Abstract
Glycoproteins with asparagine-linked (N-linked) glycans occur in all eukaryotic cells. The function of their glycan moieties is one of the central problems in contemporary cell biology. N-glycosylation may modify physicochemical and biological protein properties such as conformation, degradation, intracellular sorting or secretion. We have isolated and characterized two allelic Arabidopsis mutants, gcs1-1 and gcs1-2, which produce abnormal shrunken seeds, blocked at the heart stage of development. The mutant seeds accumulate a low level of storage proteins, have no typical protein bodies, display abnormal cell enlargement and show occasional cell wall disruptions. The mutated gene has been cloned by T-DNA tagging. It codes for a protein homologous to animal and yeast α-glucosidase I, an enzyme that controls the first committed step for N-glycan trimming. Biochemical analyses have confirmed that trimming of the α1,2- linked glucosyl residue constitutive of the N-glycan precursor is blocked in this mutant. These results demonstrate the importance of N-glycan trimming for the accumulation of seed storage proteins, the formation of protein bodies, cell differentiation and embryo development.
Keywords: Arabidopsis/α-glucosidase I/cell differentiation/N-glycosylation/seed development
Introduction
In all eukaryotes, the N-glycosylation process begins in the endoplasmic reticulum (ER) by the co-translational transfer of a precursor oligosaccharide (Glc3Man9GlcNAc2) to specific asparagine residues of nascent polypeptides. The glycosylation sites are tripeptides Asn-X-Ser/Thr, where X can be any amino acid except proline and rarely aspartic acid. N-glycan maturation occurs in the secretory pathway as the glycoproteins are transported from the ER to their final destination (for reviews see Lerouge et al., 1998; Apweiler et al., 1999; Herscovics, 1999; Moore, 1999; Parodi, 2000).
The first committed step of the maturation of the N-linked Glc3Man9GlcNAc2 oligosaccharide precursor is the trimming, by α-glucosidase I (EC, 3.2.1.1.106), of the terminal α1,2-linked glucosyl residue. This trimming occurs very early when the polypeptide is still translocating into the ER lumen. Subsequently, α-glucosidase II removes the two internal α1,3-linked glucosyl residues. In animal cells, this initial N-glycan trimming participates in the folding and quality control of newly synthesized glycoproteins mediated by calnexin and calreticulin (for a review see Hammond and Helenius, 1995). These chaperones interact with monoglucosylated glycans N-linked to misfolded proteins and retain them in the ER, until proper folding or targeting to the proteasome degradation pathway. A similar mechanism probably also occurs in plants, avoiding intracellular trafficking of misfolded or incorrectly assembled proteins (Pedrazzini et al., 1997; Herman and Larkins, 1999; Vitale and Denecke, 1999).
In addition to the control of glycoprotein folding, numerous roles have been demonstrated for N-glycans (for reviews see Olden et al., 1985; Voelker et al., 1989; Lerouge et al., 1998; Rodriguez-Boulan and Gonzalez, 1999). In general, N-glycosylation of the proteins influences their physicochemical and biological properties. For instance, N-linked oligosaccharides may contain intracellular targeting or secretion information, or may be directly involved in protein recognition, cell to cell adhesion and differentiation processes.
In plants, the role of N-glycosylation and N-glycan trimming may be essential, because all seed storage proteins are initially synthesized on the ER and several of them are N-glycosylated (Herman and Larkins, 1999; Chrispeels and Herman, 2000). So far, the importance of early steps in N-glycan trimming has been investigated essentially using cell cultures grown in the presence of glucosidase inhibitors (Lerouge et al., 1998). Thus, the available data were difficult to interpret with respect to the importance of N-glycan trimming for the accumulation of storage proteins and the formation of protein bodies during seed development.
In order to unravel molecular mechanisms underlying seed maturation, a visual screening for Arabidopsis thaliana mutants affected in seed development was carried out. In this paper, we report the isolation and characterization of two allelic Arabidopsis mutants, gcs1-1 and gcs1-2, which produce shrunken seeds. The mutant seeds accumulate a low level of storage proteins, have no typical storage protein bodies and display cell enlargement. The mutated gene has been cloned by T-DNA tagging and shown to be homologous to animal and yeast α-glucosidase I. Biochemical analyses have confirmed that N-glycan trimming is blocked in the mutants. These results demonstrate the importance of the early steps, i.e. α-glucosidase I dependent, of N-glycan trimming for the accumulation of storage proteins, the formation of protein bodies and cell differentiation, during plant embryo development.
Results
The gcs1-1 mutation affects cell differentiation and embryo development
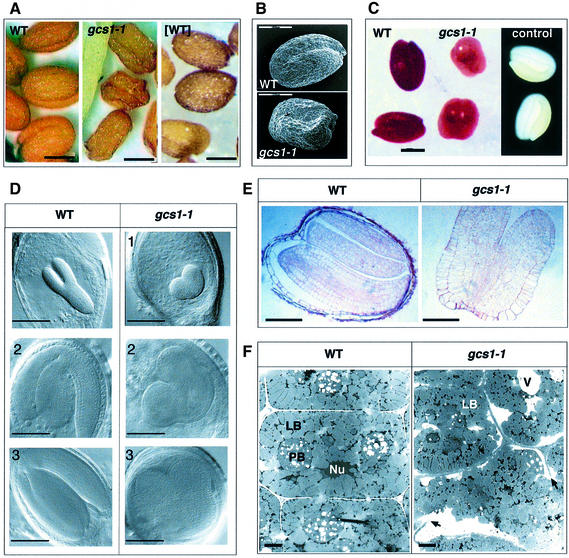
The gcs1-1 mutation was identified in a T-DNA insertion line, by visual screening of seeds (T2 generation) produced by the hemizygous primary transformant (T1). Hetero zygous plants appeared normal except for the production of wrinkled seeds (Figure 1A and B). These mature wrinkled seeds were unable to germinate after imbibition, although the embryos are metabolically active according to the tetrazolium test (Figure 1C). The morphogenesis of gcs1-1 embryos appeared to be blocked at the heart stage (Figure 1D). In contrast to wild-type embryos, the radicle and cotyledons did not elongate to form a torpedo shape. Instead, embryos increased in size by radial cell expansion, leading to the filling of the whole seed. The number of cell layers was not modified in hypocotyls and cotyledons of mutant embryos compared with wild-type embryos at the heart stage of development (Figure 1E). Attempts to rescue embryo development by growing them in vitro, in the presence of growth regulators (e.g. auxin, cytokinins or gibberellins), were unsuccessful.
Fig. 1. Microscopy analyses of wild-type and mutant seeds. (A) Light microscopy analyses of wild-type seeds (WT), gcs1 seeds selected from a heterozygous plant (gcs1-1) and seeds produced by a plant homozygous for the gcs1 mutation, which ectopically expresses GCS1 cDNA ([WT]). Scale bars, 200 µm. (B) Electron microscopy pictures of wild-type (top) and gcs1 mutant seeds (bottom). Scale bars, 200 µm. (C) Embryo viability using the tetrazolium test. Results shown are obtained with wild-type embryo (WT), gcs1-1 embryo (gcs1-1) and wild-type embryo boiled for 30 min as a negative control. Since the negative control is completely white, it is shown on a dark background. Scale bar, 200 µm. (D) Cleared seed viewed with Nomarski optics. Comparison of the development of the wild-type (WT) and gcs1 embryos after the heart stage. The radicle and cotyledons of gcs1-1 do not elongate to form the torpedo embryo (1). However, gcs1-1 embryos enlarge and fill the whole mature seed (2 and 3). Scale bars, 100 µm. (E) Light microscopy analyses of semi-thin sections stained with toluidin blue. Mature wild-type (WT) and gcs1-1 seeds. Scale bars, 80 µm. (F) Transmission electron micrographs of representative cotyledon cells from the wild-type and mutant embryos. LB, lipid bodies; Nu, nucleus; PB, protein bodies; V, vacuoles. Arrows indicate incomplete cell walls occasionally observed in the mutant cells. Scale bars, 1 µm.
The extent of cellular differentiation in arrested gcs1-1 embryos was investigated by cytological analyses. In mature wild-type embryos, protein- and oil-bodies are the most abundant organelles (Mansfield and Briarty, 1992). In mutant cells, lipid bodies looked normal (Figure 1F). On the contrary, protein bodies were absent or disorganized in mutant seeds, where vacuoles with variable amounts of electron dense material were observed. These vacuoles resembled immature protein bodies found in the cells of the young wild-type embryo. Finally, cell wall disruptions were occasionally observed in mutant cells (Figure 1F).
gcs1-1 is a T-DNA-tagged allele
Segregation analyses of kanamycin resistance (conferred by the T-DNA) and the abnormal wrinkled seed phenotype were carried out to check for the possible tagging of the mutation by a T-DNA insertion. All 110 kanamycin-resistant plants tested, from the T2 generation, segregated for both mutant seed and kanamycin-resistant phenotypes. The strict co-segregation of the phenotypes demonstrated that there was only one T-DNA insertion locus in gcs1-1, which is genetically linked to the mutation. Furthermore, the absence of homozygous plants confirmed that the mutation is lethal at the homozygous state. Southern blot analysis using T-DNA probes showed that only one T-DNA copy was present at the insertion locus (data not shown).
It was observed that <20% of the seeds (for n = 2246) presented the wrinkled phenotype, instead of 25% as expected for a normal recessive Mendelian trait. Furthermore, the ratio of seedlings resistant to kanamycin (57.4% resistant, for n = 5269 seedlings) was lower than the ratio expected for a dominant marker linked to a lethal mutation (two resistant for one sensitive seedling, 66.6%). These results suggest that the mutated allele is transmitted to the progeny at a lower frequency than the wild-type allele. Preliminary results of reciprocal crosses between wild-type and heterozygous lines indicate that the deficiency is present on both male and female sides.
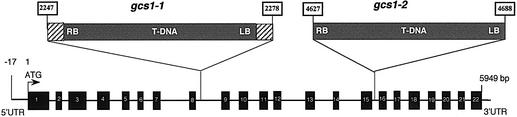
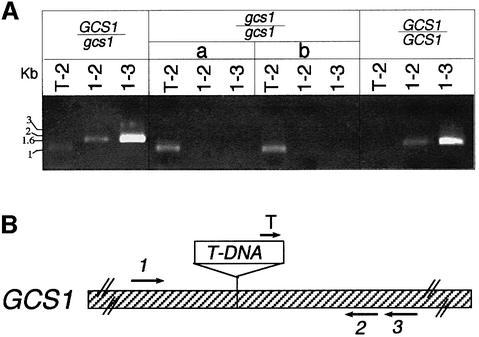
Plant genomic sequences flanking the left and right T-DNA borders were recovered by walking PCR (Devic et al., 1997) and sequenced. The two products were found to match a portion of an Arabidopsis bacterial artificial chromosome (BAC) clone. This BAC (TAMU, T1F15) mapped on chromosome 1, near the molecular marker mi185 (around cM 110). The T-DNA insertion resulted in a small deletion of genomic DNA (30 bp) and the addition of two small DNA fragments, 26 and 22 bp long, of unknown origin, between the gene and the right and left T-DNA borders, respectively. T-DNA was inserted in a putative 5949 bp gene, T1F15.4 (Figure 2). PCR analysis of the insertion locus revealed the expected polymorphism between homozygous, heterozygous and wild-type embryos (Figure 3). The coding sequence was RT–PCR amplified from 8-day-old seedling mRNA and the correct gene structure subsequently deduced (DDBJ/EMBL/GenBank accession No. AJ278990; Figure 2). The gene contains 21 introns and the coding sequence is 2559 bp long. The open reading frame differs from the hypothetical T1F15.4 protein as a result of discrepancies between the predicted and actual splice sites.
Fig. 2. Molecular analyses. Schematic representation of the gene structure at the GCS1 locus, in wild-type and gcs1 mutant genomes. The structure of the gene is deduced from the comparison between genomic and cDNA sequences (DDBJ/EMBL/GenBank accession No. AJ278990). In gcs1-1, the T-DNA was inserted between nucleotides 2247 and 2278, leading to a small deletion of 30 bp. Dashed boxes represent insertions of 26 and 22 bp of unknown origin in gcs1-1. In gcs1-2, the T-DNA was inserted between nucleotides 4629 and 4698, leading to a deletion of 58 bp. Black boxes are exons. The 5′UTR was obtained by 5′-RACE–PCR.
Fig. 3. (A) PCR analyses of the structure of the GCS1 locus. The DNA fragments corresponding to GCS1 loci were PCR amplified with genomic DNA extracted from individual embryos, after removing the seed coat. The results shown have been obtained with DNA from embryos produced by heterozygous (GCS1/gcs1) or wild-type (GCS1/GCS1) plants and with two representative selected mutant embryos (gcs1/gcs1, a and b). (B) Schematic representation of the insertion locus and location of the primers used for the analyses.
Gene expression was analysed by northern blotting and RT–PCR experiments. The mRNA was detected in all the tissues tested, at a similar level (Figure 4A). Furthermore, GCS1 mRNA accumulated at all stages of silique development (Figure 4B). These results suggested that the expression of GCS1 is constitutive.
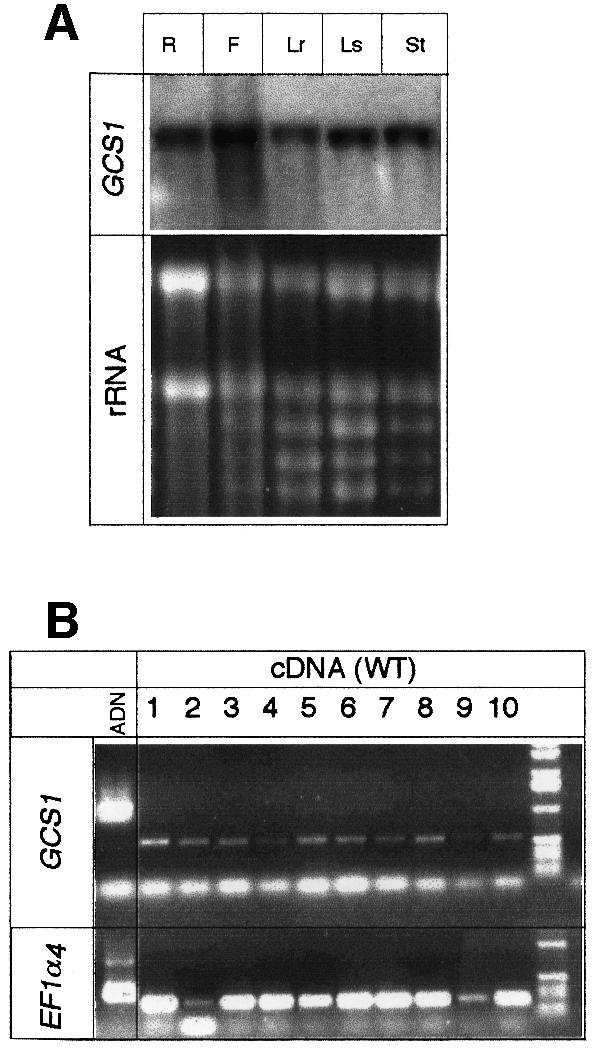
Fig. 4. Gene expression analyses. (A) Northern blot analysis of GCS1 gene expression in different tissues: roots (R), flower (F), rosette leaves (Lr), stem leaves (Ls) and stem (St). Eight micrograms of total RNA were loaded per lane. (B) RT–PCR analyses of GCS1 gene expression during silique development. Total mRNAs were extracted at different stages of flower (lane 1) and silique development (lanes 2–10). Siliques were taken in pairs from the top of the main axis, starting with the first siliques emerging from the flower (lane 2), to dry mature siliques (lane 10). The EF-1α gene (Liboz et al., 1990) was included as a control. To control that no genomic DNA contaminated PCR products, oligonucleotides were designed to amplify a region overlapping an intron. The genomic control is presented for both GCS1 and EF-1α genes.
Mutation of GCS1 is responsible for the phenotype
To provide further evidence that disruption of the GCS1 locus was responsible for the mutant phenotype, the characterization of a second allelic mutant was achieved. A T-DNA insertion line, gcs1-2, was isolated by PCR screening of the Versailles T-DNA collection (Bechtold et al., 1993). Sequencing of the mutated gene revealed that a T-DNA was inserted in the 15th intron between nucleotides 4627 and 4688 (Figure 2). Thus, the insertion gave rise to a small deletion of the genomic DNA (60 bp). As expected, gcs1-1 and gcs1-2 seeds displayed similar phenotypes and, when crossing heterozygous plants, no complementation of the phenotype was observed in F1. These results suggested that both gcs1-1 and gcs1-2 mutations were the cause of the mutant phenotype.
To confirm this result, GCS1 cDNA was ectopically expressed in gcs1-1 background, to restore the wild-type phenotype. Plants heterozygous for the mutation were transformed with a construct providing hygromycin resistance and carrying the GCS1 coding sequence under the control of a strong constitutive promoter (see Materials and methods). Out of twenty primary transformants resistant to hygromycin and able to produce seeds, two were found to be homozygous for the gcs1-1 mutation by PCR and genetic analyses of their progeny (100% kanamycin-resistant seedlings) (Figure 1A). It was concluded that ectopic expression of GCS1 cDNA was able to restore wild-type phenotype in these homozygous gcs1-1 plants.
The characterization of the two allelic mutants and restoration of the wild-type phenotype by ectopic expression of the cDNA demonstrate that gcs1 mutations cause the mutant phenotype.
GCS1 encodes a plant α-glucosidase I homologue
GCS1 mRNA encodes a predicted protein of 852 amino acids, showing strong similarities to previously identified α-glucosidase I (e.g. ∼50% to human and yeast proteins; DDBJ/EMBL/GenBank accession Nos Q13724 and AAC49157, respectively). The domain ‘ERHLDLRCW’, shown to be involved in the fixation of the substrate in the human protein (Romaniouk and Vijay, 1997), is present at a similar position in GCS1 (Figure 5A). Furthermore, sequences showing similarities to four peptides previously identified from mung bean α-glucosidase I (Zeng and Elbein, 1998) were found in GCS1 (Figure 5A). These results strongly suggest that GCS1 codes for a plant α-glucosidase I. In agreement, other structural features are also conserved in the plant protein when compared with its human homologue. For instance, a glycosylation site ‘NHT’, identified in the human protein (Kalz-Füller et al., 1995), is also present at a similar position in GCS1 (Figure 5A). GCS1 also contains an N-terminal double-arginine motif found in other type II membrane proteins of the ER (Schutze et al., 1994). Finally, analysis of the hydrophobicity profile of GCS1 revealed the presence of a highly hydrophobic region between amino acids 49 and 74 (Figure 5B). The homologous transmembrane domain found in the human protein is sufficient to mediate protein retention in the ER (Tang et al., 1997).
Fig. 5. Structure of the GCS1 protein. (A) GCS1 encodes a predicted protein of 852 amino acids. The four peptides similar to those identified from the mung bean α-glucosidase I are underlined. The N-terminal double-arginine motif found in other type II membrane proteins and the highly hydrophobic stretch of amino acids are indicated in black boxes. The putative N-glycosylation site ‘NHT’ and the site involved in fixation of the substrate appear in bold characters. (B) Analysis of the hydrophobicity profile using the method of Kyte and Doolittle (1982). The hydropathy value of amino acids was calculated over a window of three amino acid residues and was plotted as a function of amino acid positions.
N-glycan trimming is blocked and protein accumulation is affected in gcs1
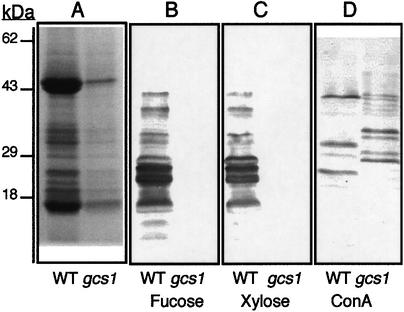
In order to investigate the consequences of GCS1 disruption on protein N-glycosylation, biochemical analyses were performed. These biochemical analyses were carried out with mutant seeds selected under a binocular microscope. Total fatty acids, sucrose and starch contents were similar in the mutant and wild-type seeds (data not shown). Polypeptides obtained from wild-type and gcs1 mutant seeds were fractionated by SDS–PAGE. Coomassie Blue staining of the gel revealed a very weak accumulation of proteins in the mutant seeds, although the profile looked similar (Figure 6A).
Fig. 6. Protein and N-glycosylation. Seed protein analysis by SDS–PAGE and Coomassie Blue staining (A), immunoblotting using antibodies specific for the β1,2-xylose (B) or α1,3-fucose (C) residues, and affinodetection of glycoproteins containing high-mannose N-glycans using ConA (D). The quantities of protein extract loaded correspond to 10 seeds in (A) and (D) and four seeds in (B) and (C).
Seed protein extracts were further analysed by immunoblotting using antibodies specific for the β1,2-xylose or the α1,3-fucose residues characteristic of plant complex N-glycans (Faye et al., 1993b) (Figure 6). Affino detection of glycoproteins containing high-mannose-type N-glycans was performed using Concanavalin A (ConA) as probe (Faye and Chrispeels, 1985) (Figure 6D). Wild-type seeds contained many different glycoproteins, with complex N-glycans reacting with anti-xylose and anti-fucose antibodies, and a few glycoproteins with high-mannose N-glycans reacting with ConA. In mutant seeds, no proteins were detected with either anti-complex glycan probe, but ConA detected additional glycoproteins of different molecular weight, as compared with the corresponding pattern obtained from wild-type seeds (Figure 6). These analyses indicated that, in the mutant, the processing of N-glycans was blocked at the first steps of N-glycan maturation, which resulted in the accumulation of ConA-reactive high-mannose-type N-glycans instead of fucose- and/or xylose-containing complex N-glycans.
Structural analyses of the N-glycans produced in the mutant
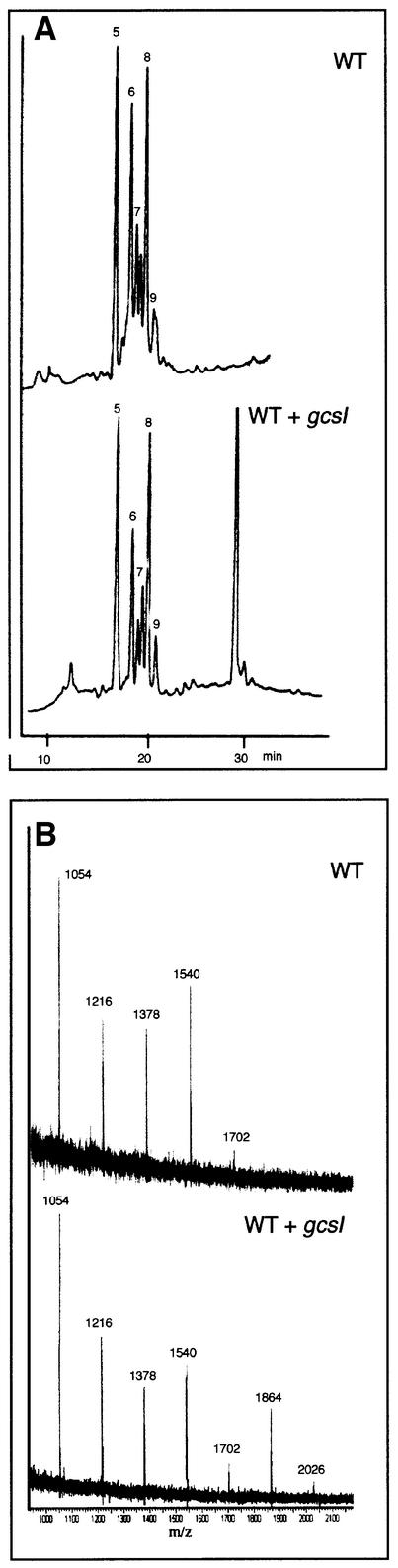
To characterize the structure of the N-glycans reacting with ConA in the mutant, N-glycans were released by endoglycosidase H treatment of seed proteins and analysed by high pH anion exchange chromatography with pulsed amperometric detection (HPAEC–PAD) and by matrix assisted laser desorption ionization time of flight (MALDI–TOF) mass spectrometry. In contrast to the analyses described above, carried out with mutant seeds selected under a binocular microscope, the higher amounts of material required for the structural analyses of N-glycans were not compatible with selection of mutant seeds. Therefore, N-linked glycans were analysed by comparing extracts from seeds produced by wild-type or heterozygous plants.
With the seeds produced by heterozygous plants, the HPAEC–PAD profile of N-glycans shows peaks between 16 and 22 min identical to those detected in the wild-type seeds, which were assigned to high-mannose glycans, from Man5GlcNAc to Man9GlcNAc (Figure 7A); however, an additional peak was observed at 29 min. The structure of the oligosaccharide(s) contained in this peak was first determined by comparison of its retention time with standard glycans. This peak was found to have the same retention time as Glc3Man7GlcNAc, which has previously been isolated from glycoproteins synthesized by sycamore cells treated with castanospermine, an inhibitor of α-glucosidase I (Lerouge et al., 1996).
Fig. 7. Structural analyses of the N-glycans. Analyses of the structure of the N-glycans released by endoglycosidase H from wild-type (WT) and a mixture of wild-type, heterozygous and mutant seeds (WT + gcs1), by HPAEC–PAD (A) and MALDI–TOF mass spectrometry (B).
The MALDI–TOF mass spectrometry analyses also showed the presence of additional structures in the N-glycans isolated from the seeds produced by heterozygous plants, when compared with wild-type seeds (Figure 7B). In addition to the (M+Na+) ions at m/z = 1054, 1216, 1378, 1540 and 1702, assigned to the sodium adducts of Man5GlcNAc to Man9GlcNAc, two ions at m/z = 1864 and 2026 were detected. These molecular ions corresponded to structures constituted of one GlcNAc residue and 10 and 11 hexose units, respectively. To confirm that such ions corresponded to the additional peak detected in the HPAEC–PAD profile, the oligosaccharides of the peak were collected and analysed by MALDI–TOF mass spectrometry. Only the ions at m/z = 1864 and 2026 were recovered, confirming that the additional peak at 29 min contained the two additional oligosaccharide species.
The data obtained by HPAEC–PAD and MALDI–TOF mass spectrometry analyses suggest that Man5GlcNAc to Man9GlcNAc oligosaccharides arose from wild-type seeds. The additional peak, assigned to the mutant seeds, contained two structures: Glc3Man7GlcNAc and Glc3Man8GlcNAc (Figure 8). The former structure was previously found to be the major glycan N-linked to glycoproteins isolated from plant cells treated with castanospermine (Lerouge et al., 1996). The Glc3Man8 GlcNAc structure probably results from a partial trimming from the oligosaccharide precursor of terminal α1,2- linked mannose residues by the Golgi α-mannosidase I.
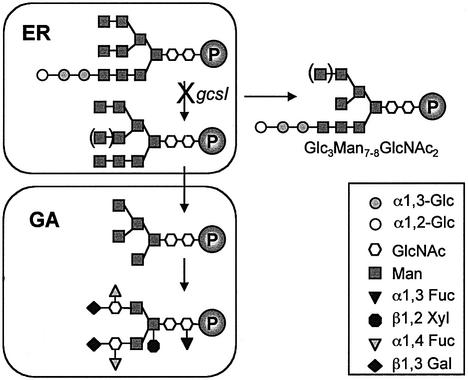
Fig. 8. Schematic representation of N-glycan trimming in WS and gcs1 seeds. As in other eukaryotes, processing of plant N-linked glycans occurs along the secretory pathway, as a glycoprotein moves from the ER and through the Golgi apparatus to its final destination. Glucosidase I removes the most external glucose of the precursor N-glycan. The glucosidase I inactivation in gcs1 mutants results in the accumulation of Glc3Man7–8GlcNAc2 glycan. ER, endoplasmic reticulum; Fuc, fucose; Gal, galactose; Glc, glucose; GA, Golgi apparatus; Man, mannose; GlcNAc, N-acetylglucosamine; P, protein; Xyl, xylose.
Discussion
GCS1 is necessary for complex N-glycan formation during embryo development
Different α-glucosidase I isoforms have been characterized after purification, from animals, plants and fungi (for review see Zeng and Elbein, 1998). They display similar enzymatic properties. The sequence similarities of GCS1 to four peptides from mung bean α-glucosidase I (Zeng and Elbein, 1998) or to the whole proteins from human (Kalz-Füller et al., 1995), mouse (Khan et al., 1999) and yeast (Romero et al., 1997) show a structural conservation of the proteins. Furthermore, biochemical analyses of N-glycans, in gcs1 mutant seeds, demonstrate that α-glucosidase I activity is affected, resulting in the accumulation of Glc3Man7–8GlcNAc2 oligosaccharides in the mutant embryos (Figure 8). Together, these results strongly support the hypothesis that GCS1 codes for an α-glucosidase I. The enzyme appears to be highly conserved throughout eukaryotic evolution, in animals, fungi and plants. This conservation is not so surprising, because glycoproteins with N-linked glycans occur in all eukaryotic cells, and the N-glycan oligosaccharide precursor with three terminal glucose residues is common to all eukaryotes (Drickamer and Taylor, 1998). Constitutive expression of animal (Khan et al., 1999) and plant (this study) genes suggests a housekeeping function for this enzyme.
In mammals, inhibition of α-glucosidase by inhibitors or in mutant cell lines never completely prevents the removal of the terminal glucosyl residue and the formation of complex oligosaccharides. This glucosidase-independent trimming is due to the presence of an alternative pathway involving an endomannosidase, which can cleave the Glc3Man fragment directly from the newly synthesized glycosylated peptide (Fujimoto and Kornfeld, 1991). Our results clearly demonstrate that complex glycans containing α1,3-fucose and β1,2-xylose are not present in the Arabidopsis gcs1 mutant seeds. This observation suggests that such an alternative pathway to glucosidase I does not occur in plants, at least during embryogenesis.
α-glucosidase I is necessary to complete seed development
In mutant seeds, the morphogenesis of the embryo is blocked at the heart stage, just before the torpedo stage characterized, in wild-type embryo, by cell expansion. At this stage, the apical–basal and radial patterns are already established (see Mayer et al., 1991; Jürgens, 1994). Thus, gcs1 cannot be considered as a pattern formation mutant.
In gcs1 embryos, cell differentiation is affected. A low amount of storage proteins accumulates and protein bodies are abnormal or absent. Cells are enlarged, and incomplete cell wall formations are occasionally observed. These results demonstrate that early steps in N-glycan trimming are important for these various aspects of plant cell differentiation. In yeast, as in mammal, glucosidase and glycosyltransferase activities do not seem to be essential for cell growth (Parodi, 2000). However, knocking out one of these enzymes may be lethal in the early steps of mammalian embryo development (Ioffe and Stanley, 1994). Similarly, our previous studies using castanospermine (Lerouge et al., 1998) and the results presented here clearly demonstrate that although α-glucosidase I is not essential for cell suspension growth, it is necessary for whole plant development. Thus, more generally, it is likely that the first step of N-glycan trimming (i.e. α-glucosidase I) is critical for the development of a multicellular organism.
In contrast, an Arabidopsis mutant affected in a later trimming step (i.e. GlcNAc transferase I activity), which accumulates Man5GlcNAc N-glycans, does not display any obvious phenotype under normal growth conditions (Von Schaewen et al., 1993). Therefore, one may hypothesize that the gcs1 phenotype could be due to a general dysfunction of the ER, due to the accumulation of abnormal N-glycoproteins, rather than to the alteration of the activity of specific proteins.
Possible causes of the cellular defects
Many proteins transported through the secretory pathway are N-glycosylated during their co-translational insertion in the ER. This early step in their maturation is often crucial for glycoprotein folding and oligomerization. This is clearly illustrated by the observation that many newly synthesized glycoproteins are rapidly degraded after their biosynthesis, when N-glycosylation is inhibited using tunicamycin (see Faye et al., 1993a for illustration in plants). Not only the presence of N-glycans but also the extent of their trimming in the ER affects the folding and transport of glycoproteins. For instance, it is well documented in mammalian cells that the inhibition of glucose trimming by glucosidase inhibitors (e.g. castanospermine or bromoconduritol) prevents or delays the folding and intracellular transport and accelerates the degradation of newly synthesized glycoproteins. Indeed, the early N-glycan processing reactions, which involve the removal of the three glucosyl residues by α-glucosidases I and II, are essential for protein folding and quality control. This control of protein quality involves two lectin-like chaperones, namely calnexin and calreticulin, through a process termed ‘the calnexin cycle’ (for review see Hammond and Helenius, 1995). Although calnexin and calreticulin have been identified in plants, it is still unclear whether they are able to act as molecular chaperones, similarly to their mammalian counterparts. However, some results suggest that similar mechanisms may occur in plants. For instance, it has been shown that mutant proteins, which are unable to form quaternary structure, are retained and degraded in the ER (Herman and Larkins, 1999). Furthermore, it has been reported that the modification of the structure of a single seed storage protein may affect the secretory pathway, leading to a reduction in storage protein synthesis and formation of abnormal protein bodies (Coleman et al., 1995). Therefore, the modification of the structure of storage proteins due to the lack of N-glycan trimming, in gcs1 embryos, may explain both the low level of storage proteins accumulated and the absence of typical protein bodies.
In plants, vesicular trafficking is essential for the expansion and elongation of cells. Indeed, it has been shown that some mutants affected in this pathway are deficient in cytokinesis, such as knolle (Lukowitz et al., 1996) or gnom/emb30 (Mayer et al., 1991; Shevell et al., 1994). Therefore, in gcs1 embryos, cytokinesis defects may be linked directly to the perturbation of the secretory pathway. Moreover, embryo arrest and defects in cytokinesis are also found in other plant mutants affected more specifically in cell wall synthesis (Nickle and Meinke, 1998; for a review see Fagard et al., 2000). As an example, the phenotype of cyt1 is similar to that of gcs1, but the mutations are not allelic [cyt1 maps on chromosome 2 (Nickle and Meinke, 1998) and gcs1 on chromosome 1]. It has been suggested that the cyt1 phenotype could be due to abnormal cellulose biosynthesis. The observation that a yeast gene homologous to GCS1 may be involved in cell wall synthesis (Simons et al., 1998) argues in favour of this hypothesis. Furthermore, during the writing of this study, we became aware that knopf mutants (Mayer et al., 1991) are also affected in GCS1 (S.Gillmore, K.Sujino, M.Palcic and C.Somerville, in preparation). These authors have shown that cellulose content is reduced in the mutant compared with wild-type embryos, demonstrating that trimming of the core N-glycans is required for cellulose biosynthesis, and giving an explanation for cell wall defects observed.
In conclusion, the genetic and biochemical characterizations of α-glucosidase I mutants reveal that N-glycan trimming is critical for the development of Arabidopsis embryos. Furthermore, cytological and biochemical analyses demonstrated that N-glycan trimming is essential for embryonic cell differentiation with respect to the accumulation of seed storage proteins, formation of protein bodies, cytokinesis and cellulose biosynthesis. These results pave the way for unravelling the underlying molecular mechanisms responsible for the different cellular defects leading to the arrest of gcs1 embryo development.
Interest of gcs1 mutants
Transgenic plants are promising systems for the production of recombinant proteins for pharmaceutical purposes (for a review on this topic see Lerouge et al., 1998). However, some N-glycan modifications, specific to plants, limit their interest for the production of mammalian glycoproteins. Different strategies may be envisaged to prevent the formation of undesirable complex-type N-glycans on recombinant proteins (Lerouge et al., 1998). The identification of gcs1 or other plant mutants defective for N-glycan trimming is a first step in the development of plants able to produce recombinant proteins with more mammalian-like N-glycans.
Materials and methods
Plant materials, growth conditions and seed viability
The wild type and mutants used were of the Wassilewskija (Ws) ecotype. The mutants are from a T-DNA insertion collection made in Versailles (Bechtold et al., 1993; Bouchez et al., 1993). Viability tests were based on the reduction of tetrazolium salts to highly coloured end products called formazans in viable seeds (Wharton, 1995). Teguments of imbibed mutant and wild-type seeds were torn and embryos were soaked in a 1% 2,3,5-triphenyl tetrazolium chloride solution (Sigma, CA). Samples were incubated for 2 days in the dark at 30°C. The development of immature embryos was investigated as described by Baus et al. (1986).
Light and electron microscopy analyses
For light microscopy, samples were fixed with 4% paraformaldehyde and 5% dimethyl sulfoxide in 0.1 M phosphate buffer pH 7, dehydrated in acetone and included in resin (Technovit 7100 kit, Heraus Kulzer, Germany), following the manufacturer’s instructions. Semi-thin sections (4–8 µm) were performed with a Jung RM 2055 microtome (Leica), stained with toluidin blue (1% w/v in 0.1 M phosphate buffer pH 7.2; Sigma, CA).
For analyses using Nomarski optics, seeds were removed from siliques and cleared for 1–24 h in a chloralhydrate solution (chloralhydrate-H2O-glycerol, 8:2:1, w:v:v) on a microscope slide. Samples were examined using a Microphot-FXA (Nikon, Japan) microscope with or without Nomarski optics. Photographs were taken using Kodak Elite 160T film.
Whole seeds were mounted on scanning electron microscopy stubs by using double-sided tape, sputter coated with carbon and gold with an Edwards 306 sputter-coater, and examined with a Philips 525 scanning electron microscope at 12 kV.
For transmission electron microscopy, embryos were fixed with 2.5% glutaraldehyde in 0.05 M phosphate buffer pH 7.2, for 2 h at 20°C. The embryos were post-fixed with 1% osmium tetroxide in the same buffer, for 2 h at 20°C. After dehydration, in a graded ethanol series, fragments were embedded in Epon’s resin. Ultrathin sections (100 nm thick) were stained with 5% aqueous uranyl acetate for 30 min then with 0.5% aqueous lead citrate for 5 min. Micrographs were taken with a Philips 420 transmission microscope at 80 kV.
Gene expression analyses
For northern analyses, total RNA was extracted from 0.5 g of plant tissue by grinding in liquid nitrogen and phenol-extracted as described by Audran et al. (1998). Eight micrograms of RNA were fractionated on a 1.2% (w/v) agarose gel containing formaldehyde in MOPS buffer and then transferred onto GeneScreen (DuPont) membranes following the manufacturer’s instructions. Blots were hybridized with a 32P-radio labelled probe made with a 300 bp PCR fragment of the 5′ region of the cDNA, and washes were carried out under high stringency.
RT–PCR experiments were performed as previously described (Dubreucq et al., 2000). Briefly, total RNA was extracted from different tissues using an RNA extraction kit (RNAeasy Plant Minikit Qiagen, Germany) supplemented with RNase-free Dnase (Qiagen, Germany) during the extraction. cDNAs were synthesized using the enhanced Avian RT–PCR Kit (Sigma, CA), with 2.5 U of PfuTurbo DNA Polymerase (Stratagene, USA). GCS1 cDNA was amplified using the primers ATG (5′-ATGACCGGAGCTAGCCGTCG) and F0 (5′-AAGTTTCGTTCCCGAAGAGG) located at the start codon and 3′-end of the cDNA, respectively (Genset, France). For gene expression analysis, a fragment of the GCS1 cDNA was amplified with the primers G5 (5′-GGATGGAAAAGTTTGTGCA) and F0. Controls were carried out with primers that amplify a constitutively expressed elongation factor ‘EF-1α’ cDNA (Liboz et al., 1990) (5′-ATGCCCCAGGACATCGTGATTTCAT and 5′-TTGGCGGCACCCTTAGCTGGATCA).
Isolation of gcs1-2
The Versailles T-DNA insertion mutants collection (Laboratoire de Génétique et Amélioration des Plantes, INRA Versailles, France) containing ∼40 000 individual lines, was screened by PCR, using primers corresponding to the right and left borders of the T-DNA and to the GCS1 gene. One positive line was detected and the position of the T-DNA within the GCS1 gene was determined by sequencing the PCR-amplified fragments.
Complementation of the gcs1 mutation
The GCS1 coding sequence placed under the control of the CaMV 35S promoter and terminator from the pLBR19 vector (Guerineau et al., 1992) was subcloned into the pBIB-HYG vector (Becker, 1990). Binary vectors were introduced into Agrobacterium tumefaciens strain C58C1 pMP90 (Koncz et al., 1984) by electroporation. T3 plants resistant to kanamycin were transformed by the in planta method (Bechtold et al., 1993), using surfactant Silwet L-77 (Clough and Bent, 1998). Transformants were selected by growing seedlings on 50 µg/ml hygromycin.
Protein methods
Arabidopsis thaliana seeds used for protein analysis were homogenized directly in a microtube containing hot denaturing buffer [20 mM Tris–HCl pH 6.8, 0.3% β-mercaptoethanol, 5% (v/v) glycerol and 1% (w/v) SDS]. The homogenate was boiled for 5 min and centrifuged for 10 min at 12 000 g. Polypeptides were separated by SDS–PAGE in 15% polyacrylamide gels under reducing conditions according to Laemmli (1970). Analytical gels were stained either in 0.25% (w/v) Coomassie Blue R-250 in 50% methanol, 5% acetic acid and destained in 25% methanol, 9% acetic acid, or with silver staining according to Blum et al. (1987). For immunodetection, polypeptides were electrophoretically transferred from the gel onto a nitrocellulose membrane. Glycoproteins were immunodetected using rabbit anti-xylose or anti-fucose serums (Faye et al., 1993b). Affinodetection of glycoproteins on the blot, using the ConA/peroxidase method, was carried out as previously described by Faye and Chrispeels (1985).
Preparation and analyses of N-glycans from A.thaliana seeds
A crude protein extract was obtained from A.thaliana seeds by homogenizing 100 mg of seeds in 10 ml of 50 mM HEPES pH 7.5, 2 mM sodium bisulfate and 0.1% SDS. Insoluble material was eliminated by centrifugation (4400 g, 15 min) at 4°C. Proteins were precipitated by the addition of 4 vols of ethanol at –20°C. After centrifugation (8000 g, 15 min), the protein pellet was then solubilized by heating for 3 min in 2 ml of 50 mM Tris–HCl pH 7.5 containing 0.1% SDS. After cooling, the pH of the protein solution was acidified to pH 5 by the addition of 1 M sodium acetate. Endoglycosidase H (0.1 U) was then added and the solution was incubated for 18 h at 37°C. Released N-glycans were purified as previously described (Bardor et al., 1999). HPAEC–PAD was performed on a Dionex DX500 system equipped with a GP50 gradient pump, an ED40 detector and a CarboPac PA1 column (4.6 × 250 mm). Elution of N-glycans was carried out using a linear gradient from 0 to 200 mM NaOAc in 100 mM NaOH at 1 ml/min, over 60 min. The additional peak detected in the mutant seed was collected and desalted on a carbograph ultra-Clean column (Packer et al., 1998) prior to mass spectrometry analysis. Mass spectra were recorded on a MALDI–TOF spectrometer ‘Tof spec E’ (Micromass, Manchester, UK), as previously described (Bardor et al., 1999).
Acknowledgments
Acknowledgements
V.Tanty, E.Harscoët and D.Garnil are acknowledged for technical assistance, A.M.Jaunet and B.Gely for helpful advice on electron microscopy, M.Bardor and V.Baltresca for help in mass spectrometry and electrophoresis analysis, G.Pelletier and N.Bechtold for providing access to the collection of T-DNA insertion mutants, D.Bouchez for his help with PCR screenings of the T-DNA lines, J.Small for correcting the manuscript and A.M.Lescure for her constant support during these past years. We are grateful to M.Miquel, S.Baud and C.Rochat for biochemical analyses of the sugars and lipids, and to G.Stewart, K.Sujino, M.Palcic and C.Somerville for sharing unpublished results. We also thank the Centre Regional Universitaire de Spectroscopie (Mont St Aignan, France) for mass spectrometry and NMR analysis, and INRA for specific support to the ‘Jeune Equipe’.
References
- Apweiler R., Hermjakob,H. and Sharon,N. (1999) On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim. Biophys. Acta, 1473, 4–8. [DOI] [PubMed] [Google Scholar]
- Audran C., Borel,C., Frey,A., Sotta,B., Meyer,C., Simmonneau,T. and Marion-Poll,A. (1998) Expression studies of the zeaxanthin epoxidase gene in Nicotiana plumbaginifolia. Plant Physiol., 118, 1021–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardor M., Faye,L. and Lerouge,P. (1999) Analysis of the N-glycosylation of recombinant glycoproteins produced in transgenic plants. Trends Plant Sci., 4, 376–380. [DOI] [PubMed] [Google Scholar]
- Baus A.D., Franzmann,L. and Meinke,D.W. (1986) Growth of arrested embryos from lethal mutants of Arabidopsis thaliana. Theor. Appl. Genet., 72, 577–586. [DOI] [PubMed] [Google Scholar]
- Bechtold N., Ellis,J. and Pelletier,G. (1993) In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Paris, 316, 1194–1199. [Google Scholar]
- Becker D. (1990) Binary vectors which allow the exchange of plant selectable markers and reporter genes. Nucleic Acids Res., 18, 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum H., Beier,H. and Gross,H.J. (1987) Improved staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis, 8, 93–99. [Google Scholar]
- Bouchez D., Camilleri,C. and Caboche,M. (1993) A binary vector based on Basta resistance for in planta transformation of Arabidopsis thaliana. C. R. Acad. Sci. De la Vie, 316, 1188–1193. [Google Scholar]
- Chrispeels M.J. and Herman,E.M. (2000) Endoplasmic reticulum-derived compartments function in storage and as mediators of vacuolar remodeling via a new type of organelle, precursor protease vesicles. Plant Physiol., 123, 1227–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.H. and Bent,A.F. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J., 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Coleman C.E., Lopes,M.A., Gillikin,J.W., Boston,R.B. and Larkins,B.A. (1995) A defective signal peptide in the maize high-lysine mutant floury 2. Proc. Natl Acad. Sci. USA, 92, 6828–6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devic M., Albert,S., Delseny,M. and Roscoe,T.J. (1997) Efficient PCR walking on plant genomic DNA. Plant Physiol. Biochem., 35, 331–339. [Google Scholar]
- Drickamer K. and Taylor,M.E. (1998) Evolving views of protein glycosylation. Trends Biochem. Sci., 23, 321–324. [DOI] [PubMed] [Google Scholar]
- Dubreucq B., Berger,N., Vincent,E., Boisson,M., Pelletier,G., Caboche,M. and Lepiniec,L. (2000) The Arabidopsis AtEPR1 extensin-like gene is specifically expressed in endosperm during seed germination. Plant J., 23, 643–652. [DOI] [PubMed] [Google Scholar]
- Fagard M., Höfte,H. and Vernhettes,S. (2000) Cell wall mutants. Plant Physiol. Biochem., 38, 15–25. [Google Scholar]
- Faye L. and Chrispeels,M.J. (1985) Characterization of N-linked oligosaccharides by affinoblotting with Concanavalin A-peroxidase and treatment of the blots with glycosidases. Anal. Biochem., 149, 218–224. [DOI] [PubMed] [Google Scholar]
- Faye L., Fitchette-Laine,A.C., Gomord,V., Chekaffi,A., Delaunay,A.M. and Driouich,A. (1993a) Detection, biosynthesis and some functions of glycans N-linked to plant secreted proteins. In Battey,N.H., Dickinson,H.G. and Hetherington,A.M. (eds), SEB Seminar Series 53: Post-translational Modifications in Plants. Cambridge University Press, Cambridge, UK, pp. 213–242.
- Faye L., Gomord,V., Fitchette-Lainé,A.C. and Chrispeels,M.J. (1993b) Affinity purification of antibodies specific for Asn-linked glycans containing α1,3-fucose or β1,2-xylose. Anal. Biochem., 209, 104–108. [DOI] [PubMed] [Google Scholar]
- Fujimoto K. and Kornfeld,R. (1991) α-Glucosidase II-deficient cells use endo α-mannosidase as a bypass route for N-linked oligosaccharide processing. J. Biol. Chem., 266, 3571–3578. [PubMed] [Google Scholar]
- Guerineau F., Lucy,A. and Mullineaux,P. (1992) Effect of two consensus sequences preceding the translation initiator codon on gene expression in plant protoplasts. Plant. Mol. Biol., 18, 815–818. [DOI] [PubMed] [Google Scholar]
- Hammond C. and Helenius,A. (1995) Quality control in the secretory pathway. Curr. Opin. Cell Biol., 7, 523–529. [DOI] [PubMed] [Google Scholar]
- Herman E.M. and Larkins,B.A. (1999) Proteins storage and vacuoles. Plant Cell, 11, 601–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herscovics A. (1999) Importance of glycosidases in mammalian glycoprotein biosynthesis. Biochim. Biophys. Acta, 1473, 96–107. [DOI] [PubMed] [Google Scholar]
- Ioffe E. and Stanley,P. (1994) Mice lacking N-acetylglucosaminidase I activity die at mid-gestation, revealing an essential role for complex or hybrid N-linked carbohydrates. Proc. Natl Acad. Sci. USA, 91, 728–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jürgens G. (1994) Pattern formation in the embryo. In Meyerowitz,E.M. and Somerville,C.R. (ed.), Arabidopsis. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 297–312.
- Kalz-Füller E., Bieberich,E. and Bause,E. (1995) Cloning and expression of glucosidase I from human hippocampus. Eur. J. Biochem., 231, 344–351. [DOI] [PubMed] [Google Scholar]
- Khan F.A., Varma,G.M. and Vijay,I.K. (1999) Genomic organization and promoter activity of glucosidase I. Glycobiology, 9, 797–806. [DOI] [PubMed] [Google Scholar]
- Koncz C., Kreuzaler,F., Kalman,Z. and Schell,J. (1984) A simple method to transfer, integrate and study expression of foreign genes, such as chicken ovalbumin and α-actin in plant tumors. EMBO J., 3, 1029–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J. and Doolittle,R.F. (1982) A simple method for displaying the hydropathic character of a protein. J. Mol. Biol., 157, 105–132. [DOI] [PubMed] [Google Scholar]
- Laemmli U. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Lerouge P., Fitchette-Lainé,A.C., Chekkafi,A., Avidgor,V. and Faye,L. (1996) Castanospermine inhibits the processing of N-glycans in sycamore cells without affecting the secretion of glycoproteins. Plant J., 10, 713–719. [DOI] [PubMed] [Google Scholar]
- Lerouge P., Cabanes-Macheteau,M., Rayon,C., Fitchette-Lainé,A.C., Gomord,V. and Faye,L. (1998) N-glycoprotein biosynthesis in plants: recent developments and future trends. Plant Mol. Biol., 38, 31–48. [PubMed] [Google Scholar]
- Liboz T., Bardet,C., Le Van Thai,A., Axelos,M. and Lescure,B. (1990) The four members of the gene family encoding the Arabidopsis thaliana translation elongation factor EF-1α are actively transcribed. Plant Mol. Biol., 14, 107–110. [DOI] [PubMed] [Google Scholar]
- Lukowitz W., Mayer,U. and Jürgens,G. (1996) Cytokinesis in the Arabidopsis embryo involves the syntaxin-related KNOLLE gene product. Cell, 84, 61–71. [DOI] [PubMed] [Google Scholar]
- Mansfield S.G. and Briarty,L.G. (1992) Cotyledon cell development in Arabidopsis thaliana during reserve deposition. Can. J. Bot., 70, 151–164. [Google Scholar]
- Mayer U., Torres-Ruiz,R.A., Berleth,T., Miséra,S. and Jurgens,G. (1991) Mutations affecting body organization in the Arabidopsis embryo. Nature, 353, 402–407. [Google Scholar]
- Moore S.E.H. (1999) Oligosaccharide transport: pumping waste from the ER into lysosomes. Trends Cell Biol., 9, 441–446. [DOI] [PubMed] [Google Scholar]
- Nickle T.C. and Meinke,D.W. (1998) A cytokinesis-defective mutant of Arabidopsis (cyt1) characterized by embryonic lethality, incomplete cell walls, and excessive callose accumulation. Plant J., 15, 321–332. [DOI] [PubMed] [Google Scholar]
- Olden K., Bernard,B.A., Humphries,M.J., Yeo,T.-K., Yeo,K.-T., White,S.L., Newton,S.A., Bauer,H.C. and Parent,J.B. (1985) Function of glycoprotein glycans. Trends Biochem. Sci., 10, 78–82. [Google Scholar]
- Packer H.N., Margaret,A.L., Jardine,D.R. and Redmond,J.W. (1998) A general approach to desalting oligosaccharides released from glycoproteins. Glycoconj. J., 15, 563–570. [DOI] [PubMed] [Google Scholar]
- Parodi A.J. (2000) Protein glucosylation and its role in protein folding. Annu. Rev. Biochem., 69, 69–93. [DOI] [PubMed] [Google Scholar]
- Pedrazzini E. et al. (1997) Protein quality control along the route to the plant vacuole. Plant Cell, 9, 1869–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Boulan E. and Gonzalez,A. (1999) Glycans in post-Golgi apical targeting: sorting signals or structural props? Trends Cell Biol., 9, 291–294. [DOI] [PubMed] [Google Scholar]
- Romaniouk A. and Vijay,I.K. (1997) Structure–function relationships in glucosidase I: amino acids involved in binding the substrate to the enzyme. Glycobiology, 7, 399–404. [DOI] [PubMed] [Google Scholar]
- Romero P.A., Dijkgraaf,G.J., Shahinian,S., Herscovics,A. and Bussey,H. (1997) The yeast CWH41 gene encodes glucosidase I. Glycobiology, 7, 997–100. [DOI] [PubMed] [Google Scholar]
- Schutze M.-P., Peterson,P.A. and Jackson,M.R. (1994) An N-terminal double-arginine motif maintains type II membrane proteins in the endoplasmic reticulum. EMBO J., 13, 1696–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevell D.E., Leu,W.M., Gillmor,C.S., Xia,G., Feldmann,K.A. and Chua,N.H. (1994) EMB30 is essential for normal cell division, cell expansion, and cell adhesion in Arabidopsis and encodes a protein that has similarity to Sec7. Cell, 77, 1051–1062. [DOI] [PubMed] [Google Scholar]
- Simons J.F., Ebersol,M. and Helenius,A. (1998) Cell wall 1,6-β-glucan synthesis in Saccharomyces cerevisiae depends on ER glucosidase I and II, and the molecular chaperone BiP/Kar2p. EMBO J., 17, 396–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang B.L., Low,S.H. and Hong,W. (1997) Endoplasmic reticulum retention mediated by the transmembrane domain of type II membrane proteins Sec12p and glucosidase I. Eur. J. Cell Biol., 73, 98–104. [PubMed] [Google Scholar]
- Vitale A. and Denecke,J. (1999) The endoplasmic reticulum-gateway of the secretory pathway. Plant Cell, 11, 615–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelker T.A., Herman,E.M. and Chrispeels,M.J. (1989) In vitro mutated phytohemagglutinin genes expressed in tobacco seeds: role of glycans in protein targeting and stability. Plant Cell, 1, 95–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Schaewen A., Sturm,A., O’Neill,J. and Chrispeels,M.J. (1993) Isolation of a mutant Arabidopsis plant that lacks N-acetyl glucosaminyl transferase I and is unable to synthesize Golgi-modified complex N-linked glycans. Plant Physiol., 102, 1109–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wharton M. (1995) The use of tetrazolium test for determining the viability of seeds of the genus Brassica. Proc. Int. Seed Test. Assoc., 20, 81–88. [Google Scholar]
- Zeng Y.-C. and Elbein,A.D. (1998) Purification to homogeneity and properties of plant glucosidase I. Arch. Biochem. Biophys., 355, 26–34. [DOI] [PubMed] [Google Scholar]