Abstract
Three snail family genes snail, escargot and worniu, encode related zinc finger transcription factors that mediate Drosophila central nervous system (CNS) development. We show that simultaneous removal of all three genes causes defective neuroblast asymmetric divisions; inscuteable transcription/translation is delayed/suppressed in the segmented CNS. Further more, defects in localization of cell fate determinants and orientation of the mitotic spindle in dividing neuroblasts are much stronger than those associated with inscuteable loss of function. In inscuteable neuroblasts, cell fate determinants are mislocalized during prophase and metaphase, yet during anaphase and telophase the great majority of mutant neuroblasts localize these determinants as cortical crescents overlying one of the spindle poles. This phenomenon, known as ‘telophase rescue’, does not occur in the absence of the snail family genes; moreover, in contrast to inscuteable mutants, mitotic spindle orientation is completely randomized. Our data provide further evidence for the existence of two distinct asymmetry-controlling mechanisms in neuroblasts both of which require snail family gene function: an inscuteable-dependent mechanism that functions throughout mitosis and an inscuteable-independent mechanism that acts during anaphase/telophase.
Keywords: insc expression/neuroblast asymmetry/snail family genes/telophase rescue
Introduction
The segmented Drosophila embryonic central nervous system (CNS) is derived from a specialized epithelial layer, the neuroectoderm (Campos-Ortega and Hartenstein, 1985; Doe, 1992; Goodman and Doe, 1993). Neural stem cells, neuroblasts (NBs), delaminate from the epithelial layer and divide asymmetrically to produce two daughter cells with different sizes. The large apical cell retains NB identity and continues to undergo successive asymmetric divisions. The small basal/lateral cell is the ganglion mother cell (GMC) and divides terminally to produce two neuron/glial cells (for a review see Lu et al., 2000). Both epithelial cells and NBs are polarized. In wild-type embryos, NB polarity required for asymmetric division is inherited from epithelial cells. Previous studies have suggested that the inheritance of this apical–basal polarity from epithelial cells is mediated through Bazooka (Baz) (Kuchinke et al., 1998; Schober et al., 1999; Wodarz et al., 1999). Baz is apically localized in the epithelial cells. When NBs delaminate from the epithelial layer, Baz and Inscuteable (Insc) (Kraut and Campos-Ortega, 1996; Kraut et al., 1996) are first concentrated in the apical stalk, maintaining the apical–basal polarity cue for NBs. Partner-of-Inscuteable (Pins) joins Baz and Insc, forming an apically localized functional complex before the NB enters mitosis (Schaefer et al., 2000; Yu et al., 2000). It has been reported that Drosophila homologue of atypical protein kinase C (DaPKC) binds to Baz, and Drosophila G-protein α-subunit binds to Pins, and these molecules might also be involved in asymmetric NB divisions (Schaefer et al., 2000; Wodarz et al., 2000).
The apical complex controls the basal localization of cell fate determinants such as Prospero (Pros) (Doe et al., 1991; Vaessin et al., 1991; Matsuzaki et al., 1992) and Numb (Uemura et al., 1989), and orients the mitotic spindle along the apical–basal axis for NB divisions (Kraut et al., 1996; Schober et al., 1999; Wodarz et al., 1999; Schaefer et al., 2000; Yu et al., 2000). Both Pros and Numb are segregated preferentially into the GMC daughter cell (Hirata et al., 1995; Knoblich et al., 1995; Spana and Doe, 1995; Spana et al., 1995). Miranda (Mir) and Partner-of-Numb (Pon), the two adaptor proteins that always co-localize with Pros (Mir) and Numb (Pon), respectively, are also segregated into the GMCs in mitosis (Ikeshima-Kataoka et al., 1997; Shen et al., 1997, 1998; Lu et al., 1998, 1999; Schuldt et al., 1998).
In mutants that disrupt apical complex formation/maintenance, cell fate determinants Pros and Numb are mislocalized in NBs and spindle orientation is affected (Kraut et al., 1996; Schaefer et al., 2000; Yu et al., 2000). It has been suggested that, in addition to the Insc/apical complex-dependent mechanism, there exists another Insc-independent asymmetry-controlling mechanism (Schober et al., 1999; Wodarz et al., 1999; Peng et al., 2000) mainly based on two observations. First, in insc null embryos, the basal cell fate determinants such as Pros and Numb are localized randomly only in the early phases of mitosis, at and prior to metaphase. Mutant NBs will redistribute these basal proteins to the basal/lateral cortex from where the future GMCs are formed in late mitotic phases (anaphase onwards); this phenomenon has been referred to as ‘telophase rescue’ (Peng et al., 2000). Consequently, most mutant GMCs inherit, at least in part, the basal cell fate determinants and adopt correct GMC identity. Secondly, spindle misorientation occurs at relatively low frequency in insc NBs. It has been reported that NBs, similarly to cells in the procephalic neurogenic region (mitotic domain 9) (Foe, 1989), rotate their mitotic spindles 90° in metaphase (Kaltschmidt et al., 2000). The major difference between the asymmetric divisions of these two types of neural stem cells is that insc is absolutely required for the spindle reorientation in mitotic domain 9 cells (Kraut et al., 1996), but appears partially dispensable in the segmented CNS. These findings suggest that when insc function is removed, a second Insc-independent asymmetry-controlling mechanism can compensate effectively for the functions of Insc in the NBs of the segmented CNS. To our knowledge, no mutations affecting the postulated Insc-independent asymmetry-controlling mechanism have been reported.
The snail (sna) mutant was identified in a genetic screen for genes involved in larval pattern formation (Nusslein-Volhard et al., 1984). The sna gene encodes a zinc finger DNA-binding transcription factor (Boulay et al., 1987; Ip et al., 1992; Kasai et al., 1992; Mauhin et al., 1993) and plays a critical role in mesoderm formation (Leptin, 1991). Before gastrulation, Sna defines the presumptive mesoderm and establishes the boundary between mesoderm and neuroectoderm by directly repressing the expression of neuroectodermal genes rhomboid and single-minded in the mesoderm (Kosman et al., 1991; Ip et al., 1992). In embryos homozygous for sna loss-of-function mutations, gastrulation does not occur and mesoderm formation is abolished (Grau et al., 1984; Nusslein-Volhard et al., 1984). Cells that normally form mesoderm will adopt a new cell fate and become part of the neuroectoderm (Rao et al., 1991).
It has been shown that sna and escargot (esg) (Whiteley et al., 1992) together with a third member of the sna gene family, worniu (wor), which also encodes a related transcription factor with Zn finger sequences, are involved in CNS development (Ashraf et al., 1999). Sna, Esg and Wor show highly homologous protein sequences and are all expressed in NBs during neurogenesis. Similarly to the Sna and Esg regulation in wing disc development (Fuse et al., 1996), Sna, Esg and Wor show functional redundancy during CNS development. It has been observed that in deficiencies that simultaneously remove sna, esg and wor, a set of GMC markers was not detected in the embryonic CNS although NB formation appeared to be normal. Ectopic expression of any one of the sna family genes alone in these deficiencies effectively rescued early embryonic CNS defects. It was suggested that Sna family proteins had essential functions during CNS development around the time of GMC formation (Ashraf et al., 1999).
We conducted a screen of the Bloomington deficiency kit looking for defects in Insc localization/expression in deficiency homozygous embryos. Our results indicate that deficiencies of the 35B–D region, uncovering all three sna family genes, showed severe NB asymmetry defects. We report here that the specific removal of the three Sna family proteins results in the down-regulation of insc transcription and translation in the NBs of the segmented embryonic CNS. Moreover, our analyses reveal the existence of two distinct asymmetry-controlling mechanisms, an Insc-dependent and an Insc-independent mechanism, both of which require the function of the sna family genes.
Results
Df(2L)TE35BC-3 embryos show defective NB asymmetry
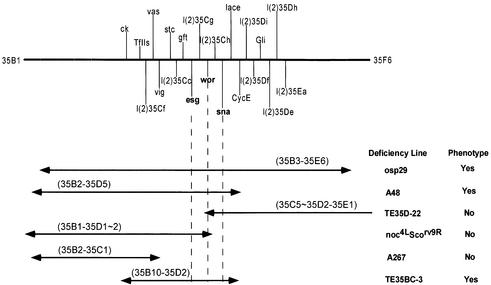
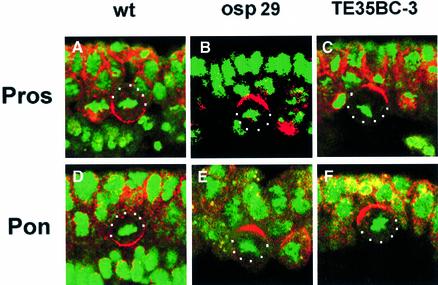
We have screened a collection of ∼170 deficiencies representing ∼70% of the fly genome from the Bloomington Stock Center, to identify possible asymmetry defects in NB divisions during early neurogenesis with anti-Pros and anti-Insc. Initially, one deficiency, Df(2L)TE35D-24 (34F5–35E1), was identified, which showed mislocalized Pros crescents and the absence of Insc expression in dividing NBs. Subsequent analyses of available deficiencies from this region (Ashburner et al., 1999) identified three additional deficiencies, Df(2L)osp29 (35B3–E6), Df(2L)A48 (35B2–35D5) and Df(2L)TE35BC-3 (35B10–35D2), all of which exhibited an identical phenotype (Figure 1). We illustrate these defects using Df(2L)osp29 and Df(2L)TE35BC-3 homozygotes. In Df(2L)osp29 embryos, Pros (Figure 2B) is no longer basally localized during NB division as seen in the wild-type embryos (Figure 2A). Since Mir is an adaptor protein for Pros localization, it is not surprising to find that Mir is also misplaced (data not shown). Basal localization of Numb and its adaptor protein Pon are also affected in Df(2L)osp29 embryos; anti-Pon antibody staining, as well as the staining with anti-Numb (data not shown), shows that the localization of the Pon/Numb crescent is no longer basal but is misplaced in mutant embryos (Figure 2E).
Fig. 1. Schematic representation of the 35B–D genomic region. Some of the deficiencies used to map the asymmetry phenotypes along with their cytological breakpoints are given. The positions of sna, wor and esg and other lethal complementation groups are shown in the diagram. Double-ended arrows represent the extent of the deficiencies.
Fig. 2. Localization of cell fate determinants is defective in deficiency lines Df(2L)osp29 and Df(2L)TE35BC-3. Confocal images of dividing NBs double-labelled with anti-Pros (red, A–C) or anti-Pon (red, D–F) and DNA staining to indicate the condensed chromosomes (green). Note that in wild-type embryos (A and D), Pros and Pon form basal crescents in dividing neuroblasts, while in the deficiency lines, although the crescents are formed, they fail to move to the basal cortex (B, C, E and F). Apical is up. NBs are outlined with white dots.
Df(2L)TE35BC-3 was the smallest deficiency avail able that exhibited these defects (Figure 2C and F). Df(2L)TE35BC-3 uncovers ∼30 known and predicted genes according to the database from the Drosophila Genome Project, including the sna family genes sna, esg and wor. Two observations suggest that the observed defects in protein localization cannot be caused by mutation of a single gene. First, Df(2L)TE35D-22 (35C5–35D2–35E1) and Df(2L)noc4LScorv9R (35B1– 35D1–2) are deficiencies with a small overlap (Ashraf et al., 1999), which together uncover 35B1–35E1, so one would expect homozygotes of one or both deficiencies to exhibit protein localization defects; yet neither of these two deficiencies when homozygous showed any protein localization defects (data not shown). Secondly, we have analysed the available lethal complementation groups in the region uncovered by Df(2L)TE35BC-3 and failed to identify the mislocalization of the Mir/Pros and Pon/Numb phenotype in any of the available mutants. These results support the notion that the observed Mir/Pros and Pon/Numb localization defects are probably multigenic in nature.
Removal of snail family genes affects Mir/Pros and Pon/Numb basal localization in mitotic NBs
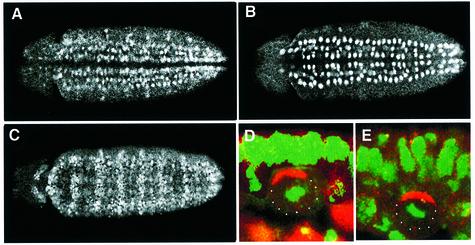
It has been reported that CNS development is abnormal in Df(2L)osp29 embryos due to deletion of Sna family proteins (Ashraf et al., 1999). Both Sna and Wor are expressed strongly in all NBs (Figure 3A and B), including those in the procephalic region, during early neurogenesis (Alberga et al., 1991; Ashraf et al., 1999). The expression of Esg is also seen in NBs (Yagi et al., 1997; Ashraf et al., 1999) and other tissues, as visualized with anti-Esg immunostaining (Figure 3C). Expression of Esg can be detected in the midline cells as well as GMCs during embryonic development. The functions of these three genes are overlapping; the early CNS defects are detected only when all three genes are removed simultaneously (Ashraf et al., 1999). In order to test whether the defects of localization of Mir/Pros and Pon/Numb seen in Df(2L)TE35BC-3 embryos are due to the absence of the three sna family genes, we examined the localization of Mir/Pros and Pon/Numb in embryos single mutant for sna, esg or wor, a double mutant for sna/esg (made by recombination; Fuse et al., 1996) and deletions that removed sna/wor or esg/wor, as well as embryos double mutant for sna/esg and further subjected to wor double-stranded RNA (RNAi; Fire et al., 1998; Kennerdell and Carthew, 1998) treatment. In single and double mutant embryos, both Mir/Pros and Pon/Numb form normal basal crescents in mitotic NBs (data not shown). Only the sna/esg double mutant embryos that have been injected with wor RNAi reproduce the phenotype found in Df(2L)TE35BC-3 embryos (Figure 3D and E).
Fig. 3. Sna family proteins are expressed in NBs and required for basal localization of Pros and Pon in dividing NBs. Confocal images of ventral views of stage 10 embryos stained with anti-Sna (A), anti-Wor (B) and anti-Esg (C). Anterior is left. (D and E) Double-labelled images of sna/esg double mutant embryos treated with wor RNAi. The NBs are stained with anti-Pros (red, D) and anti-Pon (red, E); DNA staining is green. Note that both Pros and Pon are misplaced in the sna/esg double mutant embryos with wor RNAi treatment. Apical is up.
In wild-type embryos, NBs are located between the ectoderm and mesoderm. The Df(2L)TE35BC-3 embryos lack mesoderm. Therefore, it is possible that correct NB asymmetry requires signal(s) from the mesoderm, and the asymmetry defects seen in Df(2L)TE35BC-3 could be due simply to the absence of mesoderm in these embryos. We believe this is unlikely since NB asymmetry is intact in sna embryos, which lack mesoderm and share the abnormal morphology of Df(2L)TE35BC-3 embryos. Furthermore, the partial rescue of mesoderm in Df(2L)TE35BC-3 embryos by ectopic expression (Brand et al., 1993) of the Sna protein driven by twist-gal4 does not reverse the asymmetry defects (data not shown). Thus, we conclude that mislocalization of Mir/Pros and Pon/Numb in Df(2L)TE35BC-3 embryos is due to the absence of all three sna family genes. Based on this conclusion, Df(2L)TE35BC-3 is referred to as sna/esg/wor deficient and was used in subsequent studies.
The apical complex is disrupted in sna/esg/wor-deficient embryos
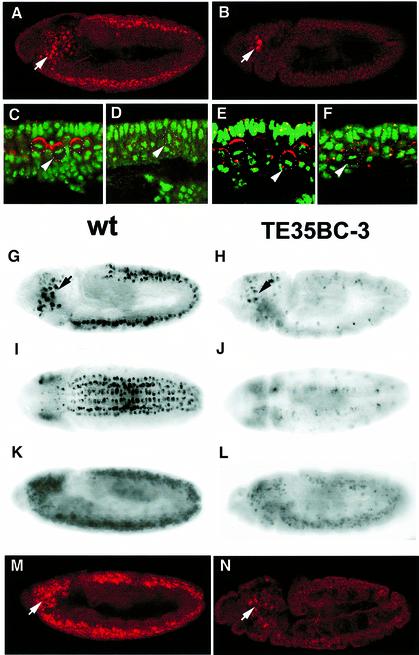
In wild-type embryos, Baz, Insc and Pins form a complex that is localized to the apical cortex of the dividing NBs (Schober et al., 1999; Wodarz et al., 1999; Schaefer et al., 2000; Yu et al., 2000). The apical complex is required for the asymmetric distribution of cell fate determinants such as Pros and Numb to the basal cortex of NBs and coordinates the orientation of the mitotic spindle along the apical–basal axis of the NB. In embryos deficient for the sna family genes, Mir/Pros and Pon/Numb are no longer concentrated to the basal cortex of mitotic NBs, indicating defects in NB asymmetry. It is possible that the asymmetry defects seen in sna/esg/wor-deficient NBs are due to the alteration of Insc expression. Anti-Insc staining indicates that Insc protein is indeed undetectable in the segmented CNS of sna/esg/wor-deficient embryos (Figure 4B and D). Although the signal intensity in the procephalic region is comparable to that in the wild-type controls, the number of cells with anti-Insc staining appears to be decreased (Figure 4B). This altered expression of Insc in the mutant embryos suggests that the mislocalization of Mir/Pros and Pon/Numb in sna/esg/wor-deficient embryos is, at least in part, due to a lack of Insc protein expression in dividing NBs (see below). As expected, Baz protein levels are low and undetectable in the great majority of mutant NBs (data not shown). The lack of easily detectable Baz in NBs is probably due to the instability of the protein when Insc is absent (Schober et al., 1999; Wodarz et al., 1999; Yu et al., 2000) since the baz mRNA levels remain unchanged in sna/esg/wor NBs (data not shown). Pins protein localization is also affected in sna/esg/wor-deficient embryos (data not shown).
Fig. 4. Dual regulation of insc expression by Sna family proteins. Lateral views of stage 10 wild-type (A) and Df(2L)TE35BC-3 (sna/esg/wor-deficient) (B) embryos stained with anti-Insc (red). Insc protein level is undetectable in sna/esg/wor-deficient NBs (D) and NBs in sna/esg double mutant embryos treated with wor RNAi (F), compared with Insc expression in NBs of wild-type (C) and sna/esg double mutant embryos (E). DNA is green. (G–L) RNA in situ hybridization images showing insc transcripts in wild-type embryos (stage 10, lateral and ventral views, G and I; and stage 11, K) and in sna/esg/wor-deficient embryos at the same developmental stages (stage 10, H and J; and stage 11, L). Insc protein expression is maintained in wild-type stage 11 embryos (M) but is undetectable in stage 11 sna/esg/wor-deficient embryos (N) even though insc transcript levels partially recover (L). Anterior is left, apical is up. Arrows indicate NBs in the procephalic region where Insc is normally expressed; NBs in the segmented CNS are indicated by arrowheads and outlined with white dots.
The down-regulation of Insc protein in NBs is also dependent on the simultaneous loss of sna, esg and wor functions. Insc expression in double mutant embryos of sna/esg was similar to that of wild-type embryos (Figure 4E). In sna/esg double mutant embryos, further removal of the third member of sna gene family, wor, with RNAi leads to the total loss of Insc protein expression (Figure 4F). Moreover, ectopic expression of any one of the sna family genes under the control of an early neural driver sca-gal4 in sna family gene mutant embryos largely restores the Insc expression in NBs (sna 79%, n = 43; esg 64%, n = 64 and wor 44%, n = 50), further indicating that Insc expression is indeed regulated by the Sna family proteins.
Dual regulation of insc expression in the segmented CNS by Sna family proteins
We further examined insc transcript levels in the sna/esg/wor-deficient embryos. In wild-type stage 9–10 embryos, insc RNA is expressed prominently in NBs of the segmented CNS and in the procephalic region (Figure 4G and I) (Kraut and Campos-Ortega, 1996). The transcript level is maintained in the segmented CNS and procephalic NBs throughout embryogenesis. In sna/esg/wor-deficient embryos, RNA in situ hybridization data indicate that the insc RNA is absent in the segmented CNS at stages 9–10 but is detectable in the procephalic NBs (Figure 4H and J). This suppression of insc RNA transcription in the segmented CNS of sna/esg/wor-deficient embryos provides evidence that the Sna family proteins are essential for insc mRNA transcription during early neurogenesis (stage 9–10). The suppression of insc transcription in the segmented CNS is transient and insc RNA can be detected, at a lower level, in late stage 11 embryos (Figure 4L). However, Insc protein in the segmented CNS of sna/esg/wor-deficient embryos remains undetectable at late stage 11 when the insc RNA levels partially recover by an unknown mechanism (Figure 4N). It is obvious that translation of insc RNA in late stage 11 embryos is inhibited in the segmented CNS of embryos deficient for sna/esg/wor. Although the inhibition mechanism is unknown, we believe that the insc 5′- and/or 3′-untranslated regions (UTRs) are involved since Insc protein (Kraut et al., 1996) can be ectopically expressed in sna/esg/wor-deficient embryos from a uas-insc transgene in which the 5′- and 3′-UTRs have been partially removed (see below). Considering that the Sna family proteins are localized to nuclei, it is unlikely that they interact directly with 5′- and/or 3′-UTRs of insc RNA. Presumably other genes regulated by the Sna family proteins mediate the observed translational effect.
The observation of delayed and decreased insc mRNA transcription and the inhibition of Insc protein synthesis in the segmented CNS of sna/esg/wor-deficient embryos suggests the dual regulation of insc expression by the Sna family proteins at both transcriptional (stage 9–10) and translational (stage 11 onwards) levels. This dual regulation mechanism is prominent in the segmented CNS but insc RNA and protein expression in the procephalic region is only partially affected in sna/esg/wor-deficient embryos. The mechanism that enables the partial restoration of insc transcription in NBs of the segmented CNS at late stage 11 in the absence of sna family gene function remains to be identified.
Insc-dependent and -independent asymmetry-controlling mechanisms are abolished in sna/esg/wor-deficient embryos
In insc22 (Burchard et al., 1995) mutant NBs, in which the apical complex required for correct asymmetric division is abolished, basal components such as Mir/Pros and Pon/Numb often form random crescents, sometimes broad and loose, from prophase to metaphase; however, Pros/Mir and Pon/Numb can eventually be redistributed to the ‘budding site’ of the future GMCs, although sometimes not as exclusively as seen in wild-type embryos, at anaphase and telophase (Figure 5B and E) even when the spindle is misorientated (Schober et al., 1999; Wodarz et al., 1999). Consequently, the great majority of all GMCs inherit, at least in part, cell fate determinants such as Pros (100%; 50/50) and adopt correct GMC fate. This phenomenon, referred to as ‘telophase rescue’ (Peng et al., 2000), does not occur in NBs lacking the three sna family genes. For example, in sna/esg/wor-deficient NBs, basal proteins Mir/Pros and Pon/Numb form a randomly localized crescent in dividing NBs but, unlike in insc embryos, these proteins are not redistributed at anaphase/telophase to the region of the cortex that gives rise to the GMC. Consequently, the great majority of the GMCs do not inherit the basal proteins Mir/Pros (Pros 90%; 45/50) and Pon/Numb (94%; 60/64) (Figure 5C and F) and thus lose their GMC identities (Broadus et al., 1998). This finding explains why GMCs were not specified correctly in Df(2L)osp29 embryos as previously reported (Ashraf et al., 1999).
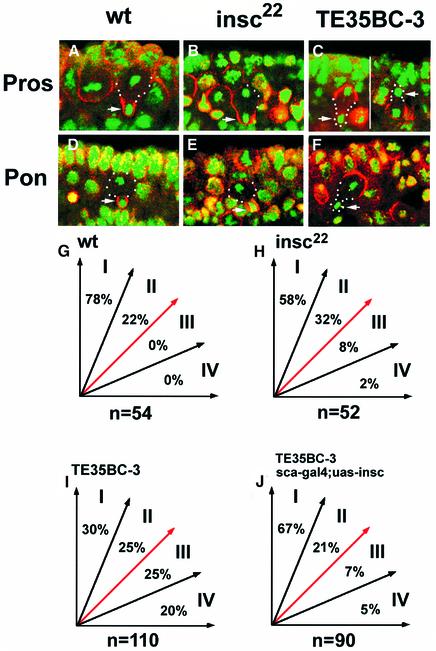
Fig. 5. Comparisons of asymmetry defects between insc22 and Df(2L)TE35BC-3 (sna/esg/wor-deficient) embryos. Confocal images of telophase NBs of wild-type (A and D), insc22 (B and E) and sna/esg/wor-deficient (C and F) embryos labelled with anti-Pros (red; A–C) or anti-Pon (red; D–F) and DNA (green). The future GMCs are indicated (arrows). The mitotic spindle orientation is sampled during late anaphase or telophase when the spindle positions are finalized in dividing NBs. Mitotic spindle orientations of NB populations are grouped arbitrarily into four sectors, depending on the angle the spindle forms with respect to the A/B axis; summary diagrams are given for wild-type (G), insc22 (H), sna/esg/wor-deficient (I) and sna/esg/wor-deficient embryos ectopically expressing Insc protein (J). The number of total NBs examined is given under each diagram. The apparent discrepancy of NB spindle orientation data in insc NBs between this study and an earlier report (Kraut et al., 1996) is most probably due to the time of sampling. We measured the NB spindle orientations in late anaphase and telophase.
Furthermore, it is known that the mitotic spindle in NBs rotates 90° during metaphase so that it is realigned along the apical–basal (A/B) axis of the embryos (Kaltschmidt et al., 2000); in insc mutants, this spindle rotation during metaphase occurs only in a small proportion (∼20%) of NBs; nevertheless, even some of these NBs are able to reorient spindles late in mitosis (Kaltschmidt et al., 2000). We measured the NB spindle orientation during anaphase or telophase in wild-type and mutant embryos and categorized them into four equal quadrants depending on the angle that the spindle forms with the A/B axis (Figure 5G–J). Based on the spindle orientation in wild-type embryos, we consider all spindles with an angle >45° relative to the A/B axis (groups III and IV) during late mitosis to be misorientated. The misoriented spindles in insc22 mutant embryos are limited (Figure 5H, 10%); the great majority of NBs (90%) have their spindles oriented within 45° of the A/B axis (groups I and II), compared with 100% in wild-type NBs. In contrast to wild-type and insc NBs, in sna/esg/wor-deficient NBs spindle orientation is completely randomized with almost equal distribution for each of the four quadrants (Figure 5I) (compare with insc22 in Figure 5H and wild-type in Figure 5G). Moreover, a small number of NBs (10%; 11/110) completely reverse their polarity, giving rise to a small apical GMC (Figure 5C; right NB), which has never been reported in any known asymmetry mutant.
These observations indicate that removal of Insc alone has only a limited effect on NB asymmetric divisions in terms of basal protein localization and spindle orientation late in mitosis, suggesting that the Insc-dependent mechanism is not the only apparatus that controls the asymmetric divisions in NBs. It appears that an Insc-independent mechanism exists that functions in parallel to coordinate the asymmetry events at later stages (anaphase onwards) of mitosis. This Insc-independent asymmetry-controlling mechanism, which is responsible for the ‘telophase rescue’ phenomenon and for prevention of random spindle orientation in insc22 embryos, is destroyed upon removal of the three sna family genes. However, one might argue that the severe asymmetry defects seen in the absence of the sna family genes might be artefactual, caused by the combination of loss of insc expression and the absence of the mesoderm. We can eliminate this possibility because in insc/sna double mutant embryos, which lack both insc and the mesoderm, NBs exhibit phenotypes that are indistinguishable from those seen in the insc single mutant (data not shown). We therefore conclude that in the absence of the sna family genes, both the Insc-dependent and -independent asymmetry-controlling mechanisms are destroyed, leading to asymmetry defects that are more severe than those seen in insc single mutants.
Ectopic expression of Insc rescues asymmetry defects in sna/esg/wor-deficient embryos
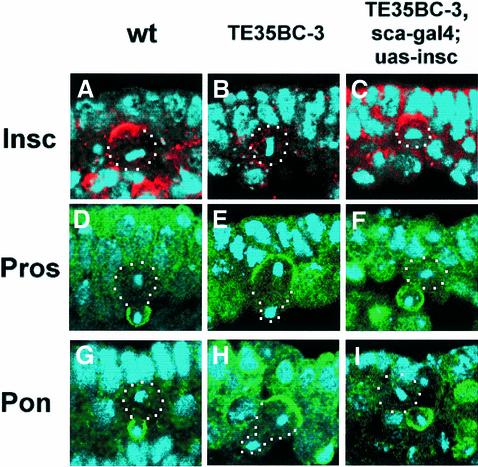
The existence of two distinct asymmetry-controlling mechanisms in wild-type NBs raises an interesting issue: how do these two mechanisms work in concert to mediate asymmetric divisions? Since embryos deficient for the sna family genes lack both mechanisms, we reasoned that by restoring the Insc-dependent mechanism in these embryos we should be able to assess the consequences of missing just the insc-independent mechanism. Ectopic expression of full-length Insc protein with an early neural driver sca-gal4 in NBs of sna family gene mutant embryos shows complete rescue of the protein localization defects described earlier. The apical complex forms normally, as indicated by the formation of apical Insc (Figure 6C) as well as Pins and Baz crescents (data not shown). The defects in basal protein localization are also completely rescued; Mir/Pros (100%, 54/54) and Pon/Numb (100%; 45/45) form tight basal crescents in mitotic NBs (Figure 6F and I). These results suggest that, with respect to protein localization, Insc protein is the only component missing in the Insc-dependent asymmetry machinery, and replacement of Insc through ectopic expression is sufficient to restore wild-type localization of the apical and basal components. Furthermore, it indicates that the Insc-independent mechanism is cryptic with respect to protein localization since it is dispensable when the Insc-dependent mechanism is in place. Either mechanism alone is able to distribute basal proteins to the cortex of the future GMC ‘budding site’ with clear temporal and efficiency differences: the Insc-dependent mechanism localizes basal proteins starting in late prophase in the form of tight crescents, while the Insc-independent mechanism is only able to redistribute, sometimes partially, mislocalized basal proteins late in mitosis (telophase rescue).
Fig. 6. Ectopic expression of Insc in Df(2L)TE35BC-3 (sna/esg/wor-deficient) embryos rescues asymmetry defects. Confocal images of metaphase NBs of wild-type (A), sna/esg/wor-deficient (B) and sna/esg/wor-deficient embryos with ectopically expressed Insc (C), double-labelled with anti-Insc (red) and DNA (blue). Images of telophase NBs of wild-type (D and G), sna/esg/wor-deficient (E and H) and sna/esg/wor-deficient embryos with ectopically expressed Insc (F and I) labelled with anti-Pros (green) or anti-Pon (green) and DNA (blue). Apical is up and telophase NBs are outlined with white dots. The future GMCs are indicated (arrows).
The spindle misorientation phenotype in sna family gene mutant embryos is also largely corrected by ectopic Insc expression. However, unlike protein localization, the rescue of mitotic spindle orientation is incomplete; the population of NBs with misoriented spindles (groups III and IV) drops from 45% to only 12% (compare Figure 5I and J). These data suggest that both the Insc-dependent and -independent mechanisms are required for correct spindle orientation in wild-type embryos since ∼10% of the mitotic spindles are misoriented in anaphase/telophase NBs defective for either mechanism. However, a complete randomization of spindle orientation is seen when both mechanisms are absent.
Discussion
Removal of the sna family genes causes defective NB asymmetric divisions
We have demonstrated that the underlying cause for the asymmetry defects associated with some deficiencies uncovering the 35B–D region of the genome, e.g. Df(2L)TE35BC-3, is the simultaneous loss of three members of the sna gene family, sna, esg and wor. All available lethal complementation groups uncovered by Df(2L)TE35BC-3, all deficiencies that remove only two out of the three sna family members and a sna/esg double mutant (Fuse et al., 1996) generated from recombination did not show any defects in any aspect of NB asymmetric division; only embryos double mutant for sna/esg, and further subjected to wor RNAi, reproduced the asymmetry defects seen in the deficiencies. These data indicate that the defects in sna/esg/wor-deficient embryos are caused by the simultaneous functional loss of all three sna family genes. The observation that the ectopic expression of sna, esg or wor in the segmented CNS of sna/esg/wor-deficient embryos reverses the asymmetry phenotypes further supports this conclusion. These conclusions are in agreement with an earlier study reporting that the sna family genes are required for CNS development (Ashraf et al., 1999).
Two parallel asymmetry mechanisms
It has been observed that in insc embryos, cell fate determinants such as Pros and Numb are mislocalized early during mitosis; however, in anaphase and telophase, the effect termed ‘telophase rescue’ causes the misplaced crescents to redistribute and overlie one spindle pole, enabling the basal cell fate determinants to segregate, exclusively or partially, to the GMCs. The insc loss-of-function alleles insc22, inscP49 and inscP72 (Burchard et al., 1995; Kraut and Campos-Ortega, 1996) all show telophase rescue. In this study, we find that essentially all NBs in insc embryos can redistribute Pros and Numb, at least partially, into GMCs. Our observations and previous studies (Schober et al., 1999; Wodarz et al., 1999; Peng et al., 2000) suggest the existence of a second asymmetry-controlling mechanism that does not require insc functions, which operates late in mitosis to coordinate protein localization with spindle orientation. These observations explain why insc mutants have minimal effect on GMC cell fate. The Insc-independent mechanism corrects the earlier errors caused by absence of Insc during anaphase/telophase, thereby enabling cell fate determinants to be inherited by the GMC. This mechanism is apparently less efficient, as shown by the fact that in some insc NBs, normally basal components form a broad and loose crescent and are only partially sequestered into GMCs. Furthermore, our observation that mitotic spindle orientation is only mildly affected in insc NBs is also consistent with an Insc-independent compensatory mechanism.
Our analysis of NB divisions in embryos deficient for the three sna family genes provides further support for the existence of an Insc-independent mechanism. In these embryos, the Insc-dependent mechanism is clearly abolished; both the transcription and the translation of insc are suppressed in the mutant NBs. In addition, telophase rescue no longer occurs; the normally basally localized components are misplaced in mitotic NBs and not redistributed to the future GMCs even at anaphase/telophase. Moreover, the spindle orientation in embryos deficient for the sna family genes becomes randomized; ∼45% of NBs exhibit misoriented spindles with an angle >45° with respect to the A/B axis at anaphase/telophase, which is not seen in wild-type NBs and is at a much higher frequency than that seen in insc22 NBs. Thus, NBs deficient for the sna family genes show two defects that are not seen in insc NB: (i) the absence of telophase rescue; and (ii) randomization of the spindle orientation late in mitosis. These observations indicate that both the Insc-dependent and -independent mechanisms require the sna family genes.
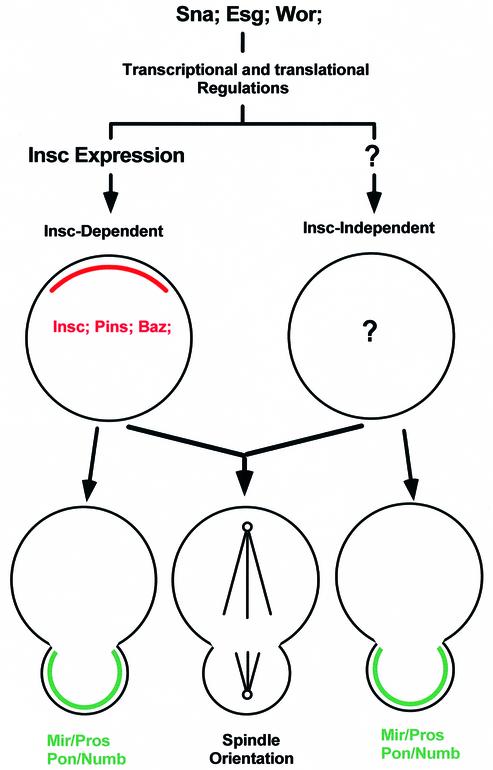
These two mechanisms can apparently function independently. In insc NBs, the Insc-independent mechanism functions in the absence of the Insc-dependent mechanism to correct the earlier (prophase to metaphase) asymmetry defects during anaphase/telophase. In sna/esg/wor-deficient NBs that have been forced to express Insc, the Insc-dependent mechanism can act in the absence of the Insc-independent mechanism to mediate the localization of the basal components from prophase to telophase, obviating the requirement for telophase rescue; however, although the Insc-dependent mechanism can reduce the extent of the mitotic spindle orientation defects seen in the sna/esg/wor NBs, it does not restore wild-type spindle orientation. Therefore, it appears that both mechanisms are required and act in concert to mediate mitotic spindle orientation. However, with respect to localization of the basal components, the effects of the Insc-independent mechanism are only visible when the Insc-dependent mechanism is absent. Figure 7 summarizes the role of the Sna family proteins and the relationship between these two asymmetry-controlling mechanisms.
Fig. 7. Relationship between the two independent asymmetry-controlling mechanisms in wild-type NBs. Sna family proteins are required for both the Insc-dependent and -independent mechanisms, most probably through transcriptional and translational regulation. Without Sna family protein functions, Insc is absent and the apical complex is not formed in NBs. Sna family proteins are also required for the Insc-independent mechanism, whose components are yet to be established. With respect to protein localization (telophase rescue), the Insc-independent mechanism is cryptic since its effects can only be observed when the Insc-dependent mechanism is absent. However, both mechanisms contribute towards the A/B orientation of the NB mitotic spindle.
Components of the asymmetry machinery in NBs
For the Insc-dependent mechanism, three components have been identified: Baz, Insc and Pins (Kraut et al., 1996; Schober et al., 1999; Wodarz et al., 1999; Schaefer et al., 2000; Yu et al., 2000) are known to form an apically localized functional complex. The function of this complex requires the participation of all members. Insc appears to be the only component of the Insc-dependent mechanism missing in sna/esg/wor-deficient embryos since ectopic expression of Insc restores its function. Little information is available on the components of the Insc-independent mechanism. Other members of asymmetry machinery identified so far in NBs are the basal components such as Mir/Pros, Pon/Numb, Stau and pros RNA (Rhyu et al., 1994; Hirata et al., 1995; Knoblich et al., 1995; Spana and Doe, 1995; Spana et al., 1995; Li et al., 1997; Broadus et al., 1998; Schuldt et al., 1998). These downstream components are controlled and coordinated by both Insc-dependent and -independent mechanisms.
Transcriptional and translational suppression of insc expression in sna/esg/wor-deficient embryos
In embryos deficient for the sna family genes, one of the major defects is the absence of Insc protein expression in the segmented CNS. RNA in situ hybridization indicates that the insc RNA transcripts are not detected in NBs of stage 9–10 embryos. Even in late stage 11 embryos when the insc RNA levels partially recover, Insc protein is never seen in the segmented CNS, indicating that the down-regulation of insc occurs at both the transcriptional and translational levels. In the procephalic region of these sna/esg/wor-deficient embryos, Insc expression is only partially affected. The 5′- and/or 3′-UTRs of the insc transcript appear to play an important role in the translational regulation of Insc expression. This is supported by two observations. First, Insc protein can be detected in sna/esg/wor embryos following ectopic expression of a cDNA construct containing the complete insc coding region but with the 5′- and 3′-UTRs partially removed. Secondly, transcripts derived from lacZ driven by a 1.2 kb insc 5′ CNS promoter sequence (our unpublished data) are not subjected to this translational repression in sna/esg/wor embryos, although their expression pattern is identical to that of Insc in the CNS. Given that the Sna family proteins are localized to nuclei, it is unlikely that they play a direct role in translational regulation. Other unknown intermediates must be involved.
To summarize, our data indicate that the sna family genes mediate two distinct asymmetry mechanisms that control wild-type NB asymmetric divisions, an Insc-dependent and an Insc-independent mechanism. These mechanisms act in parallel to effect NB asymmetric divisions. We have shown that insc expression in NBs of the segmented CNS is regulated by Sna family proteins at both the transcriptional and translational levels. With respect to protein localization, the Insc-dependent mechanism acts from prophase to telophase to localize the basal components; the Insc-independent mechanism acts during anaphase/telophase to mediate the process of ‘telophase rescue’ and its effects can only be seen when the Insc-dependent mechanism is absent. Both mechanisms contribute towards A/B orientation of the mitotic spindle.
Materials and methods
Drosophila stocks
The deficiency kit, Df(2L)osp29 and wor1 were kindly provided by the Bloomington Stock Center. The deficiencies Df(2L)TE35D-22, Df(2L)TE35BC-3 and all lethal complementation group stocks used in this study were a kind gift of John Roote (Ashburner et al., 1999). Df(2L)noc4Lscorv9R was a gift from Tony Ip. The sna1, esgG66B, sna1/esgG66B; uas-sna and uas-esg stocks were a kind gift from Shigeo Hayashi. twist-gal4 was a kind gift of Michael Bate and sca-gal4 driver was obtained from Chris Doe.
Full-length wor cDNA was amplified from a 4–8 h embryonic cDNA library (Brown and Kafatos, 1988) with specific PCR primers (5′-primer GAATTCATGCTGATTTCAACAGATGAAGG and 3′-primer GGA TCCTTAATAAATGGCCGGTGGTTGC) and subcloned into the Puast vector (Brand and Perrimon, 1993) for germline transformation.
Fusion protein and generation of anti-Wor antibody
Full-length Wor (amino acids 1–525) and N-terminal Wor (amino acids 1–305) were subcloned into the pGEX 4T-1 (Pharmacia) vector. Both glutathione S-transferase fusion proteins were purified and used to immunize mice essentially as described (Yu et al., 2000) using standard protocols.
Immunochemistry and microscopy
Embryos were collected and fixed accordingly to Yu et al. (2000). Rabbit anti-Insc (1:1000), rabbit anti-Pins (1:1000), rabbit anti-Baz (1:500; from F.Matsuzaki), rabbit anti-Pon (1:500; from Y.N.Jan), rabbit anti-Numb (1:500; from Y.N.Jan), rabbit anti-Mir (1:1000; from F.Matsuzaki), rabbit anti-β-gal (Cappel), mouse anti-Pros (1:2; from C.Q.Doe), mouse anti-Sna (1SN-5G6 and 2SN-5H6, 1:500; from Audrey Alberga and Geoff Richards), mouse anti-Esg (1:500; from Shigeo Hayashi) and mouse anti-Wor (1:1000) were used in this study. Cy3- or fluorescein isothiocyanate (FITC)-conjugated secondary antibodies were obtained from Jackson Laboratories. Stained embryos were mounted in DNA mounting medium (Lundell and Hirsh, 1994) and analysed with laser scanning confocal microscopy (Bio-Rad MRC 1024). RNA in situ hybridization experiments were carried out essentially as described in Tear et al. (1996).
Double-stranded RNA interference
The PstI fragment of wor (bases 57–597) was subcloned into pKS-ds-T7, a pBluescript (Stratagene)-derived vector with two T7 polymerase sites. The wor fragment was released with the T7 site on both ends by AscI digestion and was used for in vitro transcription with the RiboMAX™ kit (Promega). Double-stranded RNA injection was performed as described in Fire et al. (1998) and Kennerdell and Carthew (1998). After injection, embryos were aged to stage 10 at 20°C in a moist chamber then fixed and stained with antibodies.
Acknowledgments
Acknowledgements
We thank Audrey Alberga, Geoff Richards, Chris Doe, Shigeo Hayashi, Tony Ip, Yuh-Nung Jan, Fumio Matsuzaki, Kathy Matthews and the Bloomington Stock Center, and particularly John Roote, for flies and reagents, and Fook Sion Hing and Chin Tong Ong for technical assistance. W.C. would like to thank Juergen Knoblich for an interesting discussion on ‘telophase rescue’ at Keystone. We also thank the members of the Yang/Chia laboratory for discussion and help. The IMCB, Singapore, provided financial support.
References
- Alberga A, Boulay,J.L., Kempe,E., Dennefeld,C. and Haenlin,M. (1991) The snail gene required for mesoderm formation in Drosophila is expressed dynamically in derivatives of all three germ layers. Development, 111, 983–992. [DOI] [PubMed] [Google Scholar]
- Ashburner M. et al. (1999) An exploration of the sequence of a 2.9-Mb region of the genome of Drosophila melanogaster: the Adh region. Genetics, 153, 179–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashraf S.I., Hu,X., Roote,J. and Ip,T. (1999) The mesoderm determinant Snail collaborates with related zinc-finger proteins to control Drosophila neurogenesis. EMBO J., 18, 6426–6438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulay J.L., Dennefeld,C. and Alberga,A. (1987) The Drosophila developmental gene snail encodes a protein with nucleic acid binding fingers. Nature, 330, 395–398. [DOI] [PubMed] [Google Scholar]
- Brand A. and Perrimon,N. (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development, 118, 401–415. [DOI] [PubMed] [Google Scholar]
- Broadus J., Fuerstenberg,S. and Doe,C.Q. (1998) Staufen-dependent localization of prospero mRNA contributes to neuroblast daughter-cell fate. Nature, 391, 792–795. [DOI] [PubMed] [Google Scholar]
- Brown N.H. and Kafatos,F.C. (1988) Functional Drosophila cDNA libraries from Drosophila embryos. J. Mol. Biol., 203, 425–432. [DOI] [PubMed] [Google Scholar]
- Burchard S., Paululat,A., Hinz,U. and Renkawitz-Pohl,R. (1995) The mutant not enough muscles (nem) reveals reduction of the Drosophila embryonic muscle pattern. J. Cell Sci., 108, 1443–1454. [DOI] [PubMed] [Google Scholar]
- Campos-Ortega J.A. and Hartenstein,V. (1985) The Embryonic Development of Drosophila melanogaster. Spinger-Verlag, Berlin, Germany.
- Doe C.Q. (1992) Molecular markers for identified neuroblasts and ganglion mother cell in the Drosophila embryonic central nervous system. Development, 116, 855–863. [DOI] [PubMed] [Google Scholar]
- Doe C.Q., Chu-LaGraff,Q., Wright,D.M. and Scott,M.P. (1991) The prospero gene specifies cell fates in the Drosophila central nervous system. Cell, 65, 451–464. [DOI] [PubMed] [Google Scholar]
- Fire A., Xu S., Montgomery,M.K., Kostas,S.A., Driver,S.E. and Mello,C.C. (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature, 391, 806–811. [DOI] [PubMed] [Google Scholar]
- Foe V.E. (1989) Mitotic domains reveal early commitment of cells in Drosophila embryos. Development, 107, 1–22. [PubMed] [Google Scholar]
- Fuse N., Hirose,S. and Hayashi,S. (1996) Determination of wing cell fate by the escargot and snail genes in Drosophila. Development, 122, 1059–1067. [DOI] [PubMed] [Google Scholar]
- Goodman C.S. and Doe,C.Q. (1993) Embryonic development of the Drosophila central nervous system. In Bate,M. and Martinez-Arias,A. (eds), The Development of Drosophila melanogaster. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 1091–1131.
- Grau Y., Carteret,G. and Simpson,P. (1984) Mutation and chromosomal rearrangements affecting the expression of snail, a gene involved in embryonic patterning in Drosophila melanogaster. Genetics, 108, 347–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata J., Nakagoshi,H., Nabeshima,Y. and Matsuzaki,F. (1995) Asymmetric segregation of a homeoprotein, prospero, during cell division in neural and endodermal development. Nature, 377, 627–630. [DOI] [PubMed] [Google Scholar]
- Ikeshima-Kataoka H., Skeath,J.B., Nabeshima,Y., Doe,C.Q. and Matsuzaki,F. (1997) Miranda directs Prospero to a daughter cell during Drosophila asymmetric divisions. Nature, 390, 625–629. [DOI] [PubMed] [Google Scholar]
- Ip Y.T., Park,R.E., Kosman,D., Bier,E. and Levine,M. (1992) The dorsal gradient morphogen regulates stripes of rhomboid expression in the presumptive neuroectoderm of the Drosophila embryo. Genes Dev., 6, 1728–1739. [DOI] [PubMed] [Google Scholar]
- Kaltschmidt J.A., Davidson,C.M., Brown,N.H. and Brand,A.H. (2000) Rotation and asymmetry of the mitotic spindle direct asymmetric cell division in the developing central nervous system. Nature Cell Biol., 2, 7–12. [DOI] [PubMed] [Google Scholar]
- Kasai Y., Nambu,J.R., Lieberman,P.M. and Crews,S.T. (1992) Dorsal–ventral patterning in Drosophila: DNA binding of snail protein to the single-minded gene. Proc. Natl Acad. Sci. USA, 89, 3414–3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennerdell J.R. and Carthew,R.W. (1998) Use of dsRNA-mediated genetic interference to demonstrate that frizzled and frizzled 2 act in the wingless pathway. Cell, 95, 1017–1026. [DOI] [PubMed] [Google Scholar]
- Knoblich J.A., Jan,L.Y. and Jan,Y.N. (1995) Localisation of numb and prospero reveals a novel mechanism for asymmetric protein segregation during mitosis. Nature, 377, 624–627. [DOI] [PubMed] [Google Scholar]
- Kosman D., Ip,Y.T., Levine,M. and Arora,K. (1991) Establishment of the mesoderm–neuroectoderm boundary in the Drosophila embryo. Science, 254, 118–122. [DOI] [PubMed] [Google Scholar]
- Kraut R. and Campos-Ortega,J.A. (1996) inscuteable, a neural precursor gene of Drosophila, encodes a candidate for a cytoskeleton adaptor protein. Dev. Biol., 174, 65–81. [DOI] [PubMed] [Google Scholar]
- Kraut R., Chia,W., Jan,L.Y., Jan,Y.N. and Knoblich,J.A. (1996) Role of inscuteable in orienting asymmetric cell divisions in Drosophila. Nature, 383, 50–55. [DOI] [PubMed] [Google Scholar]
- Kuchinke U., Grawe,F. and Knust,E. (1998) Control of spindle orientation by the Par-3-related PDZ-domain protein Bazooka. Curr. Biol., 8, 1357–1365. [DOI] [PubMed] [Google Scholar]
- Leptin M., (1991) twist and snail as positive and negative regulators during Drosophila mesoderm development. Genes Dev., 5, 1568–1576. [DOI] [PubMed] [Google Scholar]
- Li P., Yang,X., Wasser,M., Cai,Y. and Chia,W. (1997) Inscuteable and Staufen mediate asymmetric localization and segregation of prospero RNA during Drosophila neuroblast cell divisions. Cell, 90, 437–447. [DOI] [PubMed] [Google Scholar]
- Lu B., Rothenberg,M., Jan,L.Y. and Jan,Y.N. (1998) Partner of Numb, a novel protein that colocalizes with Numb during mitosis, directs Numb asymmetric localizations in Drosophila neural and muscle progenitors. Cell, 95, 225–235. [DOI] [PubMed] [Google Scholar]
- Lu B., Ackerman,L., Jan,L.Y. and Jan,Y.N. (1999) Modes of protein movement that lead to the asymmetric localization of Partner of Numb during Drosophila neuroblast division. Mol. Cell, 4, 1–20. [DOI] [PubMed] [Google Scholar]
- Lu B., Jan,L.Y. and Jan,Y.N. (2000) Control of cell divisions in the nervous system: symmetry and asymmetry. Annu. Rev. Neurosci., 23, 531–556. [DOI] [PubMed] [Google Scholar]
- Lundell M.J. and Hirsh,J. (1994) A new visible light DNA fluorochrome for confocal microscopy. Biotechniques, 16, 434–440. [PubMed] [Google Scholar]
- Matsuzaki F., Koizumi,K., Hama,C., Yoshioka,T. and Nabeshima,Y. (1992) Cloning of the Drosophila prospero gene and its expression in ganglion mother cells. Biochem. Biophys. Res. Commun., 182, 1326–1332. [DOI] [PubMed] [Google Scholar]
- Mauhin V., Lutz,Y., Dennfeld,C. and Alberga,A. (1993) Definition of the DNA-binding site repertoire for the Drosophila transcription factor Snail. Nucleic Acids Res., 21, 3951–3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusslein-Volhard C., Wieschaus,E. and Kluding,H. (1984) Mutations affecting the pattern of the larval cuticule in Drosophila melanogaster: I. Zygotic loci on the second chromosome. Wilhelm Roux’s Arch. Dev. Biol., 193, 267–282. [DOI] [PubMed] [Google Scholar]
- Peng C.Y., Manning,L., Alberson,R. and Doe,C. (2000) The tumour-supressor genes lgl and dlg regulate basal protein targeting in Drosophila neuroblasts. Nature, 408, 596–600. [DOI] [PubMed] [Google Scholar]
- Rao Y., Vaessin,H., Jan,L.Y. and Jan,Y.N. (1991) Neuroectoderm in Drosophila embryos is dependent on the mesoderm for positioning but not for formation. Genes Dev., 5, 1577–1588. [DOI] [PubMed] [Google Scholar]
- Rhyu M.S., Yan,L.Y. and Jan,Y.N. (1994) Asymmetric distribution of Numb protein during division of the sensory organ precursor cell confers distinct fates to daughter cells. Cell, 76, 477–491. [DOI] [PubMed] [Google Scholar]
- Schaefer M., Shevchenko,A., Shevchenko,A. and Knoblich,J.A. (2000) A protein complex containing Inscuteable and the Gα-binding protein Pins orients asymmetric cell division in Drosophila. Curr. Biol., 10, 353–362. [DOI] [PubMed] [Google Scholar]
- Schober M., Schaefer,M. and Knoblich,J. (1999) Bazooka recruits Inscuteable to orient asymmetric cell divisions in Drosophila neuroblasts. Nature, 402, 548–551. [DOI] [PubMed] [Google Scholar]
- Schuldt A.J., Adams,J., Davidson,C., Micklem,D., Haseloff,J., St Johnston,D. and Brand,A.H. (1998) Miranda mediates asymmetric protein and RNA localization in the developing nervous system. Genes Dev., 12, 1847–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C.P., Jan,L.Y. and Jan,Y.N. (1997) Miranda is required for the asymmetric localization of Prospero during mitosis in Drosophila. Cell, 90, 449–458. [DOI] [PubMed] [Google Scholar]
- Shen C.P., Knoblich,J.A., Chan,Y.M., Jiang,M.M., Jan,L.Y. and Jan,Y.N. (1998) Miranda as a multidomain adapter linking apically localised Inscuteable and basally localized Staufen and Prospero during asymmetric cell division in Drosophila. Genes Dev., 12, 1837–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spana E. and Doe,C.Q. (1995) The prospero transcription factor is asymmetrically localized to the cell cortex during neuroblast mitosis in Drosophila. Development, 121, 3187–3195. [DOI] [PubMed] [Google Scholar]
- Spana E. and Doe,C.Q. (1996) Numb antagonises Notch signaling to specify sibling neuron cell fate. Neuron, 17, 21–26. [DOI] [PubMed] [Google Scholar]
- Spana E., Kopczynski,C., Goodman,C.S. and Doe,C.Q. (1995) Asymmetric localization of Numb autonomously determines sibling neuron identity in the Drosophila CNS. Development, 121, 3489–3494. [DOI] [PubMed] [Google Scholar]
- Tear G., Harris,R., Sutaria,S., Kilomanski,K., Goodman,C.S. and Seeger,M.A. (1996) Commissureless controls growth cone guidance across the CNS midline in Drosophila and encodes a novel membrane protein. Neuron, 16, 501–514. [DOI] [PubMed] [Google Scholar]
- Thummel C.S., Boulet,A.M. and Lipshitz,H.D. (1988) Vectors for Drosophila P-element-mediated transformation and tissue culture transfection. Gene, 74, 445–456. [DOI] [PubMed] [Google Scholar]
- Uemura T., Shepherd,S., Ackerman,L., Jan,L.Y. and Jan,Y.N. (1989) numb, a gene required in determination of cell fate during sensory organ formation in Drosophila embryos. Cell, 58, 349–360. [DOI] [PubMed] [Google Scholar]
- Vaessin H., Grell,E., Wolff,E., Bier,E., Jan,L.Y. and Jan,Y.N. (1991) prospero is expressed in neuronal precursors and encodes a nuclear protein that is involved in the control of axonal outgrowth in Drosophila. Cell, 67, 941–953. [DOI] [PubMed] [Google Scholar]
- Whiteley M., Noguchi,P.D., Sensabaugh,S.M., Odenwald,W.F. and Kassis,J.A. (1992) The Drosophila gene escargot encodes a zinc finger motif found in snail-related genes. Mech. Dev., 36, 117–127. [DOI] [PubMed] [Google Scholar]
- Wodarz A., Ramrath,A., Kuchinke,U. and Knust,E. (1999) Bazooka provides an apical cue for Inscuteable localization in Drosophila neuroblasts. Nature, 402, 544–547. [DOI] [PubMed] [Google Scholar]
- Wodarz A., Ramrath,A., Grimm,A. and Knust,E. (2000) Drosophila atypical protein kinase C associates with Bazooka and controls polarity of epithelia and neuroblasts. J. Cell Biol., 150, 1361–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi Y. and Hayashi,S. (1997) Role of the Drosophila EGF receptor in determination of the dorsoventral domains of escargot expression during primary neurogenesis. Genes Cells, 2, 41–53. [DOI] [PubMed] [Google Scholar]
- Yu F., Morin,X., Cai,Y., Yang,X. and Chia,W. (2000) Analysis of partner of inscuteable, a novel player of Drosophila asymmetric divisions, reveals two distinct steps in Inscuteable apical localization. Cell, 100, 399–409. [DOI] [PubMed] [Google Scholar]