Abstract
The crystallographically determined structure of the reovirus outer capsid protein σ3 reveals a two-lobed structure organized around a long central helix. The smaller of the two lobes includes a CCHC zinc-binding site. Residues that vary between strains and serotypes lie mainly on one surface of the protein; residues on the opposite surface are conserved. From a fit of this model to a reconstruction of the whole virion from electron cryomicroscopy, we propose that each σ3 subunit is positioned with the small lobe anchoring it to the protein µ1 on the surface of the virion, and the large lobe, the site of initial cleavages during entry-related proteolytic disassembly, protruding outwards. The surface containing variable residues faces solvent. The crystallographic asymmetric unit contains two σ3 subunits, tightly associated as a dimer. One broad surface of the dimer has a positively charged surface patch, which extends across the dyad. In infected cells, σ3 binds dsRNA and inhibits the interferon response. The location and extent of the positively charged surface patch suggest that the dimer is the RNA-binding form of σ3.
Keywords: capsid/dsRNA-binding protein/reovirus/σ3/X-ray crystallography
Introduction
Reoviruses are important models for the study of viral pathogenesis. They have spherical, non-enveloped particles, ∼850 Å in outer diameter (Dryden et al., 1993). The reovirus ‘core’ or ‘inner capsid particle’ (ICP), which contains five of the eight structural proteins, has an icosahedrally symmetric framework formed by the protein λ1 and stabilized by a second protein, σ2 (Reinisch et al., 2000). The λ1 shell encloses the 10 dsRNA segments of the viral genome and the components of the viral transcriptase/replicase, λ3 and µ2 (Dryden et al., 1993). The ICP bears turret-like structures, pentamers of λ2, on the 5-fold positions of its outer surface (Dryden et al., 1993; Reinisch et al., 2000). In the virion, the outer-shell proteins, µ1 and σ3, fill the surface between the projecting λ2 turrets, forming a fenestrated T = 13 (laevo) lattice (Metcalf, 1982). The remaining structural protein, σ1, inserts at the 5-fold axis into the tips of the turrets (Furlong et al., 1988; Dryden et al., 1993). The ICP can transcribe, cap and export mRNA (Shatkin and LaFiandra, 1972), and it indeed functions as a transcriptional complex in the cytoplasm of an infected cell (Borsa et al., 1981). The function of the outer-shell proteins is to introduce the ICP into the cytoplasm.
In cultured cells, the entry phase of the reovirus infection cycle includes distinct steps of attachment, endocytic uptake, outer-shell uncoating and transmembrane penetration into the cytoplasm (reviewed by Nibert et al., 1996). Attachment to cell-surface receptors is mediated by the outer capsid protein σ1 (reviewed by Lee and Gilmore, 1998). After receptor binding, virions are taken up into endocytic compartments, where endosomal proteases digest σ3 (Silverstein et al., 1970; Chang and Zweerink, 1971). This proteolytic step can be mimicked in vitro by chymotryptic digestion of purified virions (Joklik, 1972; Shatkin and LaFiandra, 1972). The infectivity of the resulting intermediate subviral particles (ISVPs) is generally similar to that of virions (Nibert et al., 1995). In a subsequent step, the ISVP perturbs the integrity of an adjacent membrane bilayer (Tosteson et al., 1993), presumably through the action of the exposed µ1 (Lucia-Jandris et al., 1993; Hooper and Fields, 1996; Chandran et al., 1999), allowing the ICPs to gain access to the cytoplasm (Borsa et al., 1981). The σ3 protein thus serves as a protective cap for µ1 in virions, and its removal by proteolysis primes the ISVP for membrane penetration.
The presentation of σ3 on the surface of a virion is critical for its function as a cap for µ1. σ3 and µ1 first associate as a hetero-oligomeric complex in the cytoplasm and then assemble onto the surface of a maturing virion (Lee et al., 1981; Shing and Coombs, 1996). The unit for cooperative assembly of µ1 and σ3 is a trimer of µ1 with three σ3 subunits bound on its periphery (K.Chandran and M.L.Nibert, unpublished; S.Liemann and S.C.Harrison, unpublished). In maps of the virion obtained by electron cryomicroscopy, (µ1)3(σ3)3 ‘heterohexamers’ occupy local and strict 3-fold positions in the T = 13 (laevo) lattice (Dryden et al., 1993). Electron cryomicroscopy also shows that the σ3 subunits project above the level of µ1, so that the virion surface appears to have rings of six knobs around the local 6-fold axes (Dryden et al., 1993). The rings surround solvent channels that extend inward toward the λ1/σ2 framework beneath. There appear to be few, if any, σ3–σ3 contacts around these rings, however.
Like structural proteins of many viruses, σ3 has an additional, regulatory function. Reoviruses activate host antiviral defense mechanisms by inducing synthesis and secretion of interferon and other cytokines, and they are believed to counteract these induced defenses through the dsRNA-binding properties of σ3 (Huismans and Joklik, 1976; Sharpe and Fields, 1982; Imani and Jacobs, 1988; Lloyd and Shatkin, 1992; Seliger et al., 1992; Beattie et al., 1995; Schmechel et al., 1997; Yue and Shatkin, 1997). Interferon induces the dsRNA-dependent protein kinase PKR, which in the presence of dsRNA phosphorylates the α-subunit of initiation factor eIF-2, thereby shutting off translational initiation (Hovanessian, 1991). Inhibition of protein synthesis in virus-infected cells (presumably those with excess, unprotected dsRNA) limits viral spread (Clemens, 1996; Mathews, 1996). Covering double-strand regions of RNA, which can occur as stems even in single-strand mRNAs, may restrict the PKR response. Inhibition of host protein synthesis by reovirus strains segregates with the S4 gene segment, which encodes σ3 (Sharpe and Fields, 1982). Consistent with the notion that σ3 prevents PKR activation by competing for binding to dsRNA (Imani and Jacobs, 1988; Giantini and Shatkin, 1989), transient or stable expression of σ3 alone does not influence cell growth or protein expression (Lloyd and Shatkin, 1992; Yue and Shatkin, 1996). The activities of the vaccinia virus E3L protein (Chang et al., 1992) and adenovirus VAI RNA (Mathews and Shenk, 1991), which both block activation of PKR, represent related strategies for inhibiting the interferon pathway. Indeed, σ3 expressed transiently from transfected cDNA of the reovirus S4 gene restores viral protein synthesis in cells infected with mutant E3L– vaccinia virus (Beattie et al., 1995) or dl31 VAI– RNA adenovirus (Lloyd and Shatkin, 1992). In these transfection experiments, both enhancement of chloramphenicol acetyltransferase reporter translation and restoration of viral protein expression correlate with the dsRNA-binding capacity of σ3 (Beattie et al., 1995; Yue and Shatkin, 1997).
The σ3 protein, which folds from a 365-residue polypeptide chain, contains a tightly bound zinc ion. σ3 interacts with dsRNA in a manner independent of RNA sequence but dependent on duplex length (Huismans and Joklik, 1976; Yue and Shatkin, 1997). Site-directed mutagenesis has implicated several basic amino acids in dsRNA binding (Denzler and Jacobs, 1994). Mutational analysis shows that loss of the zinc-binding site results not only in decreased intracellular stability, but also in loss of the ability of σ3 to interact with µ1 (Mabrouk and Lemay, 1994a; Shepard et al., 1996); loss of zinc does not, however, appear to affect dsRNA binding (Shepard et al., 1996).
We describe here the structure at 1.8 Å resolution of σ3 from reovirus type 3 Dearing (T3D). The architecture of σ3 is unlike that of other characterized proteins performing analogous functions of virus stabilization or dsRNA binding. Several features of its two-lobed structure, together with the fit into a low-resolution map of the virion obtained by electron cryomicroscopy, suggest a specific orientation for each σ3 subunit on the viral surface, with the small lobe contacting µ1 and the large lobe projecting into the surrounding solvent. The fit also suggests that to coat the virion during assembly, a σ3 subunit undergoes only subtle rearrangements from the conformation we see in the crystal. The zinc-coordinating residues of σ3 are found within the small lobe, and regions important for protease cleavage during viral entry are found in the large lobe. The presence of a tight dimer in the crystallographic asymmetric unit and the strong tendency for σ3 to dimerize in solution suggest, however, that the dsRNA-binding form of the protein is dimeric. The observed dimer has an extensive, positively charged patch on one surface, which we propose interacts with RNA.
Results
Preparation and structure determination
σ3 was expressed in Sf21 cells and purified as described in Materials and methods. The elution volume in the final Superdex-200 step suggested a dimer; dynamic light scattering and analytical ultracentrifugation supported this conclusion (data not shown). The protein crystallized in space group P212121, with two molecules in the asymmetric unit and a solvent content of ∼52%. These crystals, flash frozen in liquid nitrogen, diffracted to a resolution of 1.8 Å. We used a combination of multiple isomorphous replacement (MIR) and multiple wavelength anomalous dispersion (MAD) methods to determine phases. The entire polypeptide chain was traced from the MIR/MAD map. The two molecules in the asymmetric unit are very similar, significant differences being limited to the orientation of some of the surface side chains involved in crystallographic contacts.
Structure description
Overall outline and structure of the two lobes. The σ3 monomer is a two-lobed structure, organized around a central helix spanning the length of the protein (Figures 1 and 2). The overall dimensions of σ3 are ∼76 × 35 × 35 Å. Viewed from one side, the protein narrows at the center, separating it into a smaller and a larger lobe. Although the N-terminal residue is in the small lobe and the C-terminal residue is in the large lobe, the chain passes back and forth between lobes (Figures 2 and 3); thus, the two lobes do not strictly correspond to the sequential N- and C-terminal sections of the protein. Of the three sections of chain passing between the small and large lobes, helix B is the only one with defined secondary structure. The joint between the lobes may therefore be somewhat more flexible than the remainder of the protein. Comparison of this protein with the DALI database of protein structures (Holm and Sander, 1995), taking the small and large lobes separately or together, did not reveal any homologous structures.
Fig. 1. The folded structure of the reovirus σ3 polypeptide chain. β-strands are represented by green ribbons with arrowheads, α-helices by blue coiled ribbons and connecting loops by thin orange tubes. The zinc atom is represented by a red sphere. (A and B) Views of the structure rotated 90° with respect to each other about the long axis. View (B) is oriented such that the solvent channel within a ring of six is on the front face; the small lobe on the left would point toward the center of the virion and the large lobe, on the right, would protrude outward from the virion surface (see Figure 6).
Fig. 2. Amino acid sequence of reovirus σ3. Elements of secondary structure are color coded as in Figure 1. Helices are labeled with letters in blue, β-strands with numbers in green. Residues coordinating zinc are red, residues affecting µ1 binding are orange and residues affecting dsRNA binding are yellow. Residues 16 and 354 (discussed in the text) are underlined in black. Residues in the dimer interface are shaded gray, variable residues are underlined in red and the epitope recognized by monoclonal antibody 4F2 is boxed in green.
Fig. 3. Folding diagram for σ3. Helices are represented by blue rectangles, and β-sheets are represented by clusters of green arrows.
The small lobe contains ∼140 residues in two segments (residues 1–90 and 287–336), and contains a CCHC zinc-binding motif, which appears to be crucial for maintaining the structure of the small lobe. The small lobe contains helix A, a four-stranded anti-parallel β-sheet (strands 1–4), and the N-terminal part of the long central helix B (Figures 1 and 3). A loop from the large lobe completes the small lobe with a hairpin of bent helices F and G.
The large lobe contains ∼225 residues in two segments (residues 91–286 and 337–365). It begins with the second half of the long central helix (helix B) and, in addition to the remaining helices (C, D and E), includes two-, three- and four-stranded anti-parallel β-sheets all composed of non-consecutive strands (Figures 1 and 3). The C-terminus is neatly contained as the middle strand of the three-stranded sheet. The large lobe contains several extended loops, which may provide the sites for the proteolytic cleavages that start the disassembly process during reovirus entry (see Discussion).
Zinc site. Among known zinc-binding structures (Branden and Tooze, 1991), the CCHC zinc-binding motif of σ3 (Figure 4) most closely resembles the retroviral zinc knuckle, such as in the HIV nucleocapsid protein (De Guzman et al., 1998). Cys51 on strand 3 and Cys54 on the turn between strands 3 and 4 coordinate a zinc, together with residues His71 and Cys73. The zinc coordination in σ3 is the same as in the retroviral CCHC motif with respect to the order of liganding residues, but the α-carbon backbones of σ3 and of the retroviral CCHC zinc-binding motif do not superimpose well. Moreover, the spacing of the residues between the coordinating amino acids in σ3 (CX2CX16HX1C) is quite different from the consensus sequence of the retroviral zinc knuckle (CX2CX4HX4C). Although the retroviral zinc knuckle functions in RNA binding (De Guzman et al., 1998), the lack of similarity of the protein backbones makes an analogous mechanism for RNA binding by σ3 unlikely. Mutations in its zinc-binding residues do not affect the capacity of σ3 to bind dsRNA (Shepard et al., 1996).
Fig. 4. Zinc-binding site of σ3. The zinc is represented by a red sphere, sulfur atoms are in yellow, nitrogen atoms in blue and carbon atoms in green.
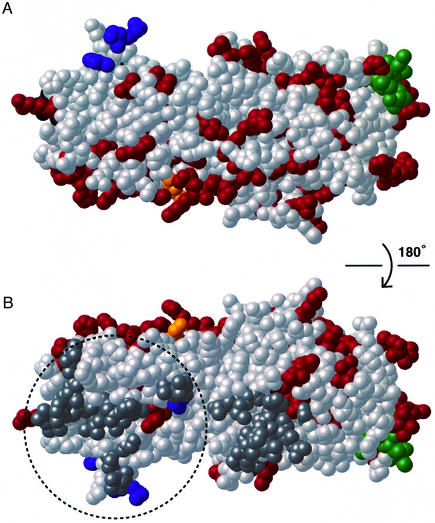
Conserved and non-conserved residues. Amino acid sequence alignments reveal that differences among σ3 proteins from 16 reovirus isolates (Kedl et al., 1995), including members of all three reovirus serotypes (Seliger et al., 1992), map mainly to one side of σ3 (Figure 5), leaving a broad conserved surface on the opposite side. This pattern suggests that the conserved side may be involved in crucial contacts with other virion proteins, primarily µ1. The non-conserved side probably corresponds to the surface facing the solvent channels in the center of the σ3 rings of six seen in reconstructions from electron cryomicroscopy (Dryden et al., 1993). The surface of the non-conserved face is composed largely of loops, while that of the conserved face is largely helical.
Fig. 5. Conserved and non-conserved residues. Residues in red are not conserved among different serotypes and isolates. Conserved residues are in gray. Residues that make up the dimer interface are in darker gray (all are conserved). Views (A) and (B) are rotated by 180° with respect to each other about the long axis. The orientation in (A) is the same as that in Figure 1B. Residues thought, based on mutational analysis, to be involved in the binding of µ1 (16, 64, 66 and 67) are shown in blue. Lys293, thought to be involved in the binding of dsRNA, is shown in yellow. Our fit of σ3 into the virion suggests that the entire conserved surface of the small lobe (dashed circle in B) may be in contact with µ1. The protease-hypersensitive loop is at the bottom right of (A) and at the top right of (B), extending roughly parallel to the long axis from the tip of the large lobe to the midpoint of the molecule.
Fit into the virion image reconstruction from electron cryomicroscopy
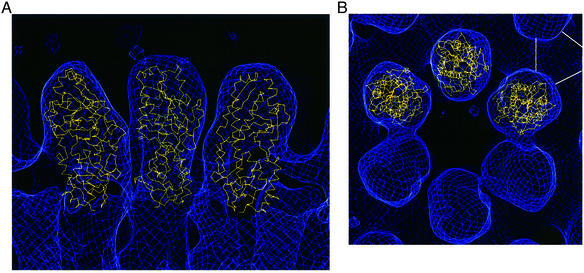
To examine the σ3 structure in the context of the mature virion, we attempted to find the best fit of our crystallographic model to an 18 Å resolution reconstruction from electron cryomicroscopy of a type 1 Lang (T1L) virion (S.B.Walter and T.S.Baker, personal communication). The model was first positioned by visual inspection into one of the knobs in a σ3 ring of six with the small lobe packed against µ1. The virion map was then restricted to contain only one σ3 molecule. Using the roughly positioned structure of σ3, an electron density map was calculated to 18 Å. This calculated map and the restricted virion map were used in a real-space maximization of the correlation of the corresponding electron densities. The correlation was calculated pixel by pixel in the overlapping maps, inside a mask derived from the structure. The optimization was performed by alternating translational and rotational searches, moving the model in small increments. The final fit has a correlation coefficient of 0.80.
Because of the relatively symmetrical shape of the σ3 monomer, there are two principal ambiguities in choosing an initial orientation for refinement: a poorly determined rotation about the long axis of the molecule and a possible flip of 180° about the midpoint, so that the large lobe packs against µ1. We tested a refinement with a starting orientation that differed from the best model by a 180° rotation about the long axis. Although it converged to a correlation of 0.79, the visual agreement of densities and the overlap of boundary outlines were poor. The refined correlation with an ‘upside-down’ starting point was 0.75. Not only is this value significantly lower than the best fit described in the preceding paragraph, but also immunological evidence strongly favors a model with the small lobe of σ3 packed contacting µ1. The monoclonal antibody 4F2 binds denatured σ3 protein in immunoblots and also binds virions (Virgin et al., 1991, 1994); thus, the epitope it recognizes must be accessible on the virion surface. This epitope has been mapped to include residue 116 in the large lobe by comparing the sequences of binding and non-binding variants of σ3 from different reovirus isolates (Kedl et al., 1995). Recent experiments with a mutant σ3, engineered with changes at positions 119 and 120, extend this mapping. Unlike wild-type protein, this mutant is no longer bound by 4F2 in immunoblots, but the altered σ3 is fully competent to recoat ISVPs (J.Jané-Valbuena, P.Jaramillo-Ferrada and M.L.Nibert, unpublished). Thus, 4F2 recognizes a sequence on helix C including residues between 116 and 120; moreover, these residues are not significant for anchoring σ3 to the virion. We therefore conclude that on the virion, the small lobe of σ3 packs against µ1, the large lobe forms the knobs visible in electron microscopy reconstructions, and the surface viewed in Figure 1B faces toward the solvent channel in the center of the σ3 rings of six (Figure 6).
Fig. 6. Fit of the structure in the low-resolution map of the virion from electron cryomicroscopy of a type 1 Lang (T1L) virion (S.B.Walter and T.S.Baker, personal communication). (A) A cross-section of a ring of six σ3 proteins on the surface of the virion, with a view tangential to the curvature of the virion, and the center of the virion below the figure. (B) A view looking down on the surface of a virion. The virion map is shown in blue, and σ3 models are displayed as α-carbon backbone traces in yellow. The orientation of the central model in (A) is the same as that in Figure 1B, with the small lobe on the bottom. The white lines in the upper right hand corner of (B) indicate a trimer of the underlying protein, µ1.
σ3 is thought to undergo a conformational change upon binding to µ1, as demonstrated by heightened protease sensitivity (Shepard et al., 1995). Successful fitting of the isolated protein model into the map of the µ1-bound form on the virion indicates that the conformational change can not involve major tertiary structural changes. The absence of a large conformational difference was suggested previously by the observation that both free and µ1-bound forms of σ3 react with conformation-dependent antibodies (Shepard et al., 1995). A more likely difference between free and µ1-bound forms of σ3 is disruption of the dimer interface.
The dimer
Figure 7 shows the σ3 dimer that constitutes the crystallographic asymmetric unit. When viewed along its 2-fold axis, the dimer has a parallelogram-like aspect, ∼76 × 70 Å, and ∼35 Å thick. The dimer interface buries a total of 2700 Å2 from each monomer, 1535 Å2 from the small lobe and 1165 Å2 from the large lobe. Interface contacts include residues 2, 4–7, 9, 10, 47, 57, 60, 62, 312, 313, 333 and 334 in the small lobe, and 164, 250, 254–257, 276–279, 338 and 342 in the large lobe. The surfaces at the dimer interface lie on the broad, conserved stripe (Figure 5), and all residues in the dimer contact are conserved among strains and serotypes.
Fig. 7. Surface representation of the σ3 dimer. Regions of positive electrostatic potential are shown in blue; regions of negative potential in red. The orientation of the top molecule in (A) is identical to the orientation in Figure 1A, and (B) is a view of the molecule rotated 180° about the long axis. Just left and right of center in (A) are positively charged patches in each monomer surrounding Lys293 (arrows), which we propose are the sites for binding dsRNA. Scale bar, 10 Å.
A map of surface electrostatic potential in the σ3 dimer (Figure 7) shows a rather even charge distribution over one of the two broad surfaces, but a large, positively charged patch that spans the two subunits on the other broad surface. This patch stands out as a likely surface for dsRNA binding. In addition to the overall positive charge, which might be involved in non-specific interaction with the backbone of an RNA duplex, this site includes Lys293 (Figure 7), which mutational analysis has implicated in dsRNA binding (Denzler and Jacobs, 1994). The two copies of Lys293 are separated by ∼30 Å, and it is thus possible for them to interact similarly with consecutive turns of an A-form dsRNA helix.
Discussion
Cleavage susceptibility
The susceptibility of σ3 to proteolytic cleavage has consequences for virion stability and initial uncoating. Although the specific sites cleaved during uncoating have yet to be identified, two independent lines of experimentation point toward a section on the large lobe as the most likely locus for initial cleavages. Many different proteases, including alkaline and acid proteases, cleave first within a hypersensitive region between residues 210 and 242 (J.Jané-Valbuena, L.A.Schiff and M.L.Nibert, unpublished). When reoviruses are present in the gut, σ3 is cleaved in the lumen of the small intestine by alkaline serine proteases (Bodkin, 1989; Bass et al., 1990), while during infection of cells via endosomes, acidic cysteine proteases are required (Sturzenbecker et al., 1987; Baer and Dermody, 1997). Reovirus strains T1L and T3D differ in their cleavage patterns and susceptibility to proteases, and the C-terminal region from residue 266 to 365 has been shown to contain the primary determinants of this difference (Jané-Valbuena et al., 1999). We note that these C-terminal residues lie near parts of the protease-hypersensitive region within the large lobe of the subunit, supporting the idea that stability of this lobe affects protease sensitivity. Studies are currently under way to identify these sequence determinants more precisely.
Viruses propagated in persistently infected cultures (PI viruses) have been shown to acquire mutations that affect viral entry. The S4 gene plays a critical role in the establishment of persistent infection (Ahmed and Fields, 1982). PI viruses can grow in the presence of ammonium chloride, a weak base that blocks acid-dependent proteolysis of viral outer capsid proteins during viral entry (Dermody et al., 1993). This trait segregates with the S1 and S4 genes, and the amino acid sequence of the S4 gene of six different PI viruses shows that they have one mutation in common: a Tyr to His substitution at position 354 (Wetzel et al., 1997). A result of this mutation is that proteolysis of PI viruses occurs with faster kinetics than that of wild-type viruses (Wetzel et al., 1997). Tyr354, located just before β-strand 12, is packed in a hydrophobic pocket that includes Tyr223, Tyr225, Leu228, Tyr238, Leu242, Val243 and Phe349. The carbonyl oxygen of Tyr354 also makes contacts with the end nitrogen of Lys234. A His at this position may destabilize the structure surrounding this pocket, rendering it more susceptible to degradation.
µ1 binding
A variety of experiments indicate that the small lobe of σ3 is the principal contact with µ1. The presence of zinc ion appears to be important for stabilizing the small lobe, and mutations of the zinc-coordinating residues Cys51 and Cys54 indeed abolish the ability of σ3 to associate with µ1 (Shepard et al., 1996). Mutations at positions 16, 64, 66 and 67, which lie on helix A and the loop between β4 and helix B, also reduce or abolish binding to µ1 (Mabrouk and Lemay, 1994a), but a mutation in the putative dsRNA-binding residue Lys293 does not (Shepard et al., 1996).
Temperature-sensitive (ts) mutants also help define the surfaces that participate in µ1 binding. Group G ts mutants (the prototype being tsG453) have a lesion in the S4 gene; they are defective in outer capsid assembly at the restrictive temperature, resulting in production of core-like particles (Fields, 1971; Morgan and Zweerink, 1974; Danis et al., 1992). Indeed, at restrictive temperature, σ3 and µ1 fail to associate, and immuno-co-precipitation has suggested that a σ3–µ1 interaction is required for their condensation onto cores (Shing and Coombs, 1996). The tsG453 σ3 differs from wild-type T3D at three positions: Asn to Lys at 16, Met to Ile at 141 and Glu to Asp at 229 (Danis et al., 1992; Shing and Coombs, 1996). The mutation at position 16 alone is sufficient to prevent σ3–µ1 interaction (Mabrouk and Lemay, 1994b; Bergeron et al., 1998). It also results in an unexplained increase in dsRNA binding. In the wild-type structure, Asn16 on helix A hydrogen-bonds with the carbonyl oxygen of Val334 (2.9 Å), but its side chain is otherwise exposed at the bottom of a deep groove. Replacement with a large side chain such as lysine might simply prevent, by steric hindrance, the binding of µ1. The interaction of Asn16 with Val334, with possible effects on σ3 dimerization or on the conformation of segments surrounding Lys293, might explain the increase in dsRNA binding associated with the Lys16 mutation.
Certain mutations in σ3 have been observed to cause extragenic suppression of ts mutations in µ1. The tsA(201) clone contains one such mutation within the µ1-encoding M2 genome segment (Mustoe et al., 1978). Two suppressors of this mutation have been mapped to the S4 genome segment in two different extragenically suppressed revertants (McPhillips and Ramig, 1984). The revertant RtsA(201)-101 contains a single mutation in σ3: a Glu to Ala substitution at amino acid 217 (Jayasuriya, 1991). The other revertant, RtsA(201)-121, has a single Gly to Ser substitution at amino acid 47 in σ3 (Jayasuriya, 1991). Assuming that the extragenic suppression of the tsA(201) phenotype is due to compensating protein interactions, these locations on σ3 are likely to mark sites of interaction with µ1. Gly/Ser47 is located on strand 3, in agreement with the proposed µ1-binding surface. Glu/Ala217 is on a large loop between strands 7 and 8 in the large lobe, and it is less clear how this residue might participate in σ3–µ1 interaction. Our fit of the σ3 model into the reconstructed electron microscopy image indicates that the whole conserved face of the small lobe (left side of the molecule as viewed in Figure 5B) may interact with µ1. Because this surface is also part of the dimer contact, µ1 binding to σ3 and σ3 dimerization are probably mutually exclusive. The apparent µ1-binding surface is more extensive than the dimerization surface, consistent with the higher affinity of the µ1–σ3 interaction.
dsRNA binding
Despite its position on the outermost surface of the virion, σ3 is a dsRNA-binding protein, binding specifically to RNA duplexes of minimal length 32–45 bp (Yue and Shatkin, 1997). This capability is tightly linked to its capacity to block the dsRNA-dependent activation of PKR and to stimulate translation (Lloyd and Shatkin, 1992; Beattie et al., 1995; Yue and Shatkin, 1997; Bergeron et al., 1998). Gel-mobility shift assays have shown that the presence of µ1 blocks dsRNA binding by σ3 (Yue and Shatkin, 1997), and co-expression of µ1 with σ3 in transfected cells negates the translation-stimulatory effect of σ3 alone (Tillotson and Shatkin, 1992). Thus, σ3 that is not complexed with µ1 appears to counter the interferon-induced shutdown of host protein synthesis in the cytoplasm of infected cells by binding dsRNA and preventing PKR activation. Consistent with this hypothesis, high levels of µ1-bound σ3 correlate with translational inhibition in reovirus-infected cells (Schmechel et al., 1997).
While agarose–polyriboinosinic acid–polyribocytidylic acid binding experiments show that the baculovirus-expressed protein we used for crystallization binds to dsRNA (data not shown), we can not yet identify the binding site on the structure with certainty. Arg236, Arg239, Lys291 and Lys293 have been implicated by mutational analysis in dsRNA binding (Denzler and Jacobs, 1994), but these residues are located on distant surfaces of the structure (helices E and F, respectively), making it difficult to incorporate all four of them into one putative binding region. Because arginines 236 and 239, located on helix E at the end of the large lobe, are completely buried, it is not immediately apparent how they could contribute to dsRNA binding. Instead, because both of these residues make extensive contacts with neighboring atoms, they are likely to be important for stabilizing the overall structure of the large lobe. They may thus influence dsRNA binding indirectly. Lysines 291 and 293 are located on helix F of the small lobe, near the central groove that separates the lobes. Lys291 is buried and makes contacts with other atoms, but Lys293 protrudes from the surface and makes no contacts with other atoms in the structure. It is the best candidate for a direct RNA contact. It is also part of the positively charged patch on the σ3 surface, as described above.
Although the dsRNA-binding surface on σ3 can not be identified with certainty, it is clear that it is not similar to the previously characterized dsRNA-binding domains (RBDs) of other dsRNA-binding proteins such as PKR (Nanduri et al., 1998), the Xenopus laevis RNA-binding protein A (Ryter and Schultz, 1998), the Drosophila staufen protein (Bycroft and Grunert, 1995) and Escherichia coli RNase III (Kharrat et al., 1995). The RBDs of these proteins are composed of two helices flanking a three-stranded anti-parallel β-sheet. Based on secondary structure predictions, it had been suggested that in σ3 the residues implicated in dsRNA binding would flank a β-sheet, and this region (amino acids 234–300) was proposed to form a domain similar to these other dsRBDs (Yue and Shatkin, 1996). In the structure presented here, we see that the residues implicated in dsRNA binding are spread out along the side of the protein and do not contact each other. Thus, they do not form any type of domain or structure on their own, even though there is in fact a β-sheet in the sequence between helix E and helix F.
We propose that the form of σ3 that binds dsRNA is the dimer seen in the crystallographic asymmetric unit. Cooperativity in binding of σ3 to dsRNA would enhance the selective coverage of double-stranded regions of RNA, once the intracellular concentration of σ3 attained a critical level. Our proposal, that σ3 binds RNA as a dimer, leads to a simple picture for this cooperative association. Contributions to cooperativity will include not only the dimer interactions seen in our structure, but also contacts between successive dimers bound with their 2-fold axes aligned along local dyads of the RNA. Thus, interactions between the ‘top’ and ‘bottom’ surfaces of the dimer, as viewed in Figure 7, could also enhance RNA affinity. If aligned as proposed, each dimer would cover approximately two turns of an RNA double helix.
Participation of the dimer in RNA binding may also allow µ1 to act as a regulatory switch. When sufficient µ1 is present, formation of µ1–σ3 complexes will compete with σ3 dimer formation and shift the usage of σ3 away from dsRNA binding.
Materials and methods
Expression and purification
σ3 protein was expressed in Sf21 cells infected with a baculovirus carrying the reovirus S4 gene (Jané-Valbuena et al., 1999). The cytoplasmic and nuclear fractions of cells were separated. Nuclei were resuspended in phosphate-buffered saline supplemented with NaCl to a final concentration of 500 mM, and DNA and cellular debris pelleted at 20 000 g for 15 min. The supernatant was diluted 6-fold in 10 mM Tris–HCl pH 7.4 and 5 mM MgCl2 to yield a final NaCl concentration of 83 mM. This solution was applied to a heparin–Sepharose column and washed with 100 mM NaCl buffer. σ3 was eluted with a step to 200 mM NaCl. The eluate was diluted 2-fold in 10 mM Tris–HCl pH 7.4 and 5 mM MgCl2 to yield a final NaCl concentration of 100 mM. This was loaded onto a Mono-Q column (Amersham Pharmacia Biotech) and eluted with a gradient to 500 mM NaCl. The σ3 peak was collected, adjusted to 500 mM NaCl and concentrated to <2.0 ml. In a final purification step, σ3 was run through a Superdex-200 size exclusion column (Amersham Pharmacia Biotech) in 500 mM NaCl, 10 mM Tris–HCl pH 8.0 and 5 mM MgCl2.
Crystallization
Crystals were grown by hanging drop vapor diffusion at 4°C in drops containing 1.5 µl of protein stock solution (15 mg/ml protein, 10 mM Tris pH 8.0, 5 mM MgCl2, 500 mM NaCl) mixed with 1.5 µl of drop solution [50 mM Tris–HCl pH 8.3, 5 mM MgCl2, 300 mM NaCl, 1–3% polyethylene glycol (PEG) 6000]. The drops were equilibrated against a well solution identical to the drop solution with the exception of having 800 mM NaCl. Crystals grew in 2–4 weeks to a size of 200 × 200 × 400 µm. The crystals were harvested into a solution of 50 mM HEPES pH 7.5, 600 mM MgSO4, 5% PEG 6000 and 10% glycerol, and left to equilibrate for several hours at 4°C for 1 h to overnight. The KAu(CN)2 derivative was obtained by soaking in the same harvest buffer supplemented with 2.0 mM KAu(CN)2 for 2 days at 4°C. Crystals were backsoaked in unsupplemented harvest buffer before freezing. The Pt(NH3)2Cl2 derivative was obtained by soaking the crystal in harvest buffer containing 2.0 mM Pt(NH3)2Cl2 for 4 days, and backsoaking before freezing. The K3UO2F5 derivative was obtained by soaking the crystal in harvest buffer with 0.2 mM K3UO2F5 for 5 days. The Au and Pt double derivative was obtained as described above for these compounds individually, but soaked together for 4 days.
Data collection and processing
Crystals belong to the space group P212121 with unit cell dimensions a = 77.9, b = 82.6, c = 132.3 Å, α = β = γ = 90°. They contain two σ3 molecules per asymmetric unit, implying a solvent content of ∼52%. Crystals were sequentially equilibrated for 30 min in harvest buffer with glycerol concentrations of 15, 20 and 25%, then flash frozen in liquid nitrogen. The data collection statistics are shown in Table I. Native data were recorded at the X25 beamline of the National Synchrotron Lightsource at Brookhaven National Laboratory using the Brandeis 4-element CCD detector. MAD data from the KAu(CN)2 derivative were recorded at the X12C beamline of the same synchrotron using the B1.2 Brandeis one-module detector. The X-ray fluorescence and transmission from the derivative crystal were measured as functions of incident X-ray energy in the vicinity of the gold L-III edge. Two energies were chosen near the absorption edge: 11 924 eV (λ = 1.03979 Å) and 11 929 eV (λ = 1.03938 Å), corresponding to the minimum f′ and maximum f′′, respectively. Two remote energies were selected at 12 203 eV (λ = 1.01600 Å) and 11 916 eV (λ = 1.04049 Å). A second data set from a KAu(CN)2 derivative was recorded at the Advanced Photon Source on a Quantum-4 detector at λ = 1.0000 Å. All other derivative data were recorded in our laboratory using a rotating anode X-ray generator coupled with a Mar345 image plate detector. Intensities were integrated and scaled using the programs DENZO and SCALEPACK (Otwinowski and Minor, 1997), respectively.
Table I. Data collection, phase determination and refinement statistics.
| Data set | Native | A | P | U | A + P | A2 |
|---|---|---|---|---|---|---|
| Wavelength (Å) | 1.1000 | 1.5418 | 1.5418 | 1.5418 | 1.5418 | 1.0000 |
| Resolution (Å) | 25–1.8 | 25–3.0 | 25–2.5 | 25–3.5 | 25–3.0 | 25–3.0 |
| Unique reflections | 79 737 | 17 747 | 30 435 | 11 373 | 17 871 | 17 586 |
| Redundancy | 5.7 | 5.6 | 4.5 | 3.8 | 5.4 | 5.0 |
| Completeness (%) | 99.1 | 99.9 | 99.1 | 87.2 | 97.3 | 97.5 |
| I/σ | 14.2 | 17.3 | 24.3 | 16.6 | 22.1 | 12.1 |
|
Rsym (%) |
5.9 (10.7) |
7.8 (17.4) |
5.3 (12.2) |
6.8 (12.8) |
6.1 (9.5) |
9.1 (18.3) |
| Phase determination (MIR) | ||||||
| Riso (%) | 17.5 | 19.4 | 12.4 | 23.7 | 19.4 | |
| No. of heavy atom sites | 3 | 9 | 4 | 5 | 3 | |
| Rcullis (centric/acentric) | 0.83/0.83 | 0.84/0.89 | 0.96/0.97 | 0.85/0.84 | 0.87/0.87 | |
| Phasing power (centric/acentric) | 0.53/0.71 | 0.58/0.74 | 0.31/0.40 | 0.49/0.64 | 0.39/0.51 | |
| Mean figure of merit |
|
0.5660 |
|
|
|
|
| Phase determination (MAD) | ||||||
| Data set |
|
f′ min |
f′′ max |
Remote I |
Remote II |
|
| Wavelength (Å) | 1.03979 | 1.03938 | 1.01600 | 1.04049 | ||
| Resolution (Å) | 25–2.4 | 25–2.4 | 25–2.4 | 25–2.4 | ||
| Rsym (%) | 5.4 (10.3) | 4.5 (9.5) | 4.7 (9.7) | 4.3 (9.2) | ||
| Ranom (%) | 2.7 (6.2) | 2.3 (5.6) | 2.7 (5.8) | 2.1 (5.6) | ||
| Unique reflections | 33 985 | 33 958 | 34 038 | 33 960 | ||
| Redundancy | 6.5 | 6.4 | 6.5 | 4.9 | ||
| Completeness | 94.8 | 94.9 | 96.5 | 90.9 | ||
| Anom. completeness | 94.6 | 94.7 | 96.2 | 90.7 | ||
| I/σ | 22.7 | 24.4 | 23.2 | 23.8 | ||
| Mean figure of merit |
|
0.5980 |
|
|
|
|
| Refinement |
25.0–1.8 Å all data |
|
|
|
|
|
| R factor (%) | 16.9 | 76 538 reflections | ||||
| Free R factor (%) | 19.5 | 2390 reflections | ||||
| R.m.s. bond length (Å) | 0.012 | |||||
| R.m.s. bond angles (°) | 1.54 | |||||
A = KAu(CN)2.
P = Pt(NH3)2Cl2.
U = K3UO2F5.
A + P = double soak in A and P.
A2 = KAu(CN)2.
Values of Rsym and Ranom in parentheses are for the outer resolution shell.
Rsym = Σ|Ii – <I>|/ΣIi
Riso = Σ||Fder| – |Fnat||/Σ|Fnat|
Ranom = Σ|<I+> – <I–>|/Σ|<I+> + <I–>|
Phasing, model building and refinement
The structure was determined by combining phases from MIR from four different derivative crystals with phases from MAD. Difference Patterson maps were calculated from all the isomorphous derivative data sets, and one of the gold atoms was located by visually solving the Patterson map for the KAu(CN)2 derivative. All other atoms both in that derivative and others were found by examination of difference Fourier maps. Phases were calculated from these derivatives by MLPHARE (CCP4, 1994), and improved by solvent flattening with the program DM. Including the anomalous occupancies in the parameters to be refined by MLPHARE allowed us to determine the correct hand. A different crystal derivatized with the same Au compound was then used to collect a MAD data set. The Remote II data set (see Table I) was used as the native against which dispersive differences were phased from the other three data sets, because of lack of confidence in having accurately targeted the minimum f′ wavelength. Anomalous differences were used for phase generation in all data sets except Remote I. MAD phases were again improved with the solvent flattening program DM. Phases from these two solutions were combined with the program SIGMAA. Two-fold averaging and phase extension allowed significant extension of the resolution of the map. The high-resolution native data were used for crystallographic refinement to 1.8 Å. The complete model was built into this experimental map. After several iterative cycles of refinement using CNS (Brünger et al., 1998) and model improvement, water molecules were placed automatically in CNS. A simulated annealing composite-omit map was calculated to check the final model. Final Rwork and Rfree values of 16.9 and 19.5% were obtained. The stereochemistry of the model was analyzed with PROCHECK (Laskowski et al., 1993); 90% of the polypeptide backbone dihedral angles were found to lie in the most favorable regions of the Ramachandran plot, with the remainder in allowed regions.
Fitting the σ3 model into the virion image reconstruction from electron cryomicroscopy
To fit our crystallographic model into an 18 Å resolution image reconstruction of the virion, the model was first positioned by visual inspection (program O; Jones et al., 1991) into one of the knobs in a σ3 ring of six, with the small lobe packed against µ1. The virion map was then restricted to contain only one σ3 molecule (with EXTEND in the CCP4 suite), and using the roughly positioned model of σ3, an 18 Å resolution electron density map was calculated (with SFALL and FFT in the CCP4 suite). This calculated map and the restricted virion map were used in a real-space maximization of the correlation of the corresponding electron densities (with MAVE in the RAVE suite; Jones, 1992). The correlation was calculated pixel by pixel in the overlapping maps, inside a mask derived from the structure. The optimization was performed by automatic alternation of translational and rotational searches. The initial translation step size was 0.5 Å. The initial rotation step size was 2°, with a convergence step size of 0.1. The value for swapping from translation to rotation was 2.0 × e–3; the criterion for correlation coefficient convergence was set to 1.0 × e–5. The sample density was 1.0 (all points used).
Preparation of figures
Figures were prepared with RIBBONS (Carson, 1991), GRASP (Nicholls et al., 1991) and O (Jones et al., 1991).
Coordinates
The atomic coordinates are being deposited in the Protein Data Bank (PDB ID 1FN9 and RCSB ID RCSB011736).
Acknowledgments
Acknowledgements
We thank M.Jacobs, X.Chen, P.Sliz and members of the Harrison and Wiley research groups for advice; R.Sweet and the staff of beamlines X25 and X12C at NSLS for assistance; and S.B.Walter and T.S.Baker for the image reconstruction of a reovirion used in Figure 6. This work was supported by NIH/NCI grant CA-13202 (to S.C.H.), NIH grant R01 A1-32139 and ACS grant RPG-98-127-01 (to L.A.S.), and DARPA research contract MDA 972-97-1-0005 (to M.L.N.). J.J.-V. was supported by a Steenbock/Wharton Fellowship from the Department of Biochemistry. M.L.N. was also supported by the Milwaukee Foundation. S.C.H. is an investigator in the Howard Hughes Medical Institute.
References
- Ahmed R. and Fields,B.N. (1982) Role of the S4 gene in the establishment of persistent reovirus infection in L cells. Cell, 28, 605–612. [DOI] [PubMed] [Google Scholar]
- Baer G.S. and Dermody,T.S. (1997) Mutations in reovirus outer-capsid protein σ3 selected during persistent infection of L cells confer resistance to protease inhibitor E64. J. Virol., 71, 4921–4928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass D.M., Bodkin,D., Dambrauskas,R., Trier,J.S., Fields,B.N. and Wolf,J.L. (1990) Intraluminal proteolytic activation plays an important role in replication of type 1 reovirus in the intestines of neonatal mice. J. Virol., 64, 1830–1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beattie E., Denzler,K., Tartaglia,J., Perkus,M., Paoletti,E. and Jacobs,B.L. (1995) Reversal of the interferon-sensitive phenotype of a vaccinia virus lacking E3L by expression of the reovirus S4 gene. J. Virol., 69, 499–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergeron J., Mabrouk,T., Garzon,S. and Lemay,G. (1998) Characterization of the thermosensitive ts453 reovirus mutant: increased dsRNA binding of σ3 protein correlates with interferon resistance. Virology, 246, 199–210. [DOI] [PubMed] [Google Scholar]
- Bodkin D.K. (1989) Proteolytic digestion of reovirus in the intestinal lumens of neonatal mice. J. Virol., 63, 4676–4681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsa J., Sargent,M.D., Lievaart,P.A. and Copps,T.P. (1981) Reovirus: evidence for a second step in the intracellular uncoating and transcriptase activation process. Virology, 111, 191–200. [DOI] [PubMed] [Google Scholar]
- Branden C. and Tooze,J. (1991) Introduction to Protein Structure. Garland Publishing, Inc., New York, NY.
- Brünger A.T. et al. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Bycroft M. and Grunert,S. (1995) NMR solution structure of a dsRNA binding domain from Drosophila staufen protein reveals homology to the N-terminal domain of ribosomal protein S5. EMBO J., 14, 3563–3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson M. (1991) Ribbons 2.0. J. Appl. Crystallogr., 24, 958–961. [Google Scholar]
- Chandran K., Walker,S.B., Chen,Y., Contreras,C.M., Schiff,L.A., Baker,T.S. and Nibert,M.L. (1999) In vitro recoating of reovirus cores with baculovirus-expressed outer-capsid proteins µ1 and σ3. J. Virol., 73, 3941–3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.-T. and Zweerink,H.J. (1971) Fate of parental reovirus in infected cells. Virology, 46, 544–555. [DOI] [PubMed] [Google Scholar]
- Chang H.-W., Watson,J. and Jacobs,B.L. (1992) The vaccinia virus E3L gene encodes a double-stranded RNA-binding protein with inhibitory activity for the interferon-induced protein kinase. Proc. Natl Acad. Sci. USA, 89, 4825–4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens M.J. (1996) Protein kinases that phosphorylate eIF2 and eIF2B and their role in eukaryotic cell translational control. In Hershey,J., Mathews,M. and Sonenberg,N. (eds), Translational Control. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 139–172.
- Collaborative Computational Project No. 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Danis C., Garzon,S. and Lemay,G. (1992) Further characterization of the ts453 mutant of mammalian orthoreovirus serotype 3 and nucleotide sequence of the mutated S4 gene. Virology, 190, 494–498. [DOI] [PubMed] [Google Scholar]
- De Guzman R.N., Wu,Z.R., Stalling,C.C., Pappalardo,L., Borer,P.N. and Summers,M.F. (1998) Structure of the HIV-1 nucleocapsid protein bound to the SL3 ψ-RNA recognition element. Science, 279, 384–388. [DOI] [PubMed] [Google Scholar]
- Denzler K.L. and Jacobs,B.L. (1994) Site-directed mutagenic analysis of reovirus σ3 protein binding to dsRNA. Virology, 204, 190–199. [DOI] [PubMed] [Google Scholar]
- Dermody T.S., Nibert,M.L., Wetzel,J.D., Tong,X. and Fields,B.N. (1993) Cells and viruses with mutations affecting viral entry are selected during persistent infections of L cells with mammalian reoviruses. J. Virol., 67, 2055–2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dryden K.A., Wang,G., Yeager,M., Nibert,M.L., Coombs,K.M., Furlong,D.B., Fields,B.N. and Baker,T.S. (1993) Early steps in reovirus infection are associated with dramatic changes in supramolecular structure and protein conformation: analysis of virions and subviral particles by cryoelectron microscopy and image reconstruction. J. Cell Biol., 122, 1023–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields B.N. (1971) Temperature-sensitive mutants of reovirus type 3: features of genetic recombination. Virology, 46, 142–148. [DOI] [PubMed] [Google Scholar]
- Furlong D.B., Nibert,M.L. and Fields,B.N. (1988) σ1 protein of mammalian reoviruses extends from the surfaces of viral proteins. J. Virol., 62, 246–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giantini M. and Shatkin,A.J. (1989) Stimulation of chloramphenicol acetyltransferase mRNA translation by reovirus capsid polypeptide σ3 in cotransfected COS cells. J. Virol., 63, 2415–2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L. and Sander,C. (1995) DALI: a network tool for protein structure comparison. Trends Biochem. Sci., 20, 478–480. [DOI] [PubMed] [Google Scholar]
- Hooper J.W. and Fields,B.N. (1996) Role of the µ1 protein in reovirus stability and capacity to cause chromium release from host cells. J. Virol., 70, 459–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovanessian A.G. (1991) Interferon-induced and double-stranded RNA-activated enzymes: a specific protein kinase and 2′,5′-oligoadenylate synthetase. J. Interferon Res., 11, 199–205. [DOI] [PubMed] [Google Scholar]
- Huismans H. and Joklik,W.K. (1976) Reovirus-coded polypeptides in infected cells: isolation of two native monomeric polypeptides with affinity for single-stranded and double-stranded RNA, respectively. Virology, 70, 411–424. [DOI] [PubMed] [Google Scholar]
- Imani F. and Jacobs,B.L. (1988) Inhibitory activity for the interferon-induced protein kinase is associated with the reovirus serotype 1 σ3 protein. Proc. Natl Acad. Sci. USA, 85, 7887–7891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jané-Valbuena J., Nibert,M.L., Spencer,S.M., Walker,S.B., Baker,T.S., Chen,Y., Centonze,V.E. and Schiff,L.A. (1999) Reovirus virion-like particles obtained by recoating infectious subvirion particles with baculovirus-expressed σ3 protein: an approach for analyzing σ3 functions during virus entry. J. Virol., 73, 2963–2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayasuriya A.K.A. (1991) Molecular characterization of the reovirus M2 gene. PhD thesis, Harvard University, Cambridge, MA.
- Joklik W.K. (1972) Studies on the effect of chymotrypsin on reoviruses. Virology, 49, 700–715. [DOI] [PubMed] [Google Scholar]
- Jones T.A. (1992) A set of averaging programs. In Dodson,E.J., Gover,S. and Wolf,W. (eds), Molecular Replacement. SERC Daresbury Laboratory, Warrington, UK, pp. 91–105.
- Jones T.A., Zou,J.Y., Cowans,S.W. and Kjeldgaard,M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Kedl R., Schmechel,S. and Schiff,L. (1995) Comparative sequence analysis of the reovirus S4 genes from 13 serotype 1 and serotype 3 field isolates. J. Virol., 69, 552–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharrat A., Macias,M.J., Gibson,T.J., Nilges,M. and Pastore,A. (1995) Structure of the dsRNA binding domain of E.coli RNase III. EMBO J., 14, 3572–3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski R., MacArthur,M., Moss,D. and Thornton,J. (1993) PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr., 26, 91–97. [Google Scholar]
- Lee P.W. and Gilmore,R. (1998) Reovirus cell attachment protein σ1: structure–function relationships and biogenesis. Curr. Top. Microbiol. Immunol., 233I, 137–153. [DOI] [PubMed] [Google Scholar]
- Lee P.W., Hayes,E.C. and Joklik,W.K. (1981) Protein σ1 is the reovirus cell attachment protein. Virology, 108, 156–163. [DOI] [PubMed] [Google Scholar]
- Lloyd R.M. and Shatkin,A.J. (1992) Translational stimulation by reovirus polypeptide σ3: substitution for VAI RNA and inhibition of phosphorylation of the α subunit of eukaryotic initiation factor 2. J. Virol., 66, 6878–6884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucia-Jandris P., Hooper,J.W. and Fields,B.N. (1993) Reovirus M2 gene is associated with chromium release from mouse L cells. J. Virol., 67, 5339–5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabrouk T. and Lemay,G. (1994a) Mutations in a CCHC zinc-binding motif of the reovirus σ3 protein decrease its intracellular stability. J. Virol., 68, 5287–5290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabrouk T. and Lemay,G. (1994b) The sequence similarity of reovirus σ3 protein to picornaviral protease is unrelated to its role in µ1 viral protein cleavage. Virology, 202, 615–620. [DOI] [PubMed] [Google Scholar]
- Mathews M.B. (1996) Interactions between viruses and the cellular machinery for protein synthesis. In Hershey,J.W.B., Mathews,M.B. and Sonenberg,N. (eds), Translational Control. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 505–548.
- Mathews M.B. and Shenk,T. (1991) Adenovirus virus-associated RNA and translational control. J. Virol., 65, 5657–5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPhillips T.H. and Ramig,R.F. (1984) Extragenic suppression of temperature-sensitive phenotype in reovirus: mapping suppressor mutations. Virology, 135, 428–439. [DOI] [PubMed] [Google Scholar]
- Metcalf P. (1982) The symmetry of the reovirus outer shell. J. Ultrastruct. Res., 78, 292–301. [DOI] [PubMed] [Google Scholar]
- Morgan E.M. and Zweerink,H.J. (1974) Reovirus morphogenesis. Corelike particles in cells infected with wild-type reovirus and temperature sensitive mutants of groups B and G. Virology, 59, 556–565. [DOI] [PubMed] [Google Scholar]
- Mustoe T.A., Ramig,R.F., Sharpe,A.H. and Fields,B.N. (1978) A genetic map of reovirus III. Assignment of the double-stranded RNA-positive mutant groups A, B and G to genome segments. Virology, 85, 545–556. [DOI] [PubMed] [Google Scholar]
- Nanduri S., Carpick,B.W., Yang,Y., Williams,B.R.G. and Qin,J. (1998) Structure of the double-stranded RNA-binding domain of the protein kinase PKR reveals the molecular basis of its dsRNA-mediated activation. EMBO J., 17, 5458–5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nibert M.L., Chappell,J.D. and Dermody,T.S. (1995) Infectious subvirion particles of reovirus type 3 Dearing exhibit a loss in infectivity and contain a cleaved σ1 protein. J. Virol., 69, 5057–5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nibert M.L., Schiff,L.A. and Fields,B.N. (1996) Reoviruses and their replication. In Fields,B.N., Knipe,D.M. and Howley,P.M. (eds), Fundamental Virology. Lippincott–Raven, Philadelphia, PA, pp. 691–730.
- Nicholls A., Sharp,K.A. and Honig,B. (1991) Protein folding and association: insights from interfacial and thermodynamics properties of hydrocarbons. Proteins, 11, 281–296. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor,W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Reinisch K.M., Nibert,M.L. and Harrison,S.C. (2000) Structure of the reovirus core at 3.6 Å resolution. Nature, 404, 960–967. [DOI] [PubMed] [Google Scholar]
- Ryter J.M. and Schultz,S.C. (1998) Molecular basis of double-stranded RNA–protein interactions: structure of a dsRNA-binding domain complexed with dsRNA. EMBO J., 17, 7505–7513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmechel S., Chute,M., Skinner,P., Anderson,R. and Schiff,L. (1997) Preferential translation of reovirus mRNA by a σ3-dependent mechanism. Virology, 232, 62–73. [DOI] [PubMed] [Google Scholar]
- Seliger L.S., Giantini,M. and Shatkin,A.J. (1992) Translational effects and sequence comparisons of the three serotypes of the reovirus S4 gene. Virology, 187, 202–210. [DOI] [PubMed] [Google Scholar]
- Sharpe A.H. and Fields,B.N. (1982) Reovirus inhibition of cellular RNA and protein synthesis: role of the S4 gene. Virology, 122, 381–391. [DOI] [PubMed] [Google Scholar]
- Shatkin A.J. and LaFiandra,A.J. (1972) Transcription by infectious subviral particles of reovirus. J. Virol., 10, 698–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard D.A., Ehnstrom,J.G. and Schiff,L.A. (1995) Association of reovirus outer capsid proteins σ3 and µ1 causes a conformational change that renders σ3 protease sensitive. J. Virol., 69, 8180–8184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard D.A., Ehnstrom,J.G., Skinner,P.J. and Schiff,L.A. (1996) Mutations in the zinc-binding motif of the reovirus capsid protein σ3 eliminate its ability to associate with capsid protein µ1. J. Virol., 70, 2065–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shing M. and Coombs,K.M. (1996) Assembly of the reovirus outer capsid requires µ1/σ3 interactions which are prevented by misfolded σ3 proteins in reovirus temperature-sensitive mutant tsG453. Virus Res., 46, 19–29. [DOI] [PubMed] [Google Scholar]
- Silverstein S.C., Schonberg,M., Levin,D.H. and Acs,G. (1970) The reovirus replicative cycle: conservation of parental RNA and protein. Proc. Natl Acad. Sci. USA, 67, 275–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturzenbecker L.J., Nibert,M., Furlong,D. and Fields,B.N. (1987) Intracellular digestion of reovirus particles requires a low pH and is an essential step in the viral infectious cycle. J. Virol., 61, 2351–2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillotson L. and Shatkin,A.J. (1992) Reovirus polypeptide σ3 and N-terminal myristoylation of polypeptide µ1 are required for site specific cleavage to µ1C in transfected cells. J. Virol., 66, 2180–2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosteson M.T., Nibert,M.L. and Fields,B.N. (1993) Ion channels induced in lipid bilayers by subvirion particles of the nonenveloped mammalian reoviruses. Proc. Natl Acad. Sci. USA, 90, 10549–10552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virgin H.W. IV, Mann,M.A., Fields,B.N. and Tyler,K.L. (1991) Monoclonal antibodies to reovirus reveal structure/function relationships between capsid proteins and genetics of susceptibility to antibody action. J. Virol., 65, 6772–6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virgin H.W. IV, Mann,M.A. and Tyler,K.L. (1994) Protective antibodies inhibit reovirus internalization and uncoating by intracellular proteases. J. Virol., 68, 6719–6729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetzel J.D., Wilson,G.J., Baer,G.S., Dunnigan,L.R., Wright,J.P., Tang,D.S.H. and Dermody,T.S. (1997) Reovirus variants selected during persistent infections of L cells contain mutations in the viral S1 and S4 genes and are altered in viral disassembly. J. Virol., 71, 1362–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Z. and Shatkin,A.J. (1996) Regulated, stable expression and nuclear presence of reovirus double-stranded RNA-binding protein σ3 in HeLa cells. J. Virol., 70, 3497–3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Z. and Shatkin,A.J. (1997) Double-stranded RNA-dependent protein kinase (PKR) is regulated by reovirus structural proteins. Virology, 234, 364–371. [DOI] [PubMed] [Google Scholar]