Abstract
The specificity of intracellular signaling events is controlled, in part, by compartmentalization of protein kinases and phosphatases. The subcellular localization of these enzymes is often maintained by protein– protein interactions. A prototypic example is the compartmentalization of the cAMP-dependent protein kinase (PKA) through its association with A-kinase anchoring proteins (AKAPs). A docking and dimerization domain (D/D) located within the first 45 residues of each regulatory (R) subunit protomer forms a high affinity binding site for its anchoring partner. We now report the structures of two D/D–AKAP peptide complexes obtained by solution NMR methods, one with Ht31(493–515) and the other with AKAP79(392–413). We present the first direct structural data demonstrating the helical nature of the peptides. The structures reveal conserved hydrophobic interaction surfaces on the helical AKAP peptides and the PKA R subunit, which are responsible for mediating the high affinity association in the complexes. In a departure from the dimer–dimer interactions seen in other X-type four-helix bundle dimeric proteins, our structures reveal a novel hydrophobic groove that accommodates one AKAP per RIIα D/D.
Keywords: AKAP/PKA/NMR/signal transduction/subcellular localization
Introduction
The molecular mechanisms that govern specificity in signal transduction events have become a topic of considerable research emphasis. Recent evidence suggests that where and when signal transduction events occur are influenced by the assembly of multiprotein complexes, which are composed of protein kinases, their effector molecules and selected substrates (Hunter, 2000). At the core of these complexes are anchoring, adapter or scaffold proteins, which provide a molecular framework on which the signaling components are assembled. These organizing proteins are often composed of modular domains that provide sites of contact with reciprocal binding surfaces on acceptor molecules (Pawson and Nash, 2000).
Compartmentalization of the cAMP-dependent protein kinase (PKA) is one well studied example of protein kinase targeting (reviewed by Colledge and Scott, 1999). The PKA holoenzyme consists of two catalytic (C) subunits and a regulatory (R) subunit dimer (Taylor et al., 1990). While phosphorylation of substrates is catalyzed by the C subunits, the subcellular location of the holoenzyme is maintained through protein–protein interactions between the R subunit dimer and A-kinase anchoring proteins (AKAPs) (Luo et al., 1990; Scott et al., 1990). The N-terminal regions of each R subunit protomer are responsible for the high affinity interaction with the AKAP (Hausken et al., 1994; Hausken and Scott, 1996). Since this region serves at least two functions: dimerization and docking to AKAPs, we have designated this module the D/D domain. Structural analysis of the type IIα D/D domain revealed a well packed, antiparallel, dimeric X-type four-helix bundle with C2 symmetry (Newlon et al., 1997, 1999).
Reciprocal studies have defined the binding surface on AKAPs that associates with the D/D domains of RII. Early mapping studies defined a primary sequence of 20 amino acids in the microtubule-associated protein 2 (MAP2) responsible for interaction with the PKA holoenzyme (Obar et al., 1989; Rubino et al., 1989). Later it was noticed that this sequence, and corresponding sequences of other AKAPs, exhibited a high probability of amphipathic helix formation (Carr et al., 1991). Although no strong consensus sequence has emerged, helical wheel alignments on over 20 AKAPs now suggest that polar and hydrophobic side chains are arranged on opposite faces of the helix (Carr et al., 1992b; Vijayaraghavan et al., 1999). Identification of an amphipathic helix is considered a defining characteristic of AKAPs, although some, such as the centrosomal anchoring protein pericentrin, appear to lack this structure (Diviani et al., 2000). A more reliable classification is to co-precipitate PKA holoenzyme or kinase activity with a candidate protein from inside cells (Colledge and Scott, 1999; Kapiloff et al., 1999; Westphal et al., 1999).
The human thyroid anchoring protein Ht31 is the prototype of the amphipathic helix model. Insertion of helix-breaking prolines at sites within the putative helix abolishes Ht31 interaction with RII in vitro and in vivo (Carr et al. 1992a,b; Hausken et al., 1996). Likewise, substitution of branched-chain hydrophobic side chains in pairs significantly reduces the PKA anchoring function (Glantz et al., 1993). Similar experiments have been performed with several AKAPs (Carr and Scott, 1992; Glantz et al., 1993; Kapiloff et al., 1999; Miki and Eddy, 1999). Peptide studies provide additional confirmation that the putative amphipathic helix of Ht31 contains PKA anchoring determinants. Peptides encompassing residues 493–515 of Ht31 bind RII and the PKA holoenzyme with nanomolar affinities (Carr et al., 1992a; Burton et al., 1997; Herberg et al., 2000). The Ht31 peptides are valuable reagents for the disruption of PKA anchoring inside cells (Carr et al., 1992a; Rosenmund et al., 1994; Burton et al., 1997). Consequently, these anchoring inhibitor reagents have been used to demonstrate a role for PKA anchoring in the regulation of a variety of cAMP-responsive events, including modulation of ion channels (Fraser and Scott, 1999), arrest of sperm motility (Vijayaraghavan et al., 1997), hormone-mediated insulin secretion (Lester et al., 1997) and phosphorylation of the transcription factor CREB (Feliciello et al., 1997).
Other well studied anchoring proteins are the AKAP79/150 family, composed of human AKAP79, the mouse ortholog AKAP150 and the bovine ortholog AKAP75 (Dodge and Scott, 2000). These are neuronal scaffolding proteins that coordinate the location of a signaling complex containing not only PKA, but also protein kinase C (PKC) and the calcium calmodulin-dependent phosphatase PP 2B (Coghlan et al., 1995; Klauck et al., 1996). The function of this signaling complex is to position two second messenger-regulated kinases and a phosphatase near to neuronal substrates at the postsynaptic densities (Faux and Scott, 1996). Recently it has been shown that the AKAP79/150 proteins are recruited into a larger signaling unit through association with the synaptic adapter protein PSD 95 or SAP-97 (Colledge et al., 2000; Husi et al., 2000). These elaborate protein–protein interactions provide a molecular bridge linking the AKAP79 signaling complex to a known substrate, the AMPA-responsive glutamate receptors. Functional studies demonstrate that assembly of the AKAP79–SAP-97 complex inside cells enhances PKA phosphorylation of serine 845 on the AMPA receptor (Colledge et al., 2000). In contrast, cAMP-responsive phosphorylation of serine 845 is prevented when a similar complex is formed with an AKAP79 mutant containing a disrupted amphipathic helix (Colledge et al., 2000).
The biochemical and functional studies performed with Ht31 and AKAP79 are based on a mechanism of PKA anchoring whereby an amphipathic helix provides an acceptor site for the D/D domain of the R subunit dimer. Yet there are no structural data to support this model. We now report the first structural details of two AKAP complexes of RIIα(1–44), one with Ht31(493–515) and the other with AKAP79(392–413). We see conserved organization of a hydrophobic interaction surface on both the AKAPs and the R subunit. These studies also identify the critical residues on both surfaces that participate in high affinity PKA anchoring interactions. Unlike other X-type four-helix bundle proteins involved in recruiting protein–protein interactions, RIIα binds AKAP in a novel mode where one AKAP helix binds at a 45° angle across the dimer interface in a well organized hydrophobic groove. Thus, despite conservation of a four-helix bundle platform, the stoichiometry and binding modes are controlled by the unique surface topology of the RIIα D/D.
Results
PKA targeting occurs through high affinity interactions of anchoring proteins with the D/D domain of the R subunit. In order to provide a molecular basis for these high affinity interactions, high resolution structural data are necessary. We employed triple-resonance, isotope filtering and three-dimensional (3D) nuclear magnetic resonance (NMR) techniques to determine the solution structures of two D/D–AKAP complexes, D/D–Ht31(493–515) and D/D–AKAP79(392–413).
Resonance assignments of the RIIα(1–44)–AKAP complexes
Sequence-specific assignments of the free and bound RIIα(1–44) D/D cross-peaks were facilitated by 13C and/or 15N incorporation. No significant RIIα protomer–protomer nuclear Overhauser enhancement (NOE) differences were observed in RIIα in the complexes compared with the free protein. The only significant changes occurred between residues that were involved in AKAP interactions. Specific protomer (protomer′) side chains in the RIIα(1–44) dimer were identified from the 3D 13C-edited (ω2)-12C-filtered (ω1)/13C-filtered (ω3) nuclear Overhauser enhancement spectroscopy (NOESY) (Ikura et al., 1992) experiment. The 3D 13C-edited (ω2)-12C-filtered (ω1)/13C-filtered (ω3) NOESY experiment also aided in the assignment of intermolecular NOEs between the D/D domain of RIIα and the AKAP peptides.
AKAP peptides bind to RIIα D/D with a stoichiometry of one peptide per RIIα dimer (Carr et al., 1992a; Newlon et al., 1999). Resonance assignments of the Ht31(493– 515) and AKAP79(392–413) peptides were determined on NMR samples of the D/D domain labeled with 15N, 13C and the natural abundance AKAP peptide. Double half-filter (DHF) experiments (Ikura and Bax, 1992; Slijper et al., 1996; Zwahlen et al., 1997) allowed nearly complete sequential 1H assignment of unlabeled Ht31(493–515) and AKAP79(392–413) in the complexes. Over 90% of the 1H resonance assignments of the 24-residue peptide of Ht31(493–515) and >85% of the 1H resonance assignments of a 22-residue peptide corresponding to AKAP79(392–413) were determined. The DHF-NOESY experiment identified either 170 or 94 intramolecular NOE cross-peaks in the Ht31 and the AKAP79 peptide, respectively.
Structures of the D/D–AKAP complexes
The structure of the D/D–AKAP complexes included 506 distance, 25 dihedral and 19 hydrogen bond restraints per monomer for RIIα(1–44). There were 170 distance and 10 hydrogen bond restraints for Ht31(493–515) and 94 distance and eight hydrogen bond restraints for AKAP79. Intermolecular restraints between the D/D and the AKAPs provided an additional 31 and 23 distance restraints for Ht31 and AKAP79, respectively.
Residues Ile5′, Leu13, Gln14, Thr17, Leu21, Leu21′ and Arg22 (where the protomers of RIIα are distinguished by the absence or presence of a ′) in RIIα show intermolecular NOEs to both Ht31 and AKAP79. There are also NOEs from residues Ile5, Leu9′, Thr10, Leu13′, Gln14′ and Val20′ in RIIα to Ht31. More NOEs were observed in the RIIα–Ht31 complex because, (i) the 13C-edited (ω2)-12C-filtered (ω1)/13C-filtered (ω3) NOESY for the RIIα–Ht31 complex had better signal to noise than that of the RIIα–AKAP79 complex, and (ii) several resonances of branched, hydrophobic protons in AKAP79 could not be assigned due to chemical shift degeneracy.
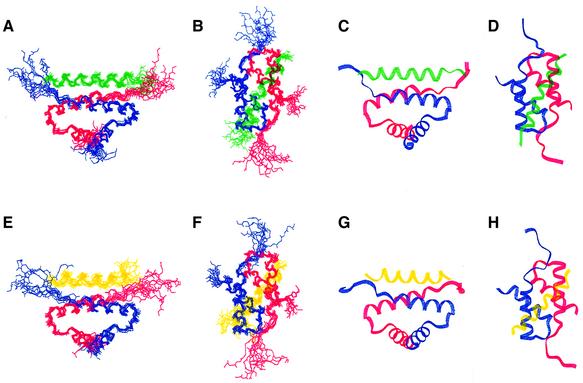
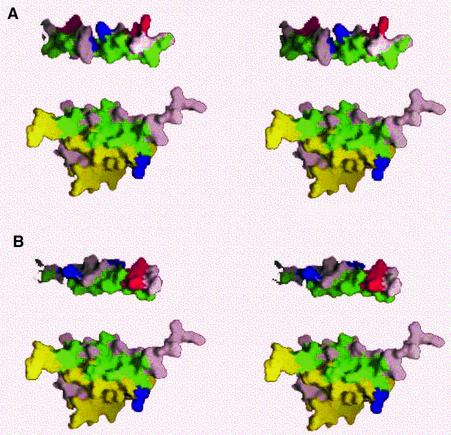
Fifty structures were generated for each complex in the final round of structure calculations. The final ensembles of the lowest energy structures for the D/D–Ht31 (493–515) complex (13 members, Figure 1A–D) and the D/D–AKAP79(392–408) complex (10 members, Figure 1E–H) were obtained with the program X-PLOR 3.851 (Brunger, 1993). Calculation of a dimer was aided by previously published protocols (Burgering et al., 1993; Nilges, 1993). All structures are well defined, with no NOE violations >0.5 Å and no dihedral angle violation >5° in either family. The lowest energy structures of the D/D–AKAP complexes were superimposed on the mean coordinate position for the backbone atoms (N, Cα, C′) of residues 9–42, 9′–42′ of the D/D and either residues 495–511 of Ht31 (Figure 1A–D) or residues 392–408 of AKAP79 (Figure 1E–H). Root mean square deviation (r.m.s.d.) values of backbone residues are presented in Table I for both complexes and reveal well defined structures with values of 0.69 and 0.64, respectively, for the backbone atoms 9–42 and 9′–42′ in RIIα in either the Ht31 or the AKAP79 complex. This precision is similar to the free D/D with r.m.s.d. values for the backbone residues 9–42 and 9′–42′ of 0.55 (Newlon et al., 1999).
Fig. 1. Backbone fold and AKAP peptide orientation of RIIα(1–44) with either Ht31(493–515) (views A–D) or AKAP79(392–413) (views E–H) bound. (A–D) Views of RIIα(1–44) (protomers in red and blue) and Ht31(493–515) in green are depicted. These views highlight two orientations of the best fit superposition of the backbone heavy atoms of the 13 lowest energy structures (A and B) or the lowest energy structure (C and D). The views (B) and (D) clarify the AKAP binding surface. (E–H) Similar views of RIIα(1–44) in red and blue and AKAP79(392–413) in yellow are shown. The best fit superposition of the 10 lowest energy structures (E and F) and the lowest energy structure (G and H) are represented. The two different views are shown in (A), (C), (E) and (G), and (B), (D), (F) and (H).
Table I. NMR structural statistics of the lowest energy structures of the RIIα(1–44)–AKAP complexes.
| NOE-derived restraints | No. of restraints |
||
|---|---|---|---|
| RIIα(1–44) (dimer) | Ht31(493–515) | AKAP79(392–408) | |
| Type of restraint | |||
| intraresidue | 370 | 63 | 29 |
| sequential | 272 | 56 | 46 |
| medium (2–5 residues) | 196 | 51 | 19 |
| long (>5 residues) | 98 | 0 | 0 |
| intermolecular (RIIA–RIIB) | 76 | ||
| intermolecular (RIIA–AKAP) | 22 or 15 | 22 | 15 |
| intermolecular (RIIB–AKAP) | 9 or 8 | 9 | 8 |
| dihedral | 50 | 0 | 0 |
| hydrogen bond | 63 | 10 | 8 |
| Total NOEs | 1046 or 1033 | 201 | 117 |
| No. NOEs per residue |
11.4 or 11.23 |
8.38 |
5.32 |
| R.m.s.ds |
Relative to mean (backbone N, Cα, C′) |
Relative to mean (heavy atoms) |
|
| Residues | |||
| Ht31(495–511) | 0.453 | ||
| AKAP79(392–408) | 1.10 | ||
| RIIα 9–42,9′–42′ | 0.635 | 0.68 | |
| RIIα 9–42,9′–42′, Ht31(495–511) | 0.693 | ||
| RIIα 9–42,9′–42′, AKAP79(392–408) |
|
1.01 |
|
| Energy (kcal/mol) |
RIIα–Ht31 complexbest, mean |
RIIα–AKAP79 complexbest, mean |
|
| Total | 235.22, 243.28 | 219.40, 236.36 | |
| Bond | 14.13, 14.12 | 12.48, 13.22 | |
| Angle | 131.56, 134.27 | 129.55, 133.06 | |
| Improper | 17.68, 17.49 | 17.74, 17.62 | |
| van der Waals | 45.66, 46.62 | 39.03, 42.05 | |
| NOE | 26.20, 30.78 | 20.61, 30.41 | |
Procheck-NMR (Laskowski et al., 1996) was used to analyze both the D/D–Ht31(493–515) and the D/D– AKAP79(392–408) families. Of all backbone Φ and ψ angles, 81.9 and 79.1% fell into the most favorable regions, and 17.1 and 18.5% were located in the additionally allowed regions of the Ramachandran plot for the Ht31 and AKAP79 complexes, respectively. Removing the disordered residues and including residues 5–44, 5′–44′ in the D/D and 493–512 in Ht31 or 392–409 in AKAP79 results in 90.2 and 87.4% of Φ, ψ angles falling in the most favorable regions, with 9.8 and 12.6% falling into additionally allowed regions on the Ramachandran plot. These regions correspond to the residues in RIIα(1–44), Ht31 and AKAP79 that have well defined secondary structure. It is possible that the low pH necessary for doing structural analysis by solution NMR has selectively destabilized residues 1–6, prior to the first helix. As a result of the expression plasmid used, there are two histidines in the N-terminus (positions –2 and 2) that are likely to be protonated at this pH and may be involved in charge–charge repulsion. Nevertheless, high affinity interactions are maintained under our experimental conditions.
Structural overview of the RIIα(1–44)–AKAP complexes
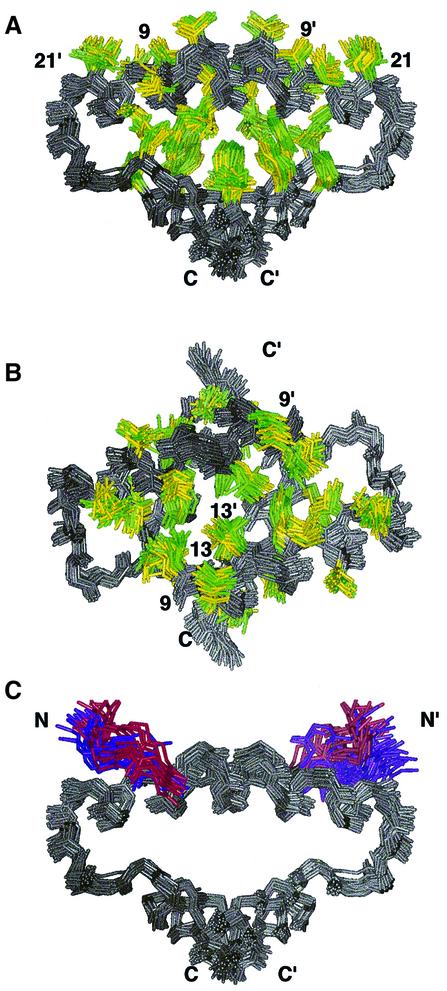
The D/D domain of RIIα in both of the AKAP complexes forms an X-type four-helix bundle with an extensive ordering of hydrophobic residues in the core as well as one exposed groove on the surface of the molecule (Figure 2A). A 90° rotation of the view in Figure 2A highlights the tight organization of the solvent-accessible hydrophobic groove (Figure 2B). The protomers of the D/D in the complexes are oriented antiparallel to each other, with helix–helix interactions alternating between antiparallel and orthogonal for the four helices around the bundle. The structural characteristics of the D/D domain of RIIα in the complexes are very similar to the free domain (Newlon et al., 1999).
Fig. 2. Representation of the hydrophobic core of RIIα(1–44) in both the free and AKAP-bound structures. The anchoring peptides have been removed for clarity and the first eight or four residues of RIIα are removed. Superposition of the 17, 13 and 10 lowest energy structures of free, Ht31(493–515)- and AKAP79(392–413)-bound RIIα(1–44), respectively. In all views, RIIα(1–44) is colored gray. Hydrophobic residues in free RIIα(1–44) are colored yellow and in both AKAP complexes are colored green. (A) This view emphasizes both the organization of the hydrophobic core and the hydrophobic groove of RIIα(1–44). (B) This view highlights the AKAP binding surface and is a 90° rotation of the view presented in (A). (C) This view is identical to (A) although the backbone atoms of residues 5–8 and 5′–8′ are indicated in either purple or red for the complex or apo-form of RIIα(1–44), respectively. Additionally, the side chain of Ile5 is also depicted and colored.
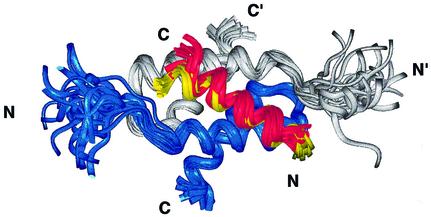
Both the 24-residue Ht31 and the 22-residue AKAP79 peptides form well defined α-helices for the N-terminal 18 residues of either peptide, respectively. These regions contain the majority of both intra- and intermolecular NOE and hydrogen bonding restraints. The C-terminal six or four residues of Ht31 and AKAP79, respectively, are defined by few NOEs. There are no observed D/D intermolecular NOE or hydrogen bonding restraints in the C-terminus of either AKAP peptide. Thus, the first 18 residues of the AKAP peptides define the D/D binding region. The overlay of the D/D and each AKAP that is shown in Figure 3 illustrates the similarity of the interactions in the two structures. The slight differences at the C-terminus of each peptide are due to helical fraying as a result of less defined interactions with the D/D, rather than true structural differences. A superposition of D/D in both complexes reveals that the first 17 residues of the two anchoring peptides, Ht31 and AKAP79, overlay quite well. Therefore, the interacting surfaces in the two peptide complexes are conserved.
Fig. 3. Similarity of the AKAP structure and binding region on RIIα(1–44) in the Ht31 and AKAP79 complexes. The superposition in RIIα(1–44), with the protomers of RIIα D/D shown in gray and blue, reveals the similarity in the backbone fold of the two complexes. The first 17 residues of the RII binding region in both Ht31(493–515) (red) and AKAP79(392–413) (yellow) are indicated, and bind to identical regions with similar helical structures to RIIα(1–44).
The stoichiometry of binding in these AKAP complexes is one RIIα dimer (two protomers) to one AKAP peptide (Carr et al., 1992a; Newlon et al., 1999). The docking of either AKAP to the D/D occurs between the surface-exposed, hydrophobic groove created by the D/D domain of RIIα and the hydrophobic face on each of the amphipathic helical AKAPs. This interaction occurs with the peptide binding at an ∼45° angle of the peptides to the symmetry axis presented by helices I and I′ in the D/D (Figure 1B and F).
Discussion
Preformed AKAP recognition surface in D/D
The position of backbone atoms of RIIα(1–44) in the AKAP–D/D complex is almost identical to that of the free structure (Newlon et al., 1999) with similar r.m.s.d. values for the ensemble of structures (Figure 1A, B, E and F). The D/D protein forms a defined antiparallel, X-type four-helix bundle motif with a five-residue N-terminal extended region. Upon binding either of the AKAP peptides, ∼10% (800 Å2) of the total surface area of the D/D becomes solvent inaccessible. This change in solvent accessibility occurs almost exclusively along the groove of helices I and I′, but also includes residues Ile3 and Ile5, which reside in the N-terminal extended region and were identified as key AKAP interaction partners in biochemical studies (Hausken et al., 1994).
The solution structures of the complexes determined in this study show that the solvent-accessible groove formed from helices I and I′ in free D/D is the AKAP recognition surface in the D/D. From the close overlay of the free and bound structures in Figure 2, it is apparent that neither the hydrophobic core nor the hydrophobic groove in RIIα is significantly affected by AKAP binding. Thus, the well ordered superposition of hydrophobic residues lining the solvent-accessible groove is a preformed recognition surface, which minimizes the change in entropy of the side chains in RIIα upon peptide binding. The largest changes that occur in the groove, or anywhere in the protein upon AKAP binding, appear at the center of the molecule, at the site of Leu13 and Leu13′. There are distinct clusters for both Leu13 and Leu13′ labeled in Figure 2B, where one of yellow (free) and one of green (complex) occupy slightly different positions in space. In either the RIIα–Ht31 or RIIα–AKAP79 complex, this residue appears to be pulled off-center and closer to the other protomer. Another residue, Leu9, also shifts in the complex structures relative to the free protein. It becomes slightly more centrally located in the hydrophobic groove, as can be seen in the two differentiated Leu9 and Leu9′ clusters of yellow and green in Figure 2B. Leu13, Leu13′, Leu9 and Leu9′ interact directly with Ht31 and AKAP79. Taken together, it appears that the RIIα D/D provides a preformed, hydrophobic, solvent-accessible groove as an interface for AKAP binding, with minor structural adjustments upon peptide binding in surface-accessible long-chain aliphatic residues (Leu9, Leu13) in helices I and I′.
Residues 5–8, on the N-terminal side of the first helices, are implicated in AKAP binding by mutational studies (Hausken et al., 1994) and are contiguous with the well placed hydrophobic residues lining a solvent-exposed crevice noted in the free RIIα D/D. In Figure 2C, we highlight backbone residues 5–8 and 5′–8′ of the D/D in purple and red for the complexed or free protein, respectively. Surprisingly, this results in only small structural changes in Ile3 and Ile5 upon complex formation, despite the fact that they are important determinants for AKAP binding (Hausken et al., 1994). This may be attributed to the selective destabilization of the region under our experimental conditions (see Results). In an interesting parallel, Wand and co-workers saw little change in backbone dynamics in calmodulin upon peptide binding (Lee et al., 2000). Indeed, they saw changes in side chain dynamics only, which may be the case for the RIIα D/D.
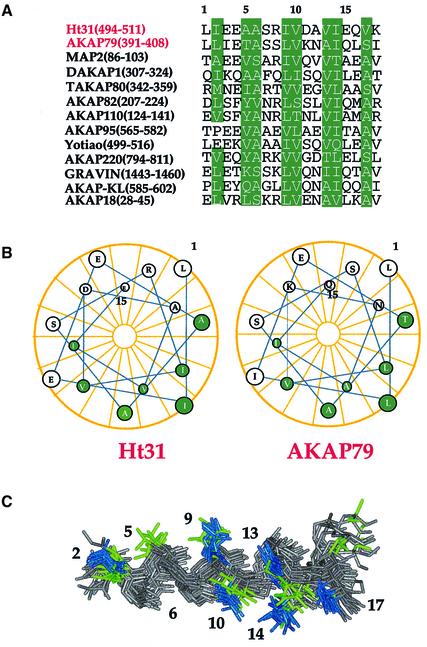
A conserved helical AKAP motif
Although sequence alignment of the binding region of many anchoring proteins reveals no conserved residues (Figure 4A), secondary structure prediction of this same region has suggested that the formation of an α-helix is required for binding (Figure 4B) (Carr et al., 1991). We present the first direct structural data demonstrating the helical nature of the peptides upon complex formation with the RIIα D/D. The DHF-NOESY data, which detect only AKAP peptide intramolecular NOEs in the D/D–AKAP complex, contain a number of helical NOEs, including many medium range dαβ,I,I+3 cross-peaks, several medium dNN cross-peaks and a few dNβ,i,i+1 cross-peaks. Amide hydrogen–deuterium exchange experiments also identified the presence of either 10 or eight slow exchanging amide protons, indicative of stable hydrogen bonds, within the helical region of the bound peptides in Ht31 and AKAP79, respectively.
Fig. 4. AKAP sequence alignment, helical wheel analysis of Ht31 and AKAP79 and structural helical overlay of Ht31 and AKAP79. (A) The alignment of the AKAP binding region (18 residues) of 13 known AKAPs is shown. The first two, Ht31 and AKAP79, are shown in red for emphasis. Conserved hydrophobic positions are shown in green. (B) Helical wheel analysis of the AKAP binding region of both Ht31 and AKAP79. The conserved hydrophobic positions as highlighted in (A) are colored green and map to a well defined space. (C) An overlay of the structures of Ht31 (red) and AKAP79 (yellow) when in RIIα are deleted for clarity. The conserved hydrophobic positions, as shown in (A) and (B), are colored either blue (Ht31) or green (AKAP79).
The backbone atoms of residues 2–17 (based on the numbering of Figure 4A) of the complexed Ht31 and AKAP79 peptides are ordered and overlay with each other well (Figure 4C). Side chains for the Ht31 peptide converge to a more defined space than those for AKAP79, because of the larger number of restraints available for the former (Figure 4C). C-terminal side chains and backbone atoms of both AKAPs are not as well defined due to fewer observed NOEs in this region for either AKAP. Hence, interactions of the C-terminal tails of the peptides with the D/D of RIIα are less defined and not as well fixed as the N-terminal residues.
Despite several amino acid differences, the arrangement of the hydrophobic face in both Ht31 and AKAP79 within the respective complex is fairly similar. An ensemble of residues 1–17 from either AKAP when bound to RIIα reveals a well ordered hydrophobic surface, comprised of residues 2, 5, 6, 9, 10, 13, 14 and 17 (residues 5 and 17 in AKAP79 are polar) based on the numbering used in the sequence alignment (Figure 4A). Free RIIα has C2 symmetry, and the result of AKAP binding is to create an asymmetric molecular complex (Figure 1A–H). It appears from our structure that the optimal AKAP sequence for tight, high affinity binding to the RIIα D/D would be a palindromic sequence of the critical hydrophobic residues, centered at residues 9/10.
The D/D–AKAP complex is formed by hydrophobic interactions
Our studies reveal that the structure of the two AKAP peptides, Ht31 and AKAP79, are helical when bound to RIIα D/D (Figures 1, 3, 4 and 6). The projection of the peptide helix axis on the I, I′ plane forms a 45° angle with I and I′ in RIIα. Each hydrophobic residue at the aligned positions 2, 5, 6, 9, 10, 13, 14 and 17 of the AKAP peptides (Figure 5) shows NOE cross-peaks to the RIIα D/D. An inspection of the R binding sequences of several AKAPs reveals that each of these positions 2, 5, 6, 9, 10, 13, 14 and 17 is a fairly conserved hydrophobic residue (Figure 4A) (Carr et al., 1991; Vijayaraghavan et al., 1999). From the stereoview of the surfaces of both complexes with the AKAP, a large hydrophobic groove created by the packing of helices I and I′ in the RIIα D/D is apparent (Figure 6). This cleft allows for docking of the conserved, hydrophobic residues in the AKAP to the RIIα D/D.7
Fig. 6. Stereoview representation of the AKAP binding surface of RIIα using Insight software (MSI, San Diego, CA). RIIα(1–44) protomers are colored yellow and pink, respectively. All hydrophobic residues in RIIa, Ht31 and AKAP79 are colored green. Acidic and basic residues are red and blue, respectively. (A) Ht31 lies above RIIα, with the hydrophobic face that contacts RIIα facing the hydrophobic face of RIIα. Additional residues in Ht31 are colored pink. (B) AKAP79 is shown in this figure with the hydrophobic face in similar orientation to RIIα to that of Ht31 in (A). Additional residues in Ht31 are colored pink.
Fig. 5. NOE contacts between RIIα and Ht31 and AKAP79. (A and B) The NOE contacts for each residue on the Ht31 (green) or AKAP79 (purple) to RIIα (A) or on RIIα to each of the AKAPs (B). (C and D) Structural view of the NOE contacts. RIIα is colored yellow and gray for each protomer. Ht31 (C) is colored green and AKAP79 (D) is colored purple. All of the residues that have observed NOE contacts are indicated.
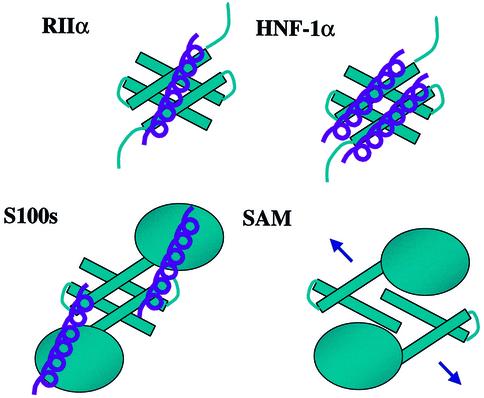
Fig. 7. Schematic diagram of protein–protein interactions mediated by X-type four-helix bundles. RIIα is unique in binding one protomer per dimer at a surface hydrophobic groove. HNF-1α presents a hydrophobic face and forms a dimer of dimers. Likewise, the stoichiometry of P53 binding to S100 is two peptides per dimer. The binding interactions with SAM are under investigation.
Mechanisms of dimerization and AKAP recognition by RII
To date, there have been a number of mutations in R subunits that alter interactions with AKAPs. These studies have indicated key residues and key differences between the R subunit types and different AKAPs. Consistently, between R isoforms, the data from GST-pulldown and RII overlay experiments have indicated both AKAP- and dimerization-specific regions within the four-helix bundle (Hausken et al., 1994, 1996; Li and Rubin, 1995; Banky et al., 1998). These functional subdomains are apparent from the structural features of the D/D domain of RIIα(1–44). Mutagenesis experiments reveal that the requirements for AKAP binding in intact dimeric proteins reside in helices I and I′ and the N-terminal extended regions of the R subunit. Indeed, our solution structures presented here reveal the intimate contact of these regions. For example, when Ile5 in RIIα or the conserved Leu21 in RIIβ is mutated to Ala, AKAP binding decreases (Li and Rubin, 1995; Hausken et al., 1996). There are intense NOE cross-peaks between both of these residues in RIIα to both of the AKAPs. Important dimer contacts involve residues in both helices I, I′ and helices II, II′ and include Ile5, Pro6, Leu9, Leu12, Leu13, Tyr16, Val20 and Leu21 in helix I, I′ (Ile5 and Pro6 are preceding and in the turn before helix I, I′, respectively), and Leu28, Val29, Ala32, Val33, Phe36, Thr37 and Leu39 in helix II, II′. Mutagenesis experiments and deletion analyses indicate that the removal of residues 1–10 (Hausken et al., 1994), or the replacement of residue 13 or 36 (Li and Rubin, 1995), abolishes dimerization and AKAP interaction. These results are consistent with the strong dimer contacts observed that involve Ile5, Pro6, Leu9, Leu13 and Phe36.
Less was known about critical AKAP residues, despite the fact that so many AKAP sequences are available. Branched, hydrophobic residues at several positions in AKAP79 are important in maintaining high affinity binding of RIIβ (Glantz et al., 1993). Potentially necessary aromatic and smaller side chains have been proposed as determinants of AKAP binding specificity between the type I and type II R isoforms (Angelo and Rubin, 1998, 2000; Miki and Eddy, 1998, 1999). We confirm the necessity of long, branched, hydrophobic residues at positions 9, 10, 13, 14, 17 and 18 in the AKAP from our solution structures of the AKAP complexes (Figures 4–6). In addition, analysis of the structure reveals additional key residues, Ala5 and Ala6 (as well as Thr5 in AKAP79), which had not been implicated previously. All of these residues show many strong NOE cross-peaks to the RIIα D/D (Figure 5).
The D/D does not bind to a conserved amino acid sequence, although a conserved amphipathic helix is predicted for this region (Carr et al., 1991). From this we can infer that the R subunit isoforms are able to recognize a variety of distinct sequence motifs that still accommodate high affinity binding. There are a number of charged side chains in both the RIIα binding region of many AKAPs and in the AKAP binding region of RIIα. However, unlike the recognition surfaces of HNF-1α and its dimerization cofactor (Rose et al., 2000a,b), we do not see any direct charge–charge interactions between either of the AKAPs and RIIα. The polar (Gln14, Thr17) and charged (Arg22) RIIα residues that show NOEs to the AKAP peptides do so through the aliphatic portions of their side chains. As all of the observed interactions between the AKAPs and the D/D domain of RIIα appear to be hydrophobic, we speculate that the conservation of charged residues, particularly in the AKAP sequences, may aid in the formation or stabilization of an amphipathic helix. The structures of the RIIα–Ht31 and RIIα–AKAP79 complexes reported here confirm the adaptability of the R subunit to recognize and bind to different AKAPs. In the present study, the AKAP binding and dimerization domain of RIIα indeed binds to two distinct AKAP sequences, AKAP79 and Ht31. At first glance, these sequences contain little sequence homology, yet bind to identical regions of the R subunit. Thus, hydrophobic interactions are the major forces that organize the molecular recognition between the R subunit and the AKAP (Figure 6).
Recruitment in signaling by unique surfaces in X-type four-helix bundles
Dimerization is a necessary prerequisite in a number of biochemical and signaling pathways, ranging from subcellular localization to DNA binding selectivity and biological activity. The X-type four-helix bundle has recently been observed as a dimerization motif in several signaling molecules (Potts et al., 1995; Rustandi et al., 1998; Newlon et al., 1999; Rose et al., 2000b) (Figure 7). An interesting feature is that these domains both aid dimerization and recruit additional protein–protein interactions with a variety of partners. Interestingly, the mode of interaction with the partner protein is dependent on the unique surface topology of exposed hydrophobic patches or grooves on the surface of the respective dimerization domain. For example, the X-type four-helix bundle D/D motif is found in the S100 family of proteins. These proteins participate in protein–protein interactions with p53 (Rustandi et al., 2000). Once Ca2+ binds, a conformational change occurs in the molecule, exposing a surface of the four-helix bundle and the E-F hand to recruit two molecules of p53 per dimer. Recently, the crystal structure of the N-terminal dimerization domain of the transcriptional activator hepatocyte nuclear factor 1α (HNF-1α) has been solved in complex with the coactivator, dimerization cofactor of HNF-1 (DCoH) to form a dimer of dimers (Rose et al., 2000a). Like the D/D of RIIα, this domain forms a stand-alone X-type four-helix bundle. Unlike what we observe for RIIα, the exposed hydrophobic patch on HNF-1α appears to have a surface ridge that facilitates formation of a dimer of dimers with two helices from DCoH binding to the exposed hydrophobic patch. A variation of the X-type four-helix bundle fold is found in the dimerization interface of the sterile α motif (SAM) domain proteins (Stapleton et al., 1999). Not a typical X-type four-helix bundle, it almost appears as though the four helices are pulled apart, which may then create a large surface for recruiting protein–protein interactions.
The X-type four-helix bundle is emerging as an adaptable, yet selective domain for supplying a platform for protein–protein interactions. In the structures of the RIIα domain complexed with either Ht31 or AKAP79 presented in this report, we observe a large hydrophobic accessible groove on one face of the X-type four-helix bundle that participates in recruitment of the AKAPs. In the S100 family, the X-type bundle contributes only partly to the protein interaction interface, with part of the interaction surface coming from the E-F hands, allowing binding of multiple (two) signaling partners (Rustandi et al., 1999). The stand-alone bundle in HNF-1α also binds multiple (two) partners; in this instance, the surface ridge in the center of the hydrophobic patch segregates the interface and allows for the packing of two additional helices with a typical helical crossing angle of –22° (Rose et al., 2000a). In a departure from this common stoichiometry of binding, RIIα presents a unique hydrophobic groove that dictates stoichiometry of one AKAP per D/D.
Our results presented here reveal, for the first time, that the molecular basis for high affinity anchoring of PKA is through discrete interactions between a preformed hydrophobic groove on the X-type four-helix bundle of the D/D with the hydrophobic face of an amphipathic helix on two different AKAPs. Comparisons with other protein structures show that the X-type bundle protein–protein interaction motif provides mechanisms for specificity in signaling events and for integration of diverse signaling pathways.
Materials and methods
Sample preparation
Recombinant RIIα(1–44) was expressed and purified from Escherichia coli as described previously (Newlon et al., 1997). AKAP79 and Ht31(493–515) were synthesized as unlabeled, biotinylated peptides and purified to >95% homogeneity by PeptidoGenic Research and Co. (Livermore, CA). In order to solubilize Ht31 or AKAP79, the pH of a 2 mM RIIα(1–44) sample in 90% H2O/10% D2O at pH 4.0 was brought up to 7.0, the sample mixed for 1 h and then the pH was slowly adjusted back to 4.0. In this way, the peptide was able to go into solution and form a complex with RIIα. As described previously (Newlon et al., 1997), the high affinity interaction with the AKAP peptides were preserved at this pH, which is optimal for structure determination.
NMR experiments
All NMR experiments were conducted on a Bruker DMX500 spectrometer at 25°C using a triple-resonance gradient probe. The majority of HN and 15N assignments of RIIα could be confirmed based on a two-dimensional titration of unlabeled AKAP peptide by following either the movement or splitting of a peak [splitting occurs because the AKAP peptides disrupt the inherent symmetry of the RIIα(1–44) dimer].
Several triple-resonance experiments were conducted on the complexes to obtain 1H and 13C chemical shift assignments of the RIIα(1–44) fragment. 13C chemical shifts that were used for Calculation Strategy Interproton (CSI) analysis were obtained by both CT-HNCA and CT-HN(CO)CA (Ikura et al., 1990) (data not shown). These experiments utilized the WATERGATE sequence (Sklenar et al., 1993) for water suppression. The 1H carrier was placed at the water frequency and then shifted to the center of the amides prior to data acquisition. The 15N and 13C carriers were placed at 117 and 57.3 p.p.m., respectively. Sixty-four real t1 (13C), 48 or 50 real t2 (15N) and 512 complex t3 (1H) points were collected with spectral widths of 3771 Hz (t1), 1266 Hz (t2) and 3005 Hz (t3). 15N decoupling was achieved by a WALTZ composite pulse scheme (Bax and Davis, 1985) during acquisition. For indirectly detected dimensions, t1 and t2, TPPI (Marion and Wuthrich, 1983) was the method used for quadrature phase detection.
Several filtered NMR experiments were conducted on a 13C,15N-labeled RIIα(1–44) complexed with either unlabeled AKAP79 or unlabeled Ht31(493–515) to obtain assignments of the unlabeled peptides, and to determine inter-peptide–protein NOEs of the bound peptide. A 1H-1H DHF-NOESY spectrum (mixing time = 150 ms) and two 1H-1H DHF-total correlation spectroscopy (TOCSY) spectra (mixing time = 30 ms, 60 ms) (Ikura and Bax, 1992; Slijper et al., 1996; Zwahlen et al., 1997) were acquired on samples in either 90% H2O/10% D2O or 100% D2O. These experiments provide intrapeptide NOE cross-peaks from unlabeled bound AKAP peptides only. Water suppression was achieved using a presaturation pulse. The spectral widths were 5501 Hz in t1 and 5482 Hz in t2. The 1H, 15N and 13C carriers were placed at 4.713, 118 and 62 p.p.m., respectively. A 3D 13C-edited (ω2)-12C-filtered (ω1)/13C-filtered (ω3) NOESY (Ikura and Bax, 1992) with a 150 ms mixing time was acquired to distinguish inter-peptide–protein NOE cross- peaks between the unlabeled AKAP peptides to the 15N, 13C labeled RIIα(1–44). This experiment was performed on a sample in 100% D2O. Carriers were placed at water for 1H, 116 p.p.m. for 15N and 40 p.p.m. for 13C nuclei. Decoupling of both 15N and 13C′ was acquired using either WALTZ16 (Bax and Davis, 1985) or SEDUCE, respectively (McCoy and Mueller, 1992a,b). Spectral widths of 5000 Hz (t1), 7546 Hz (t2) and 1024 Hz (t3) were collected with a matrix size of 128 complex points (t1), 64 complex points (t2) by 1024 complex points (t3). Quadrature phase detection was achieved using STATES in both t1 and t2 (States et al., 1982).
Additional NOEs were observed by the 3D 1H-15N NOESY-heteronuclear single quantum coherence (HSQC) experiment with a 150 ms mixing time on a 15N-labeled RIIα(1–44)/unlabeled AKAP peptide sample. Water suppression was accomplished using the gradient tailored WATERGATE sequence (Sklenar et al., 1993). Broadband 15N decoupling was achieved by employing a WALTZ16 decoupling scheme (Bax and Davis, 1985). A total of 128 real t1 (1H), 64 real t2 (15N) and 512 complex t3 (1H) points were collected with spectral widths of 4999 Hz (t1), 1773 Hz (t2) and 3005 Hz (t3). The 1H carrier was placed on the water resonance and then shifted to the center of the amides prior to detection, and the 15N carrier was centered at 117.5 p.p.m.
Hydrogen exchange experiments
The hydrogen exchange experiments were conducted on a DMX500 spectrometer with 15N-13C labeled RIIα(1–44) samples on RIIα(1–44) complexed to either AKAP79 or Ht31. A total of six DHF-NOESY (mixing time = 150 ms) experiments were conducted on each sample after the exchange of the sample from 90% H2O/10% D2O into 100% D2O. After 24 h, the HN fingerprint region for the slowly exchanging protons (those still observed after 24 h) could still be observed, but the fast exchanging (faster than 24 h) amide protons had disappeared.
Data processing
All experiments were processed using Felix 95.0 or 97.0 software (MSI, San Diego, CA). All data were apodized with a squared sinebell function of 70° phase shift in all dimensions before Fourier transformation. When needed, linear prediction to an additional one-third of the number of points collected was applied before apodization and zero filling in the indirect detected dimensions.
CSI distance restraints were obtained through measured peak volumes calibrated against known distances in elements of regular secondary structure. NOEs were classified as strong (1.8–2.7 Å), medium (1.8–3.3 Å) or ambiguous (resulting from both inter- and intramolecular contacts). Structures of the RIIα(1–44)–AKAP complexes were calculated as described by Nilges (1993) using X-PLOR 3.851 (Brunger, 1992, 1993) and target function as described (Morikis, 2000), except that the structures were subject to two rounds of refinement that included neither the non-crystallographic symmetry term nor the symmetry term. NOEs were separated into four categories: intramolecular in RIIα(1–44), intramolecular in AKAP peptide, intermolecular in RIIα(1–44) and intermolecular between RIIα(1–44) and the AKAP peptide. To avoid the inclusion of any NOEs that could have any potential contribution to multiple contacts, a conservative addition of NOE restraints was undertaken. If an NOE could arise from multiple proton resonances, it was never incorporated into the structure calculations. This was done to avoid ambiguous NOE information when orienting the AKAP onto the RIIα dimer from the 3D 13C-edited (ω2)-12C-filtered (ω1)/13C-filtered (ω3) NOESY experiment. In the final round of calculations, 50 final structures were generated and either the 10 or 13 lowest energy structures were selected for further analysis for the AKAP79 and Ht31 complexes, respectively. Procheck-NMR (Laskowski et al., 1996) was used to analyze the 17 lowest energy structures.
Acknowledgments
Acknowledgements
This work was supported, in part, by National Institutes of Health grants DK5444 (to P.A.J. and J.D.S.), DK44239 (to J.D.S.), HD36408 (to D.W.C.), CA09523 (to M.R.), and an American Heart Association pre-doctoral fellowship (to M.G.N.).
References
- Angelo R. and Rubin,C.S. (1998) Molecular characterization of an anchor protein (AKAPCE) that binds the RI subunit (RCE) of type I protein kinase A from Caenorhabditis elegans. J. Biol. Chem., 273, 14633–14643. [DOI] [PubMed] [Google Scholar]
- Angelo R.G. and Rubin,C.S. (2000) Characterization of structural features that mediate the tethering of Caenorhabditis elegans protein kinase A to a novel A kinase anchor protein. Insights into the anchoring of PKAI isoforms. J. Biol. Chem., 275, 4351–4362. [DOI] [PubMed] [Google Scholar]
- Banky P., Huang,L.J. and Taylor,S.S. (1998) Dimerization/docking domain of the type Iα regulatory subunit of cAMP-dependent protein kinase. Requirements for dimerization and docking are distinct but overlapping J. Biol. Chem., 273, 35048–35055. [DOI] [PubMed] [Google Scholar]
- Bax A. and Davis,D.G. (1985) MLEV-17-based two-dimensional homonuclear magnetization transfer spectroscopy. J. Magn. Reson., 65, 355–360. [Google Scholar]
- Brunger A.T. (1992) X-PLOR. Yale University Press, New Haven, CT.
- Brunger A.T. (1993) X-PLOR Version 3.1: A System for X-ray Crystallography and NMR. Yale University Press, New Haven, CT.
- Burgering M.J., Boelens,R., Caffrey,M., Breg,J.N. and Kaptein,R. (1993) Observation of inter-subunit nuclear Overhauser effects in a dimeric protein. Application to the Arc repressor. FEBS Lett., 330, 105–109. [DOI] [PubMed] [Google Scholar]
- Burton K.A., Johnson,B.D., Hausken,Z.E., Westenbroek,R.E., Idzerda,R.L., Scheuer,T., Scott,J.D., Catterall,W.A. and McKnight,G.S. (1997) Type II regulatory subunits are not required for the anchoring-dependent modulation of Ca2+ channel activity by cAMP-dependent protein kinase. Proc. Natl Acad. Sci. USA, 94, 11067–11072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr D.W. and Scott,J.D. (1992) Blotting and band-shifting: techniques for studying protein–protein interactions. Trends Biochem. Sci., 17, 246–249. [DOI] [PubMed] [Google Scholar]
- Carr D.W., Stofko-Hahn,R.E., Fraser,I.D.C., Bishop,S.M., Acott,T.S., Brennan,R.G. and Scott,J.D. (1991) Interaction of the regulatory subunit (RII) of cAMP-dependent protein kinase with RII-anchoring proteins occurs through an amphipathic helix binding motif. J. Biol. Chem., 266, 14188–14192. [PubMed] [Google Scholar]
- Carr D.W., Hausken,Z.E., Fraser,I.D.C., Stofko-Hahn,R.E. and Scott,J.D. (1992a) Association of the type II cAMP-dependent protein kinase with a human thyroid RII-anchoring protein. J. Biol. Chem., 267, 13376–13382. [PubMed] [Google Scholar]
- Carr D.W., Stofko-Hahn,R.E., Fraser,I.D., Cone,R.D. and Scott,J.D. (1992b) Localization of the cAMP-dependent protein kinase to the postsynaptic densities by A-kinase anchoring proteins. Characterization of AKAP 79. J. Biol. Chem., 267, 16816–16823. [PubMed] [Google Scholar]
- Coghlan V.M., Perrino,B.A., Howard,M., Langeberg,L.K., Hicks,J.B., Gallatin,W.M. and Scott,J.D. (1995) Association of protein kinase A and protein phosphatase 2B with a common anchoring protein. Science, 267, 108–112. [DOI] [PubMed] [Google Scholar]
- Colledge M. and Scott,J.D. (1999) AKAPs: from structure to function. Trends Cell Biol., 9, 216–221. [DOI] [PubMed] [Google Scholar]
- Colledge M., Dean,R.A., Scott,G.K., Langeberg,L.K., Huganir,R.L. and Scott,J.D. (2000) Targeting of PKA to glutamate receptors through a MAGUK–AKAP complex. Neuron, 27, 107–119. [DOI] [PubMed] [Google Scholar]
- Diviani D., Langeberg,L.K., Doxsey,S.J. and Scott,J.D. (2000) Pericentrin anchors protein kinase A at the centrosome through a newly identified RII-binding domain. Curr. Biol., 10, 417–420. [DOI] [PubMed] [Google Scholar]
- Dodge K. and Scott,J.D. (2000) AKAP79 and the evolution of the AKAP model. FEBS Lett., 476, 58–61. [DOI] [PubMed] [Google Scholar]
- Faux M.C. and Scott,J.D. (1996) More on target with protein phosphorylation: conferring specificity by location. Trends Biochem. Sci., 21, 312–315. [PubMed] [Google Scholar]
- Feliciello A., Li,Y., Avvedimento,E.V., Gottesman,M.E. and Rubin,C.S. (1997) A-kinase anchor protein 75 increases the rate and magnitude of cAMP signaling to the nucleus. Curr. Biol., 7, 1011–1014. [DOI] [PubMed] [Google Scholar]
- Fraser I.D. and Scott,J.D. (1999) Modulation of ion channels: a ‘current’ view of AKAPs. Neuron, 23, 423–426. [DOI] [PubMed] [Google Scholar]
- Glantz S.B., Li,Y. and Rubin,C.S. (1993) Characterization of distinct tethering and intracellular targeting domains in AKAP75, a protein that links cAMP-dependent protein kinase IIβ to the cystoskeleton. J. Biol. Chem., 268, 12796–12804. [PubMed] [Google Scholar]
- Hausken Z.E. and Scott,J.D. (1996) Properties of A-kinase anchoring proteins. Biochem. Soc. Trans., 24, 986–991. [DOI] [PubMed] [Google Scholar]
- Hausken Z.E., Coghlan,V.M., Hasting,C.A.S., Reimann,E.M. and Scott,J.D. (1994) Type II regulatory subunit (RII) of the cAMP dependent protein kinase interaction with A-kinase anchor proteins requires isoleucines 3 and 5. J. Biol. Chem., 269, 24245–24251. [PubMed] [Google Scholar]
- Hausken Z.E., Dell’Acqua,M.L., Coghlan,V.M. and Scott,J.D. (1996) Mutational analysis of the A-kinase anchoring protein (AKAP)-binding site on RII. J. Biol. Chem., 271, 29016–29022. [DOI] [PubMed] [Google Scholar]
- Herberg F.W., Maleszka,A., Eide,T., Vossebein,L. and Tasken,K. (2000) Analysis of A-kinase anchoring protein (AKAP) interaction with protein kinase A (PKA) regulatory subunits: PKA isoform specificity in AKAP binding. J. Mol. Biol., 298, 329–339. [DOI] [PubMed] [Google Scholar]
- Hunter T. (2000) Signaling—2000 and beyond. Cell, 100, 113–127. [DOI] [PubMed] [Google Scholar]
- Husi H., Ward,M.A., Choudhary,J.S., Blackstock,W.P. and Grant,S.G. (2000) Proteomic analysis of NMDA receptor–adhesion protein signaling complexes. Nature Neurosci., 3, 661–669. [DOI] [PubMed] [Google Scholar]
- Ikura M. and Bax,A. (1992) Isotope-filtered 2D NMR of a protein– peptide complex: Study of a skeletal muscle myosin light chain kinase fragment bound to calmodulin. J. Am. Chem. Soc., 114, 2433–2440. [Google Scholar]
- Ikura M., Kay,L.E. and Bax,A. (1990) A novel approach for sequential assignment of 1H, 13C and 15N spectra of proteins: heteronuclear triple-resonance three-dimensional NMR spectroscopy. Application to calmodulin. Biochemistry, 29, 4659–4667. [DOI] [PubMed] [Google Scholar]
- Ikura M., Clore,G.M., Gronenborn,A.M., Zhu,G., Klee,C.B. and Bax,A. (1992) Solution structure of a calmodulin–target peptide complex by multidimensional NMR. Science, 256, 632–637. [DOI] [PubMed] [Google Scholar]
- Kapiloff M.S., Schillace,R.V., Westphal,A.M. and Scott,J.D. (1999) mAKAP: an A-kinase anchoring protein targeted to the nuclear membrane of differentiated myocytes. J. Cell Sci., 112, 2725–2736. [DOI] [PubMed] [Google Scholar]
- Klauck T.M., Faux,M.C., Labudda,K., Langeberg,L.K., Jaken,S. and Scott,J.D. (1996) Coordination of three signaling enzymes by AKAP79, a mammalian scaffold protein. Science, 271, 1589–1592. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., Rullmannn,J.A., MacArthur,M.W., Kaptein,R. and Thornton,J.M. (1996) AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR, 8, 477–486. [DOI] [PubMed] [Google Scholar]
- Lee A.L., Kinnear,S.A. and Wand,A.J. (2000) Redistribution and loss of side chain entropy upon formation of a calmodulin–peptide complex. Nature Struct. Biol., 7, 72–77. [DOI] [PubMed] [Google Scholar]
- Lester L.B., Langeberg,L.K. and Scott,J.D. (1997) Anchoring of protein kinase A facilitates hormone-mediated insulin secretion. Proc. Natl Acad. Sci. USA, 94, 14942–14947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. and Rubin,C.S. (1995) Mutagenesis of the regulatory subunit (RIIβ) of cAMP-dependent protein kinase IIβ reveals hydrophobic amino acids that are essential for RIIβ dimerization and/or anchoring RIIβ to the cytoskeleton. J. Biol. Chem., 270, 1935–1944. [PubMed] [Google Scholar]
- Luo Z., Shafit-Zagardo,B. and Erlichman,J. (1990) Identification of the MAP2- and P75-binding domain in the regulatory subunit (RIIβ) of type II cAMP-dependent protein kinase. J. Biol. Chem., 265, 21804–21810. [PubMed] [Google Scholar]
- Marion D. and Wuthrich,K. (1983) Application of phase-sensitive two-dimensional correlated spectroscopy (COSY) for measurements of 1H-1H spin-spin coupling constants in proteins. Biochem. Biophys. Res. Commun., 113, 967–974. [DOI] [PubMed] [Google Scholar]
- McCoy M.A. and Mueller,L. (1992a) Coherence quenching induced by frequency-selective homonuclear decoupling. J. Magn. Reson., 98, 674–679. [Google Scholar]
- McCoy M.A. and Mueller,L. (1992b) Selective shaped pulse decoupling in NMR: homonuclear [13C]carbonyl decoupling. J. Am. Chem. Soc., 114, 2108–2112. [Google Scholar]
- Miki K. and Eddy,E.M. (1998) Identification of tethering domains for protein kinase A type Iα regulatory subunits on sperm fibrous sheath protein FSC1. J. Biol. Chem., 273, 34384–34390. [DOI] [PubMed] [Google Scholar]
- Miki K. and Eddy,E.M. (1999) Single amino acids determine specificity of binding of protein kinase A regulatory subunits by protein kinase A anchoring proteins. J. Biol. Chem., 274, 29057–29062. [DOI] [PubMed] [Google Scholar]
- Morikis D., Newlon,M.G. and Jennings,P.A. (2000) Structure calculations of symmetric dimers using molecular dynamics/simulated annealing and NMR restraints: the case of the RIIα subunit of protein kinase A. In Floudas,C.A. and Pardalos,P.M. (eds), Optimization in Computational Chemistry and Molecular Biology. Kluwer Academic, Dordrecht, The Netherlands, pp. 141–156.
- Newlon M.G., Roy,M., Hausken,Z.E., Scott,J.D. and Jennings,P.A. (1997) The A-kinase anchoring domain of type IIα cAMP-dependent protein kinase is highly helical. J. Biol. Chem., 272, 23637–23644. [DOI] [PubMed] [Google Scholar]
- Newlon M.G., Roy,M., Morikis,D., Hausken,Z.E., Coghlan,V., Scott,J.D. and Jennings,P.A. (1999) The molecular basis for protein kinase A anchoring revealed by solution NMR. Nature Struct. Biol., 6, 222–227. [DOI] [PubMed] [Google Scholar]
- Nilges M. (1993) A calculation strategy for the structure determination of symmetric dimers by 1H NMR. Proteins, 17, 297–309. [DOI] [PubMed] [Google Scholar]
- Obar R.A., Dingus,J., Bayley,H. and Vallee,R.B. (1989) The RII subunit of cAMP-dependent protein kinase binds to a common N-terminal domain in microtubule-associated proteins 2A, 2B and 2C. Neuron, 3, 639–645. [DOI] [PubMed] [Google Scholar]
- Pawson T. and Nash,P. (2000) Protein–protein interactions define specificity in signal transduction. Genes Dev., 14, 1027–1047. [PubMed] [Google Scholar]
- Potts B.C., Smith,J., Akke,M., Macke,T.J., Okazaki,K., Hidaka,H., Case,D.A. and Chazin,W.J. (1995) The structure of calcyclin reveals a novel homodimeric fold for S100 Ca(2+)-binding proteins [published erratum appears in Nature Struct. Biol., 1995, 2, 912]. Nature Struct. Biol., 2, 790–796. [DOI] [PubMed] [Google Scholar]
- Rose R.B., Bayle,J.H., Endrizzi,J.A., Cronk,J.D., Crabtree,G.R. and Alber,T. (2000a) Structural basis of dimerization, coactivator recognition and MODY3 mutations in HNF-1α. Nature Struct. Biol., 7, 744–748. [DOI] [PubMed] [Google Scholar]
- Rose R.B., Endrizzi,J.A., Cronk,J.D., Holton,J. and Alber,T. (2000b) High-resolution structure of the HNF-1α dimerization domain. Biochemistry, 39, 15062–15070. [DOI] [PubMed] [Google Scholar]
- Rosenmund C., Carr,D.W., Bergeson,S.E., Nilaver,G. and Scott,J.D. and Westbrook,G.L. (1994) Anchoring of protein kinase A is required for modulation of AMPA/kainate receptors on hippocampal neurons. Nature, 368, 853–856. [DOI] [PubMed] [Google Scholar]
- Rubino H.M., Dammerman,M., Shafit-Sagardo,B. and Erlichman,J. (1989) Localization and characterization of the binding site for the regulatory subunit of type II cAMP-dependent protein kinase on MAP2. Neuron, 3, 631–638. [DOI] [PubMed] [Google Scholar]
- Rustandi R.R., Drohat,A.C., Baldisseri,D.M., Wilder,P.T. and Weber,D.J. (1998) The Ca(2+)-dependent interaction of S100B(ββ) with a peptide derived from p53. Biochemistry, 37, 1951–1960. [DOI] [PubMed] [Google Scholar]
- Rustandi R.R., Baldisseri,D.M., Drohat,A.C. and Weber,D.J. (1999) Structural changes in the C-terminus of Ca2+-bound rat S100B(ββ) upon binding to a peptide derived from the C-terminal regulatory domain of p53. Protein Sci., 8, 1743–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustandi R.R., Baldisseri,D.M. and Weber,D.J. (2000) Structure of the negative regulatory domain of p53 bound to S100B(ββ). Nature Struct. Biol., 7, 570–574. [DOI] [PubMed] [Google Scholar]
- Scott J.D., Stofko,R.E., McDonald,J.R., Comer,J.D., Vitalis,E.A. and Mangeli,J. (1990) Type II regulatory subunit dimerization determines the subcellular localization of the cAMP-dependent protein kinase. J. Biol. Chem., 265, 21561–21566. [PubMed] [Google Scholar]
- Sklenar V., Piotto,M., Leppik,R. and Saudek,V. (1993) Gradient-tailored water suppression for 1H-15N HSQC experiments optimized to retain full sensitivity. J. Magn. Reson., 102, 241–245. [Google Scholar]
- Slijper M., Kaptein,R. and Boelens,R. (1996) Simultaneous 13C and 15N isotope editing of biomolecular complexes. Application to a mutant lac repressor headpiece DNA complex. J. Magn. Reson. B, 111, 199–203. [DOI] [PubMed] [Google Scholar]
- Stapleton D., Balan,I., Pawson,T. and Sicheri,F. (1999) The crystal structure of an Eph receptor SAM domain reveals a mechanism for modular dimerization. Nature Struct. Biol., 6, 44–49. [DOI] [PubMed] [Google Scholar]
- States D.J., Haberkorn,R.A. and Ruben,D.J. (1982) A two-dimensional nuclear Overhauser experiment with pure absorption phase in four quadrants. J. Magn. Reson., 48, 286–292. [Google Scholar]
- Taylor S.S., Buechler,J.A. and Yonemoto,W. (1990) cAMP-dependent protein kinase: framework for a diverse family of regulatory enzymes. Annu. Rev. Biochem., 59, 971–1005. [DOI] [PubMed] [Google Scholar]
- Vijayaraghavan S., Goueli,S.A., Davey,M.P. and Carr,D.W. (1997) Protein kinase A-anchoring inhibitor peptides arrest mammalian sperm motility. J. Biol. Chem., 272, 4747–4752. [DOI] [PubMed] [Google Scholar]
- Vijayaraghavan S., Liberty,G.A., Mohan,J., Winfrey,V.P., Olson,G.E. and Carr,D.W. (1999) Isolation and molecular characterization of AKAP110, a novel, sperm-specific protein kinase A-anchoring protein. Mol. Endocrinol., 13, 705–717. [DOI] [PubMed] [Google Scholar]
- Westphal R.S., Tavalin,S.J., Lin,J.W., Alto,N.M., Fraser,I.D., Langeberg,L.K., Sheng,M. and Scott,J.D. (1999) Regulation of NMDA receptors by an associated phosphatase–kinase signaling complex. Science, 285, 93–96. [DOI] [PubMed] [Google Scholar]
- Zwahlen C., Legault,P., Vincent,S.J.F., Greenblatt,J., Konrat,R. and Kay,L.E. (1997) Methods for measurement of intermolecular NOEs by multinuclear NMR spectroscopy: Application to a bacteriophage λ N-peptide/boxB RNA complex. J. Am. Chem. Soc., 119, 6711–6721. [Google Scholar]