Abstract
Wild-type yeast mitochondrial DNA (mtDNA) is inherited biparentally, whereas mtDNA of hypersuppressive petite mutants is inherited uniparentally in crosses to strains with wild-type mtDNA. Genomes of hypersuppressive petites contain a conserved ori sequence that includes a promoter, but it is unclear whether the ori confers a segregation or replication advantage. Fluorescent in situ hybridization analysis of wild-type and petite mtDNAs in crosses reveals no preferential segregation of hypersuppressive petite mtDNA to first zygotic buds. We identify single-stranded DNA circles and RNA-primed DNA replication intermediates in hypersuppressive petite mtDNA that are absent from non-hypersuppressive petites. Mutating the promoter blocks hypersuppressiveness in crosses to wild-type strains and eliminates the distinctive replication intermediates. We propose that promoter-dependent RNA-primed replication accounts for the uniparental inheritance of hypersuppressive petite mtDNA.
Keywords: mtDNA/replication/segregation/uniparental inheritance/yeast
Introduction
Maintaining the integrity and faithful inheritance of the mitochondrial genome is essential for respiratory function. A number of human pathologies result from the accumulation of mitochondrial DNA (mtDNA) mutations, and the distribution of wild-type and mutant mtDNAs within cells and tissues often determines the severity of the disease (Wallace, 1999). Factors that control the segregation of mtDNAs are poorly understood.
In most higher eukaryotes mtDNA is inherited uniparentally; however, this has made it difficult to study the transmission of wild-type and mutant genomes in crosses. By contrast, in the budding yeast Saccharomyces cerevisiae, mtDNA inheritance is biparental in crosses between strains with wild-type (ρ+) mitochondrial genomes (Dujon, 1981). As a consequence, this system provides unique opportunities for detailed genetic and molecular analyses of the mechanisms by which parental and recombinant mtDNAs are transmitted to progeny cells (Hermann and Shaw, 1998).
Uniparental inheritance of yeast mtDNA is observed in some crosses between ρ+ and petite (ρ–) mutant strains (Ephrussi et al., 1955). Petite strains are respiratory-deficient mutants whose mtDNAs are deleted for most of the ρ+ genome, with the remaining sequences amplified and organized as oligomeric repeats (Dujon, 1981). For most ρ– strains, some fraction of the progeny of ρ+ × ρ– crosses inherit the ρ– mtDNA and those petites are called ‘suppressive’ (Ephrussi and Grandchamp, 1965). Those petite mutants whose mtDNA is not transmitted to progeny of crosses with ρ+ strains are called ‘neutral’ petites (N ρ–). The extent of suppressiveness is generally a property of the fragment of ρ+ mtDNA retained in the petite mutant (Dujon, 1981). In the most extreme cases of hypersuppressive (HS) petites, >95% of the progeny of ρ+ × HS ρ– crosses contain exclusively the ρ– genome, despite an inherent growth advantage for ρ+ cells (Blanc and Dujon, 1980; de Zamaroczy et al., 1981).
The mtDNAs of all HS ρ– strains contain one of several ∼300 bp sequences from ρ+ mtDNA, known as ori or rep sequences (Blanc and Dujon, 1980; Faugeron-Fonty et al., 1984). Early models for hypersuppressiveness proposed that ori sequences are origins of mtDNA replication and that their high density in HS ρ– genomes provides a replicative advantage over ρ+ mtDNA. However, because many ρ– mutants lack an ori sequence, yet maintain normal levels of mtDNA in vegetatively growing cells, it is evident that ori sequences are not essential for ρ– mtDNA replication (Fangman et al., 1989).
The ρ+ mtDNAs of most strains have seven or eight ori sequences, all of which are similar in organization and primary sequence (de Zamaroczy et al., 1984; Faugeron-Fonty et al., 1984). Each ori contains a nonanucleotide promoter for the RPO41-encoded mitochondrial RNA polymerase (Greenleaf et al., 1986) and three conserved G/C-rich sequences called boxes A, B and C (Bernardi, 1982). Because only ori1, 2, 3 and 5 have been found in mtDNAs of HS ρ– strains, the others are called ‘inactive’ oris (de Zamaroczy et al., 1981). Inactive ori sequences differ from ‘active’ ones by the presence of short insertions, one of which disrupts the promoter.
Active ori elements resemble the heavy-strand origin of mammalian mtDNA, which also contains an RNA polymerase promoter and three conserved sequences, CSBI, II and III (Shadel and Clayton, 1997). Heavy-strand DNA synthesis begins with transcription from the promoter followed by a switch to DNA synthesis within CSBII. Because the C box of yeast ori elements resembles CSBII both in primary sequence and in proximity to the promoter, it has been suggested that yeast ori elements direct replication in a manner resembling that of mammalian mtDNA (Baldacci and Bernardi, 1982).
Although some biochemical evidence supports this hypothesis (Baldacci et al., 1984; Stohl and Clayton, 1992; Xu and Clayton, 1995; Graves et al., 1998), several experiments raise doubts about the relevance of RNA priming both for the replication of ρ– mtDNA (Fangman et al., 1990) and for the hypersuppressiveness of HS ρ– mitochondrial genomes (Lorimer et al., 1995). In particular, deletion of the RPO41 gene eliminates transcription of HS and N ρ– mtDNAs, but does not interfere with their replication or maintenance in vegetative cells. Moreover, the preferential transmission of HS ρ– mtDNA in a cross with an N ρ– strain is undiminished by disruption of RPO41 in both strains. Those investigators conclude that RPO41-primed replication is not necessary for hypersuppressiveness and suggest that HS ρ– genomes may have a segregational advantage over other mtDNAs.
Here, we compare the segregation and replication of mtDNA from HS ρ– and N ρ– strains. In crosses to ρ+ cells, we found no immediate segregational advantage for HS ρ– mtDNA to first medial zygotic buds compared with mtDNA of the N ρ– strain. We obtained strong evidence suggesting a unique mode of replication of HS ρ– mtDNA, which includes the formation of asymmetric single-stranded (ss) DNA circles as replication intermediates. Biolistic transformation of mitochondria was used to demonstrate that the conserved nonanucleotide promoter is necessary for hypersuppressiveness and RNA-primed DNA replication that leads to strand-specific ssDNA circles.
Results
Direct visualization of segregating mitochondrial genomes using FISH
We used fluorescent in situ hybridization (FISH) to determine whether the ori5-containing mtDNA of HS ρ– strain HS40 (Parikh et al., 1987) is preferentially transmitted to diploid buds issued from zygotes of crosses with a ρ+ strain. We analyzed in parallel a cross between the same ρ+ strain and the N ρ– strain, VAR1. These petite genomes have no sequences in common, but each contains an A/T-rich fragment of ρ+ mtDNA that includes short G/C-rich sequences (Hudspeth et al., 1984).
In these experiments, probes specific for ρ+, HS40 and VAR1 ρ– mtDNAs were generated by PCR using the fluorophore-labeled nucleotides Alexa488-5-dUTP and Alexa594-5-dUTP. Probes containing Alexa488 (green) detect ρ+ mtDNA and probes containing Alexa594 (red) detect the ρ– genomes. Control experiments confirmed that neither the HS40 nor the VAR1 probe detects ρ+ mtDNA under the conditions used for these double labeling experiments. Cells were mated synchronously for 3 h, fixed and hybridized with Alexa probes specific for the COX1 gene in ρ+ mtDNA, and specific for either of the ρ– mtDNAs (Figure 1).
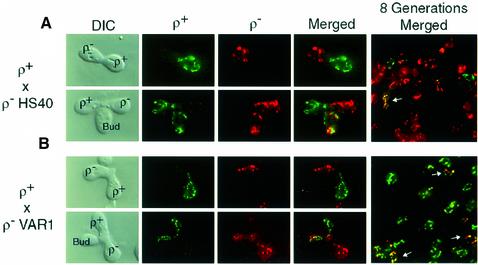
Fig. 1. FISH analysis of transmission of mtDNAs in crosses between a ρ+ strain and HS and neutral ρ– strains. mtDNAs were detected using fluorescently labeled probes containing Alexa488-dUMP (green) for ρ+ genomes and Alexa594-dUMP (red) for ρ– genomes as described in Materials and methods. (A) Representative zygotes from the ρ+ × ρ– HS40 cross. Early unbudded zygotes (top panels) and zygotes with initial medial buds (bottom panels) are shown. The last panel is a merged field of diploid progeny eight generations after mating. (B) Representative zygotes from a ρ+ × ρ– VAR1 cross. The fields are arranged as in (A).
In early zygotes, ρ+ and HS40 ρ– mtDNAs were unmixed (Figure 1A, top panels). During zygote maturation, both parental mtDNAs were transferred to the first buds (Figure 1A, bottom panels); similar results were obtained with the neutral VAR1 petite (Figure 1B). After eight generations, however, the majority of diploid progeny from the HS ρ– × ρ+ cross contained exclusively HS40 ρ– mtDNA (Figure 1A, last panel), whereas progeny from the N ρ– × ρ+ cross contained predominantly ρ+ mtDNA (Figure 1B, last panel). Even at this relatively late time point, there are still some cells with both mtDNAs (see arrows). These results suggest that the uniparental inheritance observed in HS ρ– × ρ+ crosses does not result from extremely biased segregation of HS ρ– mtDNA into the first zygotic bud.
Single-stranded molecules in mtDNA of the HS petite HS40
Because we did not find a clear segregation advantage for the mtDNA of the HS petite, we investigated whether HS ρ– mtDNA has some unique features of replication that could be related to the mechanism of hypersuppressiveness. We performed two-dimensional (2D) gel electrophoresis of mtDNA and compared the types of mtDNA molecules present in ρ– strains HS40 and VAR1. Figure 2A illustrates the resolution of linear, circular and supercoiled DNAs and various oligomeric species obtained with this method (Fangman et al., 1989). Total DNA was fractionated on each gel without prior cleavage by a restriction enzyme. The blots were hybridized with a double-stranded (ds) probe for VAR1 ρ– mtDNA (Figure 2B) and strand-specific probes for HS40 ρ– mtDNA (Figure 2C and D).
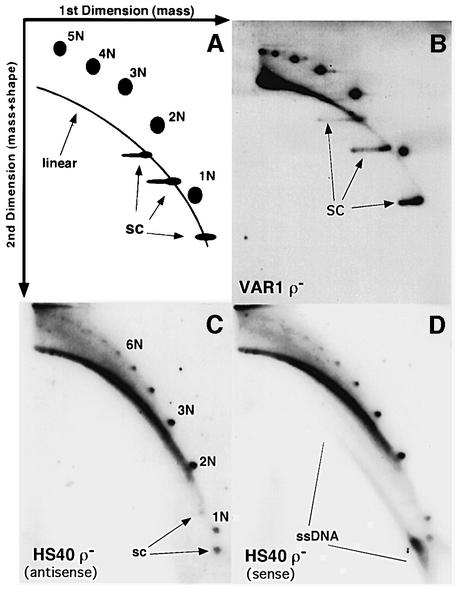
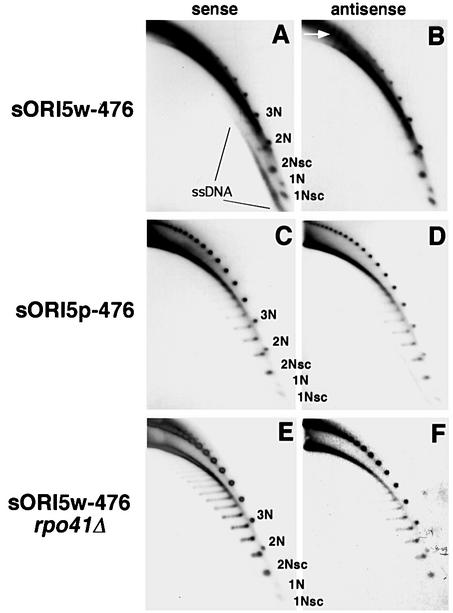
Fig. 2. Strand-specific ssDNA molecules are present in mtDNA from an HS petite. (A) Schematic diagram illustrating the types of mtDNA molecules that are resolved by this 2D gel system. The first dimension separates molecules by mass, while the second dimension resolves molecules by mass and shape. Linear dsDNA runs as a diagonal arc across the gel (linear); nicked or relaxed dsDNA circles are detected as discrete spots above the linear arc; supercoiled topoisomers migrate faster than the linear arc (SC). (B) 2D gel analysis of mtDNA from the neutral VAR1 petite. Fractionated DNA was hybridized with a random primed fragment of VAR1 DNA. (C and D) 2D gel analysis of mtDNA from the hypersuppressive petite, HS40. The gels were prepared as in (B) except that they were hybridized with 5′ end-labeled oligonucleo tides specific for the antisense (C) or sense strands (D) of HS40 mtDNA.
In each strain we observed strong signals of linear ds molecules, discrete ds circular monomers and oligomers of the repeating unit, and supercoiled molecules (Figure 2B–D). The VAR1 ρ– mtDNA (Figure 2B) contained topoisomers of supercoiled monomer, dimer and trimer circles with a range of linking numbers. The HS40 signals in Figure 2C and D differ from the VAR1 results in two ways: (i) the HS40 samples contained fewer supercoiled molecules; and (ii) there was a sense strand-specific signal below the arc of linear dsDNA (Figure 2D), where ssDNA would be found (Backert et al., 1997). These data suggest that strain HS40 ρ– produces ssDNA asymmetrically while the neutral VAR1 petite has no detectable ssDNA.
Linear and circular ss molecules in mtDNA of strain HS40
To confirm and further characterize the ssDNA in HS40 ρ– mtDNA samples, we compared the hybridization patterns of native and denatured DNA samples, as well as samples treated with specific nucleases, using the same sense strand-specific probe as in Figure 2D (Figure 3A–D). For reference, Figure 3A shows a subset of the pattern of DNA molecules shown in Figure 2D. Pre-treatment of the DNA with S1 endonuclease eliminated the ssDNA, leaving the rest of the pattern unaltered (Figure 3B). To test for the presence of ssDNA circles, we analyzed DNA that had been pre-treated with exonuclease I (ExoI). Because ExoI is a 3′–5′ single-strand-specific nuclease, only ssDNAs with a displaced 3′ end will be sensitive to treatment. ExoI treatment dramatically reduced the amount of ssDNA signal, with little effect on the pattern of duplex DNAs (Figure 3C). A fraction of the material migrating below the arc of linear dsDNA was resistant to ExoI, and that material migrated in a distinctive pattern corresponding to circular ss monomers and oligomers of the HS40 ρ– mtDNA repeating unit (arrows in Figure 3C).
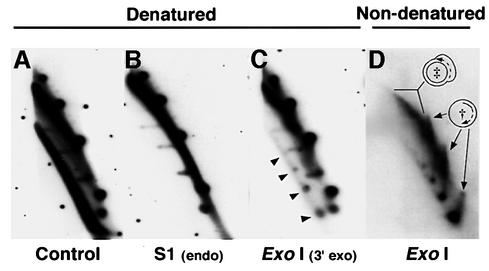
Fig. 3. Characterization of ssDNA molecules present in mtDNA of strain HS40. Shown is the part of the 2D gel pattern containing low molecular weight species of mtDNA from HS40 ρ– hybridized with the sense-strand-specific oligonucleotide, as in Figure 2D. (A) Untreated DNA, denatured before transfer. This panel resembles Figure 2D but is overexposed to improve comparison with (B)–(D). (B) DNA treated with S1 nuclease to degrade ssDNA, denatured before transfer. (C) DNA treated with ExoI to degrade ss molecules with 3′ ends, denatured before transfer. The unit length monomer, dimer and trimer ExoI-resistant ssDNA circles are indicated by arrows. (D) DNA treated with ExoI, not denatured before transfer. † indicates replication intermediates from second-strand synthesis of ssDNA circles. ‡ indicates duplex DNA with ssDNA character. These replication intermediates may represent duplex circles with expanded D-loop regions.
If the ssDNA circles present in HS40 mtDNA are replication intermediates, then as the complementary strand is made, the pattern should become more like dsDNA circles. Such molecules would be detected in this 2D gel system as a spike of signal between each ssDNA circle and the dsDNA circle of the same size. To increase the sensitivity of detecting such species, the resolved ExoI-resistant DNA molecules were transferred to nylon membranes without denaturation and hybridized using a random-primed HS40 probe. As shown in Figure 3D, ds linear and circular DNAs seen in earlier panels were virtually eliminated. However, the ssDNA circles were still present, and molecules were now evident between the position of the fully ss circle to the position of its duplex counterpart (see † in Figure 3D). The spikes stop abruptly at the duplex position because fully dsDNA no longer binds to the membrane (without prior denaturation). In addition, duplex DNA molecules with ssDNA character (see ‡ in Figure 3D) were detected as hybridization signals that increase in mass from the positions of covalently closed duplex dimer and trimer DNA circles. Although it is possible that these intermediates are sigma molecules undergoing rolling circle replication, we have been unable to detect discrete 5′ ends. Therefore, we interpret these patterns as molecules undergoing asymmetric strand displacement replication. All the ssDNA replication intermediates we observed correspond to the strand of mtDNA that would be displaced by transcription. These results, combined with the recent detection of RNA–DNA hybrids in mtDNA of an ori5 HS petite (Graves et al., 1998), suggest that the ssDNA replication intermediates may arise from RNA-primed replication.
The ori5 promoter is required for hypersuppressiveness
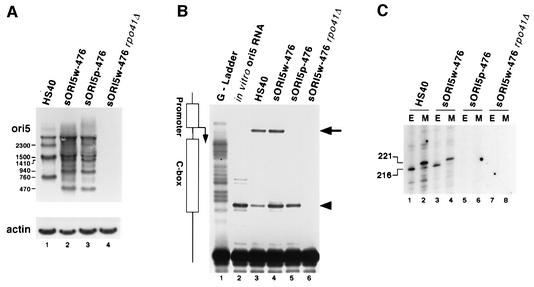
The mitochondrial RNA polymerase, encoded by the RPO41 gene, was not necessary for the preferential inheritance of HS ρ– mtDNA in crosses with an N ρ– strain (Lorimer et al., 1995). Unfortunately, it was impossible for those researchers to test the role of Rpo41p in hypersuppressiveness in ρ+ × ρ– crosses because ρ+ mtDNA is unstable in the rpo41Δ background (Greenleaf et al., 1986). We re-examined the role of transcription from the ori5 promoter in HS crosses by analyzing ρ– strains with a mutated or wild-type ori5 promoter made by reverse genetics. We constructed the 7 kb plasmid pMAC-HS40w, which contains the entire 761 bp repeating unit of the mtDNA of the HS petite HS40 (Figure 4A), and used biolistic transformation to construct strain pMIT-HS40w, which maintains the 7 kb plasmid (as oligomeric repeats) in its mitochondria (see Materials and methods). As shown in Table I, that strain was not HS in crosses with the ρ+ tester strain, even though it contains the entire HS40 repeating unit. Previous studies have shown that a short repeating unit is necessary for ori-containing ρ– strains to be HS (de Zamaroczy et al., 1981).
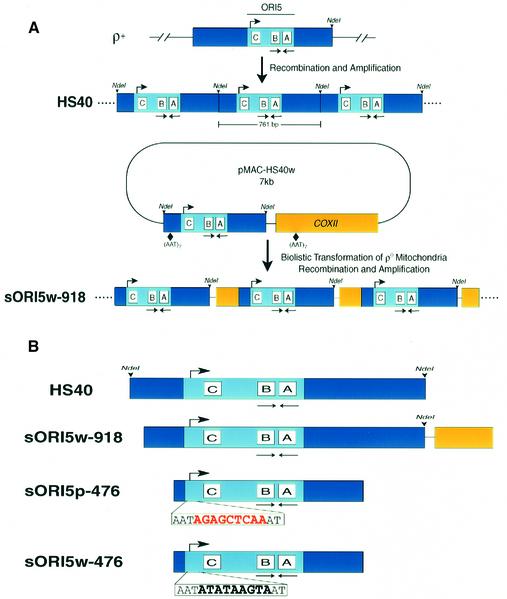
Fig. 4. Generation of recombinant petites by biolistic transformation. (A) Diagram of the 7 kb plasmid pMAC-HS40w. The 761 bp HS40 ρ– sequence is indicated by the blue box flanked by NdeI sites. The conserved ori5 sequence is shaded light blue and the promoter and A, B and C boxes are indicated. The 2.7 kb COXII fragment is indicated by the yellow box. Vector sequences are indicated by the thin line. The initial mitochondrial transformants contained the entire pMAC-HS40w as an oligomeric array of 7 kb repeats. Petite strain sORI5w-918 resulted from spontaneous recombination between (AAT)7 repeats at the indicated places in HS40 and COXII followed by amplification. (B) Summary of synthetic petite genomes obtained through biolistic transformation. The repeating unit of each petite is indicated. Petites sORI5p-476 and sORI5w-476 are identical except for the mutated nucleotides in the nonanucleotide promoter.
Table I. Suppressiveness data.
| Tester | Suppressiveness |
|||||
|---|---|---|---|---|---|---|
| HS40 ρ– | sORI5w-918 ρ– | sORI5w-476 ρ– | sORI5p-476 ρ– | pMIT-HS40w ρ– | pMIT-HS40p ρ– | |
| ρ+ | 99 | 99 | 98 | 16 | 12 | 9 |
| VAR1 ρ– | 100 | n.d. | 99 | 96 | n.d | n.d. |
| HS3324 ρ– | 58 | n.d. | 50 | 10 | n.d. | n.d. |
For each ρ+ × ρ– cross, zygote clones were selected on minimal medium and scored for the presence of ρ+ progeny by replica plating to YPGly medium. For ρ– × VAR1 ρ– crosses, zygote clones were screened by colony hybridization with probes specific to ori5 or VAR1. In each case, >200 zygote clones were scored. For ρ– × HS3324 ρ– crosses, the diploid progeny were outgrown for 2 days, total DNA was isolated and the fraction of each parental mtDNA genome was determined by restriction digestion and Southern hybridization with an ori5 probe. These values represent the average of three independent matings.
Although the 7 kb mtDNA repeats of this transformed ρ– strain were quite stable, screening identified some subclones with shorter repeating units, most of which retained HS40 sequence but were deleted for most other sequences. A number of independent isolates had a 918 bp repeating unit that contains most of the HS40 repeat including all of ori5 plus a small portion of the COXII region (sORI5w-918, see Figure 4A). DNA sequencing showed that the repeating unit of sORI5w-918 was excised from pMIT-HS40w by recombination between two (AAT)7 sequences at the positions shown in Figure 4A. Petite sORI5w-918 was HS in crosses with the ρ+ tester, yielding ∼99% zygote clones containing exclusively ρ– cells (Table I). These results establish that it is possible to generate HS strains with synthetic ρ– genomes that provide the essential control for using reverse genetics to analyze the function of the ori5 promoter.
Next, we mutated the ori5 promoter (Figure 4B, bottom) and transformed the resulting 7 kb plasmid, pMAC-HS40p, into mitochondria. Unexpectedly, the mtDNA of the resulting ρ– transformant was much more stable than ρ– pMIT-HS40w, and we were unable to obtain a subclone of pMIT-HS40p with the 918 bp repeat for comparison with sORI5w-918. Ethidium bromide treatment of strain pMIT-HS40p induced internal deletions, and screening for segregants with smaller repeating units (i.e. higher copy number of ori5 sequence) yielded strain ρ– sORI5p-476, which has a 476 bp mtDNA repeating unit that includes all of ori5 (Figure 4B).
Extensive screening did not yield any derivatives of ρ– strains with the wild-type ori5 that could be used as a matched control for strain sORI5p-476, so we had to devise an alternative strategy to construct that key control strain. We mated a MATa sORI5p-476 strain with a MATα pMIT-HS40w strain and screened for recombinants with the 476 bp repeating unit and the wild-type promoter (see Materials and methods). Several candidates (called sORI5w-476) were obtained and confirmed by sequencing. Although the two strains had the same mtDNA copy number (data not shown), test crosses showed that ρ– sORI5w-476 was 98% HS, whereas sORI5p-476 was only 16% suppressive (Table I). These results show that the ori5 promoter is necessary for hypersuppressiveness in crosses with a ρ+ tester.
That being the case, how do we explain the previous report (Lorimer et al., 1995) that RPO41 is not required for the preferential transmission of an HS ρ– genome in crosses to a neutral petite? In order to compare their study with ours, we analyzed crosses between ρ– strains sORI5w-476 and sORI5p-476 and the neutral VAR1 petite (Table I). In both crosses, the ori5-containing petite was preferentially inherited. However, in crosses between a different HS ori5-containing petite (HS3324) and sORI5p-476, there was a clear inheritance advantage for the mtDNA of the HS3324 petite. Evidently, the 16% suppressiveness observed for sORI5p-476 mtDNA in crosses to a ρ+ strain provides the genome with a sufficient advantage to outcompete the neutral VAR1 petite, but insufficient to compete with a ρ– genome containing a wild-type ori5.
Production of ssDNA requires an active ori5 promoter
Next, we determined the effect of the ori5 promoter mutation on replication intermediates identified in the HS ρ– strain HS40 (Figure 5). Aliquots of total DNA from strains sORI5w-476, sORI5p-476 and sORI5w-476 rpo41Δ were analyzed by 2D gel electrophoresis without prior restriction enzyme digestion and the blots were hybridized with strand-specific oligonucleotides as in Figure 2. As shown in Figure 5A and B, mtDNA from HS ρ– sORI5w-476 contains strand-specific ssDNA molecules below the arc of linear dsDNA. Furthermore, the ssDNA present contains ssDNA circles when treated with ExoI as in Figure 3C (data not shown).
Fig. 5. Strand-specific ssDNA depends on transcription from the ori promoter. 2D gel electrophoresis was performed on mtDNA from strains sORI5w-476 (A and B), sORI5p-476 (C and D) and sORI5w-476 rpo41Δ (E and F). Blots were first hybridized with an oligo nucleotide probe specific for the antisense strand (B, D and E), then stripped and hybridized with a sense-strand-specific oligonucleotide (A, C and E). The smaller oligomeric species are labeled in (A), (C) and (E) for reference. The arrow indicates replication intermediates migrating between linear and open-circle mtDNA.
In contrast, the mtDNA from ρ– sORI5p-476 had a dramatically reduced level of ssDNA and what little ssDNA was present was not strand specific (Figure 5C and D). Experiments using ExoI did not detect any ssDNA circles in this strain, even after long film exposure (data not shown). Also, no ssDNA was detectable in mtDNA from strain sORI5w-476 rpo41Δ (Figure 5E and F). These results indicate that transcription from the ori5 promoter is necessary for the production of strand-specific ssDNA molecules.
Several other differences, aside from ssDNA levels, were also evident in the above comparisons. First, the distribution of ds circular species was shifted toward higher oligomers in the mutants, which may be due to changes in intramolecular recombination (MacAlpine et al., 2000). A related phenotype was the marked decrease in the level of 1N molecules. Secondly, there was more broken linear dsDNA in the mtDNA samples from the ρ– strains with the wild-type promoter. Thirdly, in the strains where transcription from the promoter is impaired (sORI5p-476) or absent (sORI5w-476 rpo41Δ), the supercoiled isoforms of mtDNA were more relaxed, as evident from the increase in topological isomers (compare Figure 5B and C). This relaxation was also evident in VAR1 ρ– mtDNA (Figure 2B) and in an ori6 ρ– mutant that has a G/C-rich insertion in the promoter (not shown). Finally, mtDNA from ρ– strains with wild-type ori5 contained molecules that ran between the arc of ds linears and the ds circles (see arrow in Figure 5B). These molecules depend on transcription from the promoter element (compare Figure 5B with D and F) and were resistant to digestion with S1 and ExoI nucleases (not shown); thus, they may result from lagging strand synthesis of the displaced sense strand on large oligomeric molecules.
Transcription is not sufficient for hypersuppressiveness
To define in more detail the role of transcription in hypersuppressiveness, we analyzed northern blots of RNA isolated from strains HS40, sORI5w-476, sORI5p-476 and sORI5w-476 rpo41Δ using an oligonucleotide probe that hybridizes to transcripts from the ori5 promoter (Figure 6A). No signal was detected in control experiments with an oligonucleotide for the opposite strand (not shown). In agreement with previous findings (Baldacci and Bernardi, 1982), we observed a series of RNAs that corresponds to oligomers the length of the ρ– mtDNA repeating unit. For example, HS40 RNA contained bands ∼760, 1500 and 2300 nt long (lane 1), whereas petite sORI5w-476 contained bands ∼470, 940 and 1410 nt long (lane 2). As expected, no mitochondrial transcripts were detected in an rpo41Δ mutant strain (lane 4). Surprisingly, the petite with the mutant ori5 promoter (sORI5p-476) contained the same transcripts as did ρ– sORI5w-476, and at comparable abundance (compare lanes 2 and 3). These results suggest that the ori5 promoter is not the only site for transcription initiation in this segment of mtDNA and that it is not required for production of unit-length and oligomeric transcripts.
Fig. 6. Transcription from the nonanucleotide promoter is required for priming DNA replication. (A) Levels of transcripts from ori5-containing mtDNAs. RNA from the strains shown was balanced by hybridization with a probe for actin mRNA (bottom) and then hybridized with the ori5-specific probe to detect transcripts of those mtDNAs (top). The relative sizes of the main signals are indicated. (B) RT primer extension to map 5′ ends near ori5. The same RNAs as in (A) were hybridized with primer O969, and RT primer extension products were separated alongside a ddGTP sequencing ladder. Products with ends at the promoter element are indicated (long arrow). In vitro transcribed RNA (lane 2) was used to identify strong-stop artifacts (short arrow). (C) Detection of RNA–DNA hybrids by RNA capping. Purified mtDNA from the strains shown was digested with an excess of EcoRV or MspI, endonucleases that cut the DNA exactly 216 and 221 bp from the promoter start site, respectively, and capped with guanylyl transferase. Equal amounts of input DNA were used in each capping reaction.
Next, we used reverse transcriptase (RT) primer extension to map the 5′ ends of the transcripts present in these ρ– strains (Figure 6B). A strong band mapping to the first A in the promoter was observed in RNA from ρ– strains HS40 and sORI5w-476 (lanes 3 and 4), but no band was detected at that position in RNA from ρ– sORI5p-476 (lane 5). Those three samples all contained an additional strong primer extension product mapping to a T at the promoter-distal edge of the C box (short arrow). Because this product was also present in a control reaction using an in vitro transcribed HS40 RNA template (lane 2), we conclude that it is an artifact of primer extension. As expected, primer extensions using cellular RNA from the sORI4w-476 rpo41Δ strain yielded no signal (Figure 6B, lane 6). From these experiments, it is clear that some of the transcripts in strains HS40 and sORI5w-476 initiate at the ori5 promoter, but that similar transcripts can arise independently of the ori5 promoter. Importantly, these data suggest that specific initiation of transcription from the promoter is required for the uniparental inheritance of the mtDNA of HS petites.
In vertebrates, mtDNA replication is primed by transcription from the light-strand promoter (Shadel and Clayton, 1997). Additionally, Graves et al. (1998) demonstrated the presence of RNA–DNA hybrids that mapped to the promoter of an ori5 petite. Such RNA-primed replication intermediates can be detected by capping the RNA component with radioactive GTP (Figure 6C). Purified mtDNA was prepared from strains HS40, sORI5w-476, sORI5p-476 and sORI5w-476 rpo41Δ and digested with an excess of EcoRV or MspI, which cut the DNA exactly 216 and 221 bp away from the promoter start site, respectively. Capped intermediates mapping to the promoter were observed in petites HS40 (lanes 1 and 2) and sORI5w-476 (lanes 3 and 4), consistent with previous results (Graves et al., 1998). However, in samples from ρ– sORI5p-476 (lanes 5 and 6), no capped replication intermediates of that size were observed even though active transcription of mtDNA occurs in that strain. As expected, the sORI5w-476 rpo41Δ strain (lanes 7 and 8) lacked capped RNA–DNA hybrids. Our results demonstrate that only transcripts initiating from the ori5 promoter are capable of priming replication and make it unlikely that a specific primase activity, independent of RPO41, primes replication from the ori5 promoter.
Discussion
Competing models for the mechanism of hypersuppressiveness center on replicative or segregational advantages of HS ρ– mitochondrial genomes. Using FISH to study the transmission of ρ– genomes in crosses to a ρ+ strain, we found no preferential transfer of the HS ρ– mtDNA to the first diploid buds issued from zygotes, relative to N ρ– or ρ+ mtDNAs. These data rule out the possibility that HS ρ– mtDNA has an immediate segregational advantage early in zygote maturation. However, eight generations after mating, diploid progeny of HS40 ρ– × ρ+ crosses have predominantly ρ– mtDNAs, whereas most progeny of VAR1 N ρ– × ρ+ crosses were ρ+ at that time. Because some cells still retain ρ– and ρ+ mtDNAs up to eight generations after mating, we can also rule out the immediate destruction or inability of ρ+ genomes to replicate when in the presence of HS mtDNA.
Our molecular and genetic experiments identify promoter-primed DNA replication as a necessary factor for hypersuppressiveness. We present strong evidence for a mode of replication of HS ρ– mtDNA involving RNA priming from a nonanucleotide promoter that leads to asymmetric strand displacement. No such mode of replication was observed for mtDNA of a neutral petite. A transformed promoter mutation blocked the production of asymmetric ssDNA and inhibited hypersuppressiveness in crosses with a ρ+ strain. These observations strongly support the conclusions that initiation of replication at the ori5 promoter and asymmetric strand displacement are necessary for hypersuppressiveness. They also provide strong in vivo support for the earlier inference that ori4, 6, 7 and 8 of ρ+ mtDNA are inactive in terms of suppressiveness because their promoters are disrupted.
The ssDNAs in HS ρ– mtDNA are derived from the non-transcribed strand and include monomer and oligomeric circles as well as longer linear molecules. We also detected partially ds circular DNAs that are likely intermediates in the synthesis of the complementary strand. In the classical model for the replication of animal mtDNA, replication initiates at the heavy-strand origin, displacing the light strand asymmetrically. This unidirectional replication eventually exposes the light-strand origin at which continuous synthesis of the new heavy strand is initiated (Shadel and Clayton, 1997). Aside from asymmetric ssDNA molecules, we also detected high molecular weight replication intermediates migrating between the arc of linears and the oligomer circles that depend on promoter-primed replication (Figure 5B, arrow). Because these molecules are resistant to ExoI and S1 nucleases, we conclude that they represent molecules in which lagging strand synthesis has been initiated on the partially displaced strand. A recent report indicates that coupled leading and lagging strand synthesis occurs in animal mtDNA replication (Holt et al., 2000). Coupled leading and lagging strand synthesis must also occur in fungal mtDNAs because Y-arcs are readily detected (Han and Stachow, 1994; MacAlpine et al., 1998), even in HS petites (Lockshon et al., 1995).
Prior to this study, it was controversial whether RNA-primed replication at the ori elements was necessary for uniparental inheritance. Although earlier experiments indicated that replication from ori elements is RNA-primed (Baldacci et al., 1984; Graves et al., 1998), other studies showed that the mitochondrial RNA polymerase, RPO41, is required neither for the replication of ρ– mtDNAs nor for the preferential transmission of mtDNA of an HS ρ– strain (Fangman et al., 1990; Lorimer et al., 1995). Because ρ+ genomes are unstable in an rpo41Δ background, Lorimer et al. (1995) could not test the effect of rpo41Δ on the transmission of HS ρ– mtDNA using the standard ρ+ × HS ρ– cross.
Using reverse genetics to mutate the ori5 promoter in HS ρ– mtDNA, we have obtained compelling evidence for a role of RNA priming in HS ρ– mtDNA replication and in hypersuppressiveness. The mutated promoter in the synthetic petite sORI5p-476 did not alter the mtDNA copy number, but it dramatically reduced suppressiveness in the standard test cross to a mere 16%, as compared with 99% in the control crosses. We also analyzed these synthetic petite strains in ρ– × ρ– crosses and obtained essentially the same results as Lorimer et al. (1995): when mated to N ρ– VAR1, both sORI5p-476 and sORI5w-476 were >96% suppressive. Importantly, we found that sORI5p-476 was transmitted much less efficiently when mated to another HS ρ– strain than was sORI5w-476; these results show that the promoter mutation inhibits preferential transmission relative to an HS petite. We conclude that the mild (16%) suppressiveness of ρ– sORI5p-476 in ρ+ crosses probably explains its preferential transmission in the cross with the N ρ– strain. Further analysis is needed to determine whether the preferential transmission observed in some ρ– × ρ– crosses is due to a bias in replication or segregation.
Mutation of the promoter eliminated detectable transcripts initiating from that site, but, surprisingly, the steady-state level of transcripts was unaffected. A previous study showed that some ρ– mutants lacking a nonanucleotide promoter also accumulate transcripts due to alternative initiation sites (Zassenhaus et al., 1984). We show that those transcripts depend on Rpo41p, and therefore a hierarchy of promoters must exist in yeast mtDNA that remains to be characterized. Our finding that the promoter mutation eliminated detectable 5′ ends in transcripts, by both primer extension and capping experiments (Figure 6 and data not shown), suggests that there are probably many alternative sites for transcription initiation, at least in the absence of a canonical promoter. Our RNA blots show that transcripts from alternative promoters come from the same strand of mtDNA; if there are also alternative promoters on the other strand, then those transcripts must have a much shorter half-life.
Our finding that these alternative transcripts do not support asymmetric DNA replication suggests that this mode of replication is primed by a coordinated set of events initiated at the promoter. Consistent with this idea are the in vitro observations that stable RNA–DNA heteroduplexes occur at the C box in yeast, but only when transcription initiates from a nearby promoter (Xu and Clayton, 1995). Previous investigators have speculated that the initial transcript is cleaved to form the primer for DNA synthesis by a mitochondrial form of RNase MRP (Stohl and Clayton, 1992).
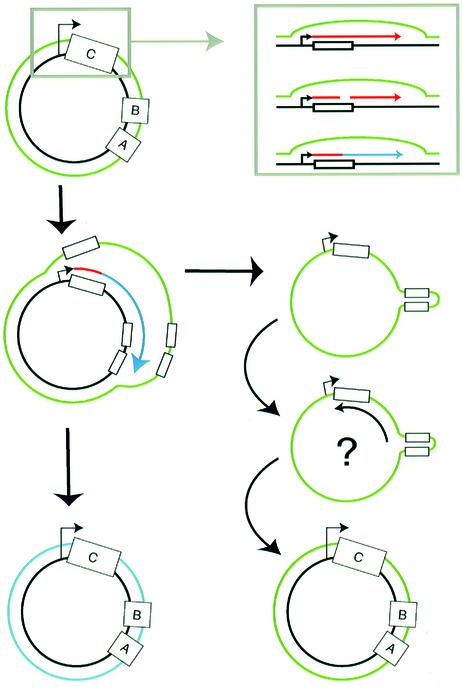
Figure 7 shows how replication initiating from the ori5 promoter would result in the asymmetric strand displacement that we observed. Transcription initiated at the promoter would generate transcripts through the C box and those transcripts would then be cleaved to yield a 3′-OH group to prime DNA replication. Elongation of the nascent strand displaces the complementary strand, resulting in the release of single strands that we observed. Events initiated on monomer or small oligomer dsDNA circles presumably result in the monomer through trimer ssDNA circles that we observed; events initiated on larger oligomers would yield even larger displaced circles that are evidently broken in our preparations, resulting in the arc of linear ssDNA detected. In the model (Figure 7), we speculate that lagging strand synthesis initiates at the hairpin formed by the A and B boxes, by analogy to the model for light-strand synthesis in mammalian mitochondria.
Fig. 7. Model for ori5-dependent mtDNA replication. Transcription initiates from the nonanucleotide promoter and transcripts (red) are processed in the C box to provide a 3′-OH end that primes DNA replication (blue) (inset). Elongation of the primed DNA strand displaces the sense strand of mtDNA (green), eventually releasing a ssDNA circle. Lagging strand synthesis of the ssDNA circle may be initiated at or near the hairpin formed by the A and B boxes.
Because maintenance of ρ– genomes requires neither transcription nor an ori element, there must be at least one other mode of mtDNA replication. Priming of replication by recombination is an attractive hypothesis for the replication of ρ– genomes that lack an ori sequence and for ori-containing genomes when RNA primers cannot be made. Recombination is very active in yeast mitochondria (Lockshon et al., 1995; MacAlpine et al., 1998), and we have previously suggested that some portion of mtDNA replication may depend on strand invasion by recombination events to make primers for DNA polymerase (Zelenaya-Troitskaya et al., 1998). Because eliminating the ori-dependent mode of replication had no effect on the steady-state level of ρ– mtDNA, it follows that mtDNA copy number is regulated by an as yet unidentified mechanism.
How then does ori-dependent replication account for the extreme inheritance advantage observed in HS petites? The high density of ori sequences in the mtDNA of HS ρ– petites relative to ρ+ strains may account for hypersuppressiveness because the ρ– mtDNAs would effectively compete for components of the replication apparatus. ori elements active in initiation of replication could provide sites for the association of mtDNA with the inner mitochondrial membrane, analogous to the membrane association of bacterial origins of replication. In bacteria, this membrane association is intimately tied to the segregation of the genome during cell division (Wheeler and Shapiro, 1997; Sharpe and Errington, 1999). Recent studies suggest that there is a segregation apparatus for mtDNA (Nunnari et al., 1997; Okamoto et al., 1998) and it will be interesting to learn whether there is a connection between this putative apparatus and active ori elements.
Materials and methods
Strains and growth conditions
The following strains were used in this study: 14 WW ρ+ (MATα ade2 trp1 ura3-52 leu2 cit1::LEU2) and its ρ– derivatives 14 WW VAR1 and 14 WW HS40; ρ0 and ρ+ derivatives of MCC109 (MATα ade2 ura3 kar1) and ID41-6/161 (MATa ade1 lys1); BS127 HS3324 ρ– (MATα ade1 ade2 leu2 trp1 ura3); JC3-TF145 coxIIΔ (MATa ade2 lys1); DBY747 ρ+ (MATa his3 leu2 trp1 ura3); HS40 ρ– and VAR1 ρ– derivatives of strain PSY142 (MATα, leu2 lys2 ura3). The mtDNA of strain ID41-6/161 ρ+ is described in Moran et al. (1995). The petite genomes HS40, HS3324 and VAR1 (also called ρ– 2-33) are described, respectively, in Lorimer et al. (1995), Parikh et al. (1987) and Hudspeth et al. (1984). DNA from strain 5657 (Research Genetics) containing a KAN disruption of RPO41 was used in a PCR to generate a cassette to knock out RPO41 in strain ID41-6/161 ρ0. All strains were grown at 30°C in YPD medium (1% yeast extract, 2% peptone and 2% dextrose).
Plasmids and mutagenesis
pGEM-H3 is a derivative of pGEM 3zf(+) (Promega) in which the polylinker SstI site was mutated to a HindIII site. MtDNA from ρ– strain HS40 was cleaved by digestion with NdeI and cloned into the SmaI site of pGEM-H3, resulting in pGEM-HS40. A 1 kb HpaII fragment of VAR1 mtDNA was cloned into pGEM 3zf(+) to yield pGEM-VAR1. pMAC-HS40w was constructed by cloning the 2.7 kb COXII PstI fragment from pPA100 (Butow et al., 1996) into the PstI site of pGEM-HS40. The ori5 promoter of pMAC-HS40w was mutated to a SacI site by oligonucleotide mutagenesis (BRL) (see Figure 4B), yielding pMAC-HS40p; the mutation was confirmed by DNA sequencing.
FISH
Two fragments of the COXI gene (Bonitz et al., 1980) were amplified from ρ+ mtDNA of strain ID41-6/161 by PCR: COXI-1 was amplified using primers FW1 (5′-AGTGGTATGGCAGGAACAGC) and RV1 (5′-GATACCTCTACCTAACGC). COXI-2 was amplified using primers FW2 (5′-ATCAGAAGCTAAAGTAACTGATC) and RV2 (5′-ACT AGTATGATGTCTAACCC). Fluorescent ρ+ probes were generated by further PCR of COXI-1 and COXI-2. Fluorescent probes for ρ– mtDNAs were generated by PCR using the universal primers SP6 and T7, pGEM-HS40 and pGEM-VAR1. In PCRs to incorporate fluorescent nucleotides, the final concentration of each dNTP was 0.2 mM. The ratio of fluorescent nucleotides Alexa488-5-dUTP (green) and Alexa594-5-dUTP (red) (Molecular Probes) to dTTP was 2.33:1 for the ρ+ fragments and 1:2.33 for the ρ– fragments. PCR was performed for 40 cycles (1 min at 94°C, 2 min at 55°C and 3 min at 72°C). These conditions maximized the specific activity of the Alexa488-5-dUTP (green) ρ+ probes. The resulting fluorescent PCR fragments were purified with QIAquick spin columns (Qiagen). To improve FISH signals, the fluorescent COXI-1, COXI-2, HS40 and VAR1 fragments were digested with HpaI and SspI, DraI and HaeII, DraI and NdeI, and AcsI, respectively. The digested fragments were precipitated and resuspended in buffer A (50% formamide, 2× SSC, 10% dextran sulfate, 1 mg/ml salmon sperm DNA).
The yeast strains DBY747 ρ+, PSY142 HS40 ρ– and PSY142 VAR1 ρ– were grown on YPD liquid medium at 30°C overnight and mated as described (Azpiroz and Butow, 1993; Okamoto et al., 1998). Samples were fixed and prepared for microscopy as described (Azpiroz and Butow, 1993). In situ hybridization was carried out using published protocols (Guacci et al., 1994; Jin et al., 1998). Briefly, spheroplasts were attached to slide wells, treated with RNase A and washed. DNA in cells was denatured, neutralized and hybridized with the mixed ρ+ probes and one ρ– probe (∼20 ng each) for 15 h. The slides were then washed to remove non-specific hybridization. Low-melting agarose was added to each slide well and slides were incubated at 4°C for 10 min prior to microscopy.
The preparations were observed using a Leica microscope (model DMRXE; Deerfield, IL) equipped for an HBO 100 W/2 mercury arc lamp, a ×100 Plan-Apochromat objective and epifluorescence. For ρ+ probes (green), the following filter sets were used: 450–490 nm band-pass excitation filter, 510 nm dichroic reflector and >515 nm long-pass emission filter. For ρ– probes (red), the following filter sets were used: 575 ± 30 nm band-pass excitation filter, 600 nm dichroic reflector and 635 ± 40 nm long-pass emission filter. Exposure time for capturing images of zygotes was 2 s for ρ+ probes (green) and 0.4 s for ρ– probes (red) to balance the fluorescence intensities of the visualized mtDNAs. Imaging was carried out as described previously (Okamoto et al., 1998).
DNA isolation and 2D gel electrophoresis
DNA was prepared from ρ– strains as described previously (MacAlpine et al., 1998) but without enrichment for molecules with some ssDNA character. Total cellular DNA (20 µg) was analyzed by 2D gel electrophoresis as described (Brewer and Fangman, 1987). Gels were transferred to nylon membranes by capillary action with or without prior denaturation. Probes of HS40 mtDNA and a 1 kb HpaII fragment of the VAR1 gene labeled by random priming in the presence of [α-32P]dATP were hybridized as described previously (MacAlpine et al., 1998). Oligonucleotides O969 (5′-GTGCTTTGTATTTATTGAATATTCTGG) and O969rc (5′-CCAGAATATTCAATAAATACAAAGCAC), which anneal to the sense and antisense strands of ori5, respectively, were 5′ end-labeled with 32P and hybridized using Rapid Hyb buffer (Amersham). The sense strand corresponds to the strand displaced by transcription from the nonanucleotide promoter. DNA was treated with ExoI for 1 h at 37°C in 50 µl reactions containing 50 µg of DNA, 67 mM glycine pH 9.5, 10 mM β-mercaptoethanol, 6.7 mM MgCl2 and 25 U ExoI (Amersham). DNA was treated with S1 endonuclease for 30 min at 37°C in 50 µl reactions containing 50 µg of DNA, 33 mM NaOAc pH 4.5, 50 mM NaCl, 30 µM ZnSO4 and 10 U of S1 (Roche).
RNA isolation and northern blots
Total RNA was isolated from mid-log phase cells and northern blotting was performed as described (Jia et al., 1997). Ori5 transcripts were detected with 5′ end-labeled O969.
Primer extension
Approximately 2 ng of 5′ end-labeled O969 were mixed with 2 µg of RNA in 1× React 2 buffer (BRL) in 10 µl, boiled for 30 s and returned slowly to room temperature. The nucleic acids were then ethanol precipitated and the pellet resuspended in 20 µl of AMV reverse transcription reaction solution [1× AMV buffer, 1 mM dNTPs and 1 U/µl AMV RT (Roche)]. The reverse transcription reactions were performed at 55°C for 1 h and stopped by the addition of 2 µl of 95% formamide containing 1 mM EDTA. The products were denatured by boiling and resolved on a sequencing gel, alongside a sequencing ladder terminated with ddGTP that was produced with cloned HS40 DNA as template and O969 as primer.
RNA capping
Total DNA was isolated as described previously (MacAlpine et al., 1998) except that pH 8.0 buffers were used (instead of pH 9.5) and RNase A was omitted. The mtDNA was purified from total DNA by banding in CsCl gradients twice. The purified mtDNA was digested with the appropriate restriction enzyme and RNA capping experiments using guanylyl transferase (USB) were carried out as described (Graves et al., 1998).
Mitochondrial transformation and strain constructions by reverse genetics
Plasmids pMAC-HS40w and pMAC-HS40p were transformed into mitochondria of strain MCC109 ρ0 as described (Butow et al., 1996). Transformed mtDNAs were transferred into strain ID41-6/161 ρ0 by cytoduction. Spontaneous recombination between (AAT)7 repeats (see Figure 4A) in the mtDNA of strain ID41-6/161 pMIT-HS40w excised a 918 bp ori5-containing fragment that was amplified in a subclone of that strain called ID41-6/161 sORI5w-918. Growth of strain ID41-6/161 ρ– pMIT-HS40p in YPD medium containing 2 µg/ml ethidium bromide led to excision and amplification of a 476 bp fragment that contains all of ori5 (sORI5p-476). Strain ID41-6/161 sORI5w-476 was obtained by mating strain MCC109 pMIT-HS40w with strain ID41-6/161 sORI5p-476 and screening for MATa recombinant ρ– strains with the 476 bp repeating unit and the wild-type promoter; several were found and strain sORI5w-476 was used here. The repeating units of these synthetic ρ– mtDNAs were confirmed by DNA sequencing.
Hypersuppressiveness assay
The suppressiveness of the petite mutant strains was measured as described (Ephrussi et al., 1955). Briefly, 2 × 107 ρ– cells of strain ID41-6/161 were mated on YPD medium for 4–5 h to 2 × 107 cells of the tester strain 14WW ρ+. The matings were diluted and zygote clones were selected on YNBD plates at 30°C. After 3 days of growth at 30°C, the zygotic clones were scored for the presence of ρ+ cells by replica plating to YPG plates or by the triphenyltetrazolium chloride (TTC) overlay method (Ogur et al., 1957). For ρ– × ρ– crosses, colony blots were used to score the transmission of petite genomes (Zinn and Butow, 1985).
Acknowledgments
Acknowledgements
This research was supported by NIH grant GM35510 to R.A.B. and P.S.P. and by research grants I-0642 and I-1211 of the Robert A.Welch Foundation to R.A.B. and P.S.P., respectively. D.M.M. was an NIH predoctoral trainee of the Division of Cell and Molecular Biology Training Program (T32 GM08203) and J.K. is an NIH predoctoral trainee of the Cardiovascular Training Program (T32 HL073670).
References
- Azpiroz R. and Butow,R.A. (1993) Patterns of mitochondrial sorting in yeast zygotes. Mol. Biol. Cell, 4, 21–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backert S., Meissner,K. and Borner,T. (1997) Unique features of the mitochondrial rolling circle-plasmid mp1 from the higher plant Chenopodium album (L.). Nucleic Acids Res., 25, 582–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldacci G. and Bernardi,G. (1982) Replication origins are associated with transcription initiation sequences in the mitochondrial genome of yeast. EMBO J., 1, 987–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldacci G., Cherif-Zahar,B. and Bernardi,G. (1984) The initiation of DNA replication in the mitochondrial genome of yeast. EMBO J., 3, 2115–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi G. (1982) The origins of replication of the mitochondrial genome of yeast. Trends Biochem. Sci., 7, 404–408. [Google Scholar]
- Blanc H. and Dujon,B. (1980) Replicator regions of the yeast mitochondrial DNA responsible for suppressiveness. Proc. Natl Acad. Sci. USA, 77, 3942–3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonitz S.G., Coruzzi,G., Thalenfield,B.E., Tzagoloff,A. and Macino,G. (1980) Assembly of the mitochondrial membrane system. Structure and nucleotide sequence of the gene coding for subunit I of cytochrome oxidase. J. Biol. Chem., 255, 11927–11941. [PubMed] [Google Scholar]
- Brewer B.J. and Fangman,W.L. (1987) The localization of replication origins on ARS plasmids in S. cerevisiae. Cell, 51, 463–471. [DOI] [PubMed] [Google Scholar]
- Butow R.A., Henke,R.M., Moran,J.V., Belcher,S.M. and Perlman,P.S. (1996) Transformation of Saccharomyces cerevisiae mitochondria using the biolistic gun. Methods Enzymol., 264, 265–278. [DOI] [PubMed] [Google Scholar]
- de Zamaroczy M., Marotta,R., Faugeron-Fonty,G., Goursot,R., Mangin,M., Baldacci,G. and Bernardi,G. (1981) The origins of replication of the yeast mitochondrial genome and the phenomenon of suppressivity. Nature, 292, 75–78. [DOI] [PubMed] [Google Scholar]
- de Zamaroczy M., Faugeron-Fonty,G., Baldacci,G., Goursot,R. and Bernardi,G. (1984) The ori sequences of the mitochondrial genome of a wild-type yeast strain: number, location, orientation and structure. Gene, 32, 439–457. [DOI] [PubMed] [Google Scholar]
- Dujon B. (1981) Mitochondrial genetics and functions. In Strathern,J.N., Jones,E.W. and Broach,J.R. (eds), The Molecular Biology of the Yeast Saccharomyces. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 505–635.
- Ephrussi B. and Grandchamp,S. (1965) Etudes sur la suppressivité des mutants à deficience respiratoire de la levure. I. Existence au niveau cellulaire de divers degrés de suppressivité. Heredity, 20, 1–7. [DOI] [PubMed] [Google Scholar]
- Ephrussi B., de Margerie-Hottinguer,H. and Roman,H. (1955) Suppressiveness: a new factor in the genetic determinism of the synthesis of respiratory enzymes in yeast. Proc. Natl Acad. Sci. USA, 41, 1065–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fangman W.L., Henly,J.W., Churchill,G. and Brewer,B.J. (1989) Stable maintenance of a 35-base pair yeast mitochondrial genome. Mol. Cell. Biol., 9, 1917–1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fangman W.L., Henly,J.W. and Brewer,B.J. (1990) RPO41-independent maintenance of ρ– mitochondrial DNA in Saccharomyces cerevisiae. Mol. Cell. Biol., 10, 10–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faugeron-Fonty G., Le Van Kim,C., de Zamarocy,M., Goursot,R. and Bernardi,G. (1984) A comparative study of the ori sequences from the mitochondrial genomes of 20 wild-type yeast strains. Gene, 32, 459–473. [DOI] [PubMed] [Google Scholar]
- Graves T., Dante,M., Eisenhour,L. and Christianson,T.W. (1998) Precise mapping and characterization of the RNA primers of DNA replication for a yeast hypersuppressive petite by in vitro capping with guanylyltransferase. Nucleic Acids Res., 26, 1309–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenleaf A.L., Kelly,J.L. and Lehman,I.R. (1986) Yeast RPO41 gene is required for transcription and maintenance of the mitochondrial genome. Proc. Natl Acad. Sci. USA, 83, 3391–3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guacci V., Hogan,E. and Koshland,D. (1994) Chromosome condensation and sister chromatid pairing in budding yeast. J. Cell Biol., 125, 517–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Z. and Stachow,C. (1994) Analysis of Schizosaccharomyces pombe mitochondrial DNA replication by two dimensional gel electrophoresis. Chromosoma, 103, 162–170. [DOI] [PubMed] [Google Scholar]
- Hermann G.J. and Shaw,J.M. (1998) Mitochondrial dynamics in yeast. Annu. Rev. Cell. Dev. Biol., 14, 265–303. [DOI] [PubMed] [Google Scholar]
- Holt I.J., Lorimer,H.E. and Jacobs,H.T. (2000) Coupled leading- and lagging-strand synthesis of mammalian mitochondrial DNA. Cell, 100, 515–524. [DOI] [PubMed] [Google Scholar]
- Hudspeth M.E.S., Vincent,R.D., Perlman,P.S., Shumard,D.S., Treisman,L.O. and Grossman,L.I. (1984) Expandable var1 gene of yeast mitochondrial DNA: in-frame insertions can explain the strain-specific protein size polymorphism. Proc. Natl Acad. Sci. USA, 81, 3148–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y., Rothermel,B., Thornton,J. and Butow,R.A. (1997) A basic helix–loop–helix zipper transcription complex functions in a signaling pathway from mitochondria to the nucleus. Mol. Cell. Biol., 17, 1110–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Q., Trelles-Sticken,E., Scherthan,H. and Loidl,J. (1998) Yeast nuclei display prominent centromere clustering that is reduced in nondividing cells and in meiotic prophase. J. Cell Biol., 141, 21–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockshon D., Zweifel,S.G., Freeman-Cook,L.L., Lorimer,H.E., Brewer,B.J. and Fangman,W.L. (1995) A role for recombination junctions in the segregation of mitochondrial DNA in yeast. Cell, 81, 947–955. [DOI] [PubMed] [Google Scholar]
- Lorimer H.E., Brewer,B.J. and Fangman,W.L. (1995) A test of the transcription model for biased inheritance of yeast mitochondrial DNA. Mol. Cell. Biol., 15, 4803–4809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlpine D.M., Perlman,P.S. and Butow,R.A. (1998) The high mobility group protein Abf2p influences the level of yeast mitochondrial DNA recombination intermediates in vivo. Proc. Natl Acad. Sci. USA, 95, 6739–6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlpine D.M., Perlman,P.S. and Butow,R.A. (2000) The numbers of individual mitochondrial DNA molecules and mitochondrial DNA nucleoids in yeast are co-regulated by the general amino acid control pathway. EMBO J., 19, 767–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran J.V., Zimmerly,S., Kennell,J.C., Lambowitz,A.M., Butow,R.A. and Perlman,P.S. (1995) Mobile group II introns of yeast mitochondrial DNA are novel site-specific retroelements. Mol. Cell. Biol., 15, 2828–2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunnari J., Marshall,W.F., Straight,A., Murray,A., Sedat,J.W. and Walter,P. (1997) Mitochondrial transmission during mating in Saccharomyces cerevisiae is determined by mitochondrial fusion and fission and the intramitochondrial segregation of mitochondrial DNA. Mol. Biol. Cell, 8, 1233–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogur M., St John,R. and Nagai,S. (1957) Tetrazolium overlay technique for populations studies of respiration deficiency in yeast genetics. Science, 125, 928–929. [DOI] [PubMed] [Google Scholar]
- Okamoto K., Perlman,P.S. and Butow,R.A. (1998) The sorting of mitochondrial DNA and mitochondrial proteins in zygotes: preferential transmission of mitochondrial DNA to the medial bud. J. Cell Biol., 142, 613–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh V.S., Morgan,M.M., Scott,R., Clements,L.S. and Butow,R.A. (1987) The mitochondrial genotype can influence nuclear gene expression in yeast. Science, 235, 576–580. [DOI] [PubMed] [Google Scholar]
- Shadel G.S. and Clayton,D.A. (1997) Mitochondrial DNA maintenance in vertebrates. Annu. Rev. Biochem., 66, 409–435. [DOI] [PubMed] [Google Scholar]
- Sharpe M.E. and Errington,J. (1999) Upheaval in the bacterial nucleoid. An active chromosome segregation mechanism. Trends Genet., 15, 70–74. [DOI] [PubMed] [Google Scholar]
- Stohl L.L. and Clayton,D.A. (1992) Saccharomyces cerevisiae contains an RNase MRP that cleaves at a conserved mitochondrial RNA sequence implicated in replication priming. Mol. Cell. Biol., 12, 2561–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace D.C. (1999) Mitochondrial diseases in man and mouse. Science, 283, 1482–1488. [DOI] [PubMed] [Google Scholar]
- Wheeler R.T. and Shapiro,L. (1997) Bacterial chromosome segregation: is there a mitotic apparatus? Cell, 88, 577–579. [DOI] [PubMed] [Google Scholar]
- Xu B.J. and Clayton,D.A. (1995) A persistent RNA–DNA hybrid is formed during transcription at a phylogenetically conserved mitochondrial DNA sequence. Mol. Cell. Biol., 15, 580–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zassenhaus H.P., Martin,N.C. and Butow,R.A. (1984) Orgins of transcripts of the yeast mitochondrial var1 gene. J. Biol. Chem., 259, 6019–6027. [PubMed] [Google Scholar]
- Zelenaya-Troitskaya O., Newman,S.M., Okamoto,K., Perlman,P.S. and Butow,R.A. (1998) Functions of the HMG box protein, Abf2p, in mitochondrial DNA segregation, recombination and copy number in Saccharomyces cerevisiae. Genetics, 148, 1763–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinn A.R. and Butow,R.A. (1985) Nonreciprocal exchange between alleles of the yeast mitochondrial 21S rRNA gene: kinetics and the involvement of a double-strand break. Cell, 40, 887–895. [DOI] [PubMed] [Google Scholar]