Abstract
Unfolding is an essential process during translocation of preproteins into mitochondria; however, controversy exists as to whether mitochondria play an active role in unfolding. We have established an in vitro system with a kinetic saturation of the mitochondrial import machinery, yielding translocation rates comparable to in vivo import rates. Preproteins with short N-terminal segments in front of a folded domain show a characteristic delay of the onset of translocation (lag phase) although the maximal import rate is similar to that of longer preproteins. The lag phase is shortened by extending the N-terminal segment to improve the accessibility to matrix heat shock protein 70 and abolished by unfolding of the preprotein. A mutant mtHsp70 defective in binding to the inner membrane prolongs the lag phase and reduces the translocation activity. A direct comparison of the rate of spontaneous unfolding in solution with that during translocation demonstrates that unfolding by mitochondria is significantly faster, proving an active unfolding process. We conclude that access of mtHsp70 to N-terminal preprotein segments is critical for active unfolding and initiation of translocation.
Keywords: Hsp70/mitochondria/protein sorting/protein unfolding/Saccharomyces cerevisiae
Introduction
A majority of mitochondrial preproteins are synthesized in the cytosol and translocated across the outer and inner mitochondrial membranes to reach their final destination in the matrix (Schatz and Dobberstein, 1996; Neupert, 1997; Pfanner et al., 1997). The preproteins are recognized by receptor proteins on the mitochondrial surface and subsequently channeled through two integral membrane translocation complexes into the mitochondrial matrix (Ryan and Pfanner, 1998; Jensen and Johnson, 1999; Koehler et al., 1999; Rassow et al., 1999; Bauer et al., 2000). The translocation of the extreme N-terminal presequence over the inner membrane is driven by an electrophoretic mechanism generated by the electrochemical potential across the inner membrane. However, the translocation of the bulk polypeptide chain requires the direct interaction with a matrix chaperone protein, the heat shock protein 70 (mtHsp70) (Kang et al., 1990; Scherer et al., 1990; Gambill et al., 1993; Voos et al., 1999). A fraction of mtHsp70 transiently binds to the inner membrane protein Tim44 in an ATP-sensitive manner (Kronidou et al., 1994; Rassow et al., 1994; Schneider et al., 1994; von Ahsen et al., 1995; Ungermann et al., 1996). By interaction with the preprotein and the membrane anchor Tim44, mtHsp70 promotes transport of the polypeptide chain into the matrix (Glick, 1995; Pfanner and Meijer, 1995; Voos et al., 1996; Voisine et al., 1999). Preproteins have to cross the membrane in an unfolded conformation since stable folded domains cannot pass through the channels of the outer and inner membrane translocases of isolated mitochondria (Eilers and Schatz, 1986; Rassow et al., 1990; Schwartz et al., 1999). In vivo studies similarly demonstrated a requirement for preprotein unfolding (Wienhues et al., 1991; Bömer et al., 1997). While it is generally accepted that mtHsp70 plays an essential role in mitochondrial protein import and that preproteins must be unfolded to be translocated across the mitochondrial membranes, the underlying mechanisms are the subject of an ongoing debate.
A central question is whether preprotein unfolding is an active or a passive process, i.e. whether or not unfolding by mitochondria is faster than spontaneous unfolding. Previous studies that correlated unfolding rates of preproteins and the corresponding mitochondrial import rates reached controversial conclusions (Matouschek et al., 1997; Gaume et al., 1998; Huang et al., 1999). One possible problem in the interpretation of the obtained data could be the use of different preprotein constructs, such as fusion proteins between a mitochondrial targeting signal and the enzyme barnase (Matouschek et al., 1997; Huang et al., 1999) or the heme-binding domain of cytochrome b2 (Gaume et al., 1998). However, in two studies, fusion proteins containing mouse dihydrofolate reductase (DHFR) were employed and still opposing conclusions concerning the mechanism of the unfolding reaction were reached (Matouschek et al., 1997; Gaume et al., 1998). In all these cases, radiolabeled preproteins have been used that were generated by in vitro translation, with the inherent problem of a very low and variable absolute amount of preproteins, impairing a direct comparison of the import assays. Since true maximal rates of the mitochondrial import reaction have not been determined to date, a quantitative analysis of import and unfolding rates has been difficult. An important advance in solving the controversy would thus be a kinetic saturation of the import machinery to assess protein unfolding by mitochondria quantitatively. So far, however, only a static saturation has been achieved by jamming the mitochondrial import sites with partially imported preproteins (Eilers and Schatz, 1986; Vestweber and Schatz, 1988; Rassow et al., 1989; Dekker et al., 1997). A kinetic saturation will require large amounts of preproteins that remain fully import competent and soluble at high concentration.
For this study, we developed an import system in isolated mitochondria with saturating amounts of preproteins that had been expressed and purified in soluble, import-competent form. The maximal import rates in this in vitro system were similar to those estimated for the in vivo reaction. We determined the quantitative contribution of the unfolding reaction to the overall import reaction and identified an initial lag phase of translocation that depended on the folding state and accessibility of the preprotein to a functional Hsp70 system in the matrix. These results directly demonstrate that mitochondria actively unfold preproteins.
Results
Saturation of the mitochondrial import machinery with a purified cleavable preprotein
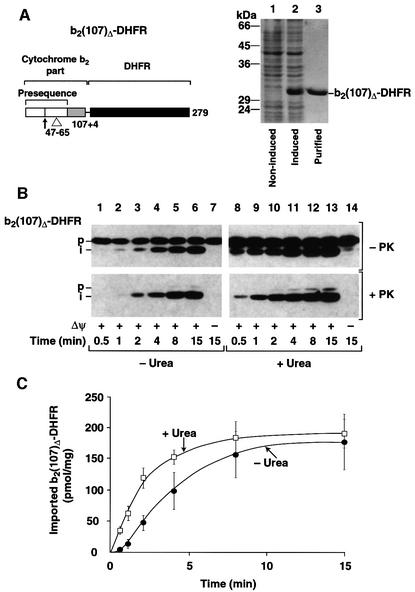
We constructed a fusion protein between an N-terminal portion of the precursor of cytochrome b2 from Saccharomyces cerevisiae and mouse DHFR. The preprotein b2(107)Δ-DHFR consists of the presequence and an initial part of cytochrome b2 up to amino acid 107, followed by a short linker and the full-length DHFR (Figure 1A, left panel). A deletion of 19 amino acids in the second part of the presequence abolished sorting into the intermembrane space and ensured translocation into the mitochondrial matrix (Koll et al., 1992; Beasley et al., 1993; Schwarz et al., 1993). Upon synthesis in Escherichia coli cells with high efficiency, b2(107)Δ-DHFR was purified to homogeneity from the soluble fraction (Figure 1A, lane 3) and incubated with isolated, energized yeast mitochondria. The fusion protein was detected by immunodecoration with affinity-purified antibodies against DHFR. Figure 1B shows that b2(107)Δ-DHFR was processed to the intermediate form by removal of the matrix-targeting presequence (upper panel, lanes 2–6). The processed form was largely protected against externally added proteinase K (Figure 1B, lower panel, lanes 3–6). Dissipation of the inner membrane potential Δψ by the potassium ionophore valinomycin completely blocked processing and transport of b2(107)Δ-DHFR to a protease-protected location (Figure 1B, lanes 7), demonstrating specific translocation across the inner mitochondrial membrane.
Fig. 1. Maximal import rate of preproteins in vitro. (A) Left panel: schematic drawing of the fusion protein cytochrome b2(107)Δ-DHFR. White, presequence; light gray, mature cytochrome b2; black, DHFR moiety. The cleavage site of the matrix processing peptidase (arrow) and the 19 amino acid deletion of the hydrophobic sorting signal (▵) are indicated. Right panel: expression and purification of b2(107)Δ-DHFR from E.coli cells. Lane 1, cell lysate of non-induced cells; lane 2, cell lysate after 2 h induction with 1 mM IPTG, lane 3, eluate from the MonoS column. The molecular mass of control proteins is indicated. (B) The import of b2(107)Δ-DHFR. Import was performed at 25°C with native (–Urea) or denatured (+Urea) preprotein for the indicated times in the presence (+Δψ) or absence (–Δψ) of an inner membrane potential. After the import, the reactions were divided and one half was treated with proteinase K (+PK). Imported preproteins were detected by western blotting using antibodies against DHFR: p, precursor form; i, matrix-targeted intermediate form. (C) Quantification of the import reaction. b2(107)Δ-DHFR was imported as described above and the absolute amount of imported preproteins in pmol/mg of mitochondrial protein was determined by a standardized western blot.
The absolute amounts of the imported preprotein were determined by standardized western blot analysis in comparison with known amounts of the purified protein. Under standard conditions, the import rates of b2(107)Δ-DHFR reached a maximal value of ∼25 pmol/min/mg of mitochondrial protein (Figure 1C, –Urea). After denaturation of the preprotein with urea, the import rate increased 2-fold (Figure 1B, right panel; and C, +Urea). In these import assays, 640 pmol of recombinant preprotein were added per mg of mitochondrial protein corresponding to 15–20 pmol import sites (Dekker et al., 1997; Sirrenberg et al., 1997), indicating that saturating amounts of preproteins were used. After 15 min incubation at 25°C, ∼25% of the added urea-denatured preprotein was imported. A further increase in the amount of added preprotein did not increase the rate of translocation (data not shown). With an estimated in vivo translocation rate of 40–60 pmol/min/mg of mitochondrial protein (see Discussion), the measured import rates thus approached the theoretical limit of what is expected for the in vivo situation.
Identification of a lag phase of preprotein translocation
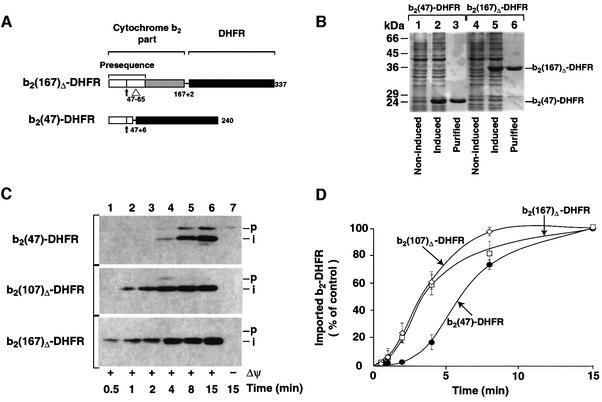
While previous studies had suggested that mitochondrial protein import followed simple first-order kinetics (Matouschek et al., 1997; Gaume et al., 1998), the import of saturating amounts of b2(107)Δ-DHFR under standard conditions (–Urea) seemed to follow a sigmoidal pattern. To characterize the import kinetics under substrate-saturating conditions in more detail, we expressed two additional fusion proteins that had different N-terminal extension lengths. The N-terminal part derived from cytochrome b2 was reduced to 47 amino acids, resulting in the fusion protein b2(47)-DHFR, which essentially consists of a matrix-targeting presequence fused to the DHFR domain (Figure 2A). b2(167)Δ-DHFR consists of the N-terminal 167 amino acids of cytochrome b2 (with the 19 residue deletion in the sorting signal) and DHFR (Figure 2A) (Koll et al., 1992; Dekker et al., 1997). After purification from the soluble fraction of E.coli (Figure 2B, lanes 3 and 6), the fusion proteins were imported into isolated mitochondria. The small preprotein b2(47)-DHFR was imported and processed efficiently to the expected intermediate form (Figure 2C, upper panel). However, in comparison with the import efficiency of the longer preproteins b2(107)Δ-DHFR and b2(167)Δ-DHFR, we found that the overall import of b2(47)-DHFR was significantly slower (Figure 2C, lanes 1–4). A quantitative analysis of the import rates of the three preproteins (Figure 2D) revealed an initial delay of ∼2–4 min in the translocation of b2(47)-DHFR, where hardly any fully imported preprotein could be detected. However, after that initial lag phase, processing and translocation rates accelerated and reached the same maximal value as was observed with the longer preproteins b2(107)Δ-DHFR and b2(167)Δ-DHFR (Figure 2D). The length of the N-terminal extension in front of DHFR is hence critical for the initial lag phase of protein import, but not of major influence for the maximal rate of preprotein translocation.
Fig. 2. Identification of an initial lag phase of the import reaction. (A) Schematic drawing of the cytochrome b2-DHFR fusion proteins used. (B) Expression and purification of b2-DHFR fusion proteins, for description see legend to Figure 1A. (C) Purified b2-DHFR preproteins were imported into wild-type mitochondria for the indicated times at 25°C in the presence (+Δψ) or absence (–Δψ) of an inner membrane potential as described in Materials and methods: p, precursor form; i, matrix-targeted intermediate form. All samples were treated with proteinase K (100 µg/ml) after import. (D) The import reactions were quantified by standardized western blotting. Values reached for each preprotein after 15 min import were set to 100% (control).
Mitochondria actively unfold preproteins with a long N-terminal extension
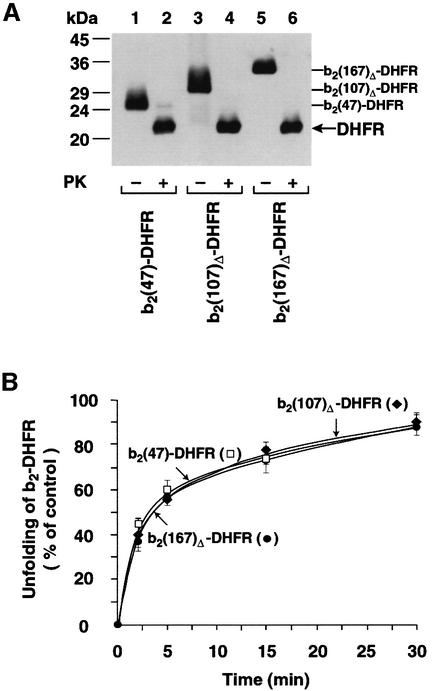
We asked whether the initial delay in import was caused by a differential effect of the N-terminal extensions on the folding state of the DHFR domain. First, we tested the folding state of the preproteins by their resistance to protease treatment. After treatment with proteinase K and analysis by western blotting, we found with all three preproteins a prominent protease-resistant band at 22 kDa that represents the folded DHFR domain (Figure 3A, lanes 2, 4 and 6). All recombinant preproteins behaved similarly in the protease digestion assay, suggesting that the DHFR domain folds independently of the respective N-terminal extensions. A native conformation of the DHFR domain in all recombinant preproteins was also demonstrated by the determination of full enzymatic activity of DHFR (Figure 3B, legend). Secondly, we asked whether the N-terminal extensions differently affected the rate of unfolding of DHFR by assaying the spontaneous loss of enzymatic activity in solution. In order to restrict the analysis to the unfolding reaction, a large excess of the bacterial chaperonin GroEL was added. GroEL binds efficiently to not fully folded DHFR and sequesters it from the solution. In the absence of ATP and the co-chaperonin GroES, GroEL stabilizes a late folding intermediate of DHFR (Viitanen et al., 1991; Goldberg et al., 1997; Beissinger and Buchner, 1998; Clark and Frieden, 1999; von Ahsen et al., 2000). Figure 3B shows that all three DHFR fusion proteins spontaneously unfolded with the identical half-time of ∼3 min. A similar value has been reported by Gaume et al. (1998).
Fig. 3. Folding states of recombinant preproteins. (A) b2-DHFR fusion proteins contain a folded DHFR domain. The indicated preproteins (40 pmol) were treated with (+PK) 50 µg/ml proteinase K at 2°C for 15 min or left untreated (–PK). After inactivation of the protease by addition of 5 mM PMSF, the samples were subjected to precipitation with trichloroacetic acid and were analyzed by SDS–PAGE and western blotting using anti-DHFR antibodies. The PK-resistant folded DHFR domains and the full-length proteins are indicated. (B) Spontaneous unfolding rates of b2-DHFR fusion proteins. Unfolding rates were determined by measuring the loss of enzymatic activity of the mouse DHFR in the presence of excess GroEL. At the start of the reaction, all preproteins showed full enzymatic activity like purified DHFR. The reduction of DHFR activity by 1 h incubation with GroEL was set to 100%. Filled circles, b2(167)Δ-DHFR; filled diamonds, b2(107)Δ-DHFR; open squares, b2(47)-DHFR.
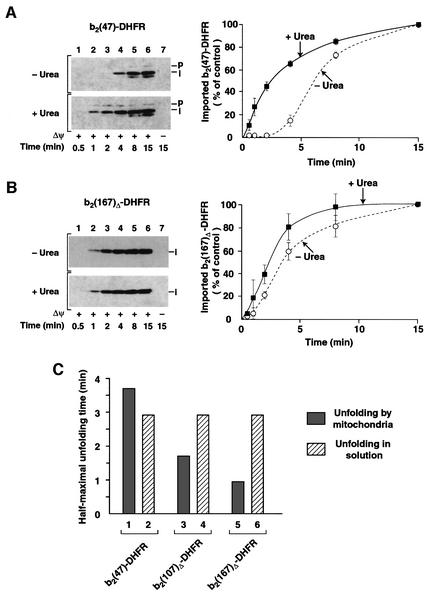
The identical folding behavior of the fusion proteins in solution suggested that the difference in import kinetics reflected an active contribution of mitochondria. We tested directly whether artificial unfolding of the fusion pro teins altered the import kinetics. b2(47)-DHFR was unfolded by treatment with urea immediately before the import reaction. Thereby the long initial lag phase was completely abolished (Figure 4A). The short lag phase of b2(167)Δ-DHFR (Figure 4B) as well as b2(107)Δ-DHFR (Figure 1C) was also abolished by denaturation of the preproteins. All three denatured preproteins followed similar import kinetics (Figures 4A and B and 1C). The only difference in the import kinetics with or without urea is the folding state of the respective preproteins. Hence, it was now possible to determine the half-time of preprotein unfolding by mitochondria by subtraction of the half-times of import of the folded preprotein from that of the denatured preprotein. The half-time for unfolding of b2(47)-DHFR by mitochondria of 3.5 min was not faster than spontaneous unfolding (Figure 4C, columns 1 and 2), i.e. no evidence for active unfolding of the short preprotein was obtained. In contrast, the half-times for mitochondrial unfolding of b2(107)Δ-DHFR and b2(167)Δ-DHFR of 1.7 and 1 min, respectively, were significantly faster than spontaneous unfolding (Figure 4C, columns 3–6). Thus, the longer the N-terminal extension of the fusion protein is, the shorter is the half-time needed for unfolding by mitochondria, demonstrating an active unfolding mechanism for the two long preproteins.
Fig. 4. Unfolding by mitochondria is faster than the spontaneous unfolding rate in solution. (A) Import of urea-denatured b2(47)-DHFR into wild-type mitochondria. The import reactions were performed as described in Figure 1B. Where indicated, the preproteins were denatured by 8 M urea (+Urea) before import. The import reactions were stopped, treated with proteinase K and imported preproteins were detected by immunodecoration with anti-DHFR antibodies. The import values reached after 15 min import were set to 100% (control). The dotted line shows import of non-denatured b2(47)-DHFR. (B) Import of urea-denatured b2(167)Δ-DHFR into wild-type mitochondria. The experiment was performed as described above. (C) Comparison of unfolding rates in solution and by mitochondria. The unfolding rates in solution were obtained by quantification of Figure 3B. The time for half-maximal unfolding during import into mitochondria was determined by subtraction of the values for the half-maximal import times of the folded preprotein from the values of half-maximal import of the urea-denatured preprotein. The values were calculated from the quantification of import reactions presented in (A) and (B) and Figure 1C.
MtHsp70 is critical for active unfolding of preproteins by mitochondria
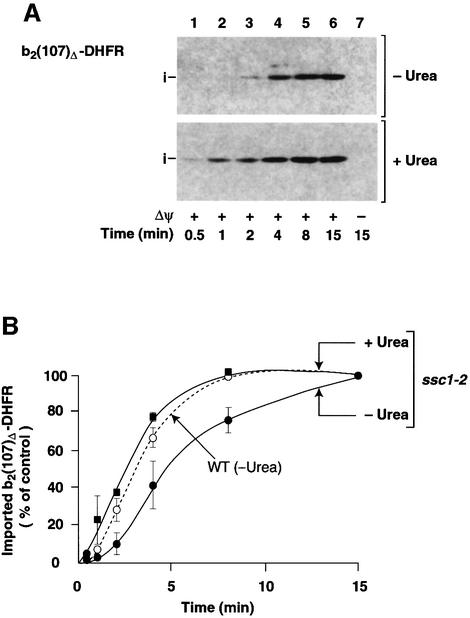
MtHsp70 has been implicated as the central component in the mitochondrial import and unfolding machinery (Glick et al., 1993; Voos et al., 1993; Matouschek et al., 1997; Gaume et al., 1998; Voisine et al., 1999). As we have shown here, the import lag phase reflects the mitochondrial unfolding activity for a given preprotein. In that case, the lag phase should also depend on the functional status of mtHsp70. In the temperature-sensitive mtHsp70 mutant form Ssc1-2, a single amino acid alteration in the peptide-binding domain of mtHsp70 causes a split phenotype: Ssc1-2 efficiently binds preproteins, but its interaction with Tim44 of the inner membrane translocase is strongly impaired (Schneider et al., 1994; von Ahsen et al., 1995; Voos et al., 1996; Voisine et al., 1999). To minimize unspecific effects of the mutation on mitochondrial and cellular growth in vivo, ssc1-2 mutant yeast cells and the corresponding wild-type cells were grown at permissive temperature. The mitochondria were isolated and pre-incubated at 37°C to induce the mutant phenotype. When b2(107)Δ-DHFR was incubated with ssc1-2 mitochondria, the lag phase of import was significantly prolonged compared with wild-type mitochondria (Figure 5A, upper panel, and B). After the initial lag phase, the import accelerated to almost wild-type efficiency (Figure 5B). When the preprotein was unfolded by urea prior to import, however, the lag phase was completely abolished (Figure 5A, lower panel, and B). Therefore, the import behavior of b2(107)Δ-DHFR in ssc1-2 versus wild-type mitochondria demonstrates that the function of mtHsp70 and the folding state of the preprotein are critical for the length of the lag phase. In ssc1-2 mitochondria, the import mechanism is shifted in the direction of passive (spontaneous) unfolding.
Fig. 5. The lag phase of import is prolonged in ssc1-2 mutant mitochondria. (A) Denatured (+Urea) or non-denatured b2(107)Δ-DHFR (–Urea) was imported into ssc1-2 mitochondria under non-permissive conditions as described in Materials and methods. All samples were treated with proteinase K after import. (B) Import kinetics of b2(107)Δ-DHFR in ssc1-2 mutant mitochondria. Imported preproteins (i-form) were quantified with anti-DHFR antibodies after SDS–PAGE and western blotting. The amount of preprotein imported after 15 min was set to 100% (control). Filled squares, import of denatured b2(107)Δ-DHFR into ssc1-2 mitochondria (ssc1-2/+Urea); filled circles, import of non-denatured b2(107)Δ-DHFR into ssc1-2 mitochondria (ssc1-2/–Urea). The dotted line shows the import rate into corresponding wild-type mitochondria as shown in Figure 2D (WT/–Urea).
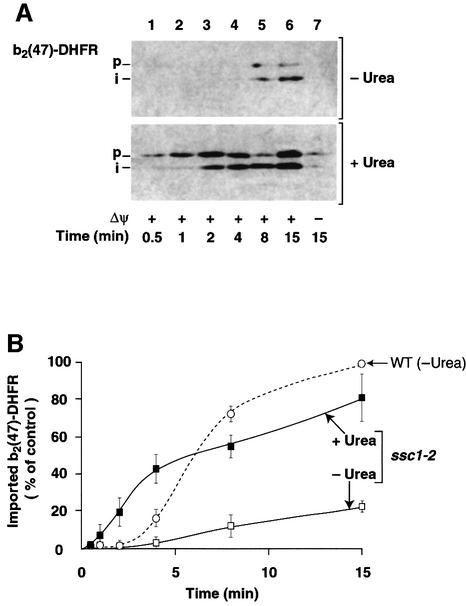
The N-terminal extension of b2(47)-DHFR is so short that it hardly spans both mitochondrial membranes and reaches to mtHsp70 in the matrix when the DHFR domain is folded, since ∼50 amino acid residues are needed to span both mitochondrial membranes (Rassow et al., 1990; Ungermann et al., 1994; Matouschek et al., 1997; Bömer et al., 1998; Gaume et al., 1998). We thus asked how the impaired interaction of Ssc1-2 with Tim44, i.e. reduced availability of mtHsp70 at the exit site of the inner membrane import channel, affected the import kinetics of b2(47)-DHFR. When b2(47)-DHFR was incubated with energized ssc1-2 mitochondria, we observed only a very low import activity (Figure 6A, upper panel, lanes 1–6). A quantification revealed that the import rate of b2(47)-DHFR into ssc1-2 mitochondria was reduced 7- to 8-fold compared with wild-type mitochondria (Figure 6B). The import rate into ssc1-2 mitochondria essentially remained very low throughout the import reaction, very similar to the rate observed during the initial lag phase in wild-type mitochondria (Figure 6B). We conclude that the strongly impaired accessibility of Ssc1-2 to the preprotein b2(47)-DHFR in transit (when the DHFR is folded) leads to a continuation of the lag phase. This effect was directly related to the folding state of the preprotein and was not caused by a general impairment of the mitochondrial import system since denaturation of b2(47)-DHFR prior to import resulted in efficient import into ssc1-2 mitochondria (Figure 6A, lower panel) with a maximal rate comparable to that in wild-type mitochondria (Figure 6B). Moreover, the lag phase was abolished by denaturation of the preprotein (Figure 6B). Taken together, our results show that the function of mtHsp70, its accessibility to a preprotein in transit and the folding state of the preproteins are crucial for active protein unfolding by mitochondria.
Fig. 6. Spontaneous unfolding of preproteins becomes rate-limiting in mtHsp70-defective mitochondria. (A) b2(47)-DHFR was imported into ssc1-2 mutant mitochondria and detected by western blotting as described in Figure 5A. Either denatured (+Urea) or non-denatured (–Urea) preprotein was used. (B) Import kinetics of b2(47)-DHFR in ssc1-2 mutant mitochondria. Import rates of non-denatured (ssc1-2/–Urea, open squares) and denatured b2(47)-DHFR (ssc1-2/+Urea, filled squares) in ssc1-2 mutant mitochondria are indicated. The dotted line shows the import rate of non-denatured b2(47)-DHFR in the corresponding wild-type mitochondria (WT/–Urea) as determined in Figure 2D. The value of imported native b2(47)-DHFR reached after 15 min import into wild-type mitochondria was set to 100% (control).
Discussion
We have solved the controversial issue of whether mitochondria play an active role in preprotein unfolding or not. Under substrate-saturating conditions, translocation starts with a lag phase that is shortened by active unfolding of preproteins. The active unfolding depends on the function of mtHsp70 and its accessibility to the preprotein in transit.
The basis of our investigation is the quantitative characterization of the mitochondrial protein translocation system using saturating amounts of soluble and fully import-competent preproteins. This import system has two major advantages over the standard system using precursor proteins generated by in vitro translation. First, import reactions can be performed under conditions where the mitochondrial translocation system is challenged with saturating amounts of preproteins. This allows the determination of the maximal rate of preprotein translocation as a characteristic parameter for the kinetic activity of the mitochondrial import system. Secondly, the absence of cofactors derived from the reticulocyte translation reaction makes it possible to study directly the effects of precursor protein conformation on the efficiency of the import reaction. With this newly established system, the measured maximal import rate in vitro for a matrix-targeted preprotein is ∼25 pmol/min/mg of mitochondrial protein. Upon denaturation of the preprotein, the import rate reached 50 pmol/min/mg. How do these rates compare with the in vivo situation (Neupert et al., 1990)? Based on a doubling time of yeast cells of 3.5 h at 25°C, an average molecular mass of 35 kDa for a mitochondrial protein (Netzer and Hartl, 1998) and taking into account that up to 50% of the mitochondrial proteins are targeted to the matrix, the import rate in vivo would reach ∼40–60 pmol/min/mg. The in vitro system presented here thus operates with an import activity closely resembling the in vivo situation. Moreover, this shows that cytosolic cofactors are not essential for an efficient activity of the mitochondrial import machinery. Cytosolic chaperones are probably involved in preventing misfolding or aggregation of some preproteins (Deshaies et al., 1988; Murakami et al., 1988; Sheffield et al., 1990; Komiya et al., 1996); the high rates of both folded and denatured preproteins in the in vitro import system demonstrate that mitochondria themselves are capable of efficient unfolding and translocation of preproteins. With a total number of import sites for cleavable preproteins in the inner membrane of 15–20 pmol/mg of mitochondrial protein (Dekker et al., 1997), about three preproteins can be translocated through one import site per minute in vitro, resulting in an average translocation velocity in the import channel of ∼9–13 amino acids/s. This velocity is slightly greater than the estimated velocity of the ribosomal translation reaction of 5–10 amino acids/s under similar conditions (Boehlke and Friesen, 1975; Bonven and Gullov, 1979).
We identified a characteristic initial delay of translocation (lag phase) that becomes longer the shorter the N-terminal extension of the DHFR fusion proteins is. The subsequently reached maximal import rate is similar for preproteins with different N-terminal extension lengths. Previous studies with radiolabeled preproteins (Matouschek et al., 1997; Gaume et al., 1998) have not resolved this initial import lag phase, yet have suggested that mitochondrial protein import follows first-order kinetics and that the overall import rate of preproteins becomes slower the shorter the N-terminal extension is. The quantitative system used here, however, shows that preprotein translocation into the mitochondrial matrix follows a sigmoidal pattern, clearly indicating that the kinetics of preprotein import are determined by a combination of polypeptide unfolding and translocation. Since in the case of a urea-denatured preprotein, the need for polypeptide unfolding is circumvented, the maximal import rate reached is higher than that of the folded preprotein. Most importantly, the lag phase is completely abolished by denaturation of the preprotein, providing the opportunity to determine directly the rate of preprotein unfolding during translocation. By comparison with unfolding of DHFR in solution (with a chaperonin trap), the controversy of active (Matouschek et al., 1997) or passive protein unfolding by mitochondria (Gaume et al., 1998) can be solved. (i) When the N-terminal extension of the DHFR fusion protein is too short to allow productive interaction with the mtHsp70 system (Rassow et al., 1990; Ungermann et al., 1994, 1996; Matouschek et al., 1997; Gaume et al., 1998), unfolding by mitochondria is not faster than spontaneous unfolding. This agrees with the observation that the outer membrane import machinery has only a low unfolding activity by trapping spontaneously unfolded preproteins (Eilers et al., 1988; Skerjanc et al., 1990; Mayer et al., 1995; Huang et al., 2000; Stan et al., 2000). (ii) DHFR fusion proteins with longer N-terminal extensions, however, are unfolded rapidly during translocation, significantly faster than spontaneous unfolding. This demonstrates an active unfolding of the preproteins by mitochondria.
MtHsp70 is of central importance for the mitochondrial unfolding activity. Impairing the function of mtHsp70 with a temperature-sensitive yeast mutant (ssc1-2) that inhibits binding of mtHsp70 to Tim44 results in a prolongation of the lag phase of translocation. The lag phase of a preprotein that is actively unfolded under wild-type conditions is shifted towards the direction of spontaneous (passive) unfolding in the mutant mitochondria, while the lag phase is almost completely abolished by denaturation of the preprotein despite the presence of the mutant form of mtHsp70. A proper interaction of mtHsp70 with Tim44 is thus of critical importance for promoting an active unfolding mechanism by mitochondria. The question arises of why Gaume et al. (1998) concluded that unfolding of DHFR by mitochondria is slower than spontaneous unfolding. The half-time for the spontaneous unfolding of DHFR in solution determined by Gaume et al. (1998) is in fact similar to the value determined by us. However, the rate constant for the translocation reaction under substrate-saturating conditions measured here with wild-type mitochondria is at least an order of magnitude higher than that obtained by Gaume et al. (1998), indicating that their import conditions with radiolabeled preproteins did not yield the maximal rates of import. Indeed, when the accessibility of mtHsp70 to the preprotein in transit was nearly blocked both by using a DHFR fusion protein with a short N-terminal extension and by employing ssc1-2 mitochondria with the impaired binding of mtHsp70 to Tim44, then we obtained import rates that are clearly lower than spontaneous unfolding. Under these limited import conditions, only trapping of spontaneously unfolded preproteins by mtHsp70 will be possible, in agreement with the view of Gaume et al. (1998) that several rounds of spontaneous unfolding of the preprotein are needed to achieve import. However, when the mitochondrial Hsp70 system works with full activity and can access the preproteins in transit, unfolding and translocation of preproteins are dramatically faster, proving the active unfolding mechanism.
In an elegant study, Huang et al. (1999) showed that mitochondria promote the unfolding of a barnase fusion protein by initiating unfolding at an N-terminal α-helix. Similarly, the heme-binding domain present in the mature part of cytochrome b2 (Xia and Mathews, 1990; Glick et al., 1993; Voos et al., 1993; Gaume et al., 1998; Voisine et al., 1999) contains an N-terminal α-helix and may thus be unraveled by a comparable mechanism. Many protein domains, however, contain an α/β fold (Branden and Tooze, 1998), where the N-terminus is buried inside the structure. The N-terminus of DHFR is present in a β-sheet that is located between two layers of α-helices. DHFR and proteins with a similar α/β fold probably require global unfolding of the domain for a release of the N-terminus. The results presented here demonstrate that mitochondria are able to actively unfold those frequently occurring domain structures.
This study resolves two kinetic phases in membrane translocation of preproteins. A first phase of active protein unfolding strongly depends on the accessibility of the preprotein to mtHsp70 and is thus influenced by the folding state of the preprotein and the functional activity of mtHsp70. In a second phase, the maximal import rate is reached that, in wild-type mitochondria, is similar for preproteins with different N-terminal extensions, i.e. is not determined by the mechanism (active or spontaneous) of the initial unfolding reaction. The maximal import rate therefore seems to represent an intrinsic property of the mitochondrial translocation motor of how fast an extended polypeptide chain is threaded through the import channels.
Materials and methods
Expression and purification of mitochondrial preproteins
Mitochondrial preproteins consisting of an N-terminal segment derived from S.cerevisiae cytochrome b2 including the mitochondrial targeting sequence (lacking residues 47–65 of the hydrophobic intermembrane space sorting signal, indicated by ▵) fused to the complete mouse DHFR were expressed from the following plasmids under control of the lac promoter. Plasmids pUHE73 [b2(167)Δ-DHFR] (Dekker et al., 1997) and pUHE96 [b2(47)-DHFR] were constructed by cloning the inserts of the respective transcription plasmids (Rassow et al., 1990; Voos et al., 1993) into pUHE. Plasmid pUHE5095 [b2(107)Δ-DHFR] was obtained by substituting the insert of pUHE73 with a PCR-generated fragment encoding the shortened segment. For expression, E.coli cells from strain BMH71-18 containing the corresponding plasmid were grown at 37°C in 500 ml of LB medium (1% tryptone, 0.5% yeast extract, 0.5% NaCl) to an OD600 of 1. After 2 h induction with 1 mM isopropyl-β-d-thiogalacto pyranoside (IPTG) at 37°C, the cells were collected by centrifugation and washed once with water. The cells were resuspended in 20 ml of pre-lysis buffer (30% sucrose, 20 mM KPi pH 8.0, 1 mM EDTA) and incubated for 10 min at 0°C. After treating the cells in 20 ml of 10 mM dithiothreitol (DTT), 1 mM phenylmethylsulfonyl fluoride (PMSF) for 10 min at 0°C, they were resuspended in 20 ml of buffer A (20 mM MOPS–KOH, 1 mM EDTA pH 8.0) containing 10 mM DTT, 1 mM PMSF and a protease inhibitor mix (Roche). Lysozyme (1 mg/ml) and 0.1% (v/v) Triton X-100 were added and the suspension was kept on ice for 10 min with occasional stiring. After sonication in a Branson Sonifier with 3× 20 pulses (40% duty cycle, micro tip setting 7), the cell lysate was centrifuged for 20 min at 15 000 g. After passing the supernatant through an acetate cellulose membrane (0.22 µm pores), the soluble proteins were loaded on a MonoS column (Pharmacia). The column was washed with 3 vols of buffer A and bound proteins were eluted with a linear salt gradient (150–450 mM NaCl in buffer A). All b2-DHFR fusion proteins eluted at ∼300 mM NaCl. The concentration of the preprotein in pooled fractions was determined by both protein concentration assays and by comparing the intensity of Coomassie Blue-stained protein bands with a set of standard proteins after SDS–PAGE. The proteins were stored in aliquots at –80°C.
Yeast strains and import of proteins into mitochondria
The following strains of S.cerevisiae were used (Gambill et al., 1993): PK82 (wild type), Matα his4-713 lys2 ura3-52 leu2-3 112 Δtrp1; and PK81 (ssc1-2), Matα ade2-101 lys2 ura3-52 leu2-3 112 Δtrp1 ssc1-2(LEU2). Mitochondria were isolated as described previously (Daum et al., 1982; Hartl et al., 1987). Import reactions consisted of 25 µg of mitochondrial protein in 100 µl assay volume. To induce the temperature-sensitive phenotype, mitochondria were pre-incubated for 15 min at 37°C in import buffer [250 mM sucrose, 3% (w/v) fatty acid-free bovine serum albumin (BSA), 80 mM KCl, 5 mM MgCl2, 20 mM MOPS–KOH pH 7.2]. After cooling to 25°C, the reactions were supplemented with 2 mM NADH and 1 mM ATP and import was started by adding 160 nM precursor protein (final concentration 640 pmol preprotein per mg of mitochondrial protein). Valinomycin (1 µM), antimycin A (8 µM) and oligomycin (20 µM) were added to control reactions to dissipate the inner membrane potential. Import reactions were stopped by cooling on ice and addition of 1 µM valinomycin. For import of unfolded precursors, preproteins were denaturated by 8 M urea, 30 mM MOPS–KOH pH 7.2, 50 mM DTT for 15 min at 25°C immediately prior to the import reaction. After the import, the reactions were treated with 100 µg/ml proteinase K where indicated, washed twice and solubilized in Laemmli sample buffer (Söllner et al., 1991). The samples were analyzed by SDS–PAGE and western blot using affinity-purified anti-DHFR antibodies. The signals were detected by the ECL system (Amersham) and quantified using the ImageMaster evaluation system (Pharmacia). SEMs were determined from at least three to five independent experiments.
Unfolding of DHFR fusion proteins
Cytochrome b2-DHFR fusion proteins (1.25 µM) were incubated with 6.5 µM GroEL (Roche) in the assay buffer (80 mM KCl, 60 mM MOPS–KOH pH 7.2) at 25°C for up to 1 h. Late folding intermediates of b2-DHFR fusion proteins were trapped by GroEL in the absence of ATP and the cofactor GroES (Clark and Frieden, 1999; von Ahsen et al., 2000). Under the conditions used, ∼70% of the total DHFR activity of the purified preproteins was abolished after 1 h incubation with GroEL. The enzymatic activity of the DHFR domain was measured at 340 nm using 150 µM dihydrofolate and 150 µM NADPH, respectively. The spontaneous unfolding rate was determined by plotting residual activity versus time.
Acknowledgments
Acknowledgements
We thank Dr E.A.Craig for the ssc1-2 mutant, Drs A.Matouschek and T.Prinz for discussion, and N.Zufall for expert technical assistance. This work was supported by the Deutsche Forschungsgemeinschaft, Sonderforschungsbereich 388, and the Fonds der Chemischen Industrie.
References
- Bauer M.F., Hofmann,S., Neupert,W. and Brunner,M. (2000) Protein translocation into mitochondria: the role of TIM complexes. Trends Cell Biol., 10, 25–31. [DOI] [PubMed] [Google Scholar]
- Beasley E.M., Müller,S. and Schatz,G. (1993) The signal that sorts yeast cytochrome b2 to the mitochondrial intermembrane space contains three distinct functional regions. EMBO J., 12, 2303–2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beissinger M. and Buchner,J. (1998) How chaperones fold proteins. Biol. Chem., 379, 245–259. [PubMed] [Google Scholar]
- Boehlke K.W. and Friesen,J.D. (1975) Cellular content of ribonucleic acid and protein in Saccharomyces cerevisiae as a function of exponential growth rate: calculation of the apparent peptide chain elongation rate. J. Bacteriol., 121, 429–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bömer U., Meijer,M., Guiard,B., Dietmeier,K., Pfanner,N. and Rassow,J. (1997) The sorting route of cytochrome b2 branches from the general mitochondrial import pathway at the preprotein translocase of the inner membrane. J. Biol. Chem., 272, 30439–30446. [DOI] [PubMed] [Google Scholar]
- Bömer U., Maarse,A.C., Martin,F., Geissler,A., Merlin,A., Schönfisch,B., Meijer,M., Pfanner,N. and Rassow,J. (1998) Separation of structural and dynamic functions of the mitochondrial translocase: Tim44 is crucial for the inner membrane import sites in translocation of tightly folded domains, but not of loosely folded preproteins. EMBO J., 17, 4226–4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonven B. and Gullov,K. (1979) Peptide chain elongation rate and ribosomal activity in Saccharomyces cerevisiae as a function of the growth rate. Mol. Gen. Genet., 170, 225–230. [DOI] [PubMed] [Google Scholar]
- Branden C. and Tooze,J. (1998) Introduction to Protein Structure. Garland, New York, NY.
- Clark A.C. and Frieden,C. (1999) The chaperonin GroEL binds to late-folding non-native conformations present in native Escherichia coli and murine dihydrofolate reductases. J. Mol. Biol., 285, 1777–1788. [DOI] [PubMed] [Google Scholar]
- Daum G., Gasser,S.M. and Schatz,G. (1982) Import of proteins into mitochondria: energy-dependent, two-step processing of the intermembrane space enzyme cytochrome b2 by isolated mitochondria. J. Biol. Chem., 257, 13075–13080. [PubMed] [Google Scholar]
- Dekker P.J.T., Martin,F., Maarse,A.C., Bömer,U., Müller,H., Guiard,B., Meijer,M., Rassow,J. and Pfanner,N. (1997) The Tim core complex defines the number of mitochondrial translocation contact sites and can hold arrested preproteins in the absence of matrix Hsp70–Tim44. EMBO J., 16, 5408–5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies R., Koch,B., Werner-Washburne,M., Craig,E.A. and Schekman,R. (1988) A subfamily of stress proteins facilitates translocation of secretory and mitochondrial precursor polypeptides. Nature, 332, 800–805. [DOI] [PubMed] [Google Scholar]
- Eilers M. and Schatz,G. (1986) Binding of a specific ligand inhibits import of a purified precursor protein into mitochondria. Nature, 322, 228–232. [DOI] [PubMed] [Google Scholar]
- Eilers M., Hwang,S. and Schatz,G. (1988) Unfolding and refolding of a purified precursor protein during import into isolated mitochondria. EMBO J., 7, 1139–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambill B.D., Voos,W., Kang,P.J., Miao,B., Langer,T., Craig,E.A. and Pfanner,N. (1993) A dual role for mitochondrial heat shock protein 70 in membrane translocation of preproteins. J. Cell Biol., 123, 109–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaume B., Klaus,C., Ungermann,C., Guiard,B., Neupert,W. and Brunner,M. (1998) Unfolding of preproteins upon import into mitochondria. EMBO J., 17, 6497–6507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick B.S. (1995) Can Hsp70 proteins act as force-generating motors? Cell, 80, 11–14. [DOI] [PubMed] [Google Scholar]
- Glick B.S., Wachter,C., Reid,G.A. and Schatz,G. (1993) Import of cytochrome b2 to the mitochondrial intermembrane space: the tightly folded heme-binding domain makes import dependent upon matrix ATP. Protein Sci., 2, 1901–1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg M.S., Zhang,J., Sondek,S., Matthews,C.R., Fox,R.O. and Horwich,A.L. (1997) Native-like structure of a protein-folding intermediate bound to the chaperonin GroEL. Proc. Natl Acad. Sci. USA, 94, 1080–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl F.-U., Ostermann,J., Guiard,B. and Neupert,W. (1987) Successive translocation into and out of the mitochondrial matrix: targeting of proteins to the intermembrane space by a bipartite signal peptide. Cell, 51, 1027–1037. [DOI] [PubMed] [Google Scholar]
- Huang S., Ratliff,K.S., Schwartz,M.P., Spenner,J.M. and Matouschek,A. (1999) Mitochondria unfold precursor proteins by unraveling them from their N-termini. Nature Struct. Biol., 6, 1132–1138. [DOI] [PubMed] [Google Scholar]
- Huang S., Murphy,S. and Matouschek,A. (2000) Effect of the protein import machinery at the mitochondrial surface on precursor stability. Proc. Natl Acad. Sci. USA, 97, 12991–12996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen R.E. and Johnson,A.E. (1999) Protein translocation: is hsp70 pulling my chain? Curr. Biol., 9, R779–R782. [DOI] [PubMed] [Google Scholar]
- Kang P.J., Ostermann,J., Shilling,J., Neupert,W., Craig,E.A. and Pfanner,N. (1990) Requirement for hsp70 in the mitochondrial matrix for translocation and folding of precursor proteins. Nature, 348, 137–143. [DOI] [PubMed] [Google Scholar]
- Koehler C.M., Merchant,S. and Schatz,G. (1999) How membrane proteins travel across the mitochondrial intermembrane space. Trends Biochem. Sci., 24, 428–432. [DOI] [PubMed] [Google Scholar]
- Koll H., Guiard,B., Rassow,J., Ostermann,J., Horwich,A.L., Neupert,W. and Hartl,F.-U. (1992) Antifolding activity of hsp60 couples protein import into the mitochondrial matrix with export to the intermembrane space. Cell, 68, 1163–1175. [DOI] [PubMed] [Google Scholar]
- Komiya T., Sakaguchi,M. and Mihara,K. (1996) Cytoplasmic chaperones determine the targeting pathway of precursor proteins to mitochondria. EMBO J., 15, 399–407. [PMC free article] [PubMed] [Google Scholar]
- Kronidou N.G., Oppliger,W., Bolliger,L., Hannavy,K., Glick,B.S., Schatz,G. and Horst,M. (1994) Dynamic interaction between Isp45 and mitochondrial hsp70 in the protein import system of the yeast mitochondrial inner membrane. Proc. Natl Acad. Sci. USA, 91, 12818–12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matouschek A., Azem,A., Ratliff,K., Glick,B.S., Schmid,K. and Schatz,G. (1997) Active unfolding of precursor proteins during mitochondrial protein import. EMBO J., 16, 6727–6736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer A., Neupert,W. and Lill,R. (1995) Mitochondrial protein import: reversible binding of the presequence at the trans side of the outer membrane drives partial translocation and unfolding. Cell, 80, 127–137. [DOI] [PubMed] [Google Scholar]
- Murakami H., Pain,D. and Blobel,G. (1988) 70-kD heat shock-related protein is one of at least two distinct cytosolic factors stimulating protein import into mitochondria. J. Cell Biol., 107, 2051–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netzer W.J. and Hartl,F.U. (1998) Protein folding in the cytosol: chaperonin-dependent and -independent mechanisms. Trends Biochem. Sci., 23, 68–73. [DOI] [PubMed] [Google Scholar]
- Neupert W. (1997) Protein import into mitochondria. Annu. Rev. Biochem., 66, 863–917. [DOI] [PubMed] [Google Scholar]
- Neupert W., Hartl,F.U., Craig,E.A. and Pfanner,N. (1990) How do polypeptides cross mitochondrial membranes? Cell, 63, 447–450. [DOI] [PubMed] [Google Scholar]
- Pfanner N. and Meijer,M. (1995) Protein sorting: pulling in the proteins. Curr. Biol., 5, 132–135. [DOI] [PubMed] [Google Scholar]
- Pfanner N., Craig,E.A. and Hönlinger,A. (1997) Mitochondrial preprotein translocase. Annu. Rev. Cell Dev. Biol., 13, 25–51. [DOI] [PubMed] [Google Scholar]
- Rassow J., Guiard,B., Wienhues,U., Herzog,V., Hartl,F.-U. and Neupert,W. (1989) Translocation arrest by reversible folding of a precursor protein imported into mitochondria. A means to quantitate translocation contact sites. J. Cell Biol., 109, 1421–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rassow J., Hartl,F.U., Guiard,B., Pfanner,N. and Neupert,W. (1990) Polypeptides traverse the mitochondrial envelope in an extended state. FEBS Lett., 275, 190–194. [DOI] [PubMed] [Google Scholar]
- Rassow J., Maarse,A.C., Krainer,E., Kübrich,M., Müller,H., Meijer,M., Craig,E.A. and Pfanner,N. (1994) Mitochondrial protein import: biochemical and genetic evidence for interaction of matrix hsp70 and the inner membrane protein MIM44. J. Cell Biol., 127, 1547–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rassow J., Dekker,P.J.T., van Wilpe,S., Meijer,M. and Soll,J. (1999) The preprotein translocase of the mitochondrial inner membrane: function and evolution. J. Mol. Biol., 286, 105–120. [DOI] [PubMed] [Google Scholar]
- Ryan M.T. and Pfanner,N. (1998) The preprotein translocase of the mitochondrial outer membrane. Biol. Chem., 379, 289–294. [PubMed] [Google Scholar]
- Schatz G. and Dobberstein,B. (1996) Common principles of protein translocation across membranes. Science, 271, 1519–1526. [DOI] [PubMed] [Google Scholar]
- Scherer P.E., Krieg,U.C., Hwang,S.T., Vestweber,D. and Schatz,G. (1990) A precursor protein partly translocated into yeast mitochondria is bound to a 70 kD mitochondrial stress protein. EMBO J., 9, 4315–4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider H.-C., Berthold,J., Bauer,M.F., Dietmeier,K., Guiard,B., Brunner,M. and Neupert,W. (1994) Mitochondrial Hsp70/MIM44 complex facilitates protein import. Nature, 371, 768–774. [DOI] [PubMed] [Google Scholar]
- Schwartz M.P., Huang,S. and Matouschek,A. (1999) The structure of precursor proteins during import into mitochondria. J. Biol. Chem., 274, 12759–12764. [DOI] [PubMed] [Google Scholar]
- Schwarz E., Seytter,T., Guiard,B. and Neupert,W. (1993) Targeting of cytochrome b2 into the mitochondrial intermembrane space: specific recognition of the sorting signal. EMBO J., 12, 2295–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffield W.P., Shore,G.C. and Randall,S.K. (1990) Mitochondrial precursor protein. Effects of 70-kD heat shock protein on polypeptide folding, aggregation and import competence. J. Biol. Chem., 265, 11069–11076. [PubMed] [Google Scholar]
- Sirrenberg C., Endres,M., Becker,K., Bauer,M.F., Walther,E., Neupert,W. and Brunner,M. (1997) Functional cooperation and stoichiometry of protein translocases of the outer and inner membranes of mitochondria. J. Biol. Chem., 272, 29963–29966. [DOI] [PubMed] [Google Scholar]
- Skerjanc I.S., Sheffield,W.P., Randall,S.K., Silvius,J.R. and Shore,G.C. (1990) Import of precursor proteins into mitochondria: site of polypeptide unfolding. J. Biol. Chem., 265, 9444–9451. [PubMed] [Google Scholar]
- Söllner T., Rassow,J. and Pfanner,N. (1991) Analysis of mitochondrial protein import using translocation intermediates and specific antibodies. Methods Cell Biol., 34, 345–358. [DOI] [PubMed] [Google Scholar]
- Stan T., Ahting,U., Dembowski,M., Künkele,K.P., Nussberger,S., Neupert,W. and Rapaport,D. (2000) Recognition of preproteins by the isolated TOM complex of mitochondria. EMBO J., 19, 4895–4902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungermann C., Neupert,W. and Cyr,D.M. (1994) The role of hsp70 in conferring unidirectionality on protein translocation. Science, 266, 1250–1253. [DOI] [PubMed] [Google Scholar]
- Ungermann C., Guiard,B., Neupert,W. and Cyr,D.M. (1996) The Δψ- and Hsp70/MIM44-dependent reaction cycle driving early steps of protein import into mitochondria. EMBO J., 15, 735–744. [PMC free article] [PubMed] [Google Scholar]
- Vestweber D. and Schatz,G. (1988) A chimeric mitochondrial precursor protein with internal disulfide bridges blocks import of authentic precursors into mitochondria and allows quantitation of import sites. J. Cell Biol., 107, 2037–2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viitanen P.V., Donaldson,G.K., Lorimer,G.H., Lubben,T.H. and Gatenby,A.A. (1991) Complex interactions between the chaperonin 60 molecular chaperone and dihydrofolate reductase. Biochemistry, 30, 9716–9723. [DOI] [PubMed] [Google Scholar]
- Voisine C., Craig,E.A., Zufall,N., von Ahsen,O., Pfanner,N. and Voos,W. (1999) The protein import motor of mitochondria: unfolding and trapping of preproteins are distinct and separable functions of matrix Hsp70. Cell, 97, 565–574. [DOI] [PubMed] [Google Scholar]
- von Ahsen O., Voos,W., Henninger,H. and Pfanner,N. (1995) The mitochondrial protein import machinery. Role of ATP in dissociation of the Hsp70⋅Mim44 complex. J. Biol. Chem., 270, 29848–29853. [DOI] [PubMed] [Google Scholar]
- von Ahsen O., Lim,J.H., Caspers,P., Martin,F., Schönfeld,H.J., Rassow,J. and Pfanner,N. (2000) Cyclophilin-promoted folding of mouse dihydrofolate reductase does not include the slow conversion of the late-folding intermediate to the active enzyme. J. Mol. Biol., 297, 809–818. [DOI] [PubMed] [Google Scholar]
- Voos W., Gambill,B.D., Guiard,B., Pfanner,N. and Craig,E.A. (1993) Presequence and mature part of preproteins strongly influence the dependence of mitochondrial protein import on heat shock protein 70 in the matrix. J. Cell Biol., 123, 119–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voos W., von Ahsen,O., Müller,H., Guiard,B., Rassow,J. and Pfanner,N. (1996) Differential requirement for the mitochondrial Hsp70–Tim44 complex in unfolding and translocation of preproteins. EMBO J., 15, 2668–2677. [PMC free article] [PubMed] [Google Scholar]
- Voos W., Martin,H., Krimmer,T. and Pfanner,N. (1999) Mechanisms of protein translocation into mitochondria. Biochim. Biophys. Acta, 1422, 235–254. [DOI] [PubMed] [Google Scholar]
- Wienhues U., Becker,K., Schleyer,M., Guiard,B., Tropschug,M., Horwich,A.L., Pfanner,N. and Neupert,W. (1991) Protein folding causes an arrest of preprotein translocation into mitochondria in vivo. J. Cell Biol., 115, 1601–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Z.-x. and Mathews,F.S. (1990) Molecular structure of flavocytochrome b2 at 2.4 Å resolution. J. Mol. Biol., 212, 837–863. [DOI] [PubMed] [Google Scholar]