Abstract
It is hypothesized that infectious prions are generated as the cellular form of the prion protein (PrPC) undergoes pronounced conformational change under the direction of an infectious PrPSc template. Conversion to the infectious conformer is particularly associated with major structural rearrangement in the central portion of the protein (residues 90–120), which has an extended flexible structure in the PrPC isoform. Using a panel of recombinant antibodies reactive with different parts of PrP, we show that equivalent major structural rearrangements occur spontaneously in this region of PrP immobilized on a surface. In contrast, regions more towards the termini of the protein remain relatively unaltered. The rearrangements occur even under conditions where individual PrP molecules should not contact one another. The propensity of specific unstructured regions of PrP to spontaneously undergo large and potentially deleterious conformational changes may have important implications for prion biology.
Keywords: conformational change/phage display/prion protein/recombinant antibodies
Introduction
Transmissible spongiform encephalopathies are a group of neurodegenerative diseases that feature the pronounced accumulation, in certain brain regions, of a misfolded isomer (PrPSc) of the cellular prion protein (PrPC) (Bolton et al., 1982; Prusiner, 1991; Weissmann et al., 1996; Prusiner et al., 1998; Weissmann, 1999). PrPC is expressed on the surface of neuronal cells, but may also be found on many other cell types (Bendheim et al., 1992). The protein contains two N-linked sugars and is anchored to the cell membrane via a glycosyl-phosphatidyl-inositol (GPI) group at the extreme C-terminus (residue 231) (Stahl et al., 1987, 1990). The physiological role of PrP is unknown. The N-terminal portion of the molecule typically contains five octarepeat regions [PHGGG(T)WGQ], which bind copper with micromolar affinity (Stockel et al., 1998; Miura et al., 1999; Viles et al., 1999). The in vivo significance of this relatively low affinity interaction, however, remains controversial (Pauly and Harris, 1998; Brown et al., 1999; Lippard, 1999; Waggoner et al., 2000). In neuronal cells, PrPC cycles rapidly between the cell surface and endocytic compartments thought to be clathrin-coated vesicles, and has an overall half-life of 4–6 h (Shyng et al., 1993, 1994; Lehmann et al., 1999). These subcellular compartments have been suggested as the site of conversion of PrPC to PrPSc and generation of prion infectivity (Caughey and Raymond, 1991; Caughey et al., 1991; Borchelt et al., 1992; Harris et al., 1996).
Early studies indicated that the physical properties of PrPC and PrPSc differ markedly. Purified PrPSc has a propensity to form aggregates that have the morphological characteristics of amyloids, binding to the birefringent dye Congo Red and possessing secondary structure content that is rich in β-sheets (Pan et al., 1993; Priola et al., 1994; Caspi et al., 1998). The aggregated protein is also partially resistant to proteolysis and is insoluble in non-ionic detergent. In contrast, PrPC contains a paucity of β-arrangements, is sensitive to protease digestion and is soluble in non-ionic detergent (Prusiner et al., 1982; Oesch et al., 1985; Prusiner, 1994).
Whereas relatively little is known of PrPSc conformation, the three-dimensional structures of recombinant mouse (Mo), Syrian hamster (SHa), bovine and human PrP folded into an α-helical conformation have been solved by nuclear magnetic resonance (NMR) (Riek et al., 1996, 1997; Donne et al., 1997; Lopez Garcia et al., 2000; Zahn et al., 2000). In each case, the C-terminal half of the protein (residues 124–231) is folded into a core comprising three α-helices, one or two short β-strands, and a disulfide bridge linking the second and third helices. The N-terminal portion of the protein, spanning residues 23–124, appears to be highly flexible and devoid of any secondary structure under the experimental conditions employed for these studies (Riek et al., 1996, 1997; Donne et al., 1997; Zahn et al., 2000). It seems likely that these recombinant molecules, despite the absence of glycosylation, closely resemble PrPC in its native environment, since antibodies bind well to recombinant PrP and PrP on the surface of living cells (Williamson et al., 1998).
The molecular events leading to the profound conformational changes in PrPC that accompany PrPSc formation are central to prion pathogenesis, but remain poorly understood. There is, however, substantial evidence to support the role of PrPSc as a template directing the fate of PrPC during prion replication (Bessen et al., 1995; Prusiner, 1997). For example, the properties distinguishing individual prion strains appear to be enciphered in distinct PrPSc conformations (Telling et al., 1996; Scott et al., 1997; Safar et al., 1998; Wadsworth et al., 1999). It follows, then, that PrPC represents a remarkably malleable substrate for infectious prion propagation. Indeed, several avenues of investigation have underlined the inherent plasticity of PrPC structure. Recombinant PrP containing residues 90–231, corresponding to the protease-resistant core of infectious PrP, has been refolded into both α-helical-rich and β-sheet-rich structures as well as various intermediates in aqueous buffers (Zhang et al., 1997; Jackson et al., 1999). Particular attention has been focussed on the properties of the 90–145 region of PrP. Studies of synthetic peptides corresponding to residues 109–121 (Gasset et al., 1992), 106–126 (Salmona et al., 1999), 109–145 and 90–145 (Zhang et al., 1995) indicate that this portion of the protein may adopt conformations rich in either α-helices or β-sheets. The β-sheet peptides can assemble into rod-shaped polymers. Significantly, synthetic peptide corresponding to MoPrP89–143 (P101L), when refolded into a β-sheet conformation, can initiate or accelerate the onset of prion pathology in transgenic mice (Kaneko et al., 2000).
Monoclonal antibody binding studies also demonstrate the innate structural flexibility of PrP (Peretz et al., 1997; Williamson et al., 1998). Antibody epitopes lying between amino acids 90–112 are found to be exposed in PrPC, but are altered or buried in infectious forms of PrP. In contrast, an epitope lying toward the C-terminus of PrP is present in both cellular and infectious forms of the protein, indicating that C-terminal regions of PrP may possess more conformational rigidity than N-terminal regions (Peretz et al., 1997; Williamson et al., 1998). In addition, an X-ray crystallographic structure of the Fab fragment of the 3F4 antibody complexed to a peptide spanning residues 104–113 of SHaPrP showed the bound peptide in a Ω-loop conformation, stabilized by two intrapeptide hydrogen bonds (Kanyo et al., 1999). This structure is a possible intermediate in the formation of a β-hairpin, and is unexpected in a region shown to be devoid of structure by NMR. One interpretation of these findings is that this portion of PrP is capable of existing either in a disordered state or in a variety of discrete but isoenergetic arrangements.
In this paper, we have extended our antibody studies to examine global and local structural changes occurring in SHaPrP23–231 over time. Using a panel of eight monoclonal Fab fragments directed against diverse linear and conformational epitopes of SHaPrP, and including two novel antibodies recognizing the N-terminal portion of the molecule (residues 29–37 and 72–86, respectively), we have followed conformational changes occurring over a 6-day period in immobilized PrP molecules using surface plasmon resonance (SPR). We report that pronounced conformational changes in the prion protein occur via distinct molecular domains, as defined by their binding to monoclonal antibody fragments. Our findings indicate that under physiological solvent conditions, monomeric PrPC is in itself an intrinsically unstable molecule prone to conformational rearrangement over time.
Results
Measuring dynamic changes in PrP conformation over time using a diverse panel of eight monoclonal antibodies
During preliminary antibody binding studies using SPR and immobilized recombinant PrP preparations, we noted that binding signals obtained with certain monoclonal antibodies decreased over time, whereas binding signals generated with other antibodies remained constant over the same period. The mass of immobilized PrP on the sensor chip was found to be invariant during the course of these experiments, seemingly precluding proteolytic degradation of PrP antigen as a facile explanation for the loss of specific epitopes. We considered the most plausible explanation for our observations was that localized regions of the PrP antigen were undergoing major structural rearrangement, while the conformations adopted by other regions of the PrP molecule remained relatively unaltered.
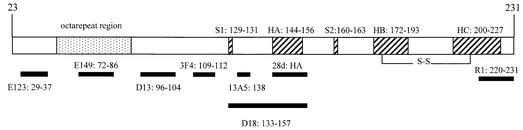
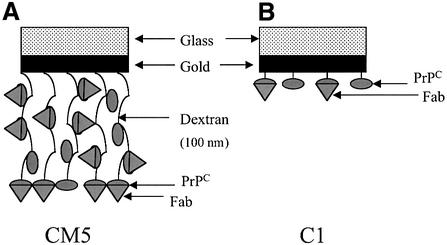
To test this hypothesis, we measured the reactivity of a diverse panel of eight PrP-specific monoclonal antibodies, the binding affinities and epitopes of which are illustrated in Table I and Figure 1, against recombin ant SHaPrP29–231 and semi-synthetic SHaPrP23–231 refolded into an α-helical conformation and immobilized by covalent linkage or via biotin, respectively, to C1 and CM5 sensor chips. As shown schematically in Figure 2, the C1 and CM5 sensor chips differ markedly in the character of their active surfaces. The carboxymethyl groups are either anchored directly to the sensor chip surface (C1) or linked to dextran strands of ∼100 nm in length, which project away from the surface of the sensor chip (CM5). Binding of each antibody was measured immediately after immobilization of the PrP antigen. The sensor chip was then incubated in physiological buffer for 4–6 days at 37°C, and antibody binding re-measured. Binding signals obtained from this set of readings were expressed as a percentage of the signal obtained at the beginning of the experiment with the same antibody.
Table I. Epitopes and kinetic constants for the binding of PrP-specific antibodies to either a synthetic peptide corresponding to SHaPrP23–98 or recombinant SHaPrP29–231 refolded into an α-helical-rich conformation, as determined by SPR.
| Fab | Epitope | kon (M–1s–1) | koff (s–1) | Kd (nM) |
|---|---|---|---|---|
| SHaPrP23–98 | ||||
| E123 | 29–37 | 1.6 ± 0.3 × 104 | 7.4 ± 0.5 × 10–4 | 46 |
| E149 | 72–86 | 2.0 ± 0.5 × 105 | 0.019 ± 0.005 | 93 |
| SHaPrP29–231 | ||||
| E123 | 29–37 | 1.1 ± 0.4 × 104 | 7.6 ± 0.4 × 10–4 | 69 |
| E149 | 72–86 | 1.0 ± 0.3 × 105 | 0.017 ± 0.002 | 170 |
| D13h | 96–104 | 5.4 ± 0.05 × 105 | 1.8 ± 0.3 × 10–4 | 3.3 |
| 3F4 | 109–112 | 1.9 ± 0.4 × 104 | 4.1 ± 0.6 × 10–4 | 21.5 |
| 13A5 | 138–141 | 2.9 ± 0.1 × 104 | 4.9 ± 0.8 × 10–4 | 17 |
| 28dh | 144–156a | 1 ± 0.3 × 106 | 0.034 ± 0.001 | 34b |
| D18h | 133–157 | 5.6 ± 0.1 × 104 | 9.1 ± 0.3 × 10–5 | 1.6 |
| R1h | 220–231 | 9.4 ± 0.2 × 104 | 1.6 ± 0.3 × 10–4 | 1.7 |
Fabs D13h, R1h, 28dh and D18h are chimeric versions of the original mouse Fabs, in which the constant regions of the antibody light and heavy (Fd portion) chain have been replaced with the corresponding human regions.
aPersonal communication from Z.Kanyo.
bThe binding curve of 28dh was fitted with the heterogeneous ligand model (see Materials and methods). The values of kon, koff and Kd are given for the majority population (89%). Fitting the part of the curve corresponding to the minority population (11%) resulted in kon = 9.0 ± 6.4 × 106 M–1s–1, koff = 1.7 ± 0.1 × 10–3 s–1 and Kd = 0.2 nM.
Fig. 1. A schematic representation of the linear sequence of SHaPrP23–231, depicting the known binding epitopes of antibodies E123, E149, D13, 3F4, 13A5, D18, 28d and R1.
Fig. 2. Schematic representation of the reactive surface of the two different SPR sensor chips CM5 (A) and C1 (B) used in this study. The sensor chips are constructed of a glass support covered with a 50 nm-thick gold film, then a linker layer. The linker layer is then either connected to the carboxymethylated surface (C1 sensor chip) or to a 100 nm-thick carboxylated dextran layer (CM5 sensor chip).
Time-dependent conformational changes in covalently immobilized SHaPrP29–231
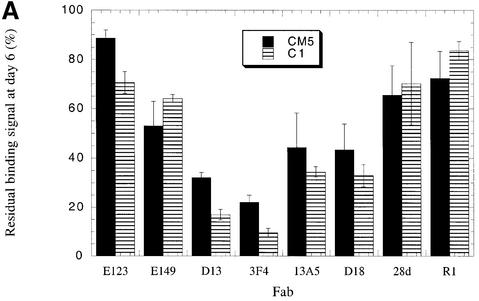
Binding measurements using the CM5 sensor chip confirmed our preliminary observation that for certain antibodies the resonance signal obtained after 6 days was dramatically lower than the value recorded immediately after antigen immobilization (Figure 3A). The greatest loss of reactivity was observed using antibodies binding epitopes lying between residues 96 and 112 of PrP. Specifically, the binding signals obtained using Fabs D13 (recognizing residues 96–104) and 3F4 (binding residues 109–112) were reduced to 32 and 22%, respectively, of those recorded immediately after immobilization of the PrP antigen. In contrast, a comparatively small reduction in binding, to 88 and 78%, respectively, of the initial values was measured for antibodies recognizing the N- and C-termini of PrP as defined by Fabs E123 (binding residues 29–37) and R1 (binding residues 220–231). Similarly, the binding of Fab 28d, which recognizes a discontinuous epitope lying within the first α-helix of PrP, diminished to 70% of the original value. However, binding of the 13A5 antibody, which recognizes sequence immediately N-terminal of helix A, was reduced to <45% of the initial level.
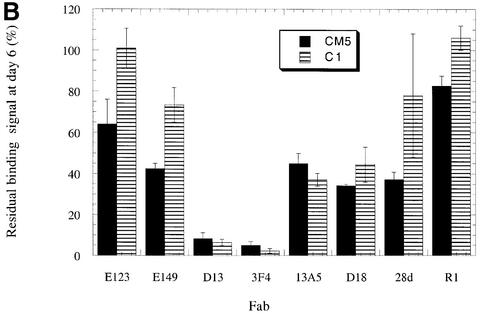
Fig. 3. Changes in antibody reactivity over time against immobilized PrP antigens. SHaPrP29–231 (A) and C-terminally biotinylated SHaPrP23–231 (B) were independently immobilized on streptavidin-coated C1 and CM5 SPR sensor chips. The reactivity of a panel of eight monoclonal antibodies was measured against freshly immobilized PrP antigen. The sensor chips were then incubated for 6 days at 37°C in HBS-P buffer, and antibody binding was re-measured. The binding signal 6 days after immobilization is given as a percentage of binding of the same antibody to freshly immobilized PrP. The data presented are the average of three independent experiments, each performed in triplicate.
When the same series of measurements was performed using the C1 sensor chip, a similar pattern of changing antibody reactivity over time was observed (Figure 3A). The binding signals obtained using Fabs D13, 3F4 and 13A5 at day 6 were reduced dramatically to 15, 10 and 27%, respectively, of the levels measured immediately after immobilization. Binding of Fabs E123, R1 and 28d, however, diminished the least over time, to 72, 62 and 71%, respectively, of the initial values. The results establish a clear disparity in the stability of epitopes located immediately N- or C-terminal to the beginning of helix A.
Equivalent conformational changes in SHaPrP23–231 immobilized via a C-terminal anchor
In its native state on the cell surface, PrPC is tethered to the cell membrane via a C-terminal GPI anchor. To replicate this arrangement more accurately in our experimental system, we constructed a hybrid SHaPrP molecule carrying a biotin group at its C-terminus. This molecule was composed of recombinant PrP containing residues 23–213, and a synthetic peptide containing residues 214–231. A lysine residue was added to the C-terminus of the synthetic peptide and a biotin moiety was covalently attached to the side chain of this amino acid via an alkyl spacer. The two recombinant and synthetic SHaPrP moieties were then linked together covalently using intein chemistry, creating a full-length SHaPrP molecule (our unpublished data). The biotinylated SHaPrP23–231 was then captured via its C-terminus to the surface of streptavidin-coated C1 and CM5 sensor chips. Each of the eight PrP-specific antibodies in our panel bound equally well to the biotinylated and recombinant PrP antigen preparations, suggesting the conformation adopted by the biotinylated molecule was equivalent to that of the recombinant PrP (data not shown).
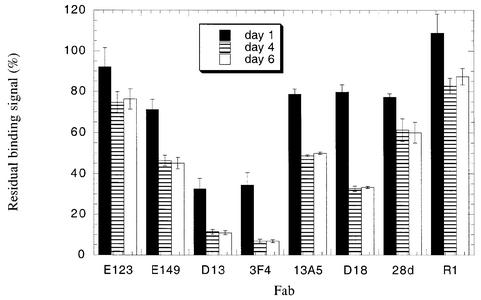
We next measured antibody reactivity against the captured SHaPrP23–231 antigen immediately after immobilization on the chip surface, and after a 1-, 4- and 6-day period. These measurements were performed using the same experimental protocols we employed for the amino-coupled recombinant PrP antigen. We found a very similar, but even more pronounced pattern of epitope loss than that observed for the amino-coupled recombinant PrP molecule (Figure 3B). Six days after immobilization on the C1 chip, only 2.3 and 6.3% of the 3F4 and D13 Fab binding sites, respectively, remained unaltered. Fab 13A5 binding was reduced to 37% of its original level. However, the binding of Fab 28d, which recognizes the first α-helix of PrP lying only six amino acids C-terminal to the 13A5 epitope, fell comparatively little, to 78% of its original value, during the same period. Moreover, antibody binding to the extreme N- and C-termini of the protein was undiminished over time.
Figure 4 illustrates a time course of conformational change in C-terminally captured PrP. Antibody reactivity was measured at 1, 4 and 6 days after antigen immobilization. The data show that after a period of only 24 h, the binding of Fabs D13 and 3F4 are reduced by ∼70%. In addition, little change in antibody reactivity is seen between days 4 and 6, indicating that the conformational rearrangement in PrP is largely complete after a 4-day period.
Fig. 4. A time course of conformational change in immobilized PrP. Antibody reactivity with SHaPrP23–231 was measured at 1, 4 and 6 days after immobilization of PrP to the sensor chip. Antibody binding is given as a percentage of the binding signal generated by the same antibody to PrP antigen immediately after immobilization. The data presented are the average of three independent experiments, each performed in triplicate.
Immobilized PrP is likely to be monomeric on the sensor chip surface
The loss of specific antibody epitopes as measured in immobilized PrP could have resulted from aggregation or oligomerization of the antigen rather than from changes in PrP conformation. To determine the likelihood of contact between individual molecules of immobilized PrP, we calculated the spatial distribution of antigen on the sensor chip surface. The maximum quantity of PrP immobilized to the sensor chip in the course of our antibody binding studies was in the order of 1000 resonance units (RU). This approximates to 1 ng, or 2.73 × 1010 molecules, of PrP per mm2 of the chip surface. Assuming an even distribution of the antigen on the sensor chip, each molecule would be located some 118 Å from its nearest neighbor, in all probability precluding the possibility of extensive association between adjacent PrP molecules. These parameters may not necessarily apply to C-terminally biotinylated PrP captured onto streptavidin-coated sensor chips, where the presence of four biotin binding sites per streptavidin molecule may allow individual PrP molecules to come within close proximity of each other. However, even in these circumstances, the relative abundance of streptavidin and PrP in our experiments dictates that there is unlikely to be more than two PrP molecules per streptavidin molecule. Moreover, since the changes in antibody binding we have recorded are seen in both covalently immobilized and C-terminally captured PrP, it is unlikely that PrP–PrP interactions on the sensor chip surface impacted significantly upon our studies.
Discussion
Conformational change in cellular PrP is a key event in the generation of prion infectivity. A detailed understanding of this process has been hampered by a lack of structural information describing the infectious form of the protein, which is highly insoluble and aggregates readily. NMR structures of recombinant PrP molecules refolded into an α-helical conformation thought to resemble PrPC reveal a highly unorthodox structure, featuring a disordered region from residues 23–128, followed by an autonomously folding C-terminal domain containing one or two short β-strands and three α-helices. Studies of recombinant PrP molecules and synthetic PrP peptides have demonstrated the ability of the protein to fold into conformations that either lack or are rich in β-arrangements. These observations hint at the conformational plasticity of some regions of PrP, and it is tempting to speculate that some portion of the disordered N-terminal half of the molecule may be capable of acquiring a greater degree of structural organization.
We have previously used PrP-specific monoclonal antibodies to compare epitope exposure in cellular and infectious PrP antigenic presentations, and have found that epitopes lying between residues 96 and 112 are available in PrPC, but lost or buried in PrP27–30 (Peretz et al., 1997). In this study, we have extended this line of investigation to examine the stability of PrPC conformation. During antibody binding experiments using recombinant PrP refolded into an α-helical conformation, we noted that with the passage of time, specific reactivity of some antibodies diminished, whereas the reactivity of other antibodies remained unchanged. To investigate this phenomenon further, we assembled a panel of eight monoclonal antibodies binding to diverse epitopes from the extreme N-terminus to the extreme C-terminus of the protein, and used SPR to track their level of reactivity over time against SHaPrP29–231 and SHaPrP23–231, immobilized on the surface of sensor chips. PrP antigens were immobilized either via covalent coupling of amino groups, or by capture onto streptavidin-coated sensor chips through a C-terminal biotin moiety.
Regardless of the mode of PrP immobilization, over time we observed a similar and reproducible pattern of changing antibody reactivity. A striking diminution in antibody binding was measured for two antibodies, D13 and 3F4, binding PrP between amino acids 96 and 112. Antibody binding and other studies had previously identified this region of PrP to be highly plastic and implicated in the conformational change associated with the acquisition of infectivity (Peretz et al., 1997; Zhang et al., 1997; Williamson et al., 1998). Comparatively little change in epitope presentation was observed for antibodies recognizing sequence either at the extreme N-terminus of the PrP molecule or C-terminal to the first α-helix. Intermediate loss of antibody binding was measured for an antibody (13A5) binding slightly N-terminal of helix A and an antibody (E149) binding in the octarepeat region of the molecule. Significantly, all of the changes in antibody reactivity were found to occur spontaneously under physiological buffer conditions in the absence of reducing or denaturing agents, and without resort to pH or buffer manipulation.
Our findings directly identify pronounced instability of specific epitopes located in regions of sequence believed to be unstructured in PrPC-like conformations. Hypo thetically, in our model system, a reduction in antibody binding may occur through a number of different mechanisms. First, aggregation of PrP molecules on the sensor chip surface could lead to some regions of the protein becoming inaccessible for antibody binding. In solution, recombinant SHaPrP29–231 refolded into an α-rich conformation and lacking β content may be converted to a more thermodynamically stable β-sheet form that is prone to aggregation and markedly less soluble. However, as described earlier, we consider it unlikely in this study that PrP molecules were immobilized on the sensor chip surface at a density sufficient to support substantive interaction between adjacent molecules. Moreover, varying the density of immobilized protein did not influence the changing pattern of antibody reactivity (data not shown).
Secondly, deamidation and racemization of asparagine 108 to isoaspartic acid has been reported to occur under some phosphate-rich solvent conditions in SHaPrP peptides containing amino acids 106–126 (Sandmeier et al., 1999). This transition could directly interfere with the binding of Fabs D13 and 3F4. To examine this possibility we performed a series of control experiments in which the binding of Fabs D13 and 3F4 were independently measured against two synthetic SHaPrP peptides composed of amino acids 96–115 and containing either Asn or isoAsp at position 108. Whereas the binding to 3F4 is abolished by the presence of the isoAsp residue, D13 binds equally well to both peptides, suggesting that even if deamidation of Asn108 occurs in a proportion of SHaPrP23–231 or SHaPrP29–231 molecules in the course of our aging experiments, this would not account for the change in binding signal observed with Fab D13. Furthermore, since the half-life of Asn108 in full-length MoPrP was measured as 33 days, it is unlikely that this conversion significantly impacted our studies, which took place over a maximum period of 6 days.
Thirdly, an apparent loss of PrP epitopes, as gauged by lower antibody binding signals, may result from a alteration in the intrinsic association and dissociation rate constants of the antibody–PrP interaction. This phenomenon may complicate the interpretation of those experiments in which maximum antibody binding (complete saturation of all available epitopes) is not realized. In these circumstances, reduced antibody binding would not necessarily reflect a loss in the number of available epitopes, but could result from diminished antibody interaction with an equal number of slightly modified epitopes. However, when the antibody binding constants to freshly immobilized and aged PrP antigen were compared, only small fluctuations were found (data not shown). These small increases and decreases in antibody affinity were factored into a binding simulation program (BIAsimulation) to determine exactly their impact on the binding signals we recorded during our experiments. In each case, differences in antibody affinity influenced our data collection only slightly, and these discrepancies were within the parameters of experimental error (see Figures 3A and B, and 4) and thus cannot account for the changing patterns of antibody reactivity we found. We therefore argue that the dynamic and localized changes in antibody binding we have observed in our studies are most readily explained by a diminution in the number of available epitopes resulting from spontaneous conformational changes in PrP.
This study identifies the 90–115 region of PrP as important in instigating conformational change in recombinant PrP molecules. Clearly, this portion of the molecule is particularly susceptible to conformational rearrangement. After an aging period of 6 days, as little as 2% of antibody binding was retained in this portion of the molecule. Indeed, after only 24 h post-immobilization, ∼70% of epitopes were lost. We infer from these data that the 90–115 region of PrP may act as a ‘nucleation’ point from which conformational change can disseminate to other parts of the protein. It is possible that strategic interventions stabilizing this portion of PrP may successfully prevent the decay of PrPC conformations.
The pattern of changing PrP conformation we describe in this report also indicates a degree of conformational heterogeneity in the rearranged protein. For example, the vast majority of immobilized PrP consistently undergoes structural transformation in the 90–115 region. Approximately 40% of these molecules retain reactivity with Fabs 13A5 and D18, indicating that the conformational rearrangement has not disseminated to the more C-terminal sequence recognized by these two antibodies. However, in the remaining 60% of the immobilized protein population, the 13A5 and D18 epitopes are lost, indicating that a more extensive structural change has occurred. The existence of distinct conformers on the sensor chip may indicate either the presence of readily distinguishable intermediates in PrP rearrangement or, alternatively, different pathways of PrP folding, some of which lead to more comprehensive structural transformation.
The role of PrP sequence between amino acids 23 and 90 in prion disease pathogenesis is uncertain. Although additional octarepeat inserts are thought to play an important role in the generation of familial Creutzfeldt–Jacob disease and Gerstmann–Sträussler– Scheinker disease (Owen et al., 1989; Goldfarb et al., 1991; Poulter et al., 1992), PrP-knockout mice overexpressing PrP with N-terminal deletions as far as, but not beyond positions 80 or 93 function normally and are capable of propagating infectious prions (Fischer et al., 1996; Shmerling et al., 1998). The conformational characteristics of amino acids 23–90 in the PrPSc conformer are unknown since structural analysis of PrPSc has been performed using the proteinase-K-resistant core of the protein, which lacks sequence N-terminal of residue 90. However, Riek et al. (1997) proposed that the N-terminal segment of PrP would adopt a β-sheet arrangement in PrPSc. We suggest from our studies that the N-terminal portion of the protein may not undergo pronounced structural change since the linear epitopes recognized by Fabs E123 and E149 are relatively well preserved during the aging process.
In summary, our studies identify intrinsic localized instability of PrP refolded into a conformation thought to closely resemble that of cell surface PrPC. We observed truly dramatic changes over time in antibody binding to a region of the molecule demonstrated to be critical in the acquisition and propagation of prion infectivity. In contrast, the conformation of other segments of the protein remained stable, as demonstrated by the relative preservation of both linear and discontinuous antibody epitopes. The distinct differences in antibody reactivity we observed between freshly immobilized and aged PrP is highly reminiscent of those differences we have described previously between PrPC and PrPSc. How closely the events we have observed relate to the conformational fate of PrPC as it is converted into PrPSc is an open question. Ongoing experiments will examine the stability of cell surface PrPC conformation. Nonetheless, our findings depict PrPC as a protein of rather pliant character. It is easy to imagine that the destiny of such a molecule could be drastically altered under the direction of a more stable and robust PrPSc template.
Materials and methods
PrP-specific antibodies
Fab fragments of monoclonal antibodies 3F4 (Kascak et al., 1987) and 13A5 (Barry and Prusiner, 1986) were prepared by papain digestion of IgG according to the manufacturer’s instructions (Pierce). Recombinant Fabs D13, D18, 28d and R1 were derived from phage libraries and have been thoroughly characterized (Williamson et al., 1996, 1998; Peretz et al., 1997). Fabs E123 and E149 were recovered from Prnp0/0 mice immunized with recombinant SHaPrP29–231, and react specifically with epitopes lying between residues 29 and 37 and 72 and 86 of SHaPrP, respectively (E.Leclerc, D.Peretz, S.B.Prusiner, D.R.Burton and R.A.Williamson, manuscript in preparation).
Surface plasmon resonance
Kinetic constants for the binding of recombinant PrP-specific Fabs were determined by SPR using the BIAcore instrument (Biacore). SHaPrP29–231 or SHaPrP23–98 (1 µg/ml) in 10 mM Na acetate pH 5.5 was immobilized to carboxymethyl groups present on CM5 and C1 sensor chips using N-hydroxysuccinimide and N-ethyl-N′-[(dimethylamino)propyl]-carbodiimidehydrochloride (Johnsson et al., 1991; Myszka, 1999). Following coupling, unreacted N-hydroxy succinimide ester groups were inactivated with 1 M ethanolamine pH 8.0 and the surface washed three times with 100 mM HCl before use. In the region of 200–500 RU were immobilized typically. The same methodology was applied to immobilize fusion protein derived from respiratory syncytial virus, which served as a reference surface. The binding of increasing concentrations of Fab (1 nM to 1 µM) was then measured in real time. Binding data was analyzed by global analysis using BiaEvaluation 3.1 software (Biacore), in which association and dissociation data for a series of Fab concentrations are fitted simultaneously (Roden and Myszka, 1996).
Measuring antibody binding to immobilized PrP over time
To study the effect of aging on PrP conformation, PrP molecules were immobilized to sensor chips using two different approaches. In the first of these, recombinant SHaPrP29–231 was immobilized on CM5 or C1 sensor chips using amine chemistry as described above (Johnsson et al., 1991). In the second approach, we created a hybrid semi-synthetic SHaPrP molecule composed of recombinant PrP (residues 23–213) coupled via intein chemistry to a synthetic peptide (residues 214–231), to which a biotinylated lysine residue was added at position 232. The resulting C-terminally biotinylated SHaPrP23–231 molecule was captured onto streptavidin-coated CM5 and C1 sensor chips. Using both these methodologies, in the region of 800–1000 RU of PrP was typically immobilized per sensor chip flow cell.
Immediately following coupling, antibody binding was measured against the immobilized PrP antigen. Fab was injected into the sensor chip at a flow rate of 50 µl/min and the increase in resonance signal above the absolute value was recorded after 180 s. After each measurement, flow cells were regenerated with 100 mM HCl. Each antibody was used at a concentration sufficient to generate 60–100% occupancy of the available sites on the sensor chip (500 nM for E123, E149 and 3F4; 400 nM for 13A5; 250 nM for D13 and 28d; 215 nM for D18; and 200 nM for R1). When the binding of all the antibodies had been measured, a 50 µl volume of HBS-P buffer (10 mM HEPES pH 7.4, 150 mM NaCl, 0.005% P20) was placed over the sensor chip, covering all the flow cells. The chip was then ‘aged’ for a period of 1, 4 or 6 days at 37°C in a high humidity environment, and binding of the anti-PrP antibody panel was evaluated as before. The binding signals measured over the course of the experiment were expressed as a percentage of the values obtained for the same antibody on day 1 of the experiment.
Acknowledgments
Acknowledgements
We are grateful to Jiri Safar, Zoltan Kanyo and Fred Cohen for their assistance with this study. This work was supported by National Institutes of Health grants HL63817, AG02132 and NS14069.
References
- Barry R. and Prusiner,S.B. (1986) Monoclonal antibodies to the cellular and scrapie prion proteins. J. Infect. Dis., 154, 518–521. [DOI] [PubMed] [Google Scholar]
- Bendheim P.E., Brown,H.R., Rudelli,R.D., Scala,L.J., Goller,N.L., Wen,G.Y., Kascsak,R.J., Cashman,N.R. and Bolton,D.C. (1992) Nearly ubiquitous tissue distribution of the scrapie agent precursor protein. Neurology, 42, 149–156. [DOI] [PubMed] [Google Scholar]
- Bessen R.A., Kocisko,D.A., Raymond,G.J., Nandan,S., Lansbury,P.T. and Caughey,B. (1995) Non-genetic propagation of strain-specific properties of scrapie prion protein. Nature, 375, 698–700. [DOI] [PubMed] [Google Scholar]
- Bolton D.C., McKinley,M.P. and Prusiner,S.B. (1982) Identification of a protein that purifies with the scrapie prion. Science, 218, 1309–1311. [DOI] [PubMed] [Google Scholar]
- Borchelt D.R., Taraboulos,A. and Prusiner,S.B. (1992) Evidence for synthesis of scrapie prion proteins in the endocytic pathway. J. Biol. Chem., 267, 16188–16199. [PubMed] [Google Scholar]
- Brown D.R., Wong,B., Hafiz,F., Clive,C., Haswell,S.J. and Jones,I.M. (1999) Normal prion protein has an activity like that of superoxide dismutase. Biochem. J., 344, 1–5. [PMC free article] [PubMed] [Google Scholar]
- Caspi S., Halimi,M., Yanai,A., Ben Sasson,S., Taraboulos,A. and Gabizon,R. (1998) The anti-prion activity of congo-red. J. Biol. Chem., 273, 3484–3489. [DOI] [PubMed] [Google Scholar]
- Caughey B. and Raymond,G.J. (1991) The scrapie-associated form of PrP is made from a cell surface precursor that is both protease- and phospholipase-sensitive. J. Biol. Chem., 266, 18217–18223. [PubMed] [Google Scholar]
- Caughey B., Raymond,G.J., Ernst,D. and Race,R.E. (1991) N-terminal truncation of the scrapie-associated form of PrP by lysosomal protease(s): implications regarding the site of conversion of PrP to the protease-resistant state. J. Virol., 65, 6597–6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donne D.G., Viles,J.H., Groth,D., Mehlhorn,I., James,T.L., Cohen,F.E., Prusiner,S.B., Wright,P.E. and Dyson,H.J. (1997) Structure of the recombinant full-length hamster prion protein PrP(29–231): the N terminus is highly flexible. Proc. Natl Acad. Sci. USA, 94, 13452–13457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer M., Rulicke,T., Raeber,A., Sailer,A., Moser,M., Oesch,B., Brandner,S., Aguzzi,A. and Weissmann,C. (1996) Prion protein (PrP) with amino-proximal deletions restoring susceptibility of PrP knockout mice to scrapie. EMBO J., 15, 1255–1264. [PMC free article] [PubMed] [Google Scholar]
- Gasset M., Baldwin,M.A., Llyod,D.H., Gabriel,J.-M., Holtzman,D.M., Cohen,F., Fletterick,R. and Prusiner,S.B. (1992) Predicted α-helical regions of the prion protein when synthesized as peptides form amyloid. Proc. Natl Acad. Sci. USA, 89, 10940–10944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfarb L.G. et al. (1991) Transmissible familial Creutzfeldt–Jakob disease associated with five, seven and eight extra octapeptide coding repeats. Proc. Natl Acad. Sci. USA, 88, 10926–10930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris D.A., Gorodinsky,A., Lehmann,S., Moulder,K. and Shyng,S.L. (1996) Cell biology of the prion protein. In Prusiner,S.B. (ed.), Prions, Prions, Prions. Springer-Verlag, Berlin, Vol. 207, pp. 77–91. [DOI] [PubMed]
- Jackson G.S. et al. (1999) Reversible conversion of monomeric human prion protein between native and fibrilogenic conformations. Science, 283, 1935–1937. [DOI] [PubMed] [Google Scholar]
- Johnsson B., Lofas,S. and Lindquist,G. (1991) Immobilization of proteins to a carboxymethyldextran-modified gold surface for biospecific interaction analysis in surface plasmon resonance sensors. Anal. Biochem., 198, 268–277. [DOI] [PubMed] [Google Scholar]
- Kaneko K. et al. (2000) A synthetic peptide initiates Gerstmann– Straussler–Scheinker (GSS) disease in transgenic mice. J. Mol. Biol., 295, 997–1007. [DOI] [PubMed] [Google Scholar]
- Kanyo Z.F., Pan,K.-M., Williamson,R.A., Burton,D.R., Prusiner,S.B., Fletterick,R.J. and Cohen,F.E. (1999) Antibody binding defines a structure for an epitope that participates in the PrPC→PrPSc conformational change. J. Mol. Biol., 293, 855–863. [DOI] [PubMed] [Google Scholar]
- Kascak R.J., Rubenstein,R., Merz,P.A., Tonna-DeMasi,M., Fersko,R., Carp,R.I., Wisnieswski,H.M. and Diringer,H. (1987) Mouse polyclonal and monoclonal antibody to scrapie-associated fibril protein. J. Virol., 61, 3688–3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann S., Milhavet,O. and Mange,A. (1999) Trafficking of the cellular isoform of the prion protein. Biomed. Pharmacother., 53, 39–46. [DOI] [PubMed] [Google Scholar]
- Lippard S.J. (1999) Free copper ions in the cell? Science, 284, 748–749. [DOI] [PubMed] [Google Scholar]
- Lopez Garcia F., Zahn,R., Riek,R. and Wuthrich,K. (2000) NMR structure of the bovine prion protein. Proc. Natl Acad. Sci USA, 97, 8334–8339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura T., Hori-i,A., Mototani,H. and Takeuchi,H. (1999) Raman spectroscopic study on the copper(II) binding mode of prion octapeptide and its pH dependence. Biochemistry, 38, 11560–11569. [DOI] [PubMed] [Google Scholar]
- Myszka D.G. (1999) Improving biosensor analysis. J. Mol. Recognit., 12, 279–284. [DOI] [PubMed] [Google Scholar]
- Oesch B. et al. (1985) A cellular gene encodes scrapie PrP 27–30 protein. Cell, 315, 331–333. [DOI] [PubMed] [Google Scholar]
- Owen F. et al. (1989) Insertion in prion protein gene in familial Creutzfeldt–Jakob disease. Lancet, 1, 51–52. [DOI] [PubMed] [Google Scholar]
- Pan K.M. et al. (1993) Conversion of α-helices into β-sheets features in the formation of the scrapie prion proteins. Proc. Natl Acad. Sci. USA, 90, 10962–10966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauly P.C. and Harris,D.A. (1998) Copper stimulates endocytosis of the prion protein. J. Biol. Chem., 273, 33107–33110. [DOI] [PubMed] [Google Scholar]
- Peretz D. et al. (1997) A conformational transition at the N terminus of the prion protein features in formation of the scrapie isoform. J. Mol. Biol., 273, 614–622. [DOI] [PubMed] [Google Scholar]
- Poulter M. et al. (1992) Inherited prion disease with 144 base pair gene insertion.1. Genealogical and molecular studies. Brain, 115, 675–685. [DOI] [PubMed] [Google Scholar]
- Priola S.A., Caughey,B., Raymond,G.J. and Chesebro,B. (1994) Prion protein and the scrapie agent: in vitro studies in infected neuroblastoma cells. Infect. Agents Dis., 3, 54–58. [PubMed] [Google Scholar]
- Prusiner S.B. (1991) Molecular biology of prions diseases. Science, 252, 1515–1522. [DOI] [PubMed] [Google Scholar]
- Prusiner S.B. (1994) Inherited prion diseases. Proc. Natl Acad. Sci. USA, 91, 4611–4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusiner S.B. (1997) Prion diseases and the BSE crisis. Science, 278, 245–251. [DOI] [PubMed] [Google Scholar]
- Prusiner S.B., Bolton,D.C., Groth,D.F., Bowman,K.A., Cochran,S.P. and McKinley,M.P. (1982) Further purification and characterization of scrapie prions. Biochemistry, 21, 6942–6950. [DOI] [PubMed] [Google Scholar]
- Prusiner S.B., Scott,M.R., DeArmond,S.J. and Cohen,F.E. (1998) Prion protein biology. Cell, 93, 337–348. [DOI] [PubMed] [Google Scholar]
- Riek R., Hornemann,S., Wider,G., Billeter,M., Glockshuber,R. and Wüthrich,K. (1996) NMR structure of the mouse prion domain PrP(121–231). Nature, 382, 180–182. [DOI] [PubMed] [Google Scholar]
- Riek R., Hornemann,S., Wider,G., Glockshuber,R. and Wüthrich,K. (1997) NMR characterization of the full-length recombinant murine prion protein, mPrP(23–231). FEBS Lett., 413, 282–288. [DOI] [PubMed] [Google Scholar]
- Roden L.D. and Myszka,D.G. (1996) Global analysis of a macromolecular interaction measured on BIAcore. Biochem. Biophys. Res. Commun., 225, 1073–1077. [DOI] [PubMed] [Google Scholar]
- Safar J., Wille,H., Itri,V., Groth,D., Serban,H., Torchia,M., Cohen,F.E. and Prusiner,S.B. (1998) Eight prion strains have PrPSc molecules with different conformations. Nature Med., 4, 1157–1165. [DOI] [PubMed] [Google Scholar]
- Salmona M. et al. (1999) Molecular determinants of the physicochemical properties of a critical prion protein region comprising residues 106–126. Biochem. J., 342, 207–214. [PMC free article] [PubMed] [Google Scholar]
- Sandmeier E., Hunziker,P., Kunz,B., Sack,R. and Christen,P. (1999) Spontaneous deamidation and isomerization of Asn 108 in prion peptide 106–126 and in full-length prion protein. Biochem. Biophys. Res. Commun., 261, 578–583. [DOI] [PubMed] [Google Scholar]
- Scott M.R., Groth,D., Tatzelt,J., Torchia,M., Tremblay,P., DeArmond,S.J. and Prusiner,S.B. (1997) Propagation of prion strains through specific conformers of the prion protein. J. Virol., 71, 9032–9044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmerling D. et al. (1998) Expression of amino terminally truncated PrP in the mouse leading to ataxia and specific cerebellar lesions. Cell, 93, 203–214. [DOI] [PubMed] [Google Scholar]
- Shyng S.L., Huber,M.T. and Harris,D.A. (1993) A prion protein cycles between the cell surface and an endocytic compartment in cultured neuroblastoma cells. J. Biol. Chem., 268, 15922–15928. [PubMed] [Google Scholar]
- Shyng S.L., Heuser,J.E. and Harris,D.A. (1994) A glycolipid-anchored prion protein is endocytosed via clathrin-coated pits. J. Cell Biol., 125, 1239–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl N., Borchelt,D.R., Hsiao,K. and Prusiner,S.B. (1987) Scrapie prion protein contains a phosphatidylinositol glycolipid. Cell, 51, 229–240. [DOI] [PubMed] [Google Scholar]
- Stahl N., Borchelt,D.R. and Prusiner,S.B. (1990) Differential release of cellular and scrapie prion proteins from cellular membranes by phosphatidylinositol-specific phospholipase C. Biochemistry, 29, 5405–5412. [DOI] [PubMed] [Google Scholar]
- Stockel J., Safar,J., Wallace,A.C., Cohen,F.E. and Prusiner,S.B. (1998) Prion protein selectively binds copper (II) ions. Biochemistry, 37, 7185–7193. [DOI] [PubMed] [Google Scholar]
- Telling G.C. et al. (1996) Evidence for the conformation of the pathologic isoform of the prion protein enciphering and propagating prion diversity. Science, 274, 2079–2082. [DOI] [PubMed] [Google Scholar]
- Viles J.H., Cohen,E.F., Prusiner,S.B., Goodin,D.B., Wright,P.E. and Dyson,H.J. (1999) Copper binding to the prion protein: structural implications of four identical cooperative binding sites. Proc. Natl Acad. Sci. USA, 96, 2042–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadsworth J.D.F., Hill,A.F., Joiner,S., Jackson,G.S. and Collinge,J. (1999) Strain-specific prion-protein conformation determined by metal ions. Nature Cell Biol., 1, 55–59. [DOI] [PubMed] [Google Scholar]
- Waggoner D.J., Drisaldi,B., Bartnikas,T.B., Casareno,R.L.B., Prohaska,J.R., Gitlin,J.D. and Harris,D.A. (2000) Brain copper content and cuproenzyme activity do not vary with prion expression level. J. Biol. Chem., 275, 7455–7458. [DOI] [PubMed] [Google Scholar]
- Weissmann C. (1999) Molecular genetics of transmissible spongiform encephalopathies. J. Biol. Chem., 274, 3–6. [DOI] [PubMed] [Google Scholar]
- Weissmann C., Fischer,M., Raeber,A., Bueler,H., Sailer,A., Shmerling,D., Rulicke,T., Brandner,S. and Aguzzi,A. (1996) The role of PrP in pathogenesis of experimental scrapie. Cold Spring Harbor Symp. Quant. Biol., 61, 511–522. [PubMed] [Google Scholar]
- Williamson R.A., Peretz,D., Smorodinsky,N., Bastidas,R., Serban,H., Mehlhorn,I., DeArmond,S.J., Prusiner,S.B. and Burton,D.R. (1996) Circumventing tolerance to generate autologous monoclonal antibodies to the prion protein. Proc. Natl Acad. Sci. USA, 93, 7279–7282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson R.A., Peretz,D., Pinilla,C., Ball,H., Bastidas,R.B., Rozenshteyn,R., Houghten,R.A., Prusiner,S.B. and Burton,D.R. (1998) Mapping the prion protein using recombinant antibodies. J. Virol., 72, 9413–9418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn R. et al. (2000) NMR solution structure of the human prion protein. Proc. Natl Acad. Sci. USA, 97, 145–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Kaneko,K., Nguyen,J.T., Livshits,T.L.B., Baldwin,M.A., Cohen,F.E., James,T.L. and Prusiner,S.B. (1995) Conformational transitions in peptides containing two putative α-helices of the prion protein. J. Mol. Biol., 250, 514–526. [DOI] [PubMed] [Google Scholar]
- Zhang H., Stockel,J., Mehlhorn,I., Groth,D., Baldwin,M.A., Prusiner,S.B., James,T.L. and Cohen,F.E. (1997) Physical studies of conformational plasticity in a recombinant prion protein. Biochemistry, 36, 3543–3553. [DOI] [PubMed] [Google Scholar]