Abstract
Isopentenyl diphosphate:dimethylallyl diphosphate (IPP:DMAPP) isomerase catalyses a crucial activation step in the isoprenoid biosynthesis pathway. This enzyme is responsible for the isomerization of the carbon–carbon double bond of IPP to create the potent electrophile DMAPP. DMAPP then alkylates other molecules, including IPP, to initiate the extraordinary variety of isoprenoid compounds found in nature. The crystal structures of free and metal-bound Escherichia coli IPP isomerase reveal critical active site features underlying its catalytic mechanism. The enzyme requires one Mn2+ or Mg2+ ion to fold in its active conformation, forming a distorted octahedral metal coordination site composed of three histidines and two glutamates and located in the active site. Two critical residues, C67 and E116, face each other within the active site, close to the metal-binding site. The structures are compatible with a mechanism in which the cysteine initiates the reaction by protonating the carbon–carbon double bond, with the antarafacial rearrangement ultimately achieved by one of the glutamates involved in the metal coordination sphere. W161 may stabilize the highly reactive carbocation generated during the reaction through quadrupole– charge interaction.
Keywords: dimethylallyl/isomerase/isopentenyl/isoprenoids/metalloproteins
Introduction
Isoprenoids comprise a family of >23 000 natural products, among them the precursors of cholesterol and taxol (Kellogg and Poulter, 1997; Sacchettini and Poulter, 1997; Edwards and Ericsson, 1999). Isoprenoids are found in all organisms (Spurgeon and Porter, 1981). There are several classes of essential molecules in this family, including sterols (Popjak, 1970), carotenoids (Goodwin, 1971), dolichols (Matsuoka et al., 1991), ubiquinones (Ashby et al., 1990) and prenylated proteins (Clarke, 1992). These extensive families of isoprenoid compounds are synthesized from two precursors: isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP). Isopentenyl diphosphate isomerase (IPP isomerase; EC 5.3.3.2) is a key enzyme for generating isoprenoid diversity and is responsible for the isomerization of the carbon–carbon double bond of IPP to create the potent electrophile DMAPP (Agranoff et al., 1960). DMAPP then alkylates other molecules, including IPP, ultimately forming the numerous isoprenoid compounds found in nature. In Eukarya and Archaea, in which the pathway is well established, IPP is synthesized from three molecules of acetyl coenzyme A by the classical mevalonate pathway (Popjak, 1970). However, in some bacteria, IPP is synthesized by a non-classical route that is not yet fully characterized (Rohmer et al., 1993). Beyond IPP, the pathway for constructing polyisoprenoid chains is similar in all three major kingdoms, and is initiated by IPP isomerase.
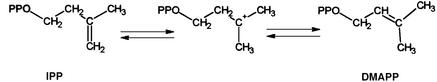
Many studies have been devoted to understanding the catalytic mechanism of IPP isomerase. IPP isomerase catalyses the interconversion of IPP and DMAPP by a stereoselective antarafacial [1.3] transposition of hydrogen (Figure 1) (Cornforth et al., 1966; Cornforth and Popjak, 1969; Poulter and Rilling, 1981). Several lines of evidence point towards a protonation–deprotonation mechanism involving an active site with two residues located on opposite faces of the allyl moiety in IPP to assist with addition and elimination of protons, with the formation of a carbonium ion during catalysis. These include proton-exchange measurements (Street et al., 1990), decreased reactivities for fluorinated analogues of IPP (Muehlbacher and Poulter, 1988) and DMAPP (Reardon and Abeles, 1986), potent-noncovalent inhibition by ammonium analogues of the putative carbocationic intermediate (Muehlbacher and Poulter, 1985, 1988; Reardon and Abeles, 1986) and irreversible inhibition by mechanism-based inhibitors containing epoxide moieties (Muehlbacher and Poulter, 1988; Lu et al., 1992).
Fig. 1. Interconversion of IPP into DMAPP by an antarafacial [1.3] transposition of hydrogen, as catalysed by IPP isomerase.
Using radioactive 3-(fluoromethyl)-3-butenyl diphosphate, an affinity label for IPP, a cysteine in position 67 (Escherichia coli numbering) was identified as a potential important amino acid and was found to be covalently modified by this substrate analogue. Subsequent site-directed mutagenesis confirmed its requirement for catalysis (Street et al., 1990). Similarly, C67 was also found to be covalently modified by the inhibitor 3-methyl-3,4-epoxybutyl diphosphate (Lu et al., 1992). Although the antarafacial stereochemistry of the isomerization suggested that IPP isomerase has two nucleophilic groups in the active site, efforts to locate a second nucleophile have been unsuccessful. These included extensive studies with active-site-directed irreversible inhibitors containing an epoxide moiety (Street et al., 1990, 1994). However, other studies with a variant of IPP isomerase where the crucial cysteine was replaced by a serine revealed that E116 was required for catalytic activity and was also modified covalently by 3-(fluoromethyl)-3-butenyl diphosphate (Street et al., 1994). Presumably, substitution of the thiol moiety at position 67 by a hydroxyl reduced the nucleophilicity of the side-chain nucleophile and allowed the E116 carboxylate to compete for the electrophilic inhibitor.
It has long been known that the enzyme requires Mg2+ or a mixture of Mn2+ and Mg2+ to show catalytic activity (Agranoff et al., 1960), and it was anticipated that these cations would participate in binding of the pyrophosphate moiety of the substrate, similarly to many enzymes acting on substrates comprising phosphate groups. However, the exact role of these cations in IPP isomerase catalytic activity has never been elucidated. Despite these numerous biochemical studies, there are no active site structures for this enzyme, and how catalysis is achieved is still a mystery.
In addition to the crucial role of IPP isomerase in the isoprenoid pathway, the catalytic mechanism of this enzyme is also of great interest, as enzymatic protonation of unactivated olefins is rarely encountered in nature. Prominent examples are Δ8–Δ7 cholesterol isomerase (Wilton et al., 1969) and enzymes catalysing proton-initiated cyclization of isoprenoids (Sacchettini and Poulter, 1997). Structural data for some cyclization reactions have been reported only fairly recently (Lesburg et al., 1997; Starks et al., 1997; Wendt et al., 1997, 1999).
Here we present the structures of IPP isomerase alone or complexed with Mn2+ or Mg2+. A key feature of the structure is a metal-binding site located within the active site, which plays a structural and catalytic role. This metal-binding site has a distorted octahedral geometry and is formed by three histidines and two glutamates. These structural analyses support a catalytic mechanism in which a cysteine initiates the reaction by protonating the carbon–carbon double bond, with the antarafacial rearrangement ultimately achieved by one of the glutamates involved in the metal coordination sphere.
Results and discussion
Overall structure
The structure of IPP isomerase alone was determined by multi-wavelength anomalous diffraction (MAD) of a seleno-methionine (SeMet) derivative and refined to 1.45 Å with an R-factor (Rfree) of 0.192 (0.212). The asymmetric unit of the P3221 crystal contains a single molecule. The structures of IPP isomerase (variant C67A) in the presence of Mn2+ or Mg2+ were subsequently determined by the molecular replacement method starting from the SeMet isomerase coordinates. These isomerase complexes crystallize, under similar solution conditions, in space group P212121, with two molecules per asymmetric unit. The Mn2+ and Mg2+ complexes have been refined to R-factors (Rfree) of 0.213 (0.249) and 0.234 (0.269) at resolutions of 2.1 and 2.7 Å, respectively.
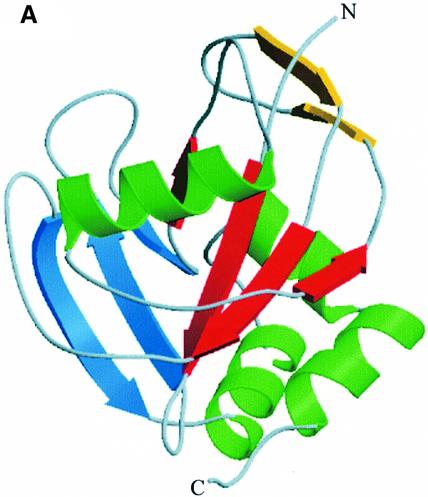
IPP isomerase is composed of 182 amino acids and folds into a compact globular protein that belongs to the class of α/β proteins, with a complex architecture. The secondary structures of IPP isomerase are illustrated in Figure 2. In the absence of metal ions, three β-sheets are identified in the structure, surrounded by four α-helices. The main β-sheet is composed of four strands (S3, S6, S7 and S10) with mixed polarities, and contains the cysteine presumably involved in the catalytic mechanism. This cysteine is located in strand S6 of the central β-sheet, and faces helix H1. The two other β-sheets are composed of three and two antiparallel strands (strands S4, S5, S11 and S8, S9, respectively) and surround the main β-sheet. The first 31 amino acids are not visible in the electron density map of the metal-free IPP isomerase at 1.45 Å resolution, revealing their great mobility. Many residues in the 103–116 region also have poorly defined electron densities, and most of these side chains are disordered. We dissolved a SeMet crystal and subjected the resulting protein to mass spectrometry and chemical sequencing analyses (results not shown). These experiments indicated the presence, in the crystallized protein, of the first N-terminal amino acids, confirming that they are highly disordered in the crystal structure.
Fig. 2. View of the overall structure of IPP isomerase by ribbon representation of secondary structure elements. (A) Metal-free structure. (B) Metal-bound structure, with two additional β-strands at the N-terminus. The location of the metal ion is shown (M). The sequence delimitation of secondary structure elements is as follows: H1, N76–L88; H2, L144–A153; H3, P160–T167; H4, R169–A177; S1, H5–L9; S2, P15–E20; S3, F35–F40; S4, L46–R51; S5, V61–N64; S6, V66–G68; S7, E96–Y99; S8, R103–T107; S9, V113–V117; S10, V120–R124; S11, V136–C142. S1 and S2 are only present in the metal-bound structures.
Metal-binding site and active site geometry
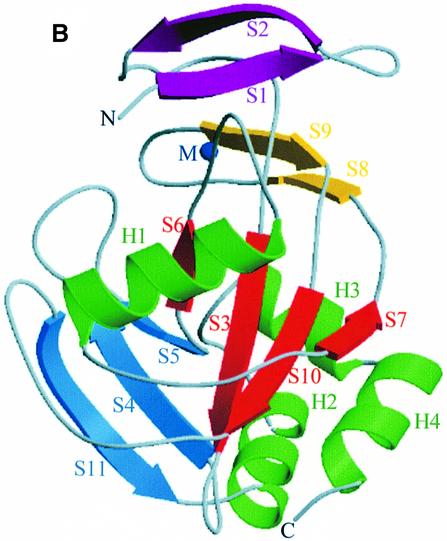
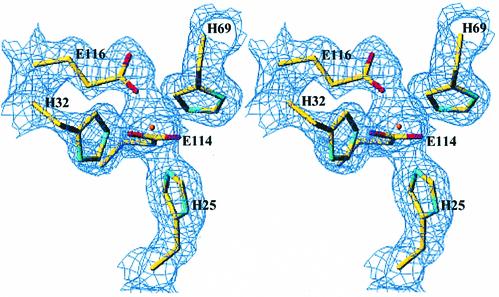
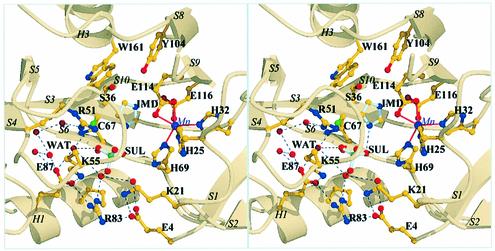
Upon metal binding, the 31 N-terminal residues fold into a small additional β-sheet of two antiparallel strands (Figure 2), partly covering a pocket comprising the putative catalytic cysteine residue, whereas residues 103–116 become well defined. These conformational changes create a distorted octahedral metal coordination site, composed of residues H25, H32, H69, E114 and E116 (Figure 3). Of these residues, only H69 is well defined in the electron density of the metal-free enzyme. Both carboxylate oxygens of E114 participate in the metal coordination. No water molecules were observed in the metal coordination sphere. These structural modifications create a deep cavity predominantly formed at its base by Y104, S36 and W161, by the metal-binding site on one side and by C67, R51 and K55 on the other side. One carboxylate oxygen of E116 is still available to form a hydrogen bond, as observed in the Mn2+ and Mg2+ complexes. In the Mg2+ complex, a water molecule is hydrogen bonded to E116. In the Mn2+ complex, we identified an imidazole molecule hydrogen bonded to E116 and stabilized within the active site by π interactions with the indole ring of W161. Although the presence of an imidazole molecule in the C67A variant is not relevant physiologically (imidazole has no effect on the activity of the wild-type enzyme), it defines a buried cavity close to the metal-binding site. The imidazole molecule was also hydrogen bonded to a well defined sulfate ion, found in the Mn2+ and Mg2+ complexes, within the active site (Figure 4).
Fig. 3. Stereo view of the metal-binding site in the Mn2+–IPP isomerase complex, refined at a resolution of 2.1 Å. The electron density map is contoured at the 1.2σ level. H25, H32, H69, E114 and E116 form a distorted octahedral coordination sphere, located in the active site.
Fig. 4. Stereo view illustrating the location, within the active site, of the sulfate ion (SUL) and the imidazole molecule (IMD). The Mn2+ ion (in purple) and its coordination site composed of H25, H32, H69 and E114 and E116 are shown, as well as S36, Y104 and W161 at the bottom of the active site. K21, K55 and R51 interact with the sulfate ion. Buried water molecules are located between an ionic cluster composed of K55, E87, R83, E4 and K21 and the catalytic C67. The water molecule hydrogen bonded to E87 is indicated (WAT).
Such observations reflect not only the availability for interactions of a carboxylate oxygen of E116 and of the indole π electrons of W161, but also the presence of a binding pocket in this protein area. The carboxylate oxygen of E116 faces the thiol group of C67, which is ∼7 Å away. Importantly, E116 and C67 are the two amino acids identified from covalent modifications of IPP isomerase by 3-(fluoromethyl)-3-butenyl diphosphate, an active-site-directed irreversible inhibitor (Street et al., 1990, 1994). The enzyme is more active when complexed with Mn2+ (Vmax = 5.87 µmol/h/mg) or, to a lesser extent, Mg2+ (Vmax = 3.97 µmol/h/mg). The binding of a transition metal to a protein with high affinity and specificity requires discrimination of the size, charge and chemical nature (Christianson and Cox, 1999). Although both metals have equivalent amino acid coordination, they may affect differently the geometry of the binding site and the precise orientation of catalytic groups such as E116 within the active site, which may explain in part the observed differences in activities. To our knowledge, such a metal coordination site for Mn2+ (and Mg2+) has never been reported. The most similar metal-binding site is the one found in manganese superoxide dismutase (Borgstahl et al., 1992), in which the Mn2+ ion is coordinated by three histidines, one carboxylate oxygen of an aspartate, and an oxygen from a water molecule, directly involved in catalysis. The coordination geometry of this cluster is trigonal bipyramidal.
Mechanistic insights into IPP isomerization
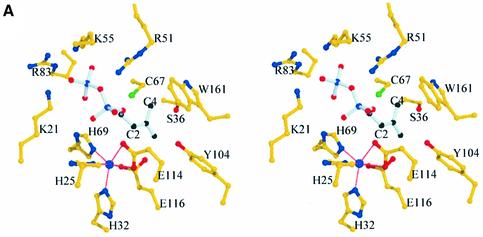
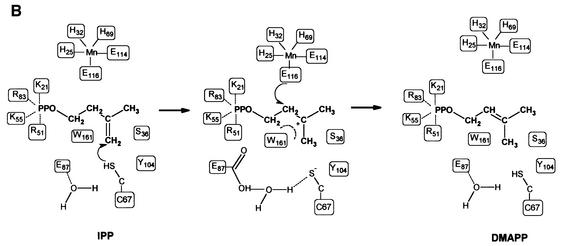
The complete active site of IPP isomerase can be mapped from the metal-bound structures and has been investigated by site-directed mutagenesis (Table I). There is a sulfate ion in the active site of the Mn2+- and Mg2+-bound isomerases, 6 Å away from the metal ion. This sulfate ion, which is stabilized by various interactions involving R51, K55 and K21 (Figure 4), may mimic the binding of the distal phosphate moiety of the substrate, helping to fit IPP within the active site. The sulfate ion does not participate in any interaction with Mn2+ or Mg2+, suggesting no direct involvement of the metal in phosphate binding. Together with biochemical studies devoted to this enzyme, the active site geometry supports a model where the allyl moiety of IPP fits in a deeply buried cavity with E116 and C67 lying on opposite faces of the substrate within the active site, and closed by W161, Y104 and S36. This cavity is occupied by the imidazole molecule in the Mn2+-bound IPP isomerase C67A variant. The presence of such a buried cavity in the active site is required to protect the highly reactive carbocation intermediate, which must be shielded from solvent. C67 may be the residue that initiates the attack on the carbon–carbon double bond of IPP, leading to the formation of an intermediate carbocation. Final deprotonation of the carbocation to form DMAPP could be accomplished via the free carboxylate oxygen of E116, for which the negative charge is stabilized via its coordination to the metal ion. The presence of such a negatively charged amino acid at this position is required for catalysis, as confirmed by the E116Q mutant, which shows no significant residual activity (Table I). C67 and E116 are the only candidates available within the active site to accomplish the protonation/deprotonation steps. A cluster of buried water molecules may help to reprotonate C67 after formation of the carbocation intermediate. This cluster is located in a region well conserved between the metal-free and -bound structures, and composed of R51, K55, R83 and E87 covering the catalytic cysteine. In the metal-free structure, E135 and E87 form salt bridges with K55 and R83, respectively. In the metal-bound structures, these residues are involved in an extended ionic network formed by K55, E87, R83, E4 and K21. These last two residues belong to the folded N-terminal regions in the metal-bound structures (Figure 4). R51, K55, E83 and K21 may interact with the phosphate moiety of IPP (Figure 5). H69 (which coordinates the metal ion) may also interact with the diphosphate group. Importantly, E87, which belongs to helix H1 (Figure 2), faces the catalytic cysteine in the opposite direction of E116. A water molecule is hydrogen bonded to E87, between this residue and C67, ∼3–3.5 Å away from the thiol group. This water molecule may be required to initiate the reprotonation of C67 after the carbocation formation. The relative importance of R51, K55, E87 and R83, which are structurally well conserved between the metal-free and -bound structures and located in the neighbourhood of the catalytic cysteine, was investigated by site-directed mutagenesis. Replacing E87 with Q completely abolished enzyme activity, providing evidence for a crucial role of the carboxylate group of E87 in the catalytic mechanism (Table I). The R51K mutant still displayed some activity, although at a low level (Table I), which indicates the importance of a positive charge at that position, but also the strict requirement for an arginine for catalytic efficiency.
Table I. Kinetic parameters for IPP isomerase mutants.
| IPP isomerase | Km (µM) | Vmax (µmol/h/mg) | Vmax/Km | Relative activity (%) |
|---|---|---|---|---|
| Wild type | 3.5 ± 0.2 | 2.1 ± 0.05 | 0.6 | 100 |
| R51K | 18.5 ± 2.0 | 0.09 ± 0.007 | 0.005 | 4.2 |
| K55R | 15.0 ± 3.6 | 0.58 ± 0.1 | 0.04 | 28 |
| K55A | 14.0 ± 1.9 | 1.386 ± 0.04 | 0.099 | 66 |
| C67A | – | – | – | 0 |
| R83K | 3.0 ± 0.5 | 2.2 ± 0.15 | 0.93 | 104 |
| E87Q | 11.5 ± 0.1 | 0.0014 ± 1 × 10–5 | 1.22 × 10–4 | 0.07 |
| E116Q | 21.0 ± 0.5 | 0.0018 ± 2 × 10–5 | 8.81 × 10–4 | 0.09 |
| W161F | 0.8 ± 0.75 | 0.017 ± 0.01 | 0.021 | 0.8 |
Fig. 5. (A) Stereo view of IPP represented within the active site. (B) Possible contacts between IPP and the Mn2+-bound enzyme, and the proposed mechanism of IPP isomerization. C67 may protonate the C4 carbon of the allyl moiety of IPP, forming a carbocation stabilized by the indole π electrons of W161. E116 is positioned to accomplish the deprotonation at carbon C2, forming DMAPP.
In our modelling study, W161 is optimally positioned deep in the active site, possibly to stabilize, through quadrupole–charge interactions, the highly reactive carbocation intermediate formed upon the nucleophilic attack of C67. This role is further supported by the W161F mutant, which shows no significant residual activity (Table I). These results suggest that the presence of a tryptophan is not only required for steric constraints at the border of the active site, but may also reflect the direct participation of the π electrons of the indole ring in the carbocation stabilization. In our model, the metal ion serves to structure the N-terminal part of the enzyme, resulting in the formation of the complete metal coordination sphere, which closes the active site to create the allyl moiety binding pocket and the diphosphate-binding sites, orienting and stabilizing E116 to accomplish the deprotonation of the carbocation intermediate (Figure 5). Biochemical data support a mechanism where the C67 is protonated, leading to an available SH group to protonate the C4 carbon of IPP. Reardon and Abeles (1986) presented evidence that IPP isomerase was inactivated by the thiol-selective reagent iodoacetamide and that the rate of inactivation depended on a single ionizable group with a pK of 9.3, which correlates with the enzyme being inactivated at pH values of >9 and suggests the presence of a cysteine in a protonated form.
In order to provide a complete mechanistic analysis, we tried to position the substrate within the active site in such a way that E116 would initiate the reaction by protonating the C4 carbon of IPP, forming a carbocation finally deprotonated by C67. Such a model could only be constructed by displacing the proximal phosphate to the observed sulfate ion position, which implies structural rearrangements in the ionic network formed by K55, E87, R83, E4 and K21 to position the distal phosphate group of IPP. In terms of shape and charge, the sulfate position is more likely to represent the distal phosphate-binding site than the proximal one. The binding geometry of the allyl group would be seriously distorted in view of the putative attack of the C4 carbon of IPP by E116. Furthermore, in such a model, there is no amino acid available to stabilize the highly reactive carbocation, an improbable situation. Finally, it should be noted that the C67S mutant still displays some activity, although at very low level (Street et al., 1994). This observation suggests that the reduced activity of the C67S mutant could result from the serine hydroxyl being less acidic than the cysteine thiol (Street et al., 1994), favouring a model where C67 is protonated.
Biological implications
The reaction catalysed by IPP isomerase involves protonation of an unactivated carbon–carbon double bond, a reaction rarely encountered in nature. Only a very few other examples of such reactions have been described at the molecular level. The structures of three cyclases found at multiple branch points in the isoprenoid pathway have been reported (Lesburg et al., 1997; Starks et al., 1997; Wendt et al., 1997, 1999). These cyclization reactions are electrophilic alkylations in which a new carbon–carbon single bond is formed by attaching a highly reactive electron-deficient carbocation to an electron-rich carbon– carbon double bond. Only one of these enzymes initiates the cyclization via the protonation of a carbon–carbon double bond; the other cyclases initiate the cyclization through the attack of the carbon–oxygen bond in the allylic diphosphate ester of the substrate. These cyclases display no structural homology to IPP isomerase and are all composed of a structural motif consisting of 10–12 mostly antiparallel α-helices that form a large catalytic cavity. The enzymes that use diphosphate-containing substrates (pentalenene synthase, epi-aristolochene synthase) also require Mg2+ for catalytic activity. They contain DDXXD sequence motifs that bind the Mg2+ ions stabilizing the diphosphate groups of the substrates. Here, the catalytic residues do not interact directly with the metal ions. Thus, the comparison of IPP isomerase with these cyclases illustrates a novel mechanism of protonation/deprotonation of the carbon–carbon bond.
The IPP isomerase structures reveal the involvement of a divalent cation in folding into the active conformation, and indicate how the initial protonation and final deprotonation of IPP occur and how the transient carbocation is stabilized. The positioning of the substrate within the active site supports a catalytic mechanism in which a cysteine initiates the formation of the transient carbocation, stabilized through quadrupole–charge interaction with the indole π electrons of W161 and deprotonated via E116, for which the carboxylate is stabilized and oriented via its coordination to the metal.
IPP isomerases have been studied in several bacteria, archaebacteria and eukaryotes, including fungi, mammals and plants (Ramos-Valdivia et al., 1997a). IPP isomerases from human, rat, hamster, Saccharomyces cerevisiae, Schizosaccharomyces pombe, Arabidopsis thaliana, Camptotheca acuminata, Clarkia breweri, Clarkia xantiana, Mycobacterium tuberculosis, Rhodobacter capsulatus, Rhodobacter sphaeroides and Streptomyces coelicolor are the best characterized enzymes (Anderson et al., 1989; Blanc and Pichersky, 1995; Hahn and Poulter, 1995; Hahn et al., 1996a,b; Campbell et al., 1997; Paton et al., 1997; Cole et al., 1998). They vary greatly in sequence length, as a result of addition of amino acids at their N-terminus. With 182 amino acids, E.coli IPP isomerase is one of the shortest sequences, and may be viewed as the basic catalytic core IPP isomerase required to perform IPP isomerization. Residues that are important for metal coordination and active site are present in all isomerase sequences, as illustrated on a structure-based sequence alignment (see Supplementary data, available at The EMBO Journal Online). For A.thaliana IPP isomerase, extensions at the N-terminus have been demonstrated not to be essential for enzymatic activity (Campbell et al., 1997). Studies with the rat and hamster enzymes have revealed the presence of the enzyme in peroxisomes (Paton et al., 1997), and some signals that target these enzymes have been identified, located in their N-terminal extensions. Escherichia coli IPP isomerase seems to represent the minimal structural core required to provide the unusual mechanism of action of IPP isomerization in its widespread distribution in nature.
Materials and methods
Cloning and in vitro mutagenesis
The idi gene encoding the E.coli IPP isomerase was previously amplified by PCR and cloned into the pET30b vector (pYL20 plasmid; Oudjama et al., 2001). The resulting protein, identified by DNA sequencing as IPP isomerase carrying a C-terminal His tag (LEHHHHHH), was overexpressed in E.coli BL21 (DE3)pLysS cells (Novagen). Mutations were introduced with the QuikChange site-directed mutagenesis kit (Stratagene). The pYL20 plasmid was used as a template in all experiments.
Protein purification
Cells expressing wild-type and mutant IPP isomerases were grown to an OD600 of 0.6 at 37°C in rich medium [1% Bacto-tryptone (Difco), 0.5% yeast extract (Difco), 0.1% glucose, 0.5% NaCl, 0.03% KH2PO4, 0.07% K2HPO4] supplemented with 30 µg/ml kanamycin. Expression of the recombinant enzymes was induced for 3 h at 37°C using 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). SeMet IPP isomerase was produced using the method described by Doublié (1997). Expression of the recombinant enzyme was induced overnight at 15°C using 1 mM IPTG. The wild-type, SeMet and mutant IPP isomerases were purified by nickel affinity chromatography as described previously (Oudjama et al., 2001). Cells from a 1 l culture were resuspended in 50 mM Tris–HCl pH 7.4 and disrupted by sonication for 20 min in a Raytheon sonic oscillator (250 W, 10 kHz). Insoluble components were removed by centrifugation at 15 000 g for 10 min. The supernatant fluid was applied to a nickel chelating Sepharose column (Amersham Pharmacia) pre-equilibrated with 50 mM Tris–HCl pH 7.4. The bound protein was eluted using a 100 ml linear gradient of 0–0.5 M imidazole in 50 mM Tris–HCl pH 7.4. The fractions containing active wild-type and mutant IPP isomerases were pooled and dialysed against 50 mM Tris–HCl pH 7.4. The SeMet enzyme was dialysed against a buffer composed of 50 mM Tris–HCl pH 7.4, 0.2 mM EDTA and 3 mM β-mercaptoethanol. The yields of the wild-type and SeMet enzymes were 224 and 70 mg/l of culture, respectively.
IPP isomerase activity assay and kinetic properties
Kinetic properties of the purified wild-type IPP isomerase carrying a C-terminal His tag were determined as described by Hahn et al. (1999) for the untagged wild-type enzyme. The apparent Km of IPP was 10 ± 0.1 µM. The maximal enzyme activity was reached by increasing metal concentrations, up to 2 and 0.2 mM for Mg2+ and Mn2+, respectively, with apparent affinity constants for Mg2+ and Mn2+ of 130 and 14 µM. No inhibition was observed with higher metal concentrations. The isomerase activity of mutant enzymes was measured as described in Ramos-Valdivia et al. (1997b).
Structure determination
SeMet IPP isomerase. Crystals of SeMet IPP isomerase (space group P3221; a = b = 71.4 Å, c = 61.8Å) were obtained as described by Oudjama et al. (2001). Crystals were cryoprotected by adding 25% glycerol to the crystallization solution, and were measured at 100K. MAD data were collected at beamline BM14 at the European Synchrotron Radiation Facility (ESRF) at the selenium absorption edge and peak (resolution 1.7 Å) and a remote wavelength was collected at beamline ID14 (resolution 1.45 Å) on the same crystal. Oscillation data were processed with the HKL suite (Otwinowski and Minor, 1997). The positions of the three selenium atoms in the asymmetric unit were determined with SOLVE (Terwilliger and Berendzen, 1999). MAD phasing in the resolution range 30–2.0 Å was performed with MLPHARE (CCP4, 1994), using the data sets collected at the selenium absorption edge and peak. Phase improvement by means of density modification was performed using DM (CCP4, 1994). Model building was accomplished using WarpNtrace (Perrakis et al., 1999) at a resolution of 1.7 Å. The structure was refined using CNS (Brunger et al., 1998) against the data set at 1.45 Å resolution using cross-validated maximum likelihood as the target function (Adams et al., 1997). The structure was inspected using TURBO-FRODO (Roussel and Cambillau, 1992). Residues 1–30 were not present in the electron density map. Residues 103–116 displayed weak electron densities for many side chains.
C67A isomerase complexes. Crystals for the C67A isomerase in the presence of 10 mM Mn2+ or Mg2+ were obtained under the conditions used to crystallize the SeMet protein, with only slight adjustments of precipitant concentrations. We used the inactive C67A variant of IPP isomerase in order to try to co-crystallize it with its substrate IPP, as no commercial inhibitor is available. We succeeded in obtaining crystals in the presence of the metals, but trials to obtain suitable crystals with a metal and the substrate have failed. The C67A variant could have a low affinity towards IPP, which prevents it from binding within the active site in the crystalline state.
Metal-bound isomerase crystals appeared within 4 days in 10–15% (w/v) polyethylene glycol (PEG) 2000 monomethylether, 10 mM MnCl2 or MgCl2, 100 mM Tris–maleate buffer pH 5.5 in the presence of 100 mM ammonium sulfate. Data sets were collected with a Mar345 imaging plate system from Marresearch equipped with Osmic optics (Osmic Inc.) and running on an FR591 rotating anode generator. The crystals belong to space group P212121, with cell parameters a = 69.3 Å, b = 72.6 Å, c = 92.5 Å. Diffraction data were processed with the MarFLM suite. The structures were determined by the molecular replacement method with the program AMoRe (Navaza, 1994), starting from the SeMet isomerase coordinates. Refinements were performed using CNS at resolutions of 2.1 and 2.7 Å for the Mn2+ and Mg2+ complexes, respectively. The N-terminal residues were readily identifiable, as were the metal ions, and were built with TURBO-FRODO. The 103–116 regions were well defined in the metal-bound structures. One metal ion was identified in each molecule of IPP isomerase, either Mn2+ or Mg2+. Both structures present a well defined sulfate ion in the active site. In the case of the Mn2+ complex, an imidazole molecule was also identified in the electron density map, interacting with the sulfate ion in a buried cavity close to the metal-binding site. Data collection statistics are given in Table II. The geometries of the structures were checked with PROCHECK (Laskowski et al., 1993). Ribbon representations of the molecules were drawn using Molscript (Kraulis, 1991) and Raster3D (Merritt and Bacon, 1997). Secondary structure definitions were accomplished using DSSP (Kabsch and Sander, 1983). The atomic coordinates and structure-factor amplitudes have been submitted to the Protein Data Bank under accession code 1HZT and 1HX3.
Table II. Data collection statistics.
| Mn2+ complex | Mg2+ complex | Se-Met λ1 | λ2 | λ3 | |
|---|---|---|---|---|---|
| Beamline | rotating anode | rotating anode | BM14 | BM14 | ID14-EH2 |
| Resolution (Å) | 2.1 | 2.7 | 1.7 | 1.7 | 1.45 |
| Wavelength (Å) | 1.5418 | 1.5418 | 0.97858 | 0.97873 | 0.93260 |
| Measurements | 118 332 | 36 432 | 231 134 | 233 816 | 278 438 |
| Unique reflections | 26 234 | 13 683 | 38 216 | 38 330 | 31 964 |
| <Redundancy> | 4.5 | 2.7 | 6.0 | 6.2 | 8.7 |
| Completenessa (%) | 94.7 (92.9) | 95.2 (99.7) | 98.6 (87.7) | 98.7 (86.5) | 98.1 (99.8) |
| Rmergea (%) | 8.7 (27.5) | 7.8 (26.9) | 3.1 (12.6) | 2.8 (12.1) | 6.9 (27.2) |
| Space group | P212121 | P212121 | P3221 | P3221 | P3221 |
aValues for the last resolution shell.
Supplementary data
Supplementary data for this paper are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
The authors thank R.Wattiez (Université de Mons-Hainaut) for the mass spectrometry analyses, and S.Wakatsuki, S.McSweeney, E.Mitchell and G.Leonard (ESRF, Grenoble) for their help and advice during data acquisition. L.D. is a research associate of the Fonds National de la Recherche Scientifique. B.C. is an undergraduate student of an Action de Recherche Concertée from the Université Libre de Bruxelles (ULB). This work was supported in part by a grant from the Fonds pour la Recherche Fondamentale Collective, by a grant from the Institut Interdisciplinaire des Sciences Nucléaires, by grants from the ULB (ARC–FER) and a grant from the Banque Nationale de Belgique.
References
- Adams P.D., Pannu,N.S., Read,R.J. and Brunger,A.T. (1997) Cross-validated maximum likelihood enhances crystallographic simulated annealing refinement. Proc. Natl Acad. Sci. USA, 94, 5018–5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agranoff B.W., Eggerer,H., Henning,U. and Lynen,F. (1960) Biosynthesis of terpenes. VII. Isopentenyl pyrophosphate isomerase. J. Biol. Chem., 235, 326–332. [PubMed] [Google Scholar]
- Anderson M.S., Muehlbacher,M., Street,I.P., Proffitt,J. and Poulter,C.D. (1989) Isopentenyl diphosphate:dimethylallyl diphosphate isomerase. An improved purification of the enzyme and isolation of the gene from Saccharomyces cerevisiae. J. Biol. Chem., 264, 19169–19175. [PubMed] [Google Scholar]
- Ashby M.N. and Edwards,P.A. (1990) Elucidation of the deficiency in two yeast coenzyme Q mutants: characterization of the structural gene encoding hexaprenyl pyrophosphate synthetase. J. Biol. Chem., 265, 13157–13164. [PubMed] [Google Scholar]
- Blanc V.M. and Pichersky,E. (1995) Nucleotide sequence of a Clarkia breweri cDNA clone of Ipi1, a gene encoding isopentenyl pyrophosphate isomerase. Plant Physiol., 108, 855–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgstahl G.E., Parge,H.E., Hickey,M.J., Beyer,W.F.,Jr, Hallewell,R.A. and Tainer,J.A. (1992) The structure of human mitochondrial manganese superoxide dismutase reveals a novel tetrameric interface of two 4-helix bundles. Cell, 71, 107–118. [DOI] [PubMed] [Google Scholar]
- Brunger A.T. et al. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Campbell M., Hahn,F.M., Poulter,C.D. and Leustek,T. (1997) Analysis of the isopentenyl diphosphate isomerase gene family from Arabidopsis thaliana. Plant Mol. Biol., 36, 323–328. [DOI] [PubMed] [Google Scholar]
- CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Christianson D.W. and Cox,D.J. (1999) Catalysis by metal-activated hydroxide in zinc and manganese metalloenzymes. Annu. Rev. Biochem., 68, 33–57. [DOI] [PubMed] [Google Scholar]
- Clarke S. (1992) Protein isoprenylation and methylation at carboxy-terminal cysteine residues. Annu. Rev. Biochem., 61, 355–386. [DOI] [PubMed] [Google Scholar]
- Cole S.T. et al. (1998) Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature, 393, 537–544. [DOI] [PubMed] [Google Scholar]
- Cornforth J.W. and Popjak,G. (1969) Chemical syntheses of substrates of sterol biosynthesis. Methods Enzymol., 15, 359–371. [Google Scholar]
- Cornforth J.W., Cornforth,R.H., Popjak,G. and Yengoyan,L. (1966) Studies on the biosynthesis of cholesterol. XX. Steric course of decarboxylation of 5-pyrophosphomevalonate and of the carbon to carbon bond formation in the biosynthesis of farnesyl pyrophosphate. J. Biol. Chem., 241, 277–281. [PubMed] [Google Scholar]
- Doublié S. (1997) Preparation of selenomethionyl proteins for phase determination. Methods Enzymol., 276, 523–530. [PubMed] [Google Scholar]
- Edwards P.A. and Ericsson,J. (1999) Sterols and isoprenoids: signalling molecules derived from the cholesterol biosynthetic pathway. Annu. Rev. Biochem., 68, 157–185. [DOI] [PubMed] [Google Scholar]
- Goodwin T.W. (1971) Biosynthesis of carotenoids and plant triterpenes: the fifth CIBA Medal lecture. Biochem. J., 123, 293–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn F.M. and Poulter,C.D. (1995) Isolation of Schizosaccharomyces pombe isopentenyl diphosphate isomerase cDNA clones by complementation and synthesis of the enzyme in Escherichia coli. J. Biol. Chem., 270, 11298–11303. [DOI] [PubMed] [Google Scholar]
- Hahn F.M., Xuan,J.W., Chambers,A.F. and Poulter,C.D. (1996a) Human isopentenyl diphosphate:dimethylallyl diphosphate isomerase overproduction, purification and characterization. Arch. Biochem. Biophys., 332, 30–34. [DOI] [PubMed] [Google Scholar]
- Hahn F.M., Baker,J.A. and Poulter,C.D. (1996b) Open reading frame 176 in the photosynthesis gene cluster of Rhodobacter capsulatus encodes idi, a gene for isopentenyl diphosphate isomerase. J. Bacteriol., 178, 619–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn F.M., Hurlburt,A.P. and Poulter,C.D. (1999) Escherichia coli open reading frame 696 is idi, a nonessential gene encoding isopentenyl diphosphate isomerase. J. Bacteriol., 181, 4499–4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. and Sander,C. (1983) Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers, 22, 2577–2637. [DOI] [PubMed] [Google Scholar]
- Kellogg B.A. and Poulter,C.D. (1997) Chain elongation in the isoprenoid biosynthetic pathway. Curr. Opin. Chem. Biol., 1, 570–578. [DOI] [PubMed] [Google Scholar]
- Kraulis P.J. (1991) MOLSCRIPT—a program to produce detailed and schematic plots of protein structures. J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- Laskowski R.A., MacArthur,M.W., Moss,D.S. and Thornton,J.M. (1993) PROCHECK: a program to check the stereochemical quality of protein structure. J. Appl. Crystallogr., 26, 283–291. [Google Scholar]
- Lesburg C.A., Zhai,G., Cane,D.E. and Christianson,D.W. (1997) Crystal structure of pentalenene synthase: mechanistic insights on terpenoid cyclization reactions in biology. Science, 277, 1820–1824. [DOI] [PubMed] [Google Scholar]
- Lu X.J., Christensen,D.J. and Poulter,C.D. (1992) Isopentenyl diphosphate isomerase: irreversible inhibition by 3-methyl-3,4-epoxybutyl diphosphate. Biochemistry, 31, 9955–9960. [DOI] [PubMed] [Google Scholar]
- Matsuoka S., Sagami,H., Kurisaki,A. and Ogura,K. (1991) Variable product specificity of microsomal dehydrodolichyl diphosphate synthase from rat liver. J. Biol. Chem., 266, 3464–3468. [PubMed] [Google Scholar]
- Merritt E.A. and Bacon,D.J. (1997) Raster3D: photorealistic molecular graphics. Methods Enzymol., 277, 505–524. [DOI] [PubMed] [Google Scholar]
- Muehlbacher M. and Poulter,D.C. (1985) Isopentenyl pyrophosphate: dimethylallylpyrophosphate isomerase. Irreversible inhibition of the enzyme by active-site directed covalent attachment. J. Am. Chem. Soc., 107, 8307–8308. [Google Scholar]
- Muehlbacher M. and Poulter,D.C. (1988) Isopentenyl diphosphate isomerase: inactivation of the enzyme with active-site-directed irreversible inhibitors and transition-state analogues. Biochemistry, 27, 7315–7328. [DOI] [PubMed] [Google Scholar]
- Navaza J. (1994) AMoRe: an automated package for molecular replacement. Acta Crystallogr. A, 50, 157–163. [Google Scholar]
- Otwinowski Z. and Minor,W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Oudjama Y., Durbecq,V., Sainz,G., Clantin,B., Tricot,C., Stalon,V., Villeret,V. and Droogmans,L. (2001) Preliminary structural studies of Escherichia coli isopentenyl diphosphate isomerase. Acta Crystallogr., 57, 287–288. [DOI] [PubMed] [Google Scholar]
- Paton V.G., Shackelford,J.E. and Krisans,S.K. (1997) Cloning and subcellular localization of hamster and rat isopentenyl diphosphate dimethylallyl diphosphate isomerase. A PTS1 motif targets the enzyme to peroxisomes. J. Biol. Chem., 272, 18945–18950. [DOI] [PubMed] [Google Scholar]
- Perrakis A., Morris,R. and Lamzin,V.S. (1999) Automated protein model building combined with iterative structure refinement. Nature Struct. Biol., 6, 458–463. [DOI] [PubMed] [Google Scholar]
- Popjak G. (1970) Natural substances formed biologically from mevalonic acid. In Goodwin,T.W. (ed.), Biochemical Symposium No. 29. Academic Press, New York, pp. 17–37.
- Poulter C.D. and Rilling,H.C. (1981) Prenyl transferases and isomerase. In Porter,J.W. and Spurgeon,S.L. (eds), Biosynthesis of Isoprenoid Compounds. Vol. 1. John Wiley and Sons, New York, NY, pp. 162–209.
- Ramos-Valdivia A.C., van der Heijden,R. and Veerpoorte,R. (1997a) Isopentenyl diphosphate isomerase: a core enzyme in isoprenoid biosynthesis. A review of its biochemistry and function. Nature Prod. Rep., 14, 591–603. [DOI] [PubMed] [Google Scholar]
- Ramos-Valdivia A.C., van der Heijden,R., Verpoorte,R. and Camara,B. (1997b) Purification and characterization of two isoforms of isopentenyl diphosphate isomerase from elicitor-treated Cinchona robusta cells. Eur. J. Biochem., 249, 161–170. [DOI] [PubMed] [Google Scholar]
- Reardon J.E. and Abeles,R.H. (1986) Mechanism of action of isopentenyl pyrophosphate isomerase: evidence for a carbonium ion intermediate. Biochemistry, 25, 5609–5616. [DOI] [PubMed] [Google Scholar]
- Rohmer M., Knani,M., Simonin,P., Sutter,B. and Sahm,H. (1993) Isoprenoid biosynthesis in bacteria: a novel pathway for the early steps leading to isopentenyl diphosphate. Biochem. J., 295, 517–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussel A. and Cambillau,C. (1992) TURBO-FRODO. Biographics, AFMB, Marseille, France.
- Sacchettini J.C. and Poulter,C.D. (1997) Creating isoprenoid diversity. Science, 277, 1788–1789. [DOI] [PubMed] [Google Scholar]
- Spurgeon S.L. and Porter,J.W. (eds) (1981) Biosynthesis of Isoprenoid Compounds. Vol. 1. John Wiley and Sons, New York, NY, pp. 1–46.
- Starks C.M., Back,K., Chappell,J. and Noel,J.P. (1997) Structural basis for cyclic terpene biosynthesis by tobacco 5-epi-aristolochene synthase. Science, 277, 1815–1820. [DOI] [PubMed] [Google Scholar]
- Street I.P., Christensen,D.J. and Poulter,C.D. (1990) Hydrogen exchange during the enzyme-catalyzed isomerization of isopentenyl diphosphate and dimethylallyl diphosphate. J. Am. Chem. Soc., 112, 8577–8578. [Google Scholar]
- Street I.P., Coffman,H.R., Baker,J.A. and Poulter,C.D. (1994) Identification of Cys139 and Glu207 as catalytically important groups in the active site of isopentenyl diphosphate:dimethylallyl diphosphate isomerase. Biochemistry, 33, 4212–4217. [DOI] [PubMed] [Google Scholar]
- Terwilliger T.C. and Berendzen,J. (1999) Automated MAD and MIR structure solution. Acta Crystallogr. D, 55, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt K.U., Poralla,K. and Schulz,G.E. (1997) Structure and function of a squalene cyclase. Science, 277, 1811–1815. [DOI] [PubMed] [Google Scholar]
- Wendt K.U., Lenhart,A. and Schultz,G.E. (1999) The structure of the membrane protein squalene-hopene cyclase at 2.0 Å resolution. J. Mol. Biol., 286, 175–187. [DOI] [PubMed] [Google Scholar]
- Wilton D.C., Rahimtula,A.D. and Akhtar,M. (1969) The reversibility of the Δ8-cholesterol–Δ7-cholesterol isomerase reaction in cholesterol biosynthesis. Biochem. J., 114, 71–73. [DOI] [PMC free article] [PubMed] [Google Scholar]