Abstract
During apoptosis, release of cytochrome c initiates dATP-dependent oligomerization of Apaf-1 and formation of the apoptosome. In a cell-free system, we have addressed the order in which apical and effector caspases, caspases-9 and -3, respectively, are recruited to, activated and retained within the apoptosome. We propose a multi-step process, whereby catalytically active processed or unprocessed caspase-9 initially binds the Apaf-1 apoptosome in cytochrome c/dATP-activated lysates and consequently recruits caspase-3 via an interaction between the active site cysteine (C287) in caspase-9 and a critical aspartate (D175) in caspase-3. We demonstrate that XIAP, an inhibitor-of-apoptosis protein, is normally present in high molecular weight complexes in unactivated cell lysates, but directly interacts with the apoptosome in cytochrome c/dATP-activated lysates. XIAP associates with oligomerized Apaf-1 and/or processed caspase-9 and influences the activation of caspase-3, but also binds activated caspase-3 produced within the apoptosome and sequesters it within the complex. Thus, XIAP may regulate cell death by inhibiting the activation of caspase-3 within the apoptosome and by preventing release of active caspase-3 from the complex.
Keywords: Apaf-1/apoptosome/caspases/XIAP
Introduction
Apoptosis is a distinct form of cell death characterized by nuclear and cytoplasmic condensation, DNA fragmentation and externalization of membrane-associated phosphatidylserine. A class of cysteine proteases, known as caspases, produce these biochemical and morphological changes by selectively cleaving a number of structural and regulatory proteins at specific aspartate residues. In a general paradigm, either receptor- or stress-induced death signals stimulate oligomerization of specific adaptor molecules, such as FADD or Apaf-1, which subsequently recruit and promote trans-activation of ‘initiator’ caspases, such as caspases-8 and -9, respectively. Initiator caspases possess long prodomains that enable them to interact with death effector domains (DEDs) or caspase-activation recruitment domains (CARDs) present in these adaptor proteins. Once activated, initiator caspases are free to activate ‘effector’ caspases, such as caspases-3 and -7, which contain short prodomains and are primarily responsible for dismantling the cell during the execution phase of apoptosis (for reviews see Cohen, 1997; Earnshaw et al., 1999; Bratton et al., 2000).
Cellular stress can stimulate the release of cytochrome c from the mitochondrial intermembrane space into the cytosol, where it interacts with the adaptor protein Apaf-1 (Green and Reed, 1998). Apaf-1 contains at least three functional domains: (i) an N-terminal CARD, which binds the prodomain of caspase-9; (ii) a CED-4 domain required for Apaf-1 self-oligomerization; and (iii) a series of C-terminal WD-40 repeats thought to mediate protein– protein interactions (Zou et al., 1997). Apaf-1, when bound to cytochrome c, apparently hydrolyzes dATP/ATP and undergoes oligomerization via its CED-4 domains (Hu et al., 1998, 1999; Srinivasula et al., 1998). Simultaneously, the CARD domain recruits and facilitates processing of procaspase-9 (Li et al., 1997; Srinivasula et al., 1998; Qin et al., 1999). This complex of cytochrome c, Apaf-1 and caspase-9 is commonly referred to as the apoptosome. Reconstitution experiments using purified recombinant proteins indicate that the apoptosome is ∼1.4 MDa in size (Saleh et al., 1999; Zou et al., 1999), whereas in native cell lysates, Apaf-1 oligomerizes into an ∼700 kDa complex and, in addition to processed caspase-9, contains fully processed caspases-3 and -7 (p17 and p12 subunits) (Cain et al., 1999). Thus, the initial processing of effector caspases by caspase-9 and their subsequent autocatalytic processing appears to take place within the apoptosome. The specific mechanisms that govern these processes remain unclear.
Caspases are inhibited by a number of viral and mammalian proteins, including inhibitor-of-apoptosis proteins (IAPs). These evolutionarily conserved proteins were first identified in baculoviruses and contain both an N-terminal tandem repeat of ∼70 amino acids, termed baculovirus IAP repeat (BIR) domain, and a C-terminal RING zinc-finger domain (Clem and Miller, 1994). Several IAP homologs containing one or both domains have been identified in mammalian cells, including X-linked IAP (XIAP or hILP), ML-IAP (livin), cellular inhibitor-of-apoptosis protein-1 (cIAP-1/hIAP-2), cIAP-2 (hIAP-1), neuronal apoptotic inhibitory protein (NAIP) and survivin (for review see Deveraux and Reed, 1999; Vucic et al., 2000). The anti-apoptotic mechanisms of some of these proteins are unknown; however, XIAP, ML-IAP, cIAP-1 and cIAP-2 appear to inhibit both stress- and death receptor-induced apoptosis through direct inhibition of distinct caspases (Deveraux et al., 1997, 1998; Vucic et al., 2000). In particular, XIAP inhibits Apaf-1-mediated activation of procaspase-9 and the activity of processed caspase-9, but not FADD-mediated activation of procaspase-8 or the activity of processed caspase-8. Perhaps more importantly, XIAP potently inhibits active caspases-3 and -7 in vitro. Thus, it is the ability of XIAP to inhibit active caspases-3 and -7 that suppresses death receptor-induced apoptosis, whereas its ability to inhibit both the apical and effector caspases protects against stress-induced apoptosis (Deveraux et al., 1997, 1998). Interestingly, XIAP inhibits active caspases-3 and -9 through distinct domains within the protein. The BIR2 domain, together with a few critical residues in the linker region between the BIR1 and BIR2 domains, is sufficient for inhibition of caspase-3, whereas the BIR3 domain inhibits caspase-9 (Takahashi et al., 1998; Sun et al., 1999, 2000).
In this study, we used a cell-free model to define the requirements for the recruitment, processing and retention of caspases within the apoptosome. We propose a multi-step process, whereby catalytically active processed or unprocessed caspase-9 is required to recruit caspase-3 to the apoptosome via an interaction between the catalytically active cysteine (C287) in caspase-9 and a critical aspartate (D175) in caspase-3 required for its processing. Caspase-3 is subsequently processed within the apoptosome to its fully mature form and remains largely associated with the complex. In addition to binding active caspases-9 and -3, XIAP also associates with oligomerized Apaf-1. Thus, by associating with the apoptosome, XIAP appears not only to influence the activation of caspase-3 by caspase-9, but also to inhibit the release of active caspase-3 from the complex.
Results and discussion
The Apaf-1 apoptosome must contain caspase-9 in order to recruit caspase-3 to the complex
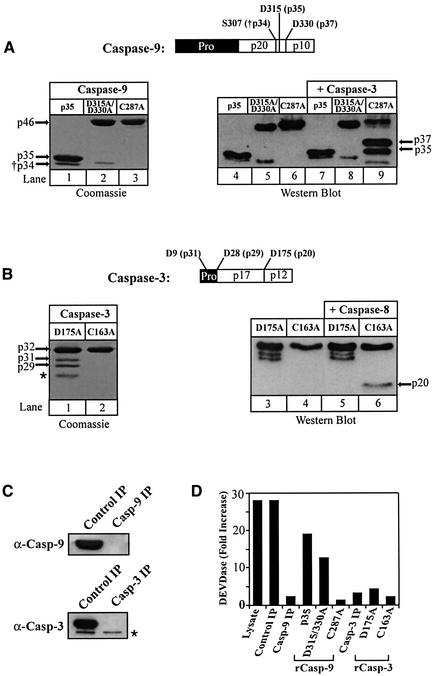
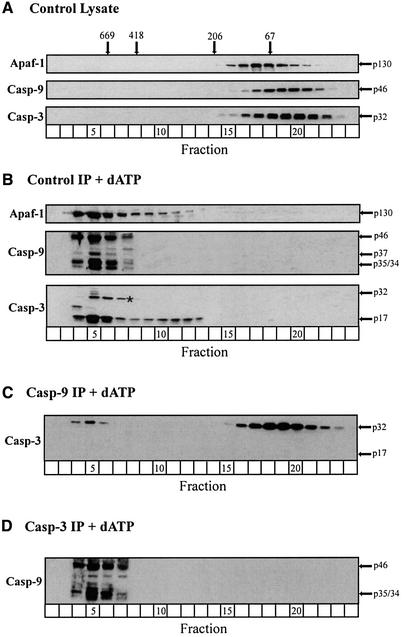
Several genetic studies using ‘knockout’ mice and in vitro studies using cytochrome c/dATP have determined the apparent order of caspase activation following mitochondrial stress (Hakem et al., 1998; Kuida et al., 1998; Woo et al., 1998; Slee et al., 1999). However, none have examined the precise order in which caspases are recruited to the apoptosome, nor the molecular determinants required for this process. By systematically immunodepleting specific caspases from the lysate (Figure 1C) and replacing them with known caspase mutants (Figure 1A and B), we wished to determine if and how the presence of a given caspase within the apoptosome might influence the recruitment/presence of other caspases. Using gel filtration chromatography we initially characterized the formation of the apoptosome in control and caspase-depleted lysates (Figure 2). In unactivated control lysates, Apaf-1 eluted primarily in its monomeric form (fractions 15–21, Figure 2A), whereas procaspases-9 and -3 eluted as homodimers (fractions 16–22, Figure 2A). Following dATP activation, Apaf-1 oligomerized into ∼700–1400 kDa apoptosome complexes, containing both unprocessed and processed caspase-9, as well as fully processed caspase-3 (fractions 4–7, Figure 2B). A second complex of active caspase-3, which we formerly termed the ‘micro-apoptosome’, was also observed (fractions 9–12, Figure 2B). These results were essentially the same as previously reported (Cain et al., 1999).
Fig. 1. Reconstitution of immunodepleted lysates with recombinant wild-type and mutant caspases-9 and -3. (A) Recombinant wild-type and fully processed caspase-9 (lane 1), non-cleavable unprocessed D315/330A caspase-9 (lane 2) and catalytically inactive C287A caspase-9 (lane 3) were isolated and purified from bacteria and their purity was determined by Coomassie Blue staining. Each caspase-9 protein was incubated with (lanes 7–9) or without (lanes 4–6) recombinant active caspase-3 (100 nM) for 1 h at 37°C. The samples were subsequently separated by SDS–PAGE and immunoblotted using a polyclonal anti-caspase-9 antibody. (B) The non-cleavable D175A caspase-3 (lanes 1, 3 and 5) and catalytically inactive C163A caspase-3 (lanes 2, 4 and 6) mutants were purified from bacteria, Coomassie-stained to assess purity and exposed to purified recombinant caspase-8 (100 nM) for 1 h at 37°C. In the D175A caspase-3 preparation, two minor internal translation products of ∼27 and ∼29 kDa were present (Fernandes-Alnemri et al., 1996), as well as a minor bacterial contaminant (*). (C) THP.1 lysates were immunodepleted of caspase-9 or -3 and (D) reconstituted with their corresponding wild-type or mutant caspases (200 nM). The reconstituted lysates were dATP-activated for 1 h at 37°C and assayed for DEVDase activity as described in Materials and methods. The anti-caspase-3 antibody recognized a non-specific band (*) in lysates, which served as a fortuitous loading control.
Fig. 2. The Apaf-1 apoptosome sequentially recruits, activates and retains caspases-9 and -3. (A) Unactivated control lysates, (B) dATP-activated immunoprecipitated (IP) control lysates, (C) dATP-activated caspase-9-depleted lysates or (D) dATP-activated caspase-3-depleted lysates were fractionated by gel filtration, as described in Materials and methods. Each fraction was mixed with 10× SDS loading buffer, separated by SDS–PAGE and immunoblotted for Apaf-1, caspase-9 and/or caspase-3. The arrows represent either the unprocessed or processed form(s) of the caspases; a non-specific band (*) was detected by the anti-caspase-3 antibody.
In order to determine whether the absence of caspase-9 had any effect on the recruitment of caspase-3 to the apoptosome, we immunodepleted lysates of procaspase-9 (Figure 1C) and activated these lysates with dATP. Apaf-1 oligomerized into an apoptosome complex (data not shown), but caspase-3 was not readily recruited to the complex and was completely unprocessed (fractions 4–7 and 16–22, compare Figure 2B and C) and inactive (Figure 1D). These data strongly suggest that Apaf-1 could oligomerize into a complex in the absence of procaspase-9, but that caspase-9 must be present to both recruit and activate caspase-3 (fractions 4–7, compare Figure 2B and C). The inability of the Apaf-1 complex to recruit caspase-3, in the absence of caspase-9, was not a consequence of inappropriate oligomerization of Apaf-1, as this complex produced significant DEVDase activity when incubated with a mixture of partially purified pro-caspases-9 and -3 obtained from THP.1 cell lysates (data not shown). We next immunodepleted lysates of procaspase-3 (Figure 1C) and activated these lysates with dATP. Removal of caspase-3 had no effect on Apaf-1 oligomerization (data not shown) or the recruitment of caspase-9 to the apoptosome (fractions 4–7, Figure 2D). However, since no active caspase-3 was present (Figure 1D), there was a slight alteration in the processing of caspase-9, in that less of the p37 form of caspase-9 was observed in the apoptosome (fractions 4–7, Figure 2D). Thus, Apaf-1 can oligomerize into a functional complex independent of caspase-9, but normally functions sequentially to recruit and activate caspases-9 and -3.
Catalytically active processed or unprocessed caspase-9 is required for recruitment of caspase-3 to the apoptosome
Since the presence of caspase-9 in the apoptosome was critical for the recruitment of caspase-3, we asked several questions: (i) does caspase-9 form a binding site through which caspase-3 must interact? (ii) does caspase-9 require processing to effectively recruit caspase-3? and (iii) does caspase-9 have to be catalytically active in order to recruit caspase-3? To address these questions, we replaced lysates depleted of endogenous procaspase-9 with wild-type caspase-9, previously processed to its p35 form in bacteria (p35 casp-9), non-cleavable procaspase-9 (D315/330A casp-9) or catalytically inactive procaspase-9 (C287A casp-9) (Figure 1A) and activated these lysates with dATP. In agreement with Stennicke et al. (1999), we found that processing of caspase-9 was not required for its activity, as D315/330A casp-9 was nearly as effective as p35 casp-9 at reconstituting DEVDase activity (Figure 1D). Moreover, the catalytically inactive C287A casp-9 mutant was completely incapable of supporting effector caspase activation (Figure 1D).
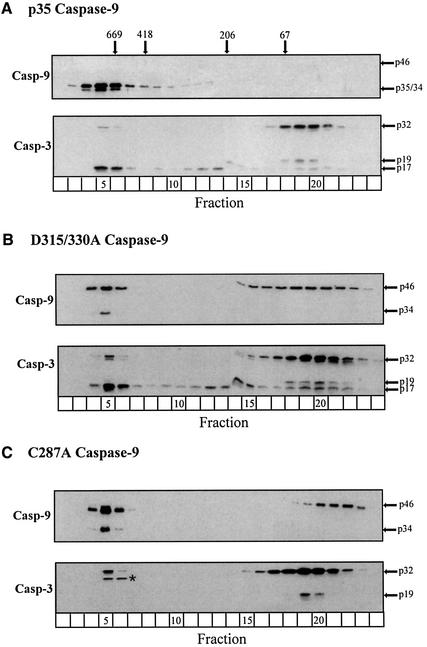
When the lysates, reconstituted with wild-type or mutant caspase-9 proteins, were examined by gel filtration, Apaf-1 was always present as an oligomerized apoptosome complex (data not shown). In the p35 casp-9 reconstitution experiments, caspase-9 readily bound to oligomerized Apaf-1 (fractions 4–7, Figure 3A), indicating that oligomerized Apaf-1 was capable of recruiting and utilizing previously processed caspase-9. The apoptosome containing recombinant p35 casp-9 also recruited and activated endogenous caspase-3, which remained largely associated with the complex (fractions 5–6, Figure 3A). Although the concentrations of p35 casp-9 used in these experiments probably exceeded the normal endogenous concentrations of caspase-9, this reconstituted system did not completely activate all of the available procaspase-3 compared with control lysates (compare Figures 3A and 2B). Therefore, potential modulators of caspase-9 activity may have been removed during immunodepletion of endogenous procaspase-9; alternatively, p35 casp-9 may not fully mimic endogenous active caspase-9. Nevertheless, the ability of p35 casp-9 to associate with Apaf-1 in the apoptosome and to recruit and process caspase-3 verified the usefulness of this in vitro model (Figure 3).
Fig. 3. Recruitment of caspase-3 to the Apaf-1 apoptosome requires catalytically active processed or unprocessed caspase-9. Caspase-9-depleted lysates were reconstituted with (A) fully processed wild-type p35 caspase-9, (B) non-cleavable D315/330A caspase-9 or (C) catalytically inactive C287A caspase-9. Following dATP activation for 1 h at 37°C, the reconstituted lysates were fractionated and immunoblotted for caspases-9 and -3, as described in Materials and methods. The arrows represent either the unprocessed or processed form(s) of the caspases; a non-specific band (*) was detected by the anti-caspase-3 antibody.
Next, we examined lysates reconstituted with the non-cleavable D315/330A casp-9 mutant. Although unprocessed caspase-9 does support DEVDase activity in dATP lysates (Stennicke et al., 1999; Figure 1D), the involvement of the apoptosome in this process has not been formally demonstrated. Indeed, we found that D315/330A casp-9 associated normally with oligomerized Apaf-1 following dATP activation, and readily recruited and activated caspase-3 (fractions 4–7, compare Figures 3B and 2B). Moreover, the active caspase-3 remained primarily associated with the apoptosome. Therefore, caspase-9 need not be processed in order to recruit and activate caspase-3 within the apoptosome.
Finally, we examined lysates reconstituted with the catalytically inactive C287A casp-9 mutant. Although this mutant could not support effector caspase activation (Figure 1D), we questioned whether C287A casp-9 was unable to recruit caspase-3 to the apoptosome or was simply unable to activate it. Following dATP activation, C287A casp-9 bound to oligomerized Apaf-1 but was incapable of effectively recruiting procaspase-3 to the apoptosome (fractions 4–6, Figure 3C). The fact that C287A casp-9 could not undergo normal autocatalytic processing to form a fully processed caspase-9 enzyme was not responsible for its lack of ability to recruit caspase-3. Indeed, both D315/330A casp-9 and C287A casp-9 are largely present within the apoptosome as unprocessed caspase-9 enzymes (fractions 4–6, Figure 3B and C), but only D315/330A casp-9 is capable of recruiting caspase-3 to the apoptosome. Thus, the catalytically active cysteine present in the active site of caspase-9 appears to be critical for recruitment of caspase-3 to the apoptosome.
Caspase-9 recruits caspase-3 to the apoptosome through recognition of a critical aspartate (D175) residue in the effector caspase
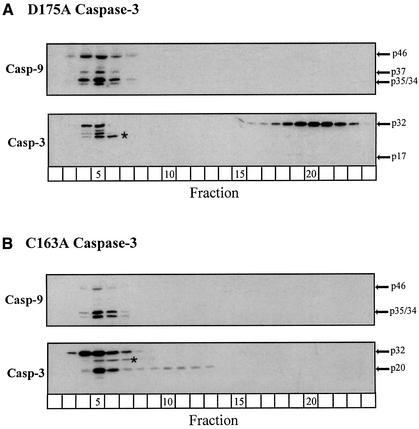
Next, we wished to determine which residues in caspase-3 might be critical for its recruitment to the complex. As caspase-3 is processed at the IETD175↓S site, located between its large and small subunits (Nicholson et al., 1995; Fernandes-Alnemri et al., 1996), we questioned whether this residue in caspase-3 might be required for its recruitment to the apoptosome. Therefore, we immunodepleted lysates of procaspase-3 (Figure 1C) and reconstituted them with D175A casp-3, which, as expected, did not support DEVDase activity following dATP activation (Figure 1D). Examination of the lysates by gel filtration revealed that Apaf-1 was oligomerized (data not shown) and caspase-9 was processed normally (fractions 4–7, Figure 4A). However, almost all the D175A casp-3 mutant remained outside the apoptosome as the unprocessed proenzyme (fractions 16–22, Figure 4A). Thus, recruitment of caspase-3 to the apoptosome ultimately requires an interaction between the active site cysteine in caspase-9 and the D175 residue in caspase-3. As with all proteases, caspases appear to bind their substrates very tightly in the transition state, but only weakly (Km in the micromolar range) in the ground state (Stennicke and Salvesen, 1999). Therefore, the absence of the C287 residue in caspase-9 or the D175 residue in caspase-3 probably inhibits formation of a normal tetrahedral intermediate necessary for substrate catalysis and, in effect, physically prohibits recruitment of procaspase-3 to the apoptosome.
Fig. 4. Caspase-9 recruits caspase-3 to the Apaf-1 apoptosome via recognition of a critical aspartate (D175) residue. Caspase-3-depleted lysates were reconstituted with (A) unprocessable D175A caspase-3 or (B) catalytically inactive C163A caspase-3. Following dATP activation for 1 h at 37°C, the reconstituted lysates were fractionated and immunoblotted for caspases-9 and -3, as described in Materials and methods. The arrows represent either the unprocessed or processed form(s) of the caspases; a non-specific band (*) was detected by the anti-caspase-3 antibody.
Both active and inactive caspase-3 remain associated with the apoptosome as long as caspase-3 is processed
At this point, all our experiments indicated that catalytically active processed or unprocessed caspase-9 was required for caspase-3 recruitment to the apoptosome, based on the presence of active, processed caspase-3 within the complex. Consequently, we questioned whether caspase-3 must be active or merely processed in order to maintain its association with the apoptosome. Therefore, we reconstituted caspase-3-depleted lysates with catalytically inactive C163A casp-3. Following dATP activation, Apaf-1 oligomerized normally (data not shown) and recruited and processed endogenous caspase-9 (fractions 4–7, Figure 4B). Most importantly, however, the C163A casp-3 mutant was efficiently recruited to the apoptosome (fractions 4–7, Figure 4B), as this mutant contained the critical D175 residue required for normal caspase-3 recruitment. Since the endogenous processed caspase-9 was present in the apoptosome, C163A casp-3 was processed to its p20 form and remained associated with the apoptosome complex (fractions 4–7, Figure 4B). However, since the p20 form of C163A casp-3 was inactive (Figure 1D), it did not remove its prodomain through autocatalytic processing to generate its p17 form (Han et al., 1997). Therefore, the processing of caspase-3, and not its activity, was required for its retention within the apoptosome.
The C163A casp-3 mutant, unlike wild-type caspase-3 or D175A casp-3, was present in significant amounts within the apoptosome as the unprocessed proform of the enzyme. The C163A casp-3 mutant was not fully processed, possibly because it had a somewhat different conformation from wild-type caspase-3, or perhaps within the context of the apoptosome, active caspase-3 participated in its own initial processing between its large and small subunits. Highly active, recombinant caspase-8 (100 nM) was also incapable of fully processing C163A casp-3 in the same period of time (lane 6, Figure 1B). Nevertheless, since the products of most enzymic reactions do not remain associated with the active site of the enzymes, we deemed it likely that the proform of C163A casp-3 in the apoptosome was associated with active processed caspase-9 and that the processed p20 form of C163A casp-3 was associated with some other protein in the complex.
Processed caspases-9 and -3 associate with Apaf-1 and XIAP in the apoptosome
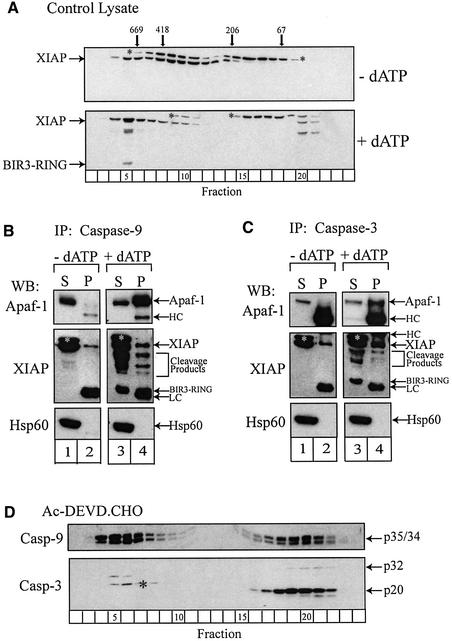
Since XIAP interacts directly with active caspases-3 and -7 in vitro and is the most potent caspase inhibitor of all known IAPs (Deveraux et al., 1998), and given that XIAP binds processed C163A casp-3 with significantly greater affinity than unprocessed C163A casp-3 (Sun et al., 1999), we speculated that XIAP might physically associate with the apoptosome and hold processed caspase-3 within the complex. Consequently, we examined normal lysates for the presence of this IAP. In unactivated lysates, XIAP was present in ∼200–700 kDa complexes. Following dATP activation, most of these complexes increased in size to ∼350–700 kDa, with the majority co-eluting with the apoptosome and some with the ‘micro-apoptosome’ complexes (Figure 5A). In activated lysates, XIAP was also cleaved into several fragments, including the BIR3-RING fragment (Deveraux et al., 1999), and each co-eluted with the apoptosome (Figure 5A).
Fig. 5. Co-immunoprecipitation of Apaf-1, caspase-9, caspase-3 and XIAP. (A) Control and dATP-activated THP.1 lysates were fractionated by gel filtration. The fractions were separated by SDS–PAGE and immunoblotted for XIAP using an antibody raised against its C-terminus. Control and dATP-activated lysates (∼15 mg/ml) were immunoprecipitated with an antibody against (B) caspase-9 or (C) caspase-3 and the resulting immuno complexes were recovered by centrifugation. The supernatants (S) and washed immunocomplexes (P) were separated by SDS–PAGE and western blotted (WB) for Apaf-1, XIAP or Hsp60. (D) Lysates were pretreated with DEVD⋅CHO (200 nM) for 1 h at 4°C and subsequently dATP-activated at 37°C for 1 h. The lysates were fractionated by gel filtration and analyzed by SDS–PAGE/immunoblotting for caspases-9 and -3, as described in Materials and methods. The arrows represent either the full-length proteins or, in the case of XIAP, various cleavage products. Bands corresponding to the light chain (LC) or heavy chain (HC) of the immunoprecipitated antibodies are also shown. Separation of the LC and the BIR3-RING fragment was difficult, but this fragment was only apparent in caspase-9 immunocomplexes obtained from dATP-activated lysates. Non-specific bands (*) were detected by the XIAP or caspase-3 antibodies.
Co-elution of Apaf-1, caspase-9, caspase-3 and XIAP in the same high molecular weight fractions suggested that they might be present within the same complex (Figures 2B and 5A). To test this hypothesis, we performed immunoprecipitation experiments using both control and dATP-activated lysates, predicting that relevant apoptosome interactions would occur primarily in activated lysates. Immunoprecipitation of caspase-9 revealed an association with both Apaf-1 and XIAP that occurred primarily in dATP-activated lysates (compare lanes 2 and 4, Figure 5B). Caspase-9 presumably associated with Apaf-1 through CARD–CARD interactions (Li et al., 1997; Qin et al., 1999) and with XIAP through active site–BIR3 interactions (Deveraux et al., 1999; Sun et al., 2000). All of the immunodetectable caspase-3/-8 cleavage products of XIAP contained the BIR3 domain and each was co-immunoprecipitated with caspase-9 (compare lanes 2 and 4, Figure 5B). This observation was significant, because all of these cleavage products, including the BIR3-RING fragment (Deveraux et al., 1999), were associated primarily with the apoptosome fractions (fraction 5, Figure 5A). Based on these experiments, we speculated that XIAP might associate with oligomerized Apaf-1, at least in part, through an interaction with processed caspase-9 (model 1, Figure 7).
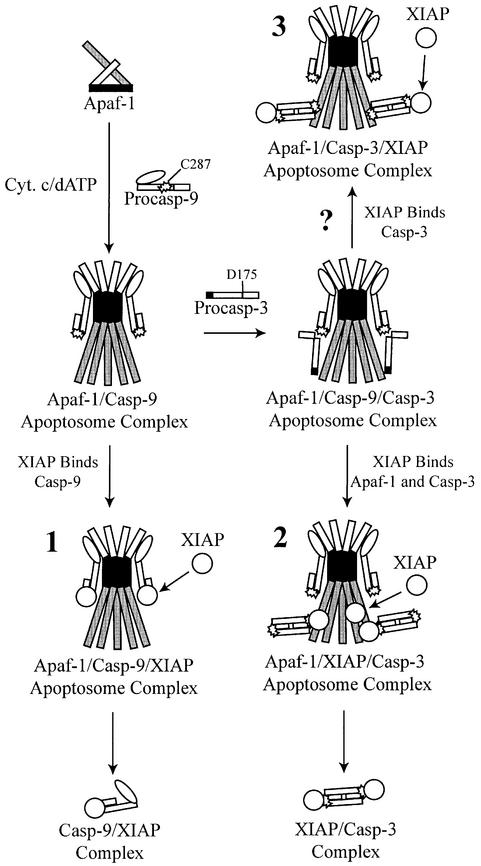
Fig. 7. Models for sequential recruitment and activation of caspases-9 and -3 and retention by XIAP. Apaf-1 undergoes cytochrome c/dATP-dependent oligomerization and recruits caspase-9 to the apoptosome through CARD–CARD interactions. Active (pro)caspase-9 then associates with XIAP (1) and/or recruits caspase-3 to the apoptosome through interactions involving the C287 residue in caspase-9 and the D175 residue in caspase-3. Following its activation, active caspase-3 is retained within the apoptosome via an interaction with XIAP, which is simultaneously associated with Apaf-1 (2).
Similarly, immunoprecipitation of caspase-3 revealed an interaction with both Apaf-1 and XIAP (compare lanes 2 and 4, Figure 5C). Since caspase-3 does not contain an obvious domain, such as a CARD, which would enable it to bind Apaf-1 directly, it was not immediately clear how caspase-3 might associate with Apaf-1. The association of caspase-3 with XIAP was more readily interpretable. Caspase-3 associated primarily with full-length XIAP in dATP-activated lysates (and possibly with undetectable BIR1–BIR2 fragments), but, in contrast to caspase-9, did not associate with the BIR3 fragments (compare lanes 2 and 4, Figure 5B and C). This was consistent with recent reports that the BIR3 domain of XIAP binds caspase-9, while the BIR2 domain and a few key residues in the linker region between the BIR1 and BIR2 domains bind caspase-3 (Takahashi et al., 1998; Sun et al., 1999, 2000). In order to verify that the interactions of active caspases-9 and -3 with Apaf-1 and XIAP were selective and not a function of non-specific adherence, we also performed Hsp27 and Hsp60 immunoblots as immunoprecipitation controls, as both proteins were highly abundant in our lysate preparation. Neither Hsp27 (data not shown) nor Hsp60 associated with caspase-3 or -9 in control or dATP-activated lysates (Figure 5B and C). Interestingly, Hsp27 and Hsp60 have been reported to interact with unprocessed and processed caspase-3, respectively (Samali et al., 1999; Pandey et al., 2000). However, we were unable to confirm these interactions under our given experimental conditions.
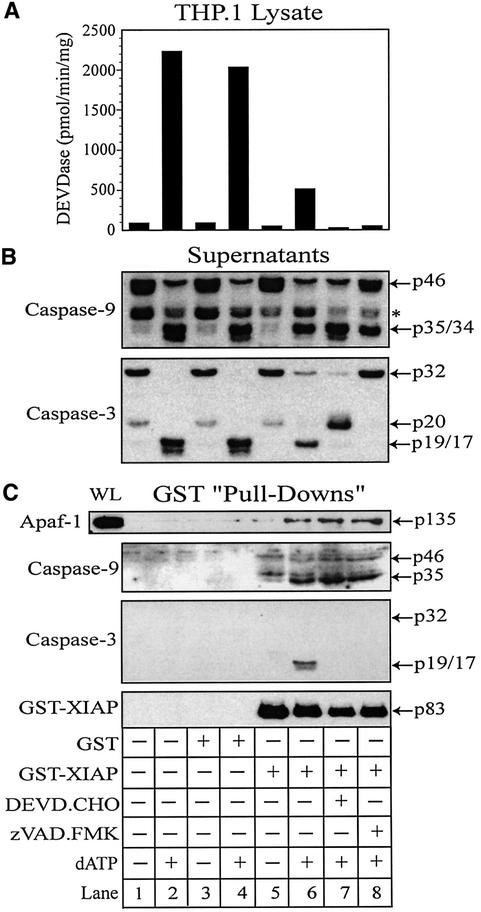
Immunoprecipitation of XIAP in control and dATP-activated lysates failed to detect the previously identified interactions of XIAP with caspase-3 or -9 (data not shown), suggesting that the XIAP antibody used probably disturbed XIAP–protein interactions. Consequently, we added endogenous amounts of recombinant purified glutathione S-transferase (GST)–XIAP (∼60 nM) to THP.1 lysates and activated them with dATP. XIAP–protein complexes were subsequently co-precipitated using glutathione (GSH)– Sepharose beads. GST–XIAP, and not GST alone, interacted with caspases-9 and -3, and with Apaf-1, but only following activation by dATP (compare lanes 3–6, Figure 6C) further supporting the notion that Apaf-1, caspase-9, caspase-3 and XIAP might exist in a single apoptosome complex.
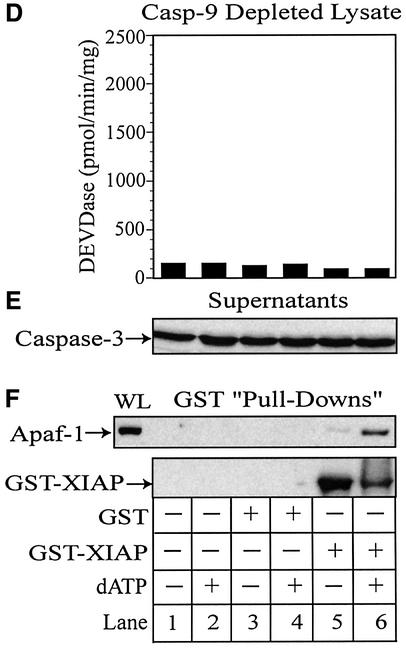
Fig. 6. XIAP directly associates with Apaf-1 and retains active caspase-3 within the apoptosome. THP.1 lysates were activated with dATP (lanes 2, 4, 6, 7 and 8) for 1 h at 37°C in the presence of GST (lanes 3 and 4) or GST–XIAP (lanes 5–8). In some treatments, the lysate was pretreated for 15 min with DEVD⋅CHO (200 nM) (lane 7) or z-VAD⋅FMK (25 µM) (lane 8). A small aliquot of each sample was immediately assayed for (A) DEVDase activity and the remainder incubated with GSH–Sepharose beads (20 µl) overnight at 4°C. Following centrifugation, supernatants were collected and the beads thoroughly washed (four times) with assay buffer. (B) Supernatants and (C) GST–XIAP/protein complexes were subsequently mixed with 2× SDS loading buffer, separated by SDS–PAGE and immunoblotted for Apaf-1, caspase-9, caspase-3 and/or XIAP. Similarly, caspase-9-depleted lysates were activated with dATP (lanes 2, 4 and 6) in the presence of GST (lanes 3 and 4) or GST–XIAP (lanes 5 and 6). A small aliquot of each sample was assayed for (D) DEVDase activity and the remainder incubated with GSH–Sepharose beads (20 µl) overnight at 4°C. After centrifugation, the resulting (E) supernatants and (F) GST–XIAP/protein complexes were separated by SDS–PAGE and immunoblotted for Apaf-1, caspase-3 and/or XIAP, as described above. All Apaf-1 western blots are presented at the same exposures; ‘WL’ represents the total amount of Apaf-1 present in each whole lysate that was available to interact with GST–XIAP.
XIAP associates directly with Apaf-1 in the apoptosome and is partially responsible for maintaining active caspase-3 within the complex
Although our experiments demonstrated an apparent complex between Apaf-1, caspase-9, caspase-3 and XIAP in dATP-activated lysates, it remained unclear whether active caspase-3 associated with the apoptosome through a direct interaction with Apaf-1 or an indirect interaction with XIAP (or another unidentified protein). We were unsuccessful in our attempts to co-immunoprecipitate active caspase-3 with an anti-caspase-9 antibody (and vice versa) in dATP-activated lysates (data not shown). Therefore, it appeared that processed caspase-3 was not associated with Apaf-1 via an interaction with either caspase-9 alone or caspase-9–XIAP complexes. Although it is formally possible that XIAP could simultaneously bind both caspases-9 and -3 through distinct BIR domains, this did not appear to be the case. Therefore, caspase-3 is not bound to the apoptosome through a sequential Apaf-1–caspase-9–XIAP–caspase-3 complex. However, it was still feasible that XIAP might directly associate with Apaf-1 and simultaneously bind and retain caspase-3 within the apoptosome (or vice versa), without any requirement for caspase-9.
Acetyl-Asp-Glu-Val-Asp aldehyde (DEVD⋅CHO) is a relatively specific caspase-3 inhibitor, which inhibits the interaction between active caspase-3 and XIAP in vitro by blocking the substrate binding site of caspase-3 (Sun et al., 1999). We reasoned that if caspase-3 was held in the apoptosome via an interaction with XIAP, DEVD⋅CHO should initiate release of caspase-3 from the complex. As expected, DEVD⋅CHO did not significantly alter Apaf-1 oligomerization (data not shown) or the recruitment and processing of caspase-9 (fractions 4–7, compare Figures 2B and 5D). In addition, DEVD⋅CHO did not significantly inhibit the activity of caspase-9, and thus allowed for processing of caspase-3 from its unprocessed to p20 form (fractions 17–22, Figure 5D). Only the autocatalytic processing of caspase-3 from its p20 to p17 form was totally inhibited, indicating that this concentration of DEVD⋅CHO selectively inhibited caspase-3 activity (fractions 17–22, Figure 5D). Significantly, however, in the presence of DEVD⋅CHO there was no longer any processed p20 associated with the apoptosome or ‘micro-apoptosome’ fractions (compare fractions 4–7 and 10–13, Figures 2B and 5D). In fact, all of the processed caspase-3 was present as free, unbound processed enzyme (fractions 17–22, Figure 5D). These data support the hypothesis that DEVD⋅CHO had selectively facilitated the release of caspase-3 from the apoptosome by disturbing an interaction between XIAP and processed caspase-3, and imply that caspase-3 might be associated with the apoptosome in a sequential Apaf-1–XIAP–caspase-3 complex (model 2, Figure 7). However, other possibilities could not be excluded, since DEVD⋅CHO could also potentially inhibit interactions between the active site of caspase-3 and additional proteins. Therefore, we examined the effects of DEVD⋅CHO and benzyloxycarbonyl-Val-Ala-Asp-(OMe) fluoromethyl ketone (z-VAD⋅FMK) on the ability of XIAP to associate with Apaf-1. In contrast to DEVD⋅CHO, z-VAD⋅FMK is a pan-caspase inhibitor that irreversibly binds the active site cysteine in all known caspases. We reasoned that if XIAP was directly associated with Apaf-1 and active caspase-3 in an Apaf-1– XIAP–caspase-3 complex (model 2, Figure 7), DEVD⋅ CHO would probably not inhibit the association of XIAP with Apaf-1, but would release caspase-3 from the apoptosome, as already demonstrated (fractions 17–22, compare Figures 2B and 5D). Alternatively, if XIAP was indirectly associated with Apaf-1 and the apoptosome via an active site interaction with caspase-3 (Apaf-1– caspase-3–XIAP complex; model 3, Figure 7) or caspase-9 (Apaf-1–caspase-9–XIAP complex; model 1, Figure 7), the presence of DEVD⋅CHO or z-VAD⋅FMK, respectively, should disrupt the association of XIAP with Apaf-1. As anticipated, both DEVD⋅CHO and z-VAD⋅FMK inhibited the interaction of XIAP with processed caspase-3 (compare lane 6 with lanes 7 and 8, Figure 6C). DEVD⋅CHO did not prevent caspase-9 from processing caspase-3 to its p20 form (compare lanes 6 and 7, Figure 6B), but did prevent XIAP from binding to p20 casp-3 by sterically hindering access of XIAP to the active site of p20 casp-3 (compare lanes 6 and 7, Figure 6C; Sun et al., 1999). In contrast, z-VAD⋅FMK precluded any interaction of XIAP with processed caspase-3 (compare lanes 6 and 8, Figure 6C), as it blocked the activity of caspase-9 and prevented formation of p20 casp-3 (compare lanes 6 and 7 with lane 8, Figure 6B). However, neither DEVD⋅CHO nor z-VAD⋅FMK disrupted the association of XIAP with Apaf-1 (compare lane 6 with lanes 7 and 8, Figure 6C), indicating that caspase-3 was probably not responsible for maintaining the presence of XIAP within the apoptosome in an Apaf-1–caspase-3– XIAP complex (model 3, Figure 7). Therefore, these data further support the idea that caspase-3 associates with the apoptosome via XIAP in an Apaf-1–XIAP–caspase-3 complex (model 2, Figure 7).
However, while z-VAD⋅FMK prevented an association between XIAP and caspase-3 by preventing activation of caspase-3, it did not disrupt the association of XIAP with processed caspase-9 (compare lanes 6 and 8, Figure 6C). Thus, in stark contrast to BIR2–caspase-3 interactions, it appeared that the BIR3 domain of XIAP might make contact with processed caspase-9 at residues outside the active site of the enzyme, or at sites not sterically hindered by the presence of z-VAD⋅FMK. However, it was also possible that z-VAD⋅FMK inhibited direct XIAP–caspase-9 interactions, but that some caspase-9 remained indirectly associated with XIAP via Apaf-1 in an XIAP–Apaf-1–caspase-9 complex. Thus, we were unable to determine whether caspase-9 was responsible for maintaining the presence of XIAP in the apoptosome or if XIAP directly associated with Apaf-1. In order to confirm the existence of an Apaf-1–XIAP–caspase-3 complex (model 2, Figure 7), we needed to establish that XIAP could bind Apaf-1 in the absence of caspase-9. As already noted, the absence of caspase-9 in lysate had no effect on Apaf-1 oligomerization (data not shown), but did prevent the recruitment, activation and association of active caspase-3 with the apoptosome (compare Figure 2B and C). Consequently, we repeated the GST–XIAP pull-down experiments with caspase-9-depleted lysates to determine whether XIAP remained associated with oligomerized Apaf-1. As anticipated, activation of caspase-9-depleted lysates with dATP did not result in processing of caspase-3 or any increase in DEVDase activity (compare lanes 1 and 2, Figure 6D and E). However, strikingly, XIAP still associated with Apaf-1 and did so almost exclusively in dATP-activated lysates (compare lanes 5 and 6, Figure 6F). Thus, XIAP was capable of selectively binding to oligomerized Apaf-1, even in the absence of caspase-9 or active caspase-3. Therefore, it appears that XIAP probably associates with the apoptosome through an interaction with Apaf-1 and is able to form an Apaf-1–XIAP–caspase-3 complex (model 2, Figure 7).
Concluding remarks
Many of the biochemical processes that take place within the cell do so within large multiprotein complexes and, not surprisingly, the activation of caspases appears no different (Bratton et al., 2000). In particular, the activity of caspase-9 is dependent upon its interaction with oligomerized Apaf-1 (Rodriguez and Lazebnik, 1999); within the context of the apoptosome, we have now demonstrated that both the active site cysteine in caspase-9 and the cleavage site aspartate (D175) in caspase-3 are required for efficient recruitment and activation of caspase-3 (Figures 2–4). Surprisingly, caspase-3 remained associated with the apoptosome following its activation, suggesting that endogenous caspase inhibitors might be associated with the apoptosome. We subsequently demonstrated for the first time that XIAP associates directly with the apoptosome and modulates the activation and retention of effector caspases within the complex (Figures 5B and C and 6). XIAP may associate with the apoptosome via an interaction with processed caspase-9 in an Apaf-1–caspase-9–XIAP complex (model 1, Figure 7). However, XIAP can bind to Apaf-1 in the absence of caspase-9 (Figure 6F) and can simultaneously bind and retain active caspase-3 within the apoptosome in an Apaf-1–XIAP–caspase-3 complex (model 2, Figure 7). Whether XIAP associates directly with Apaf-1 or does so via an additional unknown protein is not clear.
Our data do not preclude the presence of additional complexes. XIAP is present within the cell as high molecular weight complexes even in the absence of oligomerized Apaf-1 (Figure 5A) and probably binds to active caspases independent of the apoptosome (Figure 7). Conversely, not all of the active caspases present in the apoptosome are associated with XIAP. Indeed, a significant portion of active caspase-3 did not co-precipitate with GST–XIAP (lane 6, compare Figure 6B and C). Both cIAP-1 and cIAP-2 co-elute with apoptosome fractions obtained from dATP-activated lysates (data not shown) and may similarly contribute to the retention of active caspases within the apoptosome. However, we have previously demonstrated DEVDase activity within the apoptosome (Cain et al., 1999), suggesting that uninhibited, active caspase-3 is also bound to the apoptosome via additional unknown proteins.
Although processed caspase-3 remained largely associated with the apoptosome, whether it was active or bound to IAPs, some active caspase-3 was released from the apoptosome and was present in the ‘micro-apoptosome’ complexes or as the free enzyme (Figures 2–4). The factors that eventually mediate release of active caspase-3 from the apoptosome are currently under investigation, but preliminary studies suggest that ionic strength plays a role in this process. Nevertheless, it is intriguing to consider how apoptosome-bound active caspase-3 might serve an important function during apoptosis. A recent study (Faleiro and Lazebnik, 2000) suggests that apoptosome complexes are in direct contact with the nuclear membrane during apoptosis and that active caspase-9 may play an early role in the breakdown of the nuclear–cytoplasmic barrier. Therefore, one might envisage that, following inactivation of nuclear pores by caspase-9, apoptosome-bound active caspase-3 might be in an ideal position to be released and efficiently shuttled into the nucleus, where it serves to cleave a number of critical substrates (Earnshaw et al., 1999). In such a scenario, apoptosome-bound IAPs would serve to immediately inhibit caspases-9 and -3 and/or prevent release of caspase-3 from the apoptosome. After a modest stress in which only a small portion of caspases are activated, these IAPs would probably prevent apoptosis, whereas after a major insult, the activation of too many apoptosome complexes and active caspases would overcome the protective effect of IAPs, resulting in cell death.
Materials and methods
Preparation of recombinant caspases and XIAP
Several recombinant wild-type and non-cleavable or inactive mutant caspases (Figure 1) were expressed in Escherichia coli BL-21(DE3) and purified on Ni2+–Sepharose beads, as previously described (Srinivasula et al., 1996). The non-cleavable mutant caspases were incubated with appropriate active caspases to confirm the mutations in the recombinant proteins (Figure 1A and B). A minor contaminant (p34), due to bacterial processing, was present in both the p35 casp-9 and D315/330A casp-9 preparations (Figure 1A); however, this contaminant does not add significantly to their activity (Stennicke et al., 1999). GST-tagged XIAP was expressed and purified on GSH–Sepharose beads as previously described (Deveraux et al., 1999).
Preparation and activation of naive and immunodepleted cell lysates
Human monocytic THP.1 cells were grown in RPMI 1640, supplemented with 10% heat-inactivated fetal bovine serum in 5% CO2 at 37°C. Cell lysates were prepared as previously described (Cain et al., 1999). For the immunodepletion experiments, lysates were incubated for 2 h with 500 µl of protein G–Sepharose beads (50% slurry, precoated with 4% bovine serum albumin) coated with antibodies against caspase-9 or -3. Partially immunodepleted supernatants were collected by centrifugation and subjected to a second round of immunodepletion to deplete caspases completely. In vitro activation of caspases in both normal and immunodepleted lysates was initiated by incubating lysates (10 mg/ml) with dATP/MgCl2 (2 mM) at 37°C. In some experiments, wild-type or mutant caspase-9 or -3 (200 nM) proteins were added to immunodepleted lysates before activation. Exogenous cytochrome c was not required for any of these experiments, as it was released during lysate preparation. As a marker of effector caspase activation, DEVDase activity was measured in activated lysates (Cain et al., 1999).
Immunoprecipitation experiments
In some experiments, both control and dATP-activated lysates were incubated with protein G–Sepharose beads (Amersham Pharmacia Biotech) precoated with anti-caspase-9 or -3 antibodies. The resulting protein complexes were obtained by centrifugation, washed four times in activation buffer [100 mM HEPES, 0.1% (w/v) CHAPS, 10 mM dithiothreitol (DTT), 10% (w/v) sucrose pH 7.0] and immunoblotted for various components of the apoptosome, including Apaf-1, caspase-9 and caspase-3. In other experiments, purified recombinant GST–XIAP (∼60 nM) was added to lysates before activation by dATP. GST–XIAP protein complexes and their corresponding supernatants were obtained from control and dATP-activated lysates using GSH–Sepharose beads (Amersham Pharmacia Biotech) and immunoblotted for various components of the apoptosome. Both endogenous and recombinant XIAP were detected using an antibody raised against the C-terminus of the protein (amino acids 268–426) (Transduction Laboratories/Pharmingen, San Diego, CA). Consequently, this antibody recognizes the full-length protein, as well as any cleavage product containing the BIR3 domain, such as the BIR3-RING fragment (Deveraux et al., 1999), but not the corresponding BIR1–BIR2 fragment.
Analysis of the apoptosome and micro-apoptosome complexes
In order to assess the effects of various caspase mutants or caspase inhibitors (DEVD⋅CHO or z-VAD⋅FMK) on the recruitment, processing and release of various caspases from the apoptosome and micro-apopto some complexes, we used gel filtration techniques previously described (Cain et al., 1999). Briefly, activated lysate proteins (10 mg) were eluted (0.4 ml/min; 4°C) from a HiPrep 16/60 S-300 Sephacryl high-resolution column (Amersham Pharmacia Biotech) using column buffer (20 mM HEPES, 0.1% (w/v) CHAPS, 5 mM DTT, 5% (w/v) sucrose pH 7.0). Fractions (2 ml) were collected and preserved in 10× SDS loading buffer. Samples were later analyzed by western blotting for changes in the distribution or processing of Apaf-1, caspases-3 and -9 and XIAP as previously described (Cain et al., 1999). We have recently identified the presence of two apoptosome complexes in lysates, an inactive ∼1.4 MDa complex and an active ∼700 kDa complex (Cain et al., 2000). Unfortunately, resolution of these two complexes could only be achieved using Superose-6 chromatography, a technique that requires the presence of salt in the eluting buffer and consequently displaces effector caspases from the apoptosome. Both the ∼1.4 MDa and the ∼700 kDa complexes contain oligomerized Apaf-1 and processed caspase-9, and with regard to these components, appear indistinguishable. However, only the ∼700 kDa complex is capable of processing effector caspases. Thus, in the present studies, these two complexes co-elute and contain similar levels of Apaf-1 and caspase-9, but all of the effects related to recruitment and processing of effector caspases can be attributed to the ∼700 kDa complex.
Acknowledgments
Acknowledgements
An Apaf-1 polyclonal antibody was generously provided by Dr X.Wang (UT Southwestern Medical Center, Dallas, TX). Anti-caspases-3 and -9 were kind gifts from Drs D.W.Nicholson (Merck Frosst, Canada) and D.Green (La Jolla Institute for Allergy and Immunology, San Diego, CA). The authors thank D.Brown for preparation of THP.1 lysates. This work was partly funded by a European Union grant (QLG1-1999-00739) to G.M.C. and by NIH grant AG 13847 to E.S.A.
References
- Bratton S.B., MacFarlane,M., Cain,K. and Cohen,G.M. (2000) Protein complexes activate distinct caspase cascades in death receptor and stress-induced apoptosis. Exp. Cell Res., 256, 27–33. [DOI] [PubMed] [Google Scholar]
- Cain K., Brown,D.G., Langlais,C. and Cohen,G.M. (1999) Caspase activation involves the formation of the aposome, a large (∼700 kDa) caspase-activating complex. J. Biol. Chem., 274, 22686–22692. [DOI] [PubMed] [Google Scholar]
- Cain K., Bratton,S.B., Langlais,C., Walker,G., Brown,D.G., Sun,X.M. and Cohen,G.M. (2000) Apaf-1 oligomerizes into biologically active ∼700 kDa and inactive ∼1.4 MDa apoptosome complexes. J. Biol. Chem., 275, 6067–6070. [DOI] [PubMed] [Google Scholar]
- Clem R.J. and Miller,L.K. (1994) Control of programmed cell death by the baculovirus genes p35 and iap. Mol. Cell. Biol., 14, 5212–5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen G.M. (1997) Caspases: the executioners of apoptosis. Biochem. J., 326, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveraux Q.L. and Reed,J.C. (1999) IAP family proteins—suppressors of apoptosis. Genes Dev., 13, 239–252. [DOI] [PubMed] [Google Scholar]
- Deveraux Q.L., Takahashi,R., Salvesen,G.S. and Reed,J.C. (1997) X-linked IAP is a direct inhibitor of cell-death proteases. Nature, 388, 300–304. [DOI] [PubMed] [Google Scholar]
- Deveraux Q.L., Roy,N., Stennicke,H.R., Van Arsdale,T., Zhou,Q., Srinivasula,S.M., Alnemri,E.S., Salvesen,G.S. and Reed,J.C. (1998) IAPs block apoptotic events induced by caspase-8 and cytochrome c by direct inhibition of distinct caspases. EMBO J., 17, 2215–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveraux Q.L., Leo,E., Stennicke,H.R., Welsh,K., Salvesen,G.S. and Reed,J.C. (1999) Cleavage of human inhibitor of apoptosis protein XIAP results in fragments with distinct specificities for caspases. EMBO J., 18, 5242–5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw W.C., Martins,L.M. and Kaufmann,S.H. (1999) Mammalian caspases: structure, activation, substrates and functions during apoptosis. Annu. Rev. Biochem., 68, 383–424. [DOI] [PubMed] [Google Scholar]
- Faleiro L. and Lazebnik,Y. (2000) Caspases disrupt the nuclear–cytoplasmic barrier. J. Cell Biol., 151, 951–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes-Alnemri T. et al. (1996) In vitro activation of CPP32 and Mch3 by Mch4, a novel human apoptotic cysteine protease containing two FADD-like domains. Proc. Natl Acad. Sci. USA, 93, 7464–7469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green D.R. and Reed,J.C. (1998) Mitochondria and apoptosis. Science, 281, 1309–1312. [DOI] [PubMed] [Google Scholar]
- Hakem R. et al. (1998) Differential requirement for caspase 9 in apoptotic pathways in vivo. Cell, 94, 339–352. [DOI] [PubMed] [Google Scholar]
- Han Z., Hendrickson,E.A., Bremner,T.A. and Wyche,J.H. (1997) A sequential two-step mechanism for the production of the mature p17:p12 form of caspase-3 in vitro. J. Biol. Chem., 272, 13432–13436. [DOI] [PubMed] [Google Scholar]
- Hu Y., Ding,L., Spencer,D.M. and Nunez,G. (1998) WD-40 repeat region regulates Apaf-1 self-association and procaspase-9 activation. J. Biol. Chem., 273, 33489–33494. [DOI] [PubMed] [Google Scholar]
- Hu Y., Benedict,M.A., Ding,L. and Nunez,G. (1999) Role of cytochrome c and dATP/ATP hydrolysis in Apaf-1-mediated caspase-9 activation and apoptosis. EMBO J., 18, 3586–3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuida K., Haydar,T.F., Kuan,C.Y., Gu,Y., Taya,C., Karasuyama,H., Su,M.S., Rakic,P. and Flavell,R.A. (1998) Reduced apoptosis and cytochrome c-mediated caspase activation in mice lacking caspase 9. Cell, 94, 325–337. [DOI] [PubMed] [Google Scholar]
- Li P., Nijhawan,D., Budihardjo,I., Srinivasula,S.M., Ahmad,M., Alnemri,E.S. and Wang,X. (1997) Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell, 91, 479–489. [DOI] [PubMed] [Google Scholar]
- Nicholson D.W. et al. (1995) Identification and inhibition of the ICE/CED-3 protease necessary for mammalian apoptosis. Nature, 376, 37–43. [DOI] [PubMed] [Google Scholar]
- Pandey P., Farber,R., Nakazawa,A., Kumar,S., Bharti,A., Nalin,C., Weichselbaum,R., Kufe,D. and Kharbanda,S. (2000) Hsp27 functions as a negative regulator of cytochrome c-dependent activation of procaspase-3. Oncogene, 19, 1975–1981. [DOI] [PubMed] [Google Scholar]
- Qin H., Srinivasula,S.M., Wu,G., Fernandes-Alnemri,T., Alnemri,E.S. and Shi,Y. (1999) Structural basis of procaspase-9 recruitment by the apoptotic protease-activating factor 1. Nature, 399, 549–557. [DOI] [PubMed] [Google Scholar]
- Rodriguez J. and Lazebnik,Y. (1999) Caspase-9 and APAF-1 form an active holoenzyme. Genes Dev., 13, 3179–3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh A., Srinivasula,S.M., Acharya,S., Fishel,R. and Alnemri,E.S. (1999) Cytochrome c and dATP-mediated oligomerization of Apaf-1 is a prerequisite for procaspase-9 activation. J. Biol. Chem., 274, 17941–17945. [DOI] [PubMed] [Google Scholar]
- Samali A., Cai,J., Zhivotovsky,B., Jones,D.P. and Orrenius,S. (1999) Presence of a pre-apoptotic complex of pro-caspase-3, Hsp60 and Hsp10 in the mitochondrial fraction of Jurkat cells. EMBO J., 18, 2040–2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slee E.A. et al. (1999) Ordering the cytochrome c-initiated caspase cascade: hierarchical activation of caspases-2, -3, -6, -7, -8 and -10 in a caspase-9-dependent manner. J. Cell Biol., 144, 281–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasula S.M., Ahmad,M., Fernandes-Alnemri,T., Litwack,G. and Alnemri,E.S. (1996) Molecular ordering of the Fas-apoptotic pathway: the Fas/APO-1 protease Mch5 is a CrmA-inhibitable protease that activates multiple Ced-3/ICE-like cysteine proteases. Proc. Natl Acad. Sci. USA, 93, 14486–14491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasula S.M., Ahmad,M., Fernandes-Alnemri,T. and Alnemri,E.S. (1998) Autoactivation of procaspase-9 by Apaf-1-mediated oligomerization. Mol. Cell, 1, 949–957. [DOI] [PubMed] [Google Scholar]
- Stennicke H.R. and Salvesen,G.S. (1999) Catalytic properties of the caspases. Cell Death Differ., 6, 1054–1059. [DOI] [PubMed] [Google Scholar]
- Stennicke H.R., Deveraux,Q.L., Humke,E.W., Reed,J.C., Dixit,V.M. and Salvesen,G.S. (1999) Caspase-9 can be activated without proteolytic processing. J. Biol. Chem., 274, 8359–8362. [DOI] [PubMed] [Google Scholar]
- Sun C. et al. (1999) NMR structure and mutagenesis of the inhibitor-of-apoptosis protein XIAP. Nature, 401, 818–822. [DOI] [PubMed] [Google Scholar]
- Sun C., Cai,M., Meadows,R.P., Xu,N., Gunasekera,A.H., Herrmann,J., Wu,J.C. and Fesik,S.W. (2000) NMR structure and mutagenesis of the third Bir domain of the inhibitor of apoptosis protein XIAP. J. Biol. Chem., 275, 33777–33781. [DOI] [PubMed] [Google Scholar]
- Takahashi R., Deveraux,Q., Tamm,I., Welsh,K., Assa-Munt,N., Salvesen,G.S. and Reed,J.C. (1998) A single BIR domain of XIAP sufficient for inhibiting caspases. J. Biol. Chem., 273, 7787–7790. [DOI] [PubMed] [Google Scholar]
- Vucic D., Stennicke,H.R., Pisabarro,M.T., Salvesen,G.S. and Dixit,V.M. (2000) ML-IAP, a novel inhibitor of apoptosis that is preferentially expressed in human melanomas. Curr. Biol., 10, 1359–1366. [DOI] [PubMed] [Google Scholar]
- Woo M. et al. (1998) Essential contribution of caspase 3/CPP32 to apoptosis and its associated nuclear changes. Genes Dev., 12, 806–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou H., Henzel,W.J., Liu,X., Lutschg,A. and Wang,X. (1997) Apaf-1, a human protein homologous to C. elegans CED-4, participates in cytochrome c-dependent activation of caspase-3. Cell, 90, 405–413. [DOI] [PubMed] [Google Scholar]
- Zou H., Li,Y., Liu,X. and Wang,X. (1999) An APAF-1⋅cytochrome c multimeric complex is a functional apoptosome that activates procaspase-9. J. Biol. Chem., 274, 11549–11556. [DOI] [PubMed] [Google Scholar]