Abstract
Trichostatin A (TSA) and trapoxin (TPX) are potent inhibitors of histone deacetylases (HDACs). TSA is proposed to block the catalytic reaction by chelating a zinc ion in the active-site pocket through its hydroxamic acid group. On the other hand, the epoxyketone is suggested to be the functional group of TPX capable of alkylating the enzyme. We synthesized a novel TPX analogue containing a hydroxamic acid instead of the epoxyketone. The hybrid compound cyclic hydroxamic acid-containing peptide (CHAP) 1 inhibited HDAC1 at low nanomolar concentrations. The HDAC1 inhibition by CHAP1 was reversible as it was by TSA, in contrast to the irreversible inhibition by TPX. CHAP with an aliphatic chain length of five, which corresponded to that of acetylated lysine, was stronger than those with other lengths. These results suggest that TPX is a substrate mimic and that the replacement of the epoxyketone with the hydroxamic acid converted TPX to an inhibitor chelating the zinc like TSA. Interestingly, HDAC6, but not HDAC1 or HDAC4, was resistant to TPX and CHAP1, whereas TSA inhibited these HDACs to a similar extent. HDAC6 inhibition by TPX at a high concentration was reversible, probably because HDAC6 is not alkylated by TPX. We further synthesized the counterparts of all known naturally occurring cyclic tetrapeptides containing the epoxyketone. HDAC1 was highly sensitive to all these CHAPs much more than HDAC6, indicating that the structure of the cyclic tetrapeptide framework affects the target enzyme specificity. These results suggest that CHAP is a unique lead to develop isoform-specific HDAC inhibitors.
Reversible histone acetylation, which occurs at the ɛ-amino group of conserved lysine residues clustered near the amino terminus of core histones, mediates changes in nucleosome conformation, which is important in the regulation of gene expression (1). The correlation between acetylation and increased transcription has been known for many years. Highly acetylated nucleosomes are associated with transcriptionally active chromatin, whereas hypoacetylated histones are often found in inactive chromatin. Recent discovery of the enzymes controlling histone acetylation and deacetylation showed that acetylation of histones is an important step in transcription (2, 3). Acetylation and deacetylation are catalyzed by specific enzyme families, histone acetyltransferases (HATs) and histone deacetylases (HDACs), respectively. HATs were identified to be transcriptional coactivators including GCN5 (4), CREB-binding protein (CBP)/p300 (5), and p300/CBP-associated factor (PCAF; ref. 6), as well as the p160 family proteins. On the other hand, HDACs were found as yeast transcriptional regulators related to Rpd3 (7), Hda1 (8), and Sir2 (9). A number of transcriptional repressors and corepressors, such as Sin3, silencing mediator of retinoid acid and thyroid hormone receptor (SMRT), and nuclear receptor corepressor (N-CoR), were shown to recruit the HDAC complex to the promoter regions (10, 11). To date, at least nine different mammalian HDACs were described, which are classified into three classes, i.e., class I (related to Rpd3; refs. 7, 12–14), class II (related to Hda1; refs. 15–18), and the Sir2 family, the activity of which depends on nicotinamide-adenine dinucleotide (NAD; ref. 9). Specific roles of these enzymes and their target genes still remain to be elucidated.
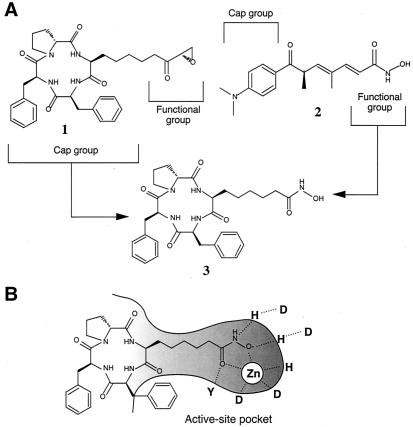
We have identified HDACs as the target of trichostatin A (TSA) and trapoxin (TPX), both of which are microbial metabolites that induce cell differentiation, cell cycle arrest, and reversal of transformed cells morphology (19). Several phytopathogenic and antifungal compounds related to TPX, such as chlamydocin and HC-toxin, have also been shown to inhibit HDAC (20). Because aberrant histone acetylation has been linked to malignant diseases in some cases, HDAC inhibitors bear great potential as new drugs because of their ability to modulate transcription and to induce differentiation and apoptosis (21). In fact, FK228 (22) and MS-275 (23), potent antitumor agents under clinical investigation, were shown to inhibit HDACs. Recent crystallographic studies (24) showed the zinc-dependent acetamide cleavage reaction by a bacterial enzyme related to HDAC [histone deacetylase-like protein (HDLP)]. Cocrystallization of this enzyme with TSA or suberoylanilide hydroxamic acid (25) demonstrated that these inhibitors mimic the substrate and that chelation of the zinc in the catalytic pocket by the hydroxamic acid group is the main mechanism of inhibition (24). Conservation of the amino acid sequences of the loops that form the active-site pocket among HDLP and class I and class II HDACs strongly suggests that HDACs present the same catalytic reaction and TSA inhibition as HDLP. In the case of TPX, we previously showed that the epoxyketone group at the terminus of the side chain of (2S,9S)-2-amino-9,10-epoxy-8-oxodecanoic acid (Aoe) is the enzyme-inhibiting group, causing irreversible inhibition (26). Because TSA can compete with TPX for binding to HDAC1 (7), it seems likely that the aliphatic chain of Aoe also acts as a substrate analogue. If this likelihood is the case, then replacement of the epoxyketone group of TPX (compound 1) with the hydroxamic acid group of TSA (compound 2) should convert it to a reversible-type inhibitor that chelates the catalytic center zinc (Fig. 1).
Figure 1.
Molecular design of a TPX/TSA hybrid HDAC inhibitor. (A) TPX (1) and TSA (2) are proposed to consist of cap groups, cyclic tetrapeptide and dimethylamino-phenyl groups, and functional groups for enzyme inhibition, an epoxyketone and a hydroxamic acid, respectively (21, 24). An inhibitor consisting of the cyclic tetrapeptide of TPX B and the hydroxamic acid of TSA (CHAP1) was synthesized (3). (B) A model for HDAC inhibition by CHAP1. The aliphatic chain with the hydroxamic acid group may be inserted into the tube-like active-site pocket of HDAC, thereby chelating the zinc ion by the hydroxamic acid group at the bottom of the pocket. The cyclic tetrapeptide portion may act as a cap to pack the molecule at the rim of the pocket. The conserved active-site residues revealed by the crystallographic studies (24) are shown as single letters.
In this paper, we show that a derivative of TPX named cyclic hydroxamic acid-containing peptide (CHAP) 1, in which Aoe in TPX B is replaced by Asu(NHOH) (Fig. 1), strongly and reversibly inhibits HDAC1. Structure-activity relationship study using HDAC1, HDAC4, and HDAC6 showed that the cyclic tetrapeptide portion affects both the enzyme inhibitory potency and specificity. CHAP1 will be a unique lead for development of specific HDAC inhibitors.
Materials and Methods
Synthesis of CHAPs.
The cyclic tetrapeptides for CHAPs were prepared according to one of the following three strategies: liquid phase synthesis of linear peptides followed by liquid phase cyclization (strategy 1); solid phase synthesis of linear peptides followed by cyclization on cleavage from resin (strategy 2); and solid phase synthesis and subsequent cleavage of linear peptides from resin, followed by liquid phase cyclization (strategy 3). The cyclic peptide precursors were finally converted to the corresponding CHAPs by side chain modification to the hydroxamic acid. The details of the procedures and experimental data concerning the synthesis of compounds 3-6 are described in the supplemental text (which is published as supplemental data on the PNAS web site, www.pnas.org).
Preparation of Recombinant HDACs and Assay for Enzyme Activity.
Cells (1–2 × 106, NIH 3T3 for HDAC1, and 293 for HDAC4 and HDAC6) were grown in a 100-mm dish for 24 h, and transfected transiently with 10 μg of each vector, pcDNA3-HD1 for human HDAC1 (27), pcDNA3.1(+)-HD4 for human HDAC4 (28), or pcDNA-mHDA2/HDAC6 for mouse HDAC6 (17), using the LipofectAmine reagent (Life Technologies, Rockville, MD). After successive cultivation in OPTI-MEM for 5 h and DMEM for 19 h, the cells were washed with PBS and lysed by sonication in lysis buffer containing 50 mM Tris⋅HCl (pH 7.5), 120 mM NaCl, 5 mM EDTA, and 0.5% Nonidet P-40. The soluble fraction collected by microcentrifugation was precleared by incubating with Protein A/G plus agarose beads (Santa Cruz Biotechnology). After the cleared supernatant had been incubated for 1 h at 4°C with 8 μg/ml of an anti-FLAG M2 antibody (Sigma) for HDAC1 and HDAC4, or an anti-HA antibody (Santa Cruz Biotechnology) for HDAC6, the agarose beads were washed three times with lysis buffer and once with HD buffer consisting of 20 mM Tris⋅HCl (pH 8.0), 150 mM NaCl, 10% glycerol, and a complete protease inhibitor mixture (Boehringer Mannheim). The bound proteins were released from the immune complex by incubating for 1 h at 4°C with 100 μg of the FLAG or the HA peptide (Sigma-Aldrich) in HD buffer (1.25 ml). The supernatant was collected after centrifugation and diluted to give 1,000–2,000 cpm in the following standard enzyme assay. The beads-bound HDAC1 and HDAC6 were used for the drug reversibility test. For the enzyme assay, 10 μl of [3H]acetyl-labeled histones (25,000 cpm/10 μg) was added to 90 μl of the enzyme fraction, and the mixture was incubated at 37°C for 15 min. The enzyme reaction was linear for at least 1 h under these conditions. The reaction was stopped by the addition of 10 μl of concentrated HCl. The released [3H]acetic acid was extracted with 1 ml of ethylacetate, and 0.9 ml of the solvent layer was taken into 5 ml of ACS (aqueous counting scintillant) II solution (Amersham Pharmacia) for determination of radioactivity. The 50% inhibitory concentrations (IC50) were determined as the means ± SD of the concentrations calculated from at least three independent dose–response curves.
Extraction of Histones and Acid/Urea/Triton Gel Electrophoresis.
Histones of cultured cells were extracted as described previously (29). The level of histone acetylation was analyzed by slab gel electrophoresis using an acid/urea/Triton (AUT) gel (1 M acetic acid, 8 M urea, 0.5% Triton X-100, 45 mM NH3, and 16% acrylamide) with an upper gel (1 M acetic acid, 6.3 M urea, and 4.4% acrylamide). After the extracted histones had been mixed with loading buffer (7.4 M urea, 1.4 M NH3, 10 mM DTT), electrophoresis was performed in 0.2 M glycine and 1 M acetic acid, and then the gels were stained with silver.
Western Blot Analysis.
Lysates from HeLa cells treated with various concentrations of TSA and CHAPs for 24 h were prepared by lysing cells with brief sonication in IP buffer (50 mM Hepes, 150 mM NaCl, 1 mM EDTA, 2.5 mM EGTA, 0.1% Tween 20, 10% glycerol, 0.1 mM PMSF, 10 mM β-glycerophosphate, 1 mM NaF, 0.1 mM Na3VO4, 10 μg/ml aprotinin, and 10 mM leupeptin, pH 7.5). Lysates were centrifuged for 10 min at 4°C, and the supernatants were frozen until analysis. An equal amount of proteins (30 μg protein/lane) was loaded and electrophoresed on SDS/polyacrylamide gels, and proteins separated were transferred onto an Immobilon-P membrane (Millipore). An anti-human Cyclin A antibody (Santa Cruz Biotechnology), an anti-human Cyclin D1 antibody (Santa Cruz Biotechnology), an anti-human Cyclin E antibody (Upstate Biotechnology, Lake Placid, NY), an anti-human p21WAF1/Cip1 antibody (Transduction Laboratories, Lexington, KY) were used, and the immune complexes were detected with an ECL Western blotting kit (Amersham Pharmacia).
MHC Class-I Molecule Up-Regulation Assay.
The activity of HDAC inhibitors to induce expression of MHC class-I molecules was evaluated by determining the concentration for 2-fold up-regulation (C×2) according to the method reported previously (30). Briefly, after 24 h of culture, B16/BL6 cells (5,000 cells/200 μl) in a 96-well microplate were incubated with various concentrations of drugs for 3 days. The expression of MHC class I molecule on the cell surface was measured by a cell ELISA method, and their C×2 values were calculated from at least three independent dose-response curves.
Flow Cytometry.
Procedures for preparation and staining of nuclear DNA with propidium iodide in HeLa cells were described previously (31).
Results
CHAP1 Is a Potent and Reversible Inhibitor of HDAC1.
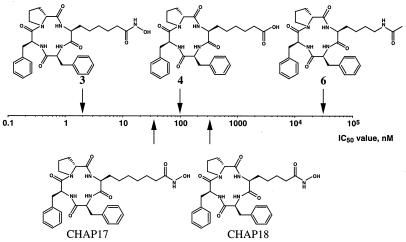
To examine the role of the epoxyketone group of TPX B [1, cyclo(Aoe-Phe-Phe-D-Pro)] in HDAC inhibition, we synthesized a series of cyclic tetrapeptide compounds containing L-Asu(NHOH) (3), L-aminosuberic acid (4), L-lysine (5), or acetylated L-lysine (6) instead of Aoe, and examined their ability to inhibit HDAC1. Compound 3 was found to be a strong inhibitor, blocking the enzyme activity at an IC50 of about 1.9 nM (Fig. 2). To our knowledge, there have been no other reports of a synthetic HDAC inhibitor stronger than TSA (Table 1). We named compound 3 CHAP1. On the other hand, compound 5 containing a free amino group was almost inactive (IC50 > 100 μM, data not shown). These results indicate that not only the epoxyketone but also the hydroxamic acid acts as a potent functional group for HDAC inhibition in the cyclic tetrapeptide antibiotic. Interestingly, compound 4 containing a carboxylic acid group showed a significant activity (IC50 = 100 ± 28 nM), whereas compound 6 containing an acetylated lysine was very weak (IC50 = 31,000 ± 4,400 nM).
Figure 2.
Effects of the structures of the functional group and aliphatic chain length on the HDAC1-inhibitory potency of CHAP1. The half-maximal inhibitory concentrations (IC50) of CHAP1 (3), cyclo(Asu-Phe-Phe-d-Pro) (4), cyclo(Lys(Ac)-Phe-Phe-d-Pro) (6), cyclo(Aaz(NHOH)-Phe-Phe-d-Pro)(Aaz = α-aminoazelaic acid) (CHAP17), and cyclo(Api(NHOH)-Phe-Phe-d-Pro) (Api = α-aminopimelic acid) (CHAP18) were plotted. The IC50 of cyclo(Lys-Phe-Phe-d-Pro) (5) was over 100 μM.
Table 1.
HDAC inhibitory activity and MHC-inducing activity of CHAPs and related compounds
| Compound | Structure | Type* | IC50
± SD, nM
|
HDAC6/HDAC1 | Cx2 ± SD, nM
|
||
|---|---|---|---|---|---|---|---|
| HDAC1 | HDAC4 | HDAC6 | MHC | ||||
| TSA | 6.0 ± 2.5 | 38 ± 4 | 8.6 ± 1.4 | 1.4 | 2.8 ± 2.0 | ||
| TPX A | cyclo(Aoe-Phe-Phe-d-Pip) | 0.82 ± 0.29 | NT† | 524 ± 240 | 640 | 3.5 ± 0.6 | |
| TPX B | cyclo(Aoe-Phe-Phe-d-Pro) | 0.11 ± 0.01 | 0.30 ± 0.03 | 360 ± 160 | 3,300 | 1.2 ± 0.9 | |
| Chlamydocin | cyclo(Aoe-Aib-Phe-d-Pro) | 0.15 ± 0.03 | NT | 1,100 ± 430 | 7,300 | 4.6 ± 2.7 | |
| Cyl-2 | cyclo(Aoe-d-Tyr(Me)-Ile-Pip) | 0.70 ± 0.45 | NT | 40,000 ± 11,000 | 57,000 | 10 ± 7 | |
| CHAP56 | cyclo(Asu(NHOH)-Phe-Phe-d-Pip) | TPX A | 6.1 ± 1.4 | NT | 150 ± 84 | 25 | 30 ± 5 |
| CHAP1 | cyclo(Asu(NHOH)-Phe-Phe-d-Pro) | TPX B | 1.9 ± 0.5 | 2.7 ± 1.3 | 19 ± 3 | 10 | 98 ± 23 |
| CHAP49 | cyclo(Asu(NHOH)-d-Tyr(Me)-Ile-Pip) | Cyl-2 | 1.2 ± 0.7 | NT | 36 ± 17 | 30 | 5.3 ± 2.2 |
| CHAP30 | cyclo(Asu(NHOH)-d-Tyr(Me)-Ile-Pro) | Cyl-1 | 4.4 ± 1.8 | NT | 110 ± 84 | 25 | 17 ± 8 |
| CHAP53 | cyclo(Asu(NHOH)-d-Phe-Leu-Pip) | WF3161 | 0.94 ± 0.33 | NT | 22 ± 10 | 23 | 20 ± 4 |
| CHAP15 | cyclo(Asu(NHOH)-Aib-Phe-d-Pro) | Chlamydocin | 0.44 ± 0.23 | NT | 38 ± 12 | 86 | 33 ± 9 |
| CHAP13 | cyclo(Asu(NHOH)-d-Pro-Ala-d-Ala) | HC-toxin | 2.9 ± 2.1 | NT | 61 ± 14 | 21 | 410 ± 86 |
Type, the natural product having the same cyclic tetrapeptide core structure as that of each CHAP.
NT, Not tested; Aib, α-aminoisobutylic acid.
We next compared the activity of CHAP compounds having different methylene chain lengths (4, 5, and 6) between their hydroxamic acid and the cyclic tetrapeptide core. As shown in Fig. 2, CHAP with the 5-carbon-long chain (3) was much more effective than 6-carbon (CHAP17) and 4-carbon (CHAP18) chains. Because the aliphatic chain length of 5 corresponds to the length between the carbonyl group and the α-carbon in acetylated lysine, it is reasonable to assume that the hydroxamic acid-containing side chain of CHAPs with a 5-carbon-long chain just fits the substrate's pocket of HDAC with which acetylated lysine interacts (Fig. 1B).
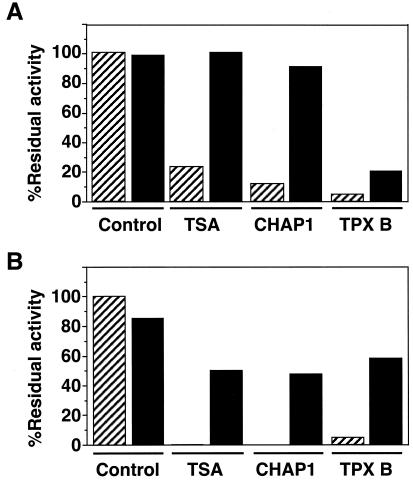
We previously reported that the inhibition of HDAC by TPX was apparently irreversible (26). This irreversible inhibition is probably due to the alkylation of the enzyme through its epoxy group, because the reduction of the epoxide caused almost complete loss of HDAC-inhibitory activity of TPX. On the other hand, TSA was a reversible inhibitor (29). To test whether the effect of CHAP1 is reversible or not, we incubated HDAC1 immobilized on the agarose beads with CHAP1, TSA, or TPX for 20 min, washed the beads thoroughly, and then analyzed the enzyme activity associated with the beads-conjugated HDAC1 in the absence of the inhibitors. As shown in Fig. 3, the activity of the enzyme that had been treated with CHAP1 was recovered to almost the initial level, whereas that of the enzyme treated with TPX B was not. These results clearly demonstrated that the replacement of the epoxyketone group with the hydroxamic acid converted the inhibitor to a reversible one.
Figure 3.
Reversibility of HDAC inhibition. (A) Effects on HDAC1. Beads-conjugated, affinity-purified HDAC1 was pretreated with 0.01% dimethyl sulfoxide (control), 100 nM TSA, 100 nM CHAP1, and 100 nM TPX B for 20 min. (B) Effects on HDAC6. Beads-conjugated, affinity-purified HDAC6 was pretreated with 0.01% dimethyl sulfoxide (control), 3 μM TSA, 3 μM CHAP1, and 100 μM TPX B for 60 min. The treated enzyme preparations were washed with drug-free HD buffer, and the residual enzyme activity was determined. The hatched and filled bars represent before and after removal of drugs, respectively.
Different Sensitivity of HDAC1 and HDAC6 to CHAP1.
Although a number of natural and synthetic compounds have been described as HDAC inhibitors, little is known about their target enzyme specificity. Class I deacetylases related to yeast Rpd3 includes HDAC1 (7), HDAC2 (12), HDAC3 (13), and HDAC8 (14), whereas class II related to Hda1 contains HDAC4 (15, 16), HDAC5 (16, 17), HDAC6 (16, 17), and HDAC7 (18). It has been reported that HDAC1 and HDAC2 are found in the same complex (12), whereas HDAC4 and HDAC5 are associated with HDAC3 (16). HDAC6, a particular isoform that contains two deacetylase domains, has not been described to associate with other known HDACs. We chose HDAC1 as a class I enzyme and HDAC4 and HDAC6 as class II deacetylases, and compared the effects of these inhibitors (Table 1). TSA inhibited all of the deacetylases tested to a similar extent, although HDAC4 was slightly resistant when compared with HDAC1 and HDAC6. TPX also strongly inhibited the enzyme activity of HDAC1 and HDAC4 at subnanomolar concentrations. Surprisingly, however, HDAC6 was highly resistant to TPX. A similar but smaller extent of resistance was also observed with CHAP1 for HDAC6. HDAC1 was most sensitive to CHAP1 among the HDACs tested, and the IC50 value for HDAC6 was ten times larger than that for HDAC1 (Table 1). These results suggest that the cyclic tetrapeptide structure is responsible, at least in part, for the weak inhibition of HDAC6 by TPX.
HDAC Inhibition by CHAPs Corresponding to Naturally Occurring Cyclic Tetrapeptide Antibiotics.
Chlamydocin, HC-toxin, Cyl-1, Cyl-2, and WF-3161 in addition to TPX A and B have been reported as cyclic tetrapeptide antibiotics containing Aoe (19). Chlamydocin and WF-3161 were described to have antifungal and antitumor activities (32, 33). HC-toxin, Cyl-1, and Cyl-2 are phytotoxic substances produced by phytopathogenic fungi, causing necrotic lesions on the leaves and sometimes serious reduction of crop yield (34–36). Of these compounds, HC-toxin and chlamydocin have been described to inhibit HDAC (20). We next synthesized CHAPs corresponding to all these naturally occurring cyclic tetrapeptide antibiotics, and their IC50 values for HDAC1 and HDAC6 were determined (Table 1). All CHAPs synthesized were potent inhibitors of HDAC1, effective at low nanomolar concentrations or subnanomolar concentrations. CHAP15 (chlamydocin-type) was the most potent inhibitor (IC50 = 0.44 nM). Again, HDAC6 was more than 10-fold resistant to these CHAPs. The difference in the IC50 between HDAC1 and HDAC6 varied with the structure of the cyclic tetrapeptide. In particular, the activity of CHAP15 to inhibit HDAC1 was 87-fold stronger than that needed to inhibit HDAC6. Interestingly, natural products containing Aoe (TPX A, TPX B, chlamydocin, and Cyl-2) showed strikingly large IC50 values for HDAC6 (360–40,000 nM), compared with those at subnanomolar levels for HDAC1 (Table 1). These results suggest that the epoxyketone group enhances the selectivity to inhibit HDAC1.
Reversible Inhibition of HDAC6 by TPX.
Because TPX has been shown to be a slow binding inhibitor (26), we tested whether the prolonged preincubation of HDAC6 with TPX results in the stronger inhibition. The inhibitory effect was almost unchanged after the 10-h incubation (data not shown). We next asked whether HDAC6 is irreversibly inhibited by TPX as was HDAC1. The affinity-purified, beads-conjugated HDAC6 was incubated with the high concentration of TSA (3 μM), CHAP1 (3 μM), or TPX B (100 μM) for 60 min, and then the residual enzyme activity was determined in the presence or absence of the drug (Fig. 3B). The activity of HDAC6 that had been treated with TPX B was recovered to levels similar to those with TSA and CHAP1 after removal of the drugs. These results suggest that TPX is a non-alkylating, reversible inhibitor for HDAC6.
CHAPs Inhibit HDAC Activity in Vivo and Affect Gene Expression and Cell Cycle Progression.
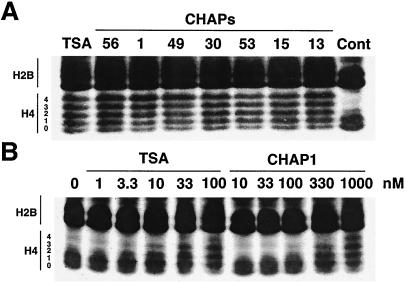
To examine the in vivo HDAC inhibition by CHAPs, we treated murine B16/BL6 melanoma cells with various concentrations of TSA and CHAPs, and the effects on histone acetylation were analyzed by AUT gel electrophoresis. AUT gel electrophoresis allows separation of each cellular histone (H1, H2A, H2B, H3, and H4) with the different extent of acetylation, because of slower migration rates of the acetylated species. Fig. 4 shows the profiles of histones H4 and H2B extracted from TSA- or CHAP-treated B16/BL6 melanoma cells. All CHAPs as well as TSA apparently induced the accumulation of highly acetylated histones, characterized by decreases in the most rapidly migrating bands of each histone species and appearance of additional slower migrating ones like a ladder on the AUT gel (Fig. 4A). These results clearly demonstrate that CHAPs inhibit HDAC not only in vitro but also in the cells. The minimal effective concentrations of TSA and CHAP1 for the increased histone acetylation were 3.3 nM and 100 nM, respectively (Fig. 4B).
Figure 4.
Effects of CHAPs on histone acetylation in vivo. (A) Acetylation by TSA and CHAPs. B16/BL6 cells were treated with TSA (1 μM), CHAP56 (1 μM), CHAP1 (10 μM), CHAP49 (1 μM), CHAP30 (1 μM), CHAP53 (1 μM), CHAP15 (1 μM), and CHAP13 (10 μM) for 6 h, and the level of histone acetylation was determined by AUT gel electrophoresis as described in Materials and Methods. (B) Dose-response. B16/BL6 cells were treated with the indicated concentrations of TSA and CHAP1 for 6 h, and the level of histone acetylation was determined.
The B16/BL6 melanoma cells are morphologically differentiated and express the MHC class-I molecule when treated with HDAC inhibitors (30). The effective concentrations of TSA and CHAP1 to induce the MHC class-I were 2.8 nM and 98 nM, respectively, which almost coincided with those required for the increased histone acetylation (Table 1). All CHAPs except CHAP13 (HC-toxin type) could induce the MHC class-I expression at the concentrations below 100 nM. The relatively weak in vivo activity of CHAP13 may be ascribed to low membrane permeability because of lack of hydrophobic aromatic amino acid residues in the cyclic tetrapeptide core.
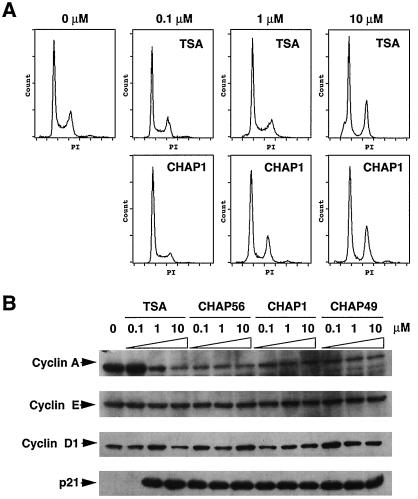
We finally analyzed the effects of CHAP compounds on the cell cycle and the expression of G1 cyclins and p21 in HeLa cells. Indeed, a marked increase in p21 expression and a decrease in cyclin A have previously been observed during the cell cycle arrest after HDAC inhibitor treatment (37–40). As shown in Fig. 5A, flow cytometric analysis showed that CHAP1 treatment caused an increase in the G1 cell population and a decrease in the S-phase cells at the concentration of 0.1 μM. Moreover, an arrest at both G1 and G2 phases was observed at the concentration of 1 μM or higher. TSA also induced an arrest of the cell cycle, but higher concentrations of the inhibitor were required to induce the cell cycle arrest in HeLa cells as described (40). Analysis of the cell cycle proteins showed that all CHAPs tested caused a down-regulation of cyclin A and a drastic induction of p21 in HeLa cells at the concentration of 0.1 μM, whereas expression of cyclin D1 and cyclin E was essentially unchanged (Fig. 5B). These effects were almost the same as those induced by TSA treatment, although the induction of p21 required 1 μM of TSA in this particular experiment, probably because of a partial degradation or inactivation of TSA in HeLa cells.
Figure 5.
Effects of CHAPs on the cell cycle and expression of cell cycle proteins. (A) HeLa cells were treated with various concentrations of TSA and CHAP1 for 24 h, and the isolated nuclei stained with propidium iodide (PI) were analyzed by flow cytometry. (B) HeLa cells were treated with the indicated concentrations of TSA, CHAP56, CHAP1, and CHAP49 for 24 h, and the amounts of cyclin A, cyclin E, cyclin D1, and p21Waf1 were determined by Western blotting.
Discussion
We showed that a series of synthetic compounds that were built from TSA and TPX-like cyclic tetrapeptide antibiotics reversibly inhibited HDAC1 at low nanomolar or subnanomolar concentrations. Crystallographic studies using the HDLP–TSA complex have shown that TSA binds by inserting its long aliphatic chain into the tube-like HDLP pocket and inhibits the enzyme activity by interacting with the zinc and active-site residues through its hydroxamic acid at one end of the aliphatic chain (24). The importance of the hydroxamic acid and the length of the aliphatic chain of CHAP1 in inhibiting HDAC1 support the idea that the side chain of Aoe in TPX is also a substrate mimic. Although the naturally occurring cyclic tetrapeptide antibiotics containing Aoe are potent in inhibiting HDAC in vitro, they show only very weak activity in the animal models (41). This weak activity is probably due to the chemical instability of the epoxyketone group in blood. CHAPs may circumvent this problem in the drug development because of the absence of the epoxyketone. In fact, the half-life of a CHAP compound {cyclo[Asu(NHOH)-D-Tyr(Me)-Ile-D-Pro]} in rat blood was about 50 min (unpublished results), whereas that of chlamydocin is 2.5 min (41).
Short-chain fatty acids such as n-butyrate are well known to inhibit HDAC. However, milimolar concentrations are required for inducing histone hyperacetylation in vivo or inhibiting HDAC in vitro (42). We showed that a TPX B analogue containing a carboxylic acid instead of the epoxyketone group (4) inhibited HDAC1 at the concentration of about 100 nM. Although this inhibitor is much weaker than CHAP1 (3), it is the most active carboxylic acid-containing inhibitor known to date. This finding indicates that the cyclic tetrapeptide structure confers the high affinity with HDAC1 onto the short-chain fatty acids, thereby potentiating the inhibitory activity. This high affinity is consistent with the model that the cyclic tetrapeptide with hydrophobic groups serves as a cap necessary for packing the inhibitor at the rim of the tube-like active-site pocket (24).
Accumulating data have suggested that each member of the HDAC family is a component of a distinct physical complex playing a distinct role in gene expression. For example, a large protein complex containing HDAC1 binds the E2F transcription factor via association with the retinoblastoma tumor suppressor protein (43, 44), whereas HDAC4 and HDAC5 associate specifically with the myocyte enhancer factor MEF2A and repress MEF2A-dependent transcription (45, 46). It is therefore likely that inhibition of a specific isoform leads to changes in transcription of a specific subset of genes. However, such an isoform-specific inhibitor has not yet been developed. TSA and suberoylanilide hydroxamic acid, simple analogues of acetylated lysine with small cap groups, may cause nonselective inhibition of class I and II HDACs. On the other hand, the cyclic tetrapeptide structure, which probably makes extensive contacts at the rim of the pocket and in the shallow grooves surrounding the pocket entrance, may mimic the substrate structure surrounding acetylated lysine (Fig. 1B). If so, it would be likely that the target enzyme specificity of CHAP can be modulated by changing amino acids in the cyclic tetrapeptide. In this study, we showed that CHAPs, as well as several cyclic tetrapeptide antibiotics containing Aoe, were less active against HDAC6 than HDAC1, supporting the idea that the cyclic tetrapeptide structure is responsible for the target enzyme specificity. HDAC6 is a unique isoform in which the catalytic domain is internally duplicated (16, 17). Furthermore, it is normally localized in the cytoplasm, and only a fraction of the protein relocalizes into the nucleus in response to stimuli for differentiation, suggesting that its natural substrates include non-histone acetylated proteins (47). The cyclic tetrapeptide structures of CHAPs synthesized in this study may be distant from the natural substrates for HDAC6.
The reversibility test showed that the effect of TPX on HDAC6, but not on HDAC1, was reversible (Fig. 3), suggesting that TPX does not alkylate HDAC6. This inability of TPX to alkylate HDAC6 is probably responsible for the higher resistance of HDAC6 to the Aoe-containing cyclic tetrapeptide antibiotics than to CHAPs (Table 1). Because the epoxide may be required for the alkylation of a catalytic pocket residue of HDAC1, presumably one of the conserved charge-relay histidines (Fig. 1B; ref. 26), it seems possible that a subtle difference in the position of the target residue results in the absence of the crosslink to HDAC6. The IC50 of these compounds for HDAC6 inhibition ranging between 100 nM and 100 μM were consistent with those for HDAC1 of non-chelating derivatives of CHAP1 such as compounds 4 and 6 (Fig. 2).
In summary, we showed potent and specific inhibition of HDAC1 by CHAPs. Our data suggest that the cyclic tetrapeptide portion affects both enzyme inhibitory potency and specificity, whereas the hydroxamic acid group acts as a potent enzyme-inhibiting group with in vivo stability better than the epoxyketone. These observations raise the possibility of developing the isoform-specific inhibitors based on CHAP. Because a marked structural diversity can be obtained by combinatorial synthesis of the cyclic tetrapeptide, CHAP is a promising framework to develop isoform-specific HDAC inhibitors. These specific HDAC inhibitors are clearly important for not only elucidating a downstream signaling step of each member of HDAC but also improving their therapeutical potential for transcription therapy and chemotherapy.
Supplementary Material
Acknowledgments
We thank Dr. E. Seto and Dr. E. Verdin for supplying HDAC cDNAs. We also thank Dr. A. Isogai for supplying Cyl-2, Dr. K. Sugita for TPX and chlamydocin, Dr. M. Nishiyama for helpful discussion, and A. Sasaki for her technical assistance. This work was supported in part by Core Research for Evolutional Science and Technology (CREST) Research Project, Japan Science and Technology Corporation, a special grant for Advanced Research on Cancer from the Ministry of Education, Culture and Science of Japan, Senri Life Science Foundation, and Takeda Science Foundation, Japan.
Abbreviations
- Aoe
(2S, 9S)-2-amino-9, 10-epoxy-8-oxodecanoic acid
- Asu
α-aminosuberic acid
- CHAP
cyclic hydroxamic-acid-containing peptide
- HDAC
histone deacetylase
- HDLP
histone deacetylase-like protein
- Pip
pipecolic acid
- TPX
trapoxin
- TSA
trichostatin A
- Tyr(Me)
O-methyltyrosine
- AUT
acid/urea/Triton
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.011405598.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.011405598
References
- 1.Grunstein M. Nature (London) 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 2.Struhl K. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 3.Kouzarides T. Curr Opin Genet Dev. 1999;9:40–48. doi: 10.1016/s0959-437x(99)80006-9. [DOI] [PubMed] [Google Scholar]
- 4.Brownell J E, Zhou J, Ranalli T, Kobayashi R, Edmondson D G, Roth S Y, Allis C D. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 5.Ogryzko V V, Schitz R L, Russanova V, Howard B H, Nakatani Y. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 6.Yang X J, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. Nature (London) 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 7.Taunton J, Hassig C A, Schreiber S L. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 8.Rundlett S E, Carmen A A, Kobayashi R, Bavykin S, Turner B M, Grunstein M. Proc Natl Acad Sci USA. 1996;93:14503–14508. doi: 10.1073/pnas.93.25.14503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Imai S, Armstrong C M, Kaeberlein M, Guarente L. Nature (London) 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 10.Kuo M H, Allis C D. BioEssays. 1998;20:615–626. doi: 10.1002/(SICI)1521-1878(199808)20:8<615::AID-BIES4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 11.Torchia J, Glass C, Rosenfeld M G. Curr Opin Cell Biol. 1998;10:373–383. doi: 10.1016/s0955-0674(98)80014-8. [DOI] [PubMed] [Google Scholar]
- 12.Laherty C D, Yang W M, Sun J M, Davie J R, Seto E, Eisenman R N. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- 13.Emiliani S, Fischle W, Van L C, Al A Y, Verdin E. Proc Natl Acad Sci USA. 1998;95:2795–2800. doi: 10.1073/pnas.95.6.2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu E, Chen Z, Fredrickson T, Zhu Y, Kirkpatrick R, Zhang G F, Johanson K, Sung C M, Liu R, Winkler J. J Biol Chem. 2000;275:15254–15264. doi: 10.1074/jbc.M908988199. [DOI] [PubMed] [Google Scholar]
- 15.Wang A H, Bertos N R, Vezmar M, Pelletier N, Crosato M, Heng H H, Th'ng J, Han J, Yang X J. Mol Cell Biol. 1999;19:7816–7827. doi: 10.1128/mcb.19.11.7816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grozinger C M, Hassig C A, Schreiber S L. Proc Natl Acad Sci USA. 1999;96:4868–4873. doi: 10.1073/pnas.96.9.4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Verdel A, Khochbin S. J Biol Chem. 1999;274:2440–2445. doi: 10.1074/jbc.274.4.2440. [DOI] [PubMed] [Google Scholar]
- 18.Kao H Y, Downes M, Ordentlich P, Evans R M. Genes Dev. 2000;14:55–66. [PMC free article] [PubMed] [Google Scholar]
- 19.Yoshida M, Horinouchi S, Beppu T. BioEssays. 1995;17:423–430. doi: 10.1002/bies.950170510. [DOI] [PubMed] [Google Scholar]
- 20.Brosch G, Ransom R, Lechner T, Walton J D, Loidl P. Plant Cell. 1995;7:1941–1950. doi: 10.1105/tpc.7.11.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marks P A, Richon V M, Rifkind R A. J Natl Cancer Inst. 2000;92:1210–1216. doi: 10.1093/jnci/92.15.1210. [DOI] [PubMed] [Google Scholar]
- 22.Nakajima H, Kim Y B, Terano H, Yoshida M, Horinouchi S. Exp Cell Res. 1998;241:126–133. doi: 10.1006/excr.1998.4027. [DOI] [PubMed] [Google Scholar]
- 23.Saito A, Yamashita T, Mariko Y, Nosaka Y, Tsuchiya K, Ando T, Suzuki T, Tsuruo T, Nakanishi O. Proc Natl Acad Sci USA. 1999;96:4592–4597. doi: 10.1073/pnas.96.8.4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Finnin M S, Donigian J R, Cohen A, Richon V M, Rifkind R A, Marks P A, Breslow R, Pavletich N P. Nature (London) 1999;401:188–193. doi: 10.1038/43710. [DOI] [PubMed] [Google Scholar]
- 25.Richon V M, Emiliani S, Verdin E, Webb Y, Breslow R, Rifkind R A, Marks P A. Proc Natl Acad Sci USA. 1998;95:3003–3007. doi: 10.1073/pnas.95.6.3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kijima M, Yoshida M, Sugita K, Horinouchi S, Beppu T. J Biol Chem. 1993;268:22429–22435. [PubMed] [Google Scholar]
- 27.Yang W M, Yao Y L, Sun J M, Davie J R, Seto E. J BiolChem. 1997;272:28001–28007. doi: 10.1074/jbc.272.44.28001. [DOI] [PubMed] [Google Scholar]
- 28.Fischle W, Emiliani S, Hendzel M J, Nagase T, Nomura N, Voelter W, Verdin E. J Biol Chem. 1999;274:11713–11720. doi: 10.1074/jbc.274.17.11713. [DOI] [PubMed] [Google Scholar]
- 29.Yoshida M, Kijima M, Akita M, Beppu T. J Biol Chem. 1990;265:17174–17179. [PubMed] [Google Scholar]
- 30.Komatsu Y, Hayashi H. J Antibiot. 1998;51:89–91. doi: 10.7164/antibiotics.51.89. [DOI] [PubMed] [Google Scholar]
- 31.Yoshida M, Beppu T. Exp Cell Res. 1988;177:122–131. doi: 10.1016/0014-4827(88)90030-4. [DOI] [PubMed] [Google Scholar]
- 32.Closse A, Hugenin R. Helv Chim Acta. 1974;57:533–545. doi: 10.1002/hlca.19740570306. [DOI] [PubMed] [Google Scholar]
- 33.Umehara K, Nakahara K, Kiyoto S, Iwami M, Okamoto M, Tanaka H, Kohsaka M, Aoki H, Imanaka H. J Antibiot. 1983;36:478–483. doi: 10.7164/antibiotics.36.478. [DOI] [PubMed] [Google Scholar]
- 34.Liesch J M, Sweeley C C, Staffeld G D, Anderson M S, Weber D J, Scheffer R P. Tetrahedron. 1982;38:45–48. [Google Scholar]
- 35.Hirota A, Suzuki A, Aizawa K, Tamura S. Agric Biol Chem. 1973;37:955–956. [Google Scholar]
- 36.Takayama S, Isogai A, Nakata M, Suzuki H, Suzuki A. Agric Biol Chem. 1984;48:839–842. [Google Scholar]
- 37.Archer S Y, Meng S, Shei A, Hodin R A. Proc Natl Acad Sci USA. 1998;95:6791–6796. doi: 10.1073/pnas.95.12.6791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vaziri C, Stice L, Faller D V. Cell Growth Differ. 1998;9:465–474. [PubMed] [Google Scholar]
- 39.Kim Y B, Lee K-H, Sugita K, Yoshida M, Horinouchi S. Oncogene. 1999;18:2461–2470. doi: 10.1038/sj.onc.1202564. [DOI] [PubMed] [Google Scholar]
- 40.Kim Y B, Ki S W, Yoshida M, Horinouchi S. J Antibiot. 2000;53:1191–1200. doi: 10.7164/antibiotics.53.1191. [DOI] [PubMed] [Google Scholar]
- 41.Stähelin H, Trippmacher A. Eur J Cancer. 1974;10:801–808. doi: 10.1016/0014-2964(74)90137-6. [DOI] [PubMed] [Google Scholar]
- 42.Cousens L S, Gallwitz D, Alberts B M. J Biol Chem. 1979;254:1716–1723. [PubMed] [Google Scholar]
- 43.Brehm A, Miska E A, McCance D J, Reid J L, Bannister A J, Kouzarides T. Nature (London) 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- 44.Magnaghi J L, Groisman R, Naguibneva I, Robin P, Lorain S, Le V J, Troalen F, Trouche D, Harel B A. Nature (London) 1998;391:601–605. doi: 10.1038/35410. [DOI] [PubMed] [Google Scholar]
- 45.Miska E A, Karlsson C, Langley E, Nielsen S J, Pines J, Kouzarides T. EMBO J. 1999;18:5099–5107. doi: 10.1093/emboj/18.18.5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lemercier C, Verdel A, Galloo B, Curtet S, Brocard M P, Khochbin S. J Biol Chem. 2000;275:15594–15599. doi: 10.1074/jbc.M908437199. [DOI] [PubMed] [Google Scholar]
- 47.Verdel A, Curtet S, Brocard M P, Rousseaux S, Lemercier C, Yoshida M, Khochbin S. Curr Biol. 2000;10:747–749. doi: 10.1016/s0960-9822(00)00542-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.