Abstract
RNA editing in higher plant chloroplasts involves C→U conversion at ∼30 specific sites. An in vitro system supporting accurate editing has been developed from tobacco chloroplasts. Mutational analysis of substrate mRNAs derived from tobacco chloroplast psbL and ndhB mRNAs confirmed the participation of cis-acting elements that had previously been identified in vivo. Competition analysis revealed the existence of site-specific trans-acting factors interacting with the corresponding upstream cis-elements. A chloroplast protein of 25 kDa was found to be specifically associated with the cis-element involved in psbL mRNA editing. Immunological analyses revealed that an additional factor, the chloroplast RNA-binding protein cp31, is also required for RNA editing at multiple sites. This combination of site-specific and common RNA-binding proteins recognizes editing sites in chloroplasts.
Keywords: chloroplast/in vitro system/RNA-binding protein/RNA editing/trans-acting factor
Introduction
Chloroplasts possess their own genome and a unique gene expression system that is regulated at the levels of transcription, RNA processing and translation, during development and in response to environmental cues (reviewed in Rochaix, 1992; Gruissem and Tonkyn, 1993; Mullet, 1993; Mayfield et al., 1996; Sugita and Sugiura, 1996). In chloroplasts of higher plants, many genes are initially transcribed as polycistronic precursors, which are subsequently processed into complex sets of overlapping transcripts including monocistronic mRNAs. During this process, some of the transcripts are also edited and/or spliced (Tanaka et al., 1987; Barkan, 1988; Westhoff and Herrmann, 1988; Neuhaus and Link, 1990; Sexton et al., 1990; Hird et al., 1991; Hoch et al., 1991). These RNA processing steps have been recognized as important regulatory steps in chloroplast gene expression (Deng and Gruissem, 1988; Barkan, 1989; Herrmann et al., 1992; Rochaix, 1992; Sugiura, 1992; Gruissem and Tonkyn, 1993; Mayfield et al., 1996; Sugita and Sugiura, 1996).
RNA editing is a process that alters nucleotide sequences of primary transcripts. It has been detected in divergent organisms including trypanosomes, slime mold, Acanthamoeba, mammals, viruses and land plants in which distinct editing mechanisms apparently operate (Benne, 1996; Simpson and Emeson, 1996; Smith et al., 1997). In land plants of all major lineages, RNA editing has been found both in chloroplast and mitochondrial transcripts. The major form of organellar editing involves C→U conversion, although rare U→C changes have also been detected (Covello and Gray, 1989; Gualberto et al., 1989; Hiesel et al., 1989, 1994; Hoch et al., 1991; Bonnard et al., 1992; Hanson et al., 1996; Maier et al., 1996; Yoshinaga et al., 1996, 1997; Freyer et al., 1997; Sugiura et al., 1998; Bock, 2000). Editing in plant organelles usually restores conserved amino acids, suggesting that it is an obligatory step in the biosynthesis of functional gene products (e.g. Bock et al., 1994; Maier et al., 1996). In chloroplasts, some transcripts undergo partial editing. In these cases, the extent of editing depends on environmental and/or developmental conditions, suggesting that editing plays a regulatory role in gene expression (Bock et al., 1993; Hirose et al., 1994, 1996, 1998; Hirose and Sugiura, 1997; Karcher and Bock, 1998). Although in land plants the chloroplast genome (∼150 genes) contains more genes than its mitochondrial counterpart (60–90 genes), the total number of edited residues is much smaller: ∼30 sites in chloroplasts (Maier et al., 1995; Wakasugi et al., 1996; Hirose et al., 1999) versus >400 sites in mitochondria (Giege and Brennicke, 1999). Using transgenic approaches in tobacco chloroplasts, the only available transformation system among plant organelles (Svab and Maliga, 1993), cis-acting elements important for the recognition of editing sites have been identified in psbL and ndhB mRNAs (Chaudhuri et al, 1995; Bock et al., 1996, 1997; Chaudhuri and Maliga, 1996, 1997; Herman and Bock, 1999). The involvement of extraplastidic site-specific trans-acting factors has also been suggested (Chaudhuri et al., 1995; Bock and Koop, 1997). Editing of the psbL mRNA in vivo creates the AUG initiation codon from ACG, resulting in the production of functional mRNA (Kudla et al., 1992). The signal directing the editing of this site is located within a 22-nucleotide-long sequence spanning the editing site (16 nt upstream and 6 nt downstream) (Chaudhuri and Maliga, 1996). In addition, an essential upstream sequence element and nucleotides critical for editing of the tobacco ndhB mRNA have also been identified (Bock et al., 1996, 1997). Recently, it has been suggested that the spacing between the upstream element and the ndhB editing site is crucial for substrate recognition (Hermann and Bock, 1999).
Here, we report the development of an accurate and efficient in vitro RNA editing system from isolated tobacco chloroplasts. Using this system, we first confirmed that the cis-acting elements, which were identified by in vivo analysis, are important for the editing of psbL and ndhB mRNAs. The involvement of site-specific trans-acting factors for each edited mRNA was determined by competition analysis. UV cross-linking experiments and immunological analysis suggested that several RNA-binding proteins, but not additional RNA factors, are involved in editing. A model for the molecular mechanism of RNA editing in chloroplasts is proposed based on our results.
Results
Development of an in vitro RNA editing system from tobacco chloroplasts
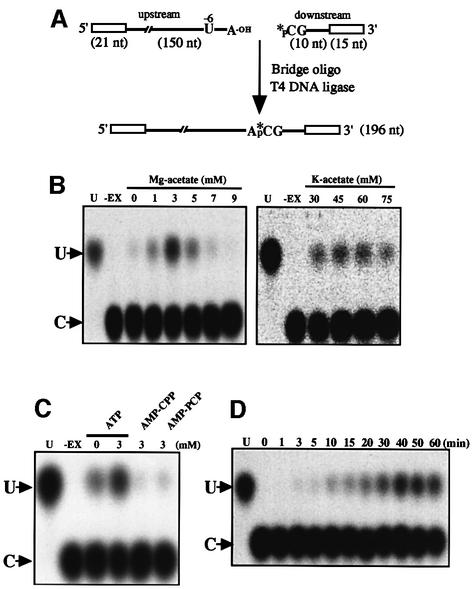
In order to dissect the biochemical mechanisms of RNA editing, we have developed an in vitro system supporting accurate editing from tobacco chloroplasts using the psbL mRNA as a model substrate. Site-specific labeling of the mRNA at the edited site led to the detection of the edited products. The upstream and downstream parts of the mRNA (with respect to the C residue to be edited) were synthesized separately (Figure 1A). The 5′ end of the downstream sequence, which constitutes the editing site, was labeled with 32P using polynucleotide kinase, and ligated to the upstream part with T4 DNA ligase in the presence of a complementary bridge DNA oligonucleotide (Moore and Sharp, 1992; Yang et al., 1995). The resulting psbL mRNA substrate was incubated with chloroplast S60 extract prepared as described below. After incubation at 28°C for the indicated times, RNA was isolated, digested with nuclease P1 and separated by cellulose thin layer chromatography (TLC) (Figure 1D).
Fig. 1. In vitro RNA editing system. (A) Synthesis of the psbL mRNA substrate labeled at the editing site. The upstream RNA (150 nt preceding the editing site with a 5′ extension of 21 nt sequence from pBluescript II) and the downstream RNA (10 nt downstream from the editing site with the 3′ extension of a 15 nt sequence from the KS primer) are ligated with T4 DNA ligase in the presence of a bridge DNA oligonucleotide. Extensions are represented by rectangular boxes. (B) [Mg+] and [K+] dependencies of the in vitro editing reaction of psbL mRNA. U, marker pU; –Ex, no chloroplast extract. pU migrates slower than pC as indicated by arrows. (C) Effect of ATP and its analogs (AMP-PCP and AMP-CPP) on the in vitro editing reaction of psbL mRNA. (D) Kinetic analysis of the in vitro editing reaction of psbL mRNA.
Intact chloroplasts were prepared from tobacco green leaves according to the procedure used previously to prepare an in vitro translation system (Hirose and Sugiura, 1996). The isolated chloroplasts were lysed with 0.2% Triton X-100 in the presence of 2 M KCl, followed by centrifugation at 60 000 g for 2 h. The resulting S60 fractions were extensively dialyzed. The TLC analysis (Figure 1B) clearly demonstrates that a fraction of the labeled C is converted to U after incubation with the chloroplast extract. The editing activity is influenced by the concentrations of magnesium (optimal at 3 mM) and potassium (optimal at 45 mM) (Figure 1B). The addition of ATP at 3 mM strikingly enhanced the editing activity, and non-hydrolyzable ATP analogs (AMP-CPP and AMP-PCP) did not substitute for ATP, suggesting that hydrolysis of ATP is required for the efficient in vitro editing reaction (Figure 1C). The C→U change proceeded linearly up to 40 min and then reached a plateau (Figure 1D). These results indicate that the α-phosphate group of the edited residue is retained during the editing reaction, suggesting that the C→U conversion in chloroplasts occurs by cytidine deamination and not by nucleotide substitution as suggested for plant mitochondria (Rajasekhar and Mulligan, 1993; Yu and Schuster, 1995).
The substrate mRNA possesses 5′ and 3′ extensions comprised of vector sequences, 22 and 15 nt, respectively (see Figure 1A). After the editing reaction, cDNA amplified from the isolated substrate mRNA by RT–PCR using primers complementary to the vector extensions (to avoid background from endogenous psbL mRNAs) yielded a single band. Sequence analysis of the cloned cDNAs revealed that C→U conversion occurred exclusively at the authentic editing site in eight cDNAs out of the 64 clones examined (56 cDNAs were not edited) (data not shown). These results confirm that the in vitro system supports accurate RNA editing of the psbL mRNA.
Cis-acting elements for the editing of psbL and ndhB mRNAs
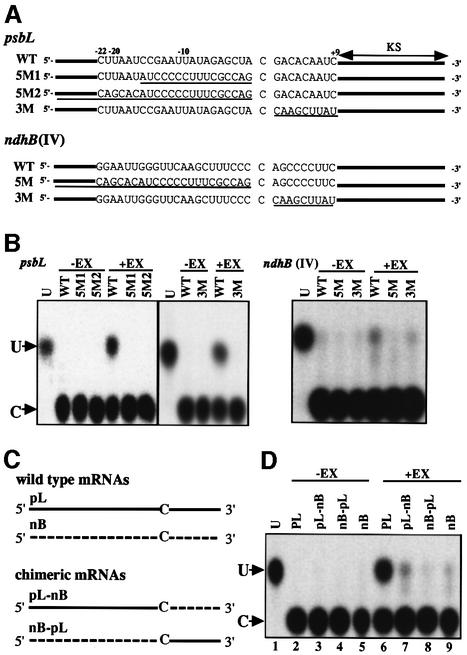
Cis-acting elements involved in editing have previously been identified using in vivo transplastome analysis of tobacco psbL and ndhB mRNAs (sites IV and V) (Bock et al., 1996; Chaudhuri and Maliga, 1996). In order to confirm whether our in vitro editing system depends on these cis-acting elements, mutational analysis of tobacco psbL and ndhB (site IV) mRNAs was performed (Figure 2A). Substitution of either the 16 nt element, the entire upstream or the 9 nt downstream sequence by a vector sequence abolished editing, indicating that both upstream and downstream regions are essential for editing of psbL mRNAs (Figure 2A and B). This result confirmed that obtained from the in vivo analysis using transplastomic tobacco plants (Chaudhuri and Maliga, 1996). A similar analysis of the ndhB mRNA showed that replacement of only the upstream, but not the downstream region inhibited editing completely. This result indicates the absolute requirement of the upstream, but not the downstream sequence for editing (Figure 2A and B), again confirming the in vivo result (Bock et al., 1996).
Fig. 2. In vitro editing using psbL and ndhB mRNAs with substitution mutations. (A) Sequences of wild-type (WT) and mutant (5M and 3M) psbL and ndhB mRNAs. Sequences of 22 nt upstream and 9 nt downstream of the respective editing sites are shown. Substitutions by a vector sequence are underlined. KS represents the sequence of the KS primer annealing region. WT sequences were from Shinozaki et al. (1986). (B) In vitro editing activity of mutant psbL and ndhB mRNAs. U, marker pU; +EX and –EX, with and without chloroplast extracts, respectively. (C) Schematic representation of chimeric mRNAs. (D) In vitro editing activity of chimeric mRNAs.
Next, to examine whether the cis-element is effective for another editing site, chimeric substrates between psbL and ndhB mRNAs were constructed: the pL-nB substrate contained the psbL upstream and the ndhB downstream sequences, and vice versa for nB-pL (Figure 2C). As shown in Figure 2D, the chimeric pL-nB mRNA was edited in vitro at a low level (∼10% of the wild-type psbL mRNA) (lane 7), whereas no editing product was observed for the nB-pL mRNA (lane 8). These results indicate that authentic combinations of the upstream and downstream sequences are required for efficient editing. It should be noted that the effects of the downstream sequence differ highly from those of an unrelated vector sequence (see Figure 2B, lane 3M).
Site-specific trans-acting factors are involved in RNA editing in chloroplasts
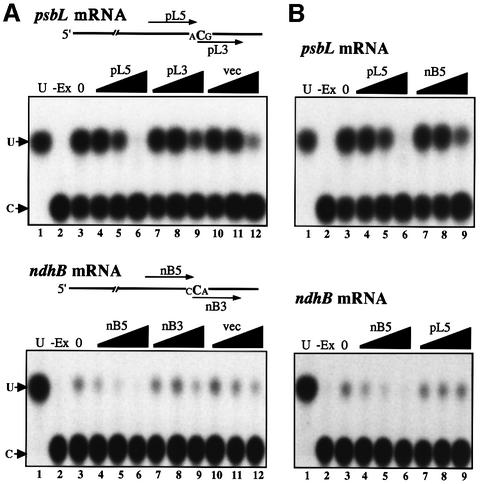
To examine the involvement of trans-acting factors in editing, competition analyses using 25 nt oligoribonucleotides corresponding to upstream and downstream regions of the editing sites in psbL or ndhB (site IV) mRNAs were carried out. As shown in Figure 3A, the editing of psbL and ndhB mRNAs was arrested by the addition of excess amounts of the upstream competitors pL5 and nB5, respectively (lanes 4–6), but not by the downstream competitors pL3 and nB3 (lanes 7–9) nor by an unrelated oligonucleotide derived from a vector sequence (lanes 10–12), although non-specific inhibition was observed in a 2000-fold excess of these competitors (lanes 9 and 12). These results strongly suggest the existence of a trans-acting factor(s) specifically interacting with the upstream region of each mRNA. The competitor RNAs were then exchanged. The editing of psbL mRNA was not arrested by an excess of nB5, the competitor for ndhB mRNA; similarly, the editing of ndhB was not inhibited by pL5 (Figure 3B, lanes 7–9), suggesting that the trans-acting factors are site specific. This result is consistent with observations using transplastomic tobacco plants where the introduction of additional copies of psbL and ndhD editing sites specifically reduced the editing efficiency of the corresponding sites in the endogenes as well as in transgenic mRNAs (Chaudhuri et al., 1995; Chaudhuri and Maliga, 1996).
Fig. 3. Competition analysis of in vitro RNA editing. (A) Increasing amounts of upstream (pL5 and nB5), downstream (pL3 and nB3) and vector (vec) oligoribonucleotides were added to in vitro editing reactions with psbL and ndhB mRNAs. pL5, pL3 and vec oligos of 1 µmol (lanes 4, 7 and 10), 10 µmol (lanes 5, 8 and 11) and 100 µmol (lanes 6, 9 and 12) were added. nB5, nB3 and vec oligos of 0.25 µmol (lanes 4, 7 and 10), 2.5 µmol (lanes 5, 8 and 11) and 25 µmol (lanes 6, 9 and 12) were added. U, authentic pU; –Ex, no chloroplast extract; 0, no competitor. (B) Analysis with heterologous competitors. nB5 (1, 10 and 100 µmol, lanes 7, 8 and 9, respectively) was added for psbL mRNA. pL5 (0.25, 2.5 and 25 µmol, lanes 7, 8 and 9, respectively) was used for ndhB mRNA.
Involvement of a 25 kDa protein in editing of psbL mRNA
The most intriguing question about trans-acting factors for RNA editing in chloroplasts is whether they contain an RNA component(s). To answer this question, the chloroplast extract was pre-treated with micrococcal nuclease. The editing activity did not decrease even though a 10 times higher concentration of the nuclease was applied than that used for the preparation of chloroplast in vitro translation systems (Hirose and Sugiura, 1996) (data not shown). Furthermore, attempts to detect RNA molecules interacting with psbL mRNAs by cross-linking in the presence of AMT (4′-aminomethyl-4,5′,8-trimethyl psoralen), which forms covalent adducts after irradiation with long-wavelength (365 nm) UV light, yielded negative results (data not shown). Although these experiments are indirect and preliminary, the trans-factor is likely to be a protein rather than an RNA.
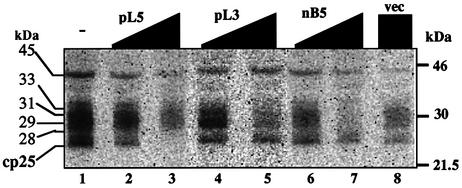
To detect proteins interacting with the upstream cis-element, UV cross-linking (254 nm) experiments were carried out using the psbL mRNA substrate labeled at the –6 residue with respect to the C to be edited (see Figure 1A). As shown in Figure 4, at least five chloroplast proteins ranging in size from 25 to 45 kDa were detected. Competition analysis revealed that binding of the 25 kDa protein (p25) to psbL mRNA was arrested by an excess amount of pL5 (lane 3), but not by pL3, nB5 or vec (lanes 5, 7 and 8). Similar experiments using the ndhB mRNA in which the –6 position was specifically labeled were carried out; however, binding of neither p25 nor any additional protein other than proteins ranging in size from 28 to 45 kDa was observed (data not shown). The characteristics of p25 binding to psbL mRNA corresponds to that of editing activity (see Figure 3), strongly suggesting that p25 is a trans-acting factor specific for the editing of psbL mRNA. On the other hand, the failure to detect any specific protein binding to the ndhB mRNA might be due to a lower amount of the factor in the chloroplast extract or co-migration in the gel with the 28 to 33 kDa bands.
Fig. 4. Detection of tobacco chloroplast proteins bound to the upstream region of psbL mRNA by UV cross-linking. The psbL mRNA with 32P-labeled U at position –6 was synthesized as in Figure 1A. The mRNA was incubated for 15 min at 28°C in the editing reaction mixture and then UV-irradiated (254 nm). The isolated RNA was digested with RNase A followed by separation of the cross-linked proteins by SDS–PAGE (lane 1). Increasing amounts (10 µmol, lanes 2, 4 and 6; 100 µmol, lanes 3, 5, 7 and 8) of competitor RNAs (see Figure 3A) were added. Protein size markers are shown at the right (Rainbow, Amersham).
A chloroplast RNA-binding protein, cp31, is a common factor for editing of psbL and ndhB mRNAs
Based on their sizes, the additional cross-linked proteins of 28–33 kDa are considered to be chloroplast ribonucleoproteins (cpRNPs: cp28, cp29A, cp29B, cp31 and cp33) previously isolated and characterized by our laboratory (Li and Sugiura, 1990; Ye et al., 1991). Each cpRNP is an abundant stromal protein that possesses two consensus-type RNA-binding domains (CS-RBD) and an N-terminal acidic domain (AD). These cpRNPs are associated with various chloroplast RNA species including mRNAs, pre-tRNAs and pre-rRNAs (Nakamura et al., 1999), suggesting that they are involved in RNA processing events and/or RNA stability/storage.
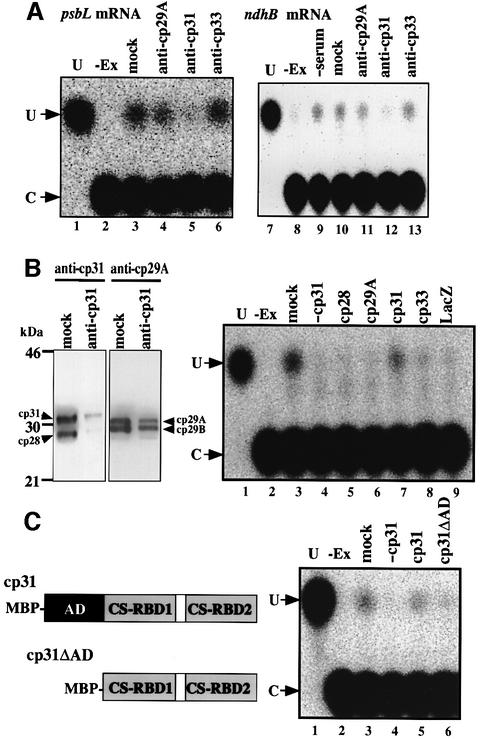
To verify the involvement of these cpRNPs in editing, we investigated the effect of antibodies against each cpRNP on the in vitro editing reactions. As shown in Figure 5A, the editing of both psbL and ndhB mRNAs was inhibited by the addition of anti-cp31, which can recognize cp28 as well as cp31 (lanes 5 and 12), while the addition of antibodies against other cpRNPs (anti-cp29A recognizes cp29A and B, while anti-cp33 recognizes only cp33) did not affect the editing activity (lanes 4, 6, 11 and 13). We then depleted cp31 from the chloroplast extract by immunoprecipitation with anti-cp31. As shown in Figure 5B (left panel), practically no cp31 (and no cp28) was present in the extract, whereas cp29A and B were still detected. Immunodepletion of cp31 from the editing extract resulted in the inhibition of psbL mRNA editing (Figure 5B, right panel, lane 4), and the addition of recombinant cp31 (0.2 µg), but not recombinant cp28, back into the cp31-depleted extract restored the editing activity (lane 7). These results indicate that cp31 is an additional essential factor involved in the editing of psbL and ndhB mRNAs, suggesting that cp31 is a common factor for RNA editing in tobacco chloroplasts. Furthermore, the addition of excess amounts (2 µg) of recombinant cp31 into the cp31-depleted extract and also into the untreated extract partially inhibited the editing of psbL mRNA (data not shown), suggesting that an appropriate concentration of cp31 is important for the editing activity.
Fig. 5. Involvement of a chloroplast ribonucleoprotein, cp31, in RNA editing. (A) Effect of antisera against tobacco chloroplast RNPs on the editing of psbL and ndhB mRNAs in vitro. (B) Immunodepletion of cp31 from the chloroplast extract (left panel) and the effect of recombinant RNPs (0.2 µg each) on editing of psbL mRNA using the cp31-depleted extract (right panel). Western blot patterns of the chloroplast extract with treatment with anti-cp31 (anti-cp31) or with pre-immune serum (mock). Detection was with anti-cp31 (left) and anti-cp29A (right) (note that anti-cp31 reacts with cp31 and 28, and anti-cp29A with cp29A and -B). (C) Effect of the AD of cp31 on editing of psbL mRNA. Structures of the recombinant cp31 and that lacking the AD (cp31ΔAD). The N-terminal extension is maltose-binding protein (MBP). cp31 and cp31ΔAD (0.2 µg each) were added to the cp31-depleted extract (lanes 5 and 6, respectively). U, authentic pU; –Ex, no chloroplast extract; mock, extracts treated with pre-immune serum; –cp31, extract treated with anti-cp31; lacZ, recombinant MBP-lacZ protein as a control.
CS-RBD1 and CS-RBD2 in cp28 and cp31 are highly conserved (85–88% identity), but their ADs are not (Li and Sugiura, 1990). Among all five cpRNPs, cp31 has the longest AD (64 amino acids). To investigate the function of the cp31 AD, recombinant cp31 lacking AD (cp31ΔAD) fused to maltose-binding protein (MBP) was prepared (Figure 5C). When cp31ΔAD was added to the cp31-depleted extract, editing was hardly detected (Figure 5C, right panel, lane 6), indicating that the AD is necessary for the function of cp31 in editing. Therefore, the AD may assist the assembly of the editing machinery.
Discussion
Development of a chloroplast in vitro RNA editing system
The RNA editing process of C→U conversion is found both in chloroplasts and mitochondria of most land plants. However, the molecular mechanism of editing in plant organella is not well understood. Two intriguing questions for the editing mechanism can be raised. (i) How are editing sites specifically recognized? (ii) What is the catalytic mechanism? In tobacco chloroplast transcripts, a total of 31 editing sites have been identified so far, and sequences surrounding these editing sites lack similarity except for the bias towards a pyrimidine residue at position –1 and an adenine residue at position +1 (Hirose et al., 1999). Therefore, it is possible that each editing site is recognized by site-specific factors. In vivo observations using transplatomic tobacco plants have already suggested the involvement of site-specific factors (Chaudhuri et al., 1995; Bock and Koop, 1997). However, chloroplast transformation techniques have their own limitations for biochemical analysis of editing processes. In plant mitochondria, an in vitro RNA editing system was described from wheat mitochondria (Araya et al., 1992). However, no further analysis has been reported.
In order to dissect the molecular mechanism of RNA editing in more detail, we have succeeded in developing a genuine in vitro system supporting accurate RNA editing from isolated tobacco chloroplasts. We utilized tobacco leaves at the defined stage (5–10 cm, grown in a growth chamber) previously selected for the preparation of our chloroplast in vitro translation system (Hirose and Sugiura, 1996). To assay editing activity, RNA substrates specifically labeled with 32P at the 5′ C, which undergoes editing, were prepared, leading to sensitive detection of the editing product without background. Detection of [5′-32P]UMP as the edited product strongly suggests that the catalytic mechanism of RNA editing in chloroplasts involves cytidine deamination as in the case of plant mitochondria (Rajasekhar and Mulligan, 1993; Yu and Schuster, 1995). The accuracy of this system was confirmed by sequencing of cDNA clones derived from the RNA substrate after an in vitro editing reaction. Among various variables for the preparation of chloroplast extracts and for the in vitro reactions, the magnesium concentration (sharp optimum at 3 mM) is one of the most important. Hydrolysis of ATP is likely to be required for efficient editing of chloroplast mRNAs (see Figure 1C). ATP is not required for the mechanism of catalysis by cytidine deaminase (Frick et al., 1989). The requirement for ATP has not been reported for the process of C→U editing of apoB mRNAs in mammals (Driscoll et al., 1989). It is therefore suggested that, in chloroplasts, ATP is utilized for editing site recognition or assembly of editing complexes rather than at the catalytic step.
Involvement of two distinct RNA-binding proteins in psbL mRNA editing
Our in vitro analysis using two different RNA editing sites revealed that site-specific trans-acting factors recognize upstream cis-acting elements of respective editing sites. In trypanosome mitochondria, editing sites are determined by sets of small RNAs, termed guide RNA (Blum et al., 1990). More than 100 small nucleolar RNAs (snoRNAs) interact with pre-rRNAs to define the sites of sugar methylation and pseudouridylation in the nucleolus of eukaryotic cells (Bachellerie et al., 1995; Tollervey, 1996; Smith and Steitz, 1997). Thus, trans-acting RNA molecules are widely utilized as guide RNAs for post-transcriptional modification of specific sites in many precursor transcripts from various organisms. However, plastid transformation tests failed to detect guide RNAs for psbL mRNA editing (Bock and Maliga, 1995). Our in vitro analyses using micrococcal nuclease treatment and psolaren cross-linking also failed to detect RNA factors involved in the RNA editing reaction. Instead, an RNA-binding protein of 25 kDa (cp25) appears to bind specifically to the cis-acting element of psbL mRNA. This result strongly suggests that a chloroplast-specific protein, but not an RNA factor, recognizes the cis-acting element and the C that undergoes conversion to a U residue.
cp31 belongs to a family of abundant chloroplast RNA-binding proteins with an N-terminal AD and two CS-RBDs (Li and Sugiura, 1990). It has been suggested to be involved in RNA processing, RNA storage and/or translation in chloroplasts (Nakamura et al., 1999). A spinach homolog of tobacco cp28 called 28RNP has been shown to be necessary for accurate 3′ end formation of chloroplast mRNAs in vitro (Schuster and Gruissem, 1991). Depletion of all five cpRNPs (cp28, 29A, 29B, 31 and 33) from tobacco chloroplast extracts did not affect the rate of 3′ processing of petD mRNA (Nakamura et al., 1999), whereas the addition of anti-cp31 (recognizing both cp28 and 31) abolished in vitro editing of both psbL and ndhB mRNAs (see Figure 5A), suggesting the involvement of either cp28 or cp31 in RNA editing. A set of experiments with depletion or addition of cp28 and cp31 to our extracts revealed that cp31, but not cp28, is required for editing. Additional experiments using the mutant cp31 lacking the AD have revealed that this domain is important for the function of cp31 in RNA editing. cp31 is a highly abundant stromal protein whose abundance per chloroplast is estimated to be higher than that of chloroplast ribosomes (T.Nakamura, M.Sugiura and M.Sugita, in preparation). On the other hand, the chloroplast transcripts possessing RNA editing sites are limited among total chloroplast RNAs, suggesting that cp31 is a multifunctional protein involved in several other post-transcriptional events as well as RNA editing. In mammalian nuclei, a set of heterogeneous nuclear ribonucleoproteins (hnRNPs) interacts with newly transcribed pre-mRNAs (Burd and Dreyfuss, 1994), and each hnRNP governs multiple pre-mRNA processing steps including RNA splicing, RNA stability and mRNA export to the cytoplasm. cp31 is thought to act as an hnRNP-like protein in chloroplasts for multiple post-transcriptional processes, including RNA editing. It is also possible that the other chloroplast RNA-binding proteins (cp28, 29A, 29B and/or 33) are involved in editing of mRNAs other than psbL and ndhB (site IV). Alternatively, cp31 may play a more indirect role in editing; namely, it may facilitate efficient editing by mediating the accessibility of editing sites to the editing machinery instead of being a part of the editing complex.
Mechanism of RNA editing in chloroplasts
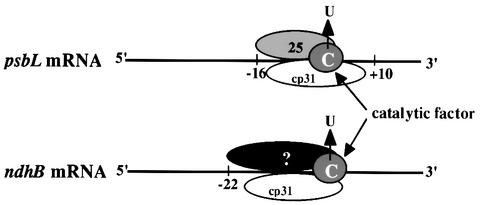
Based on our in vitro analyses, we propose a model for the mechanism of RNA editing in chloroplasts (Figure 6). A site-specific trans-acting factor interacts with the upstream cis-acting element and determines the cytidine residue to be edited. Tobacco p25 is a strong candidate for the site-specific trans-acting factor for editing of tobacco psbL mRNA. Another factor involved in editing in tobacco is an abundant RNA-binding protein, cp31, which is likely to act as a common factor for the editing of multiple sites. The catalytic factor, probably RNA cytidine deaminase, is then recruited to the editing site.
Fig. 6. Model for the mechanism of RNA editing in chloroplasts. A site-specific trans-acting factor (p25 for tobacco psbL mRNA) binds to the upstream cis-acting element of an editing site. One of the abundant chloroplast RNPs (cp31 in tobacco), probably a common factor, also binds close to every editing site. Complexes including these proteins recruit the catalytic factor of C→U conversion to the editing sites. Numbers represent cis-element positions defined by transplastomic experiments (Bock et al., 1996; Chaudhuri and Maliga, 1996).
In the case of apoB mRNA editing in mammalian nuclei, the catalytic factor apobec-1 is an RNA cytidine deaminase (Navaratnam et al., 1993, 1995; Teng et al., 1993), whose recruitment to the editing site requires a downstream ‘mooring sequence’ and a factor(s) interacting with this element (Mehta et al., 1996; Schock et al., 1996). Recently, the ‘mooring sequence’-binding factor has been purified and its cDNA cloned, leading to a model for the editing complex in which this factor (termed ACF) binds to the mooring sequence and docks apobec-1 to deaminate its target cytidine (Mehta and Driscoll, 1998; Mehta et al., 2000). In chloroplasts, the upstream sequence may correspond to the ‘mooring sequence’ of the apoB editing system. cp31 is unlikely to have RNA cytidine deaminase activity due to the lack of any known deaminase motif (Li and Sugiura, 1990), suggesting that an additional factor is required to catalyze cytidine deamination. Several genes for proteins containing the deaminase motif have been isolated from Arabidopsis; however, none of their products has been proven to be imported to chloroplasts (Faivre-Nitschke et al., 1999). Searching for additional genes encoding cytidine deaminase-like proteins in the Arabidopsis genome database could be one effective way of finding the catalytic factor.
Our in vitro experiments raised an interesting possibility that each editing site is recognized by a unique trans-acting factor. Since we previously detected at least 31 editing sites in tobacco chloroplasts, ∼31 different trans-factors would be needed, which are likely to be encoded in the nuclear genome and transported into chloroplasts. Alternatively, different combinations of abundant chloroplast RNA-binding proteins (e.g. cp31) and a limited number of trans-factors (e.g. p25) may affect the recognition of editing sites. Further in vitro analyses using other chloroplast mRNAs will provide the answers to the above questions. C→U RNA editing is also observed in plant mitochondria, where editing is 10 times more frequent. It has been reported that a mitochondrial editing site is not recognized by the chloroplast editing machinery (Sutton et al., 1995). However, the bias of the nucleotides at position –1 and position +1 at the mitochondrial editing sites is similar to that in chloroplasts, suggesting that the mechanism of editing is also similar. It will be interesting to determine how and to what extent, if any, editing machineries in both plant organelles overlap.
Materials and methods
Preparation of substrate mRNAs
The upstream RNA was synthesized using the T3 MEGASCRIPT RNA synthesis kit (Ambion), and the tobacco psbL clone containing the region from –150 to –1 and the 5′ extension of a 21 nt sequence from pBluescript II, which had been linearized with EcoT22I and blunted (see Figure 1A). The downstream RNA including the C to be edited and the 3′ extension of a 15 nt sequence from the KS primer was chemically synthesized by TaKaRa. The downstream RNA (300 pmol) was labeled with 32P at the 5′ end with T4 polynucleotide kinase and purified by passage through a Sephadex G25 spin column (Amersham Pharmacia Biotech). The labeled downstream RNA was mixed with the upstream RNA (100 pmol) and the bridge DNA oligonucleotide (200 pmol) (5′-CGGTATCGGATTGTGTCGTAGCTCTATAATTCGGATTAAG3′), and heated at 94°C for 3 min followed by cooling to room temperature for 3 h. Ligation was carried out by adding 1.4 U/µl T4 DNA ligase (TaKaRa) and incubating at 25°C overnight. The ligated RNA was purified by PAGE. The ndhB (site IV) mRNA substrate was prepared as above and includes 156 nt upstream and 10 nt downstream sequences with respect to the editing site. Chimeric mRNA substrates were prepared as above (Figure 2C, left panel).
Preparation of chloroplast S60 fractions and in vitro RNA editing reaction
Tobacco (Nicotiana tabacum var. Blight Yellow 4) was grown in a growth chamber (28°C, 16 h light and 8 h dark) for 4 weeks. Intact chloroplasts (∼500 µl packed volume) were prepared from ∼150 g of tobacco green leaves of 5–10 cm in length (stage III; Hirose and Sugiura, 1996), disrupted and extracted with 400 µl of extraction buffer [30 mM HEPES–KOH pH 7.7, 10 mM magnesium acetate, 2 M KCl and 2 mM dithiothreitol (DTT)] containing 0.2% Triton X-100, and centrifuged at 6000 g for 10 min. The transparent crude supernatant was subjected to ultra-centrifugation at 60 000 g for 2 h. The supernatant (S60 fraction) was recovered and dialyzed against dialysis buffer containing 30 mM HEPES–KOH pH 7.7, 3 mM magnesium acetate, 45 mM potassium acetate, 30 mM ammonium acetate and 10% glycerol for 12 h. All steps were carried out at 0–2°C. Reaction mixtures (25 µl) consisted of 30 mM HEPES–KOH pH 7.7, 3 mM magnesium acetate, 45 mM potassium acetate, 30 mM ammonium acetate, 3 mM ATP, 2 mM DTT, 1% polyethyleneglycol 6000, 5% glycerol, 230 U of RNase inhibitor (TaKaRa), 1× Proteinase inhibitor mixture (Complete™, Boehringer Mannheim), 50 fmol of [32P]mRNA substrate and chloroplast S60 fraction (250 µg of protein). The in vitro editing reaction was carried out at 28°C for 40 min or as indicated. The substrate mRNA was extracted with phenol–chloroform and precipitated with ethanol. The RNA was digested into 5′ mononucleotides by 1 µg of nuclease P1 in the presence of 50 mM ammonium acetate pH 4.8 at 37°C for 3 h. The resultant mononucleotides were separated on cellulose TLC plates using isopropanol:HCl:water (25:24:1) for the mobile phase. The separated [32P]mononucleotides were visualized and quantified by a Bioimaging Analyzer BAS2000 (Fuji Photo Film Co). Competitor RNA oligonucleotides of 25 nt were chemically synthesized by TaKaRa (see Figure 3A).
UV cross-linking experiments
The psbL mRNA substrate specifically labeled with 32P at U position –6 with respect to the editing site was synthesized as above (see Figure 1A). Ten femtomoles of 32P-labeled mRNA were incubated under in vitro editing conditions at 28°C for 15 min. The reaction mixture was irradiated with UV light (254 nm, 1.8 J/cm2) using Funacrosslinker (Funakoshi Co.) followed by digestion of the RNA with RNase A at 37°C for 15 min. The protein samples were precipitated with 50% acetone and dissolved in SDS–PAGE loading buffer followed by separation on 15% polyacrylamide gels containing 0.1% SDS. The separated proteins were visualized as described above.
Immunodepletion analysis
Recombinant cpRNPs with MBP and their antibodies were prepared and provided by Dr Masaru Ohta (Nakamura et al., 1999). Recombinant cp31 lacking the AD was prepared by the same procedure. Immunodepletion of cp31 (and cp28) was carried out essentially as described (Nakamura et al., 1999). The chloroplast extract (∼100 µg protein) was mixed with anti-cp31–protein A–Sepharose beads (10 mg) for 1 h, followed by dialysis against 2 l of the dialysis buffer used for S60 preparation. Depletion of cp31 was confirmed by western blot analysis using anti-cp31 and anti-cp29A (control). The recombinant cpRNPs were dialyzed against the buffer used for S60 preparation, and 0.2 µg of each cpRNP were added to the in vitro editing reaction.
Acknowledgments
Acknowledgements
We thank Dr M.Ohta for providing us with recombinant cpRNPs and their antibodies, Drs M.Sugita, T.Wakasugi and Y.Yukawa for their support and discussions, and Drs K.Tycowski and L.B.Weinstein for critical reading of the manuscript. The work was supported in part by a Grant-in-Aid from the Ministry of Education (Japan).
References
- Araya A., Domec,C., Begu,D. and Litvak,S. (1992) An in vitro system for the editing of ATP synthase subunit 9 mRNA using wheat mitochondrial extracts. Proc. Natl Acad. Sci. USA, 89, 1040–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachellerie J.-P., Michot,B., Nicoloso,M., Balakin,A., Ni,J. and Fournier,M.J. (1995) Antisense snoRNAs: a family of nucleolar RNAs with long complementaries to rRNA. Trends Biochem. Sci., 20, 261–264. [DOI] [PubMed] [Google Scholar]
- Barkan A. (1988) Proteins encoded by a complex chloroplast transcription unit are each translated from both monocistronic and polycistronic mRNAs. EMBO J., 7, 2637–2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A. (1989) Tissue-dependent plastid RNA splicing in maize: transcripts from four plastid genes are predominantly unspliced in leaf meristems and roots. Plant Cell, 1, 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benne R. (1996) RNA editing: how a message is changed. Curr. Opin. Genet. Dev., 6, 221–231. [DOI] [PubMed] [Google Scholar]
- Blum B., Bakalara,N. and Simpson,L. (1990) A model for RNA editing in kinetoplastid mitochondria: ‘Guide’ RNA molecules transcribed from maxicircle DNA provide the edited information. Cell, 60, 189–198. [DOI] [PubMed] [Google Scholar]
- Bock R. (2000) Sense from nonsense: how the genetic information of chloroplasts is altered by RNA editing. Biochimie, 82, 549–557. [DOI] [PubMed] [Google Scholar]
- Bock R. and Koop,H.U. (1997) Extraplastidic site-specific factors mediate RNA editing in chloroplasts. EMBO J., 16, 3282–3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock R. and Maliga,P. (1995) In vivo testing of a tobacco plastid DNA segment for guide RNA function in psbL editing. Mol. Gen. Genet., 247, 439–443. [DOI] [PubMed] [Google Scholar]
- Bock R., Hagemann,R., Kössel,H. and Kudla,J. (1993) Tissue- and stage-specific modulation of RNA editing of the psbF and psbL transcript from spinach plastids—a new regulatory mechanism? Mol. Gen. Genet., 240, 238–244. [DOI] [PubMed] [Google Scholar]
- Bock R., Kössel,H. and Maliga,P. (1994) Introduction of a heterologous editing site into the tobacco plastid genome: the lack of RNA editing leads to a mutant phenotype. EMBO J., 13, 4623–4628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock R., Hermann,M. and Kössel,H. (1996) In vivo dissection of cis-acting determinants for plastid RNA editing. EMBO J., 15, 5052–5059. [PMC free article] [PubMed] [Google Scholar]
- Bock R., Hermann,M. and Fuchs,M. (1997) Identification of critical nucleotide positions for plastid RNA editing site recognition. RNA, 3, 1194–1200. [PMC free article] [PubMed] [Google Scholar]
- Bonnard G., Gualberto,J.M., Lamattina,L. and Grienenberger,J.M. (1992) RNA editing in plant mitochondria. Crit. Rev. Plant Sci., 10, 503–524. [Google Scholar]
- Burd C.G. and Dreyfuss,G. (1994) Conserved structures and diversity of functions of RNA-binding proteins. Science, 265, 615–621. [DOI] [PubMed] [Google Scholar]
- Chaudhuri S. and Maliga,P. (1996) Sequences directing C to U editing of the plastid psbL mRNA are located within a 22 nucleotide segment spanning the editing site. EMBO J., 15, 5958–5964. [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri S. and Maliga,P. (1997) New insights into plastid RNA editing. Trends Plant Sci., 2, 5–6. [Google Scholar]
- Chaudhuri S., Carrer,H. and Maliga,P. (1995) Site-specific factor involved in the editing of the psbL mRNA in tobacco plastids. EMBO J., 14, 2951–2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covello P.S. and Gray,M.W. (1989) RNA editing in plant mitochondria. Nature, 341, 662–666. [DOI] [PubMed] [Google Scholar]
- Deng X.W. and Gruissem,W. (1988) Constitutive transcription and regulation of gene expression in non-photosynthetic plastids of higher plants. EMBO J., 7, 3301–3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll D.M., Wynne,J.K., Wallis,S.C. and Scott,J. (1989) An in vitro system for the editing of apolipoprotein B mRNA. Cell, 58, 519–525. [DOI] [PubMed] [Google Scholar]
- Faivre-Nitschke S.E., Grienenberger,J.M. and Gualberto,J.M. (1999) A prokaryotic-type cytidine deaminase from Arabidopsis thaliana; gene expression and functional characterization. Eur. J. Biochem., 263, 896–903. [DOI] [PubMed] [Google Scholar]
- Freyer R., Kiefer-Meyer,M.-C. and Kössel,H. (1997) Occurrence of plastid RNA editing in all major lineages of land plants. Proc. Natl Acad. Sci. USA, 94, 6285–6290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frick L., Yang,C., Marquez,V.E. and Wolfenden,R. (1989) Binding of pyrimidin-2-one ribonucleoside by cytidine deaminase as the transition-state analogue 3,4-dihydrouridine and the contribution of the 4-hydroxyl group to its binding affinity. Biochemistry, 28, 9423–9430. [DOI] [PubMed] [Google Scholar]
- Giege P. and Brennicke,A. (1999) RNA editing in Arabidopsis mitochondria effects 441 C to U changes in ORFs. Proc. Natl Acad. Sci. USA, 96, 15324–15329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruissem W. and Tonkyn,J.C. (1993) Control mechanisms of plastid gene expression. Crit. Rev. Plant Sci., 12, 19–55. [Google Scholar]
- Gualberto J.M., Lamattina,L., Bonnard,G., Weil,J.H. and Grienenberger,J.M. (1989) RNA editing in wheat mitochondria results in the conservation of protein sequences. Nature, 341, 660–662. [DOI] [PubMed] [Google Scholar]
- Hanson M.R., Sutton,C.A. and Lu,B. (1996) Plant organelle gene expression: altered by RNA editing. Trends Plant Sci., 1, 57–64. [Google Scholar]
- Hermann M. and Bock,R. (1999) Transfer of plastid RNA-editing activity to novel sites suggests a critical role for spacing in editing-site recognition. Proc. Natl Acad. Sci. USA, 96, 4856–4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann R.G., Westhoff,P. and Link,G. (1992) Biogenesis of plastids in higher plants. In Herrmann,R.G. (ed), Cell Organelles. Advances in Plant Gene Research. Springer-Verlag KG, Vienna, Austria, Vol. VI, pp. 276–349.
- Hiesel R., Wissinger,B., Schuster,W. and Brennicke,A. (1989) RNA editing in plant mitochondria. Science, 246, 1632–1634. [DOI] [PubMed] [Google Scholar]
- Hiesel R., Combettes,B. and Brennicke,A. (1994) Evidence for RNA editing in mitochondria of all major groups of land plants except the Bryophyta. Proc. Natl Acad. Sci. USA, 91, 629–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hird S.M., Webber,A.N., Wilson,R.J., Dyer,T.A. and Gray,J.C. (1991) Differential expression of the psbB and psbH genes encoding the 47 kDa chlorophyll a-protein and the 10 kDa phosphoprotein of photosystem II during chloroplast development in wheat. Curr. Genet., 19, 199–206. [DOI] [PubMed] [Google Scholar]
- Hirose T. and Sugiura,M. (1996) Cis-acting elements and trans-acting factors for accurate translation of chloroplast psbA mRNAs: development of an in vitro translation system from tobacco chloroplasts. EMBO J., 15, 1687–1695. [PMC free article] [PubMed] [Google Scholar]
- Hirose T. and Sugiura,M. (1997) Both RNA editing and RNA cleavage are required for translation of tobacco chloroplast ndhD mRNA: a possible regulatory mechanism for the expression of a chloroplast operon consisting of functionally unrelated genes. EMBO J., 16, 6804–6811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose T., Wakasugi,T., Sugiura,M. and Kössel,H. (1994) RNA editing in tobacco petB mRNAs occurs both in chloroplasts and non-photosynthetic proplastids. Plant Mol. Biol., 26, 509–513. [DOI] [PubMed] [Google Scholar]
- Hirose T., Fan,H., Suzuki,J.Y., Wakasugi,T., Tsudzuki,T., Kössel,H. and Sugiura,M. (1996) Occurrence of silent RNA editing in chloroplasts: its species specificity and the influence of environmental and developmental conditions. Plant Mol. Biol., 30, 667–672. [DOI] [PubMed] [Google Scholar]
- Hirose T., Kusumegi,T. and Sugiura,M. (1998) Translation of tobacco chloroplast rps14 mRNA depends on a Shine–Dalgarno-like sequence in the 5′-untranslated region but not on internal RNA editing in the coding region. FEBS Lett., 430, 257–260. [DOI] [PubMed] [Google Scholar]
- Hirose T., Kusumegi,T., Tsudzuki,T. and Sugiura,M. (1999) RNA editing sites in tobacco chloroplast transcripts: editing as a possible regulator of chloroplast RNA polymerase activity. Mol. Gen. Genet., 262, 462–467. [DOI] [PubMed] [Google Scholar]
- Hoch B., Maier,R.M., Appel,K., Igloi,G.L. and Kössel,H. (1991) Editing of a chloroplast mRNA by creation of an initiation codon. Nature, 353, 178–180. [DOI] [PubMed] [Google Scholar]
- Karcher D. and Bock,R. (1998) Site-selective inhibition of plastid RNA editing by heat shock and antibiotics: a role for plastid translation in RNA editing. Nucleic Acids Res., 26, 1185–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla J., Igloi,G.L., Metzlaff,M., Hagemann,R. and Kössel,H. (1992) RNA editing in tobacco chloroplasts leads to the formation of a translatable psbL mRNA by a C to U substitution within the intiation codon. EMBO J., 11, 1099–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. and Sugiura,M. (1990) Three distinct ribonucleoproteins from tobacco chloroplasts: each contains a unique amino terminal acidic domain and two ribonucleoprotein consensus motifs. EMBO J., 9, 3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier R.M., Neckermann,K., Igloi,G.L. and Kössel,H. (1995) Complete sequence of the maize chloroplast genome: gene content, hotspots of divergence and fine tuning of genetic information by transcript editing. J. Mol. Biol., 251, 614–628. [DOI] [PubMed] [Google Scholar]
- Maier R.M., Zeltz,P., Kössel,H., Bonnard,G., Gualberto,J.M. and Grienenberger,J.M. (1996) RNA editing in plant mitochondria and chloroplasts. Plant Mol. Biol., 32, 343–365. [DOI] [PubMed] [Google Scholar]
- Mayfield S.P.Y., Yohn,C.B., Cohen,A. and Danon,A. (1996) Regulation of chlorolast gene expression. Annu. Rev. Plant Physiol. Plant Mol. Biol., 46, 147–166. [Google Scholar]
- Mehta A. and Driscoll,D.M. (1998) A sequence-specific RNA-binding protein complements apobec-1 to edit apolipoprotein B mRNA. Mol. Cell. Biol., 18, 4426–4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta A., Banerjee,S. and Driscoll,D.M. (1996) Apobec-1 interacts with a 65-kDa complementing protein to edit apolipoprotein-B mRNA in vitro. J. Biol. Chem., 271, 28294–28299. [DOI] [PubMed] [Google Scholar]
- Mehta A., Kinter,M.T., Sherman,N.E. and Driscoll,D.M. (2000) Molecular cloning of apobec-1 complementation factor, a novel RNA-binding protein involved in the editing of apolipoprotein B mRNA. Mol. Cell. Biol., 20, 1846–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore M.J. and Sharp,P.A. (1992) Site-specific modification of pre-mRNA: the 2′ hydroxyl groups at the splice sites. Science, 256, 992–997. [DOI] [PubMed] [Google Scholar]
- Mullet J.E. (1993) Dynamic regulation of chloroplast transcription. Plant Physiol., 103, 309–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T., Ohta,M., Sugiura,M. and Sugita,M. (1999) Chloroplast ribonucleoproteins are associated with both mRNAs and intron-containing precursor tRNAs. FEBS Lett., 460, 437–441. [DOI] [PubMed] [Google Scholar]
- Navaratnam N., Morrison,J.R., Bhattacharya,S., Patel,D., Funahashi,T., Giannoni,F., Teng,B.B., Davidson,N.O. and Scott,J. (1993) The p27 catalytic subunit of the apolipoprotein B mRNA editing enzyme is a cytidine deaminase. J. Biol. Chem., 268, 20709–20712. [PubMed] [Google Scholar]
- Navaratnam N., Bhattacharya,S., Fujino,T., Patel,D., Jarmuz,A.L. and Scott,J. (1995) Evolutionary origins of apoB mRNA editing: catalysis by a cytidine deaminase that has acquired a novel RNA-binding motif at its active site. Cell, 81, 187–195. [DOI] [PubMed] [Google Scholar]
- Neuhaus H. and Link,G. (1990) The chloroplast psbK operon from mustard (Sinapis alba L.): multiple transcripts during seedling development and evidence for divergent overlapping transcription. Curr. Genet., 18, 377–383. [DOI] [PubMed] [Google Scholar]
- Rajasekhar V.K. and Mulligan,R.M. (1993) RNA editing in plant mitochondria: α-phosphate is retained during C-to-U conversion in mRNAs. Plant Cell, 5, 1843–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochaix J.D. (1992) Post-transcriptional steps in the expression of chloroplast genes. Annu. Rev. Cell Biol., 8, 1–28. [DOI] [PubMed] [Google Scholar]
- Schock D., Kuo,S.R., Steinburg,M.F., Bolognino,M., Sparks,J.D., Sparks,C.E. and Smith,H.C. (1996) An auxiliary factor containing a 240-kDa protein complex is involved in apolipoprotein B RNA editing. Proc. Natl Acad. Sci. USA, 93, 1097–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster G. and Gruissem,W. (1991) Chloroplast mRNA 3′ end processing requires a nuclear-encoded RNA-binding protein. EMBO J., 10, 1493–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton T.B., Christopher,D.A. and Mullet,J.E. (1990) Light-induced switch in barley psbD-psbC promoter utilization: novel mechanism regulating chloroplast gene expression. EMBO J., 9, 4485–4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K. et al. (1986) The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. EMBO J., 5, 2043–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson L. and Emeson,R.B. (1996) RNA editing. Annu. Rev. Neurosci., 19, 27–52. [DOI] [PubMed] [Google Scholar]
- Smith C.M. and Steitz,J.A. (1997) Sno storm in the nucleolus: new roles for myriad small RNPs. Cell, 89, 669–672. [DOI] [PubMed] [Google Scholar]
- Smith H.C., Gott,J.M. and Hanson,M.R. (1997) A guide to RNA editing. RNA, 3, 1105–1123. [PMC free article] [PubMed] [Google Scholar]
- Sugita M. and Sugiura,M. (1996) Regulation of gene expression in chloroplasts of higher plants. Plant Mol. Biol., 32, 315–326. [DOI] [PubMed] [Google Scholar]
- Sugiura M. (1992) The chloroplast genome. Plant Mol. Biol., 19, 149–168. [DOI] [PubMed] [Google Scholar]
- Sugiura M., Hirose,T. and Sugita,M. (1998) Evolution and mechanism of translation in chloroplasts. Annu. Rev. Genet., 32, 437–459. [DOI] [PubMed] [Google Scholar]
- Sutton C.A., Zoubenko,O.V., Hanson,M.R. and Maliga,P. (1995) A plant mitochondrial sequence transcribed in transgenic tobacco chloroplasts is not edited. Mol. Cell. Biol., 15, 1377–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svab Z. and Maliga,P. (1993) High frequency plastid transformation in tobacco by selection for a chimeric aadA gene. Proc. Natl Acad. Sci. USA, 90, 913–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M., Obokata,J., Chunwongse,J., Shinozaki,K. and Sugiura,M. (1987) Rapid splicing and stepwise processing of a transcript from the psbB operon in tobacco chloroplasts: determination of the intron sites in petB and petD. Mol. Gen. Genet., 209, 427–431. [DOI] [PubMed] [Google Scholar]
- Teng B.B., Burant,C.F. and Davidson,N.O. (1993) Molecular cloning of an apolipoprotein B messenger RNA editing protein. Science, 260, 1816–1819. [DOI] [PubMed] [Google Scholar]
- Tollervey D. (1996) Small nucleolar RNAs guide ribosomal RNA methylation. Science, 273, 1056–1057. [DOI] [PubMed] [Google Scholar]
- Wakasugi T., Hirose,T., Horihata,M., Tsudzuki,T., Kössel,H. and Sugiura,M. (1996) Creation of a novel protein-coding region at the RNA level in black pine chloroplasts: the pattern of RNA editing in the gymnosperm chloroplast is different from that in angiosperms. Proc. Natl Acad. Sci. USA, 93, 8766–8770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhoff P. and Herrmann,R.G. (1988) Complex RNA maturation in chloroplasts: the psbB operon from spinach. Eur. J. Biochem., 171, 551–564. [DOI] [PubMed] [Google Scholar]
- Yang J.H., Sklar,P., Axel,R. and Maniatis,T. (1995) Editing of glutamate receptor subunit B pre-mRNA in vitro by site-specific deamination of adenosine. Nature, 374, 77–81. [DOI] [PubMed] [Google Scholar]
- Ye L., Li,Y., Fukami-Kobayashi,K., Go,M., Konishi,T., Watanabe,A. and Sugiura,M. (1991) Diversity of a ribonucleoprotein family in tobacco chloroplasts: two new chloroplast ribonucleoproteins and a phylogenetic tree of ten chloroplast RNA-binding domains. Nucleic Acids Res., 19, 6485–6490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinaga K., Iinuma,H., Masuzawa,T. and Ueda,K. (1996) Extensive RNA editing of U to C in addition to C to U substitution in the rbcL transcripts of hornwort chloroplasts and the origin of RNA editing in green plants. Nucleic Acids Res., 24, 1008–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinaga K., Kakehi,T., Shima,Y., Iinuma,H., Masuzawa,T. and Ueno,M. (1997) Extensive RNA editing and possible double-stranded structures determining editing sites in the atpB transcripts of hornwort chloroplasts. Nucleic Acids Res., 25, 4830–4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W. and Schuster,W. (1995) Evidence for a site-specific cytidine deaminase reaction involved in C to U RNA editing of plant mitochondria. J. Biol. Chem., 270, 18227–18233. [DOI] [PubMed] [Google Scholar]