Abstract
Outer surface protein C (OspC) is a major antigen on the surface of the Lyme disease spirochete, Borrelia burgdorferi, when it is being transmitted to humans. Crystal structures of OspC have been determined for strains HB19 and B31 to 1.8 and 2.5 Å resolution, respectively. The three-dimensional structure is predominantly helical. This is in contrast to the structure of OspA, a major surface protein mainly present when spirochetes are residing in the midgut of unfed ticks, which is mostly β-sheet. The surface of OspC that would project away from the spirochete’s membrane has a region of strong negative electrostatic potential which may be involved in binding to positively charged host ligands. This feature is present only on OspCs from strains known to cause invasive human disease.
Keywords: Lyme disease/OspC/3D structure/X-ray diffraction
Introduction
Lyme disease is the most common vector-borne disease in the United States and Europe (Barbour and Fish, 1993). It is a progressive multisystem disorder which begins at the site of a tick bite, producing a primary infection that can spread to secondary sites early in infection. Secondary sites may include the nervous system, heart and joints (van der Linde et al., 1990; Steere, 1991; Hansen and Lebech, 1992). The causative agent Borrelia burgdorferi induces a strong humoral response against endoflagellar protein, p41, and a protein constituent of the protoplasmic cylinder, p93, both of which are enveloped within the outer membrane, and some outer surface lipoproteins (Osps), which are major membrane components (Burgdorfer et al., 1982; Craft et al., 1986; Schwan et al., 1995; Schwan and Piesman, 2000). Experimental OspA and OspC vaccines have limited utility since they are usually only effective against challenge by the same strain and not by heterologous strains (Fikrig et al., 1990, 1992; Simon et al., 1991; Golde et al., 1995; Gilmore et al., 1996; Probert et al., 1997).
Borrelia burgdorferi expresses OspA but not OspC when residing in the midgut of unfed ticks. However, when the tick starts feeding on mammals, OspC synthesis is induced and OspA is repressed (Schwan et al., 1995; Stevenson et al., 1995). The switch is in part due to the change in temperature; OspC is induced at 32–37°C, but not at 24°C, and this upregulation is at the transcriptional and translational levels (Tilly et al., 1997; Ramamoorthy and Philipp, 1998). Evidence suggests co-regulation of these two genes at the mRNA level (Jonsson and Bergstrom, 1995). Clearly, to survive in both hosts, spirochetes have evolved mechanisms for sensing the different host environments and responding accordingly.
The ospC gene is located on a 27 kb circular plasmid and encodes a lipoprotein of 22–23 kDa (Marconi et al., 1993; Sadziene et al., 1993). The protein is initially synthesized with an 18-amino-acid-long signal sequence which is removed during processing and lipidation at the amino proximal Cys residue. OspC proteins are highly polymorphic and this variability extends even to strains collected from a single geographical area. For example, alleles of OspC collected from a single site on Shelter Island, NY, could be clustered into 19 major groups or types (A–S) based on DNA sequence homology (Wang et al., 1999). Sequence variation within a major group is <1% but ∼15% across the major groups. Variation within a local population is comparable to the variation of similar size samples collected from the entire species. This variability has consequences in the development of OspC-based seradiagnostic antigens and anti-OspC vaccines. Of the 19 major groups, only four (A, B, I and K) contain invasive clones and cause infections of skin and extracutaneous sites, while the others are non-human pathogens or infect only the skin (Seinost et al., 1999). However, the biological function of OspC is not known, and the relationship between OspC type and pathogenicity is not understood. In order to develop an effective OspC-based vaccine, it is important to know representative three-dimensional structures of at least a few OspCs, especially those from the invasive strains. This information could be useful for rational design of an OspC-based recombinant vaccine. Here we report the crystal structure of OspC from strain HB19, a member of invasive group I, and compare it with the crystal structure of OspC from strain B31 of group A.
Results and discussion
Structure determination
The N- and C-truncated OspCs (residues 38–201) of strains HB19 and B31 of B.burgdorferi were cloned and expressed as described in Materials and methods. The, crystal structure of HB19 was solved using 2.8 Å resolution multiwavelength anomalous diffraction (MAD) data phasing with selenomethionine (SeMet)-labeled protein. Since there are four molecules per asymmetric unit, the MAD phases were further improved by non-crystallographic symmetry (NCS) averaging and density modification. The model building and initial refinements were completed with this improved set of phases. Further refinements were carried out with single wavelength 1.8 Å resolution data from a second crystal form of SeMet-labeled protein after orienting and positioning the molecule in the new unit cell by the molecular replacement method. In the final refinement cycle the NCS restraint was completely released. The model is complete except for the two N-terminal residues for which the electron density is very weak in all four monomers. The final model consists of 648 residues, 632 water molecules and six zinc ions. The crystallographic R-factor is 0.21 for 48 910 reflections in the resolution range 50–1.8 Å. The root mean square displacements (r.m.s.ds) between the NCS-related monomers are <1 Å for 162 Cα atoms.
The crystal structure of B31 was determined by the molecular replacement method using the HB19 molecule as a search model. The crystallographic R-factor for 22 087 reflections in the resolution range 50–2.5 is 0.23. The final model consists of 648 residues, 112 water molecules and one magnesium ion. As in the HB19 structure, the two N-terminal residues are not included in the model because of poor electron density.
Description of the structure
The OspC monomer is almost all helical, with four long helices plus a short fifth helix. It resembles a classical up and down α-helical bundle in agreement with the helical assignments in the previous NMR studies of OspC (Huang et al., 1999). This is in contrast to the structure of OspA, which has an unusual, elongated fold composed of 21 anti-parallel β-strands followed by a single short α-helix (Li et al., 1997). Except for helices α4 and α5 (Figure 1A and B), all helices are anti-parallel. The elongated, kidney-shaped OspC dimer is formed through a non-crystallographic 2-fold axis almost parallel to helices α1 and α1′. The interaction in the dimeric interface is almost completely hydrophobic. On dimerization, the buried surface area at the dimer interface is 4430 Å2, ∼30% of the total surface area of the dimer (14 374 Å2), suggesting that the biologically functional molecule is a dimer. Under native conditions the recombinant OspC used in these studies eluted as a dimer during gel exclusion chromatography. The dimeric interaction is mostly between helices α1 and α1′, which are parallel (Figure 1C). Additional interactions occur between helices α1 and α2′, and between the loop connecting α1 and α2, and the helix α3′. The B31 OspC dimer is essentially identical (r.m.s.d. of 322 equivalent Cα atoms = 0.88 Å). In both crystal structures, divalent cations participate in packing between dimers by coordinating with surface histidine, glutamate and/or asparagine residues. In the HB19 structure three different coordinations, i.e. tetra, penta and hexa coordinations of zinc, are observed, while the B31 structure has a single hexa-coordinated cation that is modeled as a magnesium ion, but the low refined B-value of 13 Å2 suggests that the site may be at least partially occupied by heavier trace ions. The presence of divalent ions in the crystals is probably fortuitous due to the additives used for crystallization. The N- and C-termini are on the same side of the molecule and are close to each other, the distance between the Cα atoms being 9.2 Å. This suggests that the C-terminus is also close to the membrane surface on which the lipidated N-terminus is anchored.
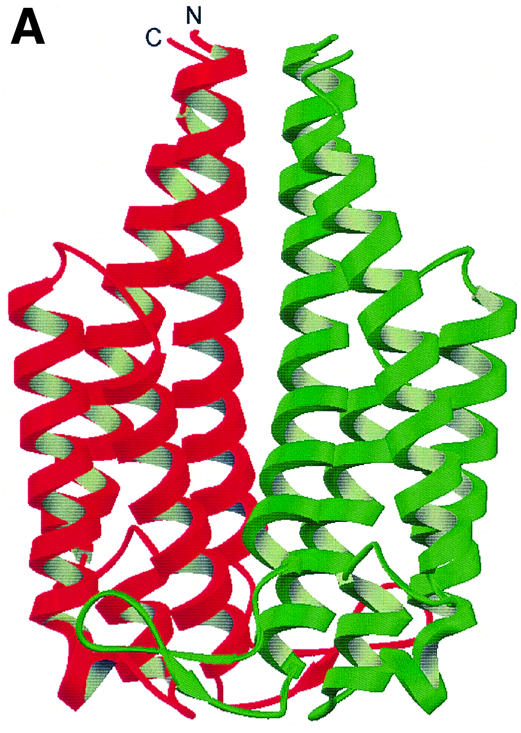
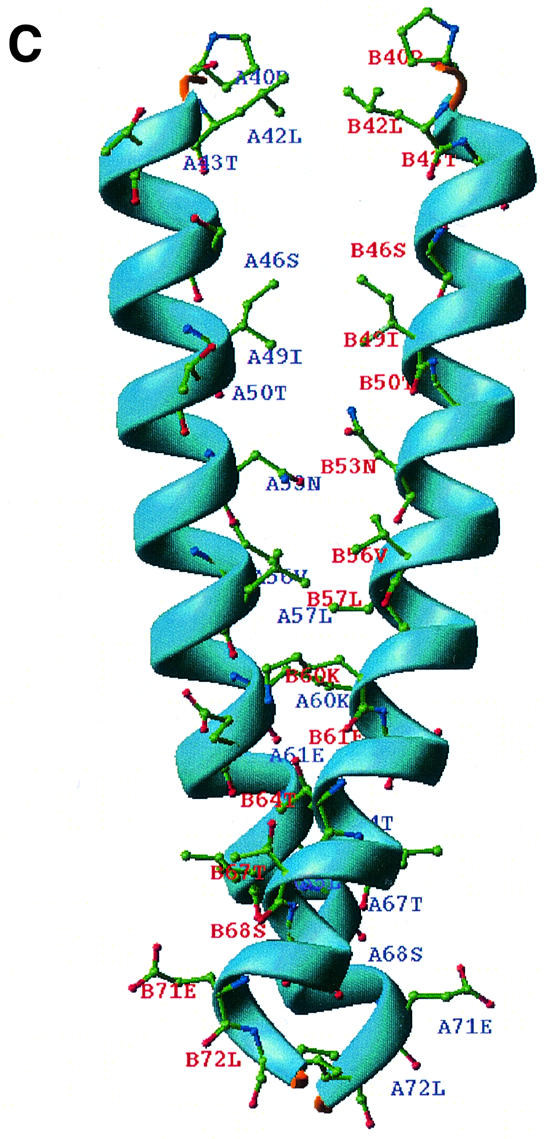
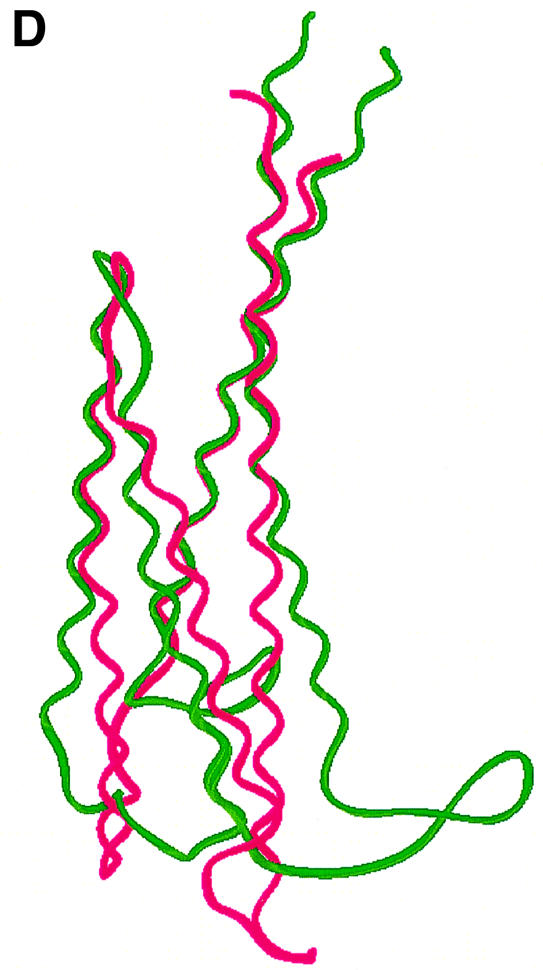
Fig. 1. (A) RIBBONS (Carson, 1991) representation of the OspC-HB19 dimer. The two monomers are colored red and green, respectively. The close proximity of the two inner most helices indicates that it is a tight dimer. (B) Topology diagram of the OspC-HB19 monomer. Red cylinders represent α-helices and green arrows β-strands. (C) The dimeric interface. Residues interacting across the 2-fold axis are shown as a ball-and-stick model. For clarity only α1 and α1′ helices are shown. (D) Superposition of Borrelia OspC-HB19 (green) and Salmonella AR (magenta) monomers. The r.m.s.d. between 122 aligned Cα atoms is 3.0 Å. Alignment was carried out with SCOP (Murzin et al., 1995).
A Dali search (Holm and Sander, 1996) indicates that OspC has similarity to the periplasmic domain of the Salmonella aspartate receptor (AR), which is also a dimer (Yeh et al., 1996). Figure 1D shows the superposition of a monomer of AR on a monomer of HB19. However, when the dimers are aligned they do not superpose, suggesting that the dimeric 2-fold axes are inclined with respect to each other. Although the three-dimensional alignment is convincing, the overall sequence homology between OspC and the periplasmic AR domain is low, with only 13% identity; thus, the similarity between the two proteins would have been missed based on sequence alone.
Putative binding site
Although four helical bundle structures have a variety of functions (Weber and Salemme, 1980), the structural similarity between AR and OspC HB19 suggests that OspC may possibly be a binding protein contributing to a fundamental biological process. Several studies have shown that B.burgdorferi has a predilection for collagenous tissue and can interact with fibronectin and cellular collagens. The spirochetes can bind to a number of different cell types, including fibroblasts. Borrelia burgdorferi can bind to a novel circulating fibroblast-like cell called the peripheral blood fibrocyte, which expresses collagen types I and III as well as fibronectin, in a process that does not require OspA or OspB (Grab et al., 1999). Interestingly, when the surface electrostatic potentials of the HB19 dimer were calculated utilizing GRASP (Nicholls et al., 1991), it was evident that the surface that projects away from the membrane (top of the molecule) has a region with strong negative electrostatic potential. It extends across the molecular 2-fold axis and is lined by carbonyl oxygens, aspartate and glutamate residues. It also has two 50 Å3 cavities ∼8 Å below the exposed surface (Figure 2). If OspC is indeed a binding protein, its binding interface may involve this region of the OspC dimer interface.
Fig. 2. Stereo view of putative binding site and cavities. The cavities as calculated by GRASP (Nicholls et al., 1991) are shown in different colors. Cavities shown in blue and green are at the top of the molecule away from the membrane surface. Residues forming these cavities are shown as a stick model in corresponding colors. Each cavity has a volume of 50 Å3 and is formed by residues Ala75, Ile76, Gly77, Lys78, Lys79, Glu89, Ala90, Asp91, His92 and Asn93 of one monomer, and Gly94, Ser95, Ser98, Gly146, Lys147 and Glu148 of the other monomer. The solvent structures in the cavities are remarkably well conserved between the HB19 and B31 molecules.
Modeling of different OspCs
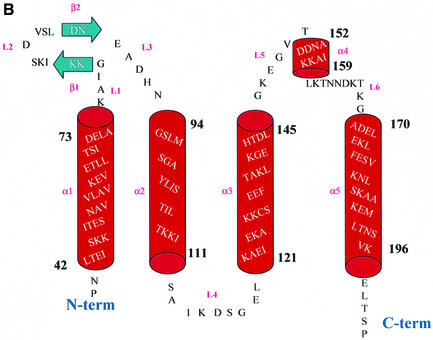
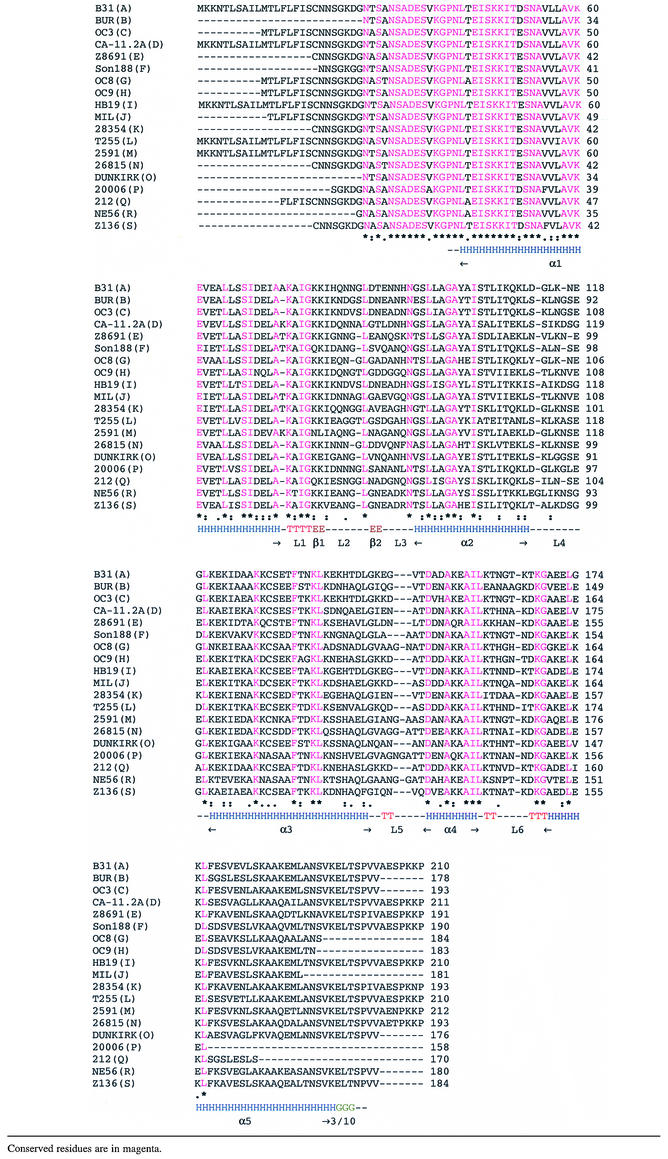
With representative structures of OspC from two different invasive groups in hand, we used this information to model one representative from each of the other 17 OspC groups (Sali and Blundell, 1993). All 17 are predicted to fold similarly to HB19 and B31 structures. A comparison at the sequence level showed that the variable regions in these sequences are mainly in the loop regions of the molecule (Table I). Presumably these variable regions constitute major antigenic sites that serve to differentiate amongst OspC antigens. The information contained in Table I will be useful for designing chimeric OspC vaccine candidates that might be effective against more than one OspC antigen.
Table I. Alignment of 19 groups (groups are given within parentheses) of OspC with their secondary structures as derived from the experimental OspC-HB19 model.
Are the structures of invasive and non-invasive subpopulations different?
It appears that only limited subpopulations of B.burgdorferi are responsible for invasive human disease, and that the OspC type can be used as a means to distinguish invasive and non-invasive spirochetes. Spirochetes with certain OspC types never cause human disease, while some others cause local infection at the primary skin site but do not cause any systematic disease (Seinost et al., 1999). Only four types (A, B, I and K) are responsible for systematic disease and are found in the secondary sites, and hence are termed invasive clones. When we compared the electrostatic potential of HB19 (group I) and B31 (group A) with similar models for the other OspCs, we noted that there is a striking distinction between OspCs associated with invasive and non-invasive spirochetes. Figure 3 shows the electrostatic potential surface viewed from the farthest point from the surface of the membrane. While the surface potential of this region is highly negative for OspCs from invasive strains, it is not so for those present on non-invasive strains. If OspC is involved in targeting the infecting spirochetes to specific host molecules, then this difference may provide a potential explanation for why some types of OspC are found associated with invasive clones. It is tempting to speculate that this region’s interaction with a positively charged ligand, such as fibronectin or a similar molecule, plays an important role in the pathogenesis of Lyme disease. Interestingly, in the crystal structure of human fibronectin, the region binding to integrin has been shown to be highly positively charged (Sharma et al., 1999).
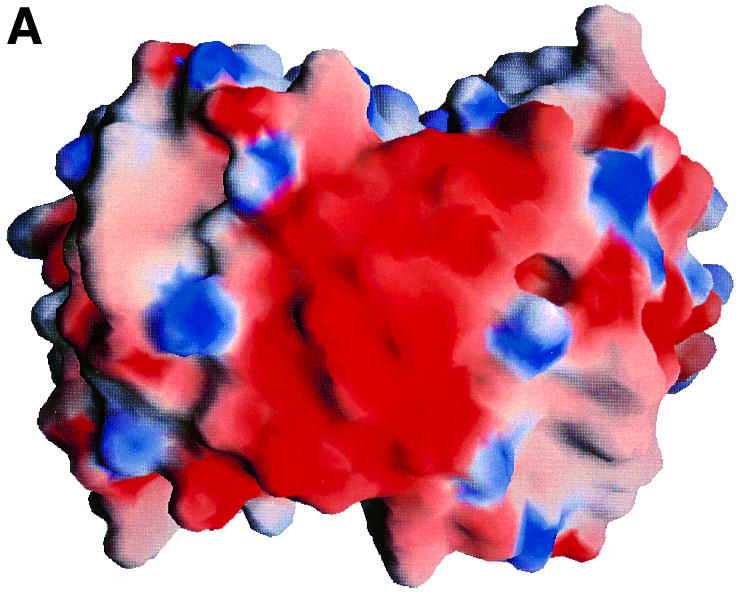
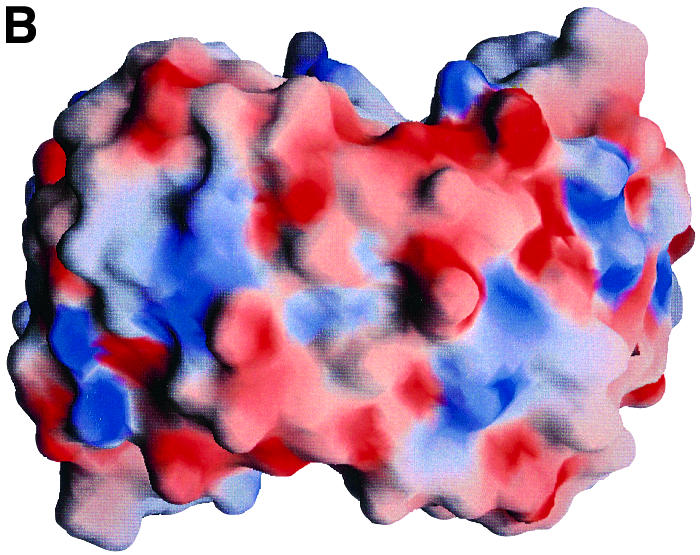
Fig. 3. Electrostatic potential surface of various OspCs. Electrostatic potential surface of (A) HB19 and (B) 212 as viewed from the top of the molecule (orthogonal to the view in Figure 1A). 212 was modeled based on HB19 coordinates. The electrostatic potential surface of B31 (not shown) is similar to that of HB19. The negative and positive potentials are shown in red and blue. The HB19 and B31 spirochetes are classified as being invasive while the 212 spirochete is from non-invasive groups. The electrostatic surface potential at the top of the OspC dimers from spirochetes in invasive groups is highly negative and clearly different from those of the non-invasive groups.
Materials and methods
Bacterial expression and purification of OspC 38–201
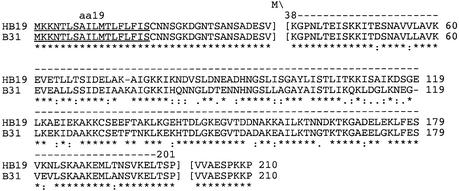
Since the NMR experiments demonstrated that the N- and C-terminal regions are highly flexible, and further showed that the N- and C-truncated proteins contain the folded core of OspC (Huang et al., 1999), OspC HB19 and B31 were synthesized in Escherichia coli BL21(DE3)/pLysS as N- and C-terminally truncated proteins. DNA fragments encoding residues 38–201 (Figure 4) were amplified by PCR using B.burgdorferi HB19 genomic DNA or a cloned B31 ospC gene as template, respectively, and two primers with different restriction sites for cloning into the kanamycin-resistant T7-based expression vector pET9c (Novagen). The 5′ primer (5′-ACAGGATCCATATGAAAGGGCCTAATCTTAC-3′) contained a NdeI site (bold). The 3′ primer (5′-CCGGGATCCTTAAGGGCTTGTAAGCTCTTTAAC-3′) contained a BamHI recognition sequence (bold). The region of each primer complementary to the Borrelia ospC sequence is underlined. PCR amplification was performed under stringent conditions using Platinum Taq DNA Polymerase High Fidelity (Gibco-BRL). The amplified fragments were cleaved with NdeI and BamHI, purified by GFX chromatography (Amersham Pharmacia Biotech, Inc.) after low-melt gel agarose electrophoresis, and inserted between the NdeI and BamHI sites of pET9c and then used for transformation of E.coli (DE3)/pLysS. Native HB19 OspC 38–201 contains only one internal Met residue, residue 190; therefore, oligonucleotide-directed mutagenesis was used to change codon 97 (ATA) I to (ATG) M using the cloned truncated gene as template.
Fig. 4. Clustal alignment (Thompson et al., 1994) of HB19 and B31 OspC proteins. The first 18 residues (underlined) function as a signal sequence for transport, proteolytic processing and lipidation. The constructs used here, aa 38–201 and aa 38–210, were made by PCR amplification of genomic DNA with primers that introduced unique restriction sites in front of aa 38 and after aa 201 or after the end of the ospC open reading frame. The 5′ primer provided the initiation codon (M). The 3′ primers had appropriately placed stop codons after residue 201. The sequences (AF337548) of the clones were confirmed by DNA sequencing. The asterisks, colons and spaces below the amino acid sequence represent complete identity, conserved changes and no homology at all, respectively.
For isolation of OspC 38–201, 0.5 l of M9TBYG medium supplemented with 50 µg/ml kanamycin and 25 µg/ml chloramphenicol were inoculated with 5 ml of an overnight culture of each transformed strain and shaken at 37°C until an OD600 of 0.4–0.6 was reached. Isopropyl-β-d-thiogalactopyranoside was added to 0.5 mM final concentration and incubation with shaking was continued for 3–4 h. Pelleted cells were resuspended in 25 ml of 20 mM Na phosphate pH 7.7, and stored frozen at –70°C. Resuspended cells were thawed to initiate lysis. After 20 min on ice, MgCl2 and DNase (Boehringer) were added at 2.5 mM and 5 µg/ml, respectively, and the lysate was incubated with occasional mixing for an additional 30 min. A cleared lysate was obtained by centrifugation at 25 000 g. The pellets were washed with 5 ml of phosphate buffer and the combined supernatants applied to a 5 ml Econo-Pac High Q cartridge (Bio-Rad) equilibrated with 10 mM Na phosphate pH 7.7, 10 mM NaCl. OspC was recovered in the flow-through, which was then dialyzed overnight at 4°C against 2× 1 l of 10 mM Na phosphate pH 6.0, 5 mM NaCl. After the dialysate was clarified by centrifugation it was applied to a 5 ml Econo-Pac High S cartridge (Bio-Rad) equilibrated with 10 mM Na phosphate pH 6.0, 5 mM NaCl. OspC was eluted with a 300 ml gradient of 5–100 mM NaCl in phosphate buffer. From a 0.5 l culture, ∼30 mg of OspC 38–201 of >99% purity could be obtained. SeMet-substituted OspC 38–201 HB19 was expressed in the E.coli 834(DE3) strain using minimal M9 media supplemented with l-SeMet and the other 19 l-amino acids (40 mg/l each). Selenomethionyl OspC 38–201 was purified using the same protocol as used for the native protein, except that egg white lysozyme (at 0.2 mg/ml) and EDTA (2.5 mM final concentration) were added to the resuspended cells to initiate lysis.
Crystallization and data collection
To determine the structure, SeMet-substituted protein was utilized. However, since there was only one internal methionine in a total of 164 residues, a mutant, Ile97Met, was generated by oligonucleotide-directed mutagenesis to increase the anomalous signal from selenium. Crystals of the SeMet OspC protein (aa 38–201) were grown with conditions similar to those for the native sample by vapor diffusion in sitting drops at 293K (Kumaran et al., 2001). Drops (6 µl) containing a 1:1 ratio of 6.9 mg/ml OspC and a precipitant containing 25% (v/v) PEG monomethyl ether 550, 10 mM zinc sulfate heptahydrate, 100 mM MES pH 6.5, were equilibrated against a reservoir containing 800 µl of the same precipitant. Long rectangular crystals with average dimensions of 0.3 × 0.3 × 1 mm3 grew in 1 week. Crystals were flash frozen after addition of 15% glycerol to the mother liquor. Diffraction data extending to 2.8 Å were collected from the frozen crystal with the B1 detector on beamline X12C at the National Synchrotron Light Source (NSLS). MAD data were collected around the selenium absorption edge (Ramakrishnan and Biou, 1997). The data were processed with DENZO and SCALEPACK (Otwinowski and Minor, 1997). Subsequently, single wavelength high-resolution data extending to 1.8 Å were collected with a better quality SeMet OspC crystal (Form II) on beamline X25 at the NSLS and were used in the later stages of refinement. Crystals of B31 OspC (aa 38–201) were grown at 4°C in 28–30% PEG 3350, 80 mM Tris–HCl pH 8.5, 100 mM MgCl2 and 18% glycerol, and diffraction was measured at NSLS beamline X12C (see Table II for diffraction data statistics).
Table II. OspC crystal, diffraction, phasing and model dataa.
| HB19 (Form I) SeMet | HB19 (Form II) SeMet | B31 native | |||
|---|---|---|---|---|---|
| Space group | P21 | P21 | P1 | ||
| Unit cell | a (Å) | 64.78 | 66.32 | 127.21 | |
| b (Å) | 46.96 | 46.25 | 33.65 | ||
| c (Å) | 110.19 | 111.78 | 47.99 | ||
| α (°) | 84.04 | ||||
| β (°) | 99.81 | 99.08 | 81.55 | ||
| γ (°) | 89.23 | ||||
| remote | edge | peak | |||
| Wavelength (Å) | 0.9300 | 0.9791 | 0.9794 | 0.9786 | 1.0920 |
| Resolution range (Å) | 50–2.8 | 50–2.8 | 50–2.8 | 50–1.8 | 50–2.5 |
| Unique reflections | 17 840 | 17 714 | 17 665 | 51 522 | 24 505 |
| Completeness (%) | 99.4 | 99.3 | 99.3 | 85.3 | 91.1 |
| R-mergeb | 0.083 | 0.079 | 0.117 | 0.087 | 0.043 |
| <I/σ(I)> | 12.2 | 14.6 | |||
| Phasing powerc | 1.2/1.4 | 1.5/1.5 | |||
| FOMc, before density modification | 0.44/0.33 | ||||
| PDB ID | 1F1Me | 1GGQ | |||
| Content of asymmetric unit | |||||
| OspC monomers (residue range) | 4 (40–201) | 4 (40–201) | |||
| solvent | 632 | 112 | |||
| ions | 6 (Zn2+) | 1 (Mg2+) | |||
| Average B-value (Å2) | |||||
| main chain | 22.1 | 43.2 | |||
| side chain | 27.0 | 45.2 | |||
| solvent | 31.5 | 30.5 | |||
| Rworkd (reflections) | 0.21 (48 910) | 0.23 (22 087) | |||
| Rfreed (reflections) | 0.24 (2612) | 0.27 (2418) | |||
| R.m.s. bonds (Å) | 0.004 | 0.005 | |||
| R.m.s. angles (°) | 1.010 | 1.130 | |||
| φ–ψ, most favored regions (%)f | 93 | 92 | |||
aHB19 and B31 were determined at Brookhaven National Laboratory and Rutgers University, respectively.
bRmerge = Σj(|Ih – <I>h|)/ΣIh, where <Ih> is the average intensity over symmetry equivalents.
cPhasing power and FOM (figure of merit) are as defined in SHARP (De La Fortelle and Bricogne, 1997); centric/acentric reflection values are shown.
dR = ΣFobs – Fcalc|/ΣFobs|; Rwork is summed over reflections used in refinement, Rfree is summed over reflections set aside for validation.
eHB19 Form II data were used for refinement.
fAs defined in PROCHECK (Laskowski et al., 1993).
Structure determination
HB19. There are four molecules per asymmetric unit and each molecule has two SeMets. The selenium positions were obtained from Patterson and difference Fourier maps with the use of the PHASES program package (Furey and Swaminathan, 1997). As described elsewhere, there are two dimers in the asymmetric unit (Kumaran et al., 2001). A self-rotation function calculation revealed a strong peak of 56% of the origin peak corresponding to κ = 180° and a relatively weak peak of 25% of the origin peak at κ = 12°. Also, a native self-Patterson showed a peak (18% of the origin peak) at (0, 0, 1/2). Accordingly, the monomers are related by a non-crystallographic 2-fold axis forming a dimer, and the dimers are related by a pseudo translation of half the unit cell along the c axis related by a small rotation of 12°. A total of eight selenium atoms was input to the SHARP program to refine the phases (De La Fortelle and Bricogne, 1997). The resulting phases were further improved by NCS averaging in DM in two steps, averaging the non-crystallographic 2-fold related monomers first and then the dimers related by pseudo translational symmetry (Cowtan, 1994). The free R-factor after NCS averaging was 0.34. The resulting electron density map was of excellent quality and revealed an almost all-helical structure. Approximately 90% of the Cα chain was traced with the baton option in ‘O’ (Jones et al., 1991). The model building was completed using the two SeMets as markers. Further refinement was carried out with the high-resolution data set. Since the cell dimensions of the Form II crystal were slightly different from those of Form I, a rigid body refinement was initially carried out. During rigid body refinement, each of the four monomers was treated as a separate rigid group and gave an R-value and free-R value of 0.39 and 0.38, respectively. 5% of the data were set apart for the free-R calculation.
B31. The B31 OspC crystal also contains two dimers in the asymmetric unit. The B31 structure was solved by the molecular replacement method with HB19 as a search model (Navaza and Saludjian, 1997).
Both models were refined by the slow-cool annealing method (Brunger et al., 1998) alternating with model building until convergence. Composite omit maps clearly showed density for divalent cations and coordinating waters. The final model of HB19 contains four monomers of 162 residues each, six zinc atoms and 632 water molecules. Individual restrained B-factors were used in the final cycles of refinement for HB19 and B31. Two N-terminal residues were not included in the model for all the monomers because of poor electron density. The final refinement statistics are given in Table II. The HB19 and B31 OspC coordinates and experimental structure factors have been deposited with the Protein Data Bank [id codes 1F1M (HB19) and 1GGQ (B31)].
Acknowledgments
Acknowledgements
We thank A.Glenn, B.Lade, H.Kycia and F.Mannino for technical assistance, and X.Yang for initial help with OspC purification and crystallization trials. We also thank X.Huang for his help. Research was supported by the National Institutes of Health (AI37256) at Brookhaven National Laboratory under contract with the US Department of Energy. B.J.L. was supported by the National Institutes of Health (#PO1-NS34092-01A2) and D.K. by the Veterans Administration Medical Center, Pittsburgh.
References
- Barbour A.G. and Fish,D. (1993) The biological and social phenomenon of Lyme disease. Science, 260, 1610–1616. [DOI] [PubMed] [Google Scholar]
- Brunger A.T. et al. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Burgdorfer W., Barbour,A.G., Hayes,S.F., Benach,J.L., Gruwaldt,E. and Davis,J.P. (1982) Lyme disease—a tick borne spirochetosis? Science, 216, 1317–1319. [DOI] [PubMed] [Google Scholar]
- Carson M. (1991) Ribbons 2.0. J. Appl. Crystallogr., 24, 958–961. [Google Scholar]
- Cowtan K. (1994) Joint CCP4 and ESF–EACBM Newsletter on Protein Crystallography, 31, 34–38. [Google Scholar]
- Craft J.E., Fischer,D.K., Shimamoto,G.T. and Steere,A.C. (1986) Antigens of Borrelia burgdorferi recognized during Lyme disease. Appearance of a new immunoglobulin M response and expansion of the immunoglobulin G response late in the illness. J. Clin. Invest., 78, 934–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Fortelle E. and Bricogne,G. (1997) Maximum-likelihood heavy atom parameter refinement in the MIR and MAD methods. Methods Enzymol., 276, 472–493. [DOI] [PubMed] [Google Scholar]
- Fikrig E., Barthold,S.W., Kantor,F.S. and Flavell,R.A. (1990) Protection of mice against the Lyme disease agent by immunizing with recombinant OspA. Science, 250, 533–536. [DOI] [PubMed] [Google Scholar]
- Fikrig E., Barthold,S.W., Persing,D.H., Sun,X., Kantor,F.S. and Flavell,R.A. (1992) Borrelia burgdorferi strain 25015: characterization of outer surface protein A and vaccination against infection. J. Immunol., 148, 2256–2260. [PubMed] [Google Scholar]
- Furey W. and Swaminathan,S. (1997) PHASES-95: a program package for the processing and analysis of diffraction data from macromolecules. Methods Enzymol., 276, 590–620. [DOI] [PubMed] [Google Scholar]
- Gilmore R.D., Kappel,K.J., Dolan,M.C., Burkot,T.R. and Johnson,B.J. (1996) Outer surface protein C (OspC), but not P39, is a protective immunogen against a tick-transmitted Borrelia burgdorferi challenge: evidence for a conformational protective epitope in OspC. Infect. Immun., 64, 2234–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golde W.T., Burkot,T.R., Piesman,J., Dolan,M.C., Capiau,C., Hauser,P., Dequesne,G. and Lobet,Y. (1995) The Lyme disease vaccine candidate outer surface protein A (OspA) in a formulation compatible with human use protects mice against natural tick transmission of B. burgdorferi. Vaccine, 13, 435–441. [DOI] [PubMed] [Google Scholar]
- Grab D.J., Lanners,H., Martin,L.N., Chesney,J., Cai,C., Adkisson,H.D. and Bucala,R. (1999) Interaction of Borrelia burgdorferi with peripheral blood fibrocytes, antigen-presenting cells with the potential for connective tissue targeting. Mol. Med., 5, 46–54. [PMC free article] [PubMed] [Google Scholar]
- Hansen K. and Lebech,A.M. (1992) The clinical and epidemiological profile of Lyme neuroborreliosis in Denmark 1985–1990: a prospective study of 187 patients with Borrelia burgdorferi specific intrathecal antibody production. Brain, 115, 399–423. [DOI] [PubMed] [Google Scholar]
- Holm L. and Sander,C. (1996) Mapping the protein universe. Science, 273, 595–603. [DOI] [PubMed] [Google Scholar]
- Huang X., Link,K., Koide,A., Dunn,J.J., Luft,B.J. and Koide,S. (1999) 1H, 13C and 15N NMR backbone assignments of 37 kDa surface antigen OspC from Borrelia burgdorferi. J. Biomol. NMR, 14, 283–284. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zou,J., Cowan,S. and Kjeldgaard,M. (1991) Improved methods in building protein models in electron density map and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Jonsson M. and Bergstrom,S. (1995) Transcriptional and translational regulation of the expression of the major outer surface proteins in Lyme disease Borrelia strains. Microbiology, 141, 1321–1329. [DOI] [PubMed] [Google Scholar]
- Kumaran D., Eswaramoorthy,S., Dunn,J.J. and Swaminathan,S. (2001) Crystallization and preliminary X-ray analysis of Borrelia burgdorferi outer surface protein C (OspC). Acta Crystallogr. D, 57, 298–300 [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., MacArthur,M.W., Moss,D.S. and Thornton,J.M. (1993) PROCHECK: a program to check the stereochemical quality for assessing the accuracy of protein structures. J. Appl. Crystallogr., 26, 283–291. [Google Scholar]
- Li H., Dunn,J.J., Luft,B.J. and Lawson,C.L. (1997) Crystal structure of Lyme disease antigen outer surface protein A complexed with an Fab. Proc. Natl Acad. Sci. USA, 94, 3584–3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marconi R.T., Samuels,D.S. and Garon,C.F. (1993) Transcriptional analyses and mapping of the ospC gene in Lyme disease spirochetes. J. Bacteriol., 175, 926–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murzin A.G., Brenner,S.E., Hubbard,T. and Chothia,C. (1995) SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol., 247, 536–540. [DOI] [PubMed] [Google Scholar]
- Navaza J. and Saludjian,P. (1997) AMoRe: an automated molecular replacement program package. Methods Enzymol., 276, 581–594. [DOI] [PubMed] [Google Scholar]
- Nicholls A., Sharp,K.A. and Honig,B. (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins, 11, 281–296. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor,W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Probert W.S., Crawford,M., Cadiz,R.B. and LeFebvre,R.B. (1997) Immunization with outer surface (Osp) A, but not OspC, provides cross-protection of mice challenged with North American isolates of Borrelia burgdorferi. J. Infect. Dis., 175, 400–405. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan V. and Biou,V. (1997) Treatment of multiwavelength anomalous diffraction data as a special case of multiple isomorphous replacement. Methods Enzymol., 276, 538–557. [PubMed] [Google Scholar]
- Ramamoorthy R. and Philipp,M.T. (1998) Differential expression of Borrelia burgdorferi proteins during growth in vitro. Infect. Immun., 66, 5119–5124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadziene A., Wilske,B., Ferdows,M.S. and Barbour,A.G. (1993) The cryptic ospC gene of Borrelia burgdorferi B31 is located on a circular plasmid. Infect. Immun., 61, 2192–2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A. and Blundell,T.L. (1993) Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol., 234, 779–815. [DOI] [PubMed] [Google Scholar]
- Schwan T.G. and Piesman,J. (2000) Temporal changes in outer surface proteins A and C of the Lyme disease-associated spirochete, Borrelia burgdorferi, during the chain of infection in ticks and mice. J. Clin. Microbiol., 38, 382–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwan T.G., Piesman,J., Golde,W.T., Dolan,M.C. and Rosa,P.A. (1995) Induction of an outer surface protein on Borrelia burgdorferi during tick feeding. Proc. Natl Acad. Sci. USA, 92, 2909–2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seinost G., Dykhuizen,D.E., Dattywyler,R.J., Golde,W.T., Dunn,J.J., Wang,I.-N., Wormser,G.P., Schriefer,M.E. and Luft,B.J. (1999) Four clones of Borrelia burgdorferi sensu stricto cause invasive infection in humans. Infect. Immun., 67, 3518–3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A., Askari,J.A., Humphries,M.J., Jones,E.Y. and Stuart,D.I. (1999) Crystal structure of a heparin- and integrin-binding segment of a human fibronectin. EMBO J., 18, 1468–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon M.M., Schaible,U.E., Kramer,M.D., Eckerskorn,C., Museteanu,C., Muller-Hermelink,K.K. and Wallich,R. (1991) Recombinant outer surface protein from Borrelia burgdorferi induces antibodies protective against spirochetal infection in mice. J. Infect. Dis., 164, 123–132. [DOI] [PubMed] [Google Scholar]
- Steere A.C. (1991) Clinical definitions and differential diagnosis of Lyme arthritis. Scand. J. Infect. Dis. Suppl., 77, 51–54. [PubMed] [Google Scholar]
- Stevenson B., Schwan,T.G. and Rosa,P.A. (1995) Temperature-related differential expression of antigens in the Lyme disease spirochete, Borrelia burgdorferi. Infect. Immun., 63, 4535–4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilly K., Casjens,S., Stevenson,B., Bono,J.L., Samuels,D.S., Hogan,D. and Rosa,P. (1997) The Borrelia burgdorferi circular plasmid cp26: conservation of plasmid structure and targeted inactivation of the ospC gene. Mol. Microbiol., 25, 361–373. [DOI] [PubMed] [Google Scholar]
- van der Linde M.R., Crijns,H.J., de Koning,J., Korstanje,J.A.H., de Graaf,J.J., Piers,D.A., van der Galien,A. and Lie,K.I. (1990) Range of atriventricular conduction disturbances in Lyme borreliosis: a report of four cases and review of other published reports. Br. Heart J., 63, 162–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang I.-N., Dykhuizen,D.E., Qui,W., Dunn,J.J., Bosler,E.M. and Luft,B.J. (1999) Genetic diversity of ospC in a local population of Borrelia burgdorferi sensu stricto. Genetics, 151, 15–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber P.C. and Salemme,F.R. (1980) Structural and functional diversity in 4-α-helical proteins. Nature, 287, 82–84. [DOI] [PubMed] [Google Scholar]
- Yeh J.I., Biemann,H.-P., Prive,G.G., Pandit,J., Koshland,D.E.,Jr and Kim,S.-H. (1996) High-resolution structures of the ligand binding domain of the wild-type bacterial aspartate receptor. J. Mol. Biol., 262, 186–201. [DOI] [PubMed] [Google Scholar]