Abstract
There are two distinct pathways for disulfide formation in prokaryotes. The DsbA–DsbB pathway introduces disulfide bonds de novo, while the DsbC–DsbD pathway functions to isomerize disulfides. One of the key questions in disulfide biology is how the isomerase pathway is kept separate from the oxidase pathway in vivo. Cross-talk between these two systems would be mutually destructive. To force communication between these two systems we have selected dsbC mutants that complement a dsbA null mutation. In these mutants, DsbC is present as a monomer as compared with dimeric wild-type DsbC. Based on these findings we rationally designed DsbC mutants in the dimerization domain. All of these mutants are able to rescue the dsbA null phenotype. Rescue depends on the presence of DsbB, the native re-oxidant of DsbA, both in vivo and in vitro. Our results suggest that dimerization acts to protect DsbC’s active sites from DsbB-mediated oxidation. These results explain how oxidative and reductive pathways can co-exist in the periplasm of Escherichia coli.
Keywords: disulfide bond/disulfide isomerase/disulfide oxidase/DsbC mutants/Escherichia coli
Introduction
The formation of disulfide bonds is one of the key steps during the folding of many secretory proteins; so important that proteins that lack their disulfides are often rapidly degraded. In bacteria, disulfide bond formation is catalyzed by the Dsb proteins (for reviews see Raina and Missiakas, 1997; Rietsch and Beckwith, 1998; Debarbieux and Beckwith, 1999). DsbA is a 21 kDa periplasmic protein with a CXXC motif in its active site. DsbA rapidly oxidizes cysteine residues in target proteins, thus leading to the formation of a disulfide bond. DsbA’s extreme oxidizing power is responsible for the oxidizing nature of the periplasm as compared with the relatively reducing environment of the cell’s cytosol (Wunderlich and Glockshuber, 1993). The inner membrane protein DsbB oxidizes DsbA efficiently (Bardwell et al., 1993; Guilhot et al., 1995). Thus, disulfide bonds flow from DsbB to DsbA and then onto folding proteins. DsbB is re-oxidized directly by ubiquinone present in the inner membrane. Electrons flow from ubiquinone via terminal cytochrome oxidases onto oxygen. DsbB provides a link between disulfide bond formation and electron transport (Kobayashi et al., 1997; Bader et al., 1999, 2000).
A second independent pathway acts on wrongly formed disulfide bonds. DsbC and DsbG act to isomerize these incorrect disulfide bonds (Rietsch et al., 1996; Darby et al., 1998; Bessette et al., 1999). Disulfide isomerization involves the nucleophilic attack of a wrong disulfide bond by the reduced catalyst forming a mixed disulfide bond between the enzyme and the target protein. The complex resolves upon formation of a disulfide bond in the target protein (Gilbert, 1997). In order to attack a disulfide bond and catalyze its rearrangement, the catalyst has to be kept in a reduced form. This is done by an inner membrane protein called DsbD, which uses the reducing power of thioredoxin to reduce DsbC (Missiakas et al., 1995; Rietsch et al., 1996, 1997).
A very important question concerning the DsbA–DsbB and DsbC–DsbD system is how these two pathways are kept separate. DsbA is found almost entirely in the oxidized form in vivo, while DsbC is entirely reduced (Joly and Swartz, 1997). This is in agreement with their divergent roles. Oxidases like DsbA need to be oxidized to be active, while isomerases need to be kept reduced. However, it begs the question of how the two systems avoid a futile cycle. Any cross-talk between the two pathways would make disulfide bond formation less efficient, as it would lead to the oxidation of the reductive pathway and vice versa. It has been shown that DsbA oxidizes DsbC very slowly in vitro, slow enough to essentially eliminate cross-talk in vivo (Darby et al., 1998). Recently, we have found that the two pathways are also kinetically separated at the level of DsbB (Bader et al., 2000). DsbB discriminates between DsbA and DsbC. The re-oxidation of DsbA by DsbB was found to be at least 500-fold faster than the re-oxidation of DsbC by DsbB, leading us to conclude that DsbB effectively oxidizes DsbA but not DsbC.
Here we provide evidence for the molecular basis of the separation of the oxidative and reductive pathways. We have isolated dsbC mutants that complement a dsbA null mutant, showing that they can actually replace dsbA in vivo. These mutants map to DsbC’s dimerization interface and fail to form stable dimers. Based on this result we targeted the dimerization interface for mutagenesis, and in doing so constructed an additional four mutants that complement the dsbA null mutation. We also found that monomeric DsbC mutants were oxidized by DsbB in vivo and in vitro. Monomerization of DsbC allows it to switch allegiances and participate in the oxidative pathway.
Results
Identification of dsbC mutants that complement dsbA
In the periplasm of Escherichia coli, DsbA is the net donor of disulfide bonds while DsbC functions as a disulfide isomerase that corrects wrongly formed disulfide bonds (Bardwell et al., 1991; Rietsch et al., 1996). To avoid inactivation of the isomerization pathway by the oxidative pathway, the two must be kept isolated. To gain insight into what makes each pathway so specific for oxidation or isomerization, we decided to convert DsbC into a net donor of disulfide bonds; in effect, allowing DsbC to act like DsbA. DsbA null strains are completely non-motile, while dsbA+ strains are motile (Dailey and Berg, 1993). Based on this phenotype, we devised a powerful genetic selection allowing the identification of dsbC variants that could complement a dsbA null mutant. Since we did not know the basis of the separation of the two pathways, we decided to randomly mutate dsbC and select for mutants that could restore motility in a dsbA null mutant strain.
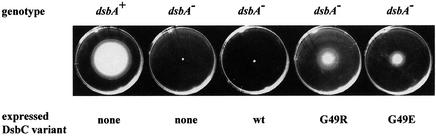
As described in Materials and methods, a plasmid library of random mutations in the dsbC gene (in which the expression of DsbC was under arabinose control) was created and transformed into a dsbA null strain. After incubation, 28 motile colonies were observed in which motility was dependent on the presence of arabinose. Figure 1 shows the motile phenotype. Plasmid DNA was prepared from these strains and used to re-transform a dsbA null mutant strain. After re-transformation, 26 of the 28 plasmids conferred motility to the previously immobile dsbA strain, confirming that, for these 26 plasmids, the motile phenotype was conferred by the plasmid. Although these mutants are not quite as motile as dsbA+ strains, they are strongly motile, whereas the starting dsbA– strain is not at all motile. Sequencing of the 26 motile mutants revealed two different mutations, both of which altered the G49 residue in DsbC. A single base substitution from GGG to AGG was found in 17 clones. This resulted in a G49 to arginine substitution. In nine clones, a G49 to glutamic acid substitution was found. The complementation depends strictly on the presence of arabinose, which induces expression of DsbC from the pBAD33a derived plasmids (data not shown). Wild-type dsbC, present as a chromosomal copy or introduced on pMB69, is not able to rescue the dsbA null phenotype under the conditions employed (Figure 1). To prove that these two mutations by themselves were sufficient to complement the dsbA null mutation, they were introduced into the dsbC gene by site-directed mutagenesis. After transformation into the dsbA null mutant strain JCB817, the mutant plasmids coding for DsbC G49R or G49E showed the same phenotype as the plasmids isolated by the selection. This confirms that the motile phenotype is due to the mutations G49R or G49E.
Fig. 1. Identification of two DsbC mutants that rescue the dsbA null phenotype. Colonies were stabbed onto M63 motility plates and grown for 24 h. A dsbA null mutation leads to a severe defect in disulfide bond formation causing the loss of motility (JCB817). This motility is not restored by the plasmid pMB69, which carries the wild-type dsbC gene. Two DsbC mutants that conferred motility were selected a number of times independently, following random mutagenesis of the dsbC gene. The two mutations were identified as dsbC G49R and G49E. The minimal medium contains 0.4% glycerol as carbon source and 0.2% arabinose to induce expression of DsbC under ara control. No rescue was observed in the absence of arabinose (data not shown).
Missiakas et al. (1994) reported that massive overexpression of wild-type DsbC in rich media could rescue a dsbA null mutant. This rescue may be due to oxidized cystine in the media oxidizing DsbC, which is then able to oxidize substrate proteins. Indeed, the simple addition of cystine and other oxidants is sufficient to rescue some phenotypes of dsbA and dsbB null mutations, in the absence of DsbC overexpression (Bardwell et al., 1991, 1993). To avoid the complicating effects of cystine and other small molecule oxidants present in rich media, we chose to perform our experiments in minimal media. Overproduction of wild-type DsbC, in strains grown in minimal media, resulted in no measurable motility. To investigate the level of DsbC overexpression from our promoter constructs, we prepared periplasmic extracts from the mutants and compared them to the starting strain. The level of expression of DsbC was unchanged in our mutants, showing that their ability to rescue dsbA null mutants was not due to enhanced DsbC expression (data not shown). Interestingly, SDS–PAGE revealed a 45 kDa protein that was present in all motile mutants and absent in both disulfide-negative strains (JCB817, JCB817 pMB69). The first 10 N-terminal amino acid residues of this band were sequenced and match exactly with the first 10 residues of E.coli alkaline phosphatase (RTPEMPVLEN). Alkaline phosphatase contains two disulfide bonds necessary for correct folding. If the disulfide bonds are not formed, the protein does not fold correctly and is degraded (Bardwell et al., 1991). This result provides additional evidence that the DsbC variants G49R and G49E are efficiently forming disulfide bonds in the absence of DsbA, the native donor of disulfide bonds in the periplasm.
DsbC G49R mutant is a monomer
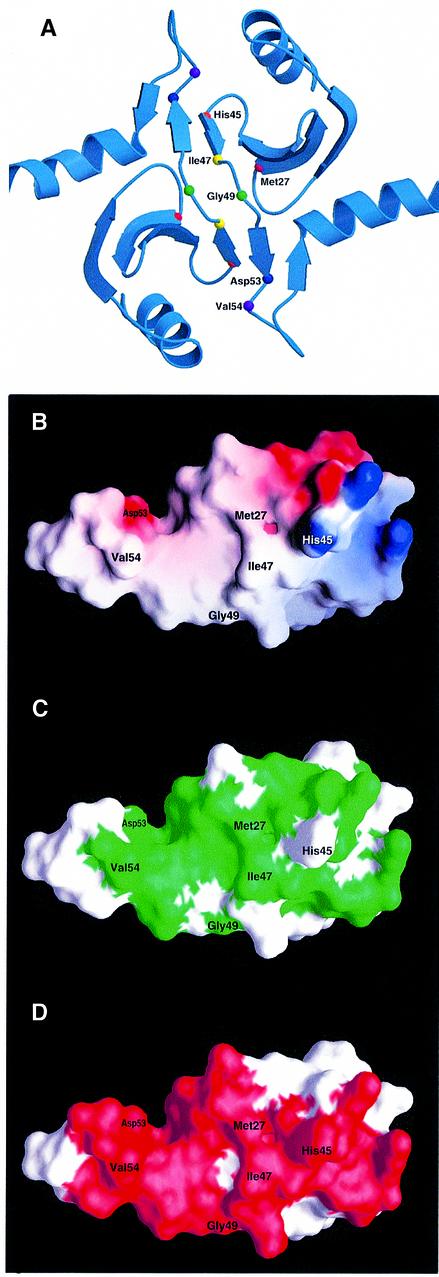
We isolated two mutants that are able to replace the function of DsbA. We conclude that both DsbC G49R and DsbC G49E are functionally more similar to DsbA than to wild-type DsbC. What distinguishes DsbC from DsbA? Although both DsbA and DsbC contain a thioredoxin fold, DsbC possesses an additional N-terminal domain, which causes DsbC to form a homodimer (Zapun et al., 1995; McCarthy et al., 2000). The N-terminal domain has also proven to be essential for DsbC’s activity as an isomerase in vitro (Sun and Wang, 2000). G49 is located in the N-terminal dimerization domain of DsbC and is a very well conserved residue. In a sequence alignment of 15 different DsbC N-terminal domains, G49 is present in 14 out of 15 DsbC proteins. It is even found in the related disulfide isomerase DsbG (data not shown). This high degree of conservation suggests that G49 is a very important residue. In the recently solved crystal structure of DsbC, G49 is located between two consecutive β-strands (Figure 3A). These two strands are the core of the dimerization interface and form an extended β-sheet with the corresponding monomer. The high conservation of the small residue G49 led us to propose that the substitution of G49 with large charged residues, such as arginine or glutamate in the isolated mutants, will act to disrupt the dimer. We therefore predict that G49 is essential for the dimerization of DsbC. Indeed, we had previously constructed the same arginine substitution at exactly the same glycine in an effort to disrupt the dimerization by rational design (see below). This represents an astonishing congruence between the in vivo genetic selection approach and rational design.
Fig. 3. Rational design of mutants in the dimerization domain of DsbC. (A) The crystal structure of DsbC is shown with the mutated residues represented as small spheres. The monomers interact via an N-terminal dimerization interface that consists of two very short consecutive β-strands from each monomer. Two extended β-sheets are formed, which consist of two strands from one monomer and four strands from the other. Two DsbC mutants that were found to complement a dsbA null mutant were mapped to G49 (drawn as a green sphere), which lies in between the interacting β-strands. These mutations replace G49 with either arginine or glutamate. An additional five residues, which were predicted to provide crucial interactions in the dimerization interface and were subject to mutagenesis, are also shown as small spheres. (B) Surface potential of DsbC’s dimerization interface. Only one monomer is shown. Values range from +7kT (blue) to 0 (white) to –7kT (red). M27, V54 and I47 were mutated to lysines. H45 forms a salt bridge with Asp53 on the other monomer (compare with Figure 3A), and was mutated to an aspartate. (C) Same as (B), except that the green color highlights conserved residues. Conserved residues were mapped according to the sequence alignment of 15 DsbC molecules. A BLAST search was performed with the entire DsbC protein sequence at the NCBI database of finished and unfinished bacterial genomes (http://www.ncbi.nlm.nih.gov/Microb_blast/unfinishedgenome.html). Fifteen microbial sequences were aligned with CLUSTAL W (Thompson et al., 1994). Identical and similar residues that are found in at least 60% of the aligned sequences are shaded in green. (D) Molecular surface of the dimerization domain showing residues involved in dimer formation. Note that most of the residues involved in dimerization are conserved.
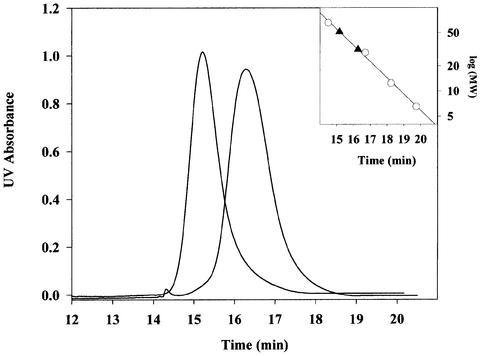
In order to investigate the biochemical properties of the mutant protein, we overexpressed and purified DsbC G49R from strain BL21. The purified protein was analyzed by Sephadex-200 gel filtration. Figure 2 shows that DsbC G49R has a significantly altered retention time as compared with wild-type DsbC. A standard curve was generated with proteins of known molecular weight. The observed retention time for DsbC G49R corresponds to an apparent mol. wt of 31.6 kDa, while wild-type DsbC runs with an apparent mol. wt of 51.4 kDa. This indicates that DsbC G49R is monomeric. Surprisingly, the loss of functional dimerization leads to a different function. The mutant protein acts as an in vivo donor of disulfide bonds rather than an isomerase.
Fig. 2. DsbC G49R is a monomer. The DsbC mutant G49R was purified and analyzed by analytical gel filtration. DsbC G49R shows a clear difference in migration on a Sephadex 200 column as compared with wild-type DsbC. The inset shows a standard curve generated with proteins of known molecular weight (open circles): bovine serum albumin (66 kDa), carbonic anhydrase (29 kDa), cytochrome c (12.4 kDa) and apoprotinin (6.5 kDa). The apparent molecular weights of wild-type DsbC and DsbC G49R (filled triangles) were determined according to this curve. An apparent mol. wt of 31.6 kDa was calculated for DsbC G49R. Wild-type DsbC runs with an apparent mol. wt of 51.4 kDa. This suggests that G49R is monomeric.
Rational design of dsbC mutants that complement a dsbA null mutation
Our results provide evidence that the dimerization of DsbC is essential for the separation of disulfide bond formation and isomerization. DsbC can be converted into a functional donor of disulfide bonds by two different in vivo selected mutations that disrupt its dimerization properties. We wanted to ascertain whether the disruption of the dimerization interface is what allows these mutants to function as oxidases. We reasoned that a rational design of mutants in the dimerization interface should reveal more DsbC variants that are able to rescue DsbA.
Substantial disruptions of the dimerization interfaces should lead to a disulfide-positive phenotype in a dsbA null mutant background. To design possible disrupting mutations, we focused on the interactions involving five residues in the interface. Figure 3 provides a view of the residues at the dimerization interface. The residues shown in Figure 3B and C are mostly hydrophobic and highly conserved. The surface area buried upon dimer formation is 1727 Å2 or 15% of the total monomer surface (Figure 3D). Residues 8, 11, 12, 23–25, 27, 40, 43–55 and 59 of each molecule are buried in the dimer interface. There are hydrophobic interactions between isoleucine 47 from molecule ‘a’ of DsbC (abbreviated as I47a) and I47b, V54a and I46b, and M27a and M27b (data not shown). In addition, there is a potential salt bridge between H45a and D53b. To disrupt these interactions, residues I47, V54 or M27 were mutated to lysine. We reasoned that the charged lysine should be very unfavorable in the hydrophobic environment provided by the residues shown in Figure 3. Residue H45 was mutated to aspartate. This was hypothesized to lead to decreased affinity of the DsbC monomers for each other since this mutation destroys a vital salt bridge that links the two monomers in the crystal structure. The positions of these mutations are shown in Figure 3A. All the designed mutations, which we predicted would disrupt the dimer interface, are able to rescue the non-motile phenotype of the dsbA null mutant (data not shown).
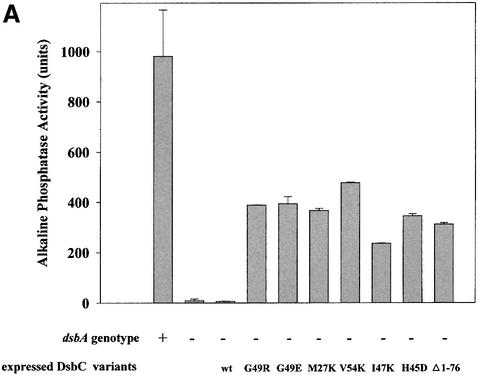
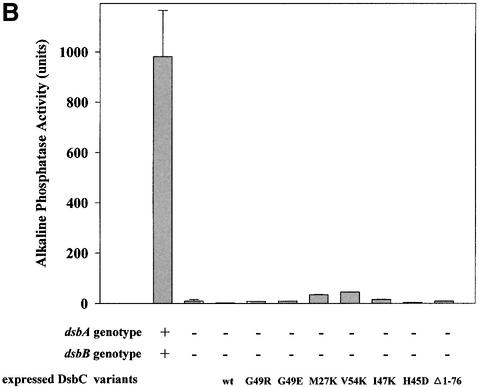
To confirm and to quantify the extent of the rescue, we performed alkaline phosphatase assays. Alkaline phosphatase is a periplasmic protein that requires two disulfide bonds for activity. The amount of alkaline phosphatase activity in a cell is often used as a measure of the extent of disulfide defect. We observed nearly full restoration of alkaline phosphatase activity in all of our mutants that were designed to disrupt the dimerization interface (Figure 4A). The simplest explanation for these results is that dimerization acts to shield DsbC from the re-oxidant DsbB. This explanation makes two predictions: (i) that the dimerization domain should be dispensable for the observed complementation; and (ii) that complementation should be DsbB dependent. To test the first prediction, we constructed a DsbC variant in which the entire dimerization domain had been deleted by site-specific mutagenesis. This variant, expressing only the thioredoxin domain of DsbC, is able to complement the dsbA null phenotype for both motility and alkaline phosphatase activity (Figure 4A).
Fig. 4. (A) All mutant DsbCs promote folding of alkaline phosphatase in vivo. Plasmids encoding DsbC or DsbC variants, respectively, were transformed into JCB817 (dsbA–). The strains were grown in M63 media supplemented with 0.4% glycerol as carbon source and 0.2% arabinose to induce expression from the pBAD33 derived plasmids. The activity of alkaline phosphatase was determined by a standard assay described in Materials and methods. JCB816 (wild type) served as a positive control. The different plasmids are listed in Table II. (B) Rescue of the dsbA null phenotype strongly depends on the presence of DsbB. Conditions were essentially as in (A), except that JCB818 (dsbA–, dsbB–) served as a strain background.
Complementation depends on the presence of DsbB
We have isolated and rationally designed mutants in the disulfide isomerase DsbC that act like the oxidase DsbA. We speculated that for these mutants to be efficient disulfide oxidases, they have to be re-oxidized by DsbB. We therefore tested whether the mutant DsbC proteins were still able to complement a disulfide-negative phenotype in a dsbA dsbB null strain. Figure 4B shows that none of the mutants can complement dsbA deficiency in the absence of dsbB. This strongly suggests that DsbB directly or indirectly is responsible for the re-oxidation of DsbC proteins containing any of the mutations G49E, G49R, H45D, I47K, V54K or M27K in the dimerization domain. We also performed AMS trapping experiments, which showed that the DsbC mutants were partially oxidized in vivo in a dsbA– background and that this oxidation was dsbB dependent (data not shown). It appears that we have been able to get DsbC to switch allegiance from being part of the isomerization pathway to becoming part of the DsbB-dependent oxidative pathway.
DsbD depletion is not sufficient for full complementation by wild-type DsbC
We considered the possibility that our DsbC mutant proteins could also be impaired in their recognition by DsbD, the in vivo reductant of DsbC. The observed phenotypes could therefore be in part due to a loss of interaction between mutant DsbC proteins and DsbD. If the entire rescue were due to lack of interaction with DsbD, we would not expect the rescue to be DsbB dependent. In addition, wild-type DsbC should be able to confer a motile phenotype if DsbD is removed by deletion. However, we did not observe restoration of alkaline phosphatase activity and motility for wild-type DsbC in a dsbA dsbD null strain (Table I). We conclude that the ability of our DsbC mutants to rescue the dsbA null phenotype can not solely be due to the loss of DsbC–DsbD interaction. In an effort to determine whether this loss of interaction may contribute in some small way to the observed phenotype, we measured the extent of rescue of these mutants in a dsbA– dsbD– strain and compared it with the extent of rescue in a dsbA– dsbD+ strain. The mutant DsbC plasmids allowed the expression of similar levels of alkaline phosphatase as a percentage of wild type independent of whether the strain background was dsbA– or dsbA– dsbD– (compare Figure 4A with Table I). The small amount of increased expression in the dsbD– strain may be due to the generally more oxidizing environment present in this background. If loss of DsbC–DsbD interaction is playing a role in DsbC acting as an oxidase, it is only playing a small one. In contrast, DsbC is unable to serve as an oxidase in a dsbB– background (Figure 4B). The most important new feature of our mutant DsbC proteins is thus their ability to serve as substrates for DsbB rather than their inability to be reduced by DsbD.
Table I. Effect of dsbD depletion on motility and alkaline phosphatase activity.
| Strain | Relevant genotype | DsbC variant expressed | Motility | Alkaline phosphatase activity (% of wt) |
|---|---|---|---|---|
| MC1000 | wt | – | + + + | 100 |
| FED126 | dsbD– | – | + + + | 109 |
| MB102 | dsbA– dsbD– | – | – | 10 |
| MB102 pMB69 | dsbA– dsbD– | wt | – | 16 |
| MB102 pMB89 | dsbA– dsbD– | G49R | + + | 76 |
| MB102 pMB90 | dsbA– dsbD– | G49E | + + | 88 |
| MB102 pMB91 | dsbA– dsbD– | M27K | + + | 82 |
| MB102 pMB92 | dsbA– dsbD– | I47K | + + | 74 |
| MB102 pMB93 | dsbA– dsbD– | V54K | + + | 73 |
| MB102 pMB94 | dsbA– dsbD– | H45D | + + | 75 |
| MB102 pMB96 | dsbA– dsbD– | Δ1–76 | + + | 44 |
DsbB re-oxidizes monomeric DsbC in vitro
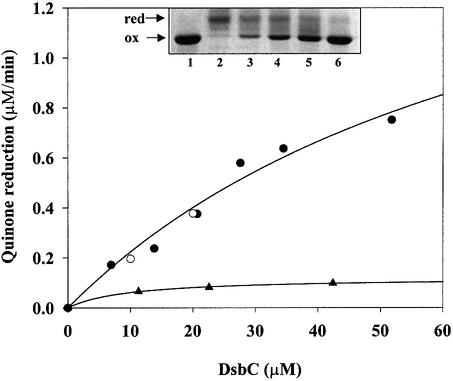
The DsbC H45D mutant was purified, its active site disulfide was reduced and its oligomerization state was analyzed by gel filtration using a Sephadex-200 column. DsbC H45D runs with a retention time of 16.90 min as compared with 15.17 min for wild-type DsbC. This corresponds to a change in apparent mol. wt from 51.4 to 37.4 kDa, suggesting that the H45D mutant is monomeric. The change is not due to degradation since the protein runs as a single band on SDS–PAGE of the same molecular weight as wild-type DsbC (data not shown). This result agrees with the prediction from the in vivo data for this mutant (Figure 4) and further supports our model. To obtain direct evidence for interaction between a mutant DsbC protein and DsbB, an enzymatic assay for the quinone reductase activity of DsbB was employed. DsbB provides the oxidative power for the periplasm by reducing ubiquinone, thus linking disulfide bond formation and electron transport (Bader et al., 1999, 2000). We mixed the reduced H45D or G49R mutants with catalytic quantities of DsbB and oxidized ubiquinone. We monitored both the reduction of ubiquinone and the oxidation of DsbC by trapping with the sulfhydryl reagent AMS. No reaction occurred when quinone alone was added. However, when catalytic quantities of DsbB were added, DsbC H45D and G49R were oxidized (Figure 5, insert). The oxidation of DsbC was accompanied by the reduction of ubiquinone, which could be quantified by following the change in absorption at 275 nm. Figure 5 shows that DsbB efficiently oxidizes both the DsbC H45D and G49R mutant proteins in vitro, but can not effectively oxidize wild-type DsbC. This suggests that monomerizing DsbC forces it to become a specific substrate for DsbB. The finding that DsbB oxidizes monomeric DsbC in vitro agrees well with the in vivo observation of dsbB dependency.
Fig. 5. DsbB re-oxidizes monomeric DsbC in vitro. DsbB’s ability to re-oxidize monomeric DsbC (H45D, filled circles; G49R, open circles) was measured by its ubiquinone reductase activity. The assay buffer consists of 100 mM sodium phosphate pH 6.0, 0.1% dodecyl-maltoside, 20 µM ubiquinone and 1 µM DsbB. The concentration of reduced DsbC H45D was steadily increased up to 50 µM. Activity of DsbB was plotted as quinone reductase activity. Reduced wild-type DsbC served as a control (filled triangles), and a much lower ubiquinone reductase activity was observed. This suggests that monomeric DsbC mutants are a superior substrate to wild-type DsbC. Insert: to visualize the redox state of DsbC after incubation with DsbB and ubiquinone. Lane 1, purified oxidized DsbC; lanes 2–6, samples taken after 0.5, 1, 2, 3 and 4 h and precipitated with 5% TCA. Free thiols were modified with AMS essentially as described before (Kobayashi et al., 1997).
Discussion
Two distinct pathways assist the proper formation of disulfide bonds within folding proteins in the periplasm (Bardwell et al., 1993; Rietsch et al., 1996). The DsbA– DsbB system drives the net introduction of disulfide bonds by providing a link to the oxidative power of the electron transport chain. DsbA is a strong oxidant that has the potential of introducing wrong disulfide bonds not normally found in the native structure of proteins (Rietsch et al., 1996; Sone et al., 1997). Efficient folding of proteins containing multiple disulfide bonds requires an enzyme that shuffles incorrectly formed disulfide bonds toward the ones found in the native state of the protein. DsbC is a disulfide isomerase that is thought to correct wrongly formed disulfide bonds by rearrangement towards the native ones. DsbC is part of the reductive pathway that is linked, via DsbD, to the thioredoxin system of the cytosol (Rietsch et al., 1997).
Although we have learned much about the flow of electrons through the two pathways, little was known about how they are able to co-exist. This work now shows that wild-type DsbC can be converted into a net donor of disulfide bonds in vivo by a single mutation at residue G49. Both DsbC G49E and G49R are able to rescue the dsbA null phenotype. These mutants are able to restore motility as well as alkaline phosphatase activity in vivo. Both alkaline phosphatase and the P ring of the flagellar motor need to be oxidized to be active (Bardwell et al., 1991; Dailey and Berg, 1993). Restoration of these two activities indicates that the mutant DsbC proteins are capable of restoring oxidative power to the periplasm. Substitution of G49 for arginine disrupts the ability of DsbC to form a stable dimer. This loss of function is accompanied by the gain of a new function: monomeric DsbC acts as a donor of disulfide bonds in vivo. The finding that loss of functional dimerization allows DsbC to act as an oxidase led us to believe that other mutations disrupting the dimerization interface may cause the same effect. The resulting mutants M27K, H45D, I47K, V54K, and one that removes the entire dimerization domain, complement a dsbA null mutant for motility and alkaline phosphatase.
The finding that different dsbC mutants were able to promote the formation of disulfide bonds raised the question of how these mutant proteins are re-oxidized. Mutant DsbC proteins do not rescue in the absence of a functional DsbB in vivo. This finding suggests that the mutant DsbC proteins are re-oxidized in vivo by DsbB, while wild-type DsbC is not. We tested this observation in vitro and found that monomeric DsbC is re-oxidized enzymatically by DsbB, whereas wild-type DsbC is not. We conclude that the failure to form a functional dimer turns DsbC into a substrate for DsbB. Hence, monomeric DsbC becomes part of the oxidative pathway. These results demonstrate how the delicate balance between the oxidative and reductive pathway is controlled by the dimerization of DsbC. It explains how DsbA and DsbC exhibit rather different redox activities in the same cellular compartment without interfering with each other.
It has been reported that thioredoxin or protein disulfide isomerase (PDI), when exported to the periplasm, is able to rescue a dsbA null phenotype (Jonda et al., 1999; Debarbieux and Beckwith, 2000). The observed rescue depends on the presence of the dsbB gene, suggesting that thioredoxin and PDI are re-oxidized by DsbB. According to our results, DsbB also re-oxidizes the thioredoxin fold of monomeric DsbC. Thus, DsbB seems to be rather promiscuous in recognizing the thioredoxin folds of DsbA, monomeric DsbC, thioredoxin and PDI. However, this promiscuity does not interfere with disulfide bond isomerization in wild-type cells because the thioredoxin fold of wild-type DsbC is not oxidized by DsbB. It is very likely that DsbC evolved from thioredoxin and that evolutionary pressure forced DsbC to find a mechanism whereby its active site is protected from DsbB. Apparently, this was achieved by dimerization of the thioredoxin domains of DsbC, thus allowing DsbC to function as an isomerase despite the presence of DsbB.
DsbC is kept in a reduced state by DsbD. In order to achieve this, DsbD must distinguish between DsbC and DsbA since any cross-talk between DsbD and DsbA would lead to inactivation of the oxidative pathway. This is very similar to the problem of how the cell avoids inactivation of the isomerization pathway due to DsbB. On the other hand, DsbD contains a thioredoxin fold, which has to be protected from DsbB-mediated oxidation. Once again, it is quite surprising how the thioredoxin folds of the various players of these systems, despite their structural similarity, seem to be protected from destructive cycles of oxidation and reduction. Very little is known about the actual mechanisms of how the isomerization pathway operates. It is therefore possible that additional barriers exist between the two pathways, which have not been identified yet.
Materials and methods
Bacterial strains and plasmid constructs
The strains and plasmids used in this study are shown in Table II. Mutations were introduced into the dsbC gene using the Stratagene QuikChange kit. The forward primers used are listed in Table III. A DsbC variant (pMB96) lacking the first 76 amino acids of the native protein was constructed following the Stratagene Exsite Protocol with some modifications. In brief, whole plasmid pMB69 was amplified by PCR using the phosphorylated primers listed in Table III. The linear PCR product was ligated in vitro, DpnI digested and transformed into XL1-blue. Transformants were screened by the loss of a BsmI site. All variants were confirmed by sequence analysis of the entire dsbC gene.
Table II. Strains and plasmids.
| Genotype | |
|---|---|
| Strain | |
| MC1000 | araD139, Δ[ara-leu]7679, galU, galK, Δ[lac]174, rpsL, thi-1 |
| JCB816 | MC1000 phoR |
| JCB817 | MC1000 phoR dsbA– |
| JCB818 | MC1000 phoR dsbA–, dsbB– |
| BL21 (DE3) | F– ompT hsdSB (rB– mB–) gal [dcm] [lon] |
| Plasmid | |
| pMB69 | pBAD33a with dsbC |
| pMB89 | pBAD33a with dsbC G49R |
| pMB90 | pBAD33a with dsbC G49E |
| pMB91 | pBAD33a with dsbC M27K |
| pMB92 | pBAD33a with dsbC I47K |
| pMB93 | pBAD33a with dsbC V54K |
| pMB94 | pBAD33a with dsbC H45D |
| pMB96 | pBAD33a with dsbC Δ1–76 |
| pMB66 | pET22 with dsbC G49R |
| pMB78 | pET28a with dsbC H45D |
Table III. Primers used in this study.
| pMB89 | 5′-AAACATATCATTCAGAGGCCTAT GTATGACGTTAGTGGCACGGC |
| pMB90 | 5′-AAACATATCATTCAGGAGCCCATGTATGACGTTAGTGGCACGGC |
| pMB91 | 5′-CGCCTGTAGCTGGCAAGAAGACAGTGTTAACTAACAGCG |
| pMB92 | 5′-GATGATGGTAAACATATCAAGCAGGGCCCAATGTATGACGTTAG |
| pMB93 | 5′-GGGGCCAATGTATGACAAGAGTGGTACCGCTCCGGTCAATGTCACC |
| pMB94/pMB78 | 5′-CATCACCGATGATGGTAAAGATATCATTCAGGGGCC |
| pMB96 | P-GAGATGATCGTTTATAAAGCGCCG |
| P-AGCCTGAGCAAAGCCTGAAAACG | |
| pMB66 | 5′-AAACATATCATTCAGAGGCCTATGTATGACGTTAGTGGCACGGC |
Selection for DsbC mutants that complement a dsbA null phenotype
Random mutagenesis of the dsbC gene was carried out in the Stratagene XL-1 Red Epicurean Coli mutator strain (Stratagene, CA). Plasmid pMB69 containing wild-type dsbC was transformed into XL-1 Red. pMB69 is derived from pBAD33a (Guzman et al., 1995), and was obtained from G.Gergiou (University of Texas). This strain lacks three key enzymes for DNA repair, which leads to a 5000-fold increase in the mutation rate. DNA extracted from ∼1500 individual colonies was pooled and electroporated into the dsbA null mutant JCB817. Mutant colonies that conferred a dsbA+ phenotype were selected by their ability to swim through 0.3% agar minimal media motility plates. Transformants from each library were pooled and serially diluted in 150 mM NaCl. Aliquots of 100 µl from these dilutions were added to 4 ml of cooling motility medium containing 0.05% arabinose and poured on top of a portion of pre-poured motility plates. Plates were incubated at 37°C until motile puffs arose that were moving into the portion of the motility plates that had not been overlaid with the mutagenized library (∼48 h). Motile puffs were streaked out onto chloramphenicol plates. Plasmids from these colonies were extracted and re-transformed into JCB817. For all of these strains, motility depended upon supplementation of the media with arabinose, suggesting that expression of DsbC was necessary for their motility. Stabbing a single colony onto minimal plates containing 0.3% agar assessed the motility of these colonies. The minimal media contained 0.4% glycerol, 0.2% arabinose, 0.2% ammonium sulfate, 1 mM magnesium sulfate, 0.1% casamino acids, 2 µg/ml biotin, 2 µg/ml nicotinamide, 0.2 µg/ml riboflavin and 2 µg/ml thiamin.
Expression and purification of proteins
DsbB was purified as described previously (Bader et al., 1999). The DsbC variant G49E was purified by anion exchange chromatography as has been described previously for wild-type DsbC (Missiakas et al., 1994). His-tagged wild-type DsbC and the variant H45D were purified over HiTrap Chelating columns (Pharmacia) charged with nickel according to the manufacturer’s manual. DsbA and DsbC were reduced with 10 mM dithiothreitol (DTT); excess DTT was removed by gel filtration on PD10 columns (Pharmacia). All proteins were >95% pure after purification.
Biochemical assays
In vivo alkaline phosphatase activity was measured using cells prepared from 25 ml overnight cultures grown in the same minimal media as described above. The OD600 for each culture was measured and a 1 ml aliquot of each overnight culture (1:10) was trapped with 0.1 M iodoacetamide for 20 min at 0°C. The cells were then washed with ice-cold 67 mM MOPS, 83 mM sodium chloride, 13 mM ammonium chloride pH 7.3, supplemented with 10 mM magnesium chloride and 10 mM iodoacetamide. The washed cells were used in the assay described previously (San Millan et al., 1989; Boyd et al., 1993). The substrate used was Sigma 104 (Sigma Diagnostics, St Louis, MO).
DsbB activity was determined by measuring its quinone reductase activity at 275 nm. In brief, 20 µM decyl-ubiquinone was incubated with reduced DsbC H45D in 100 mM sodium phosphate pH 6.0, 0.1% dodecyl-maltoside. The reaction was started by the addition of DsbB to a final concentration of 1 µM. As a control, quinone reduction was determined in the presence of wild-type DsbC. To visualize the redox state of DsbC after incubation with DsbB and ubiquinone, samples were taken and precipitated with 5% trichloroacetic acid (TCA). Free thiols were modified with 4-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid (AMS) essentially as described before (Kobayashi et al., 1997).
Analytical gel filtration of DsbC variants was performed on Sephadex 200 (Pharmacia) in 10 mM HEPES pH 7.5 and 300 mM NaCl. Wild-type DsbC served as a control. The presence of the His tag did not affect the retention time on this column as compared with the protein without a His tag. The Sigma low molecular weight gel filtration standard was used to generate a standard curve in order to obtain apparent molecular weights for DsbC variants.
Structure analysis
Residues buried in the dimer interface area were calculated using CNS (Brunger et al., 1998). Program O (Jones et al., 1991) was used to visualize the structure and to determine atom distances. Molecular surface representations were generated by GRASP (Nicholls et al., 1991). Coordinates for DsbC are available at the Protein Data Bank (accession code 1eej).
Acknowledgments
Acknowledgements
We are grateful to G.Georgiou (University of Texas) for the gift of plasmid pBAD33a-dsbC (pMB69). We would also like to acknowledge Bart Staker for many helpful suggestions. This work was supported by an NIH grant (to J.C.A.B.). P.W.H. is supported by the Boehringer Ingelheim Fonds. J.C.A.B. is a PEW scholar.
References
- Bader M., Muse,W., Ballou,D.P., Gassner,C. and Bardwell,J.C. (1999) Oxidative protein folding is driven by the electron transport system. Cell, 98, 217–227. [DOI] [PubMed] [Google Scholar]
- Bader M.W., Xie,T., Yu,C.A. and Bardwell,J.C. (2000) Disulfide bonds are generated by quinone reduction. J. Biol. Chem., 275, 26082–26088. [DOI] [PubMed] [Google Scholar]
- Bardwell J.C., McGovern,K. and Beckwith,J. (1991) Identification of a protein required for disulfide bond formation in vivo. Cell, 67, 581–589. [DOI] [PubMed] [Google Scholar]
- Bardwell J.C., Lee,J.O., Jander,G., Martin,N., Belin,D. and Beckwith,J. (1993) A pathway for disulfide bond formation in vivo. Proc. Natl Acad. Sci. USA, 90, 1038–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessette P.H., Cotto,J.J., Gilbert,H.F. and Georgiou,G. (1999) In vivo and in vitro function of the Escherichia coli periplasmic cysteine oxidoreductase DsbG. J. Biol. Chem., 274, 7784–7792. [DOI] [PubMed] [Google Scholar]
- Boyd D., Traxler,B. and Beckwith,J. (1993) Analysis of the topology of a membrane protein by using a minimum number of alkaline phosphatase fusions. J. Bacteriol., 175, 553–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger A.T. et al. (1998) Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Dailey F.E. and Berg,H.C. (1993) Mutants in disulfide bond formation that disrupt flagellar assembly in Escherichia coli. Proc. Natl Acad. Sci. USA, 90, 1043–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darby N.J., Raina,S. and Creighton,T.E. (1998) Contributions of substrate binding to the catalytic activity of DsbC. Biochemistry, 37, 783–791. [DOI] [PubMed] [Google Scholar]
- Debarbieux L. and Beckwith,J. (1999) Electron avenue: pathways of disulfide bond formation and isomerization. Cell, 99, 117–119. [DOI] [PubMed] [Google Scholar]
- Debarbieux L. and Beckwith,J. (2000) On the functional inter changeability, oxidant versus reductant, of members of the thioredoxin superfamily. J. Bacteriol., 182, 723–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert H.F. (1997) Protein disulfide isomerase and assisted protein folding. J. Biol. Chem., 272, 29399–29402. [DOI] [PubMed] [Google Scholar]
- Guilhot C., Jander,G., Martin,N.L. and Beckwith,J. (1995) Evidence that the pathway of disulfide bond formation in Escherichia coli involves interactions between the cysteines of DsbB and DsbA. Proc. Natl Acad. Sci. USA, 92, 9895–9899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman L.M., Belin,D., Carson,M.J. and Beckwith,J. (1995) Tight regulation, modulation and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol., 177, 4121–4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joly J.C. and Swartz,J.R. (1997) In vitro and in vivo redox states of the Escherichia coli periplasmic oxidoreductases DsbA and DsbC. Biochemistry, 36, 10067–10072. [DOI] [PubMed] [Google Scholar]
- Jonda S., Huber-Wunderlich,M., Glockshuber,R. and Mossner,E. (1999) Complementation of DsbA deficiency with secreted thioredoxin variants reveals the crucial role of an efficient dithiol oxidant for catalyzed protein folding in the bacterial periplasm. EMBO J., 18, 3271–3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for binding protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Kobayashi T., Kishigami,S., Sone,M., Inokuchi,H., Mogi,T. and Ito,K. (1997) Respiratory chain is required to maintain oxidized states of the DsbA–DsbB disulfide bond formation system in aerobically growing Escherichia coli cells. Proc. Natl Acad. Sci. USA, 94, 11857–11862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy A.A., Haebel,P.W., Torronen,A., Rybin,V., Baker,E.N. and Metcalf,P. (2000) Crystal structure of the protein disulfide bond isomerase, DsbC, from Escherichia coli. Nature Struct. Biol., 7, 196–199. [DOI] [PubMed] [Google Scholar]
- Missiakas D., Georgopoulos,C. and Raina,S. (1994) The Escherichia coli dsbC (xprA) gene encodes a periplasmic protein involved in disulfide bond formation. EMBO J., 13, 2013–2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missiakas D., Schwager,F. and Raina,S. (1995) Identification and characterization of a new disulfide isomerase-like protein (DsbD) in Escherichia coli. EMBO J., 14, 3415–3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls A., Sharp,K.A. and Honig,B. (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins, 11, 281–296. [DOI] [PubMed] [Google Scholar]
- Raina S. and Missiakas,D. (1997) Making and breaking disulfide bonds. Annu. Rev. Microbiol., 51, 179–202. [DOI] [PubMed] [Google Scholar]
- Rietsch A. and Beckwith,J. (1998) The genetics of disulfide bond metabolism. Annu. Rev. Genet., 32, 163–184. [DOI] [PubMed] [Google Scholar]
- Rietsch A., Belin,D., Martin,N. and Beckwith,J. (1996) An in vivo pathway for disulfide bond isomerization in Escherichia coli. Proc. Natl Acad. Sci. USA, 93, 13048–13053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietsch A., Bessette,P., Georgiou,G. and Beckwith,J. (1997) Reduction of the periplasmic disulfide bond isomerase, DsbC, occurs by passage of electrons from cytoplasmic thioredoxin. J. Bacteriol., 179, 6602–6608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Millan J.L., Boyd,D., Dalbey,R., Wickner,W. and Beckwith,J. (1989) Use of phoA fusions to study the topology of the Escherichia coli inner membrane protein leader peptidase. J. Bacteriol., 171, 5536–5541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sone M., Akiyama,Y. and Ito,K. (1997) Differential in vivo roles played by DsbA and DsbC in the formation of protein disulfide bonds. J. Biol. Chem., 272, 10349–10352. [DOI] [PubMed] [Google Scholar]
- Sun X.X. and Wang,C.C. (2000) The N-terminal sequence (residues 1–65) is essential for dimerization, activities and peptide binding of Escherichia coli DsbC. J. Biol. Chem., 275, 22743–22749. [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wunderlich M. and Glockshuber,R. (1993) Redox properties of protein disulfide isomerase (DsbA) from Escherichia coli. Protein Sci., 2, 717–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapun A., Missiakas,D., Raina,S. and Creighton,T.E. (1995) Structural and functional characterization of DsbC, a protein involved in disulfide bond formation in Escherichia coli. Biochemistry, 34, 5075–5089. [DOI] [PubMed] [Google Scholar]