Abstract
LAP2 belongs to a family of nuclear membrane proteins sharing a 43 residue LEM domain. All LAP2 isoforms have the same N-terminal ‘constant’ region (LAP2-c), which includes the LEM domain, plus a C-terminal ‘variable’ region. LAP2-c polypeptide inhibits nuclear assembly in Xenopus extracts, and binds in vitro to barrier-to-autointegration factor (BAF), a DNA-bridging protein. We tested 17 Xenopus LAP2-c mutants for nuclear assembly inhibition, and binding to BAF and BAF⋅DNA complexes. LEM domain mutations disrupted all activities tested. Some mutations outside the LEM domain had no effect on binding to BAF, but disrupted activity in Xenopus extracts, suggesting that LAP2-c has an additional unknown function required to inhibit nuclear assembly. Mutagenesis results suggest that BAF changes conformation when complexed with DNA. The binding affinity of LAP2 was higher for BAF⋅DNA complexes than for BAF, suggesting that these interactions are physiologically relevant. Nucleoplasmic domains of Xenopus LAP2 isoforms varied 9-fold in their affinities for BAF, but all isoforms supershifted BAF⋅DNA complexes. We propose that the LEM domain is a core BAF-binding domain that can be modulated by the variable regions of LAP2 isoforms.
Keywords: barrier-to-autointegration factor/emerin/Emery–Dreifuss muscular dystrophy/lamin-associated polypeptide 2 (LAP2)/nuclear envelope
Introduction
Multicellular eukaryotes express a family of proteins, termed ‘LEM domain’ proteins, at the inner nuclear membrane. The first recognized LEM domain proteins were LAP2 (Foisner and Gerace, 1993), emerin (Bione et al., 1994; Manilal et al., 1996; Clements et al., 2000) and MAN1 (Lee et al., 2000; Lin et al., 2000), which share a conserved 43 residue motif (Lin et al., 2000). Additional LEM domain proteins now include otefin (Padan et al., 1990; Goldberg et al., 1998) and Lem-3 (Lee et al., 2000). LEM domain proteins are absent from single-cell eukaryotes and plants, but are conserved among multicellular (animal) eukaryotes (reviewed by Dechat et al., 2000b; Cohen et al., 2001). LAP2 and emerin interact with lamin filaments, which underlie the inner nuclear membrane and are essential for nuclear structure and chromatin attachment to the envelope (reviewed by Gant and Wilson, 1997; Gruenbaum et al., 2000; Moir et al., 2000). The functions of lamins and LEM domain proteins have become medically relevant, because defects in emerin and A-type lamins cause Emery–Dreifuss muscular dystrophy (Bione et al., 1994; Bonne et al., 1999; Sullivan et al., 1999; reviewed by Morris and Manilal, 1999). Three additional tissue-specific diseases are caused by missense mutations that map to different regions of lamin A (Bonne et al., 2000). These discoveries, plus evidence that the lamina is essential (Lenz-Bohme et al., 1997; Liu et al., 2000) and interacts with transcriptional regulators such as Rb (reviewed by Cohen et al., 2001), suggest that lamins and lamin-binding proteins might influence gene expression (Wilson, 2000).
At least nine distinct protein isoforms of LAP2 are generated by alternative mRNA splicing in mammals (Harris et al., 1994; Furukawa et al., 1995; Berger et al., 1996) and Xenopus (Gant et al., 1999; Lang et al., 1999). The most abundant isoforms are LAP2α and LAP2β, which interact with both lamins and chromatin. LAP2β is an inner nuclear membrane protein, and binds specifically to lamin B1 (Foisner and Gerace, 1993). LAP2β is proposed to be important for nuclear reassembly and expansion (Yang et al., 1997; Gant et al., 1999), and has been implicated along with lamins as having a positive role in DNA replication competence (reviewed by Gruenbaum et al., 2000; Moir et al., 2000). The activity of LAP2β appears to be regulated, since it is differentially phosphorylated during interphase (Dreger et al., 1999) and mitosis (Foisner and Gerace, 1993). The other major isoform, LAP2α, lacks a transmembrane domain, and is found throughout the nuclear matrix during interphase (Dechat et al., 1998). LAP2α binds to A-type lamins (Vlcek et al., 1999). The binding specificities of other LAP2 isoforms towards lamins have not been tested.
We are interested in how LAP2 interacts with chromatin. All known isoforms of LAP2 have the same N-terminal region, encoded by exons 1–3 (Berger et al., 1996), which we will term the ‘constant region’ of LAP2 (LAP2-c). Deletion studies have shown that LAP2-c binds to chromatin in vitro, and that residues 1–88 of rat LAP2 are sufficient to bind to chromatin (Furukawa et al., 1998; however, see Vlcek et al., 1999). The C-terminal ‘variable’ regions, which differ between isoforms, are responsible for binding to lamins (Foisner and Gerace, 1993; Furukawa et al., 1998; Dechat et al., 2000a) and perhaps additional partners (Furukawa et al., 1997; Vlcek et al., 1999).
Experiments with Xenopus cell-free extracts demonstrate that LAP2-c can arrest nuclear envelope assembly when added to Xenopus egg extracts (Gant et al., 1999). The arrested phenotype is striking: the nuclear membranes attach to chromatin and assemble pore complexes, but the envelope fails to enclose, does not accumulate lamins and exhibits a characteristic ‘scalloped’ morphology (Gant et al., 1999), suggesting that LAP2-c may compete for binding sites important for nuclear envelope assembly. A binding partner for LAP2 on chromatin was discovered by Furukawa (1999) in a yeast two-hybrid screen, and confirmed by bead pull-down assays. This partner is a novel DNA-binding protein named BAF, the barrier-to-autointegration factor (Chen and Engelman, 1998; Lee and Craigie, 1998). [Note that there is another chromatin protein named BAF, the Brahma-associated factor (Wang et al., 1996), which is unrelated to the BAF discussed here.] BAF was discovered as a cytoplasmic factor that is acquired by retroviral pre-integration complexes and prevents viral DNA from undergoing suicidal self-integration (Lee and Craigie, 1998). It is small (89 residues; 10 kDa), and is highly conserved among multicellular eukaryotes (60% identical between humans and Caenorhabditis elegans; Cai et al., 1998). The structure of the BAF dimer has been determined by solution NMR (Cai et al., 1998) and X-ray crystallography (Umland et al., 2000). The function of BAF is unknown, but it is essential in C.elegans (Zheng et al., 2000). BAF co-localizes with chromosomal DNA during both interphase and mitosis (Furukawa, 1999). However, some BAF is also present in the cytoplasm (Lee and Craigie, 1998). BAF dimers are proposed to cross-bridge DNA restriction fragments non-covalently, forming complexes too large to enter agarose gels (Lee and Craigie, 1998). BAF binds and cross-bridges double-stranded DNA (dsDNA) in vitro, with no detectable sequence specificity (Zheng et al., 2000). When incubated with 21 bp dsDNA oligonucleotides, BAF forms a discrete nucleoprotein complex consisting of six BAF dimers and an estimated six molecules of DNA (Zheng et al., 2000), which we will denote BAF⋅DNA complexes. It is not understood how this in vitro DNA-bridging activity relates to BAF’s function(s) in vivo.
To investigate the function of the chromatin- and BAF-binding constant region of LAP2, we tested a series of alanine substitution mutations in Xenopus LAP2-c for their activity in Xenopus nuclear assembly reactions, and for binding to purified BAF dimers, DNA and BAF⋅DNA complexes.
Results
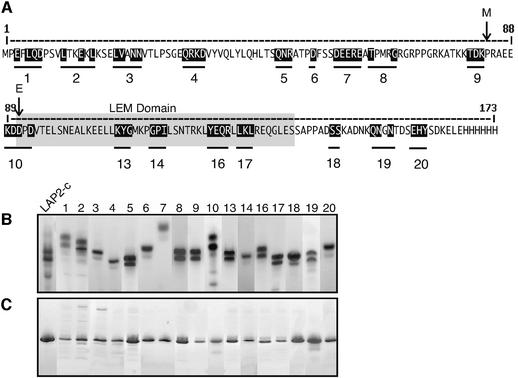
To dissect the function of the constant region of LAP2, we created 17 LAP2-c mutants by site-directed mutagenesis of Xenopus LAP2 residues 1–165 (Figure 1A; Table I). We mutated clusters of residues that were conserved between LAP2 proteins from different species, or conserved between LAP2-c and emerin (data not shown). Mutated residues were replaced by alanine, a neutral amino acid. Each His-tagged mutant LAP2-c protein was expressed in Escherichia coli and purified by affinity on Ni–agarose columns and gel filtration (see Materials and methods). Wild-type LAP2-c and all LAP2-c mutants eluted from gel filtration columns as one main peak of 40 kDa, plus a smaller peak near the void volume, which probably represented large multimers or aggregates of LAP2-c; both peaks contained LAP2-c protein when run on SDS gels or native gels (data not shown). The gel filtration results suggested two possibilities: either LAP2-c molecules might form dimers and multimers in solution, or LAP2-c monomers migrate anomalously during gel filtration. For simplicity, we assumed that LAP2-c was a monomer.
Fig. 1. Alanine substitution mutagenesis of the constant region (residues 1–165) of Xenopus LAP2 (LAP2-c). (A) The amino acid sequence of LAP2-c. Black-shaded residues were replaced by alanine; each set of mutations is indicated by a numbered bar. Mutant cluster m19 includes a tyrosine rather than alanine substitution for the second asparagine. The LEM domain (residues 91–138) is shaded gray. Arrows marked M and E indicate the start of homology to the N-termini of human MAN1 and emerin, respectively. (B) Coomassie Blue-stained native 8–25% acrylamide gel of purified wild-type LAP2-c and LAP2-c mutants, numbered as shown in (A). (C) The same proteins run on SDS-containing denaturing 8–25% acrylamide gels, stained with Coomassie Blue.
Table I. Oligonucleotides used for Xenopus LAP2 mutagenesis.
| Mutant No. | Original residues | Mutant residues | Mutagenic portion of oligonucleotide |
|---|---|---|---|
| M1 | EFLQD | AFAAA | (+15) 9GCA TTC GCA GCG GCT (+10) |
| M2 | LTKEKL | ATKAKA | (+9) 33GCT ACT AAA GCA AAG GCC (+7) |
| M3 | LVANN | AAAAA | (+9) 60GCG GCT GCG GCC GCC (+11) |
| M4 | QRKD | AAAA | (+9) 95GCG GCG GCA GCT (+10) |
| M5 | QNR | AAA | (+9) 144GCG GCC GCC (+8) |
| M6 | D | A | (+6) 162GCC (+9) |
| M7 | DEERE | AAAAA | (+9) 174GCC GCG GCG GCA GCG (+24) |
| M8 | TPMRG | APMRA | (+6) 192GCC CCT ATG AGA GCC (+7) |
| M9 | TDK | AAA | (+9) 243GCC GCC GCA (+8) |
| M10 | KDDPD | AAAPA | (+9) 267GCA GCT GCC CCA GCT (+7) |
| M13 | KYG | AAA | (+22) 324GCT GCA GCT (+14) |
| M14 | GPI | AAA | (+9) 42GCC GCA GCA (+15) |
| M16 | YEQR | AAAA | (+9) 372GCT GCG GCA GCA (+15) |
| M17 | LKL | AAA | (+9) 387GCT GCA GCA (+11) |
| M18 | SS | AA | (+9) 435GCT GCA (+12) |
| M19 | QNGN | AAGY | (+12) 456GCT GCA GGC TAC (+13) |
| M20 | EHY | AAA | (+9) 477GCG GCT GCC (+16) |
Numbers in parentheses (e.g. +15) indicate additional non-mutagenic nucleotides 5′ and 3′ to the mutagenic nucleotides. A superscript number indicates the first nucleotide of the mutagenic region with respect to the start codon A. Underlined nucleotides are mutagenic.
Wild-type and mutant LAP2-c proteins all entered native gels, consistent with but not proving that they were properly folded (Figure 1B). Each protein migrated at a characteristic position in native gels, based on its shape and net charge-to-mass ratio. Unexpectedly, wild-type LAP2-c migrated as two major bands of similar intensity in Coomassie-stained native gels (Figure 1B, LAP2-c), as did most mutants. The second band on native gels was not a degradation product, because these same proteins migrated as one major band in SDS-containing gels (Figure 1C; we speculate that LAP2-c may adapt two conformations in solution). We concluded that the purified mutant LAP2-c proteins were soluble, and thus suitable for functional testing in vitro.
Mutations both inside and outside the LEM domain disrupt the effect of LAP2-c on nuclear assembly
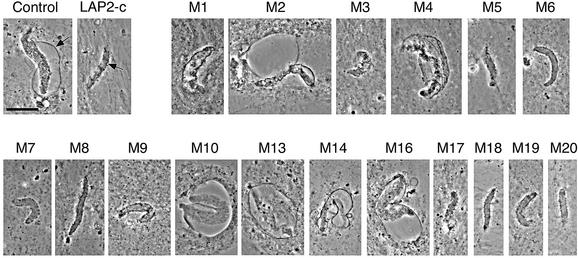
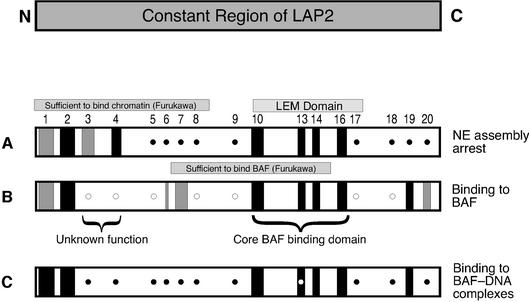
When wild-type LAP2-c is added to Xenopus cell-free nuclear assembly reactions, the forming nuclear envelopes arrest with a unique ‘scalloped’ morphology (Figure 2, ‘LAP2-c’; for transmission electron micrographs see Gant et al., 1999). To determine which residues were required for this inhibitory activity, each LAP2-c mutant was added at time zero to the Xenopus nuclear assembly reaction, which consists of membranes, chromatin, cytosol and an ATP-regenerating system (see Materials and methods). Reactions were incubated for 3–5 h to allow nuclei to assemble and reach a terminal phenotype, and then imaged by fluorescence to detect DNA (data not shown) and by phase contrast microscopy to detect nuclear envelopes (Figure 2). Many LAP2-c mutants (m5, 6, 7, 8, 9, 17, 18, 19 and 20) were indistinguishable from wild type, and inhibited nuclear assembly. However, several mutants (m2, 4, 10, 13, 14 and 16) failed to inhibit nuclear assembly (Figure 2, compare with no addition positive ‘control’). Two other mutants were partially disrupted: nuclei assembled in the presence of m1 or m3 became enclosed, but did not enlarge fully. Mutations that disrupted the effect of LAP2-c on nuclear assembly in vitro mapped to two locations: the N-terminus (mutations m1–m4) and the LEM domain (m10–m16; these results are summarized in Figure 5A).
Fig. 2. Effects of LAP2-c mutants on nuclear assembly in Xenopus egg extracts. Nuclear assembly reactions were supplemented at time zero with no additions (Control), wild-type LAP2-c protein or each LAP2-c mutant (numbered as shown in Figure 1) to a final concentration of 10 µM. Nuclei were imaged by phase contrast microscopy after 3–5 h, to allow nuclei to reach their terminal arrest phenotype; images shown are typical of at least 20 nuclei examined. The nuclear envelope is visible as a thin continuous dark line (arrow in Control). When nuclear assembly is arrested by LAP2-c, the surface area is smaller and the thin dark line is discontinuous and concave (‘scalloped’; arrow in LAP2-c; see Gant et al., 1999). Bar = 20 µm.
Fig. 5. Summary of mutagenesis results for Xenopus LAP2-c. Numbers indicate the position of each cluster of mutations in the LAP2 amino acid sequence (see Figure 1). Each row summarizes the activity of the mutant proteins in a given assay, as described in the text. (A) ability to inhibit nuclear assembly in Xenopus egg extracts; (B) ability to bind to BAF; (C) ability to supershift BAF⋅DNA complexes. Closed circles indicate that the mutant protein had the same activity as wild-type LAP2-c. Open circles indicate that the mutant protein had more activity (e.g. higher affinity for BAF) than wild-type LAP2-c. Black bars indicate loss of activity. Gray bars indicate partial loss of activity. An open circle inside a black bar for mutant m13 indicates that this mutant bound to BAF⋅DNA complexes in six out of 10 experiments. The LEM domain is indicated, as well as regions in Xenopus LAP2 that correspond to those sufficient to bind BAF in two-hybrid assays (rat residues 67–137; Furukawa, 1999), or sufficient to bind chromatin in vitro (rat residues 1–88; Furukawa et al., 1997).
Binding of LAP2-c mutants to BAF in solution
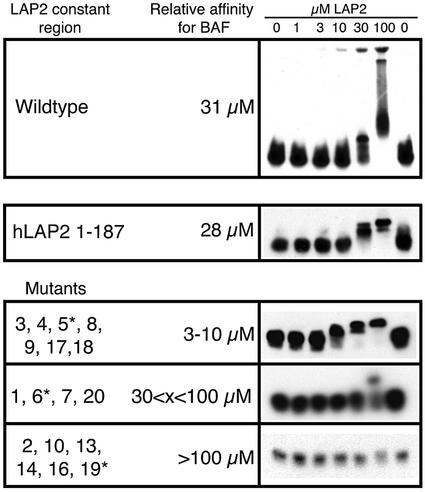
To determine which mutations disrupted LAP2-c binding to BAF, we performed solution binding assays with purified recombinant Xenopus LAP2-c protein and full-length recombinant human BAF (Cai et al., 1998), in the form of purified dimers. The formation of BAF–LAP2-c complexes was detected by changes in the migration of BAF (shifting to higher positions) in native gels immunoblotted for BAF (Figure 3; see Materials and methods). To estimate the relative binding affinity of each LAP2-c mutant for BAF, we varied the concentration of LAP2-c (wild-type or mutant) monomers from 0 to 100 µM in reactions that contained 10 µM purified BAF dimers. Densitometry was used to determine the concentration of LAP2-c at which 50% of BAF was shifted. The relative (not absolute) affinity of wild-type Xenopus LAP2-c for BAF was 31 µM (Figure 3, top), similar to that of nearly full-length Xenopus isoform 2 (see below). The LAP2-c mutants fell into three classes (Figure 3, mutants): those that bound BAF slightly better than wild-type LAP2-c (50% shifting at 3–10 µM; mutants m3, 4, 5, 8, 9, 17 and 18), those with equal or slightly lower affinity (50% shifting at 30–100 µM; mutants m1, 6, 7 and 20) and those that failed to bind BAF under these conditions (mutants m2, 10, 13, 14, 16 and 19). Mutations that severely disrupted LAP2-c binding to BAF mapped both within and outside the LEM domain. Mutations that slightly enhanced binding all mapped outside the LEM domain, as summarized in Figure 5B.
Fig. 3. Native gel shift assays for LAP2-c binding to purified recombinant human BAF dimers. BAF dimers (10 µM) were incubated with increasing concentrations (0–100 µM) of LAP2-c for 15 min to allow the proteins to bind in solution. Proteins were then resolved on native gels and immunoblotted for BAF (see Materials and methods). Relative binding affinities under these experimental conditions were estimated using densitometry scanning, to determine the LAP2-c concentration at which 50% of BAF shifted away from its original position. Results are shown for increasing concentrations of wild-type Xenopus LAP2-c (Wildtype), wild-type human LAP2-c (hLAP2 1–187; Gant et al., 1999) and Xenopus LAP2-c mutants (Mutants). LAP2-c mutants are grouped according to their relative affinities for BAF; for each group, the data shown on the right correspond to the mutant marked by an asterisk (*).
For comparison, we tested human LAP2-c binding to human BAF. The affinity of human LAP2-c (residues 1–187; Gant et al., 1999) for human BAF was similar to that of Xenopus LAP2-c for human BAF (28 versus 31 µM, respectively; Figure 3, hLAP2 1–187). This result was consistent with the high conservation of BAF protein in humans and Xenopus (84% identical; M.Segura and K.L.Wilson, unpublished findings), and suggested that our Xenopus LAP2 results are applicable to LAP2 proteins in humans.
As the concentration of LAP2-c was increased, BAF shifted to multiple positions, either forming a broad ‘smear’ or failing to enter the native gel at 100 µM LAP2-c (Figure 3, top). This reproducible result suggested that BAF and LAP2-c form a variety of higher order structures in solution. LAP2 was also detected by immunoblotting in these putative complexes (data not shown). However, LAP2 alone migrated at a variety of positions in native gels (e.g. see Figure 1B), including the positions to which BAF shifted in the presence of LAP2. Since the charge-to-mass ratio of LAP2–BAF complexes is similar to that of LAP2 alone, we were unable to detect the corresponding ‘shift’ of LAP2 into LAP2–BAF complexes.
LAP2-c interacts with BAF⋅DNA complexes
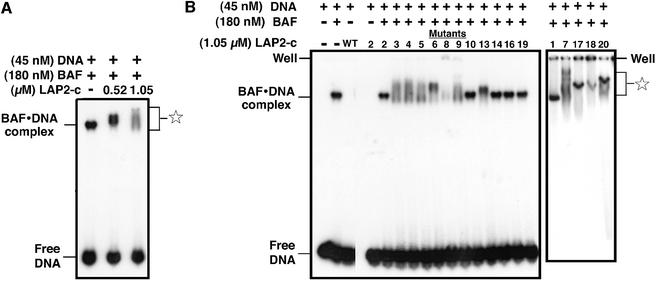
We hypothesized that LAP2-c might interact differently with BAF⋅DNA complexes than with BAF alone. To test this idea, each LAP2-c mutant was incubated with pre-formed BAF⋅DNA complexes (Materials and methods), which consist of six BAF dimers plus six molecules of DNA (Zheng et al., 2000). We used a small (21 bp) dsDNA to prevent the formation of BAF-‘bridged’ DNA aggregates (Lee and Craigie, 1998). The DNA was labeled with 32P, and BAF⋅DNA complexes were assembled for 30 min at a ratio of 4:1 (180 nM BAF dimers:45 nM DNA). Wild-type LAP2-c was then added and allowed to incubate for 30 min, and DNA-containing complexes were detected by native gel shift assay (Figure 4A; Materials and methods). The positions of free DNA and BAF⋅DNA complexes are shown (Figure 4A; Zheng et al., 2000). In reactions that contained ∼1 or ∼2 µM wild-type LAP2-c, the BAF⋅DNA complexes supershifted to successively higher positions in the gel (Figure 4, star). This result suggested that LAP2-c can bind to BAF⋅DNA complexes in vitro. Neither wild-type LAP2-c (Figure 4B, third lane, ‘wt’) nor any of the LAP2-c mutants (Figure 4B, mutant m2 and data not shown) shifted DNA in the absence of BAF. We therefore concluded that recombinant LAP2-c interacts primarily with the BAF, not the DNA, in BAF⋅DNA complexes.
Fig. 4. The constant region of LAP2 supershifts BAF⋅DNA complexes. BAF⋅DNA complexes were pre-assembled by mixing purified BAF dimers with 32P-labeled 21 bp dsDNA oligonucleotides; LAP2-c protein was then added and incubated for 30 min prior to separation on native gels and autoradiography to detect labeled DNA (see Materials and methods). (A) Autoradiograph of a native gel showing supershifting of BAF⋅DNA complexes by two concentrations of wild-type LAP2-c. (B) Autoradiograph of BAF⋅DNA complex supershifting assay with LAP2-c mutant proteins, which are numbered as shown in Figure 1. The positions of free DNA and BAF⋅DNA complexes are shown. The star indicates the positions of BAF⋅DNA complexes supershifted due to LAP2-c binding. Some supershifted complexes remained in the well, not entering the native gel.
Evidence for a conformational change in BAF when complexed with DNA: LAP2 mutant m13, which does not bind BAF, can supershift BAF⋅DNA complexes
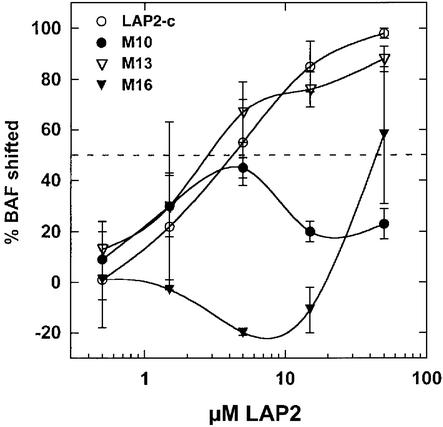
We tested the constant region mutants for binding to pre-formed BAF⋅DNA complexes. Most mutant proteins were either able to supershift BAF⋅DNA complexes (Figure 4B, mutants m3, 4, 5, 6, 7, 8, 9, 17, 18 and 20) or failed to supershift (Figure 4B, mutants m1, 2, 10, 14, 16 and 19), consistent with their ability to bind (or not bind) BAF dimers (Figure 3). These results are summarized in Figure 5C. However, there was an unexpected finding involving mutant m13. LEM domain mutant m13, which consistently failed to bind BAF dimers, was able to bind BAF⋅DNA complexes in four out of eight experiments, suggesting that it was on the threshold of a positive affinity towards BAF⋅DNA complexes (Figure 4B, supershifting by LAP2 mutant m13 is shown in the left panel). This result implied that DNA-complexed BAF might be structurally altered, such that the m13 mutations no longer mattered.
To test this unexpected result for mutant m13 further, we measured its relative binding affinity for pre-formed BAF⋅DNA complexes (Figure 6). The positive control for this experiment showed that wild-type LAP2-c prefers to interact with DNA-complexed BAF, exhibiting a 3-fold higher affinity for BAF⋅DNA complexes than for BAF alone (50% binding at ∼8 and ∼30 µM LAP2, respectively; Figures 6 and 3). This increased affinity might be due to the higher local concentration of BAF in complexes, or changes in BAF conformation that improve its binding to LAP2-c. The negative controls, mutants m10 and m16, both had low affinities for BAF and BAF⋅DNA complexes, as expected (50% binding at ≥100 µM LAP2-c; Figures 6 and 3). In two additional experiments, mutant m13 bound to pre-formed BAF⋅DNA complexes with an affinity of 6 µM (50% binding at 6 µM; Figure 6), contrasting with its undetectable affinity for BAF alone (50% binding at >100 µM; Figure 3). These relative affinities can be compared, because the results shown in Figures 3 and 6 were all obtained using the same concentration of BAF dimers (10 µM). These findings supported the hypothesis that the BAF structure might change when bound to DNA (see Discussion).
Fig. 6. LAP2-c mutant binding to pre-formed BAF⋅DNA complexes. Binding reactions were performed as described in Figure 3, with the BAF concentration held constant at 10 µM, DNA at 2.5 µM, and the concentration of LAP2-c varied from 0 to 100 µM. Binding was detected by gel shift of BAF away from its starting position (in reactions containing 2.5 µM DNA, no LAP2-c) in immunoblots of native gels probed with antibodies to BAF (not shown). The averages of densitometric scans from two experiments are shown (± SEM); the formation of BAF complexes (%BAF shifted: 100% minus %BAF remaining in original position) was plotted as a function of LAP2-c concentration. The dotted line indicates 50% binding. Mutants are numbered as in Figure 1.
LAP2 isoforms differ in their ability to bind BAF
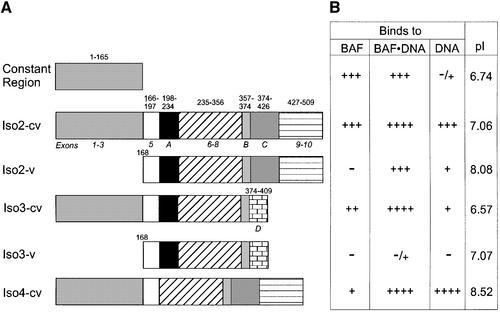
We wondered whether different variable regions of LAP2 might influence the constant region’s interactions with BAF. To test this idea, we measured the BAF-binding activity of recombinant Xenopus LAP2 isoforms 2, 3 and 4, which are related to mammalian LAP2β (Gant et al., 1999). Isoforms 2 and 4 have additional exons, and are therefore larger than LAP2β, whereas isoform 3 is shorter and has a unique C-terminal domain (see Figure 9A for diagram). In the following experiments, ‘cv’ will designate the nearly full-length isoform (constant plus variable regions), ‘c’ the constant region alone and ‘v’ the variable region alone.
Fig. 9. Summary of binding activities of different LAP2 isoforms. (A) Diagram of the constant region of Xenopus LAP2 (LAP2-c), Xenopus LAP2 isoforms 2, 3 and 4 (termed iso2-cv, iso3-cv and iso4-cv, respectively) and the variable domains of isoforms 2 and 3 (iso2-v and iso3-v). Exons are numbered according to the mouse LAP2 gene (Berger et al., 1996); letters correspond to additional exons identified in Xenopus LAP2 isoforms (Gant et al., 1999). (B) Summary of binding results in this study. Binding to BAF (relative affinities as determined in Figure 7): +++, wild-type binding to BAF; ++, 3-fold reduced binding to BAF; +, 9-fold reduced binding to BAF; –, no detectable binding to BAF. Binding to BAF⋅DNA complexes and to DNA: binding was assayed qualitatively in five independent experiments, including that shown in Figure 8. PI, isolectric point; the calculated net charges at pH 8.3 (native gel conditions) are: LAP2-c, –4.0; iso2-cv, –5.0; iso2-v, –0.6; iso3-cv, –7.7; iso3-v, –3.3; iso4-cv, +1.0.
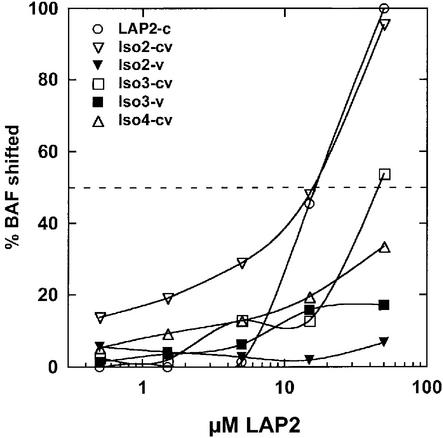
We first measured the relative affinity of BAF binding to each LAP2 isoform in the absence of DNA, by native gel shift assays and immunoblotting for BAF. The results are presented in Figure 7. BAF bound equally well to LAP2-c and iso2-cv, suggesting that the variable domain of iso form 2 had no effect on binding to BAF. Interestingly, the other isoforms bound BAF with affinities that were 3-fold lower (iso3-cv; 50% binding at ∼100 µM) or 9-fold lower (iso4-cv; 50% binding extrapolated to ∼300 µM) than isoform 2 (Figure 7). Neither of the two variable regions tested alone (iso2-v and iso3-v) bound detectably to BAF in the absence of the constant region. We concluded that the variable regions of isoform 4 and (to a lesser extent) isoform 3 can inhibit the BAF-binding activity of the constant region when linked in cis to the constant region.
Fig. 7. Different isoforms of LAP2 have different affinities for BAF dimers. Each indicated LAP2 isoform or variable region was allowed to bind BAF in solution, and LAP2–BAF complexes were detected by immunoblotting of native gels (not shown). The concentration of BAF dimers was held constant at 10 µM, and the concentration of LAP2 monomers was varied from 0 to 100 µM. We plotted the percentage of BAF that entered complexes (shifted to higher positions in the gel) as a function of the LAP2 concentration, as described for Figure 6. The dotted line indicates 50% shifted. This experiment was performed twice, with similar results.
Some but not all LAP2 isoforms can bind DNA
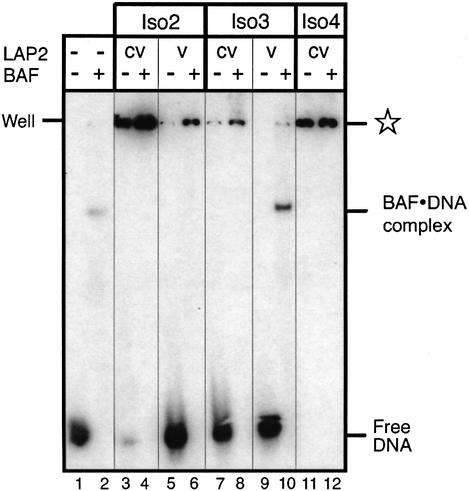
The above results suggested that LAP2 isoforms have different affinities for BAF, which might allow them to play distinct physiological roles in vivo. To explore this idea, we tested whether LAP2 isoforms differ in their ability to bind BAF⋅DNA complexes. As a control, we first tested whether they could bind to DNA alone, since mammalian LAP2β binds non-specifically to both ds- and single-stranded DNA cellulose via residues 244–296 (Furukawa et al., 1997), which are somewhat conserved (27% identical, 44% similar) in Xenopus LAP2. These experiments were carried out with 32P-labeled DNA and native gel shift assays. Control lanes showed the positions of DNA and BAF⋅DNA complexes (Figure 8, lanes 1 and 2, respectively). Both iso2-cv and iso4-cv supershifted DNA efficiently in the absence of BAF (Figure 8, lanes 3 and 11); in contrast, iso3-cv gave a low but detectable signal for DNA binding (Figure 8, lane 7). Thus, two out of three Xenopus LAP2 isoforms tested had strong affinity for DNA; this affinity did not correlate with their isoelectric point or net charge in native gels (see Figure 9B), suggesting that LAP2 isoforms can differ significantly in their DNA-binding properties. DNA-binding results from five independent experiments are summarized in Figure 9B.
Fig. 8. Native gel shift assay for binding of [32P]DNA to Xenopus LAP2 isoforms in the presence or absence of BAF. LAP2-c and the constant-plus-variable (cv) regions or variable regions (v) of LAP2 isoform 2 (iso2), isoform 3 (iso3) and isoform 4 (iso4) were incubated at a concentration of 2.8 µM with either 60 nM DNA or pre-formed BAF⋅DNA complexes (240 nM BAF plus 60 nM DNA; see Materials and methods). An autoradiograph of the native gel is shown, with the positions of free 32P-labeled DNA and BAF⋅DNA complexes indicated. The star indicates the positions of complexes supershifted due to LAP2 binding.
Variable region of isoform 2 can bind BAF⋅DNA complexes, but not BAF or DNA
In the absence of the constant region, iso2-v and iso3-v did not bind to BAF (Figure 7), and showed little or no binding to DNA (Figure 8, lanes 5 and 9). However, iso2-v, but not iso3-v, reproducibly shifted BAF⋅DNA complexes (Figure 8, lanes 6 and 10, respectively). These results suggested that the variable region of isoform 2 might bind to BAF⋅DNA complexes independently of the LEM domain.
Discussion
Our work provides biochemical evidence for interactions between LAP2 and BAF, confirming Furukawa’s (1999) discovery that BAF binds LAP2 in a yeast two-hybrid screen. Furukawa showed that residues 67–137 of rat LAP2, which includes most of the LEM domain, were sufficient to bind BAF. Our mutagenesis defines residues 89–127 of Xenopus LAP2 as a core BAF-binding region, with the important caveat that certain residues outside the LEM domain also contribute (directly or indirectly) to the interaction (see Figure 5). We further show that the LEM domain is essential for all assayed functions of LAP2-c, including its interactions with BAF and BAF⋅DNA complexes, and its ability to inhibit nuclear envelope formation competitively. Thus, the LEM domain and its associated BAF-binding activity appear to be central to LAP2 function. We do not know yet whether the LEM domain is essential for the enhancement of DNA replication by larger LAP2 polypeptides (constant plus variable domains; Gant et al., 1999).
Our work yielded the interesting result that LAP2 polypeptides prefer to bind BAF in nucleoprotein complexes with DNA (BAF⋅DNA complexes; Zheng et al., 2000), because they had higher relative affinities for the nucleoprotein complex than for BAF dimers. This finding supports the hypothesis that LAP2 binds to chromatin in vivo, and suggests that LAP2 isoforms might interact with both lamins and BAF⋅DNA complexes, thereby anchoring chromatin to the lamina and inner nuclear membrane. Surprisingly, the variable region of Xenopus LAP2 isoform 2, which had no LEM domain, also interacted with BAF⋅DNA complexes (see Figure 9). We speculate that the variable region of iso2 might have a ‘cryptic LEM-like’ domain (perhaps in exon C, exon 9 or exon 10), similar to the ‘LEM-like’ structural motif predicted for the constant region (Lin et al., 2000; see below). However, BLAST searching yielded only two regions with very weak homology to the LEM domain: one was located in exon A (residues 202–228) and the other overlapped exon A and exon 6 (residues 218–264). Since neither of these weak homologies resided in exon C, we can not yet explain the binding of iso2-v to BAF⋅DNA complexes. Isoform 4, which lacks exon A, had the lowest affinity for BAF in the absence of DNA, consistent with (but not proving) that regions of weak LEM domain homology possibly contribute to interactions with BAF. Further work is needed to test the hypothesis that multiple regions of LAP2 contribute to its affinity for BAF⋅DNA complexes.
The constant region of LAP2 has two functions
LAP2-c mutants m3 and m4 provided evidence that the constant region of LAP2 has two separable functions, because both mutants could bind BAF (slightly better than wild-type LAP2-c), but were unable to inhibit nuclear envelope assembly in Xenopus egg extracts. These mutants define a small conserved region near the N-terminus of LAP2 (residues 20–35) that is both unique to LAP2 and necessary for competitively blocking nuclear envelope assembly (see Figure 5). Interestingly, residues 1–50 are proposed to form a ‘LEM-like’ structural motif based on protein folding predictions (Lin et al., 2000), and this same region (residues 1–88) was sufficient to bind chromatin in an in vitro assay (Furukawa et al., 1998). We hypothesize that the N-terminus of LAP2 (particularly the region disrupted by mutations m3 and m4) may interact with a chromatin partner other than BAF, and we are trying to identify this putative new partner. We note that LBR, a non-LEM domain nuclear envelope protein, interacts with multiple partners including lamin B (Ye and Worman, 1994), DNA (Ye and Worman, 1994), heterochromatin (via Hp1; Ye and Worman, 1996) and chromatin (via DNA secondary structures and nucleosome linkers; Duband-Goulet and Courvalin, 2000). Thus, a new binding partner for LAP2 would not be surprising.
Variable regions linked in cis to the constant region can influence the affinity of LAP2 for BAF
Three tested isoforms of LAP2 differed 9-fold in their relative affinities for BAF. The variable region of isoform 2 was neutral when connected in cis to the constant region, having no apparent effect on the binding affinity. However, isoform 4 had 9-fold reduced affinity for BAF. This was remarkable, given that isoforms 2 and 4 differ by a single exon (Xenopus LAP2 exon A; Gant et al., 1999; see Figure 9). We speculate that residues encoded by exon A might either disrupt an inhibitory function or positively enhance binding to BAF. Isoform 3 had an intermediate (3-fold decreased) affinity for BAF: its variable region differs from that of isoform 2 by the loss of exons C, 9 and 10, and the gain of exon D (see Figure 9). The simplest model to explain the reduced affinity of isoform 3 is that residues encoded by exon D mildly inhibit binding to BAF. One main conclusion from our work is that even closely related LAP2 isoforms may have distinct biochemical properties. We do not yet know whether these differences in affinity for BAF or DNA are physiologically relevant, since the nucleoplasmic domains of all three LAP2 isoforms could supershift BAF⋅DNA complexes under our experimental conditions, and produce similar phenotypes when added to Xenopus nuclear assembly extracts (K.K.Lee, B.Lee and K.L.Wilson, unpublished data). Interestingly, the region containing exon 6 of LAP2, which is unique to LAP2β, interacts in yeast two-hybrid assays with a transcription factor named germ cell-less (mGCL; A.Simon, personal communication). This result supports our view that small domains within each LAP isoform may contribute uniquely to its binding interactions.
Evidence for conformational changes in BAF, and the formation of ‘higher order’ complexes between BAF and LAP2
The binding affinities reported here are relative, not absolute, and were used only to compare the activities of wild-type and mutant LAP2 proteins in specific assays. Because we used Xenopus LAP2 and human BAF, we expected the same results but slightly higher binding affinities for proteins from the same species (e.g. see Gant et al., 1999). Indeed, Xenopus BAF is 84% identical and 91% similar to human BAF (M.Segura and K.L.Wilson, unpublished data), and the BAF-binding affinities of Xenopus LAP2-c (31 µM) and human LAP2-c (28 µM) are similar. Other factors that might affect LAP2 binding, which were not provided under our assay conditions, include its phosphorylation status (Dreger et al., 1999), other potential post-translational modifications and additional binding partners such as lamins.
The crystal structure of the BAF dimer has been solved (Umland et al., 2000), but not the structure of the BAF⋅DNA complex. Based on the properties of mutant m13, which alters residues in the center of the LEM domain, we deduce that BAF changes its conformation at least slightly when complexed with DNA. Mutant m13 failed to bind BAF dimers, but was able to supershift BAF⋅DNA complexes. This result only makes sense if the residues mutated in LAP2-c mutant m13 (108KYG) recognize BAF differently in a BAF dimer versus a BAF⋅DNA complex. A further prediction from our work is that the stoichiometry of LAP2–BAF complexes varies as a function of the LAP2 concentration, and that binding between LAP2 and BAF might not follow simple first-order kinetics. Higher concentrations of LAP2 produced ‘mega-shifted’ complexes that failed to enter native gels, and had distinct honeycomb-like morphologies when imaged by negative-stain electron microscopy and field emission scanning electron microscopy (M.Zastrow, D.K.Shumaker and K.L.Wilson, unpublished data). Further biochemical and biophysical analysis will be important to understand these structures and determine their functional implications.
Implications for chromatin structure and nuclear envelope attachment to chromatin
One major prediction from this work is that all LEM proteins have the potential to interact with BAF dimers and BAF⋅DNA complexes. This prediction was tested recently and supported in studies with emerin; emerin binds to BAF in vitro, and mutations in the LEM domain disrupt emerin’s binding to BAF (K.K.Lee, R.S.Lee and K.L.Wilson, submitted). Thus, the function of LEM proteins is somehow linked to the function of BAF. Further studies of BAF and its interactions with LEM proteins will be important to understand what role, if any, BAF plays in linking chromatin to the nuclear lamina and nuclear envelope. Wild-type BAF added to Xenopus nuclear assembly reactions can influence nuclear envelope formation and chromatin decondensation, suggesting that BAF may be essential for nuclear assembly (M.Segura and K.L.Wilson, unpublished data). We propose that interactions between LEM proteins (e.g. LAP2, emerin and MAN1) and BAF may constitute a major mechanism for chromatin attachment to the nuclear envelope and lamina. Disruptions in the attachments between LEM proteins, lamins and BAF may also be functionally relevant for human diseases caused by defects in nuclear lamina proteins (Bonne et al., 2000; Cohen et al., 2001).
Materials and methods
Xenopus extracts and nuclear assembly reactions
Xenopus laevis eggs were fractionated by centrifugation at 200 000 g as previously described (Newmeyer and Wilson, 1991; Gant et al., 1999) to separate the cytosol and membrane fractions. Cytosol was supplemented with an ATP-regenerating system prior to freezing aliquots at –80°C. To assemble nuclei in vitro, fractions were thawed and mixed together on ice, with or without recombinant Xenopus LAP2 protein, and then supplemented with demembranated Xenopus sperm chromatin to a final concentration of 1000–1500/µl. A typical reaction contained 20 µl of cytosol, 3 µl of membranes, 1 µl of recombinant LAP2 and 1 µl of chromatin. The recombinant protein added did not exceed 9% of total volume. Nuclear assembly was initiated by incubating reactions at 22–25°C. At the indicated times, reactions were stained for DNA and fixed by adding 10 µg/ml Hoechst 33342 dye in 3.7% formaldehyde, placed under coverslips and imaged using a Nikon Microphot microscope with a Photometrics SenSys camera and IPLab software (Scanalytics).
Site-directed mutagenesis of Xenopus LAP2
The cDNA encoding the constant region of Xenopus LAP2 (residues 1–165) was mutagenized using the Quickchange site-directed mutagenesis kit (Stratagene Inc., La Jolla, CA). We started with the LAP2 cDNA in expression plasmid pET23a (Novagen Inc., Madison, WI; Gant et al., 1999), which adds a His6 tag to the C-terminus of LAP2. For each mutagenesis, the entire plasmid was replicated by PCR using a pair of complementary mutagenic oligonucleotides as primers. The altered nucleotides, which directed the replacement of the indicated wild-type residues with alanine, are shown in Table I. Each oligonucleotide also included 6–24 nucleotides of perfect homology flanking the mutagenic region, as indicated in parentheses in Table I. The original plasmid DNA strands were then destroyed by digestion with DpnI, and the remaining mutagenized plasmid DNA was transformed into Epicurian Coli XL1-blue cells. All mutations were verified by DNA sequencing (data not shown).
Expression and purification of recombinant LAP2 and BAF proteins
Wild-type human BAF was expressed in E.coli and dimers were purified as described (Cai et al., 1998). Recombinant Xenopus wild-type or mutant LAP2 proteins were purified as follows. Mutant LAP2 cDNAs were transformed into E.coli strain BL21 DE3 pLYS S (Novagen Inc.). The BL21 cells were grown to an OD600 of 0.4–0.6, and LAP2 protein expression was induced for 2–4 h using 0.4 mM isopropyl-β-d-thio galactopyranoside (IPTG; Sigma Chem. Corp., St Louis, MO). His-tagged recombinant LAP2 proteins were purified from bacteria as described (Gant et al., 1999), and concentrated by filtration on Centricon-3 units (Millipore Corp). Concentrated proteins were purified further by size fractionation on a Superdex 75 column (Pharmacia) attached to a Pharmacia FPLC System; sizes were determined relative to blue dextran (2000 kDa), catalase (232 kDa), albumin (67 kDa), ovalbumin (43 kDa), chymotrypsinogen A (25 kDa) and RNase (13.7 kDa; all from Amersham Pharmacia). The wild-type LAP2 constant region and all 17 mutant proteins eluted from gel filtration chromatography at the 40 kDa position, suggesting that they were all dimers, or that they all migrated anomalously. Consistent with the latter hypothesis, analytical centrifugation experiments suggest that LAP2 is a monomer with an extended conformation (R.Craigie, unpublished data). In samples that were frozen prior to FPLC analysis, a fraction of LAP2 protein eluted near the void volume, due to the formation of large aggregates (data not shown). Concentrations given herein are for BAF dimers (Cai et al., 1998) and LAP2 monomers.
Expression constructs for Xenopus LAP2 isoform 2 (iso2-cv; residues 1–509 of ‘clone 2’; Gant et al., 1999), isoform 3 (iso3-cv; residues 1–410 of ‘clone 3’) and isoform 4 (iso4-cv; residues 1–472 of ‘clone 4’), and corresponding variable regions iso2-v (residues 168–509) and iso3-v (168–410) were made in pET23a. The recombinant LAP2 isoform 3 used in these experiments was full length, since it naturally lacks a transmembrane domain. Iso2-cv and iso4-cv were truncated to remove the C-terminal transmembrane domain, such that their last residue corresponded to residue 509 (as numbered in isoform 2). The net charge of each protein was calculated using software available on the Web (www.scripps.edu/∼cdputnam/protcalc/html).
Native gel shift immunoblot assays for LAP2 binding to BAF and BAF⋅DNA complexes
Purified recombinant LAP2 and BAF proteins were mixed on ice in phosphate-buffered saline (PBS) plus native gel loading buffer (final concentration: 2 mM Tris pH 7.4, 0.2 mM EDTA pH 8, 0.002% bromophenol blue). Bromophenol blue did not interfere with protein binding (data not shown). Typical reactions contained 0.5–1 µl of LAP2, 1 µl of BAF, 2–2.5 µl of 1× PBS and 1 µl of native gel loading buffer. Proteins were incubated at 20–25°C for 15 min to allow binding; reactions were stopped by loading samples onto native 8–25% Phastgels and electrophoresis on a PhastSystem apparatus (Amersham Pharmacia Inc.). Some gels were stained with Coomassie R350 dye (PhastGel Blue R; Amersham). For immunoblotting, proteins were transferred to Immobilon P nitrocellulose (Millipore Inc.) in Tris–glycine buffer (25 mM Tris base, 192 mM glycine) using the semidry Phast Transfer apparatus (Amersham Pharmacia). Blots were blocked for 1 h at 20–25°C in PBS containing 0.1% Tween-20 and 10% non-fat dry milk. Subsequent incubations in primary and secondary antibodies were carried out in PBS containing 0.1% Tween-20. BAF was detected using a 1:5000 dilution of rabbit polyclonal antiserum #384 against full-length human BAF (either overnight at 4°C, or for 1 h at 20–25°C), followed by horseradish peroxidase-conjugated goat anti-rabbit antibodies (Pierce), and visualized by enhanced chemiluminescence (ECL) using either the Supersignal Dura (Pierce) or Amersham Pharmacia reagents.
Densitometry analysis of BAF immunoblots was carried out by scanning 16-bit images on a ScanMaker III (Microtech) scanner, and analyzed using IP Lab software (version 3.1.1c; Scanalytics, Vienna, VA) to determine the concentration of LAP2 at which 50% of BAF was displaced from its original position. Scanning data were plotted as spline curves using SigmaPlot software (version 4.16; Jandel Scientific, San Rafael, CA).
Native gel shift assays with [32P]DNA
The dsDNA substrate for binding assays shown in Figures 4 and 8 was made by first end labeling the single-stranded 21 base oligonucleotide RZ132 (5′-GTGTGGAAAATCTCTAGCAGT-3′) in the presence of [γ-32P]dATP using T4 polynucleotide kinase (New England Biolabs Inc.), as described (Zheng et al., 2000). The labeled oligonucleotide was then mixed with its complementary oligonucleotide RZ61 (5′-ACTGCTAGAGATTTTCCACAC-3′) and heated to 70°C. The two strands were annealed by slow cooling to 25–27°C. Annealed DNA was incubated with purified BAF dimers at the concentrations shown for 30 min at 30°C, and then subjected to electrophoresis on native 3–20% 14-cm-long acrylamide gels in TBE buffer. Gels were dried and exposed to X-ray film (Hyperfilm MP; Amersham Pharmacia Inc.). In experiments that also included LAP2, we pre-assembled BAF⋅DNA complexes by incubating 45 nM BAF dimers with 180 nM dsDNA (for Figure 4), or 240 nM BAF plus 60 nM dsDNA (Figure 8), at 30°C for 30 min. Complexes were then incubated with the indicated concentration of LAP2 for 30 min at 22–24°C, prior to native gel electrophoresis. BAF⋅DNA complexes and LAP2–BAF⋅DNA complexes were detected by changes in the migration of 32P-labeled DNA on native gels. The amount of free DNA in the binding reactions varied between experiments, as seen in the two panels of Figure 8B.
Acknowledgments
Acknowledgements
We thank Yosef Gruenbaum, Carolyn Machamer and Dan Leahy for comments on the manuscript. K.L.W. gratefully acknowledges the W.W.Smith Charitable Trust and the National Institutes of Health (RO1 GM48646) for supporting this work.
References
- Berger R. et al. (1996) The characterization and localization of the mouse thymopoietin/lamina-associated polypeptide 2 gene and its alternatively spliced products. Genome Res., 6, 361–370. [DOI] [PubMed] [Google Scholar]
- Bione S., Maestrini,E., Rivella,S., Mancini,M., Regis,S., Romeo,G. and Toniolo,D. (1994) Identification of a novel X-linked gene responsible for Emery–Dreifuss muscular dystrophy. Nature Genet., 8, 323–327. [DOI] [PubMed] [Google Scholar]
- Bonne G. et al. (1999) Mutations in the gene encoding lamin A/C cause autosomal dominant Emery–Dreifuss muscular dystrophy. Nature Genet., 21, 285–288. [DOI] [PubMed] [Google Scholar]
- Bonne G. et al. (2000) Clinical and molecular genetic spectrum of autosomal dominant Emery–Dreifuss muscular dystrophy due to mutations of the lamin A/C gene. Ann. Neurol., 48, 170–180. [PubMed] [Google Scholar]
- Cai M., Huang,Y., Zheng,R., Wei,S.Q., Ghirlando,R., Lee,M.S., Craigie,R., Gronenborn,A.M. and Clore,G.M. (1998) Solution structure of the cellular factor BAF responsible for protecting retroviral DNA from autointegration. Nature Struct. Biol., 5, 903–909. [DOI] [PubMed] [Google Scholar]
- Chen H. and Engelman,A. (1998) The barrier-to-autointegration protein is a host factor for HIV type 1 integration. Proc. Natl Acad. Sci. USA, 95, 15270–15274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements L., Manilal,S., Love,D.R. and Morris,G.E. (2000) Direct interaction between emerin and lamin A. Biochem. Biophys. Res. Commun., 267, 709–714. [DOI] [PubMed] [Google Scholar]
- Cohen M., Lee,K.K., Wilson,K.L. and Gruenbaum,Y. (2001) Transcriptional repression, apoptosis, human disease and the functional evolution of the nuclear lamina. Trends Biochem. Sci., 26, 41–47. [DOI] [PubMed] [Google Scholar]
- Dechat T., Gotzmann,J., Stockinger,A., Harris,C.A., Talle,M.A., Siekierka,J.J. and Foisner,R. (1998) Detergent–salt resistance of LAP2α in interphase nuclei and phosphorylation-dependent association with chromosomes early in nuclear assembly implies functions in nuclear structure dynamics. EMBO J., 17, 4887–4902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechat T., Korbei,B., Vaughan,O.A., Vlcek,S., Hutchison,C.J. and Foisner,R. (2000a) Lamina-associated polypeptide 2α binds intranuclear A-type lamins. J. Cell Sci., 113, 3473–3483. [DOI] [PubMed] [Google Scholar]
- Dechat T., Vlcek,S. and Foisner,R. (2000b) Review: lamina-associated polypeptide 2 isoforms and related proteins in cell cycle-dependent nuclear structure dynamics. J. Struct. Biol., 129, 335–345. [DOI] [PubMed] [Google Scholar]
- Dreger M., Otto,H., Neubauer,G., Mann,M. and Hucho,F. (1999) Identification of phosphorylation sites in native lamina-associated polypeptide 2β. Biochemistry, 38, 9426–9434. [DOI] [PubMed] [Google Scholar]
- Duband-Goulet I. and Courvalin,J.-C. (2000) Inner nuclear membrane protein LBR preferentially interacts with DNA secondary structures and nucleosomal linker. Biochemistry, 39, 6483–6488. [DOI] [PubMed] [Google Scholar]
- Foisner R. and Gerace,L. (1993) Integral membrane proteins of the nuclear envelope interact with lamins and chromosomes and binding is modulated by mitotic phosphorylation. Cell, 73, 1267–1279. [DOI] [PubMed] [Google Scholar]
- Furukawa K. (1999) LAP2 binding protein 1 (L2BP1/BAF) is a candidate mediator of LAP2–chromatin interaction. J. Cell Sci., 112, 2485–2492. [DOI] [PubMed] [Google Scholar]
- Furukawa K., Panté,N., Aebi,U. and Gerace,L. (1995) Cloning of a cDNA for lamina-associated polypeptide 2 (LAP2) and identification of regions that specify targeting to the nuclear envelope. EMBO J., 14, 1626–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa K., Glass,C. and Kondo,T. (1997) Characterization of the chromatin-binding activity of lamina-associated polypeptide (LAP) 2. Biochem. Biophys. Res. Commun., 238, 240–246. [DOI] [PubMed] [Google Scholar]
- Furukawa K., Fritze,C.E. and Gerace,L. (1998) The major nuclear envelope targeting domain of LAP2 coincides with its lamin binding region but is distinct from its chromatin interaction domain. J. Biol. Chem., 273, 4213–4219. [DOI] [PubMed] [Google Scholar]
- Gant T.M. and Wilson,K.L. (1997) Nuclear assembly. Annu. Rev. Cell Dev. Biol., 13, 669–695. [DOI] [PubMed] [Google Scholar]
- Gant T.M., Harris,C.A. and Wilson,K.L. (1999) Roles of LAP2 proteins in nuclear assembly and DNA replication: truncated LAP2β proteins alter lamina assembly, envelope formation, nuclear size and DNA replication efficiency in Xenopus laevis extracts. J. Cell Biol., 144, 1083–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg M. et al. (1998) Interactions among Drosophila nuclear envelope proteins lamin, otefin and YA. Mol. Cell. Biol., 18, 4315–4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenbaum Y., Wilson,K.L., Harel,A., Goldberg,M. and Cohen,M. (2000) Nuclear lamins: structural proteins with fundamental functions. J. Struct. Biol., 129, 313–323. [DOI] [PubMed] [Google Scholar]
- Harris C.A., Andryuk,P.J., Cline,S., Chan,H.K., Natarajan,A., Siekierka,J.J. and Goldstein,G. (1994) Three distinct human thymopoietins are derived from alternatively spliced mRNAs. Proc. Natl Acad. Sci. USA, 91, 6283–6287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang C., Paulin-Levasseur,M., Gajewski,A., Alsheimer,M., Benavente,R. and Krohne,G. (1999) Molecular characterization and developmentally regulated expression of Xenopus lamina-associated polypeptide 2 (XLAP2). J. Cell Sci., 112, 749–759. [DOI] [PubMed] [Google Scholar]
- Lee K.K., Gruenbaum,Y., Spann,P., Liu,J. and Wilson,K.L. (2000) C.elegans nuclear envelope proteins emerin, MAN1, lamin and nucleoporins reveal unique timing of nuclear envelope breakdown during mitosis. Mol. Biol. Cell, 11, 3089–3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.S. and Craigie,R. (1998) A previously unidentified host protein protects retroviral DNA from autointegration. Proc. Natl Acad. Sci. USA, 95, 1528–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz-Bohme B., Wismar,J., Fuchs,S., Reifegerste,R., Buchner,E., Betz,H. and Schmitt,B. (1997) Insertional mutation of the Drosophila nuclear lamin Dm0 gene results in defective nuclear envelopes, clustering of nuclear pore complexes and accumulation of annulate lamellae. J. Cell Biol., 137, 1001–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F., Blake,D.L., Callebaut,I., Skerjanc,I.S., McBurney,M.W., Pauline-Levasseur,M. and Worman,H.J. (2000) MAN1: an inner nuclear membrane protein that shares the LEM domain with lamina-associated polypeptide 2 and emerin. J. Biol. Chem., 275, 4840–4847. [DOI] [PubMed] [Google Scholar]
- Liu J., Ben-Shahar,T.R., Riemer,D., Treinin,M., Spann,P., Weber,K., Fire,A. and Gruenbaum,Y. (2000) Essential roles for Caenorhabditis elegans lamin gene in nuclear organization, cell cycle progression and spatial organization of nuclear pore complexes. Mol. Biol. Cell, 11, 3937–3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manilal S., Man,N.T., Sewry,C.A. and Morris,G.E. (1996) The Emery–Dreifuss muscular dystrophy protein, emerin, is a nuclear membrane protein. Hum. Mol. Genet., 5, 801–808. [DOI] [PubMed] [Google Scholar]
- Moir R.D., Spann,T.P., Lopez-Soler,R.I., Yoon,M., Goldman,A.E., Khuon,S. and Goldman,R.D. (2000) The dynamics of the nuclear lamins during the cell cycle: relationship between structure and function. J. Struct. Biol., 129, 324–334. [DOI] [PubMed] [Google Scholar]
- Morris G.E. and Manilal,S. (1999) Heart to heart: from nuclear proteins to Emery–Dreifuss muscular dystrophy. Hum. Mol. Genet., 8, 1847–1851. [DOI] [PubMed] [Google Scholar]
- Newmeyer D.D. and Wilson,K.L. (1991) Egg extracts for nuclear import and nuclear assembly reactions. Methods Cell Biol., 36, 607–634. [DOI] [PubMed] [Google Scholar]
- Padan R., Nainudel-Epszteyn,S., Goitein,R., Fainsod,A. and Gruenbaum,Y. (1990) Isolation and characterization of the Drosophila nuclear envelope otefin cDNA. J. Biol. Chem., 265, 7808–7813. [PubMed] [Google Scholar]
- Sullivan T., Escalente-Alcalde,D., Bhatt,H., Anver,M., Bhat,N., Nagashima,K., Stewart,C.L. and Burke,B. (1999) Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J. Cell Biol., 147, 913–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umland T.C., Wei,S.Q., Craigie,R. and Davies,D.R. (2000) Structural basis of DNA bridging by barrier-to-autointegration factor. Biochemistry, 39, 9130–9138. [DOI] [PubMed] [Google Scholar]
- Vlcek S.S., Just,H., Dechat,T. and Foisner,R. (1999) Functional diversity of LAP2α and LAP2β in postmitotic chromosome association is caused by an α-specific nuclear targeting domain. EMBO J., 18, 6370–6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W. et al. (1996) Purification and biochemical heterogeneity of the mammalian SWI–SNF complex. EMBO J., 15, 5370–5382. [PMC free article] [PubMed] [Google Scholar]
- Wilson K.L. (2000) The nuclear envelope, muscular dystrophy and gene expression. Trends Cell Biol., 10, 125–129. [DOI] [PubMed] [Google Scholar]
- Yang L., Guan,T. and Gerace,L. (1997) Lamin-binding fragment of LAP2 inhibits increase in nuclear volume during the cell cycle and progression into S phase. J. Cell Biol., 139, 1077–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Q. and Worman,H.J. (1994) Primary structure analysis and lamin B and DNA binding of human LBR, an integral protein of the nuclear envelope inner membrane. J. Biol. Chem., 269, 11306–11311. [PubMed] [Google Scholar]
- Ye Q. and Worman,H.J. (1996) Interaction between an integral protein of the nuclear envelope inner membrane and human chromodomain proteins homologous to Drosophila HP1. J. Biol. Chem., 271, 14653–14656. [DOI] [PubMed] [Google Scholar]
- Zheng R., Ghirlando,R., Lee,M.S., Mizuuchi,K., Krause,M. and Craigie,R. (2000) Barrier-to-autointegration factor (BAF) bridges DNA in a discrete, higher-order nucleoprotein complex. Proc. Natl Acad. Sci. USA, 97, 8997–9002. [DOI] [PMC free article] [PubMed] [Google Scholar]