Abstract
The human cytomegalovirus (HCMV) US11 polypeptide is a type I membrane glycoprotein that targets major histocompatibility complex (MHC) class I molecules for destruction in a proteasome-dependent manner. Although the US11 signal sequence appears to be a classical N-terminal signal peptide in terms of its sequence and cleavage site, a fraction of newly synthesized US11 molecules retain the signal peptide after the N-linked glycan has been attached and translation of the US11 polypeptide has been completed. Delayed cleavage of the US11 signal peptide is determined by the first four residues, the so-called n-region of the signal peptide. Its replacement with the four N-terminal residues of the H-2Kb signal sequence eliminates delayed cleavage. Surprisingly, a second region that affects the rate and extent of signal peptide cleavage is the transmembrane region close to the C-terminus of US11. Deletion of the transmembrane region of US11 (US11-180) significantly delays processing, a delay overcome by replacement with the H-2Kb signal sequence. Thus, elements at a considerable distance from the signal sequence affect its cleavage.
Keywords: ER subdomains/HCMV US11/post-translational ER processing/signal sequence cleavage/transmembrane anchor
Introduction
Membrane proteins and proteins destined for secretion are targeted to the appropriate intracellular membrane by their signal peptides (Martoglio and Dobberstein, 1998). In eukaryotes, signal peptides are 15–50 amino acids long and are usually located at the N-terminus (von Heijne, 1983). A typical signal peptide is comprised of three distinct regions: a polar N-terminal end (n-region) that may have a net positive charge, a central hydrophobic core (h-region) that consists of 6–15 hydrophobic amino acids, and a polar C-terminal (c-region) end that contains prolines and glycines (von Heijne, 1985). A signal peptide containing the consensus sequence and proper cleavage site ensures that proteins are inserted into the endoplasmic reticulum (ER) membrane and are processed properly. Mutations within the sequence immediately downstream of the signal peptide affect protein processing, and can result in both inefficient and inaccurate cleavage (Russel and Model, 1981; Folz and Gordon, 1986; Andrews et al., 1988; Wiren et al., 1988). For example, replacement of glutamic acid for leucine at the +2 position of the phage coat protein cleavage site causes inefficient removal of its signal peptide (Russel and Model, 1981). When the propeptides of human pre-pro-apolipoprotein A-II and pre-pro-parathyroid hormone are deleted, five and six residues, respectively, the generation of an improper N-terminus and a failure to direct the nascent chain to the ER properly are observed (Folz and Gordon, 1986; Wiren et al., 1988). Elements of the nascent chain at greater distances from the signal peptide are not known to affect signal peptide processing.
Shortly after its translation, the signal peptide interacts with signal recognition particle (SRP) and causes translational arrest (Walter and Blobel, 1981; Walter and Johnson, 1994). SRP is a ribonucleoprotein comprised of a 7S RNA associated with six different polypeptides (Walter and Blobel, 1980, 1982). The 54 kDa subunit of SRP interacts with the signal peptide through a hydrophobic region that promiscuously accommodates signal peptides of different lengths and sequences (Keenan et al., 1998). The SRP–nascent polypeptide chain–ribosome complex is targeted to the ER membrane where SRP binds to the SRP receptor and the ribosome weakly interacts with the translocon (mainly comprised of the Sec61p complex) (Gorlich et al., 1992; Kalies et al., 1994; for reviews see Rapoport et al., 1996; Hegde and Lingappa, 1999; Johnson and van Waes, 1999). The signal peptide is then transferred from the SRP into the channel of the translocon, where it directly associates with the Sec61α subunit of the Sec61 complex to promote tight interaction of the ribosome–nascent chain complex with the translocon (Jungnickel and Rapoport, 1995; Mothes et al., 1998; Plath et al., 1998). The signal peptide can also associate with the lipid bilayer and the TRAM protein (Martoglio et al., 1995; Voigt et al., 1996; Mothes et al., 1997), which assists in protein transport through the translocon. The interaction of the signal peptide with the Sec61 complex may also induce the removal of a ‘gating factor’, possibly BiP, from the lumenal side of the translocon, to allow access of the nascent polypeptide to the ER lumen (Crowley et al., 1994; Hamman et al., 1998). Chain elongation is re-initiated, followed by signal peptide translocation through the Sec61 channel. The hydrophobic nature of the signal peptide allows its insertion into the ER membrane, followed by signal peptidase cleavage upon lumenal exposure of the cleavage site (Blobel and Dobberstein, 1975). This cleavage site is characterized by small uncharged residues at positions –1 and –3 (von Heijne, 1990). After signal peptide cleavage, chain elongation of the nascent chain continues, while the signal peptide itself can be cleaved further by aminopeptidases or signal peptide peptidase (Lyko et al., 1995; Martoglio et al., 1997).
Signal peptidase is an endopeptidase that resembles other serine proteases (Dalbey and von Heijne, 1992) and performs a similar cleavage reaction for prokaryotic and eukaryotic signal peptidases. The crystal structure of the periplasmic domain of Escherichia coli leader peptidase (Paetzel et al., 1998) reveals important mechanistic aspects of signal peptide cleavage: the catalytic site proposed to be close to the lipid bilayer is surrounded by a hydrophobic region, explaining the requirement for small uncharged, aliphatic residues at the –1 and –3 positions of the cleavage site (Paetzel et al., 1998; von Heijne, 1998). The mammalian signal peptidase complex (SPC) is comprised of at least five subunits with molecular masses of 25, 23/22, 21, 18 and 12 kDa (Evans et al., 1986). The non-catalytic subunits of the eukaryotic SPC may function as regulatory subunits for signal peptide recognition and are located in close proximity to the translocon (Meyer and Hartmann, 1997). The Sec61p complex interacts with the 25 kDa subunit of the SPC (SPC25), which suggests a tight interaction between the SPC and the Sec61 complex (Kalies et al., 1998). This interaction may serve to recruit the SPC to the translocation site and thereby enhance the overall translocation efficiency of the nascent polypeptide.
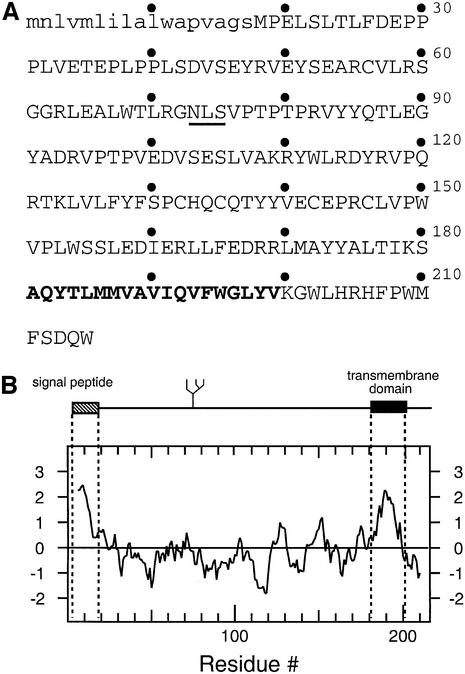
The human cytomegalovirus (HCMV) gene products US11 and US2 target the major histocompatibility complex (MHC) class I molecules for destruction by the proteasome (Wiertz et al., 1996a,b; Tortorella et al., 1998). These viral proteins associate with the class I molecules in the ER and induce the dislocation of the class I heavy chains from the ER, probably via the Sec61p complex, for degradation in the cytosol (Wiertz et al., 1996b). In all likelihood, a similar set of reactions is utilized for the removal and degradation of misfolded and abnormal ER proteins more generally (Bonifacino and Weissman, 1998). The HCMV US11 gene product is an ER-resident type I membrane glycoprotein (Figure 1), the single N-linked glycan attachment site of which is glycosylated quantitatively. The hydrophobic stretch at the N-terminus of US11 is characteristic of a signal peptide, while the hydrophobic stretch at the C-terminal end corresponds to a transmembrane/stop transfer sequence.
Fig. 1. (A) Amino acid sequence (single letter code) of HCMV US11. (B) Kyte–Doolittle hydropathy plot of US11. The predicted signal sequence is depicted in lower case. Bold face type represents the predicted transmembrane domain. The N-linked glycosylation site is underlined.
Here we report a highly unusual cleavage pattern for the US11 signal peptide. At least a fraction of the US11 signal peptide appears to be cleaved post-translationally. This trait is determined by the US11 signal peptide n-region. What cleavage occurs is also strongly influenced by the US11 transmembrane domain. Delayed cleavage of the US11 signal peptide may reflect the local ER environment in which dislocation takes place.
Results
The HCMV US11 signal peptide is cleaved post-translationally
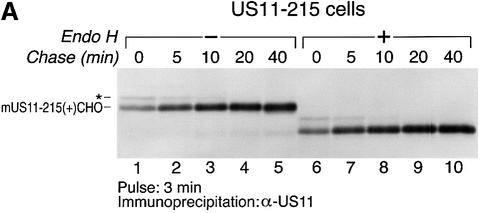
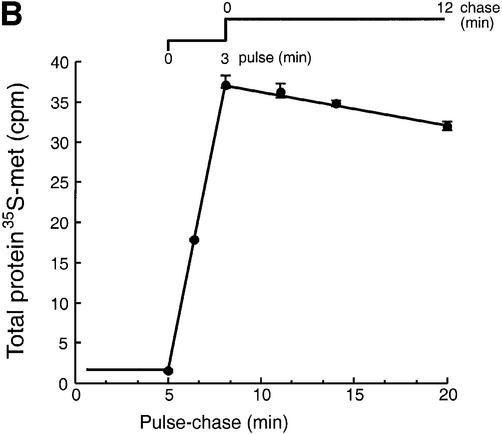
HCMV US11 is a 215 residue ER-resident protein that targets MHC class I heavy chains for destruction by the proteasome. The detailed mechanism by which the viral gene product accomplishes this is unclear, but is closely coupled to the biosynthesis of the class I and US11 products. We therefore examined whether the biosynthesis of US11 might reveal unique properties of the ER environment in which US11 normally functions. The maturation of US11 was examined in U373-MG cells stably transfected with US11 (US11-215 cells). US11-215 cells were metabolically labeled for 3 min with [35S]methionine and chased for up to 40 min. The US11 protein was recovered from cell lysates by immunoprecipitation using a polyclonal anti-US11 serum (α-US11) and analyzed by SDS–PAGE (Figure 2A). Two species of US11 of distinct mobility were recovered at early time points (Figure 2B, lanes 1 and 2). The faster moving, major species is the ER-resident, mature form of US11 (mUS11-215). It has a mobility indistinguishable from that of US11 recovered from a microsome-supplemented cell- free translation system (D.Tortorella and H.L.Ploegh, unpublished data).
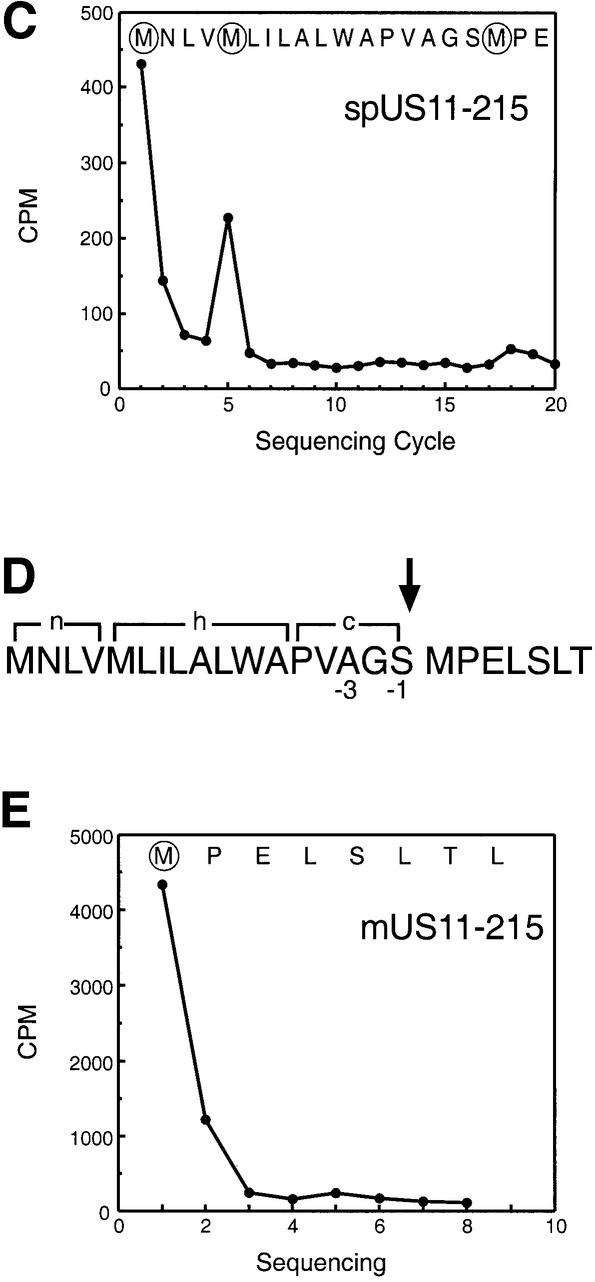
Fig. 2. Two forms of US11 exist early in biosynthesis. (A) US11-215 cells were pulsed for 3 min and chased for up to 40 min. Cells were lysed in 0.5% NP-40 and immunoprecipitated with anti-US11 serum (α-US11). The precipitates were analyzed by SDS–PAGE (12.5%). Two forms of US11 [* and mature US11-215 (+)CHO] were recovered from the US11-215 cell lysates (lanes 1–5). Half of the α-US11 precipitates were digested with Endo H (lanes 6–10). (B) Incorporation of [35S]methionine was examined during a pulse–chase experiment of US11-215 cells. TCA-precipitable radioactivity (c.p.m.) from [35S]methionine of each time point was plotted against the pulse–chase experiment. An average of three samples is represented at each value. (C) The slower moving US11 polypeptide (*) was subjected to N-terminal radiosequencing. The radioactivity (c.p.m.) from [35S]methionine of each fraction of the N-terminal radiosequencing run was plotted against Edman cycle number. (D) The n-, h- and c-regions of the US11 signal peptide are shown. The site of signal peptide cleavage is indicated by an arrow. (E) N-terminal radiosequencing of the mature form of US11 (mUS11-215) plotted as radioactivity (c.p.m.) from [35S]methionine versus Edman cycle number.
A precursor–product relationship between the two species was suggested by increased recovery at later chase points of mUS11-215 and decreased recovery of the slower moving species (*) (Figure 2A, lanes 1–4). The identity of the slower moving species (*) was unclear. Is it a distinct form of US11 or is it a protein associated with US11? Both mUS11-215 and the slower moving polypeptide (*) were recovered from SDS-denatured primary immunoprecipitates in a second round of immunoprecipitation using α-US11 serum (D.Tortorella and H.L.Ploegh, unpublished data). We therefore conclude that the slowly migrating polypeptide is a distinct form of the US11 protein.
The precursor–product conversion observed for the slower moving polypeptide (*) and mUS11-215 does not account fully for the amount of US11 recovered at early chase times. At the early time points of chase, there is a shortfall in the recovery of US11 (Figure 2A, lanes 1–3). This shortfall is not due to the continued incorporation of label during the chase (Figure 2B) and hence must result from the inability to retrieve all US11 at the early time points. Solubilization with the detergent SDS significantly improved recovery of both US11 polypeptides (* and mUS11-215) at the early time points (D.Tortorella and H.L.Ploegh, unpublished data).
Earlier experiments failed to show the presence of endoglycosidase H (Endo H)-resistant US11 and indicated that US11 was confined to the ER, as confirmed by immunoelectron microscopy (Wiertz et al., 1996a). The primary structure of US11 predicts a single N-linked glycan (CHO) attachment site at position 73 (Asn73–Leu–Ser) (Figure 1). Both polypeptides (* and mUS11-215) recovered from the US11 immunoprecipitates were susceptible to digestion by Endo H (Figure 2A, lanes 6–10). The difference between these two molecules of US11 cannot be due to an unusual modification of the N-linked glycan and, therefore, must be caused by differences in the polypeptide backbone.
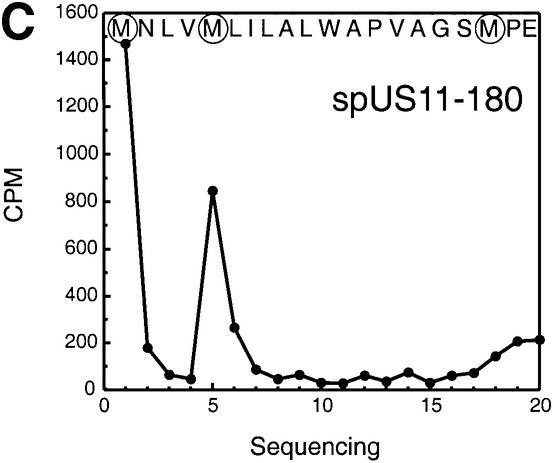
What type of modification could account for the presence of the slower moving species of US11? Based on the observed apparent molecular weight, the slowly migrating species of US11 may still contain the N-terminal signal peptide (spUS11-215). The polypeptide was isolated from [35S]methionine-labeled cells and subjected to 20 cycles of Edman degradation (Figure 2C). The observed peaks of radioactivity fit the position of the methionines at the N-terminal end of the US11 precursor sequence. These results establish that, surprisingly, the slower moving form (*) (Figure 2A) is indeed a glycosylated US11 molecule that has retained its signal peptide.
The US11 signal peptide contains a typical cleavage site
The factor known to influence signal peptide cleavage is the presence of small amino acid side chains at the –1 and –3 position relative to the cleavage site. Does the US11 signal peptide cleavage site indeed contain the consensus amino acids at the proper position? Analysis of the US11 primary sequence using the SignalP program (www.cbs.dtu.dk/services/SignalP/index.html) (Nielsen et al., 1997a,b) predicts signal peptide cleavage of US11 to occur between residues 17 and 18 (Figure 2D). Serine (17) occurs at position –1 and alanine (15) at position –3, residues that are in perfect agreement with the consensus sequence for a signal peptide cleavage site. Methionine would be the N-terminus of the processed US11 molecule. Indeed, US11 isolated from [35S]methionine-labeled US11-215 cells and subjected to eight cycles of N-terminal sequencing (Edman degradation) yielded methionine at position 1 (Figure 2E). Methionines within the N-terminal sequence of US11 occur at positions 5 and 18. Removal of only four residues from the N-terminus would not account for the mobility difference between the two forms of US11. Therefore, the methionine at position 18 must be the first residue of the mature US11 molecule. These results suggest that the unusual cleavage pattern of the US11 signal peptide is not due to an anomalous signal peptidase cleavage site.
The US11 signal peptide and the transmembrane region contribute to the delayed cleavage of the US11 signal sequence
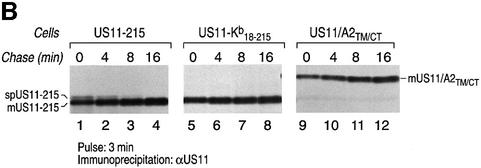
N-terminal signal peptide cleavage is presumably determined solely by the sequence of the signal peptide itself (Martoglio and Dobberstein, 1998). Changes within the n-, h- or c-region of the signal peptide and the regions directly downstream from the signal peptide affect signal peptide processing (Russel and Model, 1981; Folz and Gordon, 1986; Wiren et al., 1988; Izard and Kendall, 1994). Can the US11 signal peptide itself or regions further downstream of the US11 signal sequence, such as the US11 transmembrane region, play a role in signal peptide cleavage? We generated US11–Kb18–215 (Figure 3A), a chimeric molecule in which the US11 signal peptide was replaced with the signal peptide of the murine MHC class I heavy chain H-2Kb, a type I membrane protein. We also generated US11/A2TM/CT (Figure 3A), a chimeric molecule in which the transmembrane and cytoplasmic tail of US11 were replaced with the corresponding regions of human MHC class I heavy chain A2. Cleavage of the H-2Kb signal peptide should now generate the N-terminus of mature US11. Pulse–chase analysis of US11-215 cells shows the recovery of spUS11-215 and mUS11 at the early times points and a precursor–product relationship between the two polypeptides (Figure 3B, lanes 1–4, and C). For neither US11–Kb18–215 nor US11/A2TM/CT did we observe the presence of a signal sequence-containing precursor (Figure 3B, lanes 5–8 and 9–12). This result suggests that unique features of US11’s signal sequence and transmembrane domain contribute to the persistence of spUS11-215.
Fig. 3. The delayed cleavage of the US11 signal peptide is determined by its signal sequence and transmembrane/cytoplasmic tail region. (A) The US11 chimeric molecules US11–Kb18–215, US11/A2TM/CT and wild-type US11-215. (B) Processing of these molecules was examined in stable transfectants of MG-U373 cells using pulse–chase analysis. US11 was recovered from SDS lysates using α-US11 serum and analyzed by SDS–PAGE (12.5%). The signal peptide-containing form of US11 (spUS11-215) and the mature processed form of US11 (mUS11-215) are indicated. (C) The US11-215 molecules recovered from (B) were quantitated by a Molecular Dynamics Storm PhosphorImager. The US11 recovered at each time point is represented as percentage recovery of US11. The US11 recovered at the 8 min chase point was used as the 100% recovery value.
The recovery of mUS11-215 and US11/A2TM/CT increases with time (Figure 3B, lanes 1–4 and 9–12, and C). In contrast, recovery of US11–Kb18–215 does not significantly change during the chase (Figure 3B, lanes 5–8, and C). We therefore conclude that the US11 signal peptide is also responsible for the increased recovery of US11-215 and US11/A2TM/CT at the later time points. We suggest that the manner in which the US11 signal peptide initiates contact with the ER may contribute to its solubility properties.
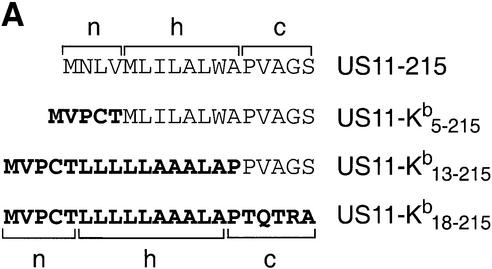
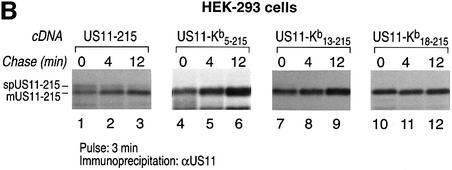
The n-, h- and c-regions of the US11 signal peptide follow the proposed consensus for a cleavable N-terminal signal peptide. However, the results obtained for the chimeric US11–Kb18–215 molecule suggest that the signal peptide itself may account for its delayed cleavage. To characterize further the segment of the US11 signal peptide that is responsible for delayed cleavage, we generated additional chimeras in which the n-region (US11–Kb5–215) or n + h-regions (US11–Kb13–215) of US11 are replaced with the corresponding regions of H-2Kb (Figure 4A). We transfected US11-215, US11–Kb5–215, US11–Kb13–215 and US11–Kb18–215 into HEK-293 cells and examined their processing by pulse– chase analysis (Figure 4B). For US11-215, a signal peptide-containing form of US11 and the mature form of US11-215 were evident at early chase times (Figure 4B, lanes 1–3). The two polypeptides showed a precursor– product relationship. For the chimeras US11–Kb5–215, US11–Kb13–215 and US11–Kb18–215, removal of the signal peptide is rapid and only the mature, cleaved form of US11 is recovered (Figure 4B, lanes 4–12). We conclude that features within the n-region of the US11 signal peptide contribute to its persistence.
Fig. 4. The n-region of the US11-215 signal peptide is responsible for its delayed cleavage. (A) The amino acid sequences of the n-, h- and c-regions of US11-215, US11–Kb5–215, US11–Kb13–215 and US11–Kb18–215. Bold letters represent the H-2Kb signal peptide. (B) US11–Kb5–215 (lanes 4–6), US11–Kb13–215 (lanes 7–9) and US11–Kb18–215 (lanes 10–12) were transfected in HEK-293 cells and analyzed by pulse–chase analysis. US11 was recovered from SDS lysates using α-US11 serum and analyzed by SDS–PAGE (12.5%). The signal peptide-containing form of US11 (spUS11-215) and the mature processed form of US11 (mUS11-215) are indicated.
During the chase, there is an increase in recovery of the mature form of US11–Kb5–215 and US11–Kb13–215 (Figure 4B, lanes 4–9), but not for US11–Kb18–215 (Figures 4B, lanes 10–12, and 3B, lanes 5–8, and C). Therefore, the c-region of the US11 signal peptide somehow contributes to recovery of mature US11. While the identity of the c-region does not affect the cleavage of the signal peptide, it does contribute to the recovery of mature US11. Perhaps the c-region is responsible for positioning nascent US11 relative to other components of the translocation machinery. This positioning may affect interactions of US11 with other ER components shortly after its completion, and hence its solubility. In contrast, the presence of the full Kb signal sequence neither delays signal peptide cleavage nor affects the recovery of US11 from cell lysates.
The US11 transmembrane region plays a role in US11 signal peptide cleavage
We next examined the role of the US11 transmembrane region in signal peptide cleavage. Such a role was suggested by the analysis of the US11/A2TM/CT chimeric construct (Figure 3). We generated a C-terminal truncation of US11 that lacks the predicted transmembrane segment and the cytoplasmic tail (US11-180) (Figure 1). The processing of wild-type US11-215 and US11-180 was examined in the appropriate U373-MG transfectants (Figure 5A). US11 recovered at the early chase times from US11-215 cells produced the usual pattern with respect to the precursor–product relationship of spUS11-215 and mUS11-215 (Figure 5A, lanes 1–4). Two major species were recovered from US11-180 cells (** and mUS11-180) (Figure 5A, lanes 5–8). A precursor–product relationship exists for the slower (**) and faster migrating species (mUS11-180) of US11-180. The two polypeptides recovered from the US11-180 transfectants represent distinct forms of the polypeptide backbone and both species of US11-180 are sensitive to Endo H (Figure 5B, compare lanes 1–4 and 5–8).

Fig. 5. Signal peptide cleavage of US11-180 is significantly delayed. (A) Processing of US11-215 and US11-180 was examined in stable transfectants of MG-U373 cells using pulse–chase analysis. US11 was recovered from SDS lysates using α-US11 serum and analyzed by SDS–PAGE (12.5%). The signal peptide-containing form of US11 (spUS11-215) and the mature processed form of US11 (mUS11-215) were immunoprecipitated from US11-215 cells (lanes 1–4). Two major species, ** and the mature processed form of US11-180 (mUS11-180), were recovered from US11-180 cells. (B) Half of the α-US11 precipitate recovered from a pulse–chase experiment of US11-180 cells was digested with Endo H (lanes 5–8). (C) The slower moving US11-180 polypeptide (**) was subjected to N-terminal radiosequencing. The radioactivity (c.p.m.) recovered at each Edman cycle is shown.
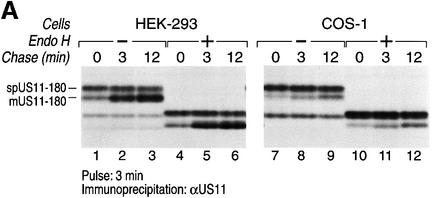
The slower moving species (**) was isolated from a US11-180 HEK-293 transfectant labeled with [35S]methionine and subjected to 20 cycles of Edman degradation (Figure 5C). The data showed persistence of the signal sequence. The absence of the transmembrane region of US11 thus strongly delays cleavage of its N-terminal signal peptide. An even more pronounced result was observed when US11-180 cDNA was transfected into HEK-293 and COS-1 cells (Figure 7).
Fig. 7. The US11 signal peptide plays a major role in processing of US11-180. US11-180 cDNA was transfected into HEK-293 and COS-1 cells. (A) Processing of US11-180 was examined by pulse–chase analysis. US11-180 was recovered from SDS lysates using α-US11 serum and analyzed by SDS–PAGE (12.5%). Half of the immunoprecipitates recovered from the respective transfectants were treated with Endo H (lanes 4–6 and 10–12). (B) The US11 signal peptide chimeric molecule US11–Kb18–180 and US11-180 were transfected in HEK-293 cells and analyzed by pulse–chase analysis. US11 was recovered from SDS lysates using α-US11 serum and analyzed by SDS–PAGE (12.5%). Half of the immunoprecipitates recovered from the respective transfectants were treated with Endo H (lanes 4–6 and 10–12). The signal peptide-containing form of US11 (spUS11-180) and the mature processed form of US11 (mUS11-180) are indicated.
mUS11-180 is a soluble protein
The Kyte–Doolittle hydropathy plot of US11 (Figure 1) suggests that the transmembrane region is located between residues 180 and 200. However, the hydrophobic nature of residues 180–200 does not ensure that it is in fact a transmembrane anchor. All attempts at proteolytic removal of the proposed cytoplasmic tail were without success. We performed Na2CO3 extractions to explore stable membrane insertion of US11-215 and US11-180 (Figure 6). US11-215 and US11-180 cells were labeled with [35S]methionine and broken with glass beads in the absence of detergent. Homogenates were then centrifuged at 1000 g to remove large debris, and the supernatant fraction was treated with 100 mM Na2CO3, followed by centrifugation at 150 000 g to sediment the extracted microsomes. US11-215 and US11-180 molecules were immunoprecipitated from detergent extracts prepared from the 1000 g pellet (Figure 6, lanes 1 and 4), the Na2CO3-treated 150 000 g soluble fraction (Figure 6, lanes 2 and 5) and the 150 000 g pellet fraction (Figure 6, lanes 3 and 6). As a soluble, lumenal control protein, we used β2-microglobulin (β2m) (Figure 6, lanes 7–12). The US11-215 polypeptide is recovered exclusively from the 150 000 g pellet fraction (Figure 6, lane 3), whereas the bulk of β2m is recovered from the 150 000 g soluble fraction (Figure 6, lane 8). These results confirm that US11-215 is a membrane protein. In contrast, the majority of US11-180 lacking its signal peptide (mUS11-180) and β2m are recovered from the 150 000 g soluble fraction (Figure 6, lanes 5 and 11). These results confirm that mUS11-180 and β2m are soluble, ER lumenal proteins.
Fig. 6. US11-180 is a soluble molecule. US11-215 and US11-180 cells were metabolically labeled for 15 min. The cells were homogenized with glass beads and centrifuged at 1000 g. The 1000 g supernatant fractions were treated with 100 mM Na2CO3, followed by centrifugation at 150 000 g. US11 molecules (lanes 1–6) and β2m (lanes 7–12) were recovered from the 1000 g (1K) pellet, 150 000 g supernatant (S) and the 150 000 g pellet (P) using α-US11 and α-β2m serum. The immunoprecipitates were analyzed by SDS–PAGE (12.5%).
A small fraction of mUS11-180 is recovered from the 150 000 g pellet fraction (Figure 6, lane 6) and may represent mUS11-180 that continues to associate with the ER membrane shortly after signal peptide cleavage and prior to its release into the ER lumen. Alternatively, a fraction of mUS11-180 may interact with an ER membrane protein in a Na2CO3-resistant manner. As might be expected, the signal peptide-containing form of US11-180 (spUS11-180) remains associated with the membrane fraction even after carbonate extraction (Figure 6, lane 6).
The identity of the signal sequence dictates delayed cleavage of the US11-180 molecule
For reasons of consistency with the data shown earlier, the experiments in Figure 5A were all conducted in U373-MG cells stably transfected with the US11-180 cDNA. The delayed cleavage of the signal peptide of US11 is not an aberration of the recipient cell line used for transfection. In fact, when we used either HEK-293 or COS-1 cells in a transient transfection protocol, the persistence of the signal peptide-containing form of both US11-215 (Figure 4B, lanes 1–3) and US11-180 (Figure 7A) was much more pronounced. The relative amount of signal sequence-containing precursor of US11-180 was increased to the extreme, such that in COS-1 cells it is in fact the predominant form of US11-180 at the end of the chase (Figure 7A, lanes 7–12). Our data show that the anomalous behavior of the US11 signal peptide is intrinsic to the US11 molecule. In transfection experiments exploiting COS-1 cells to express other type I membrane proteins, the persistence of signal peptides was not observed (Huppa and Ploegh, 1997) and to our knowledge has not been reported by others.
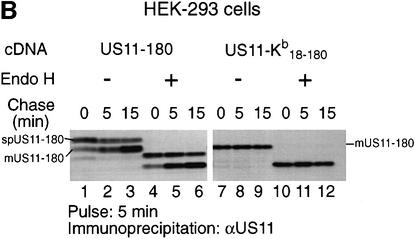
We next addressed the contribution of the signal sequence’s identity to the delayed cleavage observed for US11-180. We generated a chimeric molecule, US11–Kb18–180, in which the US11-180 signal peptide is replaced with the H-2Kb signal peptide (Figure 3A). We transfected US11-180 and US11–Kb18–180 into HEK-293 cells and examined their processing by pulse–chase analysis (Figure 7B). The immunoprecipitates were treated with Endo H to verify glycosylation and ER insertion (Figure 7B, lanes 4–6 and 10–12). For US11-180 carrying the US11 signal peptide, the signal peptide-containing form of spUS11-180 and the mature processed form of US11-180 were observed throughout the chase (Figure 7B, lanes 1–3). In contrast, a single polypeptide with a mobility similar to that of mUS11-180 is recovered from US11–Kb18–180 transfectants (Figure 7B, lanes 7–9). Delayed cleavage of the US11-180 signal peptide no longer occurs when the US11 signal peptide is replaced with the H-2Kb signal peptide. Not only the US11 transmembrane segment, but also features of the US11 signal sequence itself play a major role in US11 signal peptide cleavage.
Discussion
We describe here the unusual properties of the signal sequence of HCMV US11, a type I membrane glycoprotein. Elements contained within the signal sequence’s N-terminal segment (Met–Asn–Leu–Val) are responsible for delayed cleavage, such that a fully glycosylated, signal peptide-bearing intermediate is readily detected. In addition, the C-terminal membrane anchor also affects the rate of signal peptide cleavage; a US11 variant lacking its transmembrane/cytoplasmic tail segment (US11-180) shows an even greater delay in signal peptide cleavage than is seen for full-length US11. This effect is at its most extreme in COS-1 cells, where the glycosylated, signal peptide-containing US11-180 protein (spUS11-180) is the majority of US11 polypeptide that persists. To account for these findings, we propose an extended interaction of the signal peptide and transmembrane segment with the processing apparatus.
Conformity with the consensus parameters within the n-, h- and c-regions of the signal peptide predicts proper cleavage of an N-terminal signal peptide. The US11 signal peptide sequence fits the consensus parameters within the n-, h- and c-regions, yet fails to be cleaved efficiently from the nascent chain. Chimeric molecules in which regions (n, n + h or n + h + c) of the US11 signal peptide were replaced with the corresponding regions of the murine class I heavy chain H-2Kb signal peptide demonstrate that it is the n-region of the US11 signal sequence that is mostly responsible for the delayed cleavage of the US11 signal peptide (Figure 4). An irregular n-region has been observed to affect signal peptide processing; a surfeit of positive charges within the n-region of the HIV-1 gp-120 signal sequence probably accounts for its inefficient cleavage (Li et al., 1994, 1996). This aberrant form of gp-120 does not exit the ER and, therefore, cannot be incorporated into a nascent virion. We note that the persistence of the uncleaved signal sequence on gp-120 was never directly shown by sequence analysis.
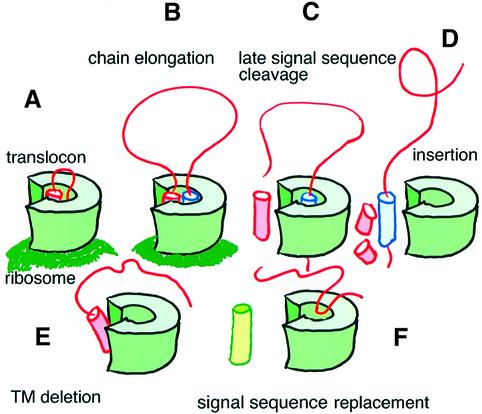
Regions outside the signal peptide can also influence its cleavage. In pre-pro-apolipoprotein A-II and pre-pro-parathyroid hormone, removal of the propeptide that is immediately downstream of the signal peptide influenced ER protein translocation and proper signal peptide processing (Russel and Model, 1981; Folz and Gordon, 1986; Andrews et al., 1988; Wiren et al., 1988). These changes mostly affect the site of cleavage, shifting it a few residues downstream, while their effect on the rate of signal peptide cleavage was not addressed in any detail. In addition, a mutation at the +2 position of the signal peptide cleavage site of phage coat protein also results in inefficient cleavage (Russel and Model, 1981). All of these mutations are localized immediately downstream of the signal peptide. In contradistinction to such signal sequence-proximal alterations, the transmembrane anchor of US11, at a considerable distance (∼160 residues) from the US11 signal sequence, strongly influences signal sequence cleavage. The rate of signal peptide cleavage for the US11 molecule lacking its transmembrane/cytoplasmic tail region (US11-180) is significantly delayed when compared with that seen for wild-type US11 (Figure 5). Replacement of the US11 signal sequence for that of H-2Kb results in rapid processing of US11 lacking the transmembrane segment, such that signal sequence-containing forms are no longer detected. The unprocessed US11-180 polypeptide is probably in an orientation unfavorable for signal peptide cleavage, and the presence of the US11 transmembrane anchor is clearly required for efficient signal peptide processing (Figure 8).
Fig. 8. Model of HCMV US11 signal peptide cleavage. (A) The signal peptide (pink) is inserted into the translocon, followed by (B) chain elongation of the US11 nascent polypeptide. (C) Upon completion of US11 translation, the US11 transmembrane segment (blue) may interact with the signal peptide to delay signal peptide cleavage. (D) Upon cleavage of the signal peptide, the US11 molecule inserts into the lipid bilayer; the signal peptide itself may be cleaved further by signal peptide peptidase. (E) The signal peptide of a truncated US11 molecule that lacks its transmembrane region and cytoplasmic tail (US11-180) is cleaved inefficiently from the nascent polypeptide. (F) Replacement of the US11 signal peptide in US11-180 with the H-2Kb signal peptide (green) results in efficient processing. The US11 transmembrane domain may position the signal peptide in an orientation favorable for cleavage.
How can the US11 transmembrane anchor accelerate removal of the US11 signal peptide? The transmembrane domain may interact with the signal peptide and position the signal peptide to facilitate access to the cleavage site. Alternatively, the transmembrane anchor may interact with the SPC and enhance recognition of the US11 signal peptide for reasons of physical proximity. While the specificity of signal peptide cleavage is appreciated in terms of the minimum sequence requirements, cleavage itself is a highly regulated process, the dynamics of which are not well understood. The non-catalytic subunits of the SPC have been cloned and isolated, yet their function remains to be determined. Our results show that regulation of signal peptide cleavage may involve cis-acting elements within the polypeptide that act at considerable distance from the actual cleavage site. Such elements could perhaps interact with the non-catalytic subunits of signal peptidase.
Immunoelectron microscopy, the maturation status of its single N-linked glycan and the kinetics with which it catalyzes accelerated destruction of class I molecules all place US11 in the ER. The ER environment of the US11 signal peptide may help determine the unusual signal peptide cleavage pattern that we observe. The site of signal peptide cleavage is in the ER and is postulated to be in close proximity to the translocon (Kalies et al., 1998). An intrinsic feature of the US11 signal peptide, more specifically the c-region of the signal peptide, may dictate an association with complexes within the ER as judged from the observed cleavage in detergent extractability (Figure 4 and D.Tortorella and H.L.Ploegh, unpublished data). Shortly after signal peptide cleavage, the recovery of the processed form of US11 increases over the chase period. We suggest that these early biosynthetic forms of US11 may reside in specialized regions of the ER.
To address an issue more peripheral to the central claims of this study: is the cleavage pattern of US11’s signal sequence related to US11-induced class I degradation? The signal peptide of the chimeric molecule US11–Kb18–215 is cleaved rapidly and this molecule readily supports class I destruction (D.Tortorella and H.L.Ploegh, unpublished data). Therefore, the identity of the US11 signal peptide itself is not essential for the ability of US11 to accelerate class I degradation. The signal peptide of the chimera US11/A2TM/CT is also cleaved rapidly, but class I heavy chains are not degraded in US11/A2TM/CT-expressing cells (D.Tortorella and H.L.Ploegh, unpublished data). Deletion of US11’s cytoplasmic tail does not abolish degradation of class I heavy chains (D.Tortorella and H.L.Ploegh, unpublished data), and consequently the identity of the transmembrane segment of US11 should be considered essential to its function.
If our interpretation is correct, then perhaps the interaction of the US11 signal peptide and US11 transmembrane segment would help keep the Sec61 complex and its accessories in a configuration that allows recruitment of the class I heavy chains to the translocon. The recorded efficiency of US11-mediated dislocation suggests that the process is tightly linked, temporally and perhaps physically, to protein translocation into the ER. Thus, close proximity of US11 to the translocation apparatus and efficient gating of the protein channel might account for the speed of the dislocation reaction. Ultimately, this aspect must be related to the properties of US11 itself. The unusual maturation of US11, as described here, may turn out be an important aspect of how the dislocation apparatus is put in place.
Materials and methods
Cell lines and antibody
U373-MG astrocytoma cells transfected with the US11-215 cDNA were prepared as described (Jones et al., 1995; Kim et al., 1995) and cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 5% fetal calf serum (FCS) and 5% calf serum. US11-201, US11-180, US11Kb18–215 and US11/A2 cells were maintained in DMEM supplemented with 5% FCS, 5% calf serum and 0.5 mg/ml geneticin (Gibco, Fredrick, MD). The human embryonic kidney cell line (HEK-293) was maintained in DMEM supplemented with 5% FCS and 5% calf serum. The anti-US11 serum was generated by immunizing rabbits with fragments of US11 (amino acids 18–36, 104–122 and 194–210) conjugated to keyhole limpet hemocyanin (Story et al., 1999). The anti-class I heavy chain serum was generated by immunizing rabbits with the bacterially expressed lumenal fragment of HLA-A2 and HLA-B27 heavy chains (Tortorella et al., 1998). The anti-β2m serum was generated by immunizing rabbits with bacterially expressed human β2m.
Metabolic labeling of cells and pulse–chase analysis
Cells were detached by trypsin treatment, followed by starvation in methionine/cysteine-free DMEM for 45 min at 37°C. Cells were metabolically labeled with 500 µCi of [35S]methionine/cysteine (1200 Ci/mmol; NEN-Dupont, Boston, MA)/ml at 37°C for the times indicated. In pulse–chase experiments, cells were radiolabeled as above and were chased for the times indicated in DMEM containing non-radiolabeled methionine (2.5 mM) and cysteine (0.5 mM). Cells were then lysed in NP-40 lysis buffer (10 mM Tris pH 7.8, 150 mM NaCl, 5 mM MgCl2, 0.5% NP-40) supplemented with 1.5 µg/ml aprotinin, 1 µM leupeptin, 2 mM phenylmethylsulfonyl fluoride (PMSF) followed by immunoprecipitation (see below). For cells lysed in 1% SDS, the SDS concentration was adjusted, prior to immunoprecipitation, to 0.063% with the NP-40 lysis mix.
Immunoprecipitation
Following cell lysis, cell debris was removed by centrifugation at 10 000 g for 10 min. Non-specific binding proteins were removed from the cell lysates by the addition of 3 µl/ml normal rabbit serum, 3 µl/ml normal mouse serum and formalin-fixed, heat-killed Staphylococcus aureus for 1 h at 4°C. Immunoprecipitation was performed by incubation with antiserum for 45 min at 4°C, followed by the addition of S.aureus for 45 min at 4°C. The pelleted S.aureus were washed four times with washing buffer (0.5% NP-40 in 50 mM Tris pH 7.4, 150 mM NaCl and 5 mM EDTA). The pellet was resuspended in SDS sample buffer (4% SDS, 5% β-mercaptoethanol, 10% glycerol, 0.025% bromophenol blue in 62.5 mM Tris pH 6.8) and the released materials were subjected to 12.5% SDS–PAGE.
cDNA, transfection and Endo H digestion
The cDNA of full-length US11 was cloned from the AD169 HCMV genome using the following primers: 5′ primer, CCGCTCCGAGCG GCGTCGACACCACCATGGAACCTTGTAATGCTTATTCTAGC; 3′ primer, GCTCTAGAGCTCACCACTGGTCCGAAAACATCCAG. The US11 cDNA was cloned into the eukaryotic expression vector pcDNA 3.1 (Invitrogen, Carlsbad, CA) using the Xho–Xba restriction site in its polylinker region. US11-180 was subcloned from US11 (pcDNA3.1). The chimeric molecules: US11/A2TM/CT [US11(amino acids 1–178)/HLA-A2(amino acids 307–365)]; US11–Kb5–215 [H-2Kb(amino acids 1–5)/US11(amino acids 5–215)]; US11–Kb13–215 [H-2Kb(amino acids 1–16)/US11(amino acids 13–215)]; US11–Kb18–215 [H-2Kb(amino acids 1–21)/US11(amino acids 18–215)]; and US11–Kb18–180 [H-2Kb(amino acids 1–21)/US11(amino acids 18–180)] were generated by initially cloning the desired fragment followed by ligation of two of the respective fragments. Using primers specific to the ends of the ligated molecule, it was recloned and inserted into pcDNA3.1. A liposome-mediated transfection (Lipofectamine, Gibco, Fredrick, MD) protocol was performed as described by the manufacturer (4 µg of DNA/20 µl of lipofectamine/10 cm dish of cells). Endo H (New England Biolabs) digestion was performed as described by the manufacturer.
Gel electrophoresis
SDS–PAGE and fluorography were performed as described (Ploegh, 1995). For N-terminal sequencing, the immunoprecipitated US11 protein was resolved by SDS–PAGE and transferred to a PVDF membrane (0.22 µm pore size) in transfer buffer (48 mM Tris-base, 39 mM glycine, 0.037% SDS, 20% methanol) using a semi-dry blotting apparatus (Buchler Instruments, Kansas, MO).
N-terminal sequence analysis
The PVDF membrane that contained the polypeptide of interest was subjected to automated Edman degradation using an Applied Biosystem Protein Sequencer, Model 477, using ATZ chemistry, at the Biopolymers Laboratory at MIT, Center for Cancer Research. The fractions from each degradation sequencing cycle were collected and counted by liquid scintillation spectrometry.
Na2CO3 treatment
US11-215 and US11-180 cells were metabolically labeled for 15 min and then washed twice in 50 mM Tris pH 7.5, 250 mM sucrose (homogenization buffer). The cells were resuspended in homogenization buffer and broken by vortexing in the presence of 106 µm glass beads. The homogenate was centrifuged at 1000 g for 5 min; the pellet fraction was resuspended in NP-40 lysis mix (see above) and the supernatant was treated with Na2CO3 (100 mM final) for 30 min at 4°C (Fujiki et al., 1982). The Na2CO3-treated samples were centrifuged at 150 000 g using a TLA 100.2 rotor in a Beckman centrifuge. The 150 000 g high pH supernatant was adjusted to pH 7 with 1 M HCl and diluted to a final 1× NP-40 lysis mix. The 150 000 g pellet was washed twice with homogenization buffer and then resuspended in 1× NP-40 lysis mix. US11 and β2m were immunoprecipitated from the 1000 g pellet, 150 000 g supernatant and the 150 000 g pellet with the respective antibody.
Acknowledgments
Acknowledgements
We thank Richard Cook of the Biopolymers Laboratory at MIT, Center for Cancer Research for protein sequencing. D.T. is a Charles A.King Trust (Boston, MA) Research Fellow. P.S. is supported by HHMI. A.R. is supported by the German Research Council (DFG). This work was funded by NIH grant 5R37-AI33456 and a grant by Boehringer-Ingelhiem.
References
- Andrews D.W., Perara,E., Lesser,C. and Lingappa,V.R. (1988) Sequences beyond the cleavage site influence signal peptide function. J. Biol. Chem., 263, 15791–15798. [PubMed] [Google Scholar]
- Blobel G. and Dobberstein,B. (1975) Transfer of proteins across membranes. II. Reconstitution of functional rough microsomes from heterologous components. J. Cell Biol., 67, 852–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino J.S. and Weissman,A.M. (1998) Ubiquitin and the control of protein fate in the secretory and endocytic pathways. Annu. Rev. Cell Dev. Biol., 14, 19–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley K.S., Liao,S., Worrell,V.E., Reinhart,G.D. and Johnson,A.E. (1994) Secretory proteins move through the endoplasmic reticulum membrane via an aqueous, gated pore. Cell, 78, 461–471. [DOI] [PubMed] [Google Scholar]
- Dalbey R.E. and von Heijne,G. (1992) Signal peptidases in prokaryotes and eukaryotes—a new protease family. Trends Biochem. Sci., 17, 474–478. [DOI] [PubMed] [Google Scholar]
- Evans E.A., Gilmore,R. and Blobel,G. (1986) Purification of microsomal signal peptidase as a complex. Proc. Natl Acad. Sci. USA, 83, 581–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folz R.J. and Gordon,J.I. (1986) Deletion of the propeptide from human preproapolipoprotein A-II redirects cotranslational processing by signal peptidase. J. Biol. Chem., 261, 14752–14759. [PubMed] [Google Scholar]
- Fujiki Y., Hubbard,A.L., Fowler,S. and Lazarow,P.B. (1982) Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J. Cell Biol., 93, 97–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlich D., Prehn,S., Hartmann,E., Kalies,K.U. and Rapoport,T.A. (1992) A mammalian homolog of SEC61p and SECYp is associated with ribosomes and nascent polypeptides during translocation. Cell, 71, 489–503. [DOI] [PubMed] [Google Scholar]
- Hamman B.D., Hendershot,L.M. and Johnson,A.E. (1998) BiP maintains the permeability barrier of the ER membrane by sealing the lumenal end of the translocon pore before and early in translocation. Cell, 92, 747–758. [DOI] [PubMed] [Google Scholar]
- Hegde R.S. and Lingappa,V.R. (1999) Regulation of protein biogenesis at the endoplasmic reticulum membrane. Trends Cell Biol., 9, 132–137. [DOI] [PubMed] [Google Scholar]
- Huppa J.B. and Ploegh,H.L. (1997) The α chain of the T cell antigen receptor is degraded in the cytosol. Immunity, 7, 113–122. [DOI] [PubMed] [Google Scholar]
- Izard J.W. and Kendall,D.A. (1994) Signal peptides: exquisitely designed transport promoters. Mol. Microbiol., 13, 765–773. [DOI] [PubMed] [Google Scholar]
- Johnson A.E. and van Waes,M.A. (1999) The translocon: a dynamic gateway at the ER membrane. Annu. Rev. Cell Dev. Biol., 15, 799–842. [DOI] [PubMed] [Google Scholar]
- Jones T.R., Hanson,L.K., Sun,L., Slater,J.S., Stenberg,R.M. and Campbell,A.E. (1995) Multiple independent loci within the human cytomegalovirus unique short region down-regulate expression of major histocompatibility complex class I heavy chains. J. Virol., 69, 4830–4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungnickel B. and Rapoport,T.A. (1995) A posttargeting signal sequence recognition event in the endoplasmic reticulum membrane. Cell, 82, 261–270. [DOI] [PubMed] [Google Scholar]
- Kalies K.U., Gorlich,D. and Rapoport,T.A. (1994) Binding of ribosomes to the rough endoplasmic reticulum mediated by the Sec61p-complex. J. Cell Biol., 126, 925–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalies K.U., Rapoport,T.A. and Hartmann,E. (1998) The β subunit of the Sec61 complex facilitates cotranslational protein transport and interacts with the signal peptidase during translocation. J. Cell Biol., 141, 887–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keenan R.J., Freymann,D.M., Walter,P. and Stroud,R.M. (1998) Crystal structure of the signal sequence binding subunit of the signal recognition particle. Cell, 94, 181–191. [DOI] [PubMed] [Google Scholar]
- Kim H.J., Gatz,C., Hillen,W. and Jones,T.R. (1995) Tetracycline repressor-regulated gene repression in recombinant human cytomegalovirus. J. Virol., 69, 2565–2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Luo,L., Thomas,D.Y. and Kang,C.Y. (1994) Control of expression, glycosylation and secretion of HIV-1 gp120 by homologous and heterologous signal sequences. Virology, 204, 266–278. [DOI] [PubMed] [Google Scholar]
- Li Y., Bergeron,J.J., Luo,L., Ou,W.J., Thomas,D.Y. and Kang,C.Y. (1996) Effects of inefficient cleavage of the signal sequence of HIV-1 gp 120 on its association with calnexin, folding and intracellular transport. Proc. Natl Acad. Sci. USA, 93, 9606–9611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyko F., Martoglio,B., Jungnickel,B., Rapoport,T.A. and Dobberstein,B. (1995) Signal sequence processing in rough microsomes. J. Biol. Chem., 270, 19873–19878. [DOI] [PubMed] [Google Scholar]
- Martoglio B. and Dobberstein,B. (1998) Signal sequences: more than just greasy peptides. Trends Cell Biol., 8, 410–415. [DOI] [PubMed] [Google Scholar]
- Martoglio B., Hofmann,M.W., Brunner,J. and Dobberstein,B. (1995) The protein-conducting channel in the membrane of the endoplasmic reticulum is open laterally toward the lipid bilayer. Cell, 81, 207–214. [DOI] [PubMed] [Google Scholar]
- Martoglio B., Graf,R. and Dobberstein,B. (1997) Signal peptide fragments of preprolactin and HIV-1 p-gp160 interact with calmodulin. EMBO J., 16, 6636–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer H.A. and Hartmann,E. (1997) The yeast SPC22/23 homolog Spc3p is essential for signal peptidase activity. J. Biol. Chem., 272, 13159–13164. [DOI] [PubMed] [Google Scholar]
- Mothes W., Heinrich,S.U., Graf,R., Nilsson,I., von Heijne,G., Brunner,J. and Rapoport,T.A. (1997) Molecular mechanism of membrane protein integration into the endoplasmic reticulum. Cell, 89, 523–533. [DOI] [PubMed] [Google Scholar]
- Mothes W., Jungnickel,B., Brunner,J. and Rapoport,T.A. (1998) Signal sequence recognition in cotranslational translocation by protein components of the endoplasmic reticulum membrane. J. Cell Biol., 142, 355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen H., Engelbrecht,J., Brunak,S. and von Heijne,G. (1997a) Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng., 10, 1–6. [DOI] [PubMed] [Google Scholar]
- Nielsen H., Engelbrecht,J., Brunak,S. and von Heijne,G. (1997b) A neural network method for identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Int. J. Neural Syst., 8, 581–599. [DOI] [PubMed] [Google Scholar]
- Paetzel M., Dalbey,R.E. and Strynadka,N.C. (1998) Crystal structure of a bacterial signal peptidase in complex with a β-lactam inhibitor. Nature, 396, 186–190. [DOI] [PubMed] [Google Scholar]
- Plath K., Mothes,W., Wilkinson,B.M., Stirling,C.J. and Rapoport,T.A. (1998) Signal sequence recognition in posttranslational protein transport across the yeast ER membrane. Cell, 94, 795–807. [DOI] [PubMed] [Google Scholar]
- Ploegh H.L. (1995) Current Protocols in Protein Science. Wiley, New York, NY.
- Rapoport T.A., Jungnickel,B. and Kutay,U. (1996) Protein transport across the eukaryotic endoplasmic reticulum and bacterial inner membranes. Annu. Rev. Biochem., 65, 271–303. [DOI] [PubMed] [Google Scholar]
- Russel M. and Model,P. (1981) A mutation downstream from the signal peptidase cleavage site affects cleavage but not membrane insertion of phage coat protein. Proc. Natl Acad. Sci. USA, 78, 1717–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Story C.M., Furman,M.H. and Ploegh,H.L. (1999) The cytosolic tail of class I MHC heavy chain is required for its dislocation by the human cytomegalovirus US2 and US11 gene products. Proc. Natl Acad. Sci. USA, 96, 8516–8521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortorella D., Story,C.M., Huppa,J.B., Wiertz,E.J., Jones,T.R., Bacik,I., Bennink,J.R., Yewdell,J.W. and Ploegh,H.L. (1998) Dislocation of type I membrane proteins from the ER to the cytosol is sensitive to changes in redox potential. J. Cell Biol., 142, 365–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt S., Jungnickel,B., Hartmann,E. and Rapoport,T.A. (1996) Signal sequence-dependent function of the TRAM protein during early phases of protein transport across the endoplasmic reticulum membrane. J. Cell Biol., 134, 25–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. (1983) Patterns of amino acids near signal-sequence cleavage sites. Eur. J. Biochem., 133, 17–21. [DOI] [PubMed] [Google Scholar]
- von Heijne G. (1985) Signal sequences. The limits of variation. J. Mol. Biol., 184, 99–105. [DOI] [PubMed] [Google Scholar]
- von Heijne G. (1990) The signal peptide. J. Membr. Biol., 115, 195–201. [DOI] [PubMed] [Google Scholar]
- von Heijne G. (1998) Life and death of a signal peptide. Nature, 396, 111–113. [DOI] [PubMed] [Google Scholar]
- Walter P. and Blobel,G. (1980) Purification of a membrane-associated protein complex required for protein translocation across the endoplasmic reticulum. Proc. Natl Acad. Sci. USA, 77, 7112–7116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P. and Blobel,G. (1981) Translocation of proteins across the endoplasmic reticulum III. Signal recognition protein (SRP) causes signal sequence-dependent and site-specific arrest of chain elongation that is released by microsomal membranes. J. Cell Biol., 91, 557–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P. and Blobel,G. (1982) Signal recognition particle contains a 7S RNA essential for protein translocation across the endoplasmic reticulum. Nature, 299, 691–698. [DOI] [PubMed] [Google Scholar]
- Walter P. and Johnson,A.E. (1994) Signal sequence recognition and protein targeting to the endoplasmic reticulum membrane. Annu. Rev. Cell Biol., 10, 87–119. [DOI] [PubMed] [Google Scholar]
- Wiertz E.J., Jones,T.R., Sun,L., Bogyo,M., Geuze,H.J. and Ploegh,H.L. (1996a) The human cytomegalovirus US11 gene product dislocates MHC class I heavy chains from the endoplasmic reticulum to the cytosol. Cell, 84, 769–779. [DOI] [PubMed] [Google Scholar]
- Wiertz E.J., Tortorella,D., Bogyo,M., Yu,J., Mothes,W., Jones,T.R., Rapoport,T.A. and Ploegh,H.L. (1996b) Sec61-mediated transfer of a membrane protein from the endoplasmic reticulum to the proteasome for destruction. Nature, 384, 432–438. [DOI] [PubMed] [Google Scholar]
- Wiren K.M., Potts,J.T.,Jr and Kronenberg,H.M. (1988) Importance of the propeptide sequence of human preproparathyroid hormone for signal sequence function. J. Biol. Chem., 263, 19771–19777. [PubMed] [Google Scholar]