Abstract
Ciliary neurotrophic factor (CNTF) is involved in the survival of a number of different neural cell types, including motor neurons. CNTF functional responses are mediated through a tripartite membrane receptor composed of two signalling receptor chains, gp130 and the leukaemia inhibitory factor receptor (LIFR), associated with a non-signalling CNTF binding receptor α component (CNTFR). CNTFR-deficient mice show profound neuronal deficits at birth, leading to a lethal phenotype. In contrast, inactivation of the CNTF gene leads only to a slight muscle weakness, mainly during adulthood, suggesting that CNTFR binds to a second ligand that is important for development. Modelling studies of the interleukin-6 family member cardiotrophin-like cytokine (CLC) revealed structural similarities with CNTF, including the conservation of a site I domain involved in binding to CNTFR. Co-expression of CLC and CNTFR in mammalian cells generates a secreted composite cytokine, displaying activities on cells expressing the gp130–LIFR complex on their surface. Correspondingly, CLC–CNTFR activates gp130, LIFR and STAT3 signalling components, and enhances motor neuron survival. Together, these observations demonstrate that CNTFR induces the secretion of CLC, as well as mediating the functional responses of CLC.
Keywords: cardiotrophin-like cytokine/CNTF receptor/gp130/LIF receptor/motor neuron
Introduction
Ciliary neurotrophic factor (CNTF) uses a multimeric receptor composed of the gp130 signal-transducing protein associated with leukaemia inhibitory factor receptor (LIFR) and a specific binding subunit, the CNTF binding receptor α component (CNTFR), which is anchored to the membrane through a glycophosphatidylinositol linkage (Hibi et al., 1990; Davis et al., 1991, 1993a,b; Gearing et al., 1991; Taga and Kishimoto, 1997). Binding of CNTF to CNTFR leads to gp130–LIFR dimerization and downstream signalling events involving, among others, the recruitment of the JAK1–STAT3 signalling pathway (Lütticken et al., 1994; Stahl et al., 1994, 1995; Heinrich et al., 1998). Gp130 and LIFR are also present in the receptor complexes for the cytokines LIF (Gearing et al., 1992), cardiotrophin-1 (CT-1) (Pennica et al., 1995), oncostatin M (OSM) (Gearing et al., 1992) and cardiotrophin-like cytokine (CLC) (Senaldi et al., 1999; Shi et al., 1999).
CNTF promotes the differentiation and survival of a wide range of cell types in the nervous system. In particular, CNTF maintains motor neuron viability in vitro, prevents the degeneration of axotomized motor neurons and attenuates motor deficits in different strains of mice with neuromuscular deficiencies (Lin et al., 1989; Stöckli et al., 1989; Oppenheim et al., 1991; Sendtner et al., 1992; Curtis et al., 1993; Mitsumoto et al., 1994). Besides its activities in the nervous system, CNTF has trophic effects on denervated skeletal muscle and is a regulator of muscular strength in aging (Helgren et al., 1994; Guillet et al., 1999).
Only a mild loss of motor neurons leading to minor muscle weakness is observed in adult CNTF–/– mice (Masu et al., 1993). Moreover, a null mutation in the human CNTF gene does not lead to neurological disease (Takahashi et al., 1994). In contrast, the CNTFR subunit is essential for survival of nearly half of all motor neurons during development, and mice harbouring a homozygous CNTFR–/– mutation die shortly after birth (DeChiara et al., 1995). These dramatic viability differences suggest that CNTFR plays a critical role during development, at a period when CNTF is almost absent, by serving as a receptor for a second, developmentally important ligand.
CLC is a new member of the CNTF/LIF family of cytokines isolated by expressed sequence tag database screening, and is also referred to as novel neurotrophin-1/B-cell stimulating factor-3 (Senaldi et al., 1999; Shi et al., 1999). CLC expressed in Escherichia coli has been shown to recruit the gp130–LIFR pathway in a neuroblastoma cell line and is able to activate NF-κB and SRE reporter constructs in myeloid cells. It also supports the survival of chicken embryonic motor and sympathetic neurons.
We recently demonstrated that CLC associates with the soluble receptor cytokine-like factor-1 (CLF) to form a heterodimeric cytokine. CLF expression is required for CLC secretion, and the CLC–CLF heterocomplex displays activities only on those cells expressing the functional CNTF receptor (Elson et al., 1998, 2000). In the present study, we show that the cellular release of functional CLC can also be mediated by CNTFR to generate a composite cytokine displaying functional activities on cells expressing the gp130 and LIFR components on their surface.
Results
Modelling of CLC
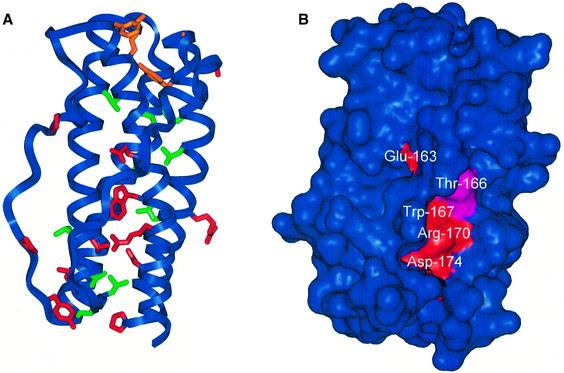
A CLC model was built based on the alignment of its sequence with those of human CNTF and murine LIF, whose crystallographic structures have been resolved (Robinson et al., 1994; McDonald et al., 1995). The alignment was obtained from multiple alignment of the interleukin (IL)-6 cytokine family using Clustal_W with only minor manual adjustments (Thompson et al., 1994) (Figure 1). The identity score between CLC and CNTF was low (15%), with only 27 residues conserved in the modelled structure (residues 7–181) (Figures 1 and 2A). However, helix D was highly conserved between CLC and CNTF, with a 14-residue-long segment (residues 160–174) displaying 50% identity. These conserved residues in CNTF are also maintained among different species (Saggio et al., 1995).
Fig. 1. Alignment of human CLC (CLCh) with human CNTF (CNTFh) and murine LIF (LIFm). Conserved residues are in bold. The residues of CNTFh and LIFm corresponding to helices A–D are in red. The residues of CNTF whose coordinates are not resolved in the crystal structure are in blue. The sequences of the leader peptides have been removed.
Fig. 2. Overall structure of CLC. (A) Ribbon drawing of modelled CLC with the side chains of residues that are conserved in CLC and CNTF. Conserved residues corresponding to the signature of LIFR binding (Tyr34, Gly39, Phe151 and Lys154) are shown in orange. Conserved leucines are shown in green. Other conserved residues are shown in red. (B) The solvent-accessible surface of CLC. The surfaces of the conserved residues located in helix D are coloured red and that of Thr166 is coloured cyan. The two views correspond to the same protein orientation.
The 27 conserved residues were not randomly located within the tri-dimensional protein structure (Figure 2A). Ten of these residues were leucines, principally located within the hydrophobic core of the protein. The four residues (Tyr34, Gly39, Phe151 and Lys154) that correspond to the ‘signature’ of LIF receptor binding, and that are conserved in CLC, LIF, CNTF, CT-1 and OSM (Grötzinger et al., 1999; Bravo and Heath, 2000), were spatially close in the tri-dimensional model of CLC. The residues equivalent to Phe151 and Lys154 in LIF (Phe156 and Lys159) and in CNTF (Phe152 and Lys155) correspond to the ‘hot spot’ residues of the site III interaction with LIFR (Inoue et al., 1995; Di Marco et al., 1996; Hudson et al., 1996).
A cluster of conserved residues was located at or near site I corresponding to the interaction site of the cytokine with its specific receptor α-chain. For CNTF, site I of CNTFR binding corresponds to the C-terminal part of helix D and the C-terminus of the AB loop (Panayotatos et al., 1995). Conserved Glu163, Trp167, Arg170 and Asp174 made a continuous stripe on the surface of helix D (Figure 2B). This stripe was partly shielded from solvent by the AB loop. Alternative loop conformations obtained by simulated annealing did not lead to significantly increased solvent exposure of the conserved residue cluster of helix D (data not shown). Receptor binding might, however, dramatically alter the positioning of the loop. Such a conformational change has been observed for human growth hormone, which belongs to the same class of four-helix-bundle cytokines (Bazan, 1990, 1991). In an unbound state, the AB loop lies on the surface of the D helix, whereas in complex with the receptor, a structural reorganization of the AB loop with an 8 Å shift in its positioning allows the direct interaction of helix D side chains with the receptor (De Vos et al., 1992).
The CNTF residue equivalent to Thr166 of CLC (Gln167) has been shown to be important for the binding of CNTF to CNTFR. When this residue was mutated to Thr, the affinity of CNTF for CNTFR was increased 10-fold (Saggio et al., 1995). This residue is located on the surface of helix D, close to the conserved cluster. The CNTF residues equivalent to Arg170 and Asp174 (Arg171 and Asp175) were crucial for the correct binding to the CNTFR chain (Inoue et al., 1997). These residues are involved in electrostatic interactions with each other, and with Glu75 and Arg72 on helix B. These electrostatic interactions might be essential for the proper positioning of the AB loop, receptor recognition, protein folding and/or structural stability of human CNTF (McDonald et al., 1995; Inoue et al., 1997). The CLC model indicates that Arg170 and Asp174 may be involved in electrostatic interactions with Asp72 and Arg168, respectively.
This striking similarity between CLC and CNTF in a region corresponding to the CNTFR binding site of CNTF (site I) prompted us to investigate the possibility that the interaction of CLC with the soluble CNTFR might allow the secretion of CLC.
CLC associates with CNTFR to form a secreted composite cytokine
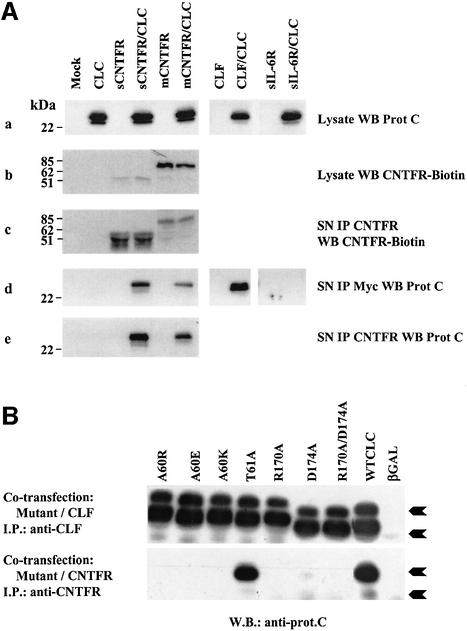
CLC cDNA was tagged with a protein C epitope coding sequence, subcloned in a mammalian expression vector and expressed in Cos-7 cells (Figure 3). Whereas the protein was readily detectable in cell lysates, secretion of the cytokine into the culture medium was not observed, in accordance with previous observations (Elson et al., 2000). Since CLC contains a putative contact site for CNTFR, the cytokine was co-expressed together with a soluble truncated form of CNTFR (sCNTFR, leading to a mature polypeptide of 316 amino acids). CLC was found to be present in the culture medium as detected by western blotting, indicating that CLC secretion can be mediated by CNTFR (Figure 3A). Interestingly, similar results were observed when co-expressing CLC together with CNTFR anchored to the membrane through a GPI linkage (Davis et al., 1991, 1993b), indicating that post-transcriptional or enzymatic processes led to the release of a soluble form of the receptor associated to CLC. In this latter case, the apparent molecular weight of the receptor portion was in the range of 75 kDa, in agreement with that described previously (Davis et al., 1993b). Similarly, CLC was secreted outside the cells when co-expressed with CLF, as we reported previously (Elson et al., 2000). In contrast, co-expression of a soluble form of IL-6 receptor (IL-6R) with CLC failed to induce secretion of the cytokine, underlining the specificity of the interaction between CLC and either CLF or CNTFR.
Fig. 3. CNTFR and CLC interact to form a secreted complex. (A) Cos-7 cells were transfected with the indicated cDNAs. (a) Expression of CLC-protC in transfected Cos-7 cell lysates detected by western blot analysis with a peroxidase-coupled anti-protC mAb. (b) Expression of CNTFR in transfected Cos-7 cell lysates detected with the biotinylated AN-E4 anti-CNTFR mAb. (c) Expression of CNTFR in transfected Cos-7 cell culture medium. Culture media were immunoprecipitated with the AN-C2 anti-CNTFR mAb (10 µg/ml). Western blot analysis of the immunoprecipitates was performed with the biotinylated AN-E4 anti-CNTFR mAb. (d) Expression of CLC-protC in transfected Cos-7 cell culture media. Culture media were immunoprecipitated with an anti-myc mAb. Western blot analysis of the immunoprecipitates was performed with the peroxidase-coupled anti-protC mAb. (e) CLC-protC is present in the culture medium as a complex with CNTFR. Culture media were immunoprecipitated with the AN-C2 anti-CNTFR mAb (10 µg/ml). Western blot analysis of the immunoprecipitates was performed with the peroxidase-coupled anti-protC mAb. (B) Physical interaction between site I CLC mutants and CLF or soluble CNTFR. Cos-7 cells were transfected with cDNAs encoding the indicated CLC mutants, wild-type CLC (positive control) or β-galactosidase (negative control), together with cDNAs encoding either CLF or sCNTFR. After 72 h, supernatants were harvested and immunoprecipitated with an anti-CLF mAb or with an anti-CNTFR mAb. Western blotting was performed with an anti-protein C mAb recognizing the wild-type as well as the mutated forms of CLC. The two differentially glycosylated forms of CLC are indicated with arrows.
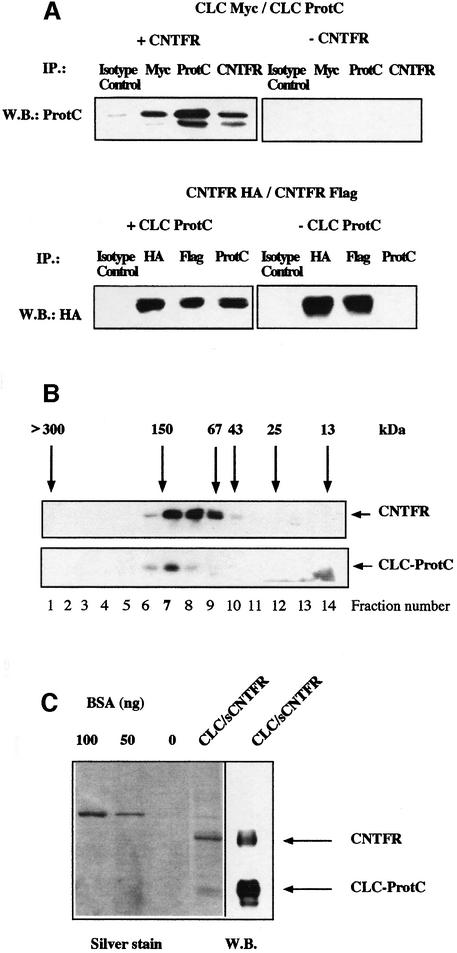
To test whether CLC and CNTFR were released as a composite cytokine, cell supernatants from Cos-7 transfected cells were subjected to immunoprecipitation with an anti-CNTFR antibody. The presence of CLC in the immunopurified fraction was revealed by western blotting using an anti-protein C antibody (Figure 3A, e). CNTFR and CLC specifically associated and could be co-precipitated, thus forming a secreted heterodimer. Similar results were also observed when the same experiments were performed in the human embryonic kidney (HEK) 293 fibroblast cell line (data not shown). The relative amount of sCNTFR engaged in the formation of the composite cytokine was determined by high-performance liquid chromatography (HPLC) gel filtration analysis of culture supernatants containing CLC–sCNTFR (see Figure 4B). Western blot analysis of the eluted fractions showed that ∼30% of soluble CNTFR was associated with CLC (an estimate of the composite cytokine concentration present in transfected fibroblast cell culture supernatants was 1–5 nM), whereas the majority of secreted CLC was retained in a complex with sCNTFR (lane 7).
Fig. 4. Stoichiometry and composition of CLC–sCNTFR cytokine. (A) Upper panel: Cos-7 cells were co-transfected with CLC-protC, CLC-myc and either sCNTFR (+CNTFR) or an empty control vector (–CNTFR). After 72 h, supernatants were immunoprecipitated with an isotype control mAb, an anti-myc mAb, an anti-protC mAb or an anti-CNTFR mAb. Western blotting analysis was performed with a biotinylated anti-protC mAb. Lower panel: Cos-7 cells were co-transfected with CNTFR-HA, CNTFR-Flag and either CLC-protC (+CLCprotC) or with an empty control vector (–CLCprotC). Supernatants were immunoprecipitated with an isotype control mAb, an anti-HA mAb, an anti-Flag mAb or an anti-protC mAb. Western blotting was performed with a peroxidase-coupled anti-HA mAb. (B) The culture supernatant of an HEK 293 cell line stably transfected with CLC–sCNTFR was size-fractionated on a Superose 12 column. Fractions were analysed by western blotting using an anti-protC mAb to detect CLC, or the AN-E4 antibody to detect sCNTFR. Column calibration was performed using standard purified proteins. (C) SDS–PAGE analysis of purified CLC–sCNTFR. CLC–sCNTFR was purified with an anti-protC column followed by QAE HPLC. Gels were silver stained and protein concentration determined using known concentrations of BSA. Western blotting analysis of a companion lane was performed using both anti-protC and anti-CNTFR mAbs for visualization.
CLC contacts CNTFR through a site I binding
Mutants of CLC were engineered to determine the binding site(s) to CNTFR. Mutations were designed to impair the putative site I of interaction with the specific receptor α-chain(s). Positions were selected based on analogy with CNTF. Two residues located at the C-terminal part of the CNTF AB loop (positions 63 and 64), equivalent to CLC Ala60 and Thr61, are known to be essential for CNTFR binding. Mutation of Gln63 of CNTF to Lys or Glu impairs CNTFR binding (Panayotatos et al., 1995), whereas mutation of the same residue to Arg creates a protein with increased activity (Panayotatos et al., 1995). Mutation of the neighbouring Trp64 to Ala also impairs CNTFR binding (Panayotatos et al., 1995). Ala60 of CLC was thus mutated to Arg, Lys and Glu, or Thr61 to Ala. Arg171 and Asp175 of CNTF, located at the C-terminal half of helix D, are also crucial for the correct binding to the CNTFR α-chain (Panayotatos et al., 1995; Inoue et al., 1997). The corresponding residues, conserved in CLC (Arg170 and Asp174), were mutated to Ala. Mutants were co-expressed in Cos-7 cells together with sCNTFR. Their ability to bind CLC and to permit the release of the CLC–sCNTFR composite cytokine was investigated (Figure 3B). Mutations introduced in the C-terminal part of helix D (positions 170 and 174) and in position 60 of the AB loop entirely abrogated the secretion of CLC when co-expressed with sCNTFR. In contrast to that observed for CNTF, the Thr61→Ala mutant conserved its capacity to bind sCNTFR and to be secreted. These results show that CLC can associate with CNTFR through a site I binding to generate a secreted composite cytokine.
Co-expression of the same set of CLC mutants with its alternate subunit, CLF (Elson et al., 2000), affected neither the CLF–CLC contact nor the secretion of the heterodimer (Figure 3B). This clearly demonstrates that CLF and sCNTFR interact with two different regions on CLC. One is identified as site I for CLC interaction with sCNTFR and the second, which remains to be identified, is required to generate the CLC–CLF composite cytokine. Interestingly, we consistently observed a preferential association of sCNTFR to a higher molecular weight form of CLC (25 kDa), whereas CLF could indifferently bind both 22 or 25 kDa forms of the molecule (Figure 3A and B). A possible explanation would be differences in the glycosylation states of CLC and in the receptor accessibility to the different glycosylated forms.
Stoichiometry of the CLC–sCNTFR cytokine
We next examined the stoichiometry and composition of the heterodimeric cytokine. In order to facilitate the detection of putative multimeric associations, CLC was tagged with either the myc or protein C (protC) epitope. Modified sCNTFR containing a Flag or haemagglutinin (HA) motif at the C-terminus were also generated. cDNAs encoding for CLC-myc and CLC-protC were simultaneously co-transfected in Cos-7 cells together with an expression vector encoding untagged sCNTFR. The culture medium was immunoprecipitated with antibodies directed against the CLC tagged motifs or CNTFR (Figure 4A). Western blotting experiments using an anti-protC monoclonal antibody (mAb) revealed that CLC-myc could be released in a dimeric form associated to CLC-protC. Additionally, the release of CLC tagged proteins depended on their co-expression with CNTFR. Similar experiments were carried out using sCNTFR labelled with Flag or HA epitopes. In contrast to that observed for CLC, secretion of the soluble receptor also occurred in the absence of CLC. Interestingly, when expressed alone, a proportion of sCNTFR spontaneously formed a stable dimer (Figure 4A and B). This was unlikely to be due to aggregate formation since protein determination by western blotting or ELISA detection (data not shown) indicated an sCNTFR concentration in the range 1–5 nM.
In order to determine the molecular weight of the CLC–sCNTFR complex, the culture supernatant of the 293 cell line stably transfected with CLC–sCNTFR was size fractionated on a Superose 12 column. Retarded fractions were studied by western blotting using mAbs directed against CNTFR or the protC epitope (Figure 4B). CLC eluted from the size-exclusion column peaked in fraction 7, corresponding to an apparent mol. wt of 120–150 kDa. With the exception of fractions surrounding fraction 7, no other significant signal could be detected in the gel filtration eluate. This indicated that fraction 7 contained the vast majority of CLC. Soluble CNTFR was recovered in fractions ranging from 50 to 150 kDa, indicating that only ∼30% of the soluble receptor was engaged in composite cytokine formation. Size-exclusion chromatography and immunoprecipitation experiments using tagged proteins (see above) support a model where at least a portion of the CLC–sCNTFR complex consists of two molecules of both CLC (mol. wt 22–25 kDa) and sCNTFR (mol. wt 50 kDa), leading to an apparent mol. wt of 140–150 kDa.
The CLC–sCNTFR complex was purified to apparent homogeneity with an anti-protein C affinity column followed by a QAE HPLC step. SDS–PAGE analysis revealed two prominent bands with apparent mol. wts of 25 and 50 kDa, respectively, identified as CLC and sCNTFR by western blotting using anti-protC and anti-CNTFR mAbs (Figure 4C).
CNTFR is required for biological responses to CLC
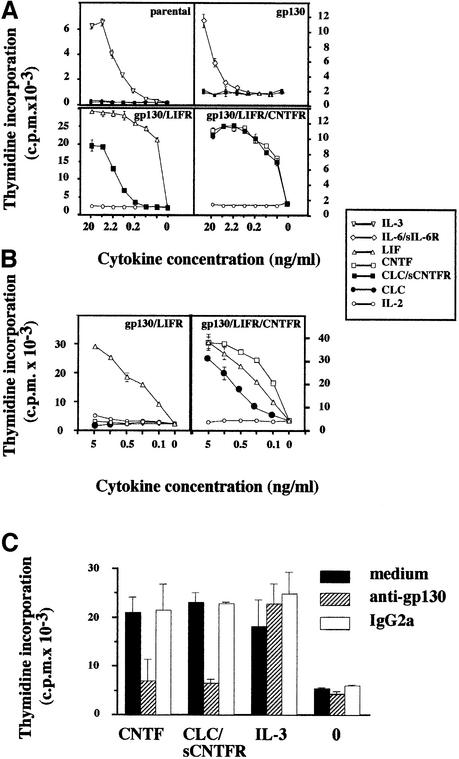
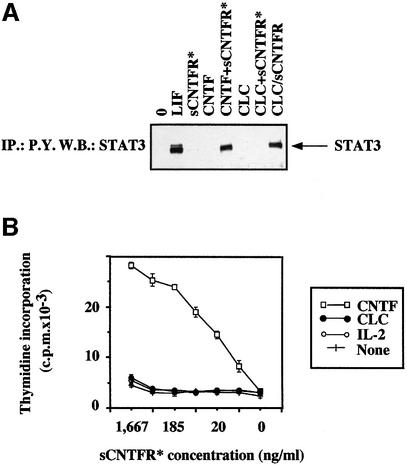
Functional properties of CLC and CLC–sCNTFR were tested in proliferation assays using derivatives of the IL-3-dependent Ba/F3 cell line (Kallen et al., 1999). Purified CLC from cell lysates or purified CLC–sCNTFR was added to cultures of Ba/F3 cell lines engineered to express the appropriate combinations of receptor subunits leading to functional IL-6-type cytokine responses (Figure 5A). The purified CLC–sCNTFR complex induced a robust proliferation of Ba/F3 cells expressing on their surface either the functional LIF receptor (gp130 and LIFR; hereafter called Ba/F3 GL) or the tripartite CNTF receptor (gp130, LIFR and CNTFR; hereafter abbreviated to Ba/F3 GLC). The respective specific activities were 2 × 105 and 5 × 107 U/mg of protein. The observed difference in specific activities was possibly due to a partial dissociation of the composite cytokine during the biological assay, rendering CLC less efficient on Ba/F GL cells, but fully active on Ba/F3 GLC cells, with the membrane-expressed tripartite CNTF receptor. In line with this observation, we recently made a fusion protein consisting of sCNTFR covalently associated to CLC and displaying a specific activity of 3 × 107 U/mg (C.Guillet, manuscript in preparation). Importantly, CLC purified from Cos-7 cell lysates triggered the proliferation of Ba/F3 GLC cells, but not of Ba/F3 GL cells, demonstrating the absolute requirement of CNTFR for generating a functional response to CLC (Figure 5B).
Fig. 5. Proliferative response of transfected Ba/F3 cell lines to the CLC–sCNTFR complex. (A) Effect of CLC–sCNTFR on parental Ba/F3 cells and cells transfected with gp130, gp130–LIFR or gp130–LIFR–CNTFR. Cells were cultured in triplicate with 3-fold dilutions of appropriate positive controls, purified CLC–sCNTFR (filled squares) or IL-2 (open circles) used as an irrelevant cytokine. After a 72 h culture period, a [3H]Tdr pulse was performed and the amount of incorporated radioactivity was determined using a β-counter. Vertical bars indicate the SEM. (B) Effect of purified CLC from Cos-7 transfected cell lysates on Ba/F3 cells transfected with gp130–LIFR and gp130–LIFR–CNTFR. Cells were cultured in triplicate with 3-fold dilutions of the indicated cytokines. (C) The AN-HH1 anti-gp130 mAb prevents the proliferative response of the Ba/F3 gp130/LIFR/CNTFR cell line to the CLC–sCNTFR complex. Transfected Ba/F3 cells were incubated in triplicate in culture medium (marked as 0) containing 1 ng/ml CNTF, CLC–sCNTFR or IL-3. The AN-HH1 antibody (hatched bars) or a control IgG2a antibody (open bars) was added at a final concentration of 30 µg/ml. After a 72 h culture period, [3H]Tdr was added for 4 h, and the incorporated radioactivity determined.
The involvement of gp130 in CLC–sCNTFR signalling was confirmed by the inhibition of proliferation following addition of a neutralizing anti-gp130 mAb to the Ba/F3 GLC cell culture (or to Ba/F3 GL cells) (Figure 5C). Furthermore, there was no detectable response to CLC–sCNTFR when Ba/F3 cells expressed gp130 alone, implicating LIFR in the functional receptor complex (Figure 5A).
Membrane binding of CLC–sCNTFR and recruitment of gp130, LIFR and STAT3 signalling proteins
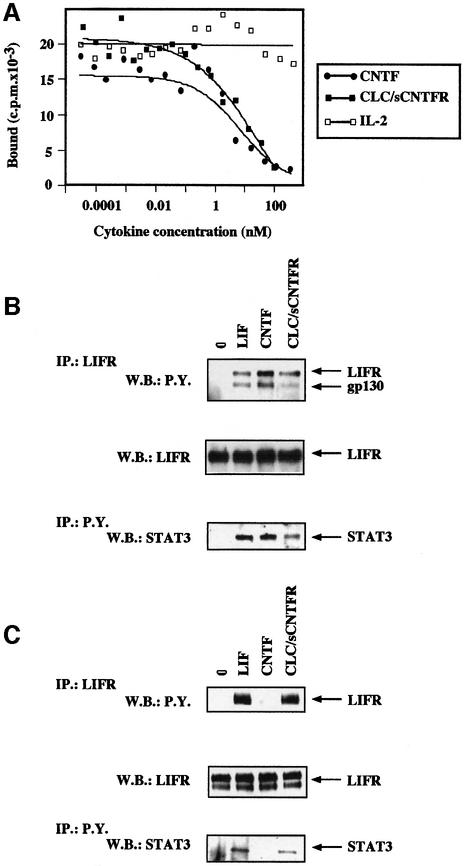
We further determined the affinity of CLC–sCNTFR for its receptor complex. Since iodination of CLC–sCNTFR completely inactivated the biological activity of the composite cytokine, affinity values were obtained by competing for CNTF binding to its functional receptor. Radiolabelled CNTF bound to its tripartite receptor expressed on the SK-N-GP neuroblastoma cell line with an affinity of 50–60 pM (data not shown), which is in agreement with our previous studies (Robledo et al., 1996, 1997). In the displacement experiments, cells were incubated in the presence of 0.6 nM iodinated CNTF and increasing concentrations of putative unlabelled competitors to reach a 100- to 200-fold excess concentration (Figure 6A). Determination of the CLC–sCNTFR affinity constant(s) from the competition experiments gave a Kd of 300 or 150 pM when considering a dimeric or a tetrameric form of the composite cytokine, respectively.
Fig. 6. Competition of CLC–sCNTFR for the binding of radiolabelled CNTF to its receptor complex. (A) The SK-N-GP cell line was incubated in the presence of 0.6 nM iodinated CNTF and increasing concentrations of unlabelled CNTF (closed circles), CLC–sCNTFR (closed squares) or IL-2 (open squares). After a 2 h incubation period at 4°C, the bound radioactivity was separated by centrifugation through an oil layer. Tyrosine phosphorylation of gp130, LIFR and STAT3 occurred in SK-N-GP neuroblastoma (gp130+LIFR+CNTFR+) (B) and in HepG-2 hepatoma cell lines (gp130+LIFR+CNTFR–) (C). The SK-N-GP and HepG2 cell lines were incubated for 10 min in the presence of 50 ng/ml LIF, CNTF, CLC–sCNTFR or without cytokine. After cell lysis in 1% Brij 96, LIFR and associated proteins were immunoprecipitated using the AN-E1 anti-LIFR antibody. Tyrosine phosphorylated proteins were analysed by SDS–PAGE followed by western blotting using an anti-phosphotyrosine mAb. After stripping, the membrane was re-stained using an anti-LIFR polyclonal antibody. For determination of tyrosine phosphorylated STAT3, cells were lysed in 1% NP-40. The proteins were then immunoprecipitated using the 4G10 anti-phosphotyrosine mAb, electrophoresed by SDS–PAGE, and western blotted with an anti-STAT3 polyclonal antibody.
The role of gp130 and LIFR as signalling components for CLC–sCNTFR was further reinforced when analysing their tyrosine phosporylation following activation by CLC–sCNTFR. Since the detection of receptor tyrosine phosphorylation is typically weak and very transient in haematopoietic cells (Chevalier et al., 1996), experiments were carried out using the SK-N-GP neuroblastoma cell line (gp130+, LIFR+, CNTFR+) and the HepG2 hepatoma cell line (gp130+, LIFR+, CNTFR–) (Kallen et al., 1999). In response to LIF, a clear induction of tyrosine phosphorylation was detected for gp130, LIFR and the STAT3 transcription factor in both cell lines studied (Figure 6B and C). CNTF induced phosphorylation of the same set of molecules only in SK-N-GP cells, as expected given that CNTFR is absent on HepG2 cells (Baumann et al., 1993). The exposure of both SK-N-GP and HepG2 cells to the CLC–sCNTFR heteromeric cytokine led to the tyrosine phosphorylation of signalling receptors and STAT3.
Additionally, CLC purified from Cos-7 transfected cell lysates was tested on a number of different cell lines (Table I). A strict correlation between STAT3 activation by CLC and surface expression of CNTFR was found. These results underlined the necessity of CNTFR involvement to generate a functional response to CLC.
Table I. STAT3 phosphorylation after stimulation with CLC purified from lysate correlates with expression of membrane CNTFR.
| Tissue origin | Cell line | CNTFR expression | STAT3 phosphorylation after cytokine stimulation |
|||
|---|---|---|---|---|---|---|
| None | LIF | CNTF | CLC | |||
| Neuroblastoma | SK-N-MC | + | – | + | + | + |
| SK-N-GP | + | – | + | + | + | |
| SK-N-SH | + | – | + | + | + | |
| IMR32 | + | – | + | + | + | |
| Glioblastoma | T98G | – | – | + | – | – |
| A172 | – | – | + | – | – | |
| CCF-STTG1 | – | – | + | – | – | |
| Epidermoid carcinoma | KB | – | – | + | – | – |
| Hepatoma | HepG2 | – | – | + | – | – |
Simultaneous cell synthesis of CLC and CNTFR is required to generate a functional composite cytokine
The addition of both CNTF and sCNTFR to the HepG2 hepatoma cell line (gp130+, LIFR+, CNTFR–) leads to STAT3 tyrosine phosphorylation (Figure 7A), as previously reported (Davis et al., 1993b). In contrast, exogenous addition of sCNTFR with purified CLC to HepG2 cells did not give rise to a response. When both CLC and sCNTFR were synthesized in the same cell, generating a secreted heterodimeric cytokine, activation of STAT3 was observed in HepG2 cells. Similarly, measurement of Ba/F3 GL cell proliferation also underlined the inability of sCNTFR to reassociate with CLC in solution to give a functional response (Figure 7B). Altogether, these experiments indicate that CLC and CNTFR need to be co-expressed in the same cell to generate a functional heterodimeric cytokine.
Fig. 7. CLC and sCNTFR co-expression in Cos-7 cells is required for functional responses. (A) STAT3 tyrosine phosphorylation in the HepG2 cell line. HepG2 cells were incubated for 10 min in the presence of LIF (50 ng/ml), purified recombinant sCNTFR (indicated with an asterisk) (1 µg/ml), CNTF (50 ng/ml), CNTF plus exogenous purified recombinant sCNTFR, purified CLC (50 ng/ml), purified CLC plus exogenous purified recombinant CNTFR, or co-synthesized CLC–sCNTFR (50 ng/ml) purified from stably transfected cells. After cell lysis in 1% NP-40, STAT3 proteins were immuno precipitated with the 4G10 anti-phosphotyrosine antibody, analysed by SDS–PAGE, and western blotted with an anti-STAT3 polyclonal antibody. (B) Proliferative response of Ba/F3 cells transfected with gp130 and LIFR to CLC plus exogenous purified recombinant sCNTFR. Cells were cultured in triplicate using 3-fold dilutions of sCNTFR and a fixed amount (20 ng/ml) of CNTF, CLC or IL-2. After a 72 h culture period, a [3H]Tdr pulse was performed and the amount of incorporated radioactivity was determined using a β-counter. Vertical bars indicate the SEM.
The CLC–sCNTFR heterodimer is a survival factor for motor neurons and neural cell lines
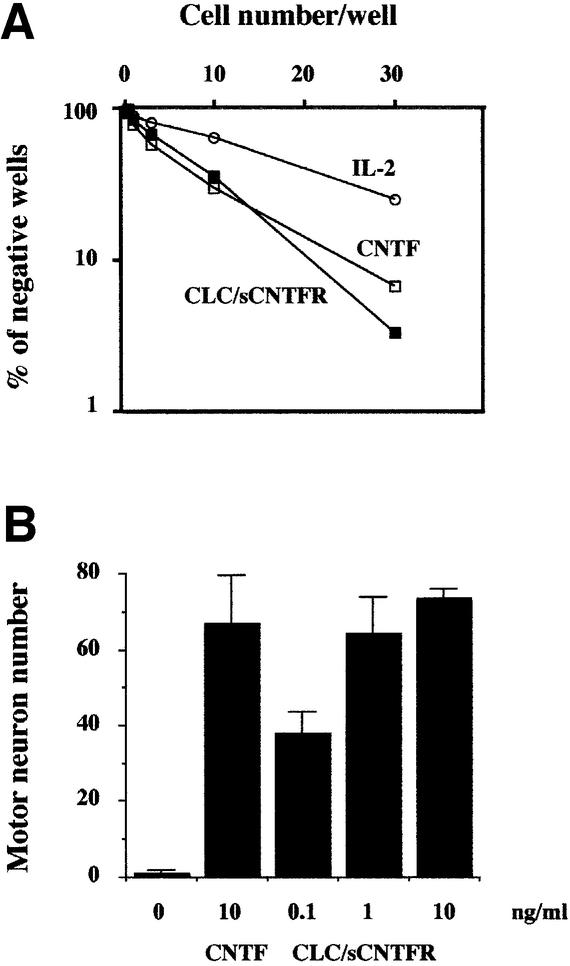
As CNTF and CLC–sCNTFR use the same signalling receptors, we studied the biological properties of CLC– sCNTFR towards cells of neural origin. Experiments were carried out using the rat Mah neuroblast cell line, whose viability can be maintained with CNTF (Ip et al., 1994). Experiments were carried out by limiting dilution and the cloning efficiency in the cultures was determined (Figure 8A). A strong increase in the frequency of growing clones was observed when Mah cells were cultured with either CLC–sCNTFR or CNTF, compared with the response observed with IL-2 used as an irrelevant cytokine.
Fig. 8. CLC–sCNTFR can substitute for CNTF on cells of neural origin. (A) Mah neuroblast cells were seeded in 60-well plates as indicated in Materials and methods. After a 5 day culture period, wells containing cells or empty wells were scored using an inverted microscope, and the cloning efficiency determined. (B) Survival effect on motor neurons. Neurons were cultured in the presence of different concentrations of purified CLC–sCNTFR. Survival values determined after 4 days of culture are means ± SEM of triplicate wells, and were compared to survival in the presence of 10 ng/ml CNTF.
We also analysed the ability of CLC–sCNTFR to support survival of rat embryonic motor neurons. A cell survival assay was performed using different concentrations of CLC–sCNTFR. After 4 days of culture, survival values were compared with those obtained using an optimal concentration of CNTF (10 ng/ml). CLC– sCNTFR was found to support the same number of motor neurons as CNTF (Figure 8B).9
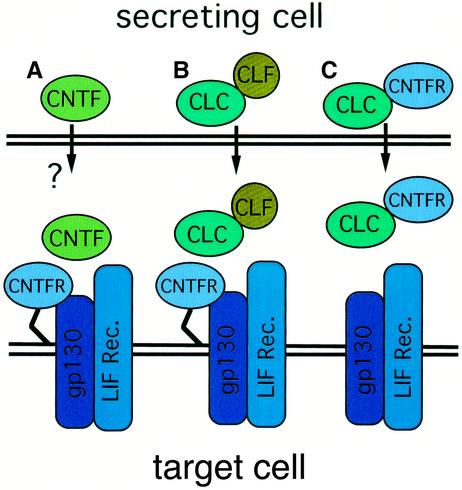
Fig. 9. Receptor complex activation by CNTF and CLC. (A) CNTF is released by trauma and activates its tripartite receptor complex. (B) CLF expression is required for CLC secretion, and the hetero complex displays activities on the tripartite CNTF receptor complex. (C) CLC associates with CNTFR within the cells before being secreted and activates the gp130–LIFR signalling complex. Stoichiometric studies indicate that in (A) and (C), at least a portion of the ligands (CLC or CNTF) dimerize and associate with two molecules of CNTFR. For CLC–CLF, the protein ratio involved in the formation of the composite cytokine is not yet known.
Discussion
CLC belongs to a constantly growing cytokine family, encompassing IL-6, CT-1, CNTF, LIF, IL-11, OSM and viral IL-6 (Bazan, 1991; Bravo and Heath, 2000). All the members of this family share the gp130 common signalling receptor, which associates with a second receptor (gp130, LIFR or OSMR, depending on the cytokine) to induce cell signalling and subsequent functional events (Taga and Kishimoto, 1997; Heinrich et al., 1998). CNTF, IL-6, IL-11 and possibly CT-1 also contact the cell membrane proteins by engaging specific α receptor components that increase the affinity and specificity of binding (Grötzinger et al., 1999; Bravo and Heath, 2000).
The secreted CLC–sCNTFR composite cytokine
Like CNTF, CLC is a non-secreted cytokine (Stöckli et al., 1989; Senaldi et al., 1999; Shi et al., 1999). Unlike CNTF, however, it contains a leader sequence, suggesting that it has the potential to be secreted via the classical secretory pathway, although it is clearly retained within the cell by an as yet unknown mechanism. We showed recently that CLC can indeed be released when co-expressed with the soluble cytokine receptor CLF, the two proteins associating to form a functional composite cytokine to activate the tripartite CNTF receptor complex (Elson et al., 2000). In the present study, we observed that CLC secretion could also be mediated by CNTFR to form an alternative secreted composite cytokine (Figure 9). The fact that CLC is capable of being secreted from cells makes it fundamentally different from CNTF, the primary structure of which lacks a peptide sequence. It has been suggested that the trophic role of CNTF is primarily pathophysiological, and it is released upon nerve injury, which causes the rupture of Schwann cells (Sendtner et al., 1997). This in turn indicates that the composite cytokines CLC–CLF and CLC–sCNTFR are secreted ligands, probably playing an important role in fetal development.
The structure of both the CLC–CLF and CLC–sCNTFR complexes resembles the IL-12 heterodimeric cytokine. The p35 subunit of IL-12 is related to the IL-6-type cytokine, whereas the p40 component belongs to the IL-6 cytokine receptor family (Yoon et al., 2000). Despite the fact that p35, like CLC, contains a signal peptide, the cytokine is only efficiently released from cells when co-expressed with the p40 soluble receptor-type subunit (D’Andrea et al., 1992). These results are therefore another example of an expanding body of evidence demonstrating that non-signalling cytokine receptor subunits not only define cytokine target cell specificity, but can also mediate cytokine secretion from producer cells.
Stoichiometric study of the CLC–sCNTFR composite cytokine revealed that a portion of the cytokine consists of two molecules each of CLC and sCNTFR. This is supported by gel filtration analysis, indicating that the complex has an apparent mol. wt of between 120 and 150 kDa. This situation is similar to that previously reported for IL-6/IL-6R, IL-11/IL-11R and CNTF– CNTFR, where receptor binding leads to hexamer formation (Ward et al., 1996; De Serio et al., 1995; Barton et al., 2000). Nevertheless, we cannot exclude the possibility that additional protein:protein ratios also co-exist for the composite cytokine. In line with this, a tetrameric model was also proposed based on the biological activities of IL-6–CNTF chimeric cytokines (Grötzinger et al., 1999; Kallen et al., 1999, 2000).
Additionally, we consistently observed the formation of dimeric soluble CNTFR in the absence of detectable ligand. This did not result from aggregate formation since the mammalian expression system we used led to an sCNTFR concentration in the range of 1–5 nM. Interestingly, expression of sCNTFR in E.coli also led to the detection, by gel filtration or non-reducing gel electrophoresis, of a receptor with an apparent molecular weight twice that observed under denaturing conditions (Panayotatos et al., 1994). This study is in line with our present observation, showing the capacity for sCNTFR to form a dimer spontaneously. Similar observations were reported previously for soluble expressed erythropoietin (EPO) receptor and for the p40 subunit of IL-12 (Ling et al., 1995; Livnah et al., 1999; Remy et al., 1999). In the case of the EPO receptor preformed dimer, the ligand binding induced a re-organization of the receptor, bringing the extracellular domains into closer proximity and allowing the subsequent phosphorylation events of the response.
The CLC–sCNTFR composite cytokine only forms within the cell
It is well known that CNTF can exert its effects on cells expressing gp130 and LIFR when soluble CNTFR is available locally (Davis et al., 1993b). CNTFR is readily cleaved from the cell surface, and its presence has been detected at physiological concentrations in cerebrospinal fluid and serum. It is released from skeletal muscle in response to peripheral injury (Davis et al., 1993b). Our observations demonstrate that the presence of recombinant soluble CNTFR in assays containing functional CLC purified from cell lysates has no potentiating effect on Ba/F3 cells expressing gp130 and LIFR. It therefore appears that CLC and CNTFR must be present either within an intracellular environment or on the cell surface for an active complex to be formed. It is noteworthy that, as demonstrated by gel filtration experiments, the vast majority of CLC remains complexed with sCNTFR in freshly isolated cell supernatants, suggesting that the complex is relatively stable in an extracellular environment. The inability of CLC to re-associate to sCNTFR in solution might be linked to a low-affinity interaction between these two proteins. Similarly, the contact strength between CNTF and CNTFR is in the 10–8 M range, and increases up to 30–50 pM when gp130 and LIFR are also engaged in CNTF tripartite receptor formation (Gearing et al., 1994; Robledo et al., 1996). A similar situation might occur for CLC binding to its tripartite membrane receptor versus soluble CNTFR alone. Additionally, we cannot exclude the possibility that association of CLC–sCNTFR inside the cells requires some particular physiological situations (local concentrations, pH values) or enzymatic processes that cannot be reproduced in solution.
We also showed that sCNTFR interacted preferentially with a CLC glycosylated form of 25 kDa in order to generate the composite cytokine. Interestingly, the importance of p35 glycosylation for IL-12 synthesis and secretion was demonstrated recently (Carra et al., 2000). Regulation of CLC–sCNTFR association and secretion might well involve a similar process. A possible explanation could also be differences in the receptor accessibility to these different glycosylated forms of CLC; this is currently under investigation in the laboratory.
CLC–CNTFR and CLC–CLF contact sites
The interaction of a cytokine with its α-chain is principally governed by a site I domain expressed on the surface of the cytokine. The site I is usually located in the terminal part of helix D, and implicates a portion of the AB loop and residues within helices A and B (Grötzinger et al., 1999; Bravo and Heath, 2000). A modelling study of CLC revealed structural similarities between CLC and CNTF, including the conservation of a site I domain involved in the binding to CNTFR. In vitro experiments indicate that CLF and CNTFR can associate simultaneously with CLC to form a multimeric complex. Furthermore, we could detect CLF associated to the CLC membrane tripartite receptor complex by immunoprecipitation experiments (Elson et al., 2000), indicating that CLF is not displaced from the cell surface following receptor engagement by CLC. This in turn suggests that CLF and CNTFR interact with two different regions on CLC, which are likely to be site I (for CNTFR) and an as yet unidentified site (for CLF). This notion is supported further by site-directed mutagenesis studies, which show that site I of CLC is indeed involved in the binding of the cytokine to CNTFR, but not with CLF. The observation that two independent non-signalling receptor subunits (CLF and CNTFR) interact with two different regions of CLC to induce its secretion makes it unlikely that CLC is retained in the cell via a retention motif, which is simply masked following interaction of the cytokine with its receptor subunit. It is also of interest to note that CLC displays a second hydrophobic region localized at amino acids 135–160, which is not a common structural feature among the cytokines, and the function of which is currently being investigated.
Biological activities of CNTFR–CLC
The CLC–sCNTFR complex induced proliferation of Ba/F3 cells expressing the tripartite CNTFR receptor complex as well as of transfectants expressing LIFR and gp130. The phosphorylation of gp130/LIFR/STAT3 signalling components was also observed in both NGP neuroblastoma cells (gp130+, LIFR+, CNTFR+) and HepG2 hepatoma cells (gp130+, LIFR+, CNTFR–). CNTFR–CLC could also mimic the biological activities of CNTF and LIF on motor neurons by supporting their survival in vitro. Both LIF and CNTF have been investigated extensively for their therapeutic potential in neurodegenerative diseases (Aebischer et al., 1996; Miller et al., 1996; Penn et al., 1997; Kurek et al., 1998; Mittoux et al., 2000). It therefore appears likely that CLC heterocomplexes have similar promise for the treatment of these disorders.
Materials and methods
Modelling studies
The alignment of human CLC with the human cytokines of the IL-6 family interacting with the LIF receptor (CNTF, LIF, CT-1 and OSM) and with murine LIF was carried out using Clustal_W (Thompson et al., 1994) and the Gonnet matrix series (Gonnet et al., 1992). The final multiple alignment required only minor manual adjustments to avoid gaps within structurally conserved regions, as determined from the crystal structure of human CNTF [Protein Data Bank (PDB) accession No. 1CNT] and mouse LIF (PDB accession No. 1LKI). CLC was modelled from residues 7 to 181 by homology with human CNTF and murine LIF. Homology modelling was carried out with MODELER (MSI, San Diego, CA) (Sali and Blundell, 1993) using the crystallographic coordinates of hCNTF as a template, except for the AB and CD loops, which were missing or partially missing in the crystal structure of CNTF. Residues 39–62 and 133–147 were modelled from the crystallographic coordinates of the equivalent residues in murine LIF. The four-residue insertion in the CD loop was modelled as an α-helix turn initiating at Pro140. The quality of the model was checked with Profile3D (MSI) (Gribskov et al., 1990). The score of 63 (out of a maximum score of 79) indicated the absence of significant misfolding. Twenty alternative conformations of the AB loop were analysed by simulated annealing and the lowest energy conformation was selected. The distance between the S atoms of Cys88 and Cys110 (located in helices B and C, respectively) was 7.4 Å and did not allow the formation of an internal disulfide bridge. The quality score was similar to the initial model. The solvent-accessible Connoly surface of the protein was calculated using a probe radius of 1.4 Å. Figure 1 was prepared using Insight (MSI).
Cells and reagents
Cos-7 and 293 fibroblast cells, and HepG2 hepatoma, KB epidermoid carcinoma, neuroblastoma and glioblastoma cell lines were routinely cultured in RPMI 1640 medium supplemented with 10% fetal calf serum (FCS). Parental and transfected Ba/F3 cells were maintained as previously described (Kallen et al., 1999). The rat neuroblast Mah cell line (Ip et al., 1994) was grown in Neurobasal medium supplemented with B7 nutrient (Life Technologies, Cergy Pontoise, France), 10% FCS and 5 µM dexamethasone. Purified recombinant LIF, IL-3 and soluble IL-6 receptor were kindly donated by Drs K.Turner and M.Stahl (Genetics Institute, Boston, MA). Human IL-6, CNTF and CNTF receptor were purchased from R&D Systems (Oxon, UK). IL-2 was a kind gift of Dr G.Zurawski (DNAX Research Insitute, CA). Polyclonal antibodies raised against STAT3 and LIFR were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). The 4G10 anti-phosphotyrosine mAb was from UBI (Lake Placid, NY). AN-HH1 (IgG2a) (anti-gp130), AN-E1 (IgG1) (anti-LIFR), anti-CNTFR (AN-E4, AN-C2) (both IgG2a) and anti-human CLF (IgG1) (AN-F-C6) were mAbs generated in the laboratory. The anti-protein C (HPC4), anti-c-myc and anti-HA antibodies were purchased from Roche Diagnostics (Roche Diagnostics, Germany). mAb recognizing the Flag epitope was from Sigma (St Louis, MO).
cDNA cloning and cell transfection
All cDNAs were cloned into the mammalian expression vector pCDNA3 (Invitrogen, San Diego, CA). cDNA encoding human CLC was amplified by PCR from lymph node cDNA. The CLC construct was modified so that the protein contained either or both the protein C (EDQVDPRLIDGK) and c-myc (EQKLISEEDL) peptide epitope at its C-terminus. cDNA encoding the extracellular portion of IL-6R was amplified from an expression construct encoding the transmembrane form of the protein by PCR (Yamasaki et al., 1988). The generation of expression vectors for membrane and soluble forms of CNTFR was described previously (Auguste et al., 1996). cDNA encoding soluble CNTFR was also modified to contain either the HA (YPYDVPDYA) or the Flag (DYKDDDDK) peptide epitope at its C-terminus. Cos-7 cells were transfected by the DEAE–dextran method as described (Robledo et al., 1997). After a 72 h culture period, the expression and function of recombinant proteins were studied. A 293 cell line stably transfected with a bicistronic expression cassette in pCDNA3 encoding both CLC (with a C-terminal protein C epitope) and sCNTFR (an internal ribosomal entry site was placed between the cDNA encoding CLCprotC and that encoding sCNTFR), allowing the simultaneous expression of CLC and sCNTFR, was generated for large scale protein production.
CLC site-directed mutagenesis
The pCDNA3 vector containing the cDNA encoding the human CLC tagged with protein C epitope was subjected to site-directed mutagenesis using the QuikChangeTM Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) following the manufacturer’s instructions. Mutations were performed on the predicted site I of CLC by replacing the original amino acid residues as indicated in Figure 3. Mutants were transfected in Cos-7 cells using the ExGen reagent (Euromedex, Souffelweyersheim, France) according to the manufacturer’s guidelines. After a 72 h culture period, cell supernatants were immunoprecipitated with either anti-CLF (AN-F-C6) or the anti-CNTFR (AN-C2) mAbs (10 µg/ml) overnight at 4°C. Samples were then treated as described for the tyrosine phosphorylation analysis and western blotted with the biotinylated anti-protein C mAb, HPC4.
CLC and CLC–sCNTFR purification
CLC-transfected Cos-7 cell lysates were obtained using the NP-40 lysate buffer (Robledo et al., 1997). Serum-free media containing the CLC–sCNTFR cytokine were collected from 293 stable transfectant culture supernatants. Samples were loaded on an anti-protein C affinity column (Roche Diagnostics). Eluted fractions were subjected further to a QAE HPLC step and tested on Ba/F3 GL and Ba/F3 GLC for biological responses. Purity and concentration determinations were carried out by SDS–PAGE and silver staining. Gel filtration HPLC was performed using a Superose 12 column (Pharmacia, Uppsala, Sweden) run in phosphate-buffered saline at a flow rate of 0.5 ml/min. Column calibration was carried out using standard purified proteins.
Radiolabelling and affinity studies
CNTF was iodinated by the two-phase method as described previously (Robledo et al., 1996). The specific activity of radiolabelled product was 3800 c.p.m./fM. Cells (3–5 × 106) were incubated with the indicated concentrations of radiolabelled ligand and increasing concentrations of unlabelled cytokine. After a 2 h incubation period at 4°C, cell-bound radioactivity was separated from the unbound fraction by centrifugation through an oil layer. Determination of affinity binding constants was performed according to Scatchard (1949).
Biological assays
For proliferation assays, Ba/F3 cell lines were seeded in 96-well plates at a concentration of 5 × 103 cells/well in RPMI 1640 medium containing 5% FCS. Serial dilutions of the cytokines tested were performed in triplicate. After a 72 h incubation period, 0.5 µCi of [3H]Tdr was added to each well for the last 4 h of the culture and the incorporated radioactivity determined by scintillation counting. Mah cell line cloning experiments were carried out in 60-well Terasaki plates. Plates were coated with poly-d-lysine and laminin, and the culture performed in 10 µl of Neurobasal medium (Life Technologies, Cergy Pontoise, France) supplemented with B7 nutrient (Life Technologies), 2.5% FCS, 5 µM dexamethasone and the indicated cytokines. After a 5 day culture period, wells containing cells and empty wells were scored using an inverted microscope. The frequency of growing cells was determined as the inverse number of seeded cells leading to 37% empty wells (Chevalier et al., 1987; Dozmorov et al., 2000). Motor neurons were isolated from E14.5 Sprague–Dawley rat ventral spinal cords as described previously (Arce et al., 1999) with slight modifications. Briefly, motor neurons were purified by a combination of metrizamide density-gradient centrifugation and indirect magnetic cell sorting with the MC192 antibody, which recognizes the p75 low-affinity NGF receptor (a specific marker for motor neurons at this stage). Purified motor neurons were plated in 4-well tissue culture dishes at a density of 800 neurons per well. Wells had been coated previously with polyornithine/laminin (Camu and Henderson, 1994). Culture medium (basal medium) was Neurobasal supplemented with B7 supplement, 2% horse serum, l-glutamine (0.5 mM), l-glutamate (25 µM) and 2 β-mercaptoethanol (25 µM). Recombinant neurotrophic factors were added at the time of cell seeding. Motor neuron survival was quantified by counting the number of large phase-bright neurons with long axonal processes in the total area of triplicate wells. Survival values were corrected for the value in basal medium, taken as no surviving motor neurons.
Tyrosine phosphorylation analysis
After a 24 h incubation period in serum-free medium, SK-N-GP and HepG2 cells were stimulated for 10 min with cytokines (Robledo et al., 1997). Cells were lysed in 10 mM Tris–HCl pH 7.6, 5 mM EDTA, 50 mM NaCl, 30 mM sodium pyrophosphate, 50 mM sodium fluoride, 1 mM sodium orthovanadate, protease inhibitors (1 µg/ml pepstatin, 2 µg/ml leupeptin, 5 µg/ml aprotinin, 1 mM phenylmethylsulfonyl fluoride) and 1% NP-40 or Brij 96. After pelleting insoluble material and protein standardization, the supernatants were immunoprecipitated overnight. Complexes were then isolated using beads coupled to protein A, subjected to SDS–PAGE and transferred onto Immobilon membranes (Millipore, Bedord, MA). The membranes were subsequently incubated with the indicated primary antibody before being incubated with the appropriate secondary antibody, or avidin labelled with peroxidase for 60 min. The reaction was visualized on X-ray film using ECL reagent (Amersham, Les Ullis, France) according to the manufacturer’s instructions. Where indicated, membranes were stripped in 0.1 M glycine pH 2.5 for 15–24 h, and neutralized in 1 M Tris–HCl pH 7.6 before reblotting.
Immunoprecipitation and western blotting
Cells were lysed in 1% Brij 96 buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 1% Brij 96) containing protease inhibitors (see above). A high-speed centrifugation step removed cell debris from both lysates and cell culture media. Tagged proteins were immunoprecipitated from samples using either anti-protein C, anti-HA or anti-Flag mAbs at a concentration of 10 µg/ml for 4 h at 4°C, and subsequently incubated with protein A– Sepharose beads for 1 h. Samples were then treated as described above for the phosphorylation analysis.
Flow cytometry
Expression of CNTFR was monitored by indirect immunofluorescence using the AN-C2 mAb following standard protocols (Kallen et al., 1999). Samples were analysed on a FACScan scanner (Becton & Dickinson, Mountain View, CA).
Acknowledgments
Acknowledgements
We thank Dr Karl-Josef Kallen for providing us with the Ba/F3 transfected cells, and Dr Christopher Henderson for his critical reading of the manuscript. H.P.-F. and C.G. were funded by grants from the city of Angers and the Departement du Maine et Loire, respectively. E.L. was funded by the Association Française contre les Myopathies. The project was supported by a grant from the Association Française contre les Myopathies.
References
- Aebischer P. et al. (1996) Intrathecal delivery of CNTF using encapsulated genetically modified xenogeneic cells in amyotrophic lateral sclerosis patients. Nature Med., 2, 696–699. [DOI] [PubMed] [Google Scholar]
- Arce V., Garces,A., de Bovis,B., Filippi,P., Henderson,C., Pettmann,B. and de Lapeyrière,O. (1999) Cardiotrophin-1 requires LIFR β to promote survival of mouse motor neurons purified by a novel technique. J. Neurosci. Res., 55, 119–126. [DOI] [PubMed] [Google Scholar]
- Auguste P., Robledo,O., Olivier,C., Froger,J., Praloran,V., Pouplard-Barthelaix,A. and Gascan,H. (1996) Alanine substitution for Thr268 and Asp269 of soluble ciliary neurotrophic factor (CNTF) receptor α component defines a specific antagonist for the CNTF response. J. Biol. Chem., 271, 26049–26056. [DOI] [PubMed] [Google Scholar]
- Barton V.A., Hall,M.A., Hudson,K.R. and Heath,J.K. (2000) Interleukin-11 signals through the formation of a hexameric receptor complex. J. Biol. Chem., 275, 36197–36203. [DOI] [PubMed] [Google Scholar]
- Baumann H., Ziegler,S.F., Mosley,B., Morella,K.K., Pajovic,S. and Gearing,D.P. (1993) Reconstitution of the response to leukemia inhibitory factor, oncostatin M and ciliary neurotrophic factor in hepatoma cells. J. Biol. Chem., 268, 8414–8417. [PubMed] [Google Scholar]
- Bazan J.F. (1990) Structural design and molecular evolution of a cytokine receptor superfamily. Proc. Natl Acad. Sci. USA, 87, 6934–6938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazan J.F. (1991) Neuropoietic cytokines in the hematopoietic fold. Neuron, 7, 197–208. [DOI] [PubMed] [Google Scholar]
- Bravo J. and Heath,J.K. (2000) Receptor recognition by gp130 cytokines. EMBO J., 19, 2399–2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camu W. and Henderson,C.E. (1994) Rapid purification of embryonic rat motoneurons: an in vitro model for studying MND/ALS pathogenesis. J. Neurol. Sci. Suppl., 124, 73–74. [DOI] [PubMed] [Google Scholar]
- Carra G., Gerosa,F. and Trinchieri,G. (2000) Biosynthesis and posttranslational regulation of human IL-12. J. Immunol., 164, 4752–4761. [DOI] [PubMed] [Google Scholar]
- Chevalier S., Lacroix,H. and Soulillou,J.P. (1987) Blood transfusion plus allograft, but not transfusion alone, induce IL-2-producing suppressor cells in Lew-1A recipients of LEW-1W heart allograft. Transplant. Proc., 19, 544–546. [PubMed] [Google Scholar]
- Chevalier S., Fourcin,M., Robledo,O., Wijdenes,J., Pouplard-Barthelaix,A. and Gascan,H. (1996) IL-6 family of cytokines induced activation of different functional sites expressed by gp130 transducing protein. J. Biol. Chem., 271, 14764–14772. [DOI] [PubMed] [Google Scholar]
- Curtis R., Adryan,K.M., Zhu,Y., Harkness,P.J., Lindsay,R.M. and DiStefano,P.S. (1993) Retrograde axonal transport of ciliary neurotrophic factor is increased by peripheral nerve injury. Nature, 365, 253–255. [DOI] [PubMed] [Google Scholar]
- D’Andrea A. et al. (1992) Production of natural killer cell stimulatory factor (interleukin 12) by peripheral blood mononuclear cells. J. Exp. Med., 176, 1387–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis S., Aldrich,T.H., Valenzuela,D.M., Wong,V.V., Furth,M.E., Squinto,S.P. and Yancopoulos,G.D. (1991) The receptor for ciliary neurotrophic factor. Science, 253, 59–63. [DOI] [PubMed] [Google Scholar]
- Davis S., Aldrich,T.H., Stahl,N., Pan,L., Taga,T., Kishimoto,T., Ip,N.Y. and Yancopoulos,G.D. (1993a) LIFR β and gp130 as heterodimerizing signal transducers of the tripartite CNTF receptor. Science, 260, 1805–1808. [DOI] [PubMed] [Google Scholar]
- Davis S. et al. (1993b) Released form of CNTF receptor α component as a soluble mediator of CNTF responses. Science, 259, 1736–1739. [DOI] [PubMed] [Google Scholar]
- DeChiara T.M. et al. (1995) Mice lacking the CNTF receptor, unlike mice lacking CNTF, exhibit profound motor neuron deficits at birth. Cell, 83, 313–322. [DOI] [PubMed] [Google Scholar]
- De Serio A., Graziani,R., Laufer,R., Ciliberto,G. and Paonessa,G. (1995) In vitro binding of ciliary neurotrophic factor to its receptors: evidence for the formation of an IL-6-type hexameric complex. J. Mol. Biol., 254, 795–800. [DOI] [PubMed] [Google Scholar]
- De Vos A.M., Ultsch,M. and Kossiakoff,A.A. (1992) Human growth hormone and extracellular domain of its receptor: crystal structure of the complex. Science, 255, 306–312. [DOI] [PubMed] [Google Scholar]
- Di Marco A., Gloaguen,I., Graziani,R., Paonessa,G., Saggio,I., Hudson,K. and Laufer,R. (1996) Identification of ciliary neurotrophic factor (CNTF) residues essential for leukemia inhibitory factor receptor binding and generation of CNTF receptor antagonists. Proc. Natl Acad. Sci. USA, 93, 9247–9252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dozmorov I., Eisenbraun,M.D. and Lefkovits,I. (2000) Limiting dilution analysis: from frequencies to cellular interactions. Immunol. Today, 21, 15–18. [DOI] [PubMed] [Google Scholar]
- Elson G.C., Graber,P., Losberger,C., Herren,S., Gretener,D., Menoud,L.N., Wells,T.N., Kosco-Vilbois,M.H. and Gauchat,J.F. (1998) Cytokine-like factor-1, a novel soluble protein, shares homology with members of the cytokine type I receptor family. J. Immunol., 161, 1371–1379. [PubMed] [Google Scholar]
- Elson G.C. et al. (2000) Cytokine-like factor-1 associates with cardiotrophin-1 like cytokine to form a functional heteromeric ligand for the CNTF receptor complex. Nature Neurosci., 3, 867–872. [DOI] [PubMed] [Google Scholar]
- Gearing D.P., Thut,C.J., VandenBos,T., Gimpel,S.D., Delaney,P.B., King,J., Price,V., Cosman,D. and Beckmann,M.P. (1991) Leukemia inhibitory factor receptor is structurally related to the IL-6 signal transducer, gp130. EMBO J., 10, 2839–2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gearing D.P. et al. (1992) The IL-6 signal transducer, gp130: an oncostatin M receptor and affinity converter for the LIF receptor. Science, 255, 1434–1437. [DOI] [PubMed] [Google Scholar]
- Gearing D.P., Ziegler,S.F., Comeau,M.R., Friend,D., Thoma,B., Cosman,D., Park,L. and Mosley,B. (1994) Proliferative responses and binding properties of hematopoietic cells transfected with low-affinity receptors for leukemia inhibitory factor, oncostatin M and ciliary neurotrophic factor. Proc. Natl Acad. Sci. USA, 91, 1119–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonnet G.H., Cohen,M.A. and Benner,S.A. (1992) Exhaustive matching of the entire protein sequence database. Science, 256, 1443–1445. [DOI] [PubMed] [Google Scholar]
- Gribskov M., Lüthy,R. and Esisenberg,D. (1990) Profile analysis. Methods Enzymol., 183, 146–159. [DOI] [PubMed] [Google Scholar]
- Grötzinger J., Kernebeck,T., Kallen,K.J. and Rose-John,S. (1999) IL-6 type cytokine receptor complexes: hexamer, tetramer or both? Biol. Chem., 380, 803–813. [DOI] [PubMed] [Google Scholar]
- Guillet C., Auguste,P., Mayo,W., Kreher,P. and Gascan,H. (1999) Ciliary neurotrophic factor is a regulator of muscular strength in aging. J. Neurosci., 19, 1257–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich P.C., Behrmann,I., Muller-Newen,G., Schaper,F. and Graeve,L. (1998) Interleukin-6 type cytokine signalling through the gp130/Jak/STAT pathway. Biochem. J., 334, 297–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helgren M. et al. (1994) Trophic effect of ciliary neurotrophic factor on denervated skeletal muscle. Cell, 76, 493–504. [DOI] [PubMed] [Google Scholar]
- Hibi M., Murakami,M., Saito,M., Hirano,T., Taga,T. and Kishimoto,T. (1990) Molecular cloning and expression of an IL-6 signal transducer, gp130. Cell, 63, 1149–1157. [DOI] [PubMed] [Google Scholar]
- Hudson K.R., Vernallis,A.B. and Heath,J.K. (1996) Characterization of the receptor binding sites of human leukemia inhibitory factor and creation of antagonists. J. Biol. Chem., 271, 11971–11978. [DOI] [PubMed] [Google Scholar]
- Inoue M., Nakayama,C., Kikuchi,K., Kimura,T., Ishige,Y., Ito,A., Kanaoka,M. and Noguchi,H. (1995) D1 cap region involved in the receptor recognition and neural cell survival activity of human ciliary neurotrophic factor. Proc. Natl Acad. Sci. USA, 92, 8579–8583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue M., Karita,H., Kikuchi,K., Nakayama,C. and Noguchi,H. (1997) V170 area plays an essential role in the biological activity of human ciliary neurotrophic factor. J. Neurochem., 68, 1436–1442. [DOI] [PubMed] [Google Scholar]
- Ip N., Boulton,T., Li,Y., Verdi,J.M., Birren,S.J., Anderson,D. and Yancopoulos,G.D. (1994) CNTF, FGF and NGF collaborate to drive the terminal differentiation of Mah cells into postmitotic neurons. Neuron, 13, 443–455. [DOI] [PubMed] [Google Scholar]
- Kallen K.J. et al. (1999) Receptor recognition sites of cytokines are organized as exchangeable modules. Transfer of the leukemia inhibitory factor receptor-binding site from ciliary neurotrophic factor to interleukin-6. J. Biol. Chem., 274, 11859–11867. [DOI] [PubMed] [Google Scholar]
- Kallen K.J., Grötzinger J. and Rose-John,S. (2000) New perspectives on the design of cytokines and growth factors. Trends Biotechnol., 18, 455–461. [DOI] [PubMed] [Google Scholar]
- Kurek J.B., Radford,A.J., Crump,D.E., Bower,J.J., Feeney,S.J., Austin,L. and Byrne,E. (1998) LIF (AM424), a promising growth factor for the treatment of ALS. J. Neurol. Sci., 160, S106–S113. [DOI] [PubMed] [Google Scholar]
- Lin L.F., Mismer,D., Lile,J.D., Armes,L.G., Butler,E.T., Vannice,J.L. and Collins,F. (1989) Purification, cloning and expression of ciliary neurotrophic factor (CNTF). Science, 246, 1023–1025. [DOI] [PubMed] [Google Scholar]
- Ling P., Gately,M.K., Gubler,A.S., Lin,P., Hollfelder,K., Su,C., Pan,Y.C. and Hakimi,J. (1995) Human IL-12 p40 homodimer binds to the IL-12 receptor but does not mediate biological activity. J. Immunol., 154, 116–127. [PubMed] [Google Scholar]
- Livnah O., Stura,E.A., Middleton,S.A., Johnson,D.L., Jolliffe,L.K. and Wilson,I.A. (1999) Crystallographic evidence for preformed dimers of erythropoietin receptor before ligand activation. Science, 283, 987–990. [DOI] [PubMed] [Google Scholar]
- Lütticken C. et al. (1994) Association of transcription factor APRF and protein kinase Jak1 with the interleukin-6 signal transducer gp130. Science, 263, 89–92. [DOI] [PubMed] [Google Scholar]
- Masu Y., Wolf,E., Holtmann,B., Sendtner,M., Brem,G. and Thoenen,H. (1993) Disruption of the CNTF gene results in motor neuron degeneration. Nature, 365, 27–32. [DOI] [PubMed] [Google Scholar]
- McDonald N.Q., Panayotatos,N. and Henrickson,W.A. (1995) Crystal structure of dimeric human ciliary neurotrophic factor determined by MAD phasing. EMBO J., 14, 2689–2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller R.G. et al. (1996) A placebo-controlled trial of recombinant human ciliary neurotrophic (rhCNTF) factor in amyotrophic lateral sclerosis. rhCNTF ALS Study Group. Ann. Neurol., 39, 256–260. [DOI] [PubMed] [Google Scholar]
- Mitsumoto H., Ikeda,K., Klinkosz,B., Cedarbaum,J.M., Wong,V. and Lindsay,R.M. (1994) Arrest of motor neurone disease in wobbler mice cotreated with CNTF and BDNF. Science, 265, 1107–1110. [DOI] [PubMed] [Google Scholar]
- Mittoux V. et al. (2000) Restoration of cognitive and motor functions by ciliary neurotrophic factor in a primate model of Huntington’s disease. Hum. Gene Ther., 11, 1177–1187. [DOI] [PubMed] [Google Scholar]
- Oppenheim R.W., Prevette,D., Qin-Wei,Y., Collins,F. and MacDonald,J. (1991) Control of embryonic motor neuron survival in vivo by ciliary neurotrophic factor. Science, 251, 1616–1618. [DOI] [PubMed] [Google Scholar]
- Panayotatos N., Everdeen,D., Liten,A., Somogyi,R. and Acheson,A. (1994) Recombinant human CNTF receptor α: production, binding, stoichiometry and characterization of its activity as a diffusable factor. Biochemistry, 33, 5813–5818. [DOI] [PubMed] [Google Scholar]
- Panayotatos N., Radziejewska,E., Acheson,A., Somogyi,R., Thadani,A., Hendrickson,W.A. and McDonald,N.Q. (1995) Localization of functional receptor epitopes on the structure of ciliary neurotrophic factor indicates a conserved, function-related epitope topography among helical cytokines. J. Biol. Chem., 270, 14007–14014. [DOI] [PubMed] [Google Scholar]
- Penn R.D., Kroin,J.S., York,M.M. and Cedarbaum,J.M. (1997) Intrathecal ciliary neurotrophic factor delivery for treatment of amyotrophic lateral sclerosis (phase I trial). Neurosurgery, 40, 94–99. [DOI] [PubMed] [Google Scholar]
- Pennica D. et al. (1995) Cardiotrophin-1. Biological activities and binding to the leukemia inhibitory factor receptor/gp130 signalling complex. J. Biol. Chem., 270, 10915–10922. [DOI] [PubMed] [Google Scholar]
- Remy I., Wilson,I.A. and Michnick,S.W. (1999) Erythropoietin receptor activation by a ligand-induced conformation change. Science, 283, 990–993. [DOI] [PubMed] [Google Scholar]
- Robinson R.C., Grey,L.M., Stauton,D., Vankelecom,H., Vernallis,A.B., Moreau,J.F., Stuart,D.I., Heath,J.K. and Jones,E.Y. (1994) The crystal structure and biological function of leukemia inhibitory factor: implications for receptor binding. Cell, 77, 1101–1116. [DOI] [PubMed] [Google Scholar]
- Robledo O., Auguste,P., Coupey,L., Praloran,V., Chevalier,S., Pouplard,A. and Gascan,H. (1996) Binding interactions of leukemia inhibitory factor and ciliary neurotrophic factor with the different subunits of their high affinity receptors. J. Neurochem., 66, 1391–1398. [DOI] [PubMed] [Google Scholar]
- Robledo O., Fourcin,M., Chevalier,S., Guillet,C., Auguste,P., Froger,J., Pouplard-Barthelaix,A., Pennica,D. and Gascan,H. (1997) Signalling of cardiotrophin-1 receptor: evidence for a third receptor component. J. Biol. Chem., 272, 4855–4863. [DOI] [PubMed] [Google Scholar]
- Saggio I., Gloaguen,I., Poiana,G. and Laufer,R. (1995) CNTF variants with increased biological potency and receptor selectivity define a functional site of receptor interaction. EMBO J., 14, 3045–3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A. and Blundell,T.L. (1993) Comparative protein modelling by satisfaction of spacial restraints. J. Mol. Biol., 234, 779–815. [DOI] [PubMed] [Google Scholar]
- Scatchard G. (1949) The attractions of proteins molecules and ions. Ann. N. Y. Acad. Sci., 52, 660–668. [Google Scholar]
- Senaldi G. et al. (1999) Novel neurotrophin-1/B cell stimulating factor-3: a cytokine of the IL-6 family. Proc. Natl Acad. Sci. USA, 96, 11458–11463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sendtner M., Schmalbruch,H., Stockli,K.A., Caroll,P., Kreutzberg,G.W. and Thoenen,H. (1992) Ciliary neurotrophic factor prevents degeneration of motor neurons in mouse mutant progressive motor neuronopathy. Nature, 358, 502–504. [DOI] [PubMed] [Google Scholar]
- Sendtner M., Gotz,R., Holtmann,B. and Thoenen,H. (1997) Endogenous ciliary neurotrophic factor is a lesion factor for axotomized motoneurons in adult mice. J. Neurosci., 17, 6999–7006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Wang,W., Yourey,P.A., Gohari,S., Zukauskas,D., Zhang,J., Ruben,S. and Alderson,R.E. (1999) Computational EST database analysis identifies a novel member of the neuropoietic cytokine family. Biochem. Biophys. Res. Commun., 262, 132–138. [DOI] [PubMed] [Google Scholar]
- Stahl N. et al. (1994) Association and activation of Jak-Tyk kinases by CNTF-LIF-OSM-IL-6 β receptor components. Science, 263, 92–95. [DOI] [PubMed] [Google Scholar]
- Stahl N., Farrugella,T.J., Boulton,T.G., Zhong,Z., Darnell,J.E. and Yancopoulos,G.D. (1995) Choice of STATs and other substrates specified by modular tyrosine-based motifs in cytokine receptors. Science, 267, 1349–1353. [DOI] [PubMed] [Google Scholar]
- Stöckli K.A., Lottspeich,F., Sendtner,M., Masiakowski,P., Caroll,P., Gotz,R., Lindholm,D. and Thoenen,H. (1989) Molecular cloning, expression and regional distribution of rat ciliary neurotrophic factor. Nature, 342, 920–923. [DOI] [PubMed] [Google Scholar]
- Taga T. and Kishimoto,T. (1997) Gp130 and the interleukin-6 family of cytokines. Annu. Rev. Immunol., 15, 797–819. [DOI] [PubMed] [Google Scholar]
- Takahashi R., Yokoii,H., Misawa,H., Hayashi,M., Hu,J. and Deguchi,T. (1994) A null mutation in the human CNTF gene is not causally related to neurological diseases. Nature Genet., 7, 79–84. [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL_W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrice choice. Nucleic Acids Res., 22, 4673–4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward L.D., Hammacher,A., Howlett,G.J., Matthews,J.M., Fabri,L., Moritz,R.L., Nice,E.C., Weinstock,J. and Simpson,R.J. (1996) Influence of interleukin-6 (IL-6) dimerization on formation of the high affinity hexameric IL-6 receptor complex. J. Biol. Chem., 271, 20138–20144. [DOI] [PubMed] [Google Scholar]
- Yamasaki K., Taga,T., Hirato,H., Kawanishi,Y., Seed,B., Taniguchi,T., Hirano,T. and Kishimoto,T. (1988) Cloning and expression of the human interleukin-6 (BSF-2/IFNβ2) receptor. Science, 241, 825–828. [DOI] [PubMed] [Google Scholar]
- Yoon C., Johnston,S.C., Tang,J., Stahl,M., Tobin,J.F. and Somers,W.S. (2000) Charged residues dominate a unique interlocking topography in the heterodimeric cytokine interleukin-12. EMBO J., 19, 3530–3541. [DOI] [PMC free article] [PubMed] [Google Scholar]