Abstract
In Chlamydomonas reinhardtii, the psaA mRNA is assembled by a process involving trans-splicing of separate transcripts, encoded at three separate loci of the chloroplast genome. At least 14 nuclear loci and one chloroplast gene, tscA, are needed for this process. We have cloned Raa3, the first nuclear gene implicated in the splicing of intron 1. The predicted sequence of Raa3 consists of 1783 amino acids and shares a small region of homology with pyridoxamine 5′-phosphate oxidases. Raa3 is present in the soluble fraction of the chloroplast and is part of a large 1700 kDa complex, which also contains tscA RNA and the first psaA exon transcript. These partners, in association with other factors, form a chloroplast RNP particle that is required for the splicing of the first intron of psaA and which may be the counterpart of eukaryotic snRNPs involved in nuclear splicing.
Keywords: Chlamydomonas reinhardtii/chloroplast/group II intron/RNA–protein complex/trans-splicing
Introduction
Group II introns have been found mainly in organellar genes from fungi, algae and higher plants, and also in eubacterial genes. Although many of these group II introns are able to catalyse their excision and exon splicing in vitro, proteins are required in vivo for splicing (Waldherr et al., 1993; Guo et al., 1997; Jenkins et al., 1997; Vogel et al., 1997; Hollander and Kück, 1999). These introns can be folded in a characteristic secondary structure with a central core and six protruding stem–loop domains, which act in cis as catalytic sites for splicing. The splicing mechanism of group II introns involves two consecutive trans-esterification reactions that result in the formation of an intermediate intron lariat–exon and, finally, in the splicing of the two exons (Michel and Ferat, 1995). The same reactions also occur during splicing of nuclear introns, except that they are catalysed by trans-acting snRNPs and splicing factors. Structural similarities exist between some of the snRNPs and the cis-acting catalytic domains of group II introns. It has therefore been proposed that group II introns represent evolutionary precursors of nuclear introns (Cech, 1986; Sharp, 1991; Weiner, 1993).
In the green alga Chlamydomonas reinhardtii, the chloroplast psaA gene, encoding one of the two major polypeptides of the photosystem I (PSI) reaction centre, consists of three exons that are separated on the circular genome (Kück et al., 1987). Each exon is flanked by group II intron sequences. Maturation of psaA mRNA depends on two separate trans-splicing reactions (Choquet et al., 1988). Splicing of the exon 1 and exon 2 transcripts also requires tscA RNA encoded by the chloroplast genome. This RNA is part of the structure of the group II intron, which is, in this case at least, tripartite (Goldschmidt-Clermont et al., 1991).
At least 14 nuclear mutants affecting trans-splicing of psaA were isolated (Girard et al., 1980; Goldschmidt-Clermont et al., 1990). These can be grouped in three classes depending on which intron fails to be spliced. Mutants of class A are defective in the splicing of exons 2 and 3, mutants of class C are defective in the splicing of exons 1 and 2, and mutants of class B are affected in the splicing of all three exons. Furthermore, one class B mutant was reported to be deficient in both tscA RNA maturation and in the splicing of the second intron (Hahn et al., 1998). Recently, the gene affected in one of the class A mutants was cloned and shown to encode a protein related to pseudouridine synthases (Perron et al., 1999). To gain further insights into the first psaA trans-splicing reaction, we have rescued a class C mutant by genomic complementation. The gene, called Raa3, for RNA maturation of psaA, encodes a predicted polypeptide of 176 kDa that does not share significant sequence identity with any other known protein, except for a small domain present in pyridoxamine 5′-phosphate oxidase (PDX). The protein is enriched in a soluble fraction of the chloroplast and is part of a 1700 kDa complex that also includes the chloroplast tscA RNA, the psaA exon 1 transcript and other unknown proteins.
Results
Cloning of the Raa3 gene
M18 is a nuclear mutant lacking PSI activity that has been shown to be deficient in the trans-splicing of the chloroplast psaA precursor RNAs (Girard et al., 1980). The mutant strain is unable to grow photoautotrophically and is highly photosensitive, in particular it is unable to grow on acetate-containing medium at 60 µE/m2s. Based on its psaA RNA phenotype, M18 is a class C mutant affected in the trans-splicing of exons 1 and 2, and lacks detectable levels of PsaA protein.
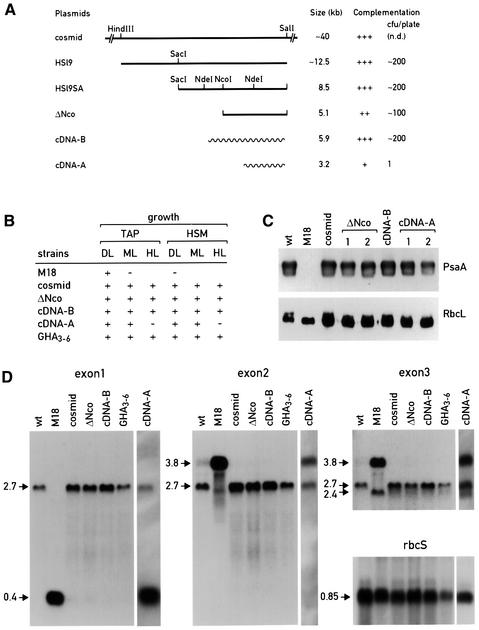
The nuclear gene Raa3 affected in M18 was isolated by genomic rescue, using an indexed cosmid library for transformation (see Materials and methods for details). A single cosmid was isolated that complements the mutant phenotype of M18 with high efficiency (200 colonies per 3 × 107 cells). The transformed cells have wild-type fluorescence transients, grow on minimal medium under all light conditions and accumulate mature psaA mRNA and PsaA protein (Figure 1). The cosmid containing nearly 40 kb of genomic DNA was digested with HindIII, SacI, NcoI or SalI. The fragments produced were still able to rescue M18, indicating that these enzymes cut outside of the Raa3 gene. A subcloned 12.5 kb SalI–HindIII fragment, HSl9, was able to complement M18. This fragment was trimmed further by digestion with SacI or NcoI. The resulting plasmids, HSl9SA and ΔNco, were still able to rescue M18. The latter fragment was the smallest genomic fragment identified able to restore the wild-type phenotype (Figure 1A).
Fig. 1. Genomic complementation of M18. (A) Mapping of Raa3. Cosmid represents the entire cosmid of ∼40 kb isolated from the indexed cosmid library that is able to rescue the M18 mutant. HSl9 is a HindIII–SalI subfragment of the cosmid. HSl9SA is the longer SacI subfragment of HSl9. ΔNco is an NcoI subclone of HSl9SA. cDNAs-B and -A are two independently isolated cDNAs (see Materials and methods). (B) Photoautotrophic growth and photosensitivity of M18 and the rescued strains obtained by transformation of M18 with the constructs described in (A). GHA3-6 is the M18 strain transformed with a HSl9SA fragment containing the entire sequence of Raa3 and an HA tag at the 3′-terminal end of the coding sequence. + and – indicate normal and absent growth, respectively. DL, dim light (6 µE/m2s); ML, medium light (60 µE/m2s); HL, high light (600 µE/m2s); TAP, Tris–acetate–phosphate medium; HSM, high salt minimal medium. (C) Immunoblot analysis. Total cell proteins (10 µg) from the indicated strains were separated by SDS–PAGE, blotted and decorated with PsaA and RbcL antibodies. (D) RNA blot analysis. The blots were hybridized with the three different psaA exon probes and RbcS as a loading control. The sizes of the major transcripts are indicated: the 2.7, 3.8, 2.4 and 0.4 kb bands correspond to the mature psaA mRNA, the exon 2–exon 3 precursor (containing the 3′ end of intron 1), the exon 3 precursor and the exon 1 precursor, respectively.
A cDNA of 3.3 kb was isolated by screening a cDNA library with a subfragment of ΔNco. When cloned into Bluescript SK, this cDNA (cDNA-A) was able to complement the M18 mutant. However, this occurred only at a low frequency (<1 transformant per 3×107 cells) and the transformants were sensitive to high light (600 µE/m2s; Figure 1B). Since the sequence of cDNA-A revealed an open reading frame (ORF) that extended beyond its 5′ end, a second screen was performed with a probe corresponding to the first 300 bp of cDNA-A to obtain a full-length cDNA. A new cDNA of 5.9 kb was isolated (cDNA-B) that fully restores the wild-type phenotype upon transformation of M18. Immunoblot analysis revealed that in all cases the rescued strains accumulate near wild-type levels of PsaA protein, even in the case of the transformants obtained with the incomplete cDNA-A (Figure 1C). Similarly, RNA blot analysis showed that all the transformants obtained accumulate the mature psaA RNA (Figure 1D). However, the transformant obtained with cDNA-A also accumulates the 0.4 kb exon 1 and the 3.8 kb exon 2–exon 3 precursor RNAs, indicating that the first psaA trans-splicing reaction is inefficient.
Southern blot analysis using >10 restriction enzymes revealed no difference between the wild-type and the mutant DNA restriction patterns when the cDNA was used as hybridization probe (data not shown). To test whether the mutant phenotype was linked to the region encoding the cDNA, M18 was crossed with the polymorphic strain S1D2, and the DNA of the progeny was examined. Analysis of four tetrads revealed co-segregation between the mutant or wild-type phenotype and the restriction fragment polymorphism of the parental strains (data not shown). This suggests that the cDNA corresponds to the gene affected in M18 or that the two are closely linked. Finally, in order to establish this correspondence without ambiguity, the Raa3 gene from the M18 allele was amplified by PCR using a proof-reading Pfu polymerase and sequenced. A small rearrangement was observed at position 1152 of the mature protein that causes a frameshift and results in a truncation of the C-terminal end of the protein. These results indicate that the isolated cosmid and cDNA correspond to the gene that is altered by the mutation of M18. This gene was named Raa3.
Raa3 sequence analysis
Both cDNAs and the genomic fragment ΔNco were sequenced. The cDNA-B contains an ORF of 1783 amino acids encoding a polypeptide of 180.5 kDa (Figure 2A). The presence of an upstream stop codon in-frame with the ATG initiation site, and the match between the size of the predicted protein and that observed by immunoblot analysis (see below), indicate that the cDNA encodes the entire protein. The 5′-untranslated region (5′-UTR) of the cDNA is very short and neither a polyadenylation signal nor a poly(A) tract is present in the 3′-UTR, suggesting that parts of the 5′- and 3′-UTRs are missing in this cDNA. A putative transit peptide for import into the chloroplast is present at the N-terminal end, with an abundance of arginine, serine and alanine residues. A potential cleavage site, Val–Ser–Gly, is present at position 41, which would give rise to a predicted mature protein of 1742 amino acids with a molecular mass of 176 kDa. Comparison of the genomic and cDNA sequences of Raa3 reveals the presence of four introns of 135, 120, 178 and 153 bp, which are confined within only 450 bp of the genomic Raa3 sequence (see Figure 2A).
Fig. 2. Raa3 sequence. (A) Deduced amino acid sequence of Raa3. The putative transit peptide is framed and the domain related to pyridoxamine 5′ phosphate oxidases is shaded. The asterisk indicates the site of the mutation of M18. The black arrow and the black inverted triangle correspond to the 5′ ends of the ΔNco fragment and of cDNA-A, respectively. The sites corresponding to the four introns are indicated by open inverted triangles. (B) Amino acid comparison of the homologous domains of pyridoxamine 5′ phosphate oxidases and Raa3. Sequences were aligned using ClustalW (gap open 10; gap ext 0.2; Blosum matrix). * = conserved residues (shaded), : = conservative substitutions, ⋅ = semi-conservative substitution (Thompson et al., 1994). Accession Nos for PDXs are Zymomonas mobilis: AF179611; yeast, P38075; Mycobacterium leprae, O33065; E.coli, P28225; Myxococcus xanthus, P21159; Synechocystis PCC 6803, P74211; O74250.
Both the cDNA-A and the genomic fragment ΔNco lack part of the 5′ end of the gene, thus truncating the N-terminal 801 and 453 amino acids of the mature protein, respectively. Since these two fragments are capable of complementing M18, although only partially in the case of cDNA-A, it appears that the N-terminal portion of Raa3 is dispensable for its function.
Database searches detected significant sequence identity between a small portion of Raa3 and one domain of PDXs (Figure 2B). These enzymes are implicated in vitamin B6 biosynthesis and are known to bind flavin mononucleotide (FMN) as immediate electron acceptor cofactor. The function of the shared domain and its significance for Raa3 function are still unknown. An unusual feature of Raa3 is the presence of several stretches of alanine, serine, proline and arginine residues, which are scattered throughout the sequence.
Raa3 is a chloroplast soluble protein
Immunoblot analysis with an antiserum raised against recombinant Raa3 protein revealed a band close to 200 kDa that is absent from M18 and present in the wild type and in the M18 strain rescued with the cosmid (Figure 3A). Since the genetic evidence indicates that Raa3 is involved in psaA trans-splicing and its sequence reveals a putative transit peptide, the protein is expected to be localized in the chloroplast. To test this hypothesis, intact chloroplasts were prepared from GHA3-6, a strain in which the Raa3 protein has been tagged with a triple haemagglutinin (HA) epitope at its C-terminal end. The tagged protein is fully functional since it rescues M18 at the same rate as the untagged version (data not shown). The protein can be identified readily with an HA antibody, which produces significantly less background than the antibody raised against Raa3. The amount of Raa3 protein was determined in total cell extracts and the chloroplast fraction, which was subdivided further into a soluble and insoluble chloroplast fraction. Figure 3B shows that the tagged Raa3 is absent from wild-type and M18 cells, present in GHA3-6, and that it is enriched in the chloroplast fraction in comparison with the total cell extract. This indicates that Raa3 is indeed a chloroplast protein. The protein is found mostly in the chloroplast soluble fraction, but small amounts can also be detected in the insoluble fraction. The absence of the cytoplasmic protein eIF4A in the chloroplast extract demonstrates that this fraction is not significantly contaminated with cytosolic proteins. The immunoblot with the chloroplast membrane protein PsaA of PSI shows that the soluble chloroplast fraction is not contaminated with membrane proteins. We conclude that Raa3 is present mostly in the chloroplast stromal phase.

Fig. 3. Accumulation and subcellular localization of Raa3. (A) Immuno blot analysis of Raa3 in wild-type and M18 rescued with the genomic (cosmid and ΔNcoI) and cDNA (cDNA-A and -B) clones. The antibody used was directed against recombinant Raa3. Dots indicate Raa3 and its truncated versions. The arrowhead indicates a non-specific band that is also present in M18. (B) Immunoblot analysis of Raa3 in whole-cell extracts from wild-type and M18 and from cell fractions of M18 transformed with the Raa3-HA construct (total cell, intact chloroplasts, soluble and insoluble chloroplast fractions) prepared as described in Materials and methods. To detect Raa3, 100 µg of proteins were loaded on a 6% polyacrylamide gel and decorated with αHA. For PsaA, RbcL and eIF4A detection, 10 µg of proteins were loaded on a 10% polyacrylamide gel and decorated with the corresponding antibodies.
A surprising result is that the genomic clone ΔNcoI and the cDNA-A construct, which lack the 5′ end of the Raa3 gene, are still able to rescue the M18 mutant. Since most chloroplast transit sequences are encoded by the 5′ end of the gene, the truncated Raa3 protein would not be expected to be imported into the chloroplast. However, earlier studies have shown that signal sequences specific for a membrane transport system lack a strict consensus and that they can be replaced by a surprisingly large number of randomly generated peptides (Schatz and Dobberstein, 1996). It is therefore possible that upon integration of the transforming DNA into the nuclear genome, transit-like sequences were fused in-frame with the truncated Raa3 sequence. Such a mechanism would predict that in each transformant, the size of the Raa3 protein is different. Immunoblots with cell extracts from the transformants obtained with ΔNcoI revealed that this was indeed the case (Figure 3A). The amount of Raa3 protein in the cDNA-A transformants was too low to be detected. These results do not exclude the alternative possibility that the chloroplast targeting sequence of Raa3 is not localized within its N-terminal part. The transformant obtained with the full-length cDNA-B produces a protein that is significantly shorter than the wild-type Raa3 protein (Figure 3A). This is most likely due to the fact that the plasmid used for the transformation lacks a nuclear promoter. Thus, the cDNA needs to be fused to a promoter and surrogate transit peptide-coding sequence in order to be expressed upon integration into the nuclear genome. Because the N-terminal region of Raa3 is dispensable, a deletion of the 5′ end of the Raa3 coding sequence can be tolerated in the transformants.
Raa3 is part of a large complex
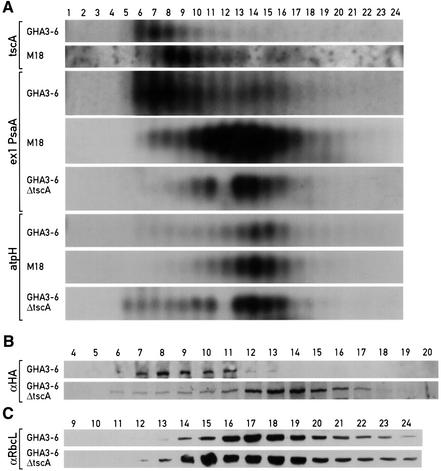
At least seven nuclear loci have been shown previously to be involved in the trans-splicing of the transcripts of exons 1 and 2 of psaA (Goldschmidt-Clermont et al., 1990). The factors encoded by these loci could potentially interact and form a catalytically active complex. To test for the presence of such a complex, soluble extracts from the Raa3-HA-tagged strain were fractionated by size exclusion chromatography and the fractions were analysed by immunoblotting. In extracts containing heparin to preserve RNA from degradation, Raa3 elutes in fractions 6–13, with its maximum in fraction 8 (Figure 4). Molecular weight markers indicate that this fraction corresponds to a size of 1700 kDa. As a protein of 176 kDa, free Raa3 would be expected to elute between fractions 19 and 20. Its larger apparent size indicates that it is part of a complex.
Fig. 4. Raa3 is part of a multimolecular complex. (A) Immunoblot analysis of cell extracts of GHA3-6 fractionated by size exclusion chromatography. A soluble fraction (see Materials and methods) was prepared in the presence of 0.5 mg/ml heparin (+heparin), or incubated on ice without heparin and with 100 µg/ml RNase (+ RNase), and subjected to size exclusion chromatography. Part of each fraction (90%) was electrophoresed on a 6% polyacrylamide gel, blotted and reacted with HA antiserum. Molecular weight size standards are included. (B) The remaining part of the fractions (10%) was electrophoresed on a 10% polyacrylamide gel, blotted and decorated with antibodies directed against RB60 and RbcL. (C) Immunoblot analysis of total cell extracts from wild type (with dilution series) and representatives from the different class C and class B psaA complementation groups probed with Raa3 and RbcL antibodies.
To determine whether RNA is associated with this complex, the same experiment was conducted in the absence of heparin, but in the presence of RNase. Under these conditions, Raa3 fractionates as a free protein, eluting in fractions 17–20 (Figure 4A). This shows that one or more RNA molecules are part of the complex. RB60, another protein known to associate with RNA (Danon and Mayfield, 1994), behaves similarly: its complex shifts to a smaller size upon treatment with RNase, as found previously (Figure 4B; Boudreau et al., 2000). A general effect of RNase on protein complex stability can be ruled out by the fact that this treatment does not alter the elution profile of Rubisco (Figure 4B).
The M18 mutation belongs to one of the seven identified nuclear loci required for the first psaA trans-splicing reaction. It is thus possible that some of the remaining six loci encode factors that are part of the Raa3 complex. Loss of any of these subunits could compromise the stability of the complex and thus of Raa3 itself. To test this possibility, protein extracts from six PSI mutants representing the different complementation groups were tested by immunoblotting. Raa3 was exclusively missing in mutants M18 and L138A, which belong to the same complementation group, but accumulated to wild-type levels in the other mutants (Figure 4C). Thus, there is no evidence for a role for these factors in the stability of the Raa3 complex. However, since the nature of the mutations has not been determined, an association of these factors with the complex remains possible.
tscA RNA and the psaA exon 1 precursor transcript are associated with the Raa3 complex
To identify the RNA associated with the Raa3 complex, an RNA blot analysis was performed on the different fractions obtained after size exclusion chromatography. One possible candidate is tscA RNA, since this small RNA, encoded by the chloroplast genome, has been postulated to represent the middle portion of the discontinuous group II intron 1 of psaA (Goldschmidt-Clermont et al., 1991). The analysis revealed that tscA RNA co-fractionates with Raa3 (Figures 4A and 5A), suggesting that both are part of the same 1700 kDa complex. In M18 extracts, tscA RNA elutes as a complex of 1500 kDa, consistent with the idea that the complex is missing a subunit (Figure 5A). The psaA exon 1 precursor transcript also co-fractionates with the Raa3 complex, but does not co-fractionate with tscA RNA in M18 (Figure 5A), suggesting that the Raa3 protein plays an important role in maintaining the stability of the large RNA–protein complex. To confirm the presence of tscA RNA in the complex, the tscA gene was deleted from GHA3-6 (see Materials and methods). The characterization of this strain shows that both Raa3 and the psaA exon 1 transcript are present in a smaller complex of 900 kDa (Figure 5A and B). The effect of the deletion appears to be specific, as the Rubisco complex preserves its normal size in the absence of tscA (Figure 5C). To test whether the co-fractionation of the RNA with the protein complex during size exclusion chromatography is specific, the elution profile of atpH mRNA, an unrelated chloroplast RNA with a size similar to that of tscA RNA, was examined. This RNA was present in fractions different from those containing tscA RNA, and the elution profile was the same in the wild-type and M18 mutant strains (Figure 5A). Taken together, these data indicate that Raa3, tscA RNA and the psaA exon 1 transcript are part of the same large RNP complex.
Fig. 5. The Raa3 complex includes tscA RNA and the psaA exon 1 transcript. (A) RNA blot analysis of cell extracts from GHA3-6 and M18 fractionated by size exclusion chromatography. Soluble extracts were prepared in the presence of heparin (0.5 mg/ml). The RNA was separated on a 1.25% formamide–agarose gel, blotted and hybridized with labelled probes of tscA, psaA exon1 and atpH. The absence of an RNA signal in fraction 12 of GHA3-6 ΔtscA is due to the accidental loss or degradation of the RNA of this strain during the lengthy preparation procedure. (B and C) Immunoblot analysis of cell extracts of the GHA3-6 strain and its derivative lacking tscA (GHA3-6 ΔtscA). Samples were handled as described in Figure 4. Blots were decorated with antibodies against HA and RbcL.
Discussion
Genetic analysis of psaA trans-splicing in the chloroplast of C.reinhardtii has revealed the existence of several nucleus-encoded splicing factors involved in this process, which fall into three classes. Some are specific for the splicing of exons 1 and 2, others are specific for the splicing of exons 2 and 3, and additional factors are required for both trans-splicing reactions. By genomic complementation of some of these psaA trans-splicing mutants, we have recently identified several of these factors, such as Raa1 and Raa2, which are both involved in the splicing of exons 2 and 3 (Perron et al., 1999; M.Goldschmidt-Clermont, unpublished results). Here we have presented the first characterization of another nucleus-encoded factor, Raa3, specifically involved in the trans-splicing of exons 1 and 2. This factor has been isolated by rescuing a nuclear mutant, M18, deficient in the first psaA trans-splicing reaction.
Unusual features of Raa3
We have localized Raa3 in the stromal phase of the chloroplast using cell fractionation and specific antibodies. Surprisingly, this factor has a different location from that of Raa2, which is associated through ionic interactions with a low density chloroplast membrane fraction that also contains RNA-binding proteins involved in translation (Zerges and Rochaix, 1998; Perron et al., 1999). The predicted precursor polypeptide encoded by the Raa3 gene consists of 1783 amino acids. Raa3 contains several long stretches of alanine, serine, proline and arginine residues and is unrelated to known proteins, with the exception of a small region that is homologous to a domain of PDX, an enzyme implicated in the biosynthesis of vitamin B6, which contains FMN as a cofactor. However, this domain does not correspond to the catalytic site of the enzyme and its role has not yet been elucidated. We reported previously that Raa2 contains two domains found in pseudouridine synthases. However, the activity of this enzyme does not appear to be required for trans-splicing (Perron et al., 1999).
Surprisingly, the N-terminal 453 residues of Raa3 are dispensable for its function, as shown by the fact that the corresponding transformant is able to grow photoautotrophically and accumulates wild-type levels of PsaA. Raa3 is still functional, at least partially, even after the removal of 801 N-terminal residues. Interestingly, in this case, the cells still accumulate near wild-type levels of PsaA, but they are sensitive to high light. One possibility, which will need to be explored further, is that PSI may be mis-assembled and that Raa3 may also have a role in this process. Similar findings have been reported for the chloroplast nucleus-encoded Nac2 factor involved in psbD mRNA stability in C.reinhardtii in which removal of the N-terminal half of the protein does not compromise its function (Boudreau et al., 2000). Since homologous recombination occurs at a very low rate in the nuclear genome of C.reinhardtii (Kindle, 1998), it is likely that upon integration into the genome, the truncated genes are fused accidentally to sequences that may act as surrogate transit sequences, a phenomenon that is well documented in other systems (Schatz and Dobberstein, 1996). The observation that the size of Raa3 varies in each transformant obtained with truncated versions of the Raa3 gene (see Figure 3A) is compatible with this proposal.
Raa3 is part of a high molecular weight complex containing tscA RNA and psaA exon 1 transcript
Raa3 is part of a high molecular weight complex of 1700 kDa. Raa1 and Raa2 are also part of a complex that is clearly distinct from that of Raa3 (K.Perron, M.Goldschmidt-Clermont and J.-D.Rochaix, unpublished results). The first intron of psaA is particularly interesting because it is tripartite, with its middle section, tscA RNA, encoded by a chloroplast locus distinct from those of exons 1 and 2. The tscA RNA is associated with the Raa3 complex based on the following observations: (i) co-fractionation of tscA RNA with the Raa3 complex during size exclusion chromatography (Figure 5); (ii) shift of the Raa3 complex to a smaller size in mutant strains lacking tscA (Figure 5); (iii) shift of the tscA RNA-containing complex to a smaller size in the Raa3 M18 mutant (Figure 5); and (iv) shift to a reduced size when the Raa3 complex is treated with RNase (Figure 4). Similar arguments can be made for the association of the psaA exon 1 transcript with the Raa3 complex.
Several soluble chloroplast proteins from higher plants have been identified that are associated with RNA (Schuster and Gruissem, 1991; Nakamura et al., 1999). Some of these proteins interact with chloroplast mRNAs as well as with intron-containing precursor tRNAs. However, the function of these protein–RNA complexes has not yet been elucidated. The existence of the Raa3 complex raises the question of how it relates to the RNA–protein particles involved in the splicing of nuclear introns, especially since group II introns are considered as the evolutionary precursors of nuclear introns (Cech, 1986). It has been proposed that the tripartite structure of the psaA intron 1 could reflect an intermediate in the evolution of nuclear introns from group II introns (Goldschmidt-Clermont et al., 1991; Sharp, 1991). Acccording to this hypothesis, the catalytic cis-acting group II intron RNA domains would have been gradually replaced by trans-acting RNA–protein particles or snRNPs. In this respect, it will be interesting to characterize the other components of the Raa3 complex fully and to test whether additional parallels can be drawn between this complex and eukaryotic snRNPs.
Materials and methods
Strains and media
Chlamydomonas reinhardtii wild-type and mutant strains were grown as described (Harris, 1989). TAP (Tris–acetate–phosphate) and HSM (high-salt minimal) media were solidified with 2% Bacto agar (Difco) for plate cultures. The mutant M18 has been described previously (Girard et al., 1980; Goldschmidt-Clermont et al., 1990).
Nucleic acid techniques
Procedures for standard molecular techniques were performed as described (Sambrook et al., 1989; Ausubel et al., 1998). The bacterial host was DH5α. Total C.reinhardtii DNA was isolated according to Rochaix et al. (1988). Total C.reinhardtii RNA was prepared with Tri reagent® according to the manufacturer’s instructions (Sigma, Fluka Chemie, Switzerland). For RNA blot experiments, 5 µg of total RNA were electrophoresed through a 1.25% agarose gel containing formaldehyde and MOPS buffer, transferred to Hybond-N+ membrane (Amersham) and probed with double-stranded DNA labelled by random priming with [α-32P]dATP. For psaA exon 1, the probe was the 400 bp HindIII fragment of R21; for exon 2, the 140 bp AvaI–KpnI fragment; and for exon 3, the 750 bp BstXI fragment in the middle of this exon.
To delete tscA, an EcoRV–BglII fragment from R12 was cloned in pUC 18. Two new restriction sites were introduced at each extremity of the tscA gene: MluI at the 3′ end and ClaI at the 5′ end using the one-tube PCR protocol (Picard et al., 1994). The recyclable aadA cassette was inserted into an NsiI site upstream of tscA (Fischer et al., 1996). This construct was reintroduced in the R12 fragment. The gene was deleted using the newly introduced sites.
Genomic complementation by transformation
The genomic indexed cosmid library used was described by Zhang et al. (1994). Cells were transformed with DNA purified from 96-well microtitre plates and selection was on TAP plates under light (60 µE/m2s). After testing 80 microtitre plates, one transformant was obtained. Cosmid DNA was prepared from the 12 columns and the eight rows of the positive plate, and tested for its ability to rescue M18 under photoautotrophic growth conditions. One rescuing cosmid (61F15) of nearly 40 kbp was isolated. This cosmid was digested with HindIII and SalI, yielding nine fragments that were tested for complementation of M18. A fragment (HSl9) of 12.5 kb able to complement M18 was digested with SacI, yielding fragments of 8.5 and 4 kb. The 8.5 kb fragment (HSl9SA) that complemented M18 was cloned in pSK (EcoRV). This plasmid was trimmed further by deleting a HindIII (polylinker)–NcoI subfragment, producing ΔNco. A NcoI–NdeI fragment of ∼2.3 kbp from HSl9SA was used to screen a cDNA library constructed by Dr Hans Sommer (Max-Planck Institute, Köln, Germany). One positive cDNA of 3.2 kb was obtained by screening 400 000 phages. When cloned into pSK, this cDNA (cDNA-A) was able to complement M18 at a low frequency. A second screening was performed using a 300 bp PCR fragment complementary to the 5′ end of ΔNco, and two cDNAs of identical size (5.8 kb) were obtained by screening 600 000 phages. When cloned into pSK, these cDNAs (cDNA-B) were able to complement M18 with a high frequency. cDNAs-A and -B as well as the genomic fragment ΔNco and part of the fragment HSl9SA were sequenced using a DNA sequencer (ABI prism, 377 DNA sequencer; Perkin Elmer). To determine the mutation affecting Raa3 in M18, gene fragments were amplified by PCR from total mutant DNA, gel purified and sequenced. The Raa3 cDNA and genomic sequences have been submitted to the DDBJ/EMBL/GenBank databases under the accession Nos AF310674 and AF310675, respectively.
HA tagging
To clone the triple HA tag, a SpeI site was introduced in the ΔNco fragment, just upstream of the stop codon, using three primers and the one-tube mutagenic PCR method (Picard et al., 1994). The HA tag was amplified using two primers each containing a SpeI site, gel purified, digested with SpeI and cloned in ΔNco. The 5′ end of Raa3 was cloned in ΔNco-HA by introducing an XhoI (polylinker)–NheI fragment from HSl9SA.
Nuclear and chloroplast transformation
The nuclear transformation was performed as described (Kindle, 1998; Stevens et al., 1996) with the following modifications. Cell wall-deficient cells (cw15) were collected by centrifugation and resuspended in TAP at a concentration of 108 cells/ml. Aliquots (0.3 ml) of the cell suspension were mixed with 300 mg of glass beads (0.4 mm; Thomas scientific) and 1.5 µg of transforming DNA, and vortexed at a maximal speed for 15 s. After addition of 0.3 ml of TAP, the contents of the tube were spread on a TAP plate, incubated in the light (60 µE/m2s) and selected for light resistance.
Deletion of tscA was achieved by chloroplast transformation (Finer et al., 1992). Transformants were restreaked four times and the homo plasmicity was verified by Southern blot analysis.
Fluorescence transients
Fluorescence transients were measured as described (Bennoun and Delepelaire, 1982; Fenton and Crofts, 1990). Cells were grown on TAP plates in the dark for 7 days.
Antiserum production and purification
The 1689 bp BamHI–BstYI fragment from ΔNco (coding for amino acids 1104–1667) was cloned into the BamHI site of pET 15b and the plasmid was introduced into Escherichia coli BL21(DE3)pLys S. For expression, transformed cells were grown at 37°C to an A600 of 0.3 in Luria–Bertani medium supplemented with ampicillin (100 mg/ml). Isopropyl-β-d- thiogalactopyranoside (IPTG; 1 mM) was added and incubation was continued for 2 h. Under these conditions, the majority of the protein was found in the inclusion bodies. Cells were collected and lysed by sonication, and the inclusion bodies were purified by four rounds of centrifugation at 12 000 g and sonication to resuspend the pellet. The final pellet was resuspended in 1× loading buffer (25 mM Tris pH 7.0, 10% glycerol, 0.2% β-mercaptoethanol, 0.01% bromophenol blue) and the proteins were loaded on an SDS–polyacrylamide gel. The gel was stained with a solution containing 0.1% Coomassie Blue in 20% ethanol and destained with a 10% ethanol solution. The band containing the protein was cut from the gel. The fragment of gel was squashed and incubated for 16 h at 4°C in a solution containing 15 mM NH4HCO3, 0.025% SDS, 1 mM dithiothreitol (DTT) and 0.1 mM phenylmethylsulfonyl fluoride (PMSF). The solution was centrifuged briefly and the supernatant conserved. This elution was repeated three times and all the supernatants were pooled and lyophilized. The protein concentration was estimated by SDS–PAGE and comparison with standard protein markers.
Polyclonal antiserum was raised in mouse by peritoneal injection of 20 µg of purified protein and Freund’s adjuvant every 20 days, with six injections in total. The antiserum was purified by affinity chromatography on Raa3–Sepharose prepared by binding recombinant protein to cyanogen bromide-activated Sepharose.
Protein extracts
A pellet of 4 × 106 cells was resuspended in lysis buffer containing protease inhibitors (50 mM Tris pH 6.8, 2% SDS, 10 mM EDTA, 5 mM ε-aminocaproic acid, 1 mM benzamidine–HCl, 25 µg/ml pepstatin A, 10 µg/ml leupeptin), incubated at 37°C for 30 min and centrifuged for 5 min in an microfuge. The resulting supernatant was used as total protein extract.
To prepare soluble and insoluble fractions, the pellet was resuspended in lysis buffer without SDS, passed through a French press (small cell, 300 p.s.i.) and centrifuged at 100 000 g for 45 min at 4°C. The resulting supernatant was the soluble fraction. The pellet was washed with 1 ml of lysis buffer without SDS and centrifuged again at 100 000 g for 15 min at 4°C to remove contaminating soluble proteins. The pellet was resuspended in lysis buffer and proteins from the insoluble fraction were extracted as described for the total protein extracts.
Chloroplast isolation was performed as described (Zerges and Rochaix, 1998) with addition of protease inhibitors (5 mM ε-aminocaproic acid, 1 mM benzamidine–HCl, 25 µg/ml pepstatin A, 10 µg/ml leupeptin, Sigma inhibitor cocktail P8849) in all solutions. Briefly, cw15 cells were broken in a Bionebulizer (Gascol Appartus Co., Terre Haute, IN) at 22 p.s.i. Broken cell extracts were fractionated on a continuous Percoll gradient (10–80%). The lower of the two bands corresponding to intact chloroplasts was collected, washed once and lysed osmotically. The soluble and insoluble fractions were prepared as described for total soluble and insoluble proteins. Protein concentration was determined with the bicinchonic acid assay (Sigma).
Immunoblot analysis
Proteins were separated by SDS–PAGE using acrylamide concentrations of 6% (w/v) (for Raa3) or 10% (w/v) (for PsaA, RbcL and eIF4A) and electroblotted to nitrocellulose membranes (Protran, 0.45 µm; Schleicher & Schuell, Inc., Keen, NH). Filters were incubated with specific antibodies, and visualized by the enhanced chemiluminescence method (SuperSignal; Pierce).
Size exclusion chromatography
The soluble protein extracts of the M18 strain complemented with the Raa3-HA protein were prepared as follows. A 100 ml culture at 2 × 106 cells/ml was centrifuged and resuspended in 0.7 ml of breaking buffer [20 mM HEPES pH 7.8, 10 mM MgCl2, 50 mM KCl, 10 mg/ml heparin, protease inhibitors (5 mM ε-aminocaproic acid, 1 mM benzamidine–HCl, 25 µg/ml pepstatin A, 10 µg/ml leupeptin, Sigma inhibitor cocktail P8849)] and lysed through the French press (small cell, 300 p.s.i.). A first centrifugation at 40 000 g for 45 min at 4°C to remove cellular debris and insoluble components was followed by a second centrifugation at 100 000 g for 45 min at 4°C. The clear supernatant was loaded on a Superose 6 PC3.2/30 column (Pharmacia Biotech, Uppsala, Sweden) using the SMART System (Pharmacia Biotech). A 50 µl aliquot (700 µg) of the proteins was injected into the column and elution was performed at 4°C with a buffer containing 20 mM HEPES–KOH pH 7.8, 50 mM KCl and 10 mM MgCl2 at a rate of 40 µl/min. Twenty-four 50 µl fractions, eluted between volumes of 600 and 1800 µl after injection, were collected. Three consecutive fractionations were performed and each pooled 150 µl fraction was precipitated by addition of 8 vols of ethanol. Proteins were pelleted by centrifugation (15 min at 15 000 g), washed with 70% ethanol and resuspended in 10 µl of loading buffer. The fractions were loaded on a 6% SDS–polyacrylamide gel. The molecular weight range was estimated by comparison with size standards included in the HMW Gel Filtration Calibration Kit (Pharmacia Biotech, Sweden).
Total RNA was prepared from pooled 150 µl fractions obtained by three consecutive runs as above. After phenol–chloroform extraction and ethanol precipitation, the pellets were resuspended in 4.5 µl of sterile water, stained with ethidium bromide and electrophoresed through a 1.5% agarose–formaldehyde denaturing gel.
Acknowledgments
Acknowledgements
We thank N.Roggli for preparing the figures. This work was supported by grant 3100-050895.97 from the Swiss National Fund.
References
- Ausubel F.A., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl,K. (1998) Current Protocols in Molecular Biology. John Wiley & Sons, New York, NY.
- Bennoun P. and Delepelaire,P. (1982) Isolation of photosynthesis mutants in Chlamydomonas. In Edelman,M., Hallick,R.B. and Chua,N.H. (eds), Methods of Chloroplast Molecular Biology. Elsevier Biomedical Press, Amsterdam, The Netherlands, pp. 25–38.
- Boudreau E., Nickelsen,J., Lemaire,S.D., Ossenbühl,F. and Rochaix,J.D. (2000) The Nac2 gene of Chlamydomonas encodes a chloroplast TPR-like protein involved in psbD mRNA stability. EMBO J., 19, 3366–3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech R.T. (1986) The generality of self-splicing RNA: relationship to nuclear mRNA splicing. Cell, 44, 207–210. [DOI] [PubMed] [Google Scholar]
- Choquet Y., Goldschmidt-Clermont,M., Girard-Bascou,J., Kuck,U., Bennoun,P. and Rochaix,J.D. (1988) Mutant phenotypes support a trans-splicing mechanism for the expression of the tripartite psaA gene in the C.reinhardtii chloroplast. Cell, 52, 903–913. [DOI] [PubMed] [Google Scholar]
- Danon A. and Mayfield,S.P. (1994) Light-regulated translation of chloroplast messenger RNAs through redox potential. Science, 266, 1717–1719. [DOI] [PubMed] [Google Scholar]
- Fenton J.M. and Crofts,A.R. (1990) Computer aided fluorescence imaging of photosynthetic systems. Photosynth. Res., 26, 59–66. [DOI] [PubMed] [Google Scholar]
- Finer J.J., Vain,P., Jones,M.W. and McMuleen,M.D. (1992) Development of the particle inflow gun for DNA delivery to plant cells. Plant Cell Rep., 11, 323–328. [DOI] [PubMed] [Google Scholar]
- Fischer N., Stampacchia,O., Redding,K. and Rochaix,J.-D. (1996) Selectable marker recycling in the chloroplast. Mol. Gen. Genet., 251, 373–380. [DOI] [PubMed] [Google Scholar]
- Girard J., Chua,N.H., Bennoun,P., Schmidt,G. and Delosme,M. (1980) Studies on mutants deficient in the photosystem I reaction centers in Chlamydomonas reinhardtii. Curr. Genet., 2, 215–221. [DOI] [PubMed] [Google Scholar]
- Goldschmidt-Clermont M., Choquet,Y., Girard-Bascou,J., Michel,F., Schirmer-Rahire,M. and Rochaix,J.D. (1991) A small chloroplast RNA may be required for trans-splicing in Chlamydomonas reinhardtii. Cell, 65, 135–143. [DOI] [PubMed] [Google Scholar]
- Goldschmidt-Clermont M., Girard-Bascou,J., Choquet,Y. and Rochaix,J.D. (1990) Trans-splicing mutants of Chlamydomonas reinhardtii. Mol. Gen. Genet., 223, 417–425. [DOI] [PubMed] [Google Scholar]
- Guo H., Zimmerly,S., Perlman,P.S. and Lambowitz,A.M. (1997) Group II intron endonucleases use both RNA and protein subunits for recognition of specific sequences in double-stranded DNA. EMBO J., 16, 6835–6848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn D., Nickelsen,J., Hackert,A. and Kück,U. (1998) A single nuclear locus is involved in both chloroplast RNA trans-splicing and 3′ end processing. Plant J., 15, 575–581. [Google Scholar]
- Harris E.H. (1989) The Chlamydomonas Sourcebook. A Comprehensive Guide to Biology and Laboratory Use. Academic Press, San Diego, CA. [DOI] [PubMed]
- Hollander V. and Kuck,U. (1999) Group II intron splicing in Escherichia coli: phenotypes of cis-acting mutations resemble splicing defects observed in organelle RNA processing. Nucleic Acids Res., 27, 2339–2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins B.D., Kulhanek,D.J. and Barkan,A. (1997) Nuclear mutations that block group II RNA splicing in maize chloroplasts reveal several intron classes with distinct requirements for splicing factors. Plant Cell, 9, 283–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindle K.L. (1998) Nuclear transformation: technology and applications. In Rochaix,J.-D., Goldschmidt-Clermont,M. and Merchant,S. (eds), The Molecular Biology of Chloroplasts and Mitochondria in Chlamydomonas. Kluwer Academic Publishers, The Netherlands.
- Kück U., Choquet,Y., Schneider,M., Dron,M. and Bennoun,P. (1987) Structural and transcription analysis of the two homologous genes for the P700 chlorophyll a-apoprotein in Chlamydomonas reinhardtii: evidence for in vivo trans-splicing. EMBO J., 6, 2185–2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel F. and Ferat,J.L. (1995) Structure and activities of group II introns. Annu. Rev. Biochem., 64, 435–461. [DOI] [PubMed] [Google Scholar]
- Nakamura T., Ohta,M., Sugiura,M. and Sugita,M. (1999) Chloroplast ribonucleoproteins are associated with both mRNAs and intron-containing precursor tRNAs. FEBS Lett., 460, 437–441. [DOI] [PubMed] [Google Scholar]
- Perron K., Goldschmidt-Clermont,M. and Rochaix,J.D. (1999) A factor related to pseudouridine synthases is required for chloroplast group II intron trans-splicing in Chlamydomonas reinhardtii. EMBO J., 18, 6481–6490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard V., Ersdal-Badju,E., Lu,A. and Bock,S.C. (1994) A rapid and efficient one-tube PCR-based mutagenesis technique using Pfu DNA polymerase. Nucleic Acids Res., 22, 2587–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochaix J.D., Mayfield,S., Goldschmidt-Clermont,M. and Erickson,J. (1988) Molecular Biology of Chlamydomonas. IRL Press, Oxford, UK.
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Schatz G. and Dobberstein,B. (1996) Common principles of protein translocation across membranes. Science, 271, 1519–1526. [DOI] [PubMed] [Google Scholar]
- Sharp P.A. (1991) Five easy pieces. Science, 254, 663. [DOI] [PubMed] [Google Scholar]
- Stevens D.R., Rochaix,J.D. and Purton,S. (1996) The bacterial phleomycin resistance gene ble as a dominant selectable marker in Chlamydomonas. Mol. Gen. Genet., 251, 23–30. [DOI] [PubMed] [Google Scholar]
- Schuster G. and Gruissem,W. (1991) Chloroplast mRNA 3′ end processing requires a nuclear-encoded RNA-binding protein. EMBO J., 10, 1493–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J., Hubschmann,T., Borner,T. and Hess,W.R. (1997) Splicing and intron-internal RNA editing of trnK–matK transcripts in barley plastids: support for MatK as an essential splice factor. J. Mol. Biol., 270, 179–187. [DOI] [PubMed] [Google Scholar]
- Waldherr M., Ragnini,A., Jank,B., Teply,R., Wiesenberger,G. and Schweyen,R.J. (1993) A multitude of supressors of group II intron-splicing defects in yeast. Curr. Genet., 24, 301–306. [DOI] [PubMed] [Google Scholar]
- Weiner A.M. (1993) mRNA splicing and autocatalytic introns: distant cousins or the products of chemical determinism? Cell, 72, 161–164. [DOI] [PubMed] [Google Scholar]
- Zerges W. and Rochaix,J.D. (1998) Low density membranes are associated with RNA-binding proteins and thylakoids in the chloroplast of Chlamydomonas reinhardtii. J. Cell Biol., 140, 101–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Herman,P.L. and Weeks,D.P. (1994) Gene isolation through genomic complementation using an indexed library of Chlamydomonas reinhardtii DNA. Plant Mol. Biol., 24, 663–672. [DOI] [PubMed] [Google Scholar]