Abstract
The VS nucleolytic ribozyme has a core comprising five helices organized by two three-way junctions. The ribozyme can act in trans on a hairpin-loop substrate, with which it interacts via tertiary contacts. We have determined that one of the junctions (2-3-6) undergoes two-stage ion-dependent folding into a stable conformation, and have determined the global structure of the folded junction using long-range distance restraints derived from fluorescence resonance energy transfer. A number of sequence variants in the junction are severely impaired in ribozyme cleavage, and there is good correlation between changes in activity and alteration in the folding of junction 2-3-6. These studies point to a special importance of G and A nucleotides immediately adjacent to helix II, and comparison with a similar junction of known structure indicates that this could adopt a guanine-wedge structure. We propose that the 2-3-6 junction organizes important aspects of the structure of the ribozyme to facilitate productive association with the substrate, and suggest that this results in an interaction between the substrate and the A730 loop to create the active complex.
Keywords: fluorescence resonance energy transfer/metal ions/RNA catalysis/RNA folding
Introduction
RNA-mediated catalysis of chemical reactions (Eckstein and Lilley, 1996) is of great importance in biology, in the processing of RNA molecules and in the translation of proteins. Moreover, RNA catalysis may have been important in the evolution of life on this planet. However, in many cases the mechanistic origin of the observed rate enhancements is imperfectly understood, and this remains a significant challenge in biological chemistry.
The relative simplicity of the small nucleolytic ribozymes provides a valuable set of systems for the dissection of RNA-mediated catalysis. In general, these ribozymes are involved in RNA processing as part of the replication cycle of circular RNA species. From studies of the hammerhead (Scott et al., 1995, 1996; Bassi et al., 1996, 1999), hairpin (Murchie et al., 1998; Walter et al., 1999) and hepatitis delta virus (Ferré-d’Amaré et al., 1998) ribozymes it is clear that ribozyme activity is closely linked to the conformation of the RNA. RNA folding is expected to bring about the correct environment for the cleavage or ligation reaction to proceed, to create the correct juxtaposition of participating groups, and to provide a local conformation of the backbone that facilitates the trajectory into an in-line transition state. The polyelectrolyte character of RNA gives a strong electrostatic component to the folding process, and interactions with divalent metal ions play a key role in the process. Site-bound metal ions may also play a direct role in the ribozyme chemistry, although some reactions can proceed in high concentrations of monovalent ions (Murray et al., 1998).
The VS ribozyme occurs in the 881 nt VS RNA found in the mitochondria of Neurospora, which is transcribed from the Varkud satellite DNA (Kennell et al., 1995). It is one of the nucleolytic ribozymes that undergo a site-specific self-cleavage reaction generating 2′,3′-cyclic phosphate and 5′-hydroxyl termini (Saville and Collins, 1990). It is, therefore, highly probable that the reaction proceeds by attack of the 2′-oxygen atom in an SN2 transesterification reaction in the same manner as the other small nucleolytic ribozymes. The ribozyme can also catalyse the ligation reaction (Saville and Collins, 1991).
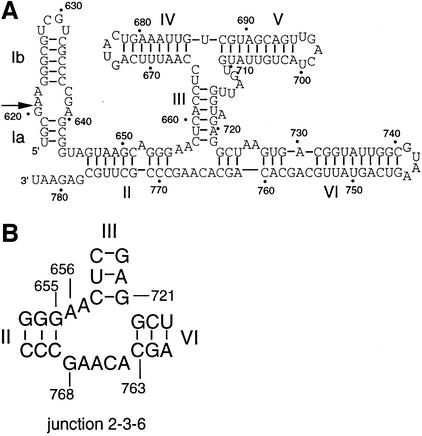
While the basic chemistry of the cleavage reaction is likely to be closely related to that of the other nucleolytic ribozymes, the VS ribozyme differs in a number of respects. First, it is larger—the basic functional element comprises 154 nt (Guo et al., 1993). Secondly, the sequence has little in common with other ribozymes, and the suggested secondary structure (Beattie et al., 1995) is quite different (Figure 1). Thirdly, although the natural form of the ribozyme functions in cis, an efficient cleavage reaction can occur in trans, whereby cleavage occurs in a stem–loop structure that is unlinked to the rest of the catalytic RNA (Guo and Collins, 1995). This suggests that, unlike other nucleolytic ribozymes, substrate recognition is achieved mainly through tertiary interactions.
Fig. 1. The sequence and secondary structure of the VS ribozyme. (A) The secondary structure comprises six helical sections, which can be considered as substrate (helix I, subdivided as Ia and Ib) and ribozyme (helices II–VI) (Beattie et al., 1995). Ribozyme cleavage occurs at the position indicated by the arrow. The conventional numbering of the nucleotides is indicated on the figure, as used throughout the text. (B) Junction 2-3-6 of the ribozyme, showing the numbering of selected nucleotides.
Trans-cleavage activity of the VS ribozyme indicates that it can be considered in two separate parts. Helix I is a stem–loop structure containing an internal loop in which the cleavage event takes place. The predicted secondary structure of the remaining part of the ribozyme shows it to be highly helical (Beattie et al., 1995), with 70% of the bases involved in conventional base pairing. This ribozyme core comprises five helices (II–VI), forming a distinctive H-shaped secondary structure that is quite unlike any other naturally occurring ribozyme. Site-directed mutagenesis experiments suggest that helices II–VI perform roles that are not sequence specific, but presumably determine the correct folding of the RNA (Beattie et al., 1995). This RNA must fold in a way that allows the substrate helix I to bind in a productive manner, via tertiary interactions. Mutations have been found in helix I where the loss of catalytic activity cannot be restored by compensatory substitutions, suggesting that this helix participates in a complex interaction. It has also been suggested that the secondary structure of the substrate is altered upon binding to the ribozyme (Andersen and Collins, 2000). A 3 bp tertiary interaction between the terminal loops of helices I and V has been found by Collins and co-workers (Rastogi et al., 1996).
It is commonly found that helical junctions are important architectural elements in functional RNA molecules, exemplified by the three- and four-way junctions of the hammerhead and hairpin ribozymes, respectively. Examination of the predicted secondary structure of the VS ribozyme core reveals two well defined three-way helical junctions. The lower 2HS5HS2 [using the IUB nomenclature of junctions (Lilley et al., 1996)] junction connects stems II, III and VI, and we refer to this as the 2-3-6 junction. The upper HS1HS5HS2 junction (the 3-4-5 junction) connects stems III, IV and V. The two three-way junctions are connected by helix III, which is a 6 bp helix containing a central bulged adenine nucleotide. We expected that these junctions would be of paramount importance in the global folding and function of the VS ribozyme, and we have therefore embarked on an analysis of the structure of the 2-3-6 junction and its effect on the catalytic activity of the VS ribozyme. We find that the junction undergoes ion-induced folding into a well defined structure, and that mutations around this junction affect both folding and catalysis. Based on a similar junction occurring in 23S rRNA, we provide an explanation for these effects and suggest how an important element in the overall structure of the VS RNA is organized by this junction to create the active ribozyme.
Results and discussion
Sequence and activity of the VS ribozyme
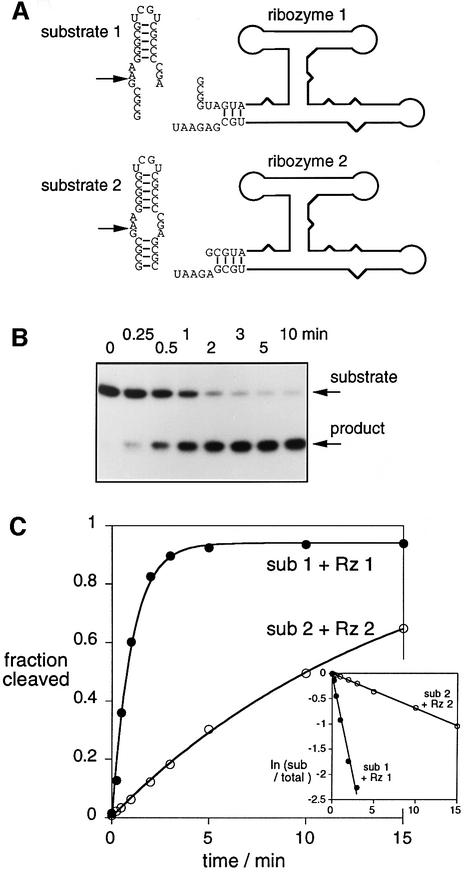
We have analysed the cleavage reaction of the VS ribozyme in trans, whereby the substrate stem–loop I is unlinked to the ribozyme consisting of stems II–VI (Guo and Collins, 1995). We have compared two different trans-acting systems (Figure 2A), in order to assess the importance of the formation of the 3 bp helix between the ribozyme and the end of the substrate (stem Ia). The sequence of ribozyme 1 plus substrate 1 allows basepairing between ribozyme and substrate, forming a 3 bp helix adjacent to the cleavage site. This intermolecular interaction is not possible with ribozyme 2 plus substrate 2.
Fig. 2. Cleavage activity of the VS ribozyme in trans. (A) The RNA has been divided into substrate and ribozyme in two ways, as indicated in this schematic. In substrate 2, helix Ia comprises four intramolecular base pairs, whereas this helix is only generated in substrate 1 by intermolecular base pairing with ribozyme 1. (B) Cleavage of radioactive 5′-32P-labelled substrate 1 by ribozyme 1 as a function of time. The substrate and 5 nt product were separated by gel electrophoresis, and an autoradiograph of the gel is shown. The reaction timepoints are written above the gel. (C) Plotted reaction progress for the cleavage of substrate (sub) 1 by ribozyme (Rz) 1 (closed circles) and substrate 2 by ribozyme 2 (open circles). Single-exponential fits are shown by the lines. The insert shows plots of ln(fraction of uncleaved substrate) versus time for the same data.
Radioactively 5′-32P-labelled substrate RNA was mixed with a large excess of core ribozyme, and incubated at 37°C. Samples were removed after certain time intervals, and the substrate and 5 nt product analysed by polyacrylamide gel electrophoresis. The result for the ribozyme 1 plus substrate 1 combination is shown in Figure 2B; site-specific cleavage is essentially complete after 5 min.
Reaction progress is plotted in Figure 2C for the two combinations of ribozyme and substrate, giving observed rate constants of 1.03 and 0.07 min–1 for systems 1 and 2, respectively. These results were not dependent on the order of mixing ribozyme, substrate and magnesium ions. We also carried out rate measurements in the alternative combinations of ribozyme and substrate (i.e. substrate 2 with ribozyme 1, and vice versa), and obtained essentially the same rates of cleavage for the substrates, irrespective of the ribozyme used. Thus, the faster rate is associated with the substrate having the 3′-deletion, but hybridization with the 5′ end of the ribozyme is evidently not necessary for this. Recently, it has been shown that the removal of stem Ia of the substrate increases the rate of cis-cleavage by a factor of 10 (Rastogi and Collins, 1998), in agreement with our results. This finding is further support for interaction between the VS ribozyme and its substrate occurring mainly through tertiary interactions.
Ion-induced folding of the 2-3-6 three-way junction observed by comparative gel electrophoresis
We expected that the conformation of the 2-3-6 helical junction (Figure 1B) would be very important in the folding and, consequently, the catalytic activity of the VS ribozyme. We have, therefore, carried out an analysis of the folding and conformation of this junction. In all our structural analysis we have removed peripheral helical discontinuities (unpaired or bulged bases) in the arms, in order to examine specifically the trajectories of the arms at the junction.
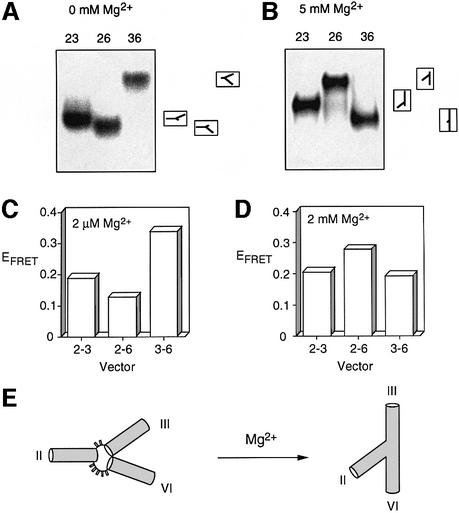
Comparative gel electrophoresis is a powerful yet simple approach for the qualitative analysis of the global structures of helical branchpoints (Lilley, 2000). For a three-way junction this requires a comparison of the electrophoretic mobility of the three species containing two long and one short helical arms. The mobility of these species is strongly affected by the angle subtended between the long arms, and thus the global structure can be deduced from the relative mobilities of these species. We have used a junction comprising a core with the sequence of the 2-3-6 junction, with arms that are extended by effectively random sequences. The central junction region (including the inner 8 bp of each helix) was RNA, while the remainder of the arms was DNA. The names of the species are taken from the identity of the long arms; for example, the species 36 has long arms III and VI, and a shortened arm II. The three long–short arm junctions were electrophoresed in 10% polyacrylamide gels, in 90 mM Tris-borate pH 8.3 in the presence and absence of 5 mM magnesium ions (Figure 3A and B). It is immediately apparent that the global structure of this junction is dependent on the divalent metal ion, since the two patterns of mobility are quite different. Ion-induced folding of VS RNA has also been indicated by chemical probing experiments (Beattie et al., 1995) showing that a large number of bases in this junction become protected from chemical modification in the presence of magnesium ions. The mobility pattern observed in the absence of added metal ions was unchanged by the addition of 25 mM NaCl, indicating that the monovalent ion is unable to induce the folding of this junction (data not shown).
Fig. 3. The global structure of the VS junction 2-3-6 studied by comparative gel electrophoresis and fluorescence resonance energy transfer (FRET). (A and B) Comparative gel electrophoresis. Three-way junctions were generated from strands in which the central 16 nt (plus the unpaired bases) were RNA and the remaining 32 nt at each end were DNA. The three possible species having two long arms of 40 bp and one shorter arm of 10 bp were created, and named according to the two long arms (e.g. species 23 has long II and III arms). The three long–short species were electrophoresed in a polyacrylamide gel in the presence of the indicated metal ion concentrations. The junctions were created from strands that were radioactively 5′-32P-labelled, and the different junction species were revealed by phosphorimaging of dried gels. The experiment was repeated in the presence of 0 (A) and 5 mM (B) magnesium ions. It is clear that the global structure is dependent on the prevailing metal ion concentration. The inter pretations of the mobility patterns are indicated to the right of the phosphorimages, corresponding to the global structures illustrated in (E). (C and D) FRET analysis. Data for species with arms of 12 bp are shown here. They are named according to the arms bearing fluorescein and Cy3, in that order. The data are presented as histograms of the efficiency of FRET (EFRET) for three end-to-end vectors, measured in the presence of 2 µM (C) or 2 mM (D) magnesium ions. (E) Global structure of the VS 2-3-6 junction in the presence and absence of magnesium ions, consistent with a qualitative interpretation of the experimental data.
The pattern of mobilities observed in low salt (Figure 3A) can be interpreted in terms of an extended structure, where the interhelical angles are directly related to the number of unpaired bases connecting them at that vertex. This is commonly found in three-way DNA junctions at low divalent metal ion concentrations (Welch et al., 1993), and it also holds true for the hammerhead ribozyme (Bassi et al., 1995, 1997). In the presence of 5 mM magnesium ions, the three long–short species migrate as narrow bands (Figure 3B). The fastest species is 36, indicating that the largest angle is subtended between helices III and VI; this would be consistent with coaxial alignment of helices III and VI, for example. The slowest species is 26, and thus the narrowest angle lies between helices II and VI. The electrophoretic data are consistent with the ion-induced change of global structure illustrated schematically in Figure 3E.
The global structure of the 2-3-6 three-way junction from FRET-derived distance data
Fluorescence resonance energy transfer (FRET) is an alternative method for the study of global conformation of branched nucleic acids in solution that can provide more quantitative data both on the global structure and the ion-induced conformational transitions (reviewed in Lilley, 2000). We compare the relative magnitude of end-to-end distances by measuring the efficiency of energy transfer (EFRET) between donor–acceptor fluorophores attached at pairs of 5′-termini. We use junctions in which the three arms have the same length, and from which helical discontinuities (unpaired or bulged bases) have been removed. We have used fluorescein and cyanine-3 (Cy3) as a donor–acceptor combination, as we have characterized them in detail (Norman et al., 2000). The fluorescent species are named according to the arms carrying the fluorescent dyes, in the order donor–acceptor. For a given junction we can study six vectors, i.e. the three possible end-to-end species, each of which may be studied in both directions (e.g. vectors 2-6 and 6-2). Efficiency of energy transfer is inversely related to the sixth power of the distance (R) between the fluorophores (Förster, 1948):
EFRET = {1 + (R/Ro)6}–1(1)
Ro is the Förster length for the fluorophores used; we have determined a value of 55.7 Å for the fluorescein-Cy3 combination (Norman et al., 2000). Each of the helices terminates with a 5′-dCpC sequence to provide a constant environment for the fluorophores (Murchie et al., 1989; Clegg et al., 1992, 1993).
Representative data for the 2-3-6 junction with arms of 12 bp are shown in Figure 3C and D. The FRET results confirm the dependence of the global structure on the concentration of the divalent ion. We have used distance information derived from FRET measurements as long-range restraints in the generation of a three-dimensional model of the global shape of the 2-3-6 junction. We are able to analyse branched RNA structures (Grainger et al., 1999) using previously defined fluorophore positions. We have previously shown by FRET and NMR that Cy3 is stacked onto the end of the helix (Norman et al., 2000), and from this we derived an effective position for fluorescein. The rotational position of fluorescein is affected by the length of the helix, because of its lateral extension. Independent distances can, therefore, be obtained by varying the lengths of the helical arms of the junction, and by reversing the fluorophore positions (e.g. vectors 2-3 and 3-2). To analyse the 2-3-6 junction we have used three different arm lengths (11, 12 and 15 bp). A single direction was used for each end-to-end distance for the species with 15 bp arms. On the basis of measurements with control helices we have concluded that FRET efficiencies <0.11 are unreliable. Discarding such values left us with a total of 14 long-range distance measurements for the junction in the presence of 2 mM magnesium ions, in the range 65.5–73.4 Å.
At the outset, nothing was known of the structure at the centre of the junction, but prediction and mutation data strongly suggested that the arms were base paired and helical for 3 bp on each arm beyond the formally unpaired core region. In order to simplify our exploration of the global shape of the 2-3-6 junction, we have used arms that are A-form helices uninterrupted by unpaired bases. The structure of the helical arms and associated fluorophores was conserved during the subsequent modelling process. The central region of the junction was incorporated into our model by means of distance restraints between the junction-proximal ends of the duplex regions that allow unpaired nucleotides to be in conformations that range from totally extended to fully A-form. We have used simulated annealing and rigid body molecular mechanics to elucidate the three-dimensional structure of the junction at high magnesium ion concentration.
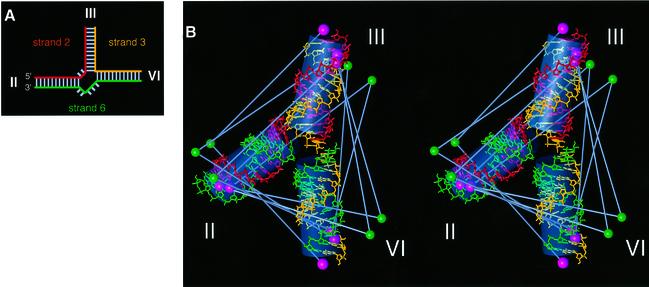
Structures having the lowest violations of experimental restraints, derived from distance-restrained molecular dynamics, fell into a single structural family (Figure 4). Due to the nature of the restraints, these low violation structures formed an extremely closely defined group. The structure obtained is almost planar, with arms III and VI being close to coaxial (158°), and arm II subtending an acute angle (65°) to arm VI and an obtuse angle (136°) to arm III. The root mean square (r.m.s.) violation for all experimental restraints was 2.6 Å, and the largest violation was 4.7 Å without any accounting for experimental error. We also used FRET to measure the inter-fluorophore distance for a simple 22 bp duplex corresponding to coaxially stacked component 11 bp helices, obtaining a value of 70.1 Å. FRET measurements on the analogous 6-3 vector of the junction gave a value of 68.7 Å. This close correspondence is confirmation of near-coaxial alignment between helices III and VI in the folded junction. The global shape of this structure is entirely consistent with the qualitative observations from comparative gel electrophoresis studies (compare Figures 4 and 3E). The structure is also in agreement with a proposal from Collins and co-workers (Rastogi and Collins, 1998), which also incorporates III–VI helical stacking.
Fig. 4. The global structure of the VS junction 2-3-6 folded in the presence of 2 mM magnesium ions, based on distance information derived from FRET measurements. (A) Schematic to illustrate the junctions used in the FRET analysis. The junctions comprise three strands, termed strands 2, 3 and 6 (taken from the arm containing the 5′ end of the strand). The colouring corresponds to that used in the structure shown in (B). (B) Stereographic representation of the low-violation structure for the junction in high magnesium ion concentration. Standard A-form helical sections are shown using a stick representation. The three strands are differentiated by colour; strand 2 is red, strand 3 is yellow and strand 6 is green. The positions of the fluorophores are indicated by single solid spheres (fluorescein, green; Cy3, pink). Grey bars indicate the FRET vectors used in the calculations. Overall helix orientations are indicated by semi-transparent grey cylinders.
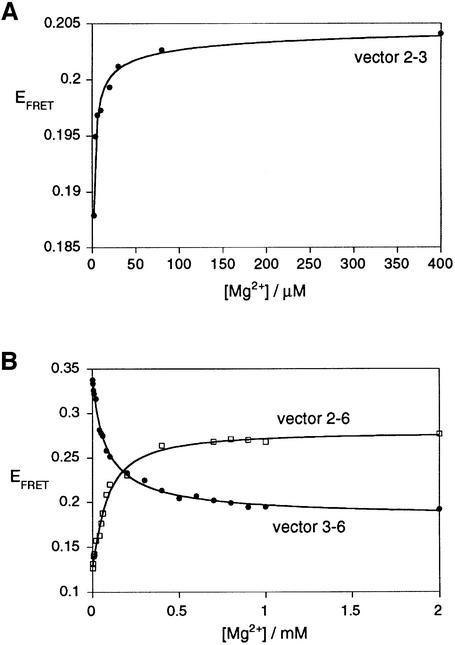
Dependence of the folding of the 2-3-6 three-way junction on magnesium ion concentration
The ion-induced folding of junction 2-3-6 of the VS ribozyme may be followed by studying the change in EFRET for a chosen end-to-end vector upon titration of magnesium ion concentration (Figure 5). The data have been fitted to a simple two-state model that assumes an all-or-none conformational transition induced by the binding of magnesium ions, with a Hill coefficient n and an apparent association constant KA. From these we can calculate the magnesium ion concentration ([Mg2+]1/2) at which the transition is 50% complete. The proportion of folded junction (α) is given by:
Fig. 5. Folding of the VS junction 2-3-6 as a function of magnesium ion concentration. Conformational transitions were studied by the change of EFRET for the three end-to-end vectors: (A) 2-3 (closed circles), (B) 2-6 (open squares) and 3-6 (closed circles). Vector 2-3 undergoes a shortening at a magnesium ion concentration lower than that inducing the shortening and lengthening of the 2-6 and 3-6 vectors, respectively. The experimental data were fitted (lines) in each case by regression to a simple two-state model where the binding of metal ions to the RNA induces a structural change.
α = KA·[Mg2+]n/(1 + KA·[Mg2+]n)(2)
For all the vectors studied, values of n were obtained that were to within experimental error of unity, indicative of non-cooperative binding. Our data suggest that two such transitions occur sequentially. Vector 2-3 exhibits a change in FRET efficiency at a very low magnesium ion concentration, with n = 1.0 ± 0.5 and [Mg2+]1/2 = 8 µM. The corresponding values for the reverse vector 3-2 are n = 1.0 ± 0.2 and [Mg2+]1/2 = 13 µM. Vectors 2-6 and 3-6 undergo a transition at a magnesium ion concentration an order of magnitude higher. The FRET efficiency for vector 2-6 increases with addition of magnesium ions, and fits to values of n = 1.0 ± 0.1 and [Mg2+]1/2 = 93 µM. For the reversed vector 6-2, values of n = 1.0 ± 0.2 and [Mg2+]1/2 = 107 µM were obtained. The FRET efficiencies for vectors 3-6 and 6-3 decrease with increased magnesium ion concentration, with values of n = 0.9 ± 0.1, [Mg2+]1/2 = 78 µM and n = 1.0 ± 0.2, [Mg2+]1/2 = 76 µM, respectively. The similarity of these values suggests that the change in end-to-end lengths of the two sets of vectors (relating helices 2 and 6, and helices 3 and 6) are responding to the same conformational transition, in the 100 µM range of magnesium ion concentration. Equivalent data have been obtained with different arm lengths. For example, using constructs with arms of 11 bp (Figure 6), we obtained values of n = 1.0 ± 0.1, [Mg2+]1/2 = 66 µM for vector 3-6.
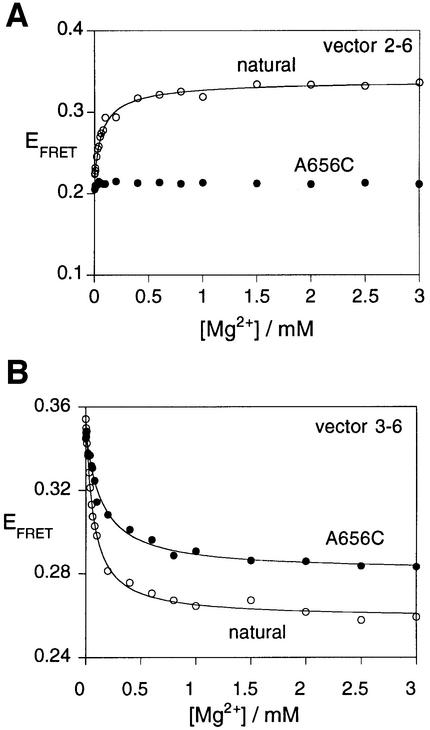
Fig. 6. Perturbed folding of a sequence variant of the VS junction 2-3-6. The folding of variant A656C was studied by FRET efficiency as a function of magnesium ion concentration. For these experiments, species with arm lengths of 11 bp were employed. Plots of EFRET as a function of magnesium ion concentration are shown for the natural and variant junctions. (A) Vector 2-6. Note that the FRET efficiency of the A656C variant (closed circles) is virtually unchanged over the complete range of ion concentration. This is markedly different from the behaviour of natural sequence junction (open circles). The data for the natural sequence junction have been fitted (line) to the two-state binding model as before. (B) Vector 3-6. This vector exhibits an ion-dependent change in FRET efficiency for the A656C variant (closed circles), but the eventual plateau value is higher than that of the natural sequence (open circles). Both sets of data have been fitted (lines) to the two-state binding model.
Thus, the FRET data indicate that the 2-3-6 junction folds in two stages, induced by the non-cooperative binding of magnesium ions in the ranges of 10 and 100 µM. The first transition corresponds to a minor alteration in structure. The second transition is more major, with significant reorientation of the helices. The lengthening of the 3-6 and 6-3 vectors in this transition indicates that the coaxial stacking of helices 3 and 6 occurs at this stage. Sood et al. (1998) observed magnesium ion-dependent protection of phosphate groups against chemical modification by ethyl nitrosourea at positions 656, 657 and 722, consistent with the ion-dependent folding of the 2-3-6 junction in the full ribozyme.
Sequence alterations in the 2-3-6 three-way junction affect VS ribozyme activity
Helical junctions exert a strong influence over the global structure of the RNA molecules in which they occur, and it therefore seemed probable that the conformation of the 2-3-6 junction would be important in the function of the VS ribozyme. While a considerable amount of structure–function data have been accumulated on this ribozyme (Guo and Collins, 1995; Rastogi et al., 1996), no mutations in the 2-3-6 junction have been reported. We have, therefore, introduced sequence changes in and around this junction in the context of the complete ribozyme, and studied the effect on ribozyme activity by measuring the rate of substrate cleavage in trans using the ribozyme 1 plus substrate 1 combination. The results are presented in Table I.
Table I. Cleavage activities of VS ribozyme sequence variants measured in trans using the ribozyme 1 + substrate 1 combination.
| Variant | kobs (min–1) |
|---|---|
| Natural | 1.03 ± 0.06 |
| G655C, C769G | 1.83 |
| C658G, G721C | 1.39 |
| G722C, C753G | 0.84 |
| A764U | 0.90 |
| C765U | 0.56 |
| A766U | 0.056 |
| A767U | 0.049 |
| G768U | 0.016 ± 0.001 |
| A656U | 0.002 ± 0.0009 |
| A656C | 0.003 |
| A657U | 0.065 ± 0.007 |
| ΔA656 | n.a. |
| ΔA767 | 0.0007 |
| ΔA656, ΔA767 | n.a. |
| G768A | 0.59 |
| A656G | 0.063 |
| A656G, G768A | 0.89 |
| A656U, G768A | 0.016 |
| A656C, G768A | n.a. |
| A656G, G768U | 0.006 |
| A656U, G768U | n.a. |
| A656C, G768U | 0.002 |
| Ribosome core | 0.096 |
The observed rate constants (kobs) are tabulated. n.a. denotes no detectable activity. Some measurements (chosen to represent a range of rates) were performed in triplicate. The standard deviations are presented in these cases; in general, errors are ∼10% of values.
Individual reversal of the terminal base pairs in the three helices flanking the junction had very small (<2-fold) effects on cleavage rates. Corresponding sequence changes had also been reported to leave ribozyme cleavage in cis unaffected (Beattie et al., 1995). The sequence of these base pairs is, therefore, not important. As a first step in the examination of the importance of the unpaired bases of the junction, we individually substituted uridine for each base. Three categories of sequence variants were found. Two species (A764U and C765U) were almost unchanged in cleavage activity, while a second class of variants (A767U, A657U and A766U) gave rate reductions of 10- to 20-fold. The largest effects were found at the bases flanking helix II, with rate reductions of 526- and 59-fold for A656U and G768U, respectively. It is, therefore, possible that an interaction between A656 and G768 could be important in determining the active conformation of the ribozyme. We therefore made a further series of single or double sequence changes in these two nucleotides, as indicated in Table I. Most changes led to large reductions in the rate of substrate cleavage, and in four cases no activity was detectable. The only changes that were relatively benign were those that preserved two purine bases at these positions. Reversing the two bases (A656G and G768A) only reduced activity by a factor of 1.2, while the single transition substitutions A656G and G768A led to reductions of 16- and 2-fold, respectively. Thus, the overall shape of the bases at this site could be important. Deletion of A656 leads to complete loss of detectable activity. The possible stereochemical role of these bases is discussed below.
Effect of mutations on the conformation of the 2-3-6 three-way junction; correlation with effects on ribozyme cleavage activity
Our results demonstrate that the 2-3-6 junction of the VS ribozyme has a precise folded structure in the presence of magnesium ions, and that a number of sequence changes in the junction have a large effect on the activity of the complete ribozyme. Since the junction is remote from the sequences that are cleaved in terms of primary sequence, it seems likely that the effect on ribozyme activity is mediated through structural effects. We have therefore examined the impact on the global structure of the 2-3-6 junction of sequence changes that have very different consequences for ribozyme activity.
The rate of cleavage by the ribozyme in which the A and G bases adjacent to stem 2 have been exchanged (A656G and G768A) is essentially unaffected in 10 mM magnesium ions. The folding of this variant junction was analysed by means of the FRET efficiencies for the 2-6 and 3-6 vectors (with 11 bp arms). This junction exhibited magnesium ion-dependent transitions (data not shown) with characteristics that were very similar to those of the natural sequence, with Hill coefficients of 1.0. The apparent affinity for magnesium ions was found to be lower ([Mg2+]1/2 = 780 µM for the 2-6 vector), but the structure formed at 10 mM magnesium ions was closely similar to that of the unmodified junction, consistent with the activity of this variant. Similarly, the sequence variants A656G and G768A (16- and 2-fold reduced activity, respectively) underwent ion-induced transitions that were very similar to the natural junction (data not shown).
By contrast, the junction containing the A656C change exhibits a cleavage rate that is reduced by >300-fold. FRET efficiencies for the 2-6 and 3-6 vectors as a function of magnesium ion concentration for this variant are compared with the corresponding data for the natural sequence in Figure 6. It is clear that the folding of this variant is markedly perturbed. Although the junction undergoes an ion-induced folding transition shown by the reduction in EFRET for the 3-6 vector (n = 1.0 ± 0.1, [Mg2+]1/2 = 143 µM), the 2-6 vector does not undergo the extent of shortening seen in the natural sequence. Moreover, the plateau EFRET value observed for the 3-6 vector (0.28) is higher than that for the natural junction (0.26), and the [Mg2+]1/2 concentration is higher. Thus, the global structure of this variant is very different from that of the natural junction. We have also examined the folding of the variant A656U (reduced in cleavage rate by ∼500-fold). The 2-6 vector of this variant also exhibits almost no change in FRET efficiency with ion concentration, and the data are closely similar to those for the A656C variant. These experiments reveal that sequence changes that affect cleavage activity in the context of the complete ribozyme also affect the conformation of the isolated 2-3-6 junction, and in particular the ion-mediated approach of helices II and VI.
The structural role of A656 and G768
The mutagenesis studies reveal that the A656 and G768 nucleotides appear to play a particularly important role in the function of the ribozyme. Sequence changes at these positions lead to the greatest impairment of cleavage activity. Replacement by purine nucleotides is relatively benign, but any other changes lead to either very low or undetectable levels of ribozyme activity. Furthermore, decreased activity correlates well with the impairment of folding of the isolated junction, providing a direct link between the structure of the 2-3-6 junction and ribozyme function.
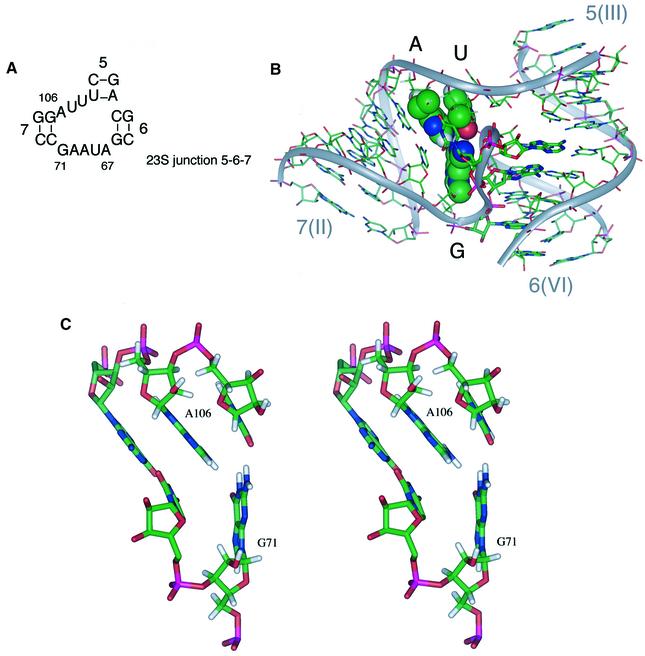
The particular sensitivity to sequence variations at A656 and G768 leads us to ask what their structural role might be, and whether these nucleotides might be interacting with one another. We have noted that a three-way junction of similar sequence (Figure 7A) is found at the 5′-terminal end of the 23S ribosomal RNA (junction 7-5-6, comprising nucleotides 54–116 in Escherichia coli). Replacement of the core sequence of the 2-3-6 junction of the VS ribozyme by the corresponding ribosomal sequence leads to significant retention of ribozyme activity; the cleavage reaction of the hybrid ribozyme proceeds to completion with a rate that is 10% that of the natural sequence (data not shown). The structure of the ribosomal junction has been determined in the context of the 50S ribosomal subunit of Haloarcula marismortui (Ban et al., 2000). Despite some differences in sequence between these two junctions, there are striking similarities in the global geometry, particularly in the coaxial stacking between arms III and VI, and in the acute angle subtended between helices II and VI (∼60° in both cases). The A⋅G nucleotides are conserved in 50 different archaeal, bacterial, mitochondrial and chloroplast 23S rRNA species (Gutell et al., 2000). These adopt an interesting geometry in the structure of the ribosomal subunit (Ban et al., 2000). G71 (corresponding to G768 in VS) is tilted upwards, where it hydrogen bonds simultaneously with A106 (corresponding to A656 in VS) and the adjacent U107 (Figure 7B and C). Molecular modelling indicates that an equivalent interaction could be made if U107 were replaced by adenine, and thus it is probable that a similar structure is formed between G768, A656 and A657 in the VS ribozyme. The result of these interactions is the formation of a wedge with the guanine base at its apex. The tilt angle between G768(71) and A656(106) sets the angle between helix II and the coaxially stacked III/VI helices. Replacement of A656 by a pyrimidine results in a large reduction in activity, together with a loss of the increased FRET efficiency for the II–VI vector on addition of magnesium ions. The impairment of folding can now be explained in terms of a disruption of the G-wedge structure, with reduced buckling at G768. The G-wedge is thus likely to be a major determinant of the global structure of the ribozyme.
Fig. 7. The structure of the ribosomal RNA junction and the pivotal role of the guanine wedge. (A) The sequence of a junction occurring near the 5′ end of 23S rRNA, whose sequence is similar to that to the VS ribozyme. The helices have been sequentially numbered from the 5′-terminus of 23S rRNA, and the nucleotide numbering corresponds to that of Haloarcula marismortui. Positions 71 and 106 correspond to G75 and A111, respectively, in E.coli. The RNA sequences of both E.coli and H.marismortui conform to this consensus. In cases where A106 is mutated, it is always replaced by guanine. (B) The structure of the three-way junction from 23S ribosomal RNA (Ban et al., 2000) consisting of the intersection of helices 7, 5 and 6 (the equivalent helices in the VS ribozyme are II, III and VI). The bases G71, A106 and U107, forming the guanine-wedge feature, are shown in space-filling representation. (C) A stereographic representation of the guanine-wedge residues and the associated first base pair of helix 7. This representation shows clearly the proposed hydrogen-bond interactions between G71-O6 and A106-HN6, G71-HN2 and U107-O2. The angle formed between the bases G71 and A106 appears to be directly related to the angle subtended between helices 7 and 6.
The importance of junction 2-3-6 in the function of the VS ribozyme
We have found that many sequence variants within the single-stranded bases located at the 2-3-6 junction have a large effect on catalytic activity of the ribozyme. A good correlation has been obtained between effects on the conformation of the isolated junction and the rate of cleavage of the full ribozyme. These results suggest that the junction plays an important role in organizing the global folding of the VS ribozyme, such that it can then bind the substrate stem–loop productively, providing the chemical environment in which the backbone cleavage reaction is accelerated.
From a functional perspective, the most significant aspect of the structure of this junction is probably the small angle subtended between helices II and VI, and, therefore, the relatively close proximity of these helices in the folded structure. This may be further influenced by the A725A726 bulge in helix VI, and deletion of both nucleotides leads to a large loss of cleavage activity (D.A.Lafontaine, unpublished data). Proximity of these two helices is consistent with the observation of a UV-induced cross-link between A652 in helix II and A761 in helix VI (D.De Abreu, A. Mittermaier and R.A.Collins, unpublished observations reported in Rastogi and Collins, 1998). In the cis-acting form of the VS ribozyme, the substrate is connected to the main body of the ribozyme via the junction-distal end of helix II. The exact manner of the tethering of the substrate appears to be unimportant, because it can be reconnected through the 3′-end of helix II with full retention of activity (Rastogi and Collins, 1998). It is, therefore, probable that the substrate helix is accommodated in the space between helices II and VI, where it is likely to make interactions with helix VI. Relative spacings suggest that this might juxtapose the substrate loop, where cleavage occurs, with the internal loop of helix VI comprising A730, C755, A756 and G757 (the A730-loop). Deletion of helix VI beyond the A730-loop has relatively small consequences for catalytic activity, but the loop itself is essential (Rastogi and Collins, 1998; D.A.Lafontaine, unpublished data). More over, the functional significance of the A730-loop is suggested by ethylnitrosourea and phosphorothioate interference, manganese-rescue experiments (Sood et al., 1998) and base modification experiments (Beattie and Collins, 1997). Analogous formation of an active site by loop–loop interaction occurs in the hairpin ribozyme (Murchie et al., 1998; Walter et al., 1998), and it is likely that contact between these loops creates the environment in which the cleavage is catalysed. In view of the probable importance of the A730-loop to VS ribozyme action we have commenced high-resolution structural studies of this element.
According to our present view, the 2-3-6 junction fulfils an essential architectural function, ensuring the correct angular disposition between helices II and VI, and thus the geometry required to facilitate the docking of the substrate helix in a productive manner. The postulated G-wedge of the junction plays an important role in determining the global stereochemistry, thus explaining the pronounced sensitivity of ribozyme cleavage to sequence changes at these positions. Evidently the global architecture of the folded RNA must be correct to allow it to function as a ribozyme.
Materials and methods
RNA synthesis and preparation of RNA constructs
DNA, RNA and mixed DNA/RNA oligonucleotides were synthesized using phosphoramidite chemistry (Beaucage and Caruthers, 1981). Synthesis and purification were performed as described in Bassi et al. (1995). Junctions were prepared by incubating stoichiometric amounts of the three appropriate RNA oligonucleotides in 90 mM Tris-borate pH 8.3, 25 mM NaCl for 10 min at 80°C, followed by slow cooling. The hybridized species were loaded onto a polyacrylamide gel and electrophoresed at 4°C for 22 h at 120 V. The buffer system contained 90 mM Tris-borate pH 8.3, 25 mM NaCl, and was recirculated at >1 l/h. Fluorescent junctions were visualized by exposure to a Dark Reader transilluminator (Clare Chemical Research). The bands were excised, the RNA electro-eluted into 8 M ammonium acetate and recovered by ethanol precipitation.
Transcription of RNA
RNA for cleavage activity experiments was synthesized by transcription using T7 RNA polymerase (Milligan et al., 1987) from double-stranded DNA templates. Templates for transcription of ribozymes were made by recursive PCR from synthetic DNA oligonucleotides. RNA was purified by electrophoresis in 8 or 20% polyacrylamide gels containing 7 M urea. RNA was recovered from crushed gel slices by elution in water at 4°C overnight. Eluted RNA was filtered, recovered by ethanol precipitation and dissolved in water.
Analysis of ribozyme cleavage
Ribozyme cleavage reactions were performed using trace concentrations (∼1 nM) of radioactively 5′-32P-labelled substrate and a large excess of ribozyme (1 µM), assumed to be single-turnover conditions. Cleavage buffer contained 50 mM Tris–HCl pH 8.0, 10 mM MgCl2, 25 mM KCl and 2 mM spermidine (Beattie et al., 1995). Substrate and ribozyme were incubated individually in cleavage buffer at 37°C for 15 min and then mixed to initiate the reaction. Two-microlitre aliquots were removed at different times, and quenched by addition of 8 µl of 95% formamide, 20 mM EDTA, 0.05% xylene cyanol FF and 0.05% bromophenol blue. Substrate and product were separated by electrophoresis in a 20% polyacrylamide gel containing 7 M urea, using 8.3 × 10.2 cm plates in a Mini-PROTEAN II Cell system (Bio-Rad). They were quantified by exposure to a storage phosphor screen and imaging (Fuji). Data were fitted to single exponential functions by non-linear regression analysis (Kalaidagraph, Abelbeck Software). Ribozyme and substrate strands were made by transcription. All transcribed RNA species begin with a 5′-GCG sequence in order to minimize the 5′ heterogeneity of the RNA population (Stage-Zimmermann and Uhlenbeck, 1998). The following species were used (all sequences written 5′ to 3′): substrate 1, GCGCGAAGGGCGUCGUCGCCCCGA; ribozyme 1, GCGGUAGUAAGCAGGGAACUCACCUCCAAUUUCAGUACUGAAAUUGUCGUAGCAGUUGACUACUGUUAUGUGAUUGGUAGAGGCUAAGUGACGGUAUUGGCGUAAGUCAGUAUUGCAGCACAGCACAAGCCCGCUUGCGAGAAU; substrate 2, GCGCGAAGGGCGUCGUCGCCCCGAGCGG; ribozyme 2, GCGUAAGCAGGGAACUCACCUCCAAUUUCAGUACUGAAAUUGUCGUAGCAGUUGACUACUGUUAUGUGAUUGGUAGAGGCUAAGUGACGGUAUUGGCGUAAGUCAGUAUUGCAGCACAGCACAAGCCCGCUUGCGAGAAU. Ribozyme strands were also prepared with the sequence changes indicated in the text.
Comparative gel electrophoresis
Comparative gel electrophoresis was carried out as described in Duckett et al. (1995). Nine DNA/RNA oligonucleotides were synthesized, of which the three longest ones had the following sequences (DNA sections are underlined): strand 2, CGCAAGCGACAGGAACCTCGAGGAAATGCGTGCAAGCGGGAACUCACCAATCAGTCTAGACTCGAGGTTCCTGTCGCTTGCG; strand 3, CGCAAGCGACAGGAACCTCGAGTCTAGACTGAUUGGUGAGGCUGUGCGGTGGAAGCTTCTCGAGGTTCCTGTCGCTTGCG; strand 6, CGCAAGCGACAGGAACCTCGAGAAGCTTCCACCGCACAGCACAAGCCCGCUUGCACGCATTTCCTCGAGGTTCCTGTCGCTTGCG. The shortened species are derived by removal of 30 nt from the 5′ or 3′ termini.
Fluorescence spectroscopy
Fluorescence spectroscopy was performed on an SLM-Aminco 8100 fluorimeter, and spectra were corrected for lamp fluctuations and instrumental variations as described in Bassi et al. (1997). Polarization artifacts were avoided by setting excitation and emission polarizers crossed at 54.74°. Values of EFRET were measured using the acceptor normalization method (Murchie et al., 1989; Clegg, 1992) described in Lilley (2000). The FRET experiments were performed using junction species constructed from one strand labelled with fluorescein, one with Cy3 and one unlabelled strand in different combinations. Junctions with three different arm lengths were analysed in these experiments. In addition, strands complementary to strand 3 (strand 3′) were made in order to make a simple duplex corresponding to a continuous III–VI helix. The junctions and duplex species were derived from the following oligonucleotide sequences (deoxyribonucleotides are underlined):
15 bp junction. Strand 2, CCAAGUGCAAGCGGGAACUCACCAAAUCACCGG; strand 3, CCGGUGAUUGGUGAGGCUGUGCGGUAUUGG; strand 6, CCAAUACCGCACAGCACAAGCCCGCUUGCACUUGG; strand 3′, CCAAUACCGCACAGCCUCACCAAUCACCGG.
12 bp junction. Strand 2, CCUGCAAGCGGGAACUCACCAAUCGG; strand 3, CCGAUUGGUGAGGCUGUGCGGUGG; strand 6, CCACCGCACAGCACAAGCCCGCUUGCAGG; strand 3′, CCACCGCACAGCCUCACCAAUCGG.
11 bp junction. Strand 2, CCGCAAGCGGGAACUCACCAAUGG; strand 3, CCAUUGGUGAGGCUGUGCGGGG; strand 6, CCCCGCACAGCACAAGCCCGCUUGCGG; strand 3′, CCCCGCACAGCCUCACCAAUGG; strand 2 (A656G), CCGCAAGCGGGGACUCACCAAUGG; strand 2 (A656C), CCGCAAGCGGGCACUCACCAAUGG; strand 2 (A656U), CCGCAAGCGGGUACUCACCAAUGG; strand 6 (G768A), CCCCGCACAGCACAAACCCGCUUGCGG.
Modelling the global structure of the 2-3-6 junction
The structure of the 2-3-6 junction was determined using molecular dynamics and mechanics within the program X-PLOR (Brünger, 1992). The junction was constructed from three 15 bp A-form RNA helices generated using InsightII (MSI), with an axial rise of 2.81 Å and a twist angle of 32.7°. The fluorophores were represented by pseudo atoms situated at positions relative to the point of attachment determined previously (Norman et al., 2000). The central unpaired nucleotides were represented by distance restraints that allowed for extended or folded conformations. FRET-derived inter-fluorophore distances were incorporated as restraints, with no allowance for experimental error. Starting structures were randomized by application of random displacement vectors to the rigid helix-fluorophore groupings. High-temperature simulated annealing and rigid-body minimization were used to find structures with low violation of experimental restraints. Fluorophores were held in position by either the use of distance restraints or use of a symmetry restraint to a reference structure. High-violation structures were discarded, leaving a highly converged family of low violation structures.
Acknowledgments
Acknowledgements
We thank Rick Collins and co-workers, and our colleagues Tim Wilson and Chris Hammann for discussion; Z.Zhao and Kaera Maxwell for chemical synthesis of RNA; the Cancer Research Campaign for financial support; and EMBO for the award of a fellowship to D.A.L.
References
- Andersen A.A. and Collins,R.A. (2000) Rearrangement of a stable RNA secondary structure during VS ribozyme catalysis. Mol. Cell, 5, 469–478. [DOI] [PubMed] [Google Scholar]
- Ban N., Nissen,P., Hansen,J., Moore,P.B. and Steitz,T.A. (2000) The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science, 289, 905–920. [DOI] [PubMed] [Google Scholar]
- Bassi G., Møllegaard,N.E., Murchie,A.I.H., von Kitzing,E. and Lilley,D.M.J. (1995) Ionic interactions and the global conformations of the hammerhead ribozyme. Nature Struct. Biol., 2, 45–55. [DOI] [PubMed] [Google Scholar]
- Bassi G.S., Murchie,A.I.H. and Lilley,D.M.J. (1996) The ion-induced folding of the hammerhead ribozyme: core sequence changes that perturb folding into the active conformation. RNA, 2, 756–768. [PMC free article] [PubMed] [Google Scholar]
- Bassi G.S., Murchie,A.I.H., Walter,F., Clegg,R.M. and Lilley,D.M.J. (1997) Ion-induced folding of the hammerhead ribozyme: a fluorescence resonance energy transfer study. EMBO J., 16, 7481–7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassi G.S., Møllegaard,N.E., Murchie,A.I.H. and Lilley,D.M.J. (1999) RNA folding and misfolding of the hammerhead ribozyme. Biochemistry, 38, 3345–3354. [DOI] [PubMed] [Google Scholar]
- Beattie T.L. and Collins,R.A. (1997) Identification of functional domains in the self-cleaving Neurospora VS ribozyme using damage selection. J. Mol. Biol., 267, 830–840. [DOI] [PubMed] [Google Scholar]
- Beattie T.L., Olive,J.E. and Collins,R.A. (1995) A secondary-structure model for the self-cleaving region of Neurospora VS RNA. Proc. Natl Acad. Sci. USA, 92, 4686–4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaucage S.L. and Caruthers,M.H. (1981) Deoxynucleoside phosphoramidites—a new class of key intermediates for deoxypolynucleotide synthesis. Tetrahedron Lett., 22, 1859–1862. [Google Scholar]
- Brünger A.T. (1992) X-PLOR (version 3.1) Manual. Yale University Press, New Haven, CT.
- Clegg R.M. (1992) Fluorescence resonance energy transfer and nucleic acids. Methods Enzymol., 211, 353–388. [DOI] [PubMed] [Google Scholar]
- Clegg R.M., Murchie,A.I.H., Zechel,A., Carlberg,C., Diekmann,S. and Lilley,D.M.J. (1992) Fluorescence resonance energy transfer analysis of the structure of the four-way DNA junction. Biochemistry, 31, 4846–4856. [DOI] [PubMed] [Google Scholar]
- Clegg R.M., Murchie,A.I.H., Zechel,A. and Lilley,D.M.J. (1993) Observing the helical geometry of double-stranded DNA in solution by fluorescence resonance energy transfer. Proc. Natl Acad. Sci. USA, 90, 2994–2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duckett D.R., Murchie,A.I.H. and Lilley,D.M.J. (1995) The global folding of four-way helical junctions in RNA, including that in U1 snRNA. Cell, 83, 1027–1036. [DOI] [PubMed] [Google Scholar]
- Eckstein F. and Lilley,D.M.J. (1996) RNA Catalysis. Springer-Verlag, Berlin, Germany.
- Ferré-d’Amaré A.R., Zhou,K. and Doudna,J.A. (1998) Crystal structure of a hepatitis delta virus ribozyme. Nature, 395, 567–574. [DOI] [PubMed] [Google Scholar]
- Förster T. (1948) Zwischenmolekulare Energiewanderung und Fluoreszenz. Ann. Phys., 2, 55–75. [Google Scholar]
- Grainger R.J., Norman,D.G. and Lilley,D.M.J. (1999) Binding of U1A protein to the 3′ untranslated region of its pre-mRNA. J. Mol. Biol., 288, 585–594. [DOI] [PubMed] [Google Scholar]
- Guo H.C.T. and Collins,R.A. (1995) Efficient trans-cleavage of a stem–loop RNA substrate by a ribozyme derived from Neurospora VS RNA. EMBO J., 14, 368–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H.C., De Abreu,D.M., Tillier,E.R., Saville,B.J., Olive,J.E. and Collins,R.A. (1993) Nucleotide sequence requirements for self-cleavage of Neurospora VS RNA. J. Mol. Biol., 232, 351–361. [DOI] [PubMed] [Google Scholar]
- Gutell R.R. et al. (2000) Comparative sequence analysis and the prediction of RNA structure, and the web. http://www.rna.icmb.utexas.edu/
- Kennell J.C., Saville,B.J., Mohr,S., Kuiper,M.T., Sabourin,J.R., Collins,R.A. and Lambowitz,A.M. (1995) The VS catalytic RNA replicates by reverse transcription as a satellite of a retroplasmid. Genes Dev., 9, 294–303. [DOI] [PubMed] [Google Scholar]
- Lilley D.M.J. (2000) Analysis of the global conformation of branched RNA species by a combined electrophoresis and fluorescence approach. Methods Enzymol., 317, 368–393. [DOI] [PubMed] [Google Scholar]
- Lilley D.M.J., Clegg,R.M., Diekmann,S., Seeman,N.C., von Kitzing,E. and Hagerman,P. (1996) Nomenclature Committee of the International Union of Biochemistry: a nomenclature of junctions and branchpoints in nucleic acids. Recommendations 1994. J. Mol. Biol., 255, 554–555. [Google Scholar]
- Milligan J.F., Groebe,D.R., Witherall,G.W. and Uhlenbeck,O.C. (1987) Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res., 15, 8783–8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchie A.I.H., Clegg,R.M., von Kitzing,E., Duckett,D.R., Diekmann,S. and Lilley,D.M.J. (1989) Fluorescence energy transfer shows that the four-way DNA junction is a right-handed cross of antiparallel molecules. Nature, 341, 763–766. [DOI] [PubMed] [Google Scholar]
- Murchie A.I.H., Thomson,J.B., Walter,F. and Lilley,D.M.J. (1998) Folding of the hairpin ribozyme in its natural conformation achieves close physical proximity of the loops. Mol. Cell, 1, 873–881. [DOI] [PubMed] [Google Scholar]
- Murray J.B., Seyhan,A.A., Walter,N.G., Burke,J.M. and Scott,W.G. (1998) The hammerhead, hairpin and VS ribozymes are catalytically proficient in monovalent cations alone. Chem. Biol., 5, 587–595. [DOI] [PubMed] [Google Scholar]
- Norman D.G., Grainger,R.J., Uhrin,D. and Lilley,D.M.J. (2000) The location of Cyanine-3 on double-stranded DNA; importance for fluorescence resonance energy transfer studies. Biochemistry, 39, 6317–6324. [DOI] [PubMed] [Google Scholar]
- Rastogi T. and Collins,R.A. (1998) Smaller, faster ribozymes reveal the catalytic core of Neurospora VS RNA. J. Mol. Biol., 277, 215–224. [DOI] [PubMed] [Google Scholar]
- Rastogi T., Beattie,T.L., Olive,J.E. and Collins,R.A. (1996) A long-range pseudoknot is required for activity of the Neurospora VS ribozyme. EMBO J., 15, 2820–2825. [PMC free article] [PubMed] [Google Scholar]
- Saville B.J. and Collins,R.A. (1990) A site-specific self-cleavage reaction performed by a novel RNA in Neurospora mitochondria. Cell, 61, 685–696. [DOI] [PubMed] [Google Scholar]
- Saville B.J. and Collins,R.A. (1991) RNA-mediated ligation of self-cleavage products of a Neurospora mitochondrial plasmid transcript. Proc. Natl Acad. Sci. USA, 88, 8826–8830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott W.G., Finch,J.T. and Klug,A. (1995) The crystal structure of an all-RNA hammerhead ribozyme: a proposed mechanism for RNA catalytic cleavage. Cell, 81, 991–1002. [DOI] [PubMed] [Google Scholar]
- Scott W.G., Murray,J.B., Arnold,J.R.P., Stoddard,B.L. and Klug,A. (1996) Capturing the structure of a catalytic RNA intermediate: the hammerhead ribozyme. Science, 274, 2065–2069. [DOI] [PubMed] [Google Scholar]
- Sood V.D., Beattie,T.L. and Collins,R.A. (1998) Identification of phosphate groups involved in metal binding and tertiary interactions in the core of the Neurospora VS ribozyme. J. Mol. Biol., 282, 741–750. [DOI] [PubMed] [Google Scholar]
- Stage-Zimmermann T.K. and Uhlenbeck,O.C. (1998) Hammerhead ribozyme kinetics. RNA, 4, 875–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter N.G., Hampel,K.J., Brown,K.M. and Burke,J.M. (1998) Tertiary structure formation in the hairpin ribozyme monitored by fluorescence resonance energy transfer. EMBO J., 17, 2378–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter N.G., Burke,J.M. and Millar,D.P. (1999) Stability of hairpin ribozyme tertiary structure is governed by the interdomain junction. Nature Struct. Biol., 6, 544–549. [DOI] [PubMed] [Google Scholar]
- Welch J.B., Duckett,D.R. and Lilley,D.M.J. (1993) Structures of bulged three-way DNA junctions. Nucleic Acids Res., 21, 4548–4555. [DOI] [PMC free article] [PubMed] [Google Scholar]