Abstract
The relaxin receptor has been recently described as a leucine-rich repeat G-protein coupled receptor and designated as LGR7. A closely related receptor, LGR8, is co-expressed by some cells. This study explored the expression of the genes for these receptors in the human fetal membranes and placenta by RT-PCR and the LGR7 protein by immunolocalization. The results showed that LGR7 was well expressed in the fetal membranes, with significantly more in the decidua (p < 0.05) than in the amnion. On the other hand, relatively low levels were expressed in the placenta. The major splice variant of LGR7 was undetectable in either the placenta or fetal membranes. Expression of LGR8 was also below the level of detectability in either tissue. Immunostaining for LGR7 was conducted with antisera to both its endomain and ectodomain, in order to seek evidence for a solubilized ectodomain. However, similar staining patterns were obtained with both antisera, with predominant staining in the cells of the amniotic epithelium, chorionic cytotrophoblast and decidua. Full-thickness fetal membranes from preterm deliveries, before and after labor or after preterm premature rupture of the membrane (PPROM) and labor were collected. In addition, membranes at term, both before and after spontaneous labor were used for analysis of LGR7 gene expression. There was significantly greater LGR7 expressed (p = 0.01) in the preterm period compared to term, indicating a potentially important role for relaxin at this time. There was a marginal decline in LGR7 gene expression after labor and delivery both at preterm and term, which did not reach significance. Immunostaining patterns showed less inter-patient variability than did gene expression, with more intense staining for LGR7 after labor and delivery.
Keywords: Human relaxin, Relaxin receptor, Fetal membranes, Placenta
INTRODUCTION
The relaxin receptor has recently been identified as a leucine-rich repeat G-protein coupled receptor known as LGR7 [1,2]. This protein contains an N-terminal low density lipoprotein (LDL) receptor cysteine-rich motif, a set of leucine-rich repeats and a seven transmembrane region with an intracellular C-terminus. It specifically binds human relaxins H1 and H2 with high affinity and causes an increase in cAMP. Thus the signaling pathway used by relaxin is different from that of insulin and the IGFs, which involve tyrosine kinase activation [2]. Another similar receptor (LGR8) was also identified and originally thought to be a second receptor for relaxin [1,2]. Subsequently its ligand has been shown to be INSL3, also known as Leydig insulin-like peptide or relaxin-like factor [3].
There are three human relaxin genes designated H1, H2 and H3 [4–6] in the human genome. Relaxin H2 is the circulating hormone, while H1 and H2 are autocrine/paracrine hormones in the decidua/fetal membranes, prostate gland and breast [7]. Relaxin H3 in rodents (relaxin-3) appears to be primarily a neuropeptide, is expressed in the brain [6] and is likely to be the ancestral peptide from which the other relaxins evolved by gene duplications. In these species, the receptor for relaxin-3 is also mainly expressed in the nervous system and is known as GPCR135, its activation decreases cAMP [8]. Currently no information on relaxin H3 is available for humans.
To date, most of the physiological studies on LGR7 have been conducted on the human and primate endometrium or on cells derived from this tissue. In two studies there appeared to be no consistent regulation of LGR7 mRNA expression with different phases of the menstrual cycle [9,10]. However, a third study showed that a significant increase in relaxin binding by autoradiography was coincident with an increase in LGR7 gene expression in the early secretory compared to the proliferative phase of the menstrual cycle [11]. Another study showed that LGR7 expression was ~50-fold higher in term decidual cells compared to endometrial cells and their treatment with relaxin H2 caused an additional increase (> 30-fold) in the expression of its mRNA, demonstrating that relaxin is capable of upregulating its receptor [12]. Most recently, expression of the relaxin receptor LGR7 has been demonstrated in pregranulosa and granulosa cells of primordial, primary and secondary human follicles both by immunocytochemistry and in situ hybridization, confirming earlier work that relaxin has a role during early follicle development [13].
Over a period of years our work has focused on relaxin expression, binding and actions in the human decidua and placenta, with a particular emphasis on the pathological up-regulation of relaxin in patients with preterm premature rupture of the fetal membranes (PPROM) [14]. We have shown that both the relaxin H1 and H2 genes and proteins are upregulated in fetal membranes free of many confounding variables: infection, labor, intrauterine growth retardation and preeclampsia [15]. The aim of the present study was to define the expression of the relaxin receptor in the fetal membranes and placenta, using semi-quantitative real-time PCR and immunolocalization of the protein to specific cell types. For this, antibodies directed to both an intracellular as well as an extracellular domain of LGR7 were used in parallel, to confirm the suggestion for a solubilized ectodomain [2].
MATERIALS AND METHODS
Tissues and explant culture
For the isolation of RNA and semi-quantitation of LGR7 gene expression, fetal membranes and placentas were collected from Kapiolani Medical Center for Women and Children (Honolulu, Hawaii, USA) with informed consent and approval from the Institutional Review Board. To study the distribution of LGR7 gene expression in the fetal membranes and placenta at term, tissues were collected after elective Ce-sarean section (n = 5) and normal spontaneous vaginal delivery (n = 3). The amnion was manually stripped from the choriodecidua and the decidua was carefully scraped from the chorion. To obtain representative samples, three pieces (100 mg each) from different areas of the membrane were pooled and used for RNA isolation. Placental samples were collected from the center of several different cotyledons avoiding the chorionic and basal plates and pooled. The expression of LGR7, its splice variant and LGR8 were studied in the placenta (n = 3) and full-thickness fetal membranes (n = 3) collected after Cesarean section at term before labor. To study the expression of LGR7 at preterm and term, full-thickness fetal membranes were collected at preterm: after elective Cesarean section for medical reasons (n = 4, 34.1–35.6 weeks), after preterm labor and delivery (n = 4, 32.1–34.0 weeks) and after preterm premature rupture of the membranes (PPROM) with labor and delivery (n = 4, 26–35.4 weeks). Samples at term were collected either before labor (n = 3, 38.1–39.3 weeks) at Cesarean section or after normal spontaneous labor and delivery (n = 3, 38.3–39.4 weeks). The tissue samples were either placed into RNAlater (Qiagen, Valencia, CA), and stored at 4°C following the manufacturer’s protocol, or flash frozen in liquid nitrogen and stored at −80°C until use.
Tissues for immunolocalization were full-thickness fetal membrane rolls collected at term after elective Cesarean section (n = 4) or after normal spontaneous labor and delivery (n = 5). Tissues were placed into Bouin’s fixative for up to 24 h and processed into blocks.
Fetal membrane explants of full-thickness tissue (n = 4) were also collected from patients after elective Cesarean section at term within 30 min of delivery. These were placed in sterile phosphate buffered saline (PBS) and dissected into 2 × 2 cm squares taken at random sites from the tissue. Two pieces per treatment were cultured in 60 mm dishes in DMEM:F12 (1:1) containing 10% fetal calf serum (FCS), 100 U/ml penicillin and 100 μg/ml streptomycin in 5% CO2 at 37°C . Explants were treated with human relaxin H2 (10–200 ng/ml) (a generous gift from BAS Medical, San Mateo, CA) for 6 and 24 h. Controls were incubated in medium alone or from unincubated tissue (To) to obtain baseline data. At the end of the incubation periods, a 1 cm diameter circle was cut from the center of each explant to avoid the original traumatized edge and immediately placed into RNAlater and stored according to the manufacturer’s instructions. The two pieces from each treatment were pooled for the isolation of RNA.
For every tissue collected, small pieces were also placed into Bouin’s fixative, processed into blocks, sectioned and stained with hematoxylin and eosin. These were examined by a pathologist and any tissue showing signs of inflammation or infection was discarded.
Isolation of RNA, end-point PCR and semi-quantitative real-time PCR
Total RNA was isolated with the RNeasy Midi kit with on-column DNase digestion (Qiagen). Tissues stored in RNAlater were removed and placed directly into RNeasy homogenization solution. Frozen tissues were pulverized in a BioPulverizer (BioSpec Products Inc., Bartlesville, OK) together with liquid nitrogen to avoid thawing. The resulting powder was mixed well and approximately 250 mg used for RNA extraction. Prior to use, each sample of RNA was assessed for quality on an Agilent Bioanalyzer 2100 (Agilent Technologies, Palo Alto, CA).
Primers for end-point PCR for detection of the splice variant of LGR7 were designed to span the splice site, such that if it were present an additional band of 100 bp would be visible on a gel, in addition to the 200 bp product corresponding to LGR7. These primers (forward: 5′-TCA GCT CCT GCA CTG TAA CG-3′; reverse: 5′-GAC CTT GGC AAA GAC ATT GCA C-3′) were synthesized by Integrated DNA Technologies Inc. (Coralville, IA). Primers for real-time PCR for LGR7, LGR8 and 18S were purchased from Applied Biosystems (ABI, Foster City, CA, Taq Man Assays on Demand).
The cDNAs were prepared using GeneAmp reagents (ABI). For end-point PCR, the reverse transcription reactions consisted of 1.0 μg total RNA, 2.0 μl of 10× PCR buffer II, 4.0 μl of 25 mM MgCl2, 8.0 μl of 10 mM dNTPs, 1.0 μl of 50 μM random hexamers, 1 μl of 20 U/μl RNase inhibitor, 1.0 μl of 50 U/μl Multiscribe reverse transcriptase, and DEPC treated water to bring the total volume to 20 μl. The reactions were incubated at 23°C for 10 min, 42°C for 15 min and inactivated at 99°C for 5 min. For real-time PCR, the reverse transcription reactions consisted of 1.0 μg total RNA, 5.0 μl of 10× PCR buffer II, 11.0 μl of 25 mM MgCl2, 10 μl of 10 mM dNTPs, 2.5 μl of 50 μM random hexamers, 1.0 μl of RNase inhibitor (20 U/μl), 1.25 μl of Multiscribe reverse transcriptase (50 U/μl), and DEPC treated water to bring the total volume to 50 μl. This was incubated at 23°C for 10 min, 48°C for 30 min and inactivated at 95°C for 5 min.
For end-point PCR, each reaction consisted of 8.0 μl of 10× PCR buffer II, 4.0 μl of 25 mM MgCl2, 0.5 μl of 5 U/μl AmpliTaq Gold DNA polymerase, 64.5 μl of deionized water and 20 μl of cDNA. These reactions with primers from Integrated DNA Technologies Inc., included 1.5 μl of each primer (10 μM) and the reactions with primers from ABI included 3.0 μl of assay mix. All reactions were incubated for one cycle at 95°C for 105 s, 35 cycles at 95°C for 15 s and 60°C for 30 s and held at 72°C for 7 min. The PCR products were run on a 1.8% agarose gel in 1× TAE buffer (40 mM Tris-acetate, 2 mM EDTA, pH 8.5) at 100 V for 50 min. For real-time PCR the ABI protocol was used: one cycle for 10 min at 95°C and 40 cycles of 15 s at 95°C and 1 min at 60°C. Each 96-well plate included a water blank and a reverse transcriptase blank. cDNA dilutions were used to generate standard curves. Real-time PCR was carried out on an MJ Research Opticon 2 Continuous Fluorescence Detector. Each reaction was performed in triplicate and the results were normalized to the expression of 18S in each sample of cDNA but in separate tubes. Real-time PCR failed to detect LGR8 expression in any of the samples of placentas or fetal membranes. Therefore, the real-time PCR products were run on a gel (as described above) together with a positive control cDNA derived from adult human uterus RNA (Stratagene, La Jolla, CA) shown previously to express LGR8 [2]. The data were analyzed as directed in the ABI Users’ Bulletin #2 and reported as relative gene expression. Data are presented as the n-fold difference relative to the group with the lowest expressed value ± SEM.
Statistical analysis was performed with ANOVA and Dunn’s Multiple Comparisons test to compare three or more data sets and with the Mann–Whitney U test to compare two sets of data. The p values < 0.05 were considered statistically significant.
Antisera and immunocytochemistry
Two rabbit antisera were used for the immunolocalization of LGR7 in the fetal membrane. One antibody was directed to a 23 amino-acid sequence (735–757) located in the intracellular domain (PDLFTYPCEMSLISQSTRLNSYS) (Phoenix Pharmaceuticals Inc., Belmont, CA). This antibody was titrated and used at 1:800 dilution and as a control, a non-immune rabbit serum was used at the same concentration. The other antiserum was made from a 13 amino-acid peptide in the extracellular LDL domain (LPQLLHCNGVDDC), designed to contain little secondary structure (Sigma-Genosys, The Woodlands, TX). The peptide was synthesized and conjugated to keyhole limpet hemocyanin to increase the antigenic response. The immunoglobulin G (IgG) was isolated and used for immunostaining at 6 μg/ml. A non-immune IgG was used as a negative control. In the event that the ectodomain was shed, immunostaining would have only detected LGR7 with the antiserum to the endodomain. Pieces of fixed tissues were embedded in paraffin blocks using conventional methods, sectioned at 7 μm and mounted onto Vectabond-treated slides (Vector Laboratories, Burlingame, CA). Tissue sections were deparaffinized and hydrated in a graded series of xylenes and ethanols and used for immunostaining with the avidin–biotin immunoperoxidase kit (Vector Labs) according to the manufacturer’s protocol. Briefly, the slides were treated with 0.3% hydrogen peroxide in methanol for 30 min to block endogenous peroxidase activity, washed in 0.015 M PBS, then incubated in 1.5% blocking serum for 20 min to saturate non-specific binding sites. Sections were incubated at room temperature with the primary antiserum for 30 min, followed by a biotinylated secondary antibody and finally incubated for 30 min with the avidin–biotin complex (ABC) reagent (Vector Laboratories). Peroxidase activity was demonstrated cytochemically by incubation with diaminobenzidine (0.5 mg/ml) and 0.03% hydrogen peroxide at room temperature for 5 min. The sections were counter-stained with hematoxylin, mounted in ProTexx (Baxter Scientific, Honolulu, HI) and viewed under brightfield microscopy.
RESULTS
The expression of LGR7 in the separated amnion, chorion, decidua and placenta, prior to labor and after spontaneous labor with vaginal delivery at term are shown in Figure 1A and B, respectively. Before labor, there was significantly greater (p < 0.05) expression of LGR7 in the decidua compared to the amnion or placenta (Figure 1A). After labor and delivery there was significantly more LGR7 gene (p < 0.05) expressed in the decidua compared to the amnion (Figure 1B). The placenta expressed relatively low levels of LGR7, both before and after labor at term (Figure 1A and B). There was marked inter-patient variability in the expression of this gene, especially in the chorion and decidua, regions of the membrane with the highest levels of expression. The non-labor and labor samples (Figure 1A and B) were not run in the same real-time PCR assay, therefore the absolute values could not be directly compared.
Figure 1.
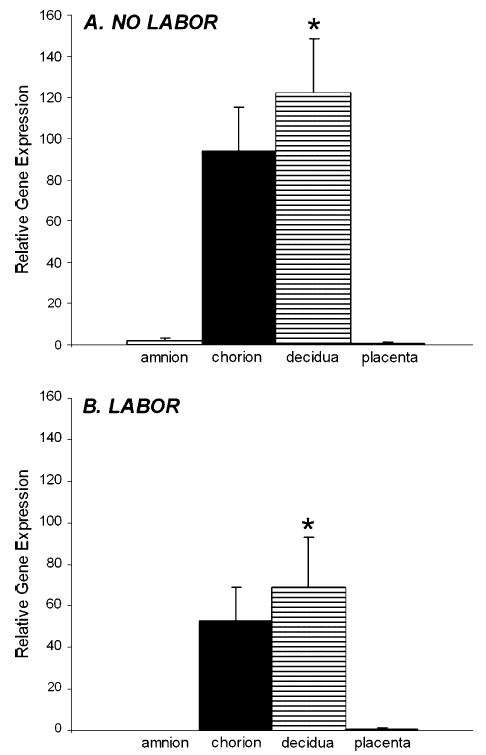
LGR7 gene expression in the amnion, chorion, decidua and placenta collected at term (A) before labor after elective Cesarean section (n = 5) and (B) after spontaneous labor and delivery (n = 3). None of these tissues had any histological evidence of inflammation or infection. Real-time PCR data were normalized to the expression of 18S in each sample and are shown as relative gene expression ± SEM compared to the expression in the placentas of each group. (A) *Significantly (p < 0.05) more LGR7 expressed in the decidua before labor compared to the amnion or placenta and (B) *significantly (p < 0.05) more in the decidua after labor compared to the amnion. There were relatively low levels of LGR7 expressed in the placenta both before and after labor.
Using real-time PCR and primers for LGR7, a single band of 100 bp was amplified in cDNAs from three different placentas (Figure 2A, lanes 1–3) and three different fetal membranes (Figure 2A, lanes 4–6). The molecular size markers are shown in lane 7. Primers were designed to detect both the LGR7 and its major splice variant by end-point PCR. A single 200 bp product corresponding to LGR7 was visualized in the RNAs from the same placental samples (Figure 2A, lanes 8–10) and fetal membranes (Figure 2A, lanes 11–13). There was no additional band visible at 100 bp, showing that the LGR7 splice variant was undetectable in these tissues. When real-time PCR primers specific for LGR8 were used with the same three placental and membrane RNAs, no PCR product was visualized (Figure 2B, lanes 1–3, placenta and lanes 4–6, membranes) although the RNA preparation from human uterus was positive for LGR8 (Figure 2B, lane 7). This shows that LGR8 is not transcribed in either the placenta or fetal membranes.
Figure 2.
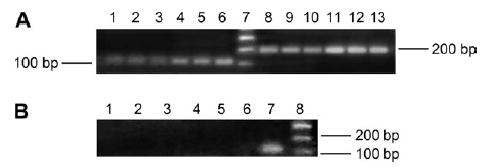
PCR products are shown on a gel when amplified with different primer sets for LGR7 (A) and LGR8 (B). Samples of RNA from three placentas and fetal membranes (different patients) were amplified with primers designed for LGR7 real-time PCR (A, lanes 1–3, placentas; lanes 4–6, membranes), a single band of 100 bp was detected in both tissues. Primers designed to detect both LGR7 and its splice variant (A, lanes 8–10, placenta; lanes 11–13 membranes) show that only LGR7 was detected at 200 bp. Lane 7 shows molecular size markers. The same RNA samples were also amplified with primers specific for LGR8 (B, lanes 1–3, placentas; lanes 4–6, membranes), its transcript was undetectable in RNAs from either tissue, while RNA from human uterus (lane 7), shows a single band at 100 bp corresponding to LGR8. Molecular size markers are shown in lane 8.
The immunostaining patterns with antisera directed to the endodomain and ectodomain of LGR7 were very similar for all tissues and reflected the pattern of gene expression shown in Figure 1. This is shown in a representative tissue collected after spontaneous term labor and delivery, which was immunostained with both antisera to LGR7 (Figure 3). The amniotic epithelial cells stained well with both antisera, directed to the endodomain (Figure 3A) and ectodomain (Figure 3B). However, the fibroblasts embedded within the extracellular matrix of the amnion and chorion sometimes stained more prominently with the antiserum to the ectodomain compared to the antiserum to the endodomain (Figure 3D and C, respectively). This was noticeable in some tissues, as in the one shown here. The strong staining in the chorionic cytotrophoblast can be readily seen in Figure 3E and F as well as in the larger decidual cells (Figure 3G and H).
Figure 3.
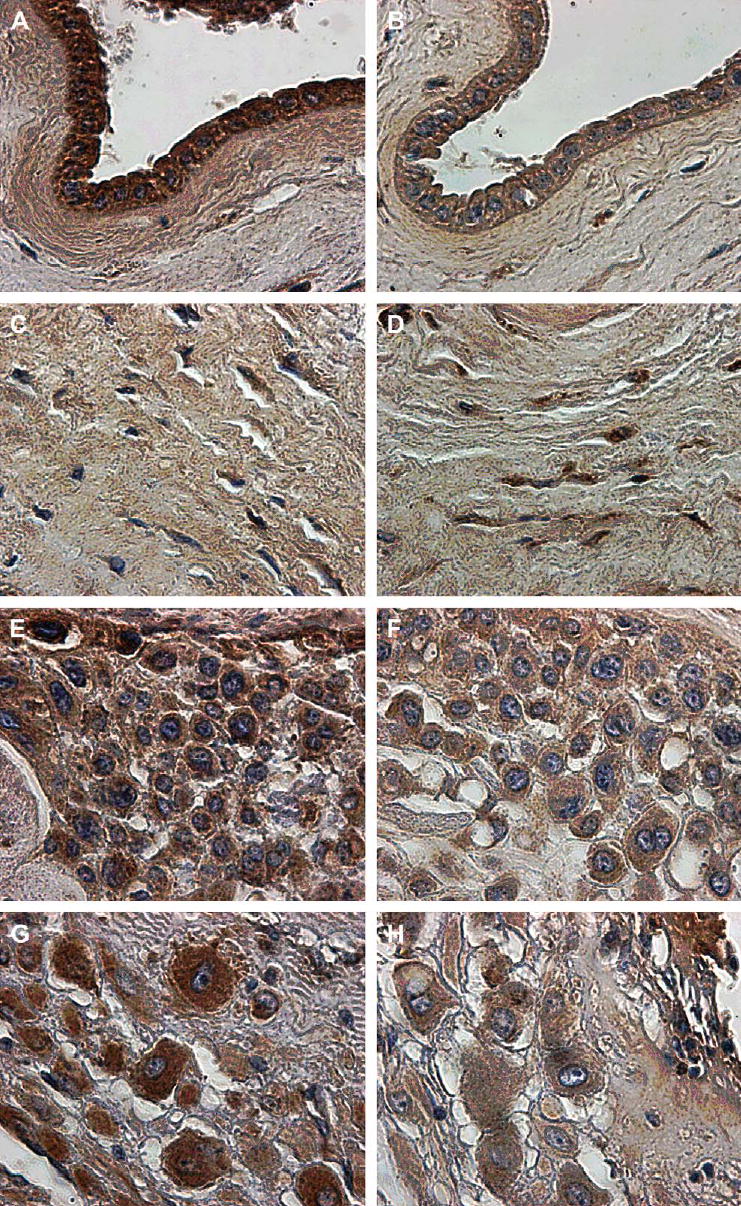
Immunostaining for LGR7 in a representative fetal membrane of a patient after spontaneous labor and delivery at term. Consecutive sections were stained with an antiserum to the endodomain (A, C, E, G) and the ectodomain (B, D, F, H) of LGR7. The amniotic epithelium (A, B) and mesenchyme of the amniotic connective tissue (C, D) showing more staining in the mesenchyme with the antiserum to the ectodomain (D) compared to the endodomain (C). The cells of the chorionic cytotrophoblast show strong cytoplasmic staining (E and F), as do those of the parietal decidua (G and H). Magnification: ×600.
The expression of the LGR7 gene was semi-quantitated in full-thickness fetal membranes collected at preterm and term. At both gestational ages, expression after labor and delivery showed a decline, although this did not reach significance at either time, labor and delivery reduced its expression by one half at preterm and caused a 5-fold reduction at term. The level of expression of the LGR7 gene in the tissues from patients with PPROM and labor was similar to that of the tissues collected at preterm prior to labor (Figure 4). These showed no decline in expression of LGR7, although all patients had experienced spontaneous labor. An especially large patient variability in LGR7 expression was evident in the pre-term tissues (Figure 4). Power analysis of these data showed that with this degree of patient variability it would be necessary to collect between 30 and 100 tissues in each of the pre-term subgroups in order to reach a level of significance. Therefore the data from all preterm tissues (n = 12) were compared to those from all the term tissues (n = 6), regardless of the mode of delivery. This showed significantly more (p = 0.01) LGR7 expressed at preterm than at term and suggested that relaxin may have an important role in the preterm period.
Figure 4.
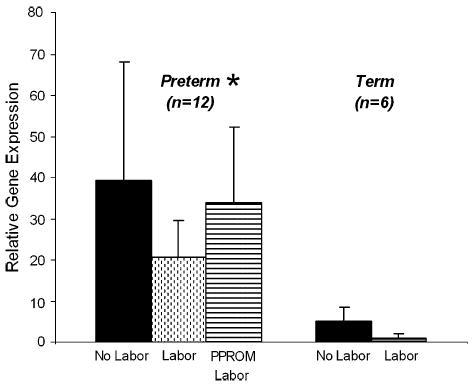
LGR7 gene expression by real-time PCR in full-thickness fetal membranes collected at preterm Cesarean section and no labor, after labor and delivery and from patients with PPROM (n = 4 of each) and at term, before and after spontaneous labor (n = 3 of each). Data are presented as the n-fold difference relative to term labor, LGR7 expression ± SEM. A 5-fold decline in expression was seen after labor at term and expression was halved after preterm labor. These were not significant due to the large inter-patient variability. However, tissues from PPROM patients remained elevated although they had experienced labor and delivery. * Disregarding the mode of delivery, there was significantly more LGR7 expressed (p = 0.01) at preterm than at term.
The intensity of LGR7 staining in tissues collected at term both before and after labor showed a different trend from the expression of the LGR7 gene. While gene expression was reduced by labor, staining for the protein was increased after labor. Although this was not quantitative, there was less variability in staining intensity than in gene expression with tissues from different patients. A tissue collected prior to labor is shown stained with the antisera to the LGR7 endodomain (Figure 5A) and ectodomain (Figure 5B). This shows lighter staining with both antisera compared to a tissue collected after spontaneous labor and delivery, shown immunostained with the antiserum to the endodomain (Figure 5C) and ectodomain (Figure 5D). Respective controls for the two antisera are shown on prelabor tissues in Figure 5E and F.
Figure 5.
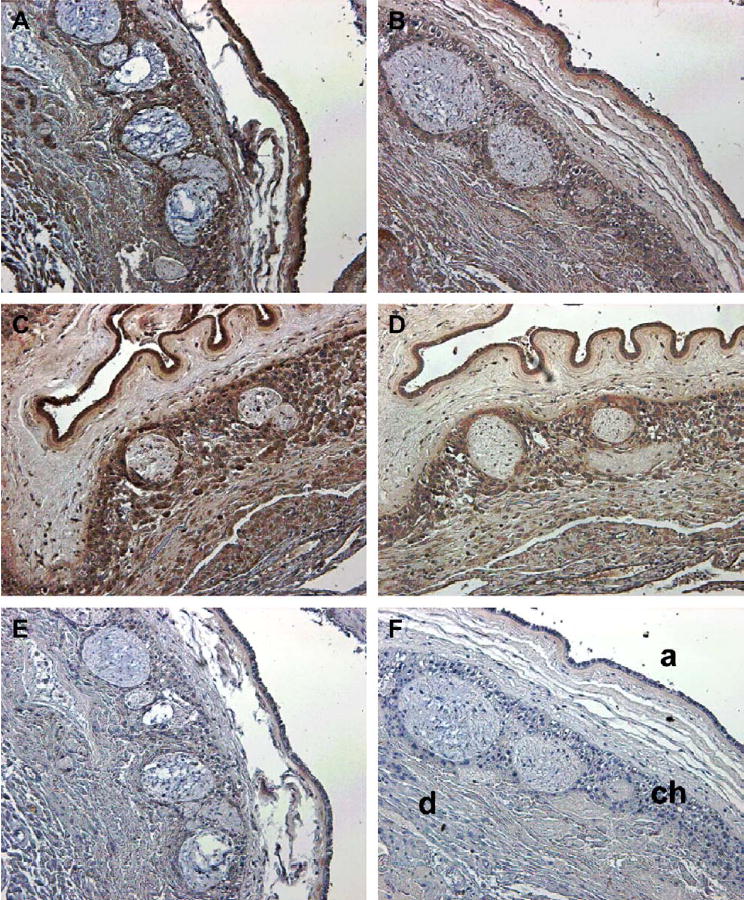
Immunostaining for LGR7 in a representative fetal membrane from a patient before labor at term, stained with an antiserum to the (A) endodomain (1:800) and (B) ectodomain (IgG: 6 μg/ml) of LGR7. In a patient after normal spontaneous labor and delivery at term stained with an antiserum to the (C) endodomain and (D) ectodomain. Controls are membranes from the patient before labor stained with (E) non-immune serum and (F) non-immune IgG, at the same concentrations as the primary antiserum/IgG. Staining patterns were similar with both antisera, but show more intense staining after (C and D) compared to before labor and delivery (A and B). Identification of the major cell layers is shown in (F); (a) = amniotic epithelium, (ch) = chorionic cytotrophoblast and (d) = decidua. Magnification: ×200.
Fetal membrane explants treated with human relaxin for 6 h showed a dose-related increase in LGR7 gene expression (Figure 6), which was not present in explants similarly treated for 24 h (data not shown). While this did not reach significance, this trend was similar to previously published data with isolated term decidual cells [12].
Figure 6.
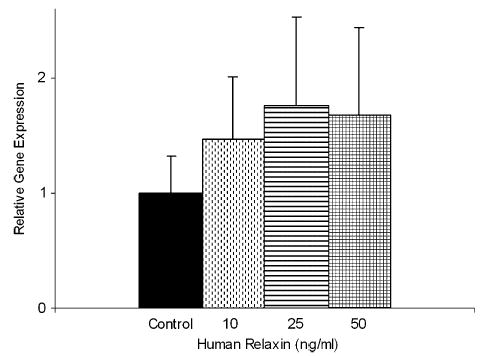
LGR7 gene expression by real-time PCR in full-thickness fetal membrane explants (n = 4 patients) treated with human relaxin for 6 h. Data presented relative to expression in the control ± SEM. A dose–response increase in the expression of LGR7 up to 25 ng/ml relaxin was evident but failed to reach significance.
DISCUSSION
This study shows that the relaxin receptor LGR7, both gene and protein, is well expressed in both the maternal and fetal cells of the fetal membrane. On the other hand, it is expressed at relatively low levels in the placenta. The splice variant of LGR7 [2] was undetectable in RNAs from both the placenta and fetal membranes. The lack of LGR8 expression in either tissue confirms that the major receptor for relaxin is LGR7 and that the actions of relaxin in the fetal membranes are mediated through this receptor [16–19]. This also suggests that INSL3, the ligand for LGR8, probably has little or no biological action in these tissues.
Previous studies from our laboratory have shown relatively high levels of relaxin gene expression in the syncytiotrophoblast of the placenta [20]. Here we found low abundance mRNA for its receptor LGR7, although by immunolocalization there was some very light positive staining with both LGR7 antisera (data not shown). The highest level of LGR7 mRNA was in the decidua and chorionic cytotrophoblast of the fetal membranes, agreeing well with our binding studies with labeled relaxins H1 and H2 [16]. The principal sites of relaxin action in the uterus are therefore in the decidua and chorion of the fetal membranes. For immunolocalization, antisera to both intracellular and extracellular domains of LGR7 were used in an attempt to identify cells which might be sequestering and therefore stain for a putative soluble extracellular domain. Alternatively, some cells might stain only with the antiserum to the endodomain, if the ectodomain were shed. Either situation could provide further evidence for a soluble extracellular domain of this receptor. However, the staining patterns with both antisera were very similar in all tissues and clearly we were unable to detect any differences in its cellular location with these two antibodies.
A high degree of variability in the expression of the LGR7 mRNA in different patients was evident; this was also noted in a study on its expression in the endometrium during the menstrual cycle [10]. This variability mandates the use of large numbers of tissues for each experiment in order to reach statistical significance. Unfortunately, this is often impractical for studies using human tissues. We eliminated all the tissues exhibiting any inflammation or infection from this study, as relaxin expression is reduced in the presence of severe chorioamnionitis [17] and we did not know any potential effects that this might have on the expression of LGR7. In order to do this, at least double the number of tissues had to be collected, as approximately half showed some degree of an inflammatory response or mild to severe chorioamnionitis.
From microarray studies of gene expression in similar tissues, collected before and after labor, published by several different laboratories, it appears that labor in general is associated more with a reduction rather than an increase in the expression of genes. This was also true for LGR7, as there was a 5-fold decrease in its expression at term after labor and a less pronounced decrease after preterm labor. On the other hand, there was more intense immunostaining after labor, suggesting more LGR7 protein was present. A more rapid decline in the LGR7 mRNA compared with its protein has been shown in a previous study, which measured mRNA levels and relaxin binding. The authors suggested that this probably reflects a more rapid turnover of RNA than the receptor protein [11] and this may also be the case here. The LGR7 protein may therefore remain highly expressed throughout the process of labor and delivery and relaxin may have a role over this period.
In three clinical situations at preterm, LGR7 expression was significantly greater than in tissues collected at term. LGR7 expression in the PPROM samples declined to less than in tissues from patients who started preterm labor with intact membranes, although these patients had also been through labor and delivery. This preliminary result needs to be confirmed by different methods and larger numbers of tissues. We have shown in several studies using different methodologies that decidual relaxin is increased in patients with PPROM [15,20]. The ability of relaxin to increase the expression of its own receptor would be particularly important in this situation. The effect of relaxin on LGR7 expression has been previously demonstrated with isolated decidual cells [12] and while our data did not reach significance, a similar trend was seen with a 6 h incubation, which was lost by 24 h. The ‘‘dilution’’ of this effect from using tissue explants, which keep the cell contacts and extracellular matrix intact was not surprising. However, this might be important, as the elevated decidual relaxin at PPROM might prevent the decline of LGR7, which occurs with labor and delivery. If this is indeed the case, a positive feed-forward effect would increase the effects of relaxin in PPROM, including an increase in the matrix metalloproteinases [18,19] as well as the more recently described induction of an inflammatory response [21]. In any event, the significantly overall higher level of LGR7 expressed over the preterm period, regardless of the type of delivery, suggests that relaxin probably has a role in these tissues at this time.
Acknowledgments
We thank the nurses and staff of the labor and delivery ward of Kapiolani Medical Center for Women and Children for their help with the tissue collection. In particular we thank Ms J. Shimoda and Mr M. Linnolt for their assistance in the identification and consenting of potential patients, and the collection of these tissues. We also acknowledge the technical help of Ms Simone Parg and BAS Medical for the generous gift of human relaxin. This work was supported by NIH grant HD-24314 and by grants to the University of Hawaii and Kapiolani Medical Center under the Research Centers in Minority Institutions Program of NIH (RR1A1-03061 and RR-11091). A.A. was supported by grant GM 07684.
References
- 1.Hsu SY, Kudo M, Chen T, Nakabayashi K, Bhalla A, van der Spek PJ, et al. The three subfamilies of leucine-rich repeat-containing G protein-coupled receptors (LGR7): identification of LGR6 and LGR7 and the signaling mechanism for LGR7. Mol Endocrinol. 2000;14:1257–71. doi: 10.1210/mend.14.8.0510. [DOI] [PubMed] [Google Scholar]
- 2.Hsu SY, Nakabayashi K, Nishi S, Kumagai J, Kudo M, Sherwood OD, et al. Activation of orphan receptors by the hormone relaxin. Science. 2002;295:671–4. doi: 10.1126/science.1065654. [DOI] [PubMed] [Google Scholar]
- 3.Kumagai J, Hsu SY, Mtsumi H, Roh JS, Fu P, Hsueh AJW. INSL3/Leydig insulin-like peptide activates the LGR8 receptor important in testis descent. J Biol Chem. 2002;277:31283–6. doi: 10.1074/jbc.C200398200. [DOI] [PubMed] [Google Scholar]
- 4.Hudson P, Haley M, Cronk M, Crawford R, Haralambidis J, Tregear G, et al. Structure of a genomic clone encoding biologically active human relaxin. Nature. 1983;301:628–31. doi: 10.1038/301628a0. [DOI] [PubMed] [Google Scholar]
- 5.Hudson P, John M, Crawford R, Haralambidis J, Scanlon D, Gorman J, et al. Relaxin gene expression in human ovaries and the predicted structure of a human preprorelaxin by analysis of cDNA clones. EMBO J. 1984;3:2333–9. doi: 10.1002/j.1460-2075.1984.tb02135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bathgate RA, Samuel CS, Burazin TC, Layfield S, Claasz AA, Reytomas IG, et al. Human relaxin gene 3 (H3) and the equivalent mouse relaxin (M3) gene. Novel members of the relaxin peptide family. J Biol Chem. 2002;277:1148–57. doi: 10.1074/jbc.M107882200. [DOI] [PubMed] [Google Scholar]
- 7.Bryant-Greenwood GD, Schwabe C. Human relaxins: chemistry and biology. Endocr Rev. 1994;15:5–26. doi: 10.1210/edrv-15-1-5. [DOI] [PubMed] [Google Scholar]
- 8.Liu C, Eriste E, Sutton S, Chen J, Roland B, Kuei C, et al. Identification of relaxin-3/INSL7 as an endogenous ligand for the orphan G-protein coupled receptor GPCR135. J Biol Chem. 2003;278:50754–64. doi: 10.1074/jbc.M308995200. [DOI] [PubMed] [Google Scholar]
- 9.Ivell R, Balvers M, Pohnke Y, Telgmann R, Bartsch O, Milde-Langosch K, et al. Immunoexpression of the relaxin receptor LGR7 in breast and uterine tissues of humans and primates. Reprod Biol Endocrinol. 2003;1:114. doi: 10.1186/1477-7827-1-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luna JJ, Riesewijk A, Horcajadas JA, de van Os R, Dominguez F, Mosselman S, et al. Gene expression pattern and immunoreactive protein localization of LGR7 receptor in human endometrium throughout the menstrual cycle. Mol Hum Reprod. 2004;10:85–90. doi: 10.1093/molehr/gah019. [DOI] [PubMed] [Google Scholar]
- 11.Bond CP, Parry LJ, Samuel CS, Gehring HM, Lederman FL, Rogers PAW, et al. Increased expression of the relaxin receptor (LGR7) in human endometrium during the secretory phase of the menstrual cycle. J Clin Endocrinol Metab. 2004;89:3477–85. doi: 10.1210/jc.2003-030798. [DOI] [PubMed] [Google Scholar]
- 12.Mazella J, Tang M, Tseng L. Disparate effects of relaxin and TGFβ1: relaxin increases, but TGFβ1 inhibits, the relaxin receptor and the production of IGFBP-1 in human endometrial stromal/decidual cells. Hum Reprod. 2004;19:1513–8. doi: 10.1093/humrep/deh274. [DOI] [PubMed] [Google Scholar]
- 13.Shirota K, Tateishi K, Koji T, Hishikawa Y, Hachisuga T, Kuroki M, et al. Early human preantral follicles have relaxin and relaxin receptor (LGR7), and relaxin promotes their development. J Clin Endocrinol Metab. 2005;90:516–21. doi: 10.1210/jc.2004-0130. [DOI] [PubMed] [Google Scholar]
- 14.Bryant-Greenwood GD, Millar LK. Human fetal membranes; their pre-term premature rupture. Biol Reprod. 2000;63:1575–9. doi: 10.1095/biolreprod63.6.1575b. [DOI] [PubMed] [Google Scholar]
- 15.Tashima LS, Yamamoto SY, Yasuda M, Millar LK, Bryant-Greenwood GD. Decidual relaxins: gene and protein up-regulation in preterm premature rupture of the membranes by complementary DNA arrays and quantitative immunocytochemistry. Am J Obstet Gynecol. 2002;187:785–97. doi: 10.1067/mob.2002.125763. [DOI] [PubMed] [Google Scholar]
- 16.Garibay-Tupas JL, Maaskant RA, Greenwood FC, Bryant-Greenwood GD. Characteristics of the binding of 32P labeled human relaxins to the human fetal membranes. J Endocrinol. 1995;145:441–8. doi: 10.1677/joe.0.1450441. [DOI] [PubMed] [Google Scholar]
- 17.Millar LK, Boesche MH, Yamamoto SY, Killeen J, De Buque RL, Chen R, et al. A relaxin mediated pathway to the preterm premature rupture of the membranes, independent of infection. Am J Obstet Gynecol. 1998;179:126–34. doi: 10.1016/s0002-9378(98)70262-5. [DOI] [PubMed] [Google Scholar]
- 18.Qin X, Garibay-Tupas J, Chua PK, Cachola L, Bryant-Greenwood GD. An autocrine/paracrine role of decidual relaxin I. Interstitial collagenase (MMP-1) and tissue plasminogen activator. Biol Reprod. 1997;56:800–11. doi: 10.1095/biolreprod56.4.800. [DOI] [PubMed] [Google Scholar]
- 19.Qin X, Chua PK, Bryant-Greenwood GD. An autocrine/paracrine role of decidual relaxin. II. Stromelysin-1 (MMP-3) and tissue inhibitor of matrix metalloproteinases (TIMP-1) Biol Reprod. 1997;56:812–20. doi: 10.1095/biolreprod56.4.812. [DOI] [PubMed] [Google Scholar]
- 20.Bogic LV, Yamamoto SY, Millar LK, Bryant-Greenwood GD. Developmental regulation of the human relaxin genes in the decidua and placenta: overexpression in the preterm premature rupture of the fetal membranes. Biol Reprod. 1997;57:908–20. doi: 10.1095/biolreprod57.4.908. [DOI] [PubMed] [Google Scholar]
- 21.Bryant-Greenwood GD, Yamamoto SY, Lowndes KM, Webster LE, Parg SS, Amano A, et al. Human decidual relaxin and preterm birth. Ann N Y Acad Sci. 2005;1041:338–44. doi: 10.1196/annals.1282.054. [DOI] [PubMed] [Google Scholar]


