Abstract
The myc oncogenes are frequently activated in human tumors, but there is no comprehensive insight into the target genes and downstream cellular pathways of these transcription factors. We applied serial analysis of gene expression (SAGE) to identify targets of N-myc in neuroblastomas. Analysis of 42 000 mRNA transcript tags in SAGE libraries of N-myc- transfected and control neuroblastoma cells revealed 114 up-regulated genes. The majority of these genes have a role in ribosome assembly and activity. Northern blot analysis confirmed up-regulation of all tested transcripts. Induction was complete within 4 h after N-myc expression. The large majority of the ribosomal proteins were induced, as well as genes controlling rRNA maturation. Cellular rRNA content was 45% induced. SAGE libraries and northern blot analysis confirmed up-regulation of many of these genes in N-myc-amplified neuroblastomas. As N-myc can functionally replace c-myc, we analyzed whether N-myc targets were induced by c-myc as well. Approximately 40% of these N-myc targets were up-regulated in a c-myc-transfected melanoma cell line. These data suggest that myc genes function as major regulators of the protein synthesis machinery.
Keywords: c-myc/N-myc/neuroblastoma/protein synthesis/ribosomes
Introduction
The members of the myc oncogene family play a prominent role in cancer. N-myc, c-myc and L-myc are rearranged, amplified, mutated and/or overexpressed in many human tumor types. The c-myc gene is expressed in a wide variety of tissues and tumors, while N-myc expression is largely restricted to embryonic tissues and neuroendocrine tumors. Approximately 20% of neuroblastomas have N-myc amplification, and these tumors follow a very aggressive course (Schwab et al., 1983; Seeger et al., 1985). Overexpression of transfected N-myc genes in neuroblastoma cell lines strongly increased proliferation rates (Bernards et al., 1986; Lutz et al., 1996). Transgenic mice overexpressing N-myc in neural crest-derived tissues showed frequent development of neuroblastomas (Weiss et al., 1997). Numerous comparable observations have implicated c-myc and L-myc in the pathogenesis of many other tumor types (Cole, 1986; Henriksson and Luscher, 1996). While c-myc and N-myc homozygous knockout mice are embryonic lethal, transgenic mice in which N-myc replaced c-myc showed a gross normal development, indicating that both proteins have largely overlapping functions (Malynn et al., 2000).
Many experiments have suggested a role for myc genes in cell cycle control, metastasis, blocking of differentiation, apoptosis and proliferation rate (Henriksson and Luscher, 1996; Dang, 1999; Schmidt, 1999). The myc family members are transcription factors with a basic/helix–loop–helix/leucine zipper (bHLHzip) domain. They form heterodimers with Max proteins and bind to the E-box motif CACGTG to activate target gene transcription (Alex et al., 1992; Blackwood et al., 1992). The limited number of initially identified targets of c-myc gave little insight into the mechanism of how c-myc induces the variety of phenotypes. Examples of myc target genes are prothymosin α, ornithine decarboxylase, CAD and the DEAD-box gene MrDb (Eilers et al., 1991; Bello-Fernandez et al., 1993; Rosenwald et al., 1993; Grandori et al., 1996; Jones et al., 1996; Boyd and Farnham, 1997). Induction of cyclins D1, E and A and cdc25A were found in some but not all model systems (reviewed in Obaya et al., 1999). Induction of cyclin D2 was consistently found and provides a direct link to the cell cycle (Bouchard et al., 1999; Perez-Roger et al., 1999; Coller et al., 2000). Furthermore, c-myc and N-myc also induce Id2 transcription, thus stimulating Rb inactivation and cell cycle progression (Lasorella et al., 2000). However, in addition to these direct effects on proliferation, several findings suggest that c-myc also promotes the growth of cells, thereby inducing an increase in cell mass that may be a prerequisite for rapid cell proliferation. Impaired in vivo expression of Drosophila dmyc results in adult flies half the normal size (Johnston et al., 1999). Both the volume and proliferation rate of their cells are reduced, and could be restored by overexpression of dmyc. B cells of transgenic mice with overexpression of c-myc show for all differentiation stages an increased cell size and increased protein synthesis rate (Iritani and Eisenman, 1999). Furthermore, fibroblasts with a homozygous inactivation of c-myc have a reduced proliferation rate as well as a reduced protein synthesis rate (Mateyak et al., 1997). Conversely, activation of c-myc in fibroblasts activates protein synthesis (Schmidt, 1999). Furthermore, a role for c-myc in protein synthesis is suggested by the finding that c-myc induces the translation initiation factors elF-4E, eIF-2-α (Rosenwald et al., 1993; Jones et al., 1996), eIF5A and eIF4G (Coller et al., 2000). Very recently, ectopic in vivo expression of c-myc was found to result in increased expression of six ribosomal protein genes in mouse liver (Kim et al., 2000). A role for myc genes in growth regulation is in line with their effect on the cell cycle. Inactivation of c-myc in fibroblasts prolonged the G1 and G2 phases of the cell cycle, but not the S phase, while high expression of myc genes accelerated transition through G1 (Steiner et al., 1995; Lutz et al., 1996). A similar effect was found for Drosophila dmyc (Johnston et al., 1999).
Here we describe the use of SAGE (serial analysis of gene expression; Velculescu et al., 1995) to identify the downstream genes that are activated by N-myc in human neuroblastoma. To date, only prothymosin α, ornithine decarboxylase and Id2 have been identified as targets of N-myc (Lutz et al., 1996; Lasorella et al., 2000). The analysis of the expression level of >40 000 transcripts identified 114 genes up-regulated in N-myc-expressing cells. Our results indicate that N-myc functions as a regulator of cell growth by stimulating genes functioning in ribosome biogenesis and protein synthesis. Several of the identified genes are induced by N-myc as well as by c-myc.
Results
SAGE libraries of N-myc-transfected neuroblastoma cell lines
To identify the downstream target genes of N-myc, we applied the SAGE technique to an N-myc-transfected neuroblastoma cell line. The SHEP cell line has no N-myc amplification and expression, or c-myc expression. A tetracycline-dependent N-myc expression vector has been introduced into these cells, resulting in the SHEP-21N clone (Lutz et al., 1996). The SHEP-21N cells have constitutive exogenous N-myc expression that can be switched off by tetracycline. N-myc expression in the SHEP-21N cells was shown to increase the rate of cell division, shorten the G1 phase of the cell cycle and render the cells more susceptible to apoptotic triggers (Lutz et al., 1996; Fulda et al., 1999). SAGE libraries were constructed from SHEP-21N cells expressing N-myc and from SHEP-2 control cells. The SHEP-2 clone was transfected with the empty expression vector. From each library, we sequenced ∼21 000 transcript tags, corresponding to 8566 (SHEP-21N) and 10 154 (SHEP-2) different transcripts (Table I). A tag for the transfected N-myc construct has a frequency of 0 and 8 in SHEP-2 and SHEP-21N, respectively. Comparison of the SAGE libraries yielded 114 significantly (P <0.01) up-regulated tags in N-myc-expressing cells, with induction levels of up to 37-fold (Tables II–IV). Another 70 tags were significantly down-regulated. Here we focused on the analysis of a series of tags induced in the N-myc-transfected cells. The transcripts corresponding to these tags were identified using the SAGEmap database (Lal et al., 1999) and our own tag assignment program (Caron et al., 2001) and checked by mRNA and expressed sequence tag (EST) sequence analyses. A comprehensive set of up-regulated genes functions in ribosome biogenesis and activity and in later steps of the protein synthesis and protein turnover machinery.
Table I. Summary of neuroblastoma SAGE libraries.
| Total tags | Different transcripts | |
|---|---|---|
| Neuroblastoma cell lines | ||
| SHEP-2 | 20 950 | 10 154 |
| SHEP-21N (N-myc transfected) | 20 938 | 8566 |
| Neuroblastoma tumors | ||
| N52 | 19 597 | 9356 |
| N159 (N-myc amplified) | 20 001 | 10 262 |
| Total | 81 486 |
Table II. Downstream targets induced by N-myc: ribosomal proteins.
| Tag sequence | SHEP-2 | SHEP-21N | Fold induction | P value | Unigene Hs. | Gene |
|---|---|---|---|---|---|---|
| GCCGAGGAAG | 1 | 37 | 37.0 | <0.001 | 82148 | ribosomal protein S12 |
| GCTTTTAAGG | 1 | 29 | 29.0 | <0.001 | 8102 | ribosomal protein S20 |
| CCCATCCGAA | 1 | 23 | 23.0 | <0.001 | 91379 | ribosomal protein L26 |
| GGCCGCGTTC | 0 | 23 | >23 | <0.001 | 5174 | ribosomal protein S17 |
| CCAGTGGCCC | 0 | 21 | >21 | <0.001 | 180920 | ribosomal protein S9 |
| GTGTTGCACA | 1 | 16 | 16.0 | <0.001 | 165590 | ribosomal protein S13 |
| GATGCTGCCA | 1 | 16 | 16.0 | <0.001 | 99914 | ribosomal protein L22 |
| CCGTCCAAGG | 2 | 31 | 15.5 | <0.001 | 80617 | ribosomal protein S16 |
| GGAGTGGACA | 1 | 14 | 14.0 | <0.001 | 75458 | ribosomal protein L18 |
| GCCTGTATGA | 2 | 27 | 13.5 | <0.001 | 180450 | ribosomal protein S24 |
| GTTCCCTGGC | 2 | 26 | 13.0 | <0.001 | 177415 | ribosomal protein Fau-S30 |
| ATGGCTGGTA | 6 | 72 | 12.0 | <0.001 | 182426 | ribosomal protein S2 |
| GTGTTAACCA | 1 | 11 | 11.0 | <0.001 | 74267 | ribosomal protein L10 |
| CACAAACGGT | 4 | 43 | 10.8 | <0.001 | 195453 | ribosomal protein S27 |
| CTCAACATCT | 3 | 32 | 10.7 | <0.001 | 73742 | ribosomal protein, large, P0 |
| GTTCGTGCCA | 2 | 18 | 9.0 | <0.001 | 179666 | ribosomal protein L35a |
| GACGACACGA | 4 | 30 | 7.5 | <0.001 | 153177 | ribosomal protein S28 |
| TCGTCTTTAT | 3 | 21 | 7.0 | <0.001 | 75538 | ribosomal protein S7 |
| GGACCACTGA | 5 | 34 | 6.8 | <0.001 | 119598 | ribosomal protein L3 |
| CCTCGGAAAA | 4 | 24 | 6.0 | <0.001 | 2017 | ribosomal protein L38 |
| AATCCTGTGG | 8 | 48 | 6.0 | <0.001 | 178551 | ribosomal protein L8 |
| ATCAAGGGTG | 4 | 21 | 5.3 | <0.001 | 157850 | ribosomal protein L9 |
| GGGCTGGGGT | 20 | 101 | 5.1 | <0.001 | 183698 | ribosomal protein L29 |
| AAGGAGATGG | 5 | 25 | 5.0 | <0.001 | 184014 | ribosomal protein L31/tag matches mitochondrial sequences |
| AAGGTGGAGG | 11 | 55 | 5.0 | <0.001 | 163593 | ribosomal protein L18a |
| TTACCATATC | 10 | 49 | 4.9 | <0.001 | 177461 | ribosomal protein L39 |
| GTGAAGGCAG | 6 | 27 | 4.5 | <0.001 | 77039 | ribosomal protein S3A |
| GAACACATCC | 4 | 18 | 4.5 | 0.002 | 75879 | ribosomal protein L19 |
| CGCCGCCGGC | 10 | 40 | 4.0 | <0.001 | 182825 | human ribosomal protein L35 mRNA |
| GCCGTGTCCG | 5 | 20 | 4.0 | 0.002 | 119213 | ribosomal protein S6 |
| AGGAAAGCTG | 13 | 52 | 4.0 | <0.001 | 76437 | ESTs, highly similar to 60S rpL36 (Rattus norvegicus) |
| CCCCAGCCAG | 7 | 27 | 3.9 | <0.001 | 75459 | ribosomal protein S3 |
| GCAGCCATCC | 13 | 48 | 3.7 | <0.001 | 4437 | ribosomal protein L28 |
| GGCAAGAAGA | 7 | 25 | 3.6 | <0.001 | 111611 | ribosomal protein L27 |
| CCCGTCCGGA | 19 | 65 | 3.4 | <0.001 | 180842 | ribosomal protein L13 |
| CGCTGGTTCC | 15 | 51 | 3.4 | <0.001 | 179943 | ribosomal protein L11 |
| TAAGGAGCTG | 9 | 30 | 3.3 | <0.001 | 77904 | ribosomal protein S26 |
| CCTTCGAGAT | 8 | 26 | 3.3 | 0.001 | 76194 | ribosomal protein S5 |
| GGATTTGGCC | 33 | 103 | 3.1 | <0.001 | 119500 | ribosomal protein, large, P2 |
| AGGCTACGGA | 20 | 63 | 3.2 | <0.001 | 119122 | 60S ribosomal protein L13A |
| CTGCTATACG | 7 | 22 | 3.1 | <0.004 | 180946 | ribosomal protein L5 |
| TGTGCTAAAT | 12 | 35 | 2.9 | <0.001 | 179779 | ribosomal protein L37 |
| GAGGGAGTTT | 34 | 97 | 2.9 | <0.001 | 76064 | ribosomal protein L27a |
| AAGAAGATAG | 8 | 22 | 2.8 | 0.007 | 184776 | ribosomal protein L23a |
| ACATCATCGA | 17 | 46 | 2.7 | <0.001 | 182979 | ribosomal protein L12 |
| CTGTTGGTGA | 12 | 31 | 2.6 | 0.003 | 3463 | ribosomal protein S23 |
| AAGACAGTGG | 26 | 63 | 2.4 | <0.001 | 184109 | ribosomal protein L37a |
| TTGGTCCTCT | 46 | 108 | 2.3 | <0.001 | 108124 | ribosomal protein L41 |
| CTCCTCACCT | 12 | 28 | 2.3 | 0.008 | 119122 | 60S ribosomal protein L13A |
| AATAGGTCCA | 22 | 50 | 2.3 | <0.001 | 113029 | ribosomal protein S25 |
| ACTCCAAAAA | 23 | 46 | 2.0 | 0.004 | 133230 | ribosomal protein S15 |
| CTGGGTTAAT | 47 | 87 | 1.9 | <0.001 | 126701 | ribosomal protein S19 |
| TCAGATCTTT | 38 | 74 | 1.9 | <0.001 | 75344 | ribosomal protein S4, X-linked |
| AGCTCTCCCT | 38 | 56 | 1.9 | 0.003 | 82202 | ribosomal protein L17 |
| TAATAAAGGT | 45 | 78 | 1.7 | 0.002 | 118690 | ribosomal protein S8 |
| TTCAATAAAA | 56 | 89 | 1.6 | 0.004 | 177592 | ribosomal protein, large, P1 |
| Additional tags with P values >0.01 and ≤0.05 | ||||||
| AAGGTCGAGC | 1 | 8 | 8.0 | 0.022 | 184582 | ribosomal protein L24 |
| CTCGAGGAGG | 0 | 6 | >6 | 0.016 | 3254 | ribosomal protein L23-like |
| GCTCCGAGCG | 0 | 5 | >5 | 0.028 | 80617 | ribosomal protein S16 |
| TACAAGAGGA | 5 | 16 | 3.2 | 0.014 | 174131 | ribosomal protein L6 |
| CCATTGCACT | 7 | 17 | 2.4 | 0.032 | 53798 | ESTs, highly similar to 60S RP L18A |
| CGCCGGAACA | 12 | 27 | 2.3 | 0.012 | 286 | ribosomal protein L4 |
| ATTATTTTTC | 8 | 18 | 2.3 | 0.037 | 153 | ribosomal protein L7 |
| CAATAAATGT | 34 | 53 | 1.6 | 0.027 | 179779 | ribosomal protein L37 |
| CCAGAACAGA | 40 | 60 | 1.5 | 0.030 | 111222 | ribosomal protein L30 |
| GCATAATAGG | 38 | 55 | 1.4 | 0.048 | 184108 | ribosomal protein L21 |
Data obtained from the comparison of the SAGE libraries of the transfected neuroblastoma cell lines (SHEP-2 versus SHEP-21N). The transcripts are ordered by fold induction. P values were calculated by Monte Carlo simulations according to the SAGE 300 software package (see Materials and methods; Velculescu et al., 1995). Tag frequencies are given for the total SAGE libraries of SHEP-2 and SHEP-21N, ∼21 000 tags each.
Table IV. Downstream targets induced by N-myc: glycolysis.
| Tag sequence | SHEP-2 | SHEP-21N | Fold induction | P value | Unigene Hs. | Gene |
|---|---|---|---|---|---|---|
| GCGACCGTCA | 1 | 14 | 14.0 | <0.001 | 183760 | aldolase A fructose-bisphosphate (ALDOA) |
| TAGCTTCTTC | 0 | 7 | >7 | 0.008 | 76392 | aldehyde dehydrogenase 1, solublea |
| TCTGCTTGTC | 0 | 5 | >5 | 0.028 | 76392 | aldehyde dehydrogenase 1, solublea |
| TGGCCCCACC | 3 | 18 | 6.0 | <0.001 | 198281 | pyruvate kinase |
| TGAGGGAATA | 4 | 21 | 5.3 | <0.001 | 83848 | triosephosphate isomerase 1 (TPI1) |
| TACCATCAAT | 17 | 59 | 3.5 | <0.001 | 195188 | glyceraldehyde 3-phosphate dehydrogenase (GAPDH) |
| Additional tags with P value >0.01 and ≤0.05 | ||||||
| TGACTGAAGC | 0 | 5 | >5 | 0.028 | 3343 | 3-phosphoglycerate dehydrogenase mRNA |
| CGGCTGAATT | 0 | 5 | >5 | 0.028 | 75888 | ESTs, highly similar to 6-P-gluconate dehydrogenase, decarboxylating |
| ACCTTGTGCC | 0 | 5 | >5 | 0.028 | 878 | sorbitol dehydrogenase |
Transcripts are listed by fold induction.
aTwo reliable tags were found for this gene.
Induction of genes involved in ribosome biogenesis and protein synthesis
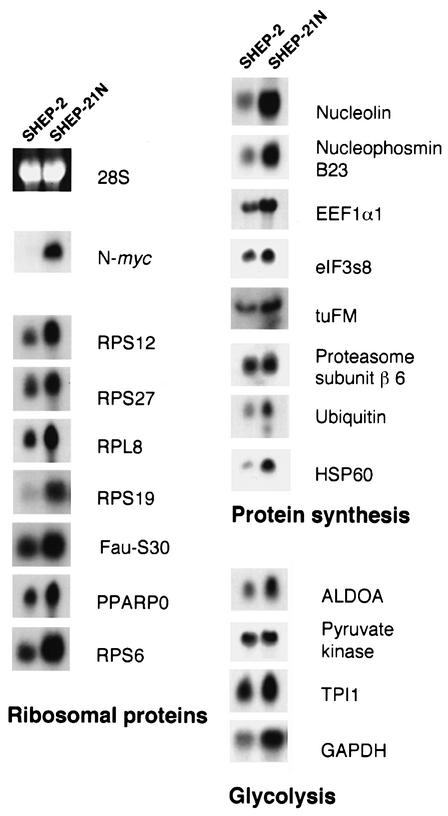
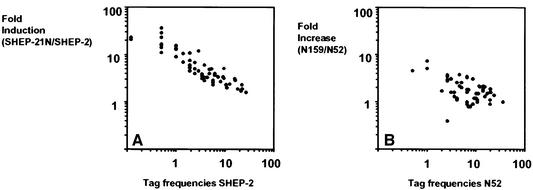
The first functional group consists of 56 ribosomal protein genes, which were induced up to 37-fold (P <0.01; Table II). Tags for 10 more ribosomal protein genes were less strongly up-regulated (0.01> P >0.05), with induction levels from 1.4- to 8-fold (Table II, lower part). These 66 tags correspond to 82% of the human ribosomal proteins (Wool and Chan, 1996). Northern blot analysis of SHEP-2 and SHEP-21N with probes for seven ribosomal protein genes (S12, S27, Fau-S30, L8, S6, S19 and PPARP0) confirmed their up-regulation (Figure 1). The fraction of tags for ribosomal protein mRNAs was increased from 4.1% in SHEP-2 to 12.6% in SHEP-21N. The level of induction of individual ribosomal protein genes is a function of their basal expression levels in SHEP-2. Highly expressed genes are less induced than genes with a low basic expression in SHEP-2 (Figure 2).
Fig. 1. Northern blot analysis of N-myc downstream target genes. Equal amounts of total RNA from exponentially growing SHEP-2 and SHEP-21N cells were loaded. Northern blots were hybridized with probes for the 19 indicated N-myc targets. RNA quantification was checked by ethidium bromide staining; the 28S band is shown.
Fig. 2. Level of induction of the 56 ribosomal protein genes identified as N-myc targets (P <0.01) in SHEP-21N cells. (A) Fold induction by N-myc in SHEP-21N cells as a function of the basic expression levels in SHEP-2. x-axis, basic expression level in SHEP-2 cells normalized per 10 000 tags; y-axis, fold induction in SHEP-21N cells. (B) Increase in the same 56 ribosomal protein genes in the N-myc-amplified neuroblastoma N159 as a function of the basic expression level in N-myc single-copy neuroblastoma N52. x-axis, expression level in N52 normalized per 10 000 tags; y-axis, fold increase in N159 relative to N52.
A second functional group of genes up-regulated in SHEP-21N consists of two nucleolar protein genes (Table III). Nucleophosmin (or B23) is induced from 26 to 55 tags (P <0.001), which was confirmed by northern blot analysis (Figure 1). Nucleophosmin is an abundant nucleolar protein that processes rRNA by cleavage of the 5′ end of the 5.8S pre-rRNA (Zhang et al., 1997) and functions in assembly and nuclear–cytoplasmic shuttling of pre-ribosomal particles (Borer et al., 1989; Olson, 1991). Transcription of another nucleolar protein is also induced: nucleolin, which has two tags due to alternative transcripts, is induced from 4 to 12 tags in total (P = 0.035). This induction was confirmed by northern blot analysis (Figure 1). Nucleolin processes pre-rRNA to mature 18S rRNA (Ginisty et al., 1998, 1999). It binds to nucleophosmin and is also involved in the assembly of pre-ribosomal particles and their nucleo-cytoplasmic transport. The induction of nucleolin and nucleophosmin in SHEP-21N suggests that rRNA and ribosome biosynthesis are targets of N-myc stimulation.
Table III. Downstream targets induced by N-myc: protein synthesis, protein degradation and ribosome biogenesis.
| Tag sequence | SHEP-2 | SHEP-21N | Fold induction | P value | Unigene Hs. | Gene |
|---|---|---|---|---|---|---|
| GCATAGGCTG | 0 | 12 | >12 | <0.001 | 198304 | Tu translation elongation factor, mitochondrial (tufM) |
| GCCCAGCTGG | 1 | 11 | 11.0 | 0.003 | 223241 | translation elongation factor 1δ (EEF1δ) |
| TGTGTTGAGA | 12 | 111 | 9.3 | <0.001 | 181165 | eukaryotic translation elongation factor 1α1 (EEF1α1) |
| TGGGCAAAGC | 5 | 39 | 7.8 | <0.001 | 2186 | eukaryotic translation elongation factor 1γ (EEF1γ) |
| CAGTCTAAAA | 0 | 8 | >8 | 0.004 | 76118 | ubiquitin C-terminal esterase L1 (ubiquitin thiolesterase) |
| GAGCGGGATG | 0 | 8 | >8 | 0.004 | 77060 | proteasome subunit 6 (β type) |
| GGCTCCCACT | 3 | 16 | 5.3 | 0.002 | 74335 | 90-kDa heat-shock protein (HSP90) |
| GCATTTAAAT | 86 | 138 | 1.6 | <0.001 | 261802 | eukaryotic translation elongation factor 1β (EEF1β) |
| AGCACCTCCA | 22 | 66 | 3.0 | <0.001 | 75309 | eukaryotic translation elongation factor 2 |
| TGAAATAAAA | 26 | 55 | 2.1 | <0.001 | 173205 | nucleophosmin (B23) |
| Additional tags with P values > 0.01 and ≤0.05 | ||||||
| CTGGCGAGCG | 1 | 9 | 9.0 | 0.011 | 174070 | human ubiquitin carrier protein (E2-EPF) |
| GGGGCAGGGC | 1 | 8 | 8.0 | 0.022 | 119140 | eukaryotic translation initiation factor 5A |
| GGCCCTGAGC | 2 | 11 | 5.5 | 0.012 | 71618 | human RNA polymerase II subunit (hsRPB10) |
| TACCAGTGTA | 0 | 5 | >5 | 0.028 | 79037 | 60-kDa heat-shock protein 1 (HSP60) |
| TGGCTAGTGT | 2 | 10 | >5 | 0.019 | 118065 | proteasome subunit, β type, 7 |
| TACAAAACCA | 1 | 4 | 4.0 | 0.202 | 79110 | nucleolina |
| GTTTTTGCTT | 3 | 8 | 2.7 | 0.110 | 79110 | nucleolina |
| CAGATCTTTG | 3 | 11 | 3.7 | 0.029 | 119502 | proteasome subunit, α type, 7 |
| TCACAAGCAA | 4 | 15 | 3.8 | 0.010 | 146763 | αNAC mRNA |
| CGCCGCGGTG | 5 | 16 | 3.2 | 0.014 | 4835 | eukaryotic translation initiation factor 3, subunit 8 (eIFs8) |
| GTGACAGAAG | 5 | 13 | 2.6 | 0.047 | 129673 | eukaryotic translation initiation factor 4A, isoform 1 |
| CCATTGCACT | 7 | 17 | 2.4 | 0.032 | 173694 | ESTs, highly similar to probable ubiquitin C-terminal hydrolase |
| AACTAAAAAA | 71 | 97 | 1.4 | 0.028 | 3297 | ubiquitin |
Genes are listed by fold induction.
aTwo reliable tags were found for this gene due to alternative polyadenylation.
Additionally, tags corresponding to nine translation initiation and elongation factors were induced (Table III). They are eukaryotic translation initiation factors eIF3s8, eIF4A and eIF5A, and the subunits α, β, γ and δ of translation elongation factor 1 (EEF1). Furthermore, elongation factor 2 and the mitochondrial elongation factor Tu (tuFM) are up-regulated. Northern blot analysis of SHEP-21N and SHEP-2 confirmed the induction of eIF3s8, EEF1α1 and tuFM (Figure 1). These data further support a role for N-myc as a regulator of protein synthesis.
Genes involved in routing, folding and degradation of proteins were also up-regulated. The nascent polypeptide-associated complex α (NAC) mRNA was induced (Table III, lower part). NAC protects nascent cytosolic proteins from translocation to the endoplasmatic reticulum (Wiedmann et al., 1994). Induction of the chaperones HSP60 (from 0 to 5 tags) and HSP90 (from 3 to 12 tags) further suggested an increased cellular capacity for protein folding and maturation. Additionally, the cellular capacity for protein degradation was possibly induced. Three ubiquitin pathway proteins (ubiquitin, ubiquitin C-terminal esterase L1 and ubiquitin carrier protein) and three proteasome subunits (β type 6, β type 7 and α type 7) showed increased tag frequencies. Northern blot analysis confirmed induction of HSP60, proteasome subunit β type 6 and ubiquitin in SHEP-21N (Figure 1).
Up-regulation of glycolysis genes
Another functional group of N-myc-induced genes encoded key enzymes in the glycolytic pathway (Table IV, upper part). Tags for aldolase A fructose-bisphosphate (ALDOA), triosephosphate isomerase 1 (TPI1), glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and pyruvate kinase are all increased (Table IV). A series of other metabolic enzymes was also induced. Northern blot analysis confirmed the mRNA induction of ALDOA, pyruvate kinase, TPI1 and GAPDH (Figure 1).
N-myc activates downstream targets within 4 h
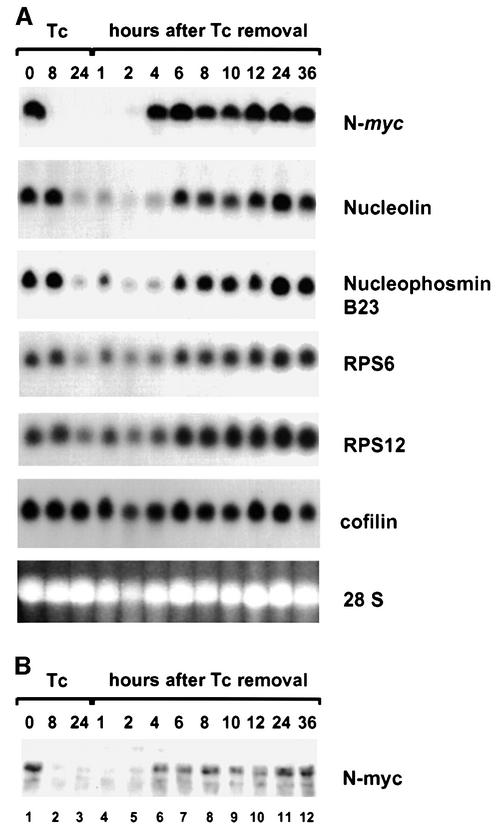
In several systems it was observed that c-myc can induce cell growth and cell mass. The finding that many genes with a role in protein synthesis are induced in SHEP-21N therefore raises the question of whether their up-regulation is an indirect and late effect consequent on N-myc-induced growth or whether these genes are early targets of induction by N-myc and the cause of myc-mediated cell growth. We therefore tested in a time course experiment whether the genes of the protein synthesis machinery are early or late targets of N-myc-mediated induction. N-myc expression can be switched off reversibly in SHEP-21N cells by tetracycline. SHEP-21N cells were treated for 24 h with tetracycline. Northern blot analysis showed that the N-myc mRNA expression is switched off within 8 h of tetracycline treatment (Figure 3A, lanes 1–3). After 24 h, cells were washed and grown for an additional 2–36 h without tetracycline. N-myc mRNA expression is restored between 2 and 4 h after tetracycline removal (Figure 3A, lanes 5 and 6). Western blot analysis showed that N-myc protein expression closely follows N-myc mRNA expression (Figure 3B). The northern blot filter was hybridized with probes for the N-myc downstream targets nucleolin, nucleophosmin and the ribosomal protein genes RPS6 and RPS12 (Figure 3A). After repression of N-myc by tetracycline, the mRNA levels of these genes were unaffected at 0 and 8 h, but were reduced to low basic levels at 24 h. Within 2–4 h after re-expression of N-myc mRNA and protein, the mRNA expression of all four genes was strongly re-induced (Figure 3B, lanes 6 and 7). Similar results were obtained for EEF1A1, TPI1 and eIF3s8 (data not shown). The expression level of cofilin that we used as a control does not change significantly during the time course. To exclude a direct effect of tetracycline on nucleolin or nucleophosmin expression, we conducted the same experiment with SHEP-2 cells, but no effect on gene expression was observed (data not shown). Fluorescence activated cell sorting (FACS) analysis of SHEP-21N at 0 and 24 h of tetracycline treatment and at 7.5 h after tetracycline removal showed no change in cell volume, formally excluding the possibility that induction of the genes of the protein synthesis machinery is a result of increased cell volume (data not shown). These results show that the genes of the protein synthesis machinery are early targets in the N-myc downstream pathway, although not necessarily direct targets of N-myc. Induction of these genes by N-myc is highly versatile: expression drops after N-myc abrogation and is restored swiftly after N-myc re-expression.
Fig. 3. Time course analysis of N-myc and downstream target gene induction in SHEP-21N cells. SHEP-21N cells were treated for 24 h with 10 ng/ml tetracycline, washed and grown for an additional 36 h without tetracycline. Cells were harvested at 0, 8 and 24 h of tetracycline treatment. Subsequent samples were taken at 1, 2, 4, 8, 10, 12, 24 and 36 h after removal of the antibiotic. (A) Northern blot analysis of total RNA at the indicated time points. (B) Western blot analysis of N-myc protein at the indicated time points. A 10 µg aliquot of total protein samples of the time course experiment were fractionated through a 10% SDS–polyacrylamide gel, blotted on an Immobilon membrane and probed with a monoclonal anti-N-myc antibody.
Effect at the protein level and induction of rRNA synthesis
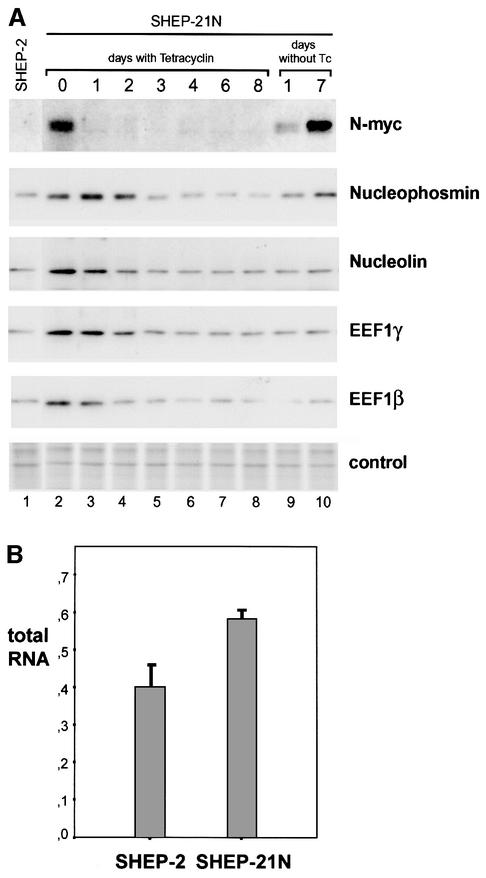
To analyze further the effect of N-myc on the protein synthesis machinery, we analyzed the protein levels of nucleolin, nucleophosmin, EEF1γ and EEF1β by western blotting. All four proteins are more strongly expressed in SHEP-21N cells compared with SHEP-2 (Figure 4, lanes 1 and 2). As a further test that these expression levels are controlled by N-myc, we treated the SHEP-21N cells for 1–8 days with tetracycline, which suppressed N-myc expression (Figure 4, lanes 3–8). After 2–3 days, the high protein expression levels of the four N-myc-induced genes dropped to the basic expression level observed in SHEP-2 cells. Regulation of the mRNA level of these genes by N-myc is therefore effective at the protein level. Finally, when tetracycline was washed away after 8 days and cells were cultured for 1 or 7 days without tetracycline, N-myc expression was restored and expression levels of the four target proteins were increased (Figure 4, lanes 9 and 10).
Fig. 4. Expression of N-myc, nucleolin, nucleophosmin, translation elongation factors EEF1γ and EEF1β and total RNA content of SHEP-2 and SHEP-21N cells. (A) Western blot analysis. Total cell extracts (10 µg) were fractionated through an acrylamide gel, blotted and probed with monoclonal antibodies against N-myc and nucleo phosmin, and with polyclonal antibodies against nucleolin, EEF1γ and EEF1β. SHEP-21N cells were treated for 0–8 days with tetracycline (lanes 2–8) and subsequently cultured for 1 or 7 days without tetracycline (lanes 9 and 10). A Coomassie Blue staining is shown as control for loading. (B) Total RNA content of SHEP-2 and SHEP-21N cells. RNA was isolated from 10 samples of 106 cells of each cell line and analyzed spectrophotomerically. Error bars give the SD.
The induction by N-myc of nucleolin and nucleophosmin, two genes with a key role in rRNA processing and ribosome biogenesis, urged us to analyze whether SHEP-21N cells have a higher rRNA content than SHEP-2 cells. Total RNA was isolated from 10 samples of 106 cells of SHEP-2 and SHEP-21N. Spectrophotometric analysis showed that SHEP-21N cells on average have a 45% higher yield than SHEP-2 cells (P <0.001, Student’s t-test for independent samples; Figure 4B). Triplicate experiments on independently cultured cells gave the same results. Densitometric quantification of the 18S and 28S rRNA bands fractionated on agarose gels confirmed that this increase is caused by rRNA (data not shown).
We also measured protein content and the rate of protein synthesis. Lysates of 106 SHEP-2 and SHEP-21N cells contained equivalent amounts of total protein (data not shown). Protein synthesis rates were analyzed by [35S]methionine incorporation, but no differences were observed between SHEP-2 and SHEP-21N cells. Also, when N-myc expression was switched off by a 48 h tetracycline treatment, no differences in incorporation could be observed (data not shown). N-myc therefore strongly induces the rRNA content of SHEP-21N cells, but not the protein synthesis rate. The protein synthesis in SHEP-21N cells may be limited by a factor not induced by N-myc, or may have been maximal already in the SHEP-2 cells and beyond a level that can be boosted by N-myc.
SAGE libraries of neuroblastomas with and without amplification of endogenous N-myc
To analyze whether genes of the protein synthesis machinery are also induced in neuroblastomas with N-myc amplification, we generated SAGE libraries of two neuroblastomas. Neuroblastoma N159 has N-myc amplification and expression, and neuroblastoma N52 is an N-myc single-copy tumor without N-myc expression (Figure 5B, lanes 9 and 10). We sequenced 39 598 tags of the two libraries (Table I). The tag frequencies were normalized per 20 000 tags and compared. N-myc was represented by 16 tags in N159 and 0 tags in N52. There are 52 tags differentially expressed (P <0.01) in the libraries. We analyzed which of the N-myc target genes identified in the SHEP cells correlated with N-myc in the two tumors. The 56 significantly (P <0.01) induced ribosomal protein genes detected in SHEP-21N cells produce a total of 988 tags in N52 and 1600 tags in N159 (per 20 000 tags). The N-myc-amplified N159 tumor therefore has 62% higher ribosomal protein gene expression (Figure 2B). This strongly suggests that N-myc induces ribosomal protein gene expression in vivo. Other genes functioning in protein synthesis are also up-regulated. Increased expression in N159 compared with N52 is seen for nucleophosmin (from 4 to 19 tags), nucleolin (from 3 to 9 tags), eukaryotic translation initiation factor 4A, isoform 1 (from 4 to 8 tags) and the translation elongation factors EEF1α1 (from 50 to 98 tags) and EEF1γ (from 18.4 to 31 tags). There is almost no induction of the genes involved in glycolysis. The expression levels of nucleolin, nucleophosmin and ribosomal protein S6 were confirmed by hybridization of northern blots with total RNA from N159 and N52 (Figure 5B). These results show that the expression levels of many of the N-myc target genes identified in SHEP-21N cells also correlate in vivo with N-myc amplification and overexpression.
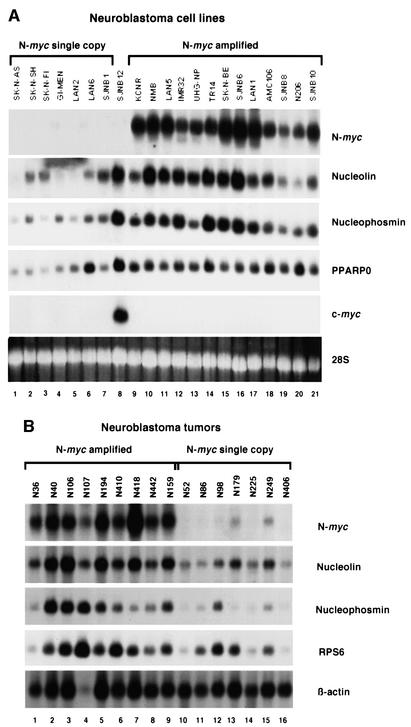
Fig. 5. Northern blot analysis of total RNA from neuroblastoma cell lines and tumors. Filters were hybridized with the indicated probes. RNA quantification was checked by ethidium bromide staining; the 28S band is shown. (A) Panel of 21 neuroblastoma cell lines. (B) Panel of 16 fresh tumors. Tumors in lanes 1–9 are N-myc amplified.
N-myc target gene expression in neuroblastoma cell lines and tumors
We further analyzed the expression of N-myc downstream genes in a panel of neuroblastoma cell lines and tumors. Hybridization of a northern blot of total RNA from 21 neuroblastoma cell lines showed a fair correlation between expression of N-myc, nucleolin, nucleophosmin and the ribosomal protein PPARP0 (Figure 5A). Cell line SJNB12 shows high expression of the N-myc target genes (Figure 5A, lane 7). This cell line has no N-myc expression, but has c-myc amplification and overexpression (Cheng et al., 1995), suggesting that c-myc may induce the same target genes as N-myc (see below).
As cell lines are not fully representative of neuroblastoma tumors in vivo, we analyzed 16 fresh neuroblastomas of all stages. A northern blot analysis showed a rather good overall correlation between expression of N-myc, nucleolin and nucleophosmin (Figure 5B). There are some exceptions, but the results suggest that nucleolin and nucleophosmin are also in vivo targets of N-myc induction. Ribosomal protein S6 (RPS6) expression showed a less consistent relationship with N-myc, indicating that besides N-myc, other factors may also modulate its expression.
Several N-myc target genes are also induced by c-myc
N-myc belongs to the same family of proto-oncogenes as c-myc. Since N-myc can replace c-myc in transgenic mice without inducing gross phenotypic defects (Malynn et al., 2000), and since both myc proteins share the same target recognition sequence, we analyzed whether the N-myc downstream targets identified in this study are also induced by c-myc. We analyzed the melanoma cell line IGR39D and a c-myc-transfected clone of this cell line (clone 3; Versteeg et al., 1988). Northern blots with total RNA of these cell lines were hybridized with the 19 probes tested on the SHEP-2 and SHEP-21N cells. Eight of the N-myc targets appeared to be induced by c-myc as well (Figure 6). They are the ribosomal protein genes S12, S27, S19 and S6, and nucleolin, nucleophosmin, ubiquitin and GAPDH. The remaining 11 genes showed no induction by c-myc. Therefore, c-myc and N-myc share >40% of their target genes in the cell systems tested here. Interestingly, nucleophosmin, nucleolin and most ribosomal protein genes are among them.
Fig. 6. Northern blot analysis of induction of N-myc target genes in a c-myc-transfected melanoma cell line. Clone 3 is a c-myc-transfected clone of the IGR39D melanoma cell line. Equal amounts of total RNA of IGR39D and clone 3 were loaded. Filters were hybridized with the indicated probes.
Discussion
One of the surprising aspects of myc oncogenes is their multitude of phenotypic effects. They are known to induce growth, cell division, metastasis and apoptosis. A series of target genes of myc transcription factors has been identified, some of which can be related to specific phenotypes. However, our knowledge of myc target genes is probably still fragmentary and insufficient to explain the full range of phenotypes. As a step towards a complete inventory of the myc downstream pathway, we applied the SAGE technology to N-myc-transfected cells. We have chosen to compare SHEP-21N with SHEP-2 cells, as we aimed to identify all genes that are up-regulated in a situation of stable and enduring N-myc expression, rather than in a transition period after induction of N-myc. SAGE provides an integral gene expression profile of a tissue or cell line. Comparison of the SAGE libraries of the N-myc-expressing SHEP-21N and control SHEP-2 cells identified 114 genes significantly (P <0.01) induced in SHEP-21N. Moreover, since SAGE is quantitative, the libraries permit the analysis of the full myc-induced transcription shift. N-myc turns out to have a massive effect on genes with a role in protein synthesis. Approximately 80% of ribosomal protein genes turned out to be enhanced, as well as some key genes in rRNA maturation and ribosome assembly. Furthermore, expression of many translation initiation and elongation factors is considerably enhanced. We detected induction of 89 genes involved in protein synthesis (Tables II and III). Together, they produced 1119 of the 20 950 sequenced tags in SHEP-2. These 89 genes therefore contributed 5.3% of the total number of transcripts in SHEP-2. In SHEP-21N, these genes give rise to 3327 transcript tags, or 15.9% of all transcripts. Two of the up-regulated genes, nucleolin and nucleophosmin, function in rRNA maturation and ribosome assembly (reviewed in Ginisty et al., 1999). We therefore analyzed whether N-myc expression results in higher rRNA levels. We found a striking 45% higher rRNA content in SHEP-21N than in SHEP-2 cells on a per cell basis. Somewhat surprisingly, there was no overall increase in the rate of protein synthesis in SHEP-21N cells. One interpretation is that some rate-limiting components of the protein synthesis machinery are not induced in SHEP-21N cells. Alternatively, protein synthesis may already have been maximal in SHEP-2, beyond a level that can be boosted further.
Crucial to the interpretation of these data is the finding that genes of the protein synthesis machinery are early targets of the N-myc pathway. Activation of N-myc results within 2–4 h in full induction of expression of the tested target genes, amongst which were nucleolin, nucleophosmin, two ribosomal protein genes, a translation initiation factor and a translation elongation factor. This leads us to the conclusion that the massive induction of genes of the protein synthesis pathway is an early effect of N-myc. However, we have not addressed the question of whether these genes are direct targets of N-myc or, in contrast, part of a hierarchical pathway with myc at the top. Although nucleolin was identified previously as a direct target of c-myc (Greasley et al., 2000), the time course experiments do not exclude the possibility that other genes of the protein synthesis machinery are induced by an intermediary transcription factor that is up-regulated by N-myc. The quantitative character of SAGE enabled a further analysis of the induction. All N-myc downstream targets have a fair basal expression level in SHEP-2 cells, which is not surprising in view of their essential role in protein synthesis. Interestingly, the ribosomal protein genes do not show an equal overall induction by, for example, a factor of two or three, but induction appears to be related to the basic level of expression in SHEP-2. The strongest induction is observed for genes with the weakest basal expression level (Figure 2). This could suggest that the ribosomal protein genes in SHEP-2 cells are restricted in their expression to a variable extent and that N-myc can relieve this restriction.
Induction of genes of the protein synthesis machinery is likely to be a general effect of N-myc in neuroblastomas. Comparison of SAGE libraries of neuroblastoma cell lines with and without N-myc amplification shows a 62% increase of ribosomal protein gene transcripts in N-myc-expressing cells. Moreover, northern blot analysis of 37 neuroblastomas and neuroblastoma cell lines showed an overall induced expression level of nucleolin, nucleophosmin and ribosomal protein genes in N-myc-amplified cases (Figure 5). In addition, c-myc was found to induce a series of N-myc target genes as well. Of the 19 targets of N-myc that we tested on northern blots, eight were induced in melanoma cells with ectopic expression of c-myc. Amongst them are the ribosomal protein genes S12, S27, S19 and S6, and nucleolin and nucleophosmin. These data suggest that induction of the protein synthesis machinery is a major function of both c-myc and N-myc.
Myc genes are general inducers of the protein synthesis machinery
Our data are well in line with some of the recently identified target genes of c-myc and with phenotypic effects of myc observed in vivo and in vitro. Early analyses identified two translation initiation factors as targets of c-myc (Rosenwald et al., 1993; Jones et al., 1996), while recent microarray analyses revealed induction by c-myc of two more translation initiation factors, nucleolin and one ribosomal protein gene (RPS11) (Coller et al., 2000). Nucleolin was also identified as a c-myc target by Greasley et al. (2000). The microarray analyses of Coller et al. (2000) do not reveal induction of 32 other ribosomal protein genes that were represented on their chips. However, it was observed recently that 4 days after in vivo transduction of a c-myc-expressing retrovirus in mouse, liver cells expressing ectopic c-myc are greatly enlarged and have increased expression of six ribosomal protein genes as well as nucleolin and nucleophosmin (Kim et al., 2000). Together with our SAGE analyses in which the qualitative and quantitative induction of genes of the protein synthesis machinery were established, these data implicate the protein synthesis machinery as a major target of induction by myc proteins. These findings are in agreement with phenotypic effects of myc genes observed in several experiments (Schmidt, 1999). Rat fibroblasts with inactivated c-myc alleles showed a slower growth rate and a reduced protein synthesis rate (Mateyak et al., 1997), while induction of protein synthesis in fibroblasts was observed after c-myc activation (Schmidt, 1999). B cells with ectopic c-myc expression in transgenic mice are larger at any stage of differentiation and have an increased protein synthesis rate (Iritani and Eisenman, 1999). Drosophila with a mutated dmyc grow more slowly and only attain a tiny body volume (Johnston et al., 1999). The data in animal model systems, in normal fibroblasts and in neuroblastoma tumor cells all suggest that induction of the protein synthesis machinery is a major function of myc genes. In addition to their direct effect on the cell cycle by inducing cyclin D2 and Id2, this induction of the protein synthesis machinery may provide the increase in cell mass required to keep the cell volume in step with proliferation.
Materials and methods
Cell lines
Neuroblastoma cell lines and culture conditions were as described before (Cheng et al., 1995). The melanoma cell lines IGR39D and clone 3 were described earlier (Versteeg et al., 1988). The SHEP cell lines were maintained in RPMI 1640 medium supplemented with 10% fetal calf serum, 4 mM l-glutamine, 100 U/ml penicillin and 100 µg/ml streptomycin (Lutz et al., 1996). Tetracycline (Sigma) was used at a concentration of 10 ng/ml to inhibit N-myc expression.
Generation of SAGE libraries
SAGE was performed as described (Velculescu et al., 1995) with minor adaptations. Total RNA was extracted by guanidium thiocyanate (Chomczynski and Sacchi, 1987). Poly(A)+ RNA was isolated using the MessageMaker kit (Gibco-BRL) according to the manufacturer’s instructions. SAGE libraries were generated using minimally 4 µg of poly(A)+ RNA. The cDNA was synthesized according to the Superscript Choice System (Gibco-BRL), digested with NlaIII and bound to streptavidin-coated magnetic beads (Dynal). Linkers containing recognition sites for BsmFI were ligated to the cDNA. Linker tags including a cDNA tag were released by BsmFI digestion, ligated to one another and amplified. The PCR products were heated for 5 min at 65°C before preparative analysis on a polyacrylamide gel. After the ligation into concatamers, a second heating step was included (15 min at 65°C) and fragments between 800 and 1500 bp were purified and cloned in pZero-1 (Invitrogen). Colonies were screened with PCR using M13 forward and reverse primers. Inserts >300 bp were sequenced with a BigDye terminator kit and analyzed on a 377 ABI automated sequencer (Perkin Elmer).
Analysis of the SAGE database
The SAGE libraries were analyzed using the SAGE 300 program software package (Velculescu et al., 1997). P values were calculated using Monte Carlo simulations. Transcripts were identified by comparison of the tags in the database with the ‘tag to gene map’ (SAGEmap) from the Cancer Genome Anatomy Project at the NCBI (http://www.ncbi.nlm.nih.gov/SAGE). This database links Unigene clusters to SAGE tags (Lal et al., 1999). The gene assignments were subsequently checked by hand for sequencing errors causing incorrect tags and for erroneous gene assignments based on hybrid Unigene clusters. Other database analyses and generation of specific primers utilized the Wisconsin GCG package software.
Northern blot analysis
Total RNA (20 µg per lane) was electrophoresed through a 0.8% agarose gel in the presence of 6.7% formaldehyde and blotted on Hybond N membranes (Amersham) in 10× SSC. Hybridization was carried out overnight in 0.5 M NaHPO4 pH 7.0, 7% SDS, 1 mM EDTA at 65°C. Filters were washed in 40 mM NaHPO4, 1% SDS at 65°C. Probes were labeled by random priming of sequence-verified PCR products. A complete list of all the primers used in RT–PCRs is available on request.
Total protein content
Exponentially growing cells were harvested and cell number was determined using a Coulter counter. Cells (1 × 106) were lysed in 20 mM Tris–HCl pH 8.0, 137 mM NaCl, 10% glycerol, 1% NP-40 and protease inhibitors (protease cocktail, Roche). Samples were assayed with the Bio-Rad Protein assay. Assays were performed at least in duplicate.
Western blots
Cell lysates were separated on an SDS–polyacrylamide gel and electroblotted onto Immobilon-P transfer membrane (Millipore). Blocking of the membrane and incubation with antibodies involved standard procedures. Proteins were visualized using the ECL detection system (Amersham). Anti-nucleophosmin monoclonal antibody was a gift of Dr P.K.Chan (Baylor College of Medicine). The antibody against nucleolin was a gift of Dr P.Bouvet (CNRS, IPBS, Toulouse, France). Anti-N-myc was obtained from PharmIngen (Clone B8.4.B). Rabbit anti-human EEF1γ and anti-human EEF1β antibodies (Sanders et al., 1996) were a gift of Dr J.Dijk (Sylvius Laboratories, LUMC, Leiden, The Netherlands).
Total rRNA content
Total RNA of 1 × 106 exponentially growing cells was extracted by guanidium isothiocyanate (Chomczynski and Sacchi, 1987) and quantified spectrophotometrically. Results of 10 isolations of each of the cell lines SHEP-2 and SHEP-21N were statistically analyzed with the Student’s t-test for independent samples. Aliquots on a per cell basis were subjected to agarose gel electrophoresis and stained with ethidium bromide. The relative fluorescence of the rRNA bands was quantified using the Kodak Digital Science 1D Image Analysis Software package (EDAS 120).
FACS analysis
SHEP-21N cells treated or not treated with tetracycline were trypsinized, stained with propidium iodine and analyzed on a Beckman FACScan flow cytometer. Forward scatter (FSC) was used as a means for cell mass. FSC was measured for the total cell population or for the G0/G1 fraction, and did not differ for SHEP-21N cells treated for 0 or 24 h with tetracycline and for cells subsequently cultured for 7.5 h without tetracycline.
Acknowledgments
Acknowledgements
We thank Dr Pui K.Chan and Dr Phillipe Bouvet for their kind gifts of anti-nucleophosmin and anti-nucleolin antibodies, respectively, and Dr Jan Dijk for antibodies to translation elongation factors. We thank Adam Benham and Ineke Braakman for their kind help in the protein synthesis experiments and for their hospitality, and Alvin Chan, Jan Molenaar and Danielle Veenma for help in some of the experiments. This research was supported by grants from the Stichting Kindergeneeskundig Kankeronderzoek (SKK), the Dutch Cancer Foundation (NKB/KWF) and the A.Meelmeijer Fund.
References
- Alex R., Sozeri,O., Meyer,S. and Dildrop,R. (1992) Determination of the DNA sequence recognized by the bHLH-zip domain of the N-Myc protein. Nucleic Acids Res., 20, 2257–2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bello-Fernandez C., Packham,G. and Cleveland,J.L. (1993) The ornithine decarboxylase gene is a transcriptional target of c-Myc. Proc. Natl Acad. Sci. USA, 90, 7804–7808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernards R., Dessain,S.K. and Weinberg,R.A. (1986) N-myc amplification causes down-modulation of MHC class I antigen expression in neuroblastoma. Cell, 47, 667–674. [DOI] [PubMed] [Google Scholar]
- Blackwood E.M., Kretzner,L. and Eisenman,R.N. (1992) Myc and Max function as a nucleoprotein complex. Curr. Opin. Genet. Dev., 2, 227–235. [DOI] [PubMed] [Google Scholar]
- Borer R.A., Lehner,C.F., Eppenberger,H.M. and Nigg,E.A. (1989) Major nucleolar proteins shuttle between nucleus and cytoplasm. Cell, 56, 379–390. [DOI] [PubMed] [Google Scholar]
- Bouchard C. et al. (1999) Direct induction of cyclin D2 by Myc contributes to cell cycle progression and sequestration of p27. EMBO J., 18, 5321–5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd K.E. and Farnham,P.J. (1997) Myc versus USF: discrimination at the cad gene is determined by core promoter elements. Mol. Cell. Biol., 17, 2529–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron H. et al. (2001) The human transcriptome map: clustering of highly expressed genes in chromosomal domains. Science, 291, 1289–1292. [DOI] [PubMed] [Google Scholar]
- Cheng N.C., Van Roy,N., Chan,A., Beitsma,M., Westerveld,A., Speleman,F. and Versteeg,R. (1995) Deletion mapping in neuroblastoma cell lines suggests two distinct tumor suppressor genes in the 1p35–36 region, only one of which is associated with N-myc amplification. Oncogene, 10, 291–297. [PubMed] [Google Scholar]
- Chomczynski P. and Sacchi,N. (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction. Anal. Biochem., 162, 156–159. [DOI] [PubMed] [Google Scholar]
- Cole M.D. (1986) The myc oncogene: its role in transformation and differentiation. Annu. Rev. Genet., 20, 361–384. [DOI] [PubMed] [Google Scholar]
- Coller H.A., Grandori,C., Tamayo,P., Colbert,T., Lander,E.S., Eisenman,R.N. and Golub,T.R. (2000) Expression analysis with oligonucleotide microarrays reveals that MYC regulates genes involved in growth, cell cycle, signaling and adhesion. Proc. Natl Acad. Sci. USA, 97, 3260–3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang C.V. (1999) c-Myc target genes involved in cell growth, apoptosis and metabolism. Mol. Cell. Biol., 19, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilers M., Schirm,S. and Bishop,J.M. (1991) The MYC protein activates transcription of the α-prothymosin gene. EMBO J., 10, 133–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulda S., Lutz,W., Schwab,M. and Debatin,K.M. (1999) MycN sensitizes neuroblastoma cells for drug-induced apoptosis. Oncogene, 18, 1479–1486. [DOI] [PubMed] [Google Scholar]
- Ginisty H., Amalric,F. and Bouvet,P. (1998) Nucleolin functions in the first step of ribosomal RNA processing. EMBO J., 17, 1476–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginisty H., Sicard,H., Roger,B. and Bouvet,P. (1999) Structure and functions of nucleolin. J. Cell Sci., 112, 761–772. [DOI] [PubMed] [Google Scholar]
- Grandori C., Mac,J., Siebelt,F., Ayer,D.E. and Eisenman,R.N. (1996) Myc–Max heterodimers activate a DEAD box gene and interact with multiple E box-related sites in vivo. EMBO J., 15, 4344–4357. [PMC free article] [PubMed] [Google Scholar]
- Greasley P.J., Bonnard,C. and Amati,B. (2000) Myc induces the nucleolin and BN51 genes: possible implications in ribosome biogenesis. Nucleic Acids Res., 28, 446–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriksson M. and Luscher,B. (1996) Proteins of the Myc network: essential regulators of cell growth and differentiation. Adv. Cancer Res., 68, 109–182. [DOI] [PubMed] [Google Scholar]
- Iritani B.M. and Eisenman,R.N. (1999) c-Myc enhances protein synthesis and cell size during B lymphocyte development. Proc. Natl Acad. Sci. USA, 96, 13180–13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston L.A., Prober,D.A., Edgar,B.A., Eisenman,R.N. and Gallant,P. (1999) Drosophila myc regulates cellular growth during development. Cell, 98, 779–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones R.M., Branda,J., Johnston,K.A., Polymenis,M., Gadd,M., Rustgi,A., Callanan,L. and Schmidt,E.V. (1996) An essential E box in the promoter of the gene encoding the mRNA cap-binding protein (eukaryotic initiation factor 4E) is a target for activation by c-myc. Mol. Cell. Biol., 16, 4754–4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Li,Q., Dang,C.V. and Lee,L.A. (2000) Induction of ribosomal genes and hepatocyte hypertrophy by adenovirus-mediated expression of c-Myc in vivo. Proc. Natl Acad. Sci. USA, 97, 11198–11202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal A. et al. (1999) A public database for gene expression in human cancers. Cancer Res., 59, 5403–5407. [PubMed] [Google Scholar]
- Lasorella A., Noseda,M., Beyna,M. and Iavarone,A. (2000) Id2 is a retinoblastoma protein target and mediates signalling by Myc oncoproteins. Nature, 407, 592–598. [DOI] [PubMed] [Google Scholar]
- Lutz W., Stohr,M., Schurmann,J., Wenzel,A., Lohr,A. and Schwab,M. (1996) Conditional expression of N-myc in human neuroblastoma cells increases expression of α-prothymosin and ornithine decarboxylase and accelerates progression into S-phase early after mitogenic stimulation of quiescent cells. Oncogene, 13, 803–812. [PubMed] [Google Scholar]
- Malynn B.A., de Alboran,I.M., O’Hagan,R.C., Bronson,R., Davidson,L., DePinho,R.A. and Alt,F.W. (2000) N-myc can functionally replace c-myc in murine development, cellular growth and differentiation. Genes Dev., 14, 1390–1399. [PMC free article] [PubMed] [Google Scholar]
- Mateyak M.K., Obaya,A.J., Adachi,S. and Sedivy,J.M. (1997) Phenotypes of c-Myc-deficient rat fibroblasts isolated by targeted homologous recombination. Cell Growth Differ., 8, 1039–1048. [PubMed] [Google Scholar]
- Obaya A.J., Mateyak,M.K. and Sedivy,J.M. (1999) Mysterious liaisons: the relationship between c-Myc and the cell cycle. Oncogene, 18, 2934–2941. [DOI] [PubMed] [Google Scholar]
- Olson M.O. (1991) The role of protein in nucleolar structure and function. In Strauss,R. and Wilson,S.H. (eds), The Eukaryotic Nucleus. Telford Press, Caldwell, NJ, pp. 541–546.
- Perez-Roger I., Kim,S.H., Griffiths,B., Sewing,A. and Land,H. (1999) Cyclins D1 and D2 mediate myc-induced proliferation via sequestration of p27(Kip1) and p21(Cip1). EMBO J., 18, 5310–5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenwald I.B., Rhoads,D.B., Callanan,L.D., Isselbacher,K.J. and Schmidt,E.V. (1993) Increased expression of eukaryotic translation initiation factors eIF-4E and eIF-2α in response to growth induction by c-myc. Proc. Natl Acad. Sci. USA, 90, 6175–6178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders J., Brandsma,M., Janssen,G.M., Dijk,J. and Moller,W. (1996) Immunofluorescence studies of human fibroblasts demonstrate the presence of the complex of elongation factor-1βγδ in the endoplasmic reticulum. J. Cell Sci., 109, 1113–1117. [DOI] [PubMed] [Google Scholar]
- Schmidt E.V. (1999) The role of c-myc in cellular growth control. Oncogene, 18, 2988–2996. [DOI] [PubMed] [Google Scholar]
- Schwab M., Alitalo,K., Klempnauer,K.H., Varmus,H.E., Bishop,J.M., Gilbert,F., Brodeur,G., Goldstein,M. and Trent,J. (1983) Amplified DNA with limited homology to myc cellular oncogene is shared by human neuroblastoma cell lines and a neuroblastoma tumour. Nature, 305, 245–248. [DOI] [PubMed] [Google Scholar]
- Seeger R.C., Brodeur,G.M., Sather,H., Dalton,A., Siegel,S.E., Wong,K.Y. and Hammond,D. (1985) Association of multiple copies of the N-myc oncogene with rapid progression of neuroblastomas. N. Engl. J. Med., 313, 1111–1116. [DOI] [PubMed] [Google Scholar]
- Steiner P., Philipp,A., Lukas,J., Godden-Kent,D., Pagano,M., Mittnacht,S., Bartek,J. and Eilers,M. (1995) Identification of a Myc-dependent step during the formation of active G1 cyclin–cdk complexes. EMBO J., 14, 4814–4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velculescu V.E., Zhang,L., Vogelstein,B. and Kinzler,K.W. (1995) Serial analysis of gene expression. Science, 270, 484–487. [DOI] [PubMed] [Google Scholar]
- Velculescu V.E., Zhang,L., Zhou,W., Vogelstein,J., Basrai,M.A., Bassett,D.E.,Jr, Hieter,P., Vogelstein,B. and Kinzler,K.W. (1997) Characterization of the yeast transcriptome. Cell, 88, 243–251. [DOI] [PubMed] [Google Scholar]
- Versteeg R., Noordermeer,I.A., Kruse-Wolters,M., Ruiter,D.J. and Schrier,P.I. (1988) c-myc down-regulates class I HLA expression in human melanomas. EMBO J., 7, 1023–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss W.A., Aldape,K., Mohapatra,G., Feuerstein,B.G. and Bishop,J.M. (1997) Targeted expression of MYCN causes neuroblastoma in transgenic mice. EMBO J., 16, 2985–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedmann B., Sakai,H., Davis,T.A. and Wiedmann,M. (1994) A protein complex required for signal-sequence-specific sorting and trans location. Nature, 370, 434–440. [DOI] [PubMed] [Google Scholar]
- Wool I., Chan,Y.L. and Glück,A. (1996) Mammalian ribosomes: the structure and the evolution of proteins. In Hershey,J., Mathews,M. and Sonenberg,N. (eds), Translational Control. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 685–732.
- Zhang L., Zhou,W., Velculescu,V.E., Kern,S.E., Hruban,R.H., Hamilton,S.R., Vogelstein,B. and Kinzler,K.W. (1997) Gene expression profiles in normal and cancer cells. Science, 276, 1268–1272. [DOI] [PubMed] [Google Scholar]