Abstract
A crucial step in transcription is the recruitment of RNA polymerase to promoters. In the transcription of human rRNA genes by RNA Polymerase I (Pol I), transcription factor SL1 has a role as the essential core promoter binding factor. Little is known about the mechanism by which Pol I is recruited. We provide evidence for an essential role for hRRN3, the human homologue of a yeast Pol I transcription factor, in this process. We find that whereas the bulk of human Pol I complexes (Iα) are transcriptionally inactive, hRRN3 defines a distinct subpopulation of Pol I complexes (Iβ) that supports specific initiation of transcription. Human RRN3 interacts directly with TAFI110 and TAFI63 of promoter-selectivity factor SL1. Blocking this connection prevents recruitment of Pol I β to the rDNA promoter. Furthermore, hRRN3 can be found in transcriptionally autonomous Pol I holoenzyme complexes. We conclude that hRRN3 functions to recruit initiation-competent Pol I to rRNA gene promoters. The essential role for hRRN3 in linking Pol I to SL1 suggests a mechanism for growth control of Pol I transcription.
Keywords: growth control/holoenzyme/nucleolus/rRNA/transcription
Introduction
Much of the regulation of eukaryotic gene expression is at the level of initiation of transcription. RNA polymerases themselves lack sequence-specific DNA-binding properties. A fundamental step in gene activation, therefore, is the assembly of transcription pre-initiation complexes at the gene promoter that ultimately lead to the recruitment of the enzyme. How is specificity of RNA polymerase recruitment achieved?
In prokaryotes, the interaction between a distinct σ factor and bacterial core RNA polymerase confers promoter selectivity to the resulting holoenzyme (Gross et al., 1992). In eukaryotes, three nuclear RNA polymerases (Pol I, II and III) are responsible for the expression of distinct sets of genes. The mechanisms of specific recruitment are known for Pol II and Pol III. In the transcription of protein-coding genes by Pol II, the general transcription factor TFIIB was found to be essential in linking Pol II to the core promoter binding factor TFIID, a TBP–TAF (TBP-associated factor) complex (for review see Orphanides et al., 1996). It was subsequently recognized that the general transcription factor TFIIIB, also an assembly of TBP and TAFs, determines Pol III recruitment and, interestingly, contains a TFIIB-related protein (BRF) which contacts subunits of Pol III (Chedin et al., 1998; Kumar et al., 1998). Archaea also possess a homologue of the eukaryotic TFIIB, termed TFB, which functions together with the archaeal TBP to direct transcription by RNA polymerase (Bell and Jackson, 1998). So, what determines recruitment of Pol I to the rRNA gene promoters in mammalian cells?
In human cell-free systems, two transcription factors have been identified that are required for high levels of accurate initiation of transcription from the rRNA gene promoter: (i) selectivity factor SL1, a TBP–TAFI complex essential for transcription initiation (Learned et al., 1985; Bell et al., 1988; Comai et al., 1992, 1994; Zomerdijk et al., 1994; Beckmann et al., 1995; Zomerdijk and Tjian, 1998) and (ii) upstream binding factor (UBF), a multiple HMG box-containing architectural protein and activator of Pol I transcription (Learned et al., 1986; Bell et al., 1988; Jantzen et al., 1990; Reeder et al., 1995). These two transcription factors were found to interact cooperatively, and support efficient recruitment and initiation of transcription by Pol I (Bell et al., 1988). Given the genic and functional conservation of the TFIIB proteins in transcriptional mechanisms in Archaea and eukaryotes, it came as somewhat of a surprise not to find polypeptides with homology to TFIIB in the cloned subunits of SL1 (Comai et al., 1994; Zomerdijk et al., 1994). Therefore, it remained unclear whether the crucial link between SL1 and Pol I was direct or perhaps mediated by another general transcription factor.
Here, we have identified and functionally defined a factor essential for the recruitment of Pol I to rRNA gene promoters. Human RRN3 is the homologue of a transcription factor essential for Pol I-dependent transcription in Saccharomyces cerevisiae. In yeast, Rrn3p is associated with a small fraction of Pol I complexes that is competent for specific initiation of transcription, and is involved in growth control of Pol I transcription (Yamamoto et al., 1996; Keener et al., 1998; Milkereit and Tschochner, 1998). Recent complementation studies in yeast with a human homologue of RRN3 have shown that RRN3 is functionally conserved between yeast and humans (Moorefield et al., 2000). However, the function of RRN3 remained unclear. We demonstrate that hRRN3 interacts directly with SL1 and, in addition, defines a distinct subpopulation of transcriptionally active and initiation-competent Pol I. Thus, hRRN3 connects the polymerase to the promoter-selectivity factor SL1.
Results
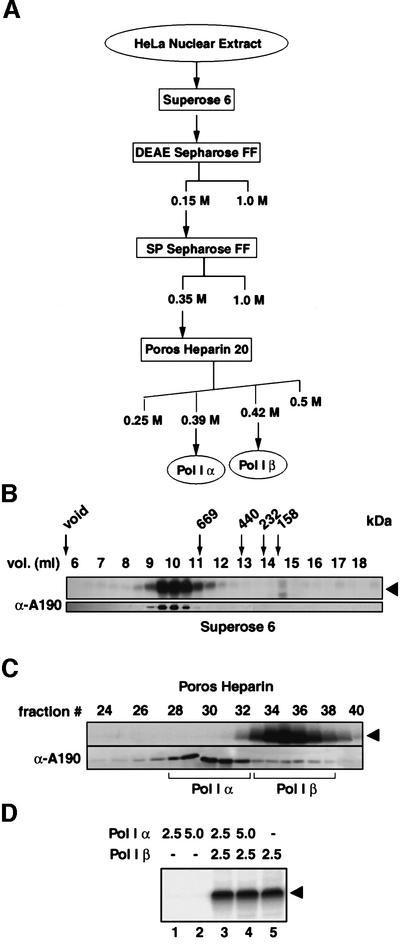
Human Pol I is found in a complex of >1 MDa
In the human cell-free system used for this study, the transcription factors SL1 and UBF were well defined and characterized, while the molecular composition of Pol I and possible associated factors have remained elusive. In order to study and understand the essential interactions during recruitment of Pol I by SL1, we extensively purified Pol I from HeLa cell nuclear extracts as outlined in Figure 1A. The Superose 6 step in the purification separates the three components that reconstitute accurate and efficient transcription initiation from the rDNA promoter. The majority of Pol I activity, as assayed in a reconstituted transcription assay with rDNA promoter, SL1 and UBF, eluted at 9.5–11 ml from the Superose 6 size-exclusion column (Figure 1B, top panel). The presence of Pol I activity in the fractions correlated with detection of the largest subunit of human Pol I by a specific antiserum (Figure 1B, bottom panel). The Superose 6 fractions did not support transcription in the absence of SL1. Extrapolating from the size standards run in parallel, we estimate that the majority of Pol I is in a complex of >1 MDa. No significant alteration in the elution profile could be detected in experiments where we fractionated the nuclear extract at salt concentrations as high as 0.5 M KCl, and the Pol I retention time at different flow rates remained constant (data not shown), suggesting that Pol I is part of a relatively stable and ‘globular’ complex.
Fig. 1. Two functionally distinct forms of human Pol I. (A) Schematic outline of the procedure for the purification of Pol I from HeLa cell nuclear extract. (B) Pol I size fractionated as a complex of >1 MDa. Samples from fractions (0.5 ml) of the Superose 6 column were tested in a specific transcription assay with the rDNA promoter, supplemented with SL1 and UBF, and transcripts were detected by S1 nuclease protection (arrowhead). The largest subunit of human Pol I, hA190, was detected in the same elution volumes from the Superose 6 column in immunoblots with anti-A190 antibodies. Size standards for the Superose 6 column are indicated above the lanes. (C) Poros Heparin columns separate Pol I α and β. The bulk of Pol I, Pol I α, is in fractions 28–32 as revealed with anti-A190 antibodies on immunoblots (bottom panel). The second peak of Pol I, Pol I β in fractions 34–36, constitutes a minor fraction of the total ‘soluble’ Pol I, yet in a reconstituted transcription assay with rDNA, SL1 and UBF support specific initiation of transcription (top panel; arrowhead). Pol I α is inactive in that same assay. (D) Mixing of Pol I α and β peak fractions. Pooled peak fractions of Pol I α and β (in µl) from the Poros Heparin column were tested, separately (lanes 1, 2 and 5) or mixed (lanes 3 and 4) as indicated above the lanes, for their ability to support specific initiation of transcription with SL1, UBF and rDNA (arrowhead).
Two functionally distinct Pol I enzyme complexes
The peak fractions from the Superose 6 column that displayed Pol I activity were pooled and subjected to chromatography on DEAE and SP Sepharose ion- exchange columns. When subsequently subjected to affinity chromatography on Poros Heparin, Pol I fractionated in two closely eluting forms, named Pol I α and β. Pol I β (in fractions 33–38 at 0.42 M KCl) supported specific initiation of transcription on the rDNA promoter when supplemented with UBF and SL1 (Figure 1C, top panel). Immunodetection of the largest subunit of Pol I indicated that Pol I β constituted <10% of the total soluble pool of Pol I (Figure 1C, bottom panel), and similarly the non-specific transcription activity for Pol I β was ∼10% of that of total Pol I (data not shown). The majority of Pol I, Pol I α (in fractions 28–32 at 0.39 M KCl), did not support specific initiation on a rRNA gene promoter template (Figure 1C, top panel), yet displayed non-specific RNA synthesis activity on sheared calf thymus DNA. We propose that the Pol I α fraction is distinct from the DNA-bound polymerases engaged in elongation of transcription, which appear not to extract from the nuclei under the conditions used. Mixing of Pol I α with Pol I β neither inhibited the activity of Pol I β nor activated Pol I α (Figure 1D), suggesting that the inability of Pol I α to support specific initiation is not due either to the presence of an excess of a trans-acting inhibitor in the Pol I α fractions or to the lack of a trans-activator.
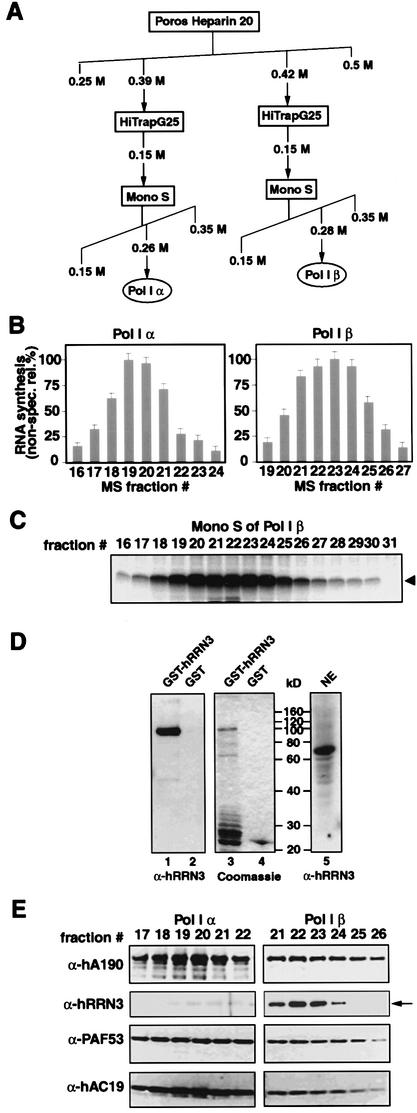
hRRN3 co-fractionates with the initiation-competent form of Pol I, Pol I β
We cloned the human homologue of RRN3 (see Materials and methods), which shows no sequence similarity to any other known gene in the databases, and studied its function in Pol I-dependent transcription. We demonstrate that hRRN3 is associated exclusively with the initiation-competent form of human Pol I, Pol I β.
The two forms of human Pol I from the Poros Heparin column were purified further by subjecting them separately to chromatography on a Mono S column (Figure 2A). Pol I elution was followed in a non-specific transcription assay with fractions from this column (Figure 2B). A peak of Pol I α activity appeared in fractions 18–21 (at 260 mM KCl; Figure 2B, left panel), and of Pol I β in fractions 21–24 (at 280 mM KCl; Figure 2B, right panel). It should be noted that the ratio of Pol I α and β, as found in the earlier steps of the purification (see Figure 1B), was the same at each chromatographic step, with Pol I α ∼10 times more abundant than Pol I β, indicating that this purification did not convert one form into the other. As we have shown for the cruder fractions of Pol I α (Figure 1C), the Mono S fractions for this polymerase complex were inactive on the rDNA promoter in a reconstituted transcription assay containing SL1 and UBF (data not shown). In contrast, the Pol I β fractions from the Mono S column support high levels of specific initiation of transcription (Figure 2C).
Fig. 2. hRRN3 is specifically associated with promoter- and initiation-competent Pol I β. (A) Schematic outline of the purification of Pol I α and β. (B) Mono S (MS) fractions were assayed in a non-specific transcription assay with calf thymus DNA, and the activities were expressed as a percentage of maximal activity for each form of Pol I. Note that the ratio of Pol I α and β remained the same throughout the purification procedure, with Pol I α ∼10 times more abundant than Pol I β. (C) Fractions from the Mono S column for Pol I β were assayed in a reconstituted transcription assay with SL1, UBF and rDNA promoter (arrowhead). (D) Affinity-purified anti-peptide antibodies raised against hRRN3 react in immunoblots with both an Escherichia coli-expressed recombinant GST–hRRN3 fusion protein of 100 kDa (lane 1) and a single protein of 74 kDa, the predicted molecular weight for hRRN3, in HeLa cell nuclear extract (lane 5). (E) hRRN3 is specific for Pol I β, and is lacking in Pol I α. Immunoblots of peak fractions for Pol I α and Pol I β from the Mono S columns were probed with anti-human A190 (Pol I), anti-human RRN3, anti-mouse PAF53 and anti-human AC19 (subunit shared by Pol I and III) antibodies.
Anti-peptide antibodies to hRRN3 were raised, peptide affinity purified and tested for specificity (Figure 2D). The antibodies specifically bind to a 74 kDa protein in HeLa nuclear extracts, the predicted size for hRRN3 (Figure 2D, lane 5). The antibodies also recognize a recombinant glutathione S-transferase (GST)–hRRN3 fusion protein (Figure 2D, compare lanes 1 and 3). Having validated their specificity, protein blots of peak fractions from the Mono S column of Pol I α and β were probed for the presence of hRRN3. Clearly, a fraction of the cellular hRRN3 co-fractionated precisely and exclusively with Pol I β (Figure 2E). Although Pol I α and β can be separated on Heparin and Mono S columns, they elute at salt concentrations very close to each other. It is, therefore, not surprising to find a weak signal for hRRN3 from the ‘tail’ of the Pol I β peak in the side-fractions of Pol I α (Figure 2E, left panel). The presence of subunits of the core polymerase in the Pol I fractions was confirmed with an antibody specific for the largest subunit of human Pol I (A190), an antibody specific for mouse polymerase associated factor 53 (PAF53), which cross-reacts with human PAF53, and with an antibody that we have raised against human AC19, a small polymerase subunit shared by Pol I and Pol III (Figure 2E).
Note that we have named the two Pol I forms Pol I α and β to distinguish them from the previously described Pol I species in mammalian cells (Pol IA and Pol IB) (Gissinger et al., 1974; Schwartz and Roeder, 1974; Matsui et al., 1976) for two reasons. First, Pol I α and β were recovered from salt-extracted nuclei without nuclear disintegration, and therefore were likely to represent primarily the ‘free’ pool of Pol I, whereas in the previous studies nuclei and nucleoli were sonicated in the presence of salt, and under those conditions both ‘free’ and ‘transcriptionally engaged’ polymerases had been recovered. Secondly, Pol IA, when compared with Pol IB, lacked the third largest subunit, PAF53 (Hanada et al., 1996), whereas PAF53 is present in both Pol I α and β (see Figure 2E).
Hence, the co-fractionation of hRRN3 through multiple chromatographic steps with the initiation-competent form of Pol I, Pol I β, and not with Pol I α, which shares many of the same core polymerase subunits, suggests a specific and tight association of hRRN3 with the Pol I β complex and, therefore, a role for this factor in accurate transcription initiation on the rDNA promoter.
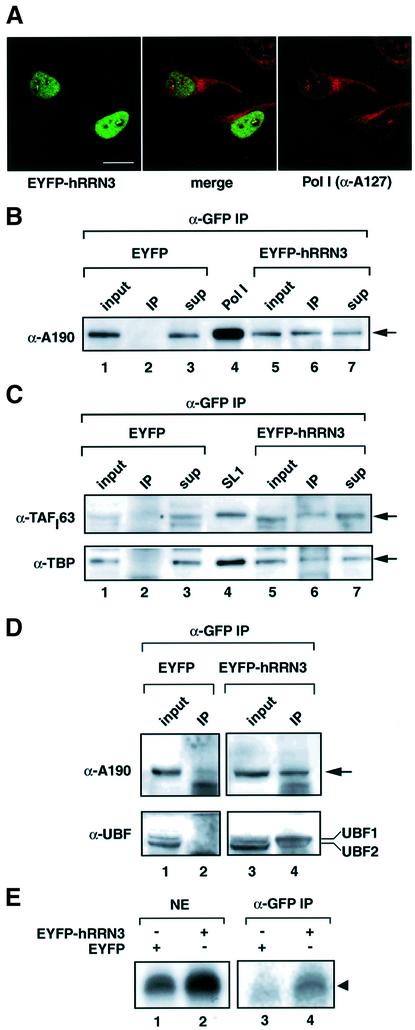
hRRN3 in sub-nucleolar structures co-localizes with Pol I
Pol I-dependent transcription is confined to a specific subcompartment of the nucleus, the nucleolus. To investigate the localization of hRRN3 in vivo, hRRN3 was expressed as an enhanced yellow fluorescent protein (EYFP) fusion in HeLa cells. EYFP–hRRN3 was observed as bright dots in nucleoli and as a diffuse signal in the nucleoplasm (Figure 3A, left panel). An antibody specific for the second largest subunit of Pol I (A127) detected both forms of the enzyme, and a co-localization of nucleolar hRRN3 with a fraction of Pol I in substructures in the nucleolus can be observed (Figure 3A, right and middle panel).
Fig. 3. hRRN3 co-localizes and co-immunoprecipitates with Pol I and is found in a complex with SL1 and UBF. (A) Imaging of EYFP–hRRN3 expression (green) in transfected HeLa cells and of the Pol I second largest subunit (A127, red), reveals co-localization in sub-nucleolar structures (merged image in the middle, where yellow indicates co-localization). Scale bar, 10 µm. (B) Extracts (2.7 mg) from HEK293 cells transfected with EYFP (lanes 1–3) and EYFP–hRRN3 (lanes 5–7) expression constructs were immunoprecipitated with anti-GFP antibodies. Immunocomplexes were subjected to SDS–PAGE, immunoblotted and probed with an antibody specific for human A190 (Pol I largest subunit). Forty micrograms of the input (lanes 1 and 5) and supernatant (sup) after the immunoprecipitations (lanes 3 and 7) were loaded, and these, therefore, represent ∼1.5% of the total protein subjected to immunoprecipitation. As a marker, we loaded highly purified Pol I (lane 4). Essentially the same immunoprecipitation results were obtained (data not shown) when the precipitations were performed in the presence of high concentrations (200 µg/ml) of ethidium bromide (Lai and Herr, 1992). (C) The immunoprecipitates of (B) were analysed for SL1 subunits with antibodies specific for TAFI63 (Comai et al., 1994; Zomerdijk et al., 1994) and a mouse monoclonal antibody, SL39, against human TBP, both at 1:1000. Input and supernatant are as in (B). Highly purified SL1 was loaded as a marker (lane 4). Note the slightly different mobilities of SL1 subunits in the relatively pure protein samples (lanes 4 and 6) compared with those in complex protein mixtures (lanes 5 and 7). (D) The immunoprecipitates of (B) were analysed in parallel for Pol I largest subunit A190, and UBF1 and 2. Anti-UBF antibodies (1:1000) were in a rabbit polyclonal serum. (E) Nuclear extracts prepared from HEK293 cells transfected with EYFP and EYFP–hRRN3 expression plasmids were tested for specific transcription initiation activity on the rDNA promoter (lanes 1 and 2). Anti-GFP-immunoprecipitated complexes from these nuclear extracts were transcriptionally active upon the addition of rRNA gene promoter template DNA and ribonucleoside triphosphates (lane 4). In the absence of added rDNA template, no transcription was observed (data not shown) and the immunoprecipitate from the EYFP-transfected cells was inactive (lane 3).
hRRN3 in Pol I holoenzymes including SL1 and UBF
When EYFP–hRRN3 in extracts from transfected HEK293 cells was immunoprecipitated with an antibody specific for the fluorescent protein tag, a small fraction of Pol I (∼1.5%) was found associated with hRRN3 (Figure 3B, lane 6). In the control experiment with EYFP-transfected cells, this co-immunoprecipitation was not observed (Figure 3B, lane 2). Therefore, these results are in agreement with the in vivo co-localization and the chromatographic co-fractionation of hRRN3 with Pol I, and taken together the data suggest strongly that hRRN3 is tightly associated with Pol I. Interestingly, a small fraction of SL1 co-immunoprecipitated with hRRN3 (Figure 3C, compare lanes 6 and 2), and UBF1, though little of UBF2, was also found in a complex with hRRN3 (Figure 3D, compare lanes 4 and 2). The transfected EYFP–hRRN3 fusion protein stimulated Pol I transcription in extracts derived from these cells (up to 5-fold stimulation), suggesting a positive function for hRRN3 in transcription by Pol I (Figure 3E, lanes 1 and 2). Remarkably, the anti-green fluorescent protein (GFP)-immunoprecipitated complexes from the EYFP–hRRN3-transfected cells were able to support specific initiation of transcription when rDNA and nucleotides were provided to these complexes, which were still bound to immobilized antibodies (Figure 3E, compare lanes 4 and 3). Thus, EYFP–hRRN3 is functional, and these results suggest the presence of immunopurified, transcriptionally autonomous protein assemblies, which are characteristic for Pol I holoenzyme complexes (SaezVasquez and Pikaard, 1997; Seither et al., 1998; Albert et al., 1999; Hannan et al., 1999).
hRRN3 interacts with SL1
Next we asked whether or not hRRN3 could bind SL1 and/or UBF directly. To study such interactions, we used an affinity resin of GST–hRRN3 purified on glutathione– Sepharose from Escherichia coli extracts (Figure 2D, lane 3). No detectable direct interaction between hRRN3 and highly purified and recombinant UBF1 could be observed (data not shown). Interestingly, highly purified human SL1 (see Materials and methods; J.K.Friedrich and J.C.B.M.Zomerdijk, unpublished data) was retained specifically on this affinity resin, suggesting a direct interaction between hRRN3 and SL1 in the absence of Pol I (Figure 4A, compare lanes 4 and 2). The direct interaction between hRRN3 and SL1 was further substantiated in an experiment where we first immunoprecipitated SL1 with anti-TBP monoclonal antibodies from the already highly purified SL1 fraction, and used this as an affinity resin to capture FLAG-peptide affinity-purified, radiolabelled hRRN3 produced in reticulocyte lysates. Indeed, hRRN3 showed a significant interaction with the anti-TBP resin pre-incubated with SL1 (Figure 4B, compare lanes 4 and 3), under conditions where no specific interaction between radiolabelled luciferase and immunocomplexed SL1 was detectable (Figure 4B, lane 5). Incubation of radiolabelled hRRN3 with renatured SL1 on a PVDF membrane revealed an interaction between hRRN3 and two polypeptides in the SL1 fraction. These proteins were identified with SL1 subunit-specific antibodies as TAFI110 and TAFI63 (Figure 4C). Consistent with this observed direct interaction, GST affinity chromatography showed binding of these TAFI subunits specifically to the GST–hRRN3 fusion protein.
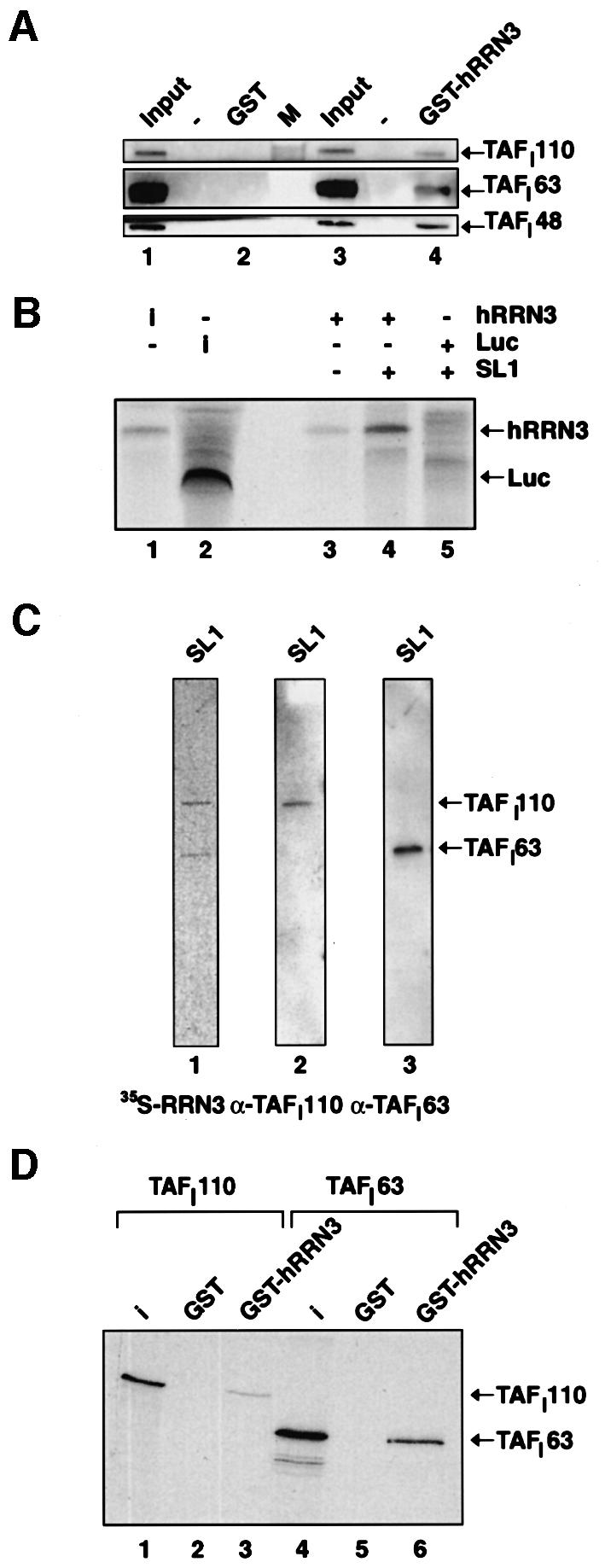
Fig. 4. hRRN3 interacts with SL1. (A) Highly purified SL1 (see Materials and methods) specifically interacts with recombinant and purified GST–hRRN3, as revealed by immunoblotting of the relevant strips of the immunoblot with antibodies specific for three subunits of SL1, TAFI110, TAFI63 and TAFI48. (B) hRRN3 interacts with SL1, which had been immunoprecipitated with antibodies specific for TBP. FLAG-epitope affinity-purified, 35S-radiolabelled hRRN3 (10% of input in lane 1) and luciferase (10% of input, lane 2) were incubated with SL1 immobilized via a TBP antibody to protein G–Sepharose beads (lanes 4 and 5). As an additional control, hRRN3 was added to antibody-loaded beads without SL1 (lane 3). Bound proteins were subjected to SDS–PAGE. The gel was fixed, dried and subjected to autoradiography. (C) FLAG-tag affinity-purified [35S]hRRN3 specifically interacts with two subunits of SL1, TAFI110 and TAFI63 in a far-western blot of highly purified SL1 (lane 1). The blot was probed with antibodies specific for TAFI110 (lane 2) and TAFI63 (lane 3), confirming their identity. (D) GST–hRRN3 interacts with two subunits of SL1. GST (lane 2 and 5) and GST–hRRN3 (lanes 3 and 6) on glutathione beads were incubated with in vitro translated [35S]methionine-labelled TAFI110 and TAFI63, and after extensive washing the resulting protein complexes were resolved by SDS–PAGE and autoradiography. Ten per cent of the TAFI110 and TAFI63 inputs are shown in lanes 1 and 4, respectively.
hRRN3 is essential for the recruitment of Pol I by SL1 to the rDNA promoter
Recently, in a yeast two-hybrid analysis, an interaction between yeast Rrn3 and Rrn6, a component of yeast core factor (CF), has been described (Peyroche et al., 2000). CF fulfils a function similar to metazoan SL1, and therefore, taken together with the data presented here, the functionally conserved RRN3 protein (Moorefield et al., 2000) may serve to recruit Pol I to the promoter by interacting with a promoter-selective transcription factor in yeast and human. Here, we have tested this directly for mammalian Pol I-dependent transcription. We asked whether or not antibodies specific for hRRN3 could inhibit the recruitment of Pol I by SL1 on an immobilized rDNA promoter template. No significant Pol I binding to the template could be observed in the absence of SL1 or with Pol I α (data not shown). In a comparison with control antibodies (Figure 5A, lane 1), affinity-purified anti-hRRN3 antibodies clearly blocked the recruitment of Pol I β to SL1-bound promoter DNA in a dose-dependent manner (Figure 5A, lanes 2 and 3). The loss of recruitment of Pol I β to promoter-bound SL1 could be a consequence of the hRRN3 antibody interfering with the interaction between hRRN3 and SL1. To study this, we affinity purified radiolabelled hRRN3 and tested it for its ability to bind promoter-bound SL1. A specific interaction between promoter-bound SL1 and hRRN3 was observed (Figure 5B, lanes 3 and 4). This is consistent with the interaction we observed between GST–hRRN3 and free SL1 (see Figure 4A, lane 4), and between affinity-purified, radiolabelled hRRN3 and immunoprecipitated SL1 (see Figure 4B, lane 4). Human RRN3 did not bind DNA in the absence of SL1 (Figure 5B, lane 5). Importantly, whereas control antibodies had no effect (Figure 5B, lane 6), anti-hRRN3 antibodies completely blocked the binding of hRRN3 to SL1 at the promoter (Figure 5B, lane 7). Protein blots confirmed that SL1 was present on the immobilized promoter template (Figure 5C, lanes 2 and 3) and that the antibody-induced loss of hRRN3 binding to SL1 on the immobilized template was not due to loss of SL1 from the template (Figure 5C, lanes 4 and 5). Taken together, these results strongly suggest an essential role for hRRN3 in the recruitment of Pol I by SL1, as it provides a direct link between these components.
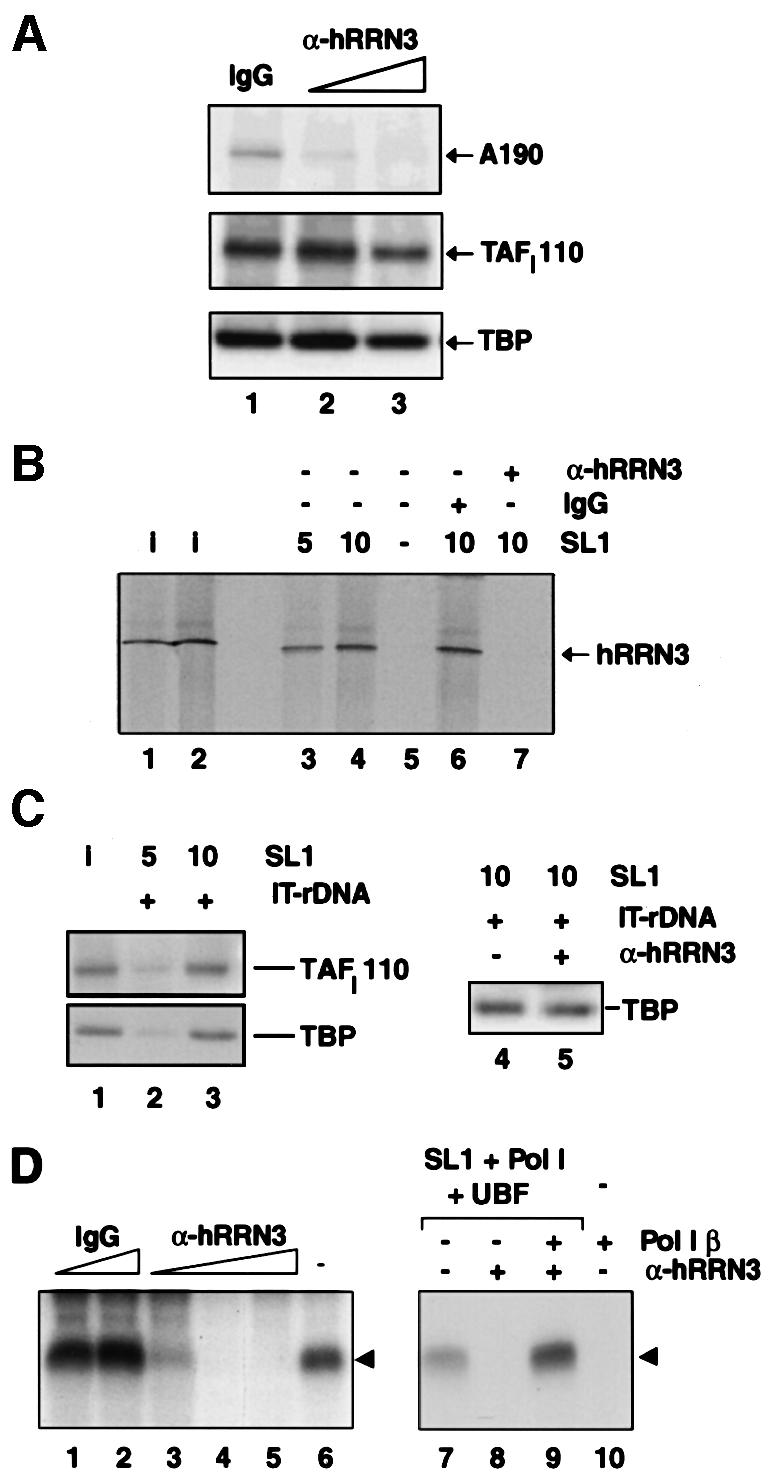
Fig. 5. hRRN3 is essential for the SL1-mediated recruitment of Pol I to the rDNA promoter. (A) Affinity-purified antibodies against hRRN3, in a dose-dependent manner, prevent the recruitment of Pol I β to SL1, which was pre-bound to the rDNA promoter. Immobilized rDNA promoter template DNA (IT-DNA) was pre-incubated for 30 min with highly purified SL1 and, in parallel, a 0.2 M KCl fraction from DEAE columns, named D0.2, and containing UBF and initiation-competent Pol I (Comai et al., 1992), was pre-incubated for 30 min with affinity-purified anti-hRRN3 antibodies (4 and 8 µg, lanes 2 and 3, respectively) or control sheep IgG antibodies (8 µg, lane 1). The immobilized templates were washed in TM10/0.05 M KCl to remove unbound SL1, and then added to the UBF/Pol I/antibody mixture. This reaction was incubated for a further 20 min, after which the immobilized templates were washed in TM10/0.05 M KCl. Template-bound proteins were eluted in 10 M urea at room temperature and analysed by immunoblotting with antibodies specific for human A190, TAFI110 (Comai et al., 1994; Zomerdijk et al., 1994) and TBP. (B) Antibodies specific for hRRN3 block the binding of highly purified hRRN3 to SL1 pre-bound to the rDNA promoter. Immobilized rDNA promoter template DNA was pre-incubated for 20 min with either 5 or 10 µl of highly purified SL1 (lanes 3 and 4, respectively) or without SL1 (lane 5). Templates were washed and mixed with FLAG-epitope affinity-purified, 35S-radiolabelled hRRN3. In a separate experiment, the purified 35S-radiolabelled hRRN3 had been pre-incubated for 20 min with peptide affinity-purified anti-hRRN3 antibodies (8 µg, lane 7) or IgG (8 µg, lane 6) before addition to promoter-bound SL1. The reactions were incubated for 20 min and then the templates were washed. The template-bound proteins were eluted in urea and subjected to SDS–PAGE. The gel was fixed, dried and subjected to autoradiography to reveal [35S]FLAG-hRRN3. Lanes 1 and 2 are 10 and 20%, respectively, of the input of [35S]FLAG-hRRN3. In the absence of SL1, hRRN3 did not bind the promoter DNA template (lane 5). (C) The binding of SL1 to the immobilized template (IT-rDNA) from the experiment described in (B) was verified by immunoblotting the reactions of lane 3 and 4 of (B) with antibodies specific for two subunits of SL1, TAFI110 and TBP (lanes 2 and 3). SL1 input (5 µl) is shown (lane 1). Antibodies specific for hRRN3 did not displace SL1 from the immobilized rDNA promoter template (lanes 4 and 5). Reactions were performed as described in (B) for lanes 6 and 7. (D) Inhibition of Pol I transcription with affinity-purified anti-hRRN3 antibodies (2, 4 and 8 µg, lanes 3–5, respectively), but not with control IgG (4 and 8 µg, lanes 1 and 2, respectively). The experimental procedure was as outlined in (C), except that template-bound proteins were not eluted, but rather tested for their ability to support specific initiation of transcription upon addition of ribonucleoside triphosphates in a 30 min reaction. Transcripts were detected in an S1 nuclease protection assay (arrowhead). Lane 6 is a control transcription reaction in the absence of antibodies. Supplementing a reaction with 2.5 µl of Pol I β recovers anti-hRRN3 antibody-induced inhibition of transcription. Lane 8 illustrates the inhibition of transcription with 4 µg of affinity-purified anti-hRRN3 antibodies, and add-back of Pol I β restores transcription (lane 9). Control reactions were loaded in lane 7 (no antibodies) and lane 10 (no SL1 and UBF in the reaction), illustrating that Pol I β by itself does not support specific initiation of transcription.
The anti-hRRN3 antibodies, therefore, should affect specific transcription initiation. Indeed, antibodies specific for hRRN3 inhibit Pol I transcription (Figure 5D, lanes 3–5), but do not affect non-specific transcription (data not shown). In agreement with targeting of Pol I β by the hRRN3 antibodies and interference with the Pol I recruitment, an excess of Pol I β (which by itself cannot support transcription; Figure 5D, lane 10) restored transcription initiation in the antibody-inhibited transcription reaction (Figure 5D, compare lane 9 with 8).
Discussion
The essential role of hRRN3 in the SL1-mediated recruitment of Pol I to rRNA gene promoters
We provide several lines of evidence for an essential function of the Pol I transcription factor hRRN3 in the SL1-mediated recruitment of Pol I to rRNA gene promoters. We have shown a stable, specific and selective association of hRRN3 with a subpopulation of Pol I, Pol I β, which supported SL1-dependent and promoter-directed initiation of transcription. Our results, and those in the mouse, Acanthamoeba and S.cerevisiae systems, therefore illustrate an apparent evolutionary conservation in eukaryotes of the apportionment of Pol I into two functionally distinct forms (Bateman and Paule, 1986; Tower and Sollner-Webb, 1987; Milkereit and Tschochner, 1998). In extracts from transfected cells, a tagged version of the hRRN3 protein co-immunoprecipitated with SL1, UBF1 and Pol I, and these immunopurified complexes supported accurate initiation of transcription from rDNA promoter templates. Furthermore, we have demonstrated an interaction between SL1 and hRRN3, as demonstrated in several protein–protein interaction assays where either of the interacting partners was used as an affinity resin. For example, hRRN3 was retained specifically on highly purified SL1 tethered by anti-TBP antibodies and on SL1 pre-bound to an immobilized promoter DNA template, and conversely SL1 interacted with tethered recombinant hRRN3 GST fusion proteins on glutathione–agarose. Far-western analyses supported the notion of a direct interaction of hRRN3 with SL1, since renatured TAFI110 and TAFI63 subunits of SL1 interacted with radiolabelled hRRN3, and indeed these same TAFIs interacted with GST–hRRN3 in pull-down assays. Antibodies specific for hRRN3 prevented the interaction between hRRN3 and SL1, and an excess of Pol I–hRRN3 complex (Pol I β) when added back reversed this inhibition. These experiments explain at a molecular level how the hRRN3-specific antibody both inhibits transcription initiation and abolishes the recruitment of Pol I to an SL1-engaged promoter rDNA template. Therefore, we propose that the interaction of hRRN3 with SL1 is essential for the recruitment of mammalian Pol I by SL1 at the rRNA gene promoter. This correlates well with the suggested role for yeast Rrn3p in bridging polymerase to CF (Peyroche et al., 2000), and taken together these results signify an apparent similarity in the mechanism of Pol I recruitment in yeast and metazoans.
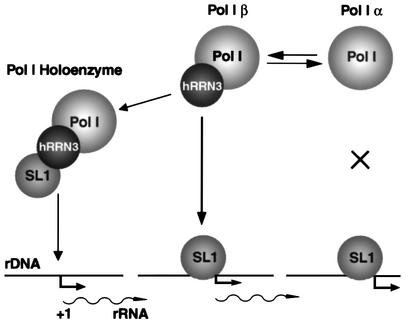
Stepwise assembly and Pol I holoenzyme recruitment
Our results are compatible with two pathways for the assembly of transcription pre-initiation complexes, as outlined in Figure 6. In one scenario, Pol I β is directly recruited to the rDNA promoter as a result of a productive interaction between polymerase-associated hRRN3 and promoter-bound SL1. In the second, an interaction between SL1 and hRRN3 in Pol I β leads to the formation of a holoenzyme complex, which may include UBF, and this pre-assembled complex then binds the rDNA promoter. Since SL1 remains at the promoter following escape by Pol I from the rDNA promoter (Panov et al., 2001), the frequent re-initiations of Pol I transcription are likely to follow the former pathway. Indeed, the predominant form of the initiation-competent form of Pol I in the nucleus is Pol I β, with only very low levels of it in holoenzyme complexes, which were detectable only after affinity purification from cell extracts that overexpressed tagged hRRN3 (Figure 3). The transcriptional autonomy of the affinity-purified complexes fits with the proper definition of polymerase holoenzymes, which support accurate initiation of transcription from promoter DNA templates in the absence of accessory factors.
Fig. 6. A model for the role of hRRN3 in productive Pol I pre-initiation complex formation at the rRNA gene promoters. hRRN3 has an essential function in linking Pol I to SL1 at the rDNA promoter and in Pol I holoenzyme complex assembly. hRRN3, specifically associated with the initiation-competent form of Pol I, Pol I β, interacts with SL1. Pol I α lacks hRRN3 and is not competent for productive interaction with SL1. The interaction of hRRN3 with SL1 may occur in solution, leading to the formation of a Pol I holoenzyme complex that displays promoter selectivity (during ‘de novo’ pre-initiation complex assembly), and/or may take place at the rDNA promoter, where pre-bound SL1 recruits Pol I β via a crucial connection with hRRN3 (during re-initiation of transcription).
Regulation of Pol I transcription by interconversion of Pol I α and β
There is evidence in S.cerevisiae to suggest a role for RRN3 in growth control, and the proposed molecular mechanism intimated a differential association of RRN3 with the initiation-competent form of Pol I (Milkereit and Tschochner, 1998). The mouse homologue of yeast RRN3 has recently been identified as TIF-IA (Bodem et al., 2000), a factor whose activity or abundance appears to be regulated under varying growth conditions (Buttgereit et al., 1985). In addition, in both mammals and yeast, a large fraction of the polymerase in the nucleus, here named Pol I α, is unable to support specific initiation, despite its ability to synthesize RNA from non-specific templates. The function for hRRN3 defined here allows us to explain at a molecular level the role of murine transcription factor TIF-IA (Buttgereit et al., 1985; Schnapp et al., 1990, 1993), otherwise known as Factor C* (Tower and Sollner-Webb, 1987; Brun et al., 1994), in growth control of Pol I-dependent transcription. We propose a comprehensive model for the regulation of rRNA gene transcription at the level of polymerase recruitment, and suggest that growth control of Pol I transcription in mammalian cells involves the regulated interconversion between the initiation-competent Pol I β and an inactive Pol I complex that lacks hRRN3, perhaps Pol I α (Figure 6). In this respect, it is interesting to note that we found a large fraction of the total cellular hRRN3 not associated with polymerases (data not shown); the nature of this hRRN3 and, perhaps, its associated factors is currently under investigation.
Stimulation of transcription initiation by recombinant hRRN3 can be observed in a nuclear extract, yet recombinant hRRN3 cannot simply convert highly purified Pol I α into a promoter-competent Pol I β (data not shown). This suggests that in mammalian cells the interconversion of Pol I α to β may involve additional factors or, perhaps, modifications in addition to and/or required for the incorporation of hRRN3 into the polymerase complex. We note that other contacts between SL1 and polypeptides in Pol I β, and between UBF and Pol I (Schnapp et al., 1994; Hanada et al., 1996), are likely to occur, but nevertheless the interaction between hRRN3 and SL1 is essential for the stable recruitment of the enzyme. Given the essential role for hRRN3 in linking the polymerase to SL1 in rRNA gene expression, the interactions between hRRN3 and SL1, and between hRRN3 and Pol I, are likely to be major targets of regulatory pathways that control ‘de novo’ assembly of Pol I pre-initiation complexes at rDNA promoters, and Pol I recruitment during re-initiation of transcription.
hRRN3, like TFIIB, provides the bridge between polymerase and the promoter-selectivity factor
A fundamental step in gene activation is the recruitment of RNA polymerase by promoter-bound factors, and general transcription factors with domains homologous to TFIIB function in this process, both in Archaea and eukaryotes. No TFIIB-like polypeptides were found in SL1 (Comai et al., 1994; Zomerdijk et al., 1994), and human and yeast RRN3 primary amino acid sequence analyses also failed to reveal homology with conserved domains in TFIIB. Yet hRRN3 emerges as functionally related to TFIIB, TFB and the BRF subunit of TFIIIB, since all of them are critically important in connecting polymerase to promoter-selectivity factors. Thus, we may define hRRN3 as the ‘missing link’ in Pol I-dependent transcription.
Materials and methods
Cloning and expression vectors of hRRN3
The original full-length cDNA clone of hRRN3 was isolated from a HeLa λTriplEx cDNA library (Clontech) and subcloned for GST, EYFP and FLAG-tagged expression (for details of identification and cloning of hRRN3 see Supplementary data, available at The EMBO Journal Online).
Antibodies, immunoblotting, immunoprecipitation and immunofluorescence
Peptides corresponding to the following sequences of hRRN3 were coupled to bovine serum albumin (BSA) (Merck BDH) and keyhole limpet haemocyanin (Calbiochem) to raise sheep antisera (Diagnostics Scotland): MAAPLLHTRLPGDAC, EKFPVRKSERTLEC and CGS PPVLYMQPSPL. The same peptides were used to affinity purify the antibodies.
Primary antibodies used were: anti-A190 (human Pol I largest subunit) antibodies (1:250), peptide-affinity purified from sheep immunized with a mixture of three peptides derived from the human A190 Pol I largest subunit (K.I.Panov and J.C.B.M.Zomerdijk, unpublished data); anti-hRRN3 affinity-purified sheep antibodies (1:1000); anti-PAF53 antibodies (1:1000) (Hanada et al., 1996); anti-AC19 antibodies (1:1000), a polyclonal sheep serum raised against recombinant human AC19 small subunit shared between Pol I and Pol III (G.Miller and J.C.B.M.Zomerdijk, unpublished data); anti-TAFI110 (1:1000) and anti-TAFI63 (1:2000) (Zomerdijk et al., 1994). Appropriate secondary antibodies conjugated to horseradish peroxidase (Jackson ImmunoResearch) were used to detect immunocomplexes on the blots by chemiluminescence (ECL; Amersham Pharmacia Biotech).
Immunoprecipitations were carried out from either whole-cell lysates or nuclear extracts. Human embryonic kidney (HEK) 293 cells were transfected using calcium phosphate with either EYFP (pEYFP-C1; Invitrogen) or EYFP–hRRN3 (pEYFP-C1–hRRN3; see Supplementary data) expression vectors. Cells were lysed 20 h post-transfection in 0.5 ml (per 10 cm plate) of 50 mM Tris–HCl pH 7.5, 0.5 M NaCl, 1% NP-40, 1% deoxycholate, 1% SDS, 2 mM EDTA, EDTA-free complete protease inhibitor cocktail (Roche). Each whole-cell lysate (2.7 mg) was pre-cleared with 10 µl of protein G–Sepharose beads (Amersham Pharmacia) for 30 min at 4°C, and then used in immunoprecipitation experiments with 10 µg of anti-GFP antibodies (Roche) bound to 25 µl of protein G–Sepharose beads for 2 h at 4°C. The beads were washed four times in 1 ml of 50 mM Tris–HCl pH 7.5, 0.25 M NaCl, and precipitates were analysed by immunoblotting. Immunoprecipitations from the nuclear extracts were performed with 50 µg of each nuclear extract, which had been pre-cleared for 30 min with 10 µl of protein G–Sepharose beads, with 4 µg of anti-GFP antibodies bound to 7.5 µl of protein G–Sepharose beads in TM10/0.4 M KCl and 0.05% NP-40 for 3 h at 4°C. TM10 buffer is: 50 mM Tris–HCl pH 7.9, 12.5 mM MgCl2, 1 mM EDTA, 10% glycerol, 1 mM sodium metabisulfite and 1 mM dithiothreitol (DTT). Beads were washed four times in 1 ml of TM10/0.4 M KCl buffer and used in transcription assays.
EYFP–hRRN3 was transfected into HeLa cells using the effectene method (Qiagen). The cells were incubated for 20 h, washed in phosphate-buffered saline (PBS), then fixed in 4% paraformaldehyde in PBS for 10 min, permeabilized with 0.1% Tween-20 in PBS for 10 min, and incubated with anti-A127 antibodies (1:50; specific for the second largest subunit of rat Pol I; Hannan et al., 1998) for 1 h at room temperature (RT). Subsequently, the slides were washed four times for 5 min in PBS at RT, incubated with anti-rabbit Texas Red secondary antibodies (1:250; Jackson ImmunoResearch) for 30 min at RT, and then washed in PBS, mounted in Mowiol/Dabco and allowed to dry. Cells were viewed using a Zeiss LSM 410 confocal laser scanning microscope.
GST-affinity chromatography
Escherichia coli BL21 (DE3) cells were transformed with pGEX-4T-3-hRRN3 (see Supplementary data) or pGEX-4T-3 (Amersham Pharmacia). Cells were grown to an OD600 nm of 0.6 and induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside for 3 h at 37°C. Cells were harvested, resuspended in buffer T/0.15 M NaCl (buffer T: 50 mM Tris–HCl pH 7.9, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride), sonicated and centrifuged at 15 000 g for 25 min at 4°C. The bacterial lysate was bound to 1 ml of glutathione–Sepharose beads (Amersham Pharmacia Biotech) for 1 h at 4°C. Beads were washed three times at 4°C for 10 min with 20 ml of buffer T/0.15 M NaCl, and then three times for 10 min with 20 ml of buffer T/1.0 M NaCl. Twenty microlitres of beads were used as affinity resin in 600 µl of TM10/0.15 M KCl. The beads were blocked with 20 µg of BSA for 1 h, and then highly purified SL1 was added and incubated for 2 h at 4°C. The beads were then washed in 600 µl of TM10/0.2 M KCl for 5 min at 4°C, followed by washes in TM10/0.25, TM10/0.3 and TM10/0.35 M KCl buffers. Bound proteins were analysed by immunoblotting.
TAFI110 and TAFI63 were in vitro translated (Comai et al., 1994) and independently incubated with GST– or GST–hRRN3–beads. The pull-down with GST or GST–hRRN3 was carried out in 20 mM Tris–HCl pH 7.9, 100 mM KCl, 20% glycerol, 0.2 mM EDTA, 0.5% NP-40 for 2 h, followed by four 10-min washes with the same buffer. Bound proteins were analysed by SDS–PAGE and autoradiography.
In vitro translation and affinity purification of [35S]FLAG-hRRN3
[35S]FLAG-hRRN3 was synthesized from plasmid pcDNA3-FLAG-hRRN3 (see Supplementary data; pcDNA3 was from Invitrogen) in a total reaction volume of 300 µl, in the TNT-coupled rabbit reticulocyte lysate system (Promega). Reactions were diluted with 1.2 ml of TM10/0.2 M KCl buffer. To affinity purify the protein, 25 µl of anti-FLAG M2-beads (Kodak) were added and incubated, with shaking, overnight at 4°C. The beads were washed twice in 500 µl volumes of TM10/0.05, TM10/0.3, TM10/0.6 and TM10/0.1 M KCl buffers, and eluted in 50 µl of 1 mg/ml FLAG peptide in TM10/0.1 M KCl at 4°C for 4 h.
Far-western analysis
Highly purified SL1 was subjected to SDS–PAGE and transferred to an Immobilon-P (Millipore) membrane.The membrane was probed with FLAG-purified, [35S]methionine-labelled hRRN3, as described (Kaelin et al., 1992), and with antibodies specific for TAFI110 and TAFI63 (Zomerdijk et al., 1994).
In vitro transcription assays
In vitro transcription reactions were performed as described previously (Learned et al., 1986; Bell et al., 1988) at a final salt concentration of 50–70 mM KCl. Supercoiled prHu3 plasmid DNA, which contains the human rRNA gene promoter from –515 to +1548 (Learned and Tjian, 1982), or immobilized linear DNA fragments were used as templates in the transcription reaction. The resulting transcripts were analysed in an S1 nuclease protection assay after annealing the RNA to a 5′-end-labelled oligonucleotide, which was identical to the region between –20 and +40 of the promoter template strand (Bell et al., 1988). For the non-specific transcription assay and preparation of the immobilized template see Supplementary data.
Purification of Pol I complexes and SL1 from HeLa cell nuclear extracts
Pol I was purified from HeLa cell nuclei (Figures 1 and 2) by passage over Superose 6, DEAE–Sepharose and SP Sepharose, and separated into Pol I α and β on Poros Heparin and Mono S columns. For a detailed protocol see Supplementary data. Pol I α and β were free from SL1 and UBF.
SL1 was purified from HeLa nuclear extract on heparin–agarose and S-Sepharose columns (Comai et al., 1994), followed by chromatography on Poros Heparin and Superose 6 columns (see Supplementary data). SL1 fractions were identified by in vitro transcription assays. SL1 was free from Pol I and UBF. In Figure 4B, SL1 was immunopurified with anti-TBP monoclonal antibodies (SL39; kindly provided by N.Hernandez). Protein G–Sepharose-pre-cleared SL1 was incubated overnight at 4°C with the protein G-bound, anti-TBP antibody. Beads were washed extensively with several changes of 1 ml TM10/0.3 before equilibration to TM10/0.05.
Supplementary data
Supplementary data for this paper are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank L.Rothblum, M.Muramatsu, B.McStay and N.Hernandez for kindly providing us with antibodies specific for rat second largest Pol I subunit (A127), mouse PAF53, human UBF and human TBP, respectively; C.Clark for technical assistance; G.Bloomberg (University of Bristol, UK) for peptide synthesis; and the National Cell Culture Center (Minneapolis, MN, USA) for HeLa cells. We thank our colleagues in the Zomerdijk laboratory and T.Owen-Hughes, N.D.Perkins, S.G.E.Roberts and J.Russell for advice and critical reading of the manuscript. G.M. received a Wellcome Trust Prize PhD studentship. L.T-.M. is supported by a Biotechnology and Biological Sciences Research Council grant to A.I.L. A.I.L. is a Principal Research Fellow of the Wellcome Trust. J.C.B.M.Z. is a Wellcome Trust Senior Research Fellow in the Basic Biomedical Sciences.
References
- Albert A.C., Denton,M., Kermekchiev,M. and Pikaard,C.S. (1999) Histone acetyltransferase and protein kinase activities copurify with a putative Xenopus RNA polymerase I holoenzyme self-sufficient for promoter-dependent transcription. Mol. Cell. Biol., 19, 796–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman E. and Paule,M.R. (1986) Regulation of eukaryotic ribosomal RNA transcription by RNA polymerase modification. Cell, 47, 445–450. [DOI] [PubMed] [Google Scholar]
- Beckmann H., Chen,J.L., O’Brien,T. and Tjian,R. (1995) Coactivator and promoter-selective properties of RNA polymerase I TAFs. Science, 270, 1506–1509. [DOI] [PubMed] [Google Scholar]
- Bell S.D. and Jackson,S.P. (1998) Transcription in Archaea. Cold Spring Harb. Symp. Quant. Biol., 63, 41–51. [DOI] [PubMed] [Google Scholar]
- Bell S.P., Learned,R.M., Jantzen,H.M. and Tjian,R. (1988) Functional cooperativity between transcription factors UBF1 and SL1 mediates human ribosomal RNA synthesis. Science, 241, 1192–1197. [DOI] [PubMed] [Google Scholar]
- Bodem J., Dobreva,G., Hoffmann-Rohrer,U., Iben,S., Zentgraf,H., Delius,H., Vingron,M. and Grummt,I. (2000) TIF-IA, the factor mediating growth-dependent control of ribosomal RNA synthesis, is the mammalian homolog of yeast Rrn3p. EMBO rep., 1, 171–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brun R.P., Ryan,K. and Sollner-Webb,B. (1994) Factor C*, the specific initiation component of the mouse RNA polymerase I holoenzyme, is inactivated early in the transcription process. Mol. Cell. Biol., 14, 5010–5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttgereit D., Pflugfelder,G. and Grummt,I. (1985) Growth-dependent regulation of rRNA synthesis is mediated by a transcription initiation factor (TIF-IA). Nucleic Acids Res., 13, 8165–8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chedin S., Ferri,M.L., Peyroche,G., Andrau,J.C., Jourdain,S., Lefebvre,O., Werner,M., Carles,C. and Sentenac,A. (1998) The yeast RNA polymerase III transcription machinery: a paradigm for eukaryotic gene activation. Cold Spring Harb. Symp. Quant. Biol., 63, 381–389. [DOI] [PubMed] [Google Scholar]
- Comai L., Tanese,N. and Tjian,R. (1992) The TATA-binding protein and associated factors are integral components of the RNA polymerase I transcription factor, SL1. Cell, 68, 965–976. [DOI] [PubMed] [Google Scholar]
- Comai L., Zomerdijk,J.C., Beckmann,H., Zhou,S., Admon,A. and Tjian,R. (1994) Reconstitution of transcription factor SL1: exclusive binding of TBP by SL1 or TFIID subunits. Science, 266, 1966–1972. [DOI] [PubMed] [Google Scholar]
- Gissinger F., Kedinger,C. and Chambon,P. (1974) Animal DNA-dependent RNA polymerases. 10. General enzymatic properties of purified calf thymus RNA polymerases AI and B. Biochimie, 56, 319–333. [DOI] [PubMed] [Google Scholar]
- Gross C.A., Lonetto,M. and Losick,R. (1992) Bacterial sigma factors. In McKnight,S.L. and Yamamoto,K.R. (eds), Transcriptional Regulation. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 129–176. [Google Scholar]
- Hanada K., Song,C.Z., Yamamoto,K., Yano,K., Maeda,Y., Yamaguchi,K. and Muramatsu,M. (1996) RNA polymerase I associated factor 53 binds to the nucleolar transcription factor UBF and functions in specific rDNA transcription. EMBO J., 15, 2217–2226. [PMC free article] [PubMed] [Google Scholar]
- Hannan R.D., Hempel,W.M., Cavanaugh,A., Arino,T., Dimitrov,S.I., Moss,T. and Rothblum,L. (1998) Affinity purification of mammalian RNA polymerase I. Identification of an associated kinase. J. Biol. Chem., 273, 1257–1267. [DOI] [PubMed] [Google Scholar]
- Hannan R.D., Cavanaugh,A., Hempel,W.M., Moss,T. and Rothblum,L. (1999) Identification of a mammalian RNA polymerase I holoenzyme containing components of the DNA repair/replication system. Nucleic Acids Res., 27, 3720–3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jantzen H.M., Admon,A., Bell,S.P. and Tjian,R. (1990) Nucleolar transcription factor hUBF contains a DNA-binding motif with homology to HMG proteins. Nature, 344, 830–836. [DOI] [PubMed] [Google Scholar]
- Kaelin W.G. Jr et al. (1992) Expression cloning of a cDNA encoding a retinoblastoma-binding protein with E2F-like properties. Cell, 70, 351–364. [DOI] [PubMed] [Google Scholar]
- Keener J., Josaitis,C.A., Dodd,J.A. and Nomura,M. (1998) Reconstitution of yeast RNA polymerase I transcription in vitro from purified components. TATA-binding protein is not required for basal transcription. J. Biol. Chem., 273, 33795–33802. [DOI] [PubMed] [Google Scholar]
- Kumar A., Grove,A., Kassavetis,G.A. and Geiduschek,E.P. (1998) Transcription factor IIIB: the architecture of its DNA complex and its roles in initiation of transcription by RNA polymerase III. Cold Spring Harb. Symp. Quant. Biol., 63, 121–129. [DOI] [PubMed] [Google Scholar]
- Lai J.S. and Herr,W. (1992) Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proc. Natl Acad. Sci. USA, 89, 6958–6962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Learned R.M. and Tjian,R. (1982) In vitro transcription of human ribosomal RNA genes by RNA polymerase I. J. Mol. Appl. Genet., 1, 575–584. [PubMed] [Google Scholar]
- Learned R.M., Cordes,S. and Tjian,R. (1985) Purification and characterization of a transcription factor that confers promoter specificity to human RNA polymerase I. Mol. Cell. Biol., 5, 1358–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Learned R.M., Learned,T.K., Haltiner,M.M. and Tjian,R.T. (1986) Human rRNA transcription is modulated by the coordinate binding of two factors to an upstream control element. Cell, 45, 847–857. [DOI] [PubMed] [Google Scholar]
- Matsui T., Onishi,T. and Muramatsu,M. (1976) Nucleolar DNA-dependent RNA polymerase from rat liver. 2. Two forms and their physiological significance. Eur. J. Biochem., 71, 361–368. [DOI] [PubMed] [Google Scholar]
- Milkereit P. and Tschochner,H. (1998) A specialized form of RNA polymerase I, essential for initiation and growth-dependent regulation of rRNA synthesis, is disrupted during transcription. EMBO J., 17, 3692–3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorefield B., Greene,E.A. and Reeder,R.H. (2000) RNA polymerase I transcription factor Rrn3 is functionally conserved between yeast and human. Proc. Natl Acad. Sci. USA, 97, 4724–4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orphanides G., Lagrange,T. and Reinberg,D. (1996) The general transcription factors of RNA polymerase II. Genes Dev., 10, 2657–2683. [DOI] [PubMed] [Google Scholar]
- Panov K.I., Friedrich,J.K. and Zomerdijk,J.C.B.M. (2001) A step subsequent to pre-initiation complex assembly at the rDNA promoter is rate limiting for human RNA Polymerase I-dependent transcription. Mol. Cell. Biol., 21, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyroche G., Milkereit,P., Bischler,N., Tschochner,H., Schultz,P., Sentenac,A., Carles,C. and Riva,M. (2000) The recruitment of RNA polymerase I on rDNA is mediated by the interaction of the A43 subunit with Rrn3. EMBO J., 19, 5473–5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeder R.H., Pikaard,C.S. and McStay,B. (1995) UBF, an architectural element for RNA polymerase I promoters. In Eckstein,F. and Lilley,D.M.J. (eds), Nucleic Acids and Molecular Biology. Springer-Verlag, Berlin, Germany, pp. 251–263.
- SaezVasquez J. and Pikaard,C.S. (1997) Extensive purification of a putative RNA polymerase I holoenzyme from plants that accurately initiates rRNA gene transcription in vitro. Proc. Natl Acad. Sci. USA, 94, 11869–11874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnapp A., Pfleiderer,C., Rosenbauer,H. and Grummt,I. (1990) A growth-dependent transcription initiation factor (TIF-IA) interacting with RNA polymerase I regulates mouse ribosomal RNA synthesis. EMBO J., 9, 2857–2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnapp A., Schnapp,G., Erny,B. and Grummt,I. (1993) Function of the growth-regulated transcription initiation factor TIF-IA in initiation complex formation at the murine ribosomal gene promoter. Mol. Cell. Biol., 13, 6723–6732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnapp G., Santori,F., Carles,C., Riva,M. and Grummt,I. (1994) The HMG box-containing nucleolar transcription factor UBF interacts with a specific subunit of RNA polymerase I. EMBO J., 13, 190–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz L.B. and Roeder,R.G. (1974) Purification and subunit structure of deoxyribonucleic acid-dependent ribonucleic acid polymerase I from the mouse myeloma, MOPC 315. J. Biol. Chem., 249, 5898–5906. [PubMed] [Google Scholar]
- Seither P., Iben,S. and Grummt,I. (1998) Mammalian RNA polymerase I exists as a holoenzyme with associated basal transcription factors. J. Mol. Biol., 275, 43–53. [DOI] [PubMed] [Google Scholar]
- Tower J. and Sollner-Webb,B. (1987) Transcription of mouse rDNA is regulated by an activated subform of RNA polymerase I. Cell, 50, 873–883. [DOI] [PubMed] [Google Scholar]
- Yamamoto R.T., Nogi,Y., Dodd,J.A. and Nomura,M. (1996) RRN3 gene of Saccharomyces cerevisiae encodes an essential RNA polymerase I transcription factor which interacts with the polymerase independently of DNA template. EMBO J., 15, 3964–3973. [PMC free article] [PubMed] [Google Scholar]
- Zomerdijk J.C.B.M. and Tjian,R. (1998) Structure and assembly of human selectivity factor SL1. In Paule,M.R. (ed.), Transcription of Eukaryotic Ribosomal RNA Genes by RNA Polymerase I. Springer-Verlag, Austin, TX, pp. 67–73.
- Zomerdijk J.C.B.M., Beckmann,H., Comai,L. and Tjian,R. (1994) Assembly of transcriptionally active RNA polymerase I initiation factor SL1 from recombinant subunits. Science, 266, 2015–2018. [DOI] [PubMed] [Google Scholar]