Abstract
The tra-1 and tra-2 sex-determining genes promote female fates in Caenorhabditis elegans. Classical genetic studies placed tra-1 as the terminal regulator of the pathway with tra-2 acting upstream as a regulator of regulators of tra-1. Here we report the surprising result that the TRA-1 transcription factor binds the intracellular domain of the TRA-2 membrane protein. This binding is dependent on the MX regulatory domain, a region of the TRA-2 intracellular domain shown previously to be critical for the onset of hermaphrodite spermatogenesis. The functional importance of the TRA-1–TRA-2 physical interaction is supported by genetic interactions between tra-1(0) and tra-2(mx) mutations: a reduction of tra-1 gene dose from two copies to one copy enhances the tra-2(mx) feminization phenotype, but has no apparent somatic effect. In Caenorhabditis briggsae, we also find an MX-dependent interaction between Cb-TRA-1 and Cb-TRA-2, but intriguingly, no cross-species interactions are seen. The conservation of the TRA-1– TRA-2 interaction underscores its importance in sex determination.
Keywords: Caenorhabditis briggsae/Caenorhabditis elegans/sex determination/TRA-1/TRA-2
Introduction
Caenorhabditis elegans XX animals are self-fertilizing hermaphrodites, making sperm first and then oocytes; XO animals are male. Specification as hermaphrodite or male relies initially on the X:A ratio to control a pathway of sex-determining genes to direct sexual cell fates (reviewed in Meyer, 1997). The transient generation of sperm in an otherwise female XX animal is regulated by modulating the activities of sex-determining genes that direct male or female development (Puoti et al., 1997). Of particular importance to this paper are two sex-determining genes, tra-1 and tra-2, which specify female development (Hodgkin and Brenner, 1977). We have found an unexpected physical interaction between the TRA-1 and TRA-2 proteins, which suggests a functional relationship between these two regulators that was not predicted by previous analyses.
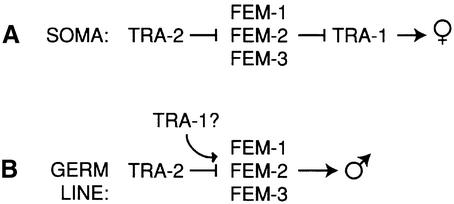
Figure 1 shows an abbreviated sex determination pathway that highlights functional relationships among genes at the end of the pathway. In somatic tissues, the pathway ultimately controls activity of TRA-1 (Hodgkin and Brenner, 1977; Hodgkin, 1986) (Figure 1A). The tra-1 gene encodes two proteins: TRA-1A with five zinc fingers and TRA-1B with only two zinc fingers (Zarkower and Hodgkin, 1992). TRA-1A is homologous to Drosophila cubitus interuptus (ci) and vertebrate GLI proteins, and is essential for tra-1 activity; no role is known for TRA-1B (Zarkower and Hodgkin, 1992). TRA-1A functions as a transcription factor (Conradt and Horvitz, 1999; Chen and Ellis, 2000), and also promotes transport of tra-2 mRNA to the cytoplasm (Graves et al., 1999). In tra-1 XX null mutants, somatic tissues are masculinized, and the germ line makes a reduced number of sperm (Hodgkin and Brenner, 1977; Hodgkin, 1987; Schedl et al., 1989). Therefore, wild-type TRA-1 promotes female development in somatic tissues and abundant spermatogenesis in the germ line.
Fig. 1. Genetic regulation of sex determination in C.elegans. Only genes essential to this paper are presented; for a full description of the pathway see Meyer (1997). For references see the text. (A) Somatic tissues. TRA-1 is the terminal regulator and directs female development. TRA-2 promotes female development by negatively regulating the FEM proteins. (B) Germ line. TRA-1 is not the terminal regulator and promotes spermatogenesis, perhaps by positively regulating activity of the FEM proteins.
The tra-2 gene encodes multiple tra-2 transcripts to generate TRA-2A, a membrane protein with similarity to Drosophila and vertebrate patched, and the oocyte-specific TRA-2B, which corresponds to the intracellular domain (ic) of TRA-2A (Kuwabara et al., 1992, 1998). In addition, TRA-2A can be cleaved by the TRA-3 protease to free the TRA-2ic from its membrane attachment (Sokol and Kuwabara, 2000). Genetic studies have shown that tra-2 negatively regulates the fem genes and promotes female development in both somatic and germ-line tissues (Hodgkin and Brenner, 1977; Hodgkin, 1986) (Figure 1).
TRA-2ic, whether generated as a cleavage product of TRA-2A (Sokol and Kuwabara, 2000) or as the separate translation product TRA-2B (Kuwabara et al., 1998), carries the feminizing activity of TRA-2 (Kuwabara and Kimble, 1995). One aspect of that feminizing activity resides in the N-terminal portion, which binds and inhibits FEM-3 (Mehra et al., 1999). Intriguingly, the C-terminal region contains a 22-amino-acid MX region that is also critical for sex determination (Kuwabara et al., 1998). This MX region was defined by a series of tra-2(mx) missense mutations, which have both gain-of-function and loss-of-function character and were designated ‘mx’ for mixed character. The germ lines of both tra-2(mx)/+ and tra-2(mx) XX animals are feminized, albeit with greater penetrance in the homozygotes; in contrast, tra-2(mx) XO males make sperm continuously (Doniach, 1986; Schedl and Kimble, 1988). In addition, the somatic tissues of tra-2(mx) mutants can be weakly masculinized (Doniach, 1986; Schedl and Kimble, 1988). Therefore, the TRA-2(MX) region has a major role in the onset of hermaphrodite spermatogenesis and a minor role in somatic tissues.
In this paper, we report the unexpected finding that TRA-1A binds TRA-2ic. This binding occurs in the C-terminal portion of TRA-2ic and depends on the TRA-2(MX) regulatory region. TRA-1–TRA-2ic binding was discovered and analyzed using yeast two-hybrid assays, and confirmed as a direct physical interaction in vitro. To test its biological function, we examined the effect of reducing tra-1 gene dose on the tra-2(mx) phenotype. We find that tra-1(0) is a dominant enhancer of tra-2(mx)/+ feminization in the germ line, but have not detected a similar enhancement of tra-2(mx) masculinization of somatic tissues. Finally, we demonstrate that the TRA-1–TRA-2ic interaction is conserved: Caenorhabditis briggsae TRA-1 binds the intracellular domain of C.briggsae TRA-2 in an MX-dependent manner. This conservation over millions of years supports the idea that TRA-1–TRA-2ic binding plays a key role in sex determination.
Results
Identification of TRA-1 in a yeast two-hybrid screen for TRA-2 interactors
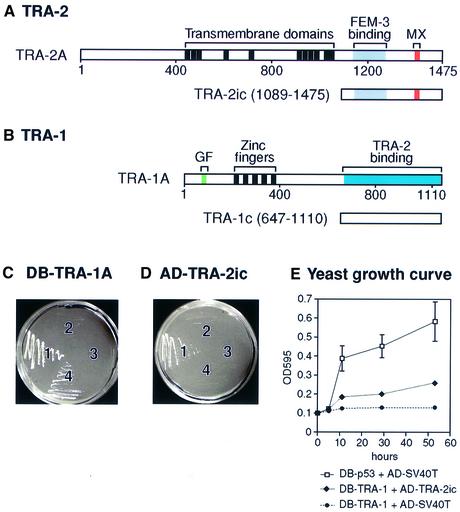
To search for proteins that regulate or are regulated by TRA-2, we performed a yeast two-hybrid screen using a TRA-2ic fragment as bait (Figure 2A). From 1.2 million transformants, we identified one positive clone that encodes the C-terminal portion [amino acids (aa) 647–1110] of TRA-1A; we call this fragment TRA-1c (Figure 2B). As GAL4 DNA-binding domain (DB) fusions, TRA-2ic has weak activating activity and TRA-1c has strong activity (data not shown). By contrast, full-length DB–TRA-1A protein possesses little activation activity (Figure 2C, sector 3). We therefore used DB–TRA-1A for subsequent analyses of the TRA-1– TRA-2 interaction (Figure 2C and D).
Fig. 2. Caenorhabditis elegans TRA-1 interacts with C.elegans TRA-2ic in yeast. (A) The tra-2 gene encodes two products: TRA-2A is a transmembrane protein and TRA-2ic corresponds to the TRA-2A cytoplasmic region (aa 1089–1475, with numbering from the TRA-2A sequence) (Kuwabara et al., 1992). The region that binds FEM-3 extends from aa 1133 to 1273 (Mehra et al., 1999); the MX domain is defined as aa 1392–1413 (Kuwabara et al., 1998). TRA-2ic was derived from a TRA-2A cDNA, and is equivalent to TRA-2B. (B) The tra-1 gene encodes two products: the longer TRA-1A shown here contains five zinc fingers; the shorter TRA-1B possesses only the two N-terminal fingers and lacks the region corresponding to TRA-1c (Zarkower and Hodgkin, 1992). The GF region of TRA-1 was defined by missense gain-of-function mutations clustered in a region encoding aa 73–88 (de Bono et al., 1995); it should be noted that the TRA-1(GF) region resides in the N-terminal part of TRA-1A, whereas the TRA-2-binding region resides in the C-terminal part of TRA-1A. TRA-1c extends from aa 647 to 1110. (C and D) Yeast were cultured on –His/–Trp/–Leu + 2.5 mM 3-AT medium at 30°C for 4–5 days. A two-hybrid interaction was scored as positive by growth on this selective medium, and as negative by a lack of growth. (C) Yeast were transformed with plasmid pairs and streaked in four sectors: sector 1, pDB-p53 and AD–SV40T; sector 2, pDB-TRA-1A and pAD-SV40T; sector 3, pDB-TRA-1A and pAD-vector; sector 4, pDB-TRA-1A and pAD-TRA-2ic. (D) Yeast were transformed with plasmid pairs and streaked in four sectors: sector 1, pDB-TRA-1A and pAD-TRA-2ic; sector 2, pDB-vector and pAD-TRA-2ic; sector 3, pDB-p53 and pAD-TRA-2ic; and sector 4, pDB-LAM5′ and pAD-TRA-2ic. (E) Yeast growth curves in liquid media SD/–His/–Trp/–Leu + 2 mM 3-AT. Each point is an average of three cultures; standard deviations are shown for the DB-p53 + AD-SV40T curve; standard deviations for other curves are within the size of the triangles/dots used for the graphic and are therefore not visible.
Our yeast two-hybrid assays relied on growth on selective medium (see Materials and methods); growth was scored as positive for the interaction and lack of growth scored as negative. By this assay, yeast transformed with DB–p53, and activation domain (AD)–SV40 T-antigen fusions grew on this medium (Figure 2C, sector 1). DB–TRA-1A did not interact with SV40 T antigen fused to an AD (Figure 2C, sector 2) and did not self-activate (Figure 2C, sector 3), but DB–TRA-1A did interact with AD–TRA-2ic (Figure 2C, sector 4). Conversely, AD–TRA-2ic interacted with TRA-1A (Figure 2D, sector 1), but did not self-activate (Figure 2D, sector 2) or interact with p53 or LAM5 (Figure 2D, sectors 3 and 4). The relative strength of the TRA-1–TRA-1ic interaction compared with that of p53/SV40 T antigen was determined by measuring yeast growth rates (Figure 2E). These results suggest that TRA-1A interacts specifically with the cytoplasmic portion of TRA-2.
Distinct TRA-2 regions bind FEM-3 and TRA-1
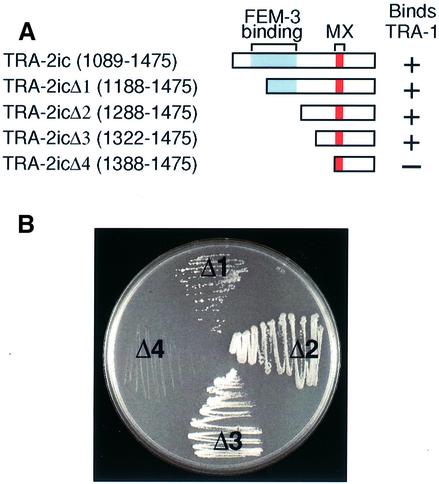
The FEM-3-binding region of TRA-2 extends from aa 1133 to 1273 (Mehra et al., 1999) (Figures 2A and 7A). To ask where TRA-1A binds, we employed a series of N-terminal deletions of AD–TRA-2ic (Figure 3A). Specifically, TRA-2icΔ1 removed 100 amino acids, TRA-2icΔ2 removed 200 amino acids, TRA-2icΔ3 removed 233 amino acids and TRA-2icΔ4 removed 300 amino acids. We found that removal of 100, 200 or 233 amino acids from the TRA-2ic N-terminus had little or no effect on interactions with TRA-1A (Figure 3B). However, deletion of 300 amino acids disrupted the interaction (Figure 3B, sector Δ4). Therefore, the TRA-1–TRA-2ic interaction does not require the FEM-3-binding domain (aa 1133–1273), but instead requires a region of at most 154 amino acids (aa 1322–1475), which contains the MX regulatory region (aa 1392–1413).
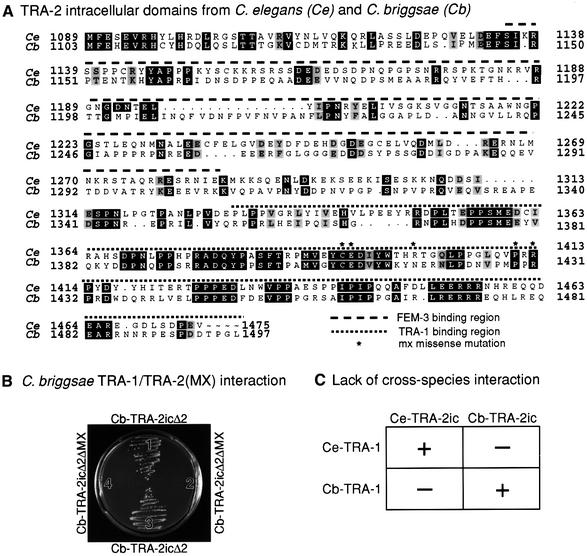
Fig. 7. Conservation of TRA-1–TRA-2 interaction. (A) Alignment of amino acid sequences of TRA-2ic for C.elegans (Ce) and C.briggsae (Cb). Numbering for C.elegans TRA-2 is from Kuwabara et al. (1992); numbering for C.briggsae TRA-2 is from Kuwabara (1996). Identical amino acids are black; similar amino acids are gray; the FEM-3-binding region is indicated by a dashed line over the sequence (Mehra et al., 1999); the TRA-1-binding region by a dotted line over the sequence (this work); amino acids altered by tra-2(mx) missense mutations are marked by an asterisk (Kuwabara et al., 1998). (B) Yeast two-hybrid results. Yeast were co-transformed with pDB-Cb-TRA-1, a plasmid encoding C.briggsae TRA-1A as a GAL4 DNA-binding hybrid, and either of two plasmids as AD hybrids: pAD-Cb-TRA-2icΔ2 encodes a fragment of C.briggsae TRA-2ic deleted for its N-terminal 200 amino acids; pCb-TRA-2icΔ2ΔMX encodes a form of Cb-TRA-2icΔ2 that also deletes the MX region. Yeast were streaked on SD/–His/–Trp/–Leu + 5 mM 3-AT plates and scored for growth after 4 days. Sectors 1 and 3, Cb-TRA-2icΔ2 interacts with pDB-Cb-TRA-1; sectors 2 and 4, Cb-TRA-2icΔ2ΔMX does not interact with pDB-Cb-TRA-1. (C) Summary of yeast two-hybrid results to test cross-species interactions between TRA-1 and TRA-2. See text for explanation.
Fig. 3. The part of TRA-2ic that binds TRA-1 includes the MX region and does not overlap with the FEM-3-binding region. (A) TRA-2 N-terminal deletions used to define the TRA-1 binding domain; amino acid numbering and motif colors are the same as in Figure 2. (B) Yeast were transformed with one plasmid encoding DB–TRA-1A and one plasmid bearing an N-terminal deletion as depicted in (A). All yeast were cultured on –His/–Trp/–Leu + 2.5 mM 3-AT at 30°C for 4–5 days and then scored for growth. Yeast growth was robust for two of the deletions (TRA-2icΔ2 and TRA-2icΔ3) and no growth was observed for TRA-2icΔ4. For TRA-2icΔ1, growth was less robust than for TRA-2icΔ2 and TRA-2icΔ3, but was still easily detectable. In repeats of this experiment, growth was always easily detectable using TRA-2icΔ1, TRA-2icΔ2 and TRA-2icΔ3, and never detectable using TRA-2icΔ4.
The TRA-2 MX region is required for the TRA-1–TRA-2 interaction
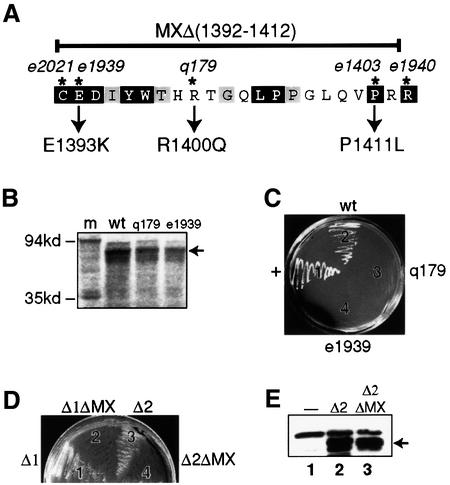
To ask whether the TRA-2 MX region is required for TRA-1 binding, we engineered mx mutations into TRA-2ic (Figure 4A). The MXΔ deletion removes 21 of the 22 MX amino acids, and three point mutations, E1393K, R1400Q and P1411L, were made corresponding to tra-2(mx) alleles e1939, q179 and e1403, respectively (Figure 4A). The size of each MX mutant protein fused with the GAL4 AD was confirmed by examination of in vitro translation products (Figure 4B and data not shown); in addition, expression of the TRA-2(MX) mutant proteins in yeast was verified by western blotting (data not shown). The interaction of TRA-1A with wild-type TRA-2ic and the TRA-2(MX) mutant proteins was assayed in yeast. In contrast to wild-type TRA-2ic, which interacted well with TRA-1 (Figure 4C, sector 2), the three TRA-2(MX) mutants all failed to interact (Figure 4C, sectors 3 and 4 and data not shown). We conclude that the MX region of TRA-2 is critical for the TRA-1–TRA-2 interaction.
Fig. 4. The TRA-2 MX region is required for the TRA-1–TRA-2 interaction. (A) Amino acid sequence of the TRA-2 MX region. The MX region extends from the cysteine at aa 1392 to the arginine at aa 1413. Amino acids that are identical in C.elegans, C.briggsae and C.remanei are shown in black; amino acids identical in two of these species, including C.elegans, are shown in gray. The amino acids changed in tra-2(mx) mutants are marked by asterisks and include: tra-2(e2021), a C to Y transition at aa 1392; tra-2(e1939), an E to K transition at aa 1393; tra-2(q179) or tra-2(e2019), an R to Q transition at aa 1400; tra-2(e1403), a P to L transition at aa 1411; and tra-2(e1940), an R to Q transition at aa 1413 (Kuwabara et al., 1998). The MX mutants employed in this study are: MXΔ, which removes aa 1392–1412; MX-E1393K, the tra-2(e1939) change; MX-R1400Q, the tra-2(q179) change; and MX-P1411L, the tra-2(e1403) change. Data are presented only for MX-E1393K and MX-R1400Q. (B) MX mutant proteins. Clones encoding GAL4 AD fusion proteins with either wild-type (+) TRA-2ic or one of three mutant TRA-2ic proteins were tested by expression in vitro for generation of protein of the correct size (see Materials and methods). Lanes are as follows: m, molecular weight markers; wt, wild-type AD–TRA-2ic fusion; q179, AD–TRA-2icR1400Q fusion; e1939, AD–TRA-2icE1393K fusion. Molecular weights are indicated on the left. (C) Two-hybrid results with TRA-2ic-MX mutants. Yeast were co-transformed with one plasmid encoding TRA-1A as a GAL4 DNA-binding hybrid and one of three plasmids encoding TRA-2ic or a TRA-2ic-MX mutant as AD hybrids; they were then cultured on SD/–His/–Trp/–Leu with 2.5 mM 3-AT medium at 30°C for 4–5 days. Sector 1, pDB-p53 and pAD-SV40T; sector 2, AD–TRA-2ic; sector 3, AD–TRA-2ic(q179); sector 4, AD–TRA-2ic(e1939). (D) Two-hybrid results with TRA-2icΔ and TRA-2icΔ-MXΔ proteins. Yeast were co-transformed with one plasmid encoding TRA-1A as a GAL4 DNA-binding hybrid and one of four plasmids encoding AD–TRA-2icΔ1 (sector 1), AD–TRA-2icΔ1ΔMX (sector 2), AD–TRA-2icΔ2 (sector 3) or AD–TRA-2icΔ2ΔMX (sector 4); the yeast were then cultured on –His/–Trp/–Leu + 2.5 mM 3-AT medium for 4–5 days. (E) Expression of TRA-2 mutant proteins in yeast. Immunoblot of yeast proteins probed with mouse anti-HA antibody. Lane 1, yeast CG1945; lane 2, yeast CG1945 transformed with pAD-TRA-2icΔ2; lane 3, yeast CG1945 transformed with pAD-TRA-2icΔ2ΔMX. The GAL4AD–TRA-2ic fusion proteins (either wild type or deletion) are indicated by the arrow.
We tested further the importance of the MX region for TRA-1–TRA-2 binding using deletions TRA-2icΔ1 and TRA-2icΔ2 (Figure 3A). To this end, the MXΔ deletion was engineered into each of two constructs to generate AD–TRA-2icΔ1ΔMX and AD–TRA-2icΔ2ΔMX. We found that yeast co-transformed with plasmids encoding DB–TRA-1A, AD–TRA-2icΔ1 and AD–TRA-2icΔ2 grew well (Figure 4D, sectors 1 and 3), whereas those co-transformed with plasmids encoding DB–TRA-1A, AD–TRA-2icΔ1ΔMX and AD–TRA-2icΔ2ΔMX did not grow (Figure 4D, sectors 2 and 4). Expression of TRA-2icΔ2 and TRA-2bΔ2ΔMX proteins in yeast was verified by western blotting (Figure 4E, arrow). This result substantiates the finding that the TRA-2(MX) region is required for the TRA-1–TRA-2 interaction.
TRA-1 and TRA-2 interact in vitro
To confirm the TRA-1–TRA-2 interaction in vitro, a fragment encoding TRA-2icΔ2 (Figure 3A) was fused in-frame to a sequence encoding a T7 tag; this frag ment either harbored the wild-type MX region or the MXΔ deletion (Figure 4A). The resultant proteins, T7-TRA-2icΔ2 and T7-TRA-2icΔ2ΔMX, were expressed in Escherichia coli and purified with T7 antibody-conjugated agarose beads (Figure 5A). To tag TRA-1, the TRA-1c fragment (Figure 2B) was fused in-frame to an S-tag. The in vitro translated, S-tagged TRA-1c was added to either T7-TRA-2icΔ2 (Figure 5B, lane 2) or T7-TRA-2icΔ2ΔMX (Figure 5B, lane 3) and incubated to permit binding. Subsequently, beads were washed and pelleted, and associated proteins were analyzed by western blotting using alkaline phosphatase conjugated to S-protein (see Materials and methods). We found that T7-TRA-2icΔ2 protein pulled down S-tagged TRA-1c (Figure 5B, lane 4), but that T7-TRA-2icΔ2ΔMX did not (Figure 5B, lane 5). We conclude that TRA-1 and TRA-2 bind each other in vitro and that this binding requires the MX region.
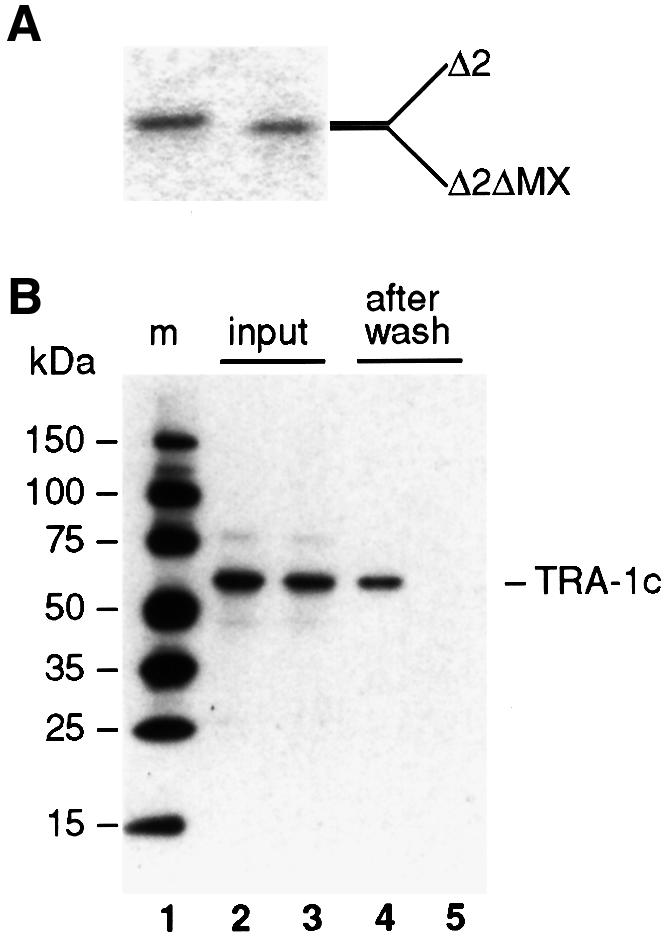
Fig. 5. TRA-1 interacts with TRA-2ic in vitro. (A) The TRA-2 proteins used in the in vitro assay were purified T7-tagged TRA-2icΔ2 and T7-tagged TRA-2icΔ2ΔMX. TRA-2icΔ2 lacks the FEM-3-binding domain, but possesses the MX domain, as shown in Figure 3A; TRA-2icΔ2ΔMX is an N-terminal deletion corresponding to TRA-2icΔ2, but it also has the deletion of MX, MXΔ, as shown in Figure 4A. (B) S-tagged TRA-1c was incubated with T7-tagged TRA-2ic proteins. Lane 1, molecular weight markers; lane 2, input S-tagged TRA-1c incubated with T7-tagged TRA-2icΔ2; lane 3, input S-tagged TRA-1c incubated with T7-tagged TRA-2icΔ2ΔMX; lane 4, the same as lane 2 but after washing; lane 5, the same as lane 3 but after washing.
A functional test for tra-1/tra-2(mx) interactions
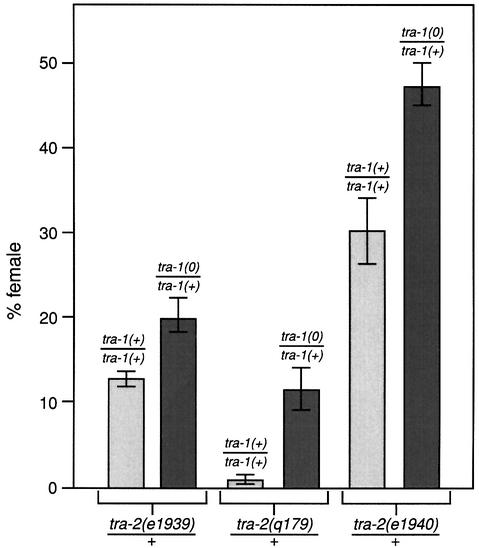
Normally, tra-2(mx)/+ XX animals are either hermaphrodite or female with the percentage of females characteristic of an individual allele (Doniach, 1986; Schedl et al., 1989); tra-1(0)/+ XX animals are all hermaphrodite (Hodgkin and Brenner, 1977). To test the in vivo importance of the TRA-1–TRA-2 interaction, we compared the percentage females among tra-2(mx)/+ XX animals with that among tra-2(mx)/+; tra-1(0)/+ XX animals. For each of three different alleles, tra-2(e1939mx), tra-2(q179mx) and tra-2(e1940mx), XX animals bearing only one copy of wild-type tra-1 were significantly more feminized than those with two copies of wild-type tra-1 (Figure 6). In contrast to the effect of tra-1(0) on the tra-2(mx) mutations, tra-1(0)/+ did not feminize fem-3(0)/+ XX animals (data not shown). Therefore, tra-1(0) is a dominant enhancer of tra-2(mx)/+, consistent with the idea that TRA-1–TRA-2ic binding promotes spermatogenesis in wild-type animals (see Discussion).
Fig. 6. Enhancement of tra-2(mx) by removal of one copy of tra-1. The percentage of females among XX animals of genotype tra-2(mx)/+ is compared with the percentage of females among XX animals of genotype tra-2(mx)/+; tra-1(0)/+. Each column represents results from at least three matings. The experiments compared were performed in the same incubator and in the same box of plates. For tra-2(e1939mx)/+, 13 ± 1% females were observed (n = 100) and for tra-2(e1939mx)/+; tra-1(0)/+, 20 ± 2% females were found (n = 175). For tra-2(q179)/+, 1.5 ± 0.4% females were observed (n = 486), and for tra-2(q179mx)/+; tra-1(0)/+, 11 ± 3.5% females were found (n = 303). For tra-2(e1940mx)/+, 31 ± 4% females were observed (n = 100) and for tra-2(e1940mx)/+; tra-1(0)/+, 47 ± 3% females were found (n = 104).
To examine the interaction between tra-1 and tra-2(mx) in somatic tissues, we examined tra-2(q179mx) homozygotes that were either tra-1(0)/+ or tra-1(0) homozygotes. Animals of these genotypes were distinguished by the presence or absence of green fluorescent protein (GFP) expressed in the pharynx, which was carried by the hT2[qIs48] chromosome used to balance tra-1(0). We found that tra-1(0)/+; tra-2(mx) XX animals retained their hermaphrodite morphology: tails had the whip-like morphology typical of hermaphrodites and vulvas appeared normal (n = 342). In tra-1(0); tra-2(mx) double homozygotes, XX animals were male, as expected for tra-1(0) homozygotes: no vulva or hermaphrodite-like tail was observed (n = 120). To see some more subtle effects of tra-2(mx) on tra-1(0) males, we examined them at the L4 or early adult stage by Nomarski microscopy. The somatic gonad was not feminized: no hermaphrodite-like somatic gonads were found in tra-2(q179); tra-1(0) double mutants (n = 31). We therefore found no effect of tra-2(mx) on somatic tissues, either when the tra-1 dose was reduced by half or when tra-1 was removed entirely.
TRA-1–TRA-2ic binding is conserved
The C.briggsae tra-1 and tra-2 genes are functionally similar to their C.elegans relatives (Kuwabara and Shah, 1994; de Bono and Hodgkin, 1996) and the MX region is conserved (Kuwabara and Shah, 1994) (Figure 7A). If the TRA-1–TRA-2 interaction is critical for sex determination, that interaction should also be conserved. We therefore tested a nearly full-length TRA-1A from C.briggsae for interactions with Cb-TRA-2ic, a fragment equivalent to Ce-TRA-2ic (Figure 2A). Whereas yeast transformed with plasmids encoding DB–Cb-TRA-1 and AD–Cb-TRA-2ic were able to grow on selective medium, those transformed with pDB-Cb-TRA-1 and a pAD empty vector or pAD-SV40 T antigen failed to grow on the same medium (data not shown). Furthermore, yeast transformed with plasmids encoding DB–p53 or the DB empty vector together with pAD-CbTRA-2ic could not grow on this medium (data not shown). Therefore, C.briggsae TRA-1 interacts with C.briggsae TRA-2c specifically in yeast.
To ask whether the Cb-TRA-1–Cb-TRA-2 interaction relied on the MX region, we tested N-terminal deletions of TRA-2ic similar to those described above for C.elegans TRA-2ic and obtained similar results: deletions of either 100 or 200 amino acids from the N-terminus of C.briggsae TRA-2ic had no effect, but an N-terminal deletion of 300 amino acids disrupted the interaction (data not shown). Finally, we deleted the MX region from Cb-TRA-2icΔ2 (a deletion analogous to Ce-TRA-2icΔ2; Figure 3A). The Cb-MX deletion was equivalent to Ce-MXΔ (Figure 4A). Whereas yeast transformed with pAD-Cb-TRA-2icΔ2 and pDB-Cb-TRA-1 grew well on selective medium (Figure 7B, sectors 1 and 3), those transformed with pAD-Cb-TRA-2icΔ2ΔMX and pDB-Cb-TRA-1 did not grow on the same medium (Figure 7B, sectors 2 and 4). Expression in yeast of the AD–Cb-TRA-2icΔ2 and AD–Cb-TRA-2icΔ2ΔMX fusion proteins was verified by western blotting (data not shown). We conclude that the C.briggsae TRA-1–TRA-2 interaction requires the MX region.
Finally, we asked whether C.elegans TRA-1 could interact with C.briggsae TRA-2ic and vice versa. Experiments were carried out similarly to those described above and are summarized in Figure 7C. We found that the C.elegans DB–TRA-1 fusion protein did not interact with the C.briggsae AD–TRA-2ic fusion protein, and conversely that the C.briggsae DB–TRA-1 fusion protein did not interact with the C.elegans AD–TRA-2ic fusion protein (data not shown). We conclude that the C.elegans and C.briggsae proteins have diverged too far to interact.
Discussion
In this paper, we report three conclusions about nematode sex determination. First, the TRA-1 transcription factor binds the C-terminal region of TRA-2 in an MX-dependent manner. Secondly, a tra-1 null mutation dominantly enhances the semi-dominant tra-2(mx)/+ feminization of germ-line tissues. In contrast, no enhancement of the recessive tra-2(mx) weak masculinization of somatic tissues was detected. And thirdly, the TRA-1–TRA-2 interaction is conserved in C.briggsae. In the following sections, we discuss the functional significance of the TRA-1–TRA-2 interaction in controlling sexual fates and possible mechanisms by which it may do so.
The TRA-1–TRA-2 interaction is critical for promoting spermatogenesis
We have found that TRA-1 binds TRA-2ic in a region that includes the MX regulatory region. Furthermore, TRA-1 does not bind TRA-2(MX) mutant proteins. The precise site of TRA-1 binding within TRA-2ic is not known, but must involve the MX region either directly or indirectly. The TRA-1–TRA-2ic interaction occurs between two proteins that were previously predicted to reside in distinct subcellular compartments (Kuwabara et al., 1992; Zarkower and Hodgkin, 1992). However, the TRA-1 transcription factor has been detected in both nucleus and cytoplasm (Graves et al., 1999), and an overexpressed TRA-2ic::GFP fusion protein is nuclear (Lum et al., 2000). Because TRA-2ic can be generated by translation of the smaller tra-2 transcript (Kuwabara et al., 1998) or by cleavage of the TRA-2A membrane protein (Sokol and Kuwabara, 2000), TRA-2ic may indeed be nuclear in wild-type animals. Therefore, TRA-1 and TRA-2 are present in the same subcellular compartments within C.elegans and are likely to interact there as well.
The TRA-2 MX region is critical for control of sexual fates: the germ line of tra-2(mx) XX mutants is feminized, making no sperm and only oocytes, and somatic tissues are weakly masculinized (Doniach, 1986; Schedl and Kimble, 1988). To explore the importance of TRA-1–TRA-2 binding in C.elegans, we asked whether genetic interactions between tra-1 null and tra-2(mx) mutations could be observed. This type of experiment assumes a correlation between gene dosage and amount of protein made. Normally, one tra-1 copy is sufficient for proper sex determination (Hodgkin and Brenner, 1977; Hodgkin, 1986). Therefore, the wild-type TRA-2 protein is not sensitive to a reduction of tra-1 gene dose. However, we reasoned that a tra-2(mx) mutation might provide a more sensitized background, which might be dependent on tra-1 gene dosage. Indeed, the tra-2(mx)/+; tra-1(0)/+ germ line was more often feminized than that of tra-2(mx)/+ animals. An effect on somatic tissues, however, was not observed in animals of the same genotype. We conclude that the tra-2(mx) mutant protein is sensitive to tra-1 gene dose, consistent with an in vivo role for the TRA-1–TRA-2 interaction in germ-line sex determination.
tra-1 and germ-line sex determination
TRA-1 is not the terminal regulator of sexual fates in the germ line (Hodgkin, 1986) (see Figure 1). In tra-1 null mutants, somatic tissues are strongly masculinized, but the germ line makes fewer sperm than are made in wild-type males (Hodgkin and Brenner, 1977; Hodgkin, 1987; Schedl et al., 1989). Therefore, TRA-1 promotes female development in somatic tissues and abundant spermatogenesis in males. In tra-1 gain-of-function mutants, both somatic and germ-line tissues are feminized, suggesting that TRA-1 may also direct oogenesis (Hodgkin, 1987; de Bono et al., 1995). We propose a third role for TRA-1 in germ-line sex determination. tra-2(mx) mutants are defective in the onset of hermaphrodite spermatogenesis, and tra-1 is a dominant enhancer of that defect. Therefore, in addition to its previously suggested roles, TRA-1 participates in controlling the onset of hermaphrodite spermatogenesis. This third role may be mechanistically similar to the role of tra-1 in promoting continued spermatogenesis in males.
How does the TRA-1–TRA-2 interaction influence sex determination?
The binding of TRA-1 to TRA-2ic in an MX-dependent manner provides evidence for a new mechanism by which TRA-1 and TRA-2 act together to influence sex determination. Molecular analyses of tra-2(mx) mutations led to the idea that a repressor might bind the TRA-2 MX region and thereby inhibit the feminizing activity of TRA-2 to promote male development (Kuwabara et al., 1998). One possibility is that TRA-1 is that repressor; alternatively, TRA-2 might repress TRA-1. How might repression of TRA-1 be consistent with a requirement for TRA-1 in promoting spermatogenesis? One mechanism by which TRA-1 controls germ-line sexual fates appears to be transcriptional. The fog-3 gene is required for specification of sperm (Ellis and Kimble, 1995), and harbors a series of TRA-1 binding sites in its promoter (Chen and Ellis, 2000). Intriguingly, these TRA-1 sites can mediate either repression or activation of the fog-3 promoter (Chen and Ellis, 2000). One simple idea is that TRA-1 activates fog-3 at one level of expression or in one state which exists in males and larval hermaphrodites as sperm are specified, and represses fog-3 at a second level or state which exists during oogenesis. The binding of TRA-2 to TRA-1 might promote the TRA-1 level or state critical for spermatogenesis. For example, sequestration of TRA-1 by TRA-2 could decrease available TRA-1 and keep TRA-1 at a level appropriate for fog-3 activation.
An alternative scenario brings in FEM-3, a protein required for male development in both somatic and germ-line tissues (Hodgkin, 1986; Barton et al., 1987). The N-terminal region of TRA-2ic binds FEM-3 and inhibits its ability to direct male development (Mehra et al., 1999). TRA-1 binding to the C-terminal region of TRA-2ic may activate FEM-3. Possible mechanisms for such activation include enhancing a modification required for activity, promoting assembly into an active complex, or competing with FEM-3 binding and thereby freeing FEM-3 to promote male development. Such a complex relationship among regulatory proteins would be difficult to unravel by conventional genetic tests.
Does the TRA-1–TRA-2ic interaction affect somatic sex determination?
The TRA-1 and TRA-2 proteins are present in somatic tissues and direct female development there (Hodgkin and Brenner, 1977). Does the TRA-1–TRA-2(MX) interaction play a role in somatic sex determination? The somatic effects of tra-2(mx) mutants are extremely weak: for three alleles (e1939, q179 and e1940), somatic masculinization is observed only in tra-2(mx)/tra-2(0) heterozygotes; for the other two alleles (e2021 and e1403), masculinization is so weak in homozygotes that most XX animals are self-fertile (Doniach, 1986; R.Edgar and T.Schedl, personal communication; S.Wang and J.Kimble, unpublished). Such weak masculinization might result from a specific effect on the TRA-1–TRA-2 interaction or from a non-specific defect, such as reduced stability of the TRA-2 mutant protein.
In an attempt to observe a role of the TRA-1–TRA-2 interaction in somatic sex determination, we examined somatic tissues in tra-2(mx); tra-1(0)/+ mutants. We used tra-2(q179), an allele that is strongly feminized in the germ line (Schedl et al., 1989), which is reduced in its ability to bind TRA-1 (this work) and which exhibits no somatic masculinization in XX homozygotes. Because this allele was clearly sensitive to the dose of tra-1 in the germ line, we reasoned that it might also be sensitive in somatic tissues. However, tra-2(q179); tra-1(0)/+ XX animals showed no somatic masculinization. In particular, the tail had the typical whip-like morphology of the hermaphrodite. Since the tail is a particularly sensitive monitor of sexual fate, a cautious interpretation is that the TRA-1–TRA-2 interaction has no major role in somatic sexual fates.
Conservation of the TRA-1–TRA-2 interaction
The tra-1 and tra-2 genes are among the most divergent genes in C.elegans and C.briggsae (de Bono and Hodgkin, 1996; Kuwabara, 1996). Each C.elegans protein is only ∼40% identical to its C.briggsae homolog. Nonetheless, the TRA-1–TRA-2 interaction has been conserved (this work), which argues strongly that it plays a critical role in sex determination. Within TRA-2, the TRA-2(MX) region is highly conserved between C.elegans and C.briggsae (Kuwabara, 1996), and we have found that this region is required for the interaction between TRA-1 and TRA-2 in both species (this work). Nonetheless, the C.elegans proteins do not interact with their C.briggsae partners. This lack of interaction between proteins of different species underscores the divergence between the sex-determining proteins of these two species, and argues that the TRA-1–TRA-2 interaction is sufficiently critical that it has been retained by co-evolution of the tra-1 and tra-2 genes.
In C.elegans, the TRA-1–TRA-2 interaction has been implicated in the onset of hermaphrodite spermatogenesis, and a similar function in C.briggsae seems plausible. An intriguing question is whether the TRA-1–TRA-2 interaction has been conserved in Caenorhabditis remanei, a female/male species. Analysis of that interaction awaits cloning of the Cr-tra-1 gene. However, we do know that the TRA-2 MX region has been conserved in C.remanei (Haag and Kimble, 2000). If the TRA-1–TRA-2 interaction has been similarly conserved, it is likely to play a more general role in nematode sex determination that is not limited to hermaphrodite/male species. Possibilities include continued spermatogenesis in XO males or control of sexual fates in somatic tissues.
Materials and methods
DNA manipulation
Most DNA manipulations were performed according to Sambrook et al. (1989). Primers and their position in the respective genes used in this study are listed in Table I.
Table I. Primers and their positions.
| Primer sequence | Annealing start position |
|---|---|
| P1: 5′-CGCGGATCCGTATGTTTGAAAGTGAAGTTCGACAC-3′ | Ce-tra-2, nt 3301–3324 |
| P2: 5′-CGCGGATCCGTGGAAATGGTGATAACACTGAAC-3′ | Ce-tra-2, nt 3601–3621 |
| P3: 5′-CGCGGATCCGTTCGCAAGAAAATTTGGACAAAG-3′ | Ce-tra-2, nt 3901–3922 |
| P4: 5′-CGCGGATCCGTCCTGCCAACCTTCCAGTTGA-3′ | Ce-tra-2, nt 4000–4019 |
| P5: 5′-CGCGGATCCGTGTTGAATATTGCGAAGATA-3′ | Ce-tra-2, nt 4201–4219 |
| P6: 5′-CCGCTCGAGTTAAACCTCTGGGTCTGATAGGTC-3′ | Ce-tra-2, nt 4464–4441 |
| P7: 5′-ACGCGTCGACGGAGTACTTGAAGTCCTGGAGGTAGC-3′ | Ce-tra-2, nt 4268–4245 |
| P8: 5′-ACGCGTCGACGGGGTACTTGAAGTCCTGGAGGTAGCTGTCCAGTTTGGTGTGTCCAGTAAATATC-3′ | Ce-tra-2, nt 4268–4245 |
| P9: 5′-ACGCGTCGACGGGGTACTTGAAGTCCTGGAGGTAGCTGTCCAGTTCGGTGTGTCCAGTAAATATCTTTGCAATATTCAACCATTGGACGG-3′ | Ce-tra-2, nt 4268–4245 |
| P10: 5′-ACGCGTCGACGATATTCAACCATTGGACGGG 3′ | Ce-tra-2, nt 4208–4190 |
| P11: 5′-CGGGATCCGTATGTTCGAACACGAAGTTCG-3′ | Cb-tra-2, nt 3339–3358 |
| P12: 5′-CCGCTCGAGCTAAAGACCAGGAGTGTC-3′ | Cb-tra-2, nt 4526–4509 |
| P13: 5′-CGCGGATCCGGATTCGGTGGATCGGGATCG-3′ | Ce-tra-1, nt 2098–2108 |
| P14: 5′-ATAAGAATGCGGCCGCTTAAAATTGATGACGTGGCTTTTTGGG-3′ | Ce-tra-1, nt 3492–3466 |
| P15: 5′-GGAATTCTCGCAAGAAAATTTGGACAAAG-3′ | Ce-tra-2, nt 3901–3922 |
| P16: 5′-ATAAGAATGCGGCCGCTTAAACCTCTGGGTCTGATAGGTC-3′ | Ce-tra-2, nt 4464–4441 |
| P17: 5′-CATGCCATGGAGATGTACCCATACGACGTCCCAGACTACGCTACCAGTCATGGAGAAGAGACT-3′ | Cb-tra-1, nt 841–861 |
| P18: 5′-GGAATTCTTAAAAACTGCGTGGCTTC-3′ | Cb-tra-1, nt 3705–3687 |
| P19: 5′-CGGGATCCGAGAAGTTCGGAAGAAAGTACA-3′ | Cb-tra-2, nt 3939–3958 |
| P20: 5′-AAGGCCTGTAACCAACCATTGCCGGTG-3′ | Cb-tra-2, nt 4259–4240 |
| P21: 5′-CTATCTATTCGATGATGAAG-3′ | – |
| P22: 5′-ACAGTTGAAGTGAACTTGCG-3′ | – |
The numbering in C.elegans tra-2 was according to Kuwabara et al. (1992). The numbering in C.briggsae was according to Kuwabara (1996). The numbering of C.elegans tra-1 was according to Zarkower and Hodgkin (1992). The numbering of C.briggsae tra-1 was according to de Bono and Hodgkin (1996). Restriction enzymes used in cloning are italicized; nucleotide changes associated with tra-2(mx) mutations are underlined.
To construct pAD-TRA-2ic, a C-terminal fragment corresponding to nucleotides (nt) 3301–4464 in the tra-2 gene was PCR amplified from pJK349 (Kuwabara and Kimble, 1995) with primers P1 and P6. The resulting PCR fragment was digested with BamHI and XhoI and ligated to pACTII (Clontech) digested with the same restriction enzymes. A PCR-based strategy was used to make deletions at the N-terminus of TRA-2ic. pAD-TRA-2icΔ1 was constructed by PCR amplifying a Ce-tra-2 cDNA fragment from nt 3601–4464 with primers P2 and P6, and then cloned into BamHI- and XhoI-cut pACTII. pAD-TRA-2icΔ2, pAD-TRA-2icΔ3 and pAD-TRA-2icΔ4 were constructed in the same way, except that primer sets P3 and P6, P4 and P6 or P5 and P6 were used, respectively.
To make Ce-TRA-2ic mx mutants, mutations were introduced in primers, and Ce-tra-2 cDNA amplified and cloned into an intermediate vector, pGEM-TRA-2ic. pGEM-TRA-2ic was made by ligating a BamHI–XhoI fragment from pAD-TRA-2ic into pGEM7zf(+) (Promega) digested with the same enzymes. To construct pAD-TRA-2icMX-E1393K, PCR product amplified from Ce-tra-2ic with P1 and P8 was digested with BamHI and SalI and then cloned into pGEM-TRA-2ic digested with the same enzymes. Then the BamHI–XhoI fragment of TRA-2ic was cloned into pACTII. pAD-TRA-2ic R1400Q and pAD-TRA-2ic P1411L were constructed with the same strategy, but with primer pairs P1 and P9, or P1 and P7 to amplify tra-2ic cDNA. To construct pAD-TRA-2icΔ1ΔMX, a PCR product amplified from pAD-TRA-2icMXΔ(1392–1412) by primer pair P2 and P6 was cloned into BamHI- and XhoI-digested pACTII. pAD-TRA-2icΔ2ΔMX was built with the same strategy as pAD-TRA-2icΔ1ΔMX except that primer pair P3 and P6 was used to amplify from pAD-TRA-2cMXΔ(1392–1412).
To make pET-TRA-2icΔ2 and pET-TRA-2icΔ2ΔMX, primers P15 and P16 were used to amplify the tra-2ic (nt 3901–4464) fragment with pAD-TRA-2icΔ2 and pAD-TRA-2icΔ2ΔMX as template, respectively. PCR products were digested with EcoRI and NotI and ligated to pET28a(+) (Novagen), to make pET-TRA-2icΔ2 and pET-TRA-2icΔ2ΔMX, respectively. Expression in E.coli produced T7-tagged TRA-2icΔ2Δ and TRA-2icΔ2ΔMX proteins.
To construct pCITE-TRA-1c, a 1.4 kb PCR product (nt 2098–3492) amplified from pDZ120 (a gift from D.Zarkower) with primers P13 and P14 was digested with BamHI and NotI and ligated to pCITE4a(+) (Novagen) digested with the same enzymes. In vitro translation in TNT Quick System (Promega) produced an S-tagged TRA-1c protein.
pAD-Cb-TRA-2ic was constructed in two steps. First, a 1.2 kb PCR fragment (nt 3339–4526) amplified from a C.briggsae cDNA library by primers P11 and P12 was digested with BamHI and XhoI (a BamHI site exists at nt 3573), and the released 1 kb fragment was ligated to pACTII digested with BamHI and XhoI, to make pAD-Cb-TRA-2icΔN. Then, the same PCR fragment was digested with BamHI and the released 0.2 kb fragment was ligated to BamHI-digested pAD-Cb-TRA-2cNΔ to make pAD-Cb-TRA-2ic.
To make pDB-Cb-TRA-1, restriction sites were added at the 5′ end of primers P17 and P18, which were used to amplify C.briggsae tra-1 cDNA. The 3 kb PCR fragment (C.briggsae tra-1 cDNA sequence nt 841–3705) was digested with NcoI and EcoRI and ligated to pAS2-1 cut with the same enzymes.
To delete the N-terminus from Cb-TRA-2ic, primers P11 and P20 were used to amplify a C-terminus of Cb-TRA-2ic; the resulting PCR fragment was digested with BamHI and XhoI and ligated into pACTII digested with the same enzymes, to make pAD-Cb-TRA-2icΔ2. To delete the MX region from Cb-TRA-2Δ2, primers P12 and P20 were used to amplify the Cb-TRA-2ic fragment; the PCR product was digested with StuI and BamHI, and then ligated into pAD-Cb-TRA-2Δ2 digested with the same enzymes, to make pAD-Cb-TRA-2icΔ2ΔMX.
Yeast two-hybrid assays
pDB-TRA-2ic was used as bait to screen the ACT-RB-2 C.elegans cDNA library (Kraemer et al., 1999). The large-scale yeast transformation procedure followed Clontech protocol. Because pDB-TRA-2c had weak self-activation, the screen was performed on medium SD/–His/–Trp/–Leu supplemented with 20 mM 3-aminotriazole (3-AT). Approximately 200 yeast colonies grew on this medium from 1.2 million transformants, and 19 were analyzed by sequencing the inserts. Yeast plasmid DNA was isolated and transformed into E.coli strain KC-10 according to the manufacturer’s instructions (Clontech). Inserts were PCR amplified with primers P21 and P22, sequenced, and used to search the C.elegans genomic sequence database with the BLAST program.
Yeast two-hybrid assays were carried out by introducing relevant plasmids into yeast strain CG1945 (Clontech) and plated on SD/–His/–Trp/–Leu medium supplemented with 2.5–5 mM 3-AT.
Yeast growth rate in liquid media was determined by inoculating a colony in the SD/–His/–Trp/–Leu + 2 mM 3-AT and monitoring the OD at 595 nm at indicated time points.
Purification of recombinant protein from E.coli
pET-TRA-2icΔ2 and pET-TRA-2icΔ2ΔMX were transformed in E.coli strain λDE3(LysS) (Novagen) and grown in 5 ml of Luria–Bertani (LB) medium + 50 µg/ml ampicillin overnight at 37°C. Bacteria were diluted to 100 ml of + 50 µg/ml medium and cultured for a further 3 h; after adding 1 mM isopropyl-β-d-thiogalactopyranoside, they were shaken at 37°C for 2 h. Bacteria were collected by centrifugation, resuspended in 5 ml of phosphate-buffered saline (PBS), sonicated, and centrifuged to remove cell debris. T7 antibody-conjugated agarose beads (100 µl) were mixed with supernatant, washed and resuspended according to the manufacturer’s protocol (Novagen). Purified proteins were examined on 4–20% gradient polyacrylamide gels (Bio-Rad) with Coomassie Blue staining.
Immunoblot hybridization
Yeast was grown overnight in 5 ml of culture at 30°C, and collected by spinning in a microfuge for 30 s. Preparation of yeast extract was modified from Clontech’s protocol. Briefly, yeast pellet was resuspended in 100 µl of disruption buffer (Clontech), mixed with glass beads and vortexed for 1 min. The crude extract was spun in a microfuge for 30 s, and 50 µl of supernatant transferred to a fresh tube. Clear extract (20 µl) was mixed with loading buffer and boiled for 2 min. Boiled samples were examined on a 4–20% gradient polyacrylamide precast gel (Bio-Rad). Protein was transferred to a polyvinylidene difluoride (PVDF) membrane according to the manufacturer’s instructions (Amersham). The membrane was washed briefly with PBS buffer and incubated in 5% non-fat milk in PBS solution for 2 h with rotation. Then, the membrane was transferred to 5% non-fat milk in PBS + 0.25% Tween-20 solution containing anti-HA antibody (Boehringer) (1:1000 dilution), and incubated for 1 h with rotation. After the membrane was washed three times with PBS + 0.25% Tween-20 solution, each for 10 min with rotation, it was incubated in 5% non-fat milk in PBS + 0.25% Tween-20 solution containing horseradish peroxidase-conjugated, anti-mouse secondary antibody for 1 h with rotation. Then the membrane was washed a further three times before exposure to X-ray film for 0.5–5 min.
In vitro transcription and in vitro translation
pGEM-AD-TRA-2ic, pGEM-AD-TRA-2icMX-P1411L, pGEM-AD-TRA-2icMX-E1393K and pGEM-AD-TRA-2icMXR1400Q were digested with SmaI and transcribed using an SP6 Megascript kit (Ambion). RNA (0.1 µg) was used for in vitro translation in wheat germ extract (Promega) in the presence of [35S]methionine (Amersham) according to the manufacturer’s instructions. Translation product (5 µl) was subjected to SDS–PAGE analysis on 10% separation polyacrylamide gel. The gel was treated with Amplify® (Amersham) before drying and exposure to X-ray film (Kodak).
In vitro pull-down assay
S-tagged TRA-1c was synthesized in TNT Quick® System (Promega) in a total volume of 50 µl according to the manufacturer’s instructions. Translation product (10 µl) was mixed with ∼0.1 µg (10 µl) of purified T7-tagged TRA-2icΔ2, either wild-type or MX deletion, in a total volume of 50 µl, and incubated at room temperature for 30 min. The mixture (10 µl) was removed as an inoculation amount control. The rest of the mixture was washed five times with 1× binding/washing buffer from the T7 purification kit (Novagen). After the final spin, the sample was resuspended in 10 µl of 1× binding/washing buffer mixed with 10 µl of loading buffer. The sample was boiled for 5 min before loading onto 4–20% gradient precast polyacrylamide gel (Bio-Rad) for SDS–PAGE analysis. Protein was transferred to PVDF membrane according to the manufacturer’s instruction. Alkaline phosphatase-conjugated S-protein (Novagen) was used to detect S-tagged TRA-1c according to the manufacturer’s instruction.
Genetics
Strains. The nonsense mutant tra-1(e1099) was used for tra-1(0). The nonsense mutant fem-3(e1996) was used for fem-3(0). The tra-2(mx) strains tra-2(e1939mx), tra-2(q179mx) and tra-2(e1940mx) were maintained as homozygotes. The double mutant strain, tra-2(q179mx); tra-1(e1099)/hT2[qIs48](I;III) was made by standard methods; tra-1(e1099)/hT2[qIs48](I;III) was used for controls. The hT2 translocation chromosome balances tra-1; the qIs48 insertion onto hT2 allows tra-1(e1099)/+ heterozygotes to be distinguished from tra-1(e1099) homozygotes by the presence or absence of the GFP marker inserted onto hT2.
Assays. tra-1(e1099) males or N2 males were mated to tra-2(e1939), tra-2(e1940) or tra-2(q179) females at 20°C. To assess an animal as hermaphrodite or female, L4s were picked individually onto separate plates, and scored as self-fertile (hermaphrodite) or sterile with stacked oocytes and an otherwise female morphology (female). To assess the effect of tra-2(mx) on somatic sexual characters, green and non-green adult progeny from a tra-2(q179mx); tra-1(e1099)/hT2[qIs48](I;III) parent were scored by dissecting scope for vulva and tail morphology. In addition, tra-2(q179mx); tra-1(e1099) and tra-1(e1099) homozygotes were compared by Nomarski microscopy for effects on somatic gonadal morphology; these males were either L4 larvae or young adults within 1 day after their L4 to adult molt.
Acknowledgments
Acknowledgements
We are grateful to Dr David Zarkower for the full-length tra-1 cDNA, to Dr Alex Puoti for the C.briggsae cDNA library and to Dr Laura Mathies for generation of the hT2[qIs48] green balancer. We are also grateful to David Lum and Andrew Spence for communicating results prior to publication. We thank Betsy Goodwin, Eric Haag, Laura Mathies, Dave Rudel and Kellee Siegfried for their insightful comments on the manuscript. J.K. is an investigator with the Howard Hughes Medical Institute.
References
- Barton M.K., Schedl,T.B. and Kimble,J. (1987) Gain-of-function mutations of fem-3, a sex-determination gene in Caenorhabditis elegans. Genetics, 115, 107–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P.-J. and Ellis,R.E. (2000) TRA-1A regulates transcription of fog-3, which controls germ cell fate in C. elegans. Development, 127, 3119–3129. [DOI] [PubMed] [Google Scholar]
- Conradt B. and Horvitz,H.R. (1999) The TRA-1A sex determination protein of C. elegans regulates sexually dimorphic cell deaths by repressing the egl-1 cell death activator gene. Cell, 98, 317–327. [DOI] [PubMed] [Google Scholar]
- de Bono M. and Hodgkin,J. (1996) Evolution of sex determination in Caenorhabditis: unusually high divergence of tra-1 and its functional consequences. Genetics, 144, 587–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bono M., Zarkower,D. and Hodgkin,J. (1995) Dominant feminizing mutations implicate protein–protein interactions as the main mode of regulation of the nematode sex-determining gene tra-1. Genes Dev., 9, 155–167. [DOI] [PubMed] [Google Scholar]
- Doniach T. (1986) Activity of the sex-determining gene tra-2 is modulated to allow spermatogenesis in the C. elegans hermaphrodite. Genetics, 114, 53–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis R.E. and Kimble,J. (1995) The fog-3 gene and regulation of cell fate in the germ line of Caenorhabditis elegans. Genetics, 139, 561–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves L.E., Segal,S. and Goodwin,E.B. (1999) TRA-1 regulates the cellular distribution of the tra-2 mRNA in C. elegans. Nature, 399, 802–805. [DOI] [PubMed] [Google Scholar]
- Haag E.S. and Kimble,J. (2000) Regulatory elements required for development of Caenorhabditis elegans hermaphrodites are conserved in the tra-2 homologue of C. remanei, a male/female sister species. Genetics, 155, 105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin J. (1986) Sex determination in the nematode C. elegans: analysis of tra-3 suppressors and characterization of fem genes. Genetics, 114, 15–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin J. (1987) A genetic analysis of the sex determining gene, tra-1, in the nematode Caenorhabditis elegans. Genes Dev., 1, 731–745. [DOI] [PubMed] [Google Scholar]
- Hodgkin J.A. and Brenner,S. (1977) Mutations causing transformation of sexual phenotype in the nematode Caenorhabditis elegans. Genetics, 86, 275–287. [PMC free article] [PubMed] [Google Scholar]
- Kraemer B., Crittenden,S., Gallegos,M., Moulder,G., Barstead,R., Kimble,J. and Wickens,M. (1999) NANOS-3 and FBF proteins physically interact to control the sperm–oocyte switch in Caenorhabditis elegans. Curr. Biol., 9, 1009–1018. [DOI] [PubMed] [Google Scholar]
- Kuwabara P.E. (1996) Interspecies comparison reveals evolution of control regions in the nematode sex-determining gene tra-2. Genetics, 144, 597–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwabara P.E. and Kimble,J. (1995) A predicted membrane protein, TRA-2A, directs hermaphrodite development in Caenorhabditis elegans. Development, 121, 2995–3004. [DOI] [PubMed] [Google Scholar]
- Kuwabara P.E. and Shah,S. (1994) Cloning by synteny: identifying C. briggsae homologues of C. elegans genes. Nucleic Acids Res., 22, 4414–4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwabara P.E., Okkema,P.G. and Kimble,J. (1992) tra-2 encodes a membrane protein and may mediate cell communication in the Caenorhabditis elegans sex determination pathway. Mol. Biol. Cell, 3, 461–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwabara P.E., Okkema,P.G. and Kimble,J. (1998) Germ-line regulation of the Caenorhabditis elegans sex-determining gene tra-2. Dev. Biol., 204, 251–262. [DOI] [PubMed] [Google Scholar]
- Lum D.H., Kuwabara,P.E., Zarkower,D. and Spence,A.M. (2000) Direct protein–protein interaction between the intracellular domain of TRA-2 and the transcription factor TRA-1A modulates feminizing activity in Caenorhabditis elegans. Genes Dev., 14, 3153–3165. [PMC free article] [PubMed] [Google Scholar]
- Mehra A., Gaudet,J., Hick,L., Kuwabara,P.E. and Spence,A.M. (1999) Negative regulation of male development in Caenorhabditis elegans by a protein–protein interaction between TRA-2A and FEM-3. Genes Dev., 13, 1453–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer B.J. (1997) Sex determination and X chromosome dosage compensation. In Riddle,D.L., Blumenthal,T., Meyer,B.J. and Priess,J.R. (eds), C. elegans II. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 209–240. [PubMed]
- Puoti A., Gallegos,M., Zhang,B., Wickens,M.P. and Kimble,J. (1997) Controls of cell fate and pattern by 3′ untranslated regions: the Caenorhabditis elegans sperm/oocyte decision. Cold Spring Harb. Symp. Quant. Biol., 62, 19–24. [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Schedl T. and Kimble,J. (1988) fog-2, a germ-line-specific sex determination gene required for hermaphrodite spermatogenesis in Caenorhabditis elegans. Genetics, 119, 43–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schedl T., Graham,P.L., Barton,M.K. and Kimble,J. (1989) Analysis of the role of tra-1 in germline sex determination in the nematode Caenorhabditis elegans. Genetics, 123, 755–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol S.B. and Kuwabara,P.E. (2000) Proteolysis in Caenorhabditis elegans sex determination: cleavage of TRA-2A by TRA-3. Genes Dev., 14, 901–906. [PMC free article] [PubMed] [Google Scholar]
- Zarkower D. and Hodgkin,J. (1992) Molecular analysis of the C. elegans sex-determining gene tra-1: a gene encoding two zinc finger proteins. Cell, 70, 237–249. [DOI] [PubMed] [Google Scholar]