Abstract
A cloverleaf structure at the 5′ terminus of poliovirus RNA binds viral and cellular proteins. To examine the role of the cloverleaf in poliovirus replication, we determined how cloverleaf mutations affected the stability, translation and replication of poliovirus RNA in HeLa S10 translation–replication reactions. Mutations within the cloverleaf destabilized viral RNA in these reactions. Adding a 5′ 7-methyl guanosine cap fully restored the stability of the mutant RNAs and had no effect on their translation. These results indicate that the 5′ cloverleaf normally protects uncapped poliovirus RNA from rapid degradation by cellular nucleases. Preinitiation RNA replication complexes formed with the capped mutant RNAs were used to measure negative-strand synthesis. Although the mutant RNAs were stable and functional mRNAs, they were not active templates for negative-strand RNA synthesis. Therefore, the 5′ cloverleaf is a multifunctional cis-acting replication element required for the initiation of negative-strand RNA synthesis. We propose a replication model in which the 5′ and 3′ ends of viral RNA interact to form a circular ribonucleoprotein complex that regulates the stability, translation and replication of poliovirus RNA.
Keywords: poliovirus/protein synthesis/ribonucleoprotein/RNA replication/RNA stability
Introduction
Poliovirus genomic RNA is a multifunctional molecule that serves as an mRNA for viral protein synthesis and as a template for negative-strand RNA synthesis. The RNA contains cis-active elements, which regulate its translation and replication, and a single open reading frame that encodes the viral capsid and replication proteins (Figure 1A). The multifunctional nature of this RNA molecule necessitates that regulatory mechanisms, mediated by cis-active RNA replication elements and trans-active viral and cellular proteins, control its many activities.
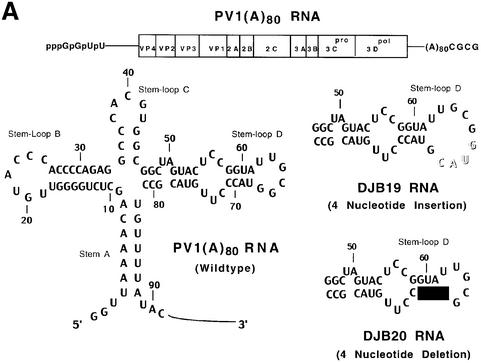
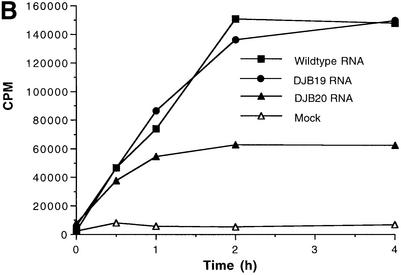
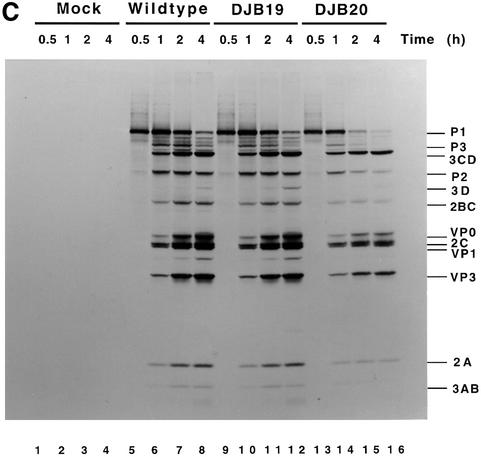
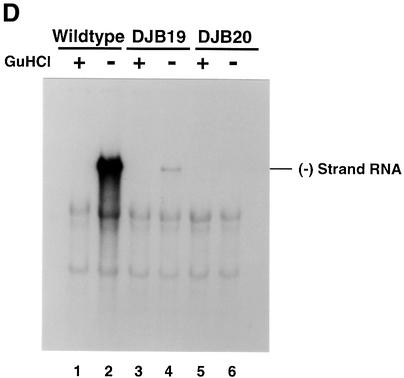
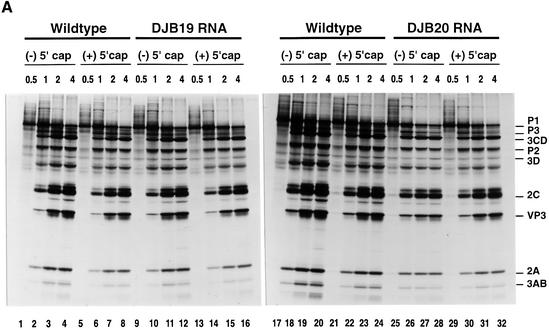
Fig. 1. Effect of 5′ cloverleaf mutations on viral protein synthesis and negative-strand RNA synthesis. (A) Diagram of poliovirus transcript RNA, T7-PV1(A)80 RNA, and 5′ cloverleaf structure. The wild-type cloverleaf structure, the GUAC insertion in stem–loop D in DJB19 RNA and the GUAC deletion (black box) in stem–loop D in DJB20 RNA are shown. (B) Viral protein synthesis was measured in HeLa S10 in vitro translation–replication reactions containing either T7-PV1(A)80 RNA (wild type), DJB19 RNA or DJB20 RNA. The translation reactions contained [35S]methionine (1.2 mCi/ml) and 50 µg/ml RNA as indicated. At the indicated times, 1 µl samples were removed from each reaction. The labeled viral proteins were precipitated in 5% trichloroacetic acid, collected on filters and quantitated by scintillation counting. The amount of labeled protein synthesized in each reaction was plotted as a function of reaction time. (C) SDS–PAGE analysis of the labeled viral proteins. At the indicated times, 4 µl samples were removed from each translation reaction. The labeled viral proteins were solubilized in 50 µl of SDS sample buffer, denatured at 100°C for 3 min and 20 µl portions were separated by electrophoresis in an SDS–9–18% polyacrylamide gel. The gel was fixed and fluorographed. (D) Negative-strand RNA synthesis was measured using preinitiation RNA replication complexes isolated from HeLa S10 translation–RNA replication reactions containing guanidine HCL and each of the indicated RNAs using the procedures described in Materials and methods. Reactions containing the preinitiation RNA replication complexes were incubated at 37°C for 32 min and 32P-labeled negative-strand RNA was fractionated by CH3HgOH–agarose gel electrophoresis. The position of poliovirus negative-strand RNA is indicated.
Following translation, poliovirus genomic RNA acts as a template for negative-strand RNA synthesis. Once poliovirus RNA templates are cleared of translating ribosomes, the RNA replication cycle begins with the initiation of negative-strand RNA synthesis. This requires a membrane-associated replication complex of viral and cellular proteins and a viral RNA template that contains the cis-active replication elements needed for negative-strand synthesis (Caliguiri and Tamm, 1969, 1970; Bienz et al., 1987, 1992, 1994; Egger et al., 2000). Conserved sequences and structures in the 3′ untranslated region (UTR) in poliovirus RNA are needed for efficient negative-strand synthesis (Sarnow et al., 1986; Rohll et al., 1995; Pilipenko et al., 1996; Melchers et al., 1997; Mirmomeni et al., 1997; J.Wang et al., 1999). Although needed for efficient replication, there appears to be a significant degree of flexibility in the sequences and structures needed for this function since chimeric polioviruses, which contain 3′ UTRs from other picornaviruses are viable (Rohll et al., 1995). The 3′ terminal poly(A) tail is also essential for poliovirus replication. It is known that the length of the poly(A) tail is an important factor in the virus replication cycle since removing most of the poly(A) tail from poliovirion RNA or using transcripts with a poly(A)12 tail reduces the specific-infectivity of viral RNA by >10-fold (Spector and Baltimore, 1974; Sarnow, 1989). A comparison of poliovirus RNA transcripts with poly(A)12 and poly(A)80 tails in HeLa S10 translation–replication reactions showed that a short poly(A) tail has no significant effect on translation but severely inhibits negative-strand RNA synthesis (Barton et al., 1996). Therefore, the 3′ UTR and the associated poly(A) tail serve as important cis-active determinants in the initiation of negative-strand synthesis.
A conserved stem–loop structure in the 2C coding sequence and a 5′ terminal cloverleaf structure are also important determinants in the replication of poliovirus RNA. The stem–loop structure in the 2C coding sequence is reported to be a cis-active replication element (CRE) that is required for negative-strand synthesis in cells transfected with viral RNA (Goodfellow et al., 2000; Rieder et al., 2000) and in the synthesis of VPg-pUpU in vitro (Paul et al., 2000; Rieder et al., 2000). The 2C CRE is conserved in the Enterovirus genus and appears to be functionally related to a similar replication element in the P1-coding region of human rhinovirus type 14 (McKnight and Lemon, 1998) and cardioviruses (Lobert et al., 1999). The 5′ cloverleaf structure is also conserved in other enterovirus genomes (Zell and Stelzner, 1997). Disrupting this structure can severely inhibit poliovirus RNA replication in infected cells (Andino et al., 1990a, 1993; Xiang et al., 1995; Zhao et al., 2000) and block the association of viral and cellular proteins with this RNA in binding assays (Andino et al., 1990a, 1993; Xiang et al., 1995). Based on these and other observations, it was suggested that the 5′ cloverleaf forms a functional ribonucleoprotein complex that is required in trans for the initiation of synthesis of a new positive-strand RNA molecule at the 3′ end of negative-strand RNA templates (Andino et al., 1993). A similar model was also proposed for the replication of the positive-strand RNA plant virus, brome mosaic virus (Progue and Hall, 1992).
In this study, we investigated the role of the 5′ cloverleaf in the initiation of poliovirus negative-strand RNA synthesis. We show that 5′ cloverleaf mutations decreased the stability of poliovirus RNA and dramatically inhibited negative-strand RNA synthesis in preinitiation replication complexes. A 5′ 7-methyl guanosine cap stabilized the mutant RNAs but did not restore their ability to serve as templates for negative-strand synthesis. The results of this study indicate that the 5′ cloverleaf structure is a cis-acting replication element in poliovirus genomic RNA that is needed for both stability and negative-strand RNA synthesis. We propose a model in which the 5′ and 3′ ends of poliovirus RNA interact to form a circular ribonucleoprotein complex that is required for the initiation of negative-strand RNA synthesis.
Results
Effect of 5′ cloverleaf mutations on protein synthesis and negative-strand RNA synthesis
To investigate the role of the 5′ cloverleaf structure in poliovirus RNA replication, we compared the translation and replication of two mutant RNAs with that of T7-PV1(A)80 RNA (wild type) using HeLa S10 translation–RNA replication reactions and preinitiation RNA replication complexes. Small insertion and deletion mutations were engineered in stem–loop D of the 5′ cloverleaf to form DJB19 RNA [T7-PV1(A)80 G66+(GUAC) RNA] and DJB20 RNA [T7-PV1(A)80 ΔG67-C70 RNA], respectively (Figure 1A). In agreement with previous studies (Trono et al., 1988), DJB20 RNA was non-infectious in transfected cells and DJB19 RNA resulted in the formation of virus with a small-plaque phenotype (data not shown) (Kuge and Nomoto, 1987; Andino et al., 1990b).
Viral protein synthesis and polyprotein processing were quantitatively normal for DJB19 RNA relative to wild-type RNA (Figure 1B and C, lanes 5–12). Protein synthesis was reduced by about half in reactions containing DJB20 RNA although polyprotein processing was normal (Figure 1B and C, lanes 13–16). The initial rate of DJB20 RNA translation was similar to wild-type RNA, but was significantly inhibited after 30 min (Figure 1B). The reduced level of protein synthesis found with DJB20 RNA was unexpected since the 5′ cloverleaf is not required for translation initiation by the internal ribosome entry site (IRES) in the 5′ UTR of poliovirus RNA (Wimmer et al., 1993).
Negative-strand synthesis was measured using preinitiation RNA replication complexes that were resuspended in reaction mixes containing puromycin and [α-32P]CTP as described in Materials and methods. The presence of puromycin in these reactions displaced translating ribosomes from viral RNA templates and provided optimal conditions for negative-strand RNA synthesis (Barton et al., 1999). We and others have previously shown that only labeled negative-strand RNA is synthesized in reactions containing preinitiation replication complexes formed with transcript RNAs containing two non-viral 5′ terminal Gs (Figure 1A) (Barton et al., 1996, 1999; Herold and Andino, 2000). The presence of the two extra 5′ Gs has no effect on negative-strand synthesis but reduces positive-strand synthesis below detectable levels. As expected, full-length labeled negative-strand RNA was synthesized in preinitiation RNA replication complexes containing wild-type RNA transcripts (Figure 1D, lane 2). In marked contrast, negative-strand RNA synthesis was severely inhibited in preinitiation complexes containing DJB19 RNA (Figure 1D, lane 4). With DJB20 RNA, negative-strand synthesis was inhibited below detectable levels (Figure 1D, lane 6). Therefore, disruption of the 5′ cloverleaf structure resulted in a dramatic inhibition of negative-strand synthesis.
Mutations in 5′ cloverleaf destabilize poliovirus RNA
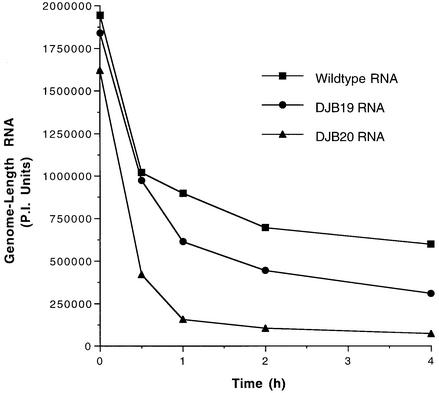
Because defects in viral RNA stability would ultimately lead to defects in translation and RNA replication, we compared the stability of the mutant RNAs with wild-type poliovirus RNA in HeLa S10 translation–RNA replication reactions. The stability of viral RNAs added to these reactions was determined by measuring the amount of 32P-labeled full-length RNA that was recovered as a function of incubation time. Since the HeLa S10 translation–replication reactions were programmed with excess amounts of input RNA to obtain maximum rates of protein synthesis, a significant amount of this RNA is degraded during the first 30 min of the reaction (Figure 2). The majority of the remaining input RNA, however, was stable for the balance of the reaction. DJB19 RNA and DJB20 RNA were both degraded more rapidly than wild-type RNA in these reactions (Figure 2). DJB20 RNA was very unstable and was completely degraded during the first hour of the reaction (Figure 2). DJB19 RNA exhibited an intermediate level of stability, and the amount of this RNA that was detected at 4 h was about half the amount of wild-type RNA (Figure 2). These results indicate that the 5′ cloverleaf plays a crucial role in maintaining the stability of poliovirus RNA.
Fig. 2. Mutations in 5′ cloverleaf structure decrease the stability of poliovirus RNA. Four micrograms of 32P-labeled T7-PV1(A)80 RNA, DJB19 RNA or DJB20 RNA (∼580 000 c.p.m./µg) were added to 50 µl of HeLa S10 translation–RNA replication reactions containing 2 mM guanidine HCl. The reactions were incubated at 34°C and 7 µl samples were removed at the indicated times and added to 300 µl of 0.5% SDS buffer. The labeled RNA samples were phenol extracted, ethanol precipitated and analyzed by electrophoresis in a CH3HgOH–agarose gel. The amount of full-length labeled viral RNA detected at each time point was quantitated using PhosphorImager (PI) analysis. Arbitrary PI units of full-length viral RNA were plotted versus incubation time.
The results of the stability assays appear to be generally consistent with the amount of labeled viral protein that was synthesized in the same reactions. When viral protein synthesis was measured using wild-type RNA, labeled proteins were found to accumulate in a linear manner during the first 2 h of the reaction (Figure 1B). In contrast, protein synthesis was inhibited after ∼30 min in reactions containing DJB20 RNA (Figure 1B). The rapid degradation of DJB20 RNA in this reaction was consistent with the early termination of protein synthesis and the inhibition of negative-strand synthesis. The reduced stability of DJB19 RNA might also be expected to have some effect on the translation of this mutant RNA. However, the fact that excess input RNA was added at the start of the HeLa S10 translation–RNA replication reactions may explain why the reduced stability of DJB19 RNA did not appear to have a measurable effect on translation. Although viral protein synthesis was not inhibited by the mutation in DJB19 RNA, negative-strand synthesis was severely inhibited (Figure 1D, lane 4). The difference observed between translation and negative-strand synthesis could have resulted in part from the fact that the majority of the viral proteins were synthesized during the first 2 h of the reaction. In contrast, the preinitiation RNA replication complexes were isolated after 4 h of incubation. Therefore, the instability of the mutant RNAs might be expected to have a greater effect on negative-strand synthesis than on translation.
Effect of 5′cap on the translation, stability and replication of DJB19 and DJB20 RNAs
As part of a separate study, we discovered that the presence of a 5′ terminal 7-methyl guanosine cap restored the stability of a mutant poliovirus RNA that contained a large (30 nt) deletion in the 5′ cloverleaf (data not shown). This suggested that the instability observed with 5′ cloverleaf mutations was due to an increased susceptibility of these RNAs to degradation by nucleases present in the HeLa S10 extracts. By overcoming defects in RNA stability by adding a 5′ cap, we were able to determine whether the 5′ cloverleaf mutations in DJB19 and DJB20 RNA affected subsequent steps in the viral replication cycle independent of their effect on RNA stability.
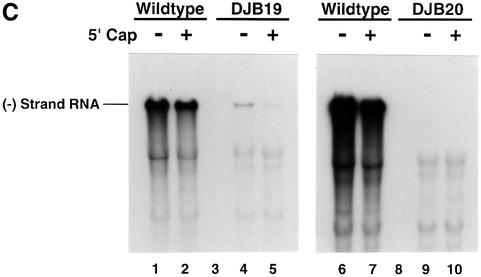
As expected, a 5′ cap had no significant effect on the translation (Figure 3A, lanes 1–8 and 17–24), stability (Figure 3B, lanes 1, 2, 5 and 6) or replication (Figure 3C, lanes 1, 2, 6 and 7) of wild-type RNA. Capped DJB19 RNA (Figure 3A, lanes 13–16) was translated with equal efficiency with wild-type RNA (Figure 3A, lanes 5–8) and was stable in preinitiation RNA replication complexes (Figure 3B, lanes 3 and 4). Capped DJB19 RNA, however, did not serve as a functional template for negative-strand RNA synthesis (Figure 3C, lane 5). In contrast to wild-type RNA, only a trace amount of labeled negative-strand RNA was synthesized in a 30 min reaction. Therefore, negative-strand synthesis was severely inhibited with this mutant RNA although it was stable and translated normally.
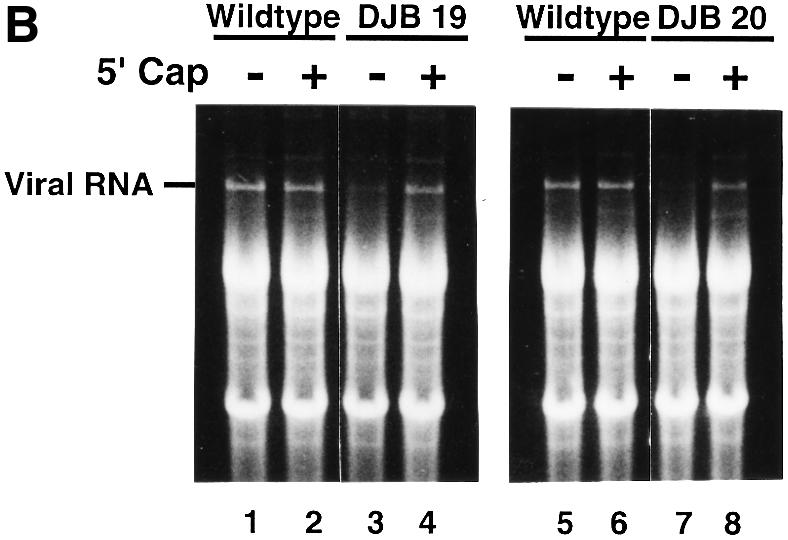
Fig. 3. Capped DJB19 and DJB20 RNAs are stable and translate at wild-type or slightly reduced levels but are not functional templates for negative-strand synthesis. (A) Labeled viral protein synthesis was measured as described in Figure 1 using HeLa S10 translation–RNA replication reactions that contained T7-PV1(A)80 RNA (wild type), DJB19 RNA or DJB20 RNA (50 µg/ml) with or without a 5′ cap as indicated. Labeled viral proteins synthesized at each time point were analyzed by SDS–PAGE as described in Figure 1C. (B and C) The stability of the input RNAs and negative-strand RNA synthesis was measured in reactions containing preinitiation RNA replication complexes that were isolated from HeLa S10 translation–RNA replications containing guanidine HCl and the indicated viral RNAs. The reactions were incubated at 37°C for 30 min and the total RNA and the labeled product RNAs were analyzed by CH3HgOH–agarose gel electrophoresis as described in Materials and methods. (B) UV light visualization of total RNA within the gels after being stained with ethidium bromide is shown. The position of the input viral RNA in the gel is indicated. (C) 32P-labeled negative-strand RNA detected by autoradiography of the dried gels. The position of 32P-labeled poliovirus negative-strand RNA is marked.
The presence of a 5′ cap also restored the stability of DJB20 RNA in preinitiation RNA replication complexes (Figure 3B, lanes 7 and 8). A slight increase in translation was observed with capped DJB20 RNA (Figure 3A, compare lanes 25–28 and 29–32), but viral protein synthesis remained at ∼60% of the level observed with wild-type RNA (Figure 3A, lanes 17–24). This result was similar to a previous report where a six-nucleotide insertion mutation in the cloverleaf was reported to partially inhibit the translation of poliovirus RNA in infected cells (Simoes and Sarnow, 1991). Although capped DJB20 RNA was stable and was translated to produce all of the viral replication proteins, albeit in reduced amounts, negative-strand RNA synthesis was completely inhibited in reactions containing this RNA (Figure 3C, lanes 9 and 10). Taken together, the results indicate that the 5′ cloverleaf mutations inhibited negative-strand RNA synthesis independently of their effect on viral RNA stability or translation.
In vitro complementation assays for the replication of DJB2 and DJB18 RNAs
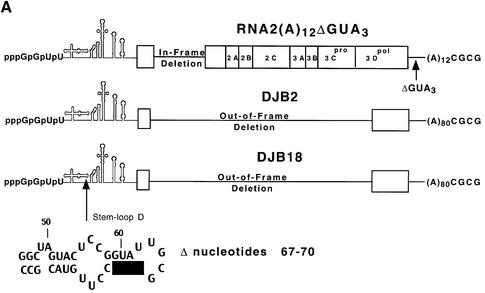
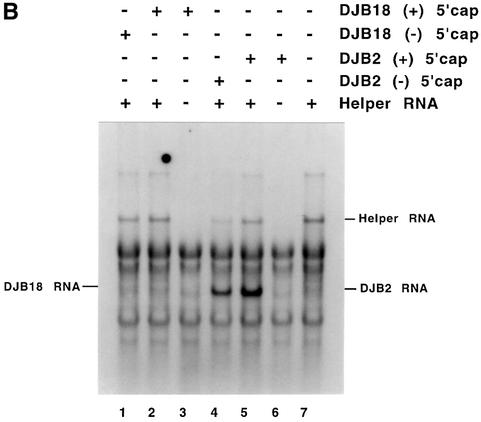
Since the decreased level of translation that was observed with DJB20 RNA might result in less than optimal conditions for measuring negative-strand synthesis, we measured negative-strand synthesis in complementation assays where the viral replication proteins were provided in trans. In this experiment, DJB2 RNA (T7-PV1(A)80ΔC869-T6011 RNA) was used as the positive control for measuring RNA replication (Figure 4A). This subgenomic RNA transcript did not encode any of the viral replication proteins itself, but it acted as a functional template for negative-strand synthesis when co-translated with a helper RNA. Therefore, DJB2 RNA contained the minimum set of the cis-active sequences that were required for negative-strand RNA synthesis in vitro. DJB18 RNA (T7-PV1(A)80ΔC869-T6011ΔG67-C70 RNA) was used as the mutant RNA template in this experiment (Figure 4A). DJB18 RNA was identical to DJB2 RNA except that it contained a 4 nt deletion mutation in the 5′ cloverleaf. This deletion was identical to the mutation in DJB20 RNA (Figure 4A). As described in Materials and methods, RNA2(A)12ΔGUA3 (Figure 4A) was used as the helper RNA in these experiments to provide all of the viral replication proteins in trans.
Fig. 4. Complementation analysis of DJB2 and DJB18 RNA replication in vitro. (A) Diagram of RNA2(A)12ΔGUA3, the helper RNA used in this experiment. The co-translation of this helper RNA provided the viral replication proteins in trans in reactions containing viral transcript RNAs that contained a deletion in the coding sequence for the viral replication proteins. DJB2 RNA does not encode any of the viral replication proteins but was shown in a separate study in our laboratory to contain all of the cis-active replication elements required to act as a template for negative-strand RNA synthesis. DJB18 RNA is identical to DJB2 RNA except for the deletion of poliovirus nucleotides 67–70 in the 5′ cloverleaf. (B) HeLa S10 translation–RNA replication reactions (100 µl) that contained 2 mM guanidine HCl and the indicated RNA (±) 5′ cap were incubated for 4 h at 34°C. The helper RNA was added at a 2:1 molar ratio relative to DJB2 RNA or DJB18 RNA, and the total RNA concentration was maintained at 100 µg/ml in each reaction. Negative-strand RNA synthesis was measured in preinitiation RNA replication complexes that were isolated from these reactions as described in Materials and methods. Reactions containing the preinitiation RNA replication complexes were incubated at 37°C for 25 min, and labeled negative-strand synthesis was analyzed by CH3HgOH–agarose gel electrophoresis. The mobility of DJB2 RNA, DJB18 RNA and the helper RNAs is shown. Control experiments showed that equivalent amounts of the viral replication proteins were synthesized in each reaction that contained the helper RNA (data not shown).
Negative-strand synthesis was measured in preinitiation replication complexes isolated from HeLa S10 translation–replication reactions containing the helper RNA and either DJB2 RNA (±) 5′ cap or DJB18 RNA (±) 5′ cap as indicated (Figure 4B). As expected, DJB2 RNA (±) 5′ cap was an active template for negative-strand synthesis in the presence of the helper RNA (Figure 4B, lanes 4–6). In contrast, capped DJB18 RNA, although stable, was not an active template for negative-strand synthesis even in the presence of the viral replication proteins (Figure 4B, lanes 1–3). Because DJB18 RNA and DJB2 RNA were identical except for the cloverleaf mutation, these results indicate that the 5′ cloverleaf was a cis element that was required for negative-strand RNA synthesis.
Discussion
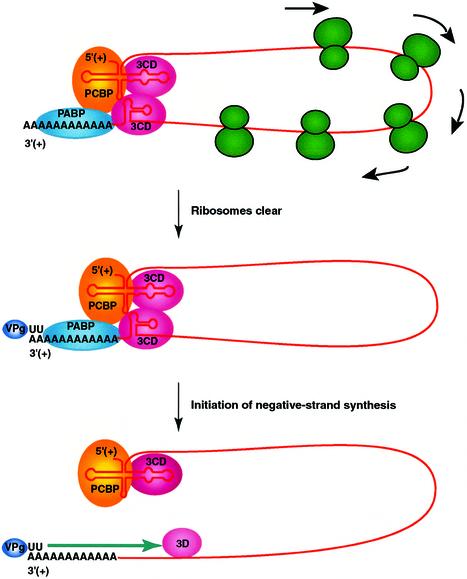
This paper shows that disrupting the structure of the 5′ cloverleaf affects the stability, translation and replication of poliovirus RNA. The addition of a 5′ cap to the mutant RNAs restored their stability to wild-type levels. The restoration of stability, however, did not overcome all of the observed effects of the cloverleaf mutations on translation and negative-strand synthesis. A key finding of this study is that the 5′ cloverleaf is a cis-acting replication element in the poliovirus genome that is required for stability and the initiation of negative-strand synthesis. We propose a replication model in which the 5′ and 3′ ends of poliovirus RNA interact to form a circular ribonucleoprotein (RNP) complex that regulates poliovirus RNA replication (Figure 5).
Fig. 5. Model of circular RNP complex that is used to initiate negative-strand RNA synthesis. Viral proteins 3CD and VPg, poly(A) binding protein (PABP) and poly(rC) binding protein (PCBP) interact with each other and the ends of the viral RNA to form a circular RNP complex. The inhibition of translation initiation allows for the clearance of the viral RNA over time by the elongation of translating ribosomes to the 3′ end of the coding sequence (top). Once the template RNA is cleared of translating ribosomes, VPg-pUpU (or VPg) associates with the 3′ end of the viral RNA template and completes the formation of the circular preinitiation RNA replication complex (middle). Negative-strand synthesis initiates by the elongation of the VPg-primer by the viral polymerase, 3Dpol (bottom). Additional viral and cellular proteins, proteolytic processing events and cellular membranes are also required for viral RNA replication but for clarity are not depicted in this model. See text for additional details.
Role of 5′ cloverleaf in maintaining RNA stability
VPg is a small viral protein that is covalently linked to the 5′ end of poliovirus RNA (Flanegan et al., 1977; Nomoto et al., 1977). This protein is removed from the RNA by a cellular enzyme upon its entry into the cytoplasm of infected cells or HeLa S10 extracts (Hewlett et al., 1976; Nomoto et al., 1976, 1977; Ambros et al., 1978; Ambros and Baltimore, 1980). The resulting RNA possesses a 5′ terminal phosphate rather than a 5′ cap like most eukaryotic mRNAs. Removal of the 5′ cap from cellular mRNAs leads to their rapid degradation by cellular nucleases (Decker and Parker, 1994). In contrast, poliovirus RNA although lacking a 5′ cap is relatively stable in the cytoplasm of infected cells and in HeLa S10 extracts. As demonstrated in this paper, disrupting the structure of the 5′ cloverleaf caused a dramatic reduction in RNA stability. The presence of a 5′ cap on these RNAs, however, restored their stability to wild-type levels. These results suggest that the 5′ cloverleaf, most likely in the form of an RNP complex, functions like a 5′ cap and protects poliovirus RNA from rapid degradation by cellular nucleases. Because significant levels of the viral proteins were not expressed in vitro until after 30 min, a 5′ cloverleaf–RNP complex containing only cellular proteins was apparently responsible for initially protecting the viral RNA from degradation. This in vitro situation mimics what must also occur in vivo when poliovirus RNA is released from virions into the cytoplasm of infected cells. Poly(rC) binding proteins 1 and 2 (PCBP1 and PCBP2) are known to bind to the 5′ cloverleaf (Gamarnik and Andino, 1997; Parsley et al., 1997). The interaction of PCBP and perhaps other cellular proteins with the 5′ cloverleaf may mediate the initial stability of poliovirus RNA prior to the synthesis of the viral replication proteins. This possibility appears to be related to the known ability of PCBP (also described as αCP1 and αCP2) to mediate the stability of α-globin mRNA by binding to a cis-acting stability element in the 3′ UTR and to poly(A) binding protein (PABP) bound to the poly(A) tail (Kiledjian et al., 1995; Z.Wang et al., 1999; Wang and Kiledjian, 2000). Therefore, the interaction of PCBP and perhaps other cellular proteins with the 5′ cloverleaf may help mediate the stability of poliovirus RNA after the removal of VPg and prior to the synthesis of viral replication proteins.
Switch from translation to RNA replication
After being translated to synthesize the viral proteins, poliovirus RNA is cleared of translating ribosomes and then copied to synthesize negative-strand RNA. The molecular mechanisms that regulate this process are not well defined. The viral polymerase is not able to displace translating ribosomes from viral RNA templates and directly mediate the switch from translation to negative-strand synthesis (Barton et al., 1999). Therefore, a mechanism apparently exists to prevent the continuous translation of a molecule of viral RNA, or the replication of this RNA would be inhibited. One or more of the viral proteins may play a role in a feedback pathway to inhibit translation initiation. Previous studies have established that the RNA phages use this type of mechanism to inhibit translation initiation and to clear viral RNAs of translating ribosomes (Kolakofsky and Weissmann, 1971; Weber et al., 1972). For poliovirus, protein 3CD binds the 5′ cloverleaf in association with PCBP (Andino et al., 1990a,b, 1993; Gamarnik and Andino, 1997; Parsley et al., 1997). PCBP also binds to the IRES in the 5′ UTR of poliovirus RNA and is required for translation initiation (Blyn et al., 1996, 1997; Gamarnik and Andino, 1998; Silvera et al., 1999; Walter et al., 1999). It has been proposed that the preferential binding of PCBP to the 5′ cloverleaf in the presence of 3CD might inhibit translation initiation, and play a role in the switch from translation to RNA replication (Gamarnik and Andino, 1998). Although questions remain concerning the functional relevance of this and other models, it seems likely that the 5′ cloverleaf is involved in this process. Therefore, it is possible that 5′ cloverleaf mutations could affect the feedback inhibition of translation initiation and as a result indirectly inhibit negative-strand synthesis (Gamarnik and Andino, 1998). It is important to note that this was not responsible for the inhibition of negative-strand synthesis observed in this study, since negative-strand synthesis was always measured in the presence of puromycin (Barton et al., 1999). Therefore, we were able to determine how 5′ cloverleaf mutations affected negative-strand synthesis independent of any effect they may have had on the mechanism regulating the normal switch from translation to replication.
Disrupting the 5′ cloverleaf structure inhibited negative-strand synthesis
Only trace amounts of labeled negative-strand RNA were synthesized in preinitiation RNA replication complexes containing capped DJB19 RNA. Since this RNA was stable and translated at wild-type levels, we concluded that the 4 nt insertion mutation in the 5′ cloverleaf of this RNA was directly responsible for the dramatic decrease in negative-strand RNA synthesis observed. In experiments with capped DJB20 RNA, we showed that negative-strand synthesis was completely inhibited by the 4 nt deletion in the 5′ cloverleaf. Although protein synthesis was partially inhibited in these reactions, the results suggest that the structural change in the 5′ cloverleaf of this RNA was responsible for the inhibition in negative-strand synthesis. This was confirmed in an in vitro complementation assay where we showed that the 4 nt deletion mutation was directly responsible for the observed inhibition in negative-strand synthesis. These results demonstrate that the 5′ cloverleaf was required in cis for the initiation of negative-strand RNA synthesis.
The 5′ cloverleaf structure appears to be functionally different from the 2C CRE that was recently reported to be needed for negative-strand synthesis in transfected cells (Goodfellow et al., 2000; Rieder et al., 2000). In the in vitro replication assays described here, we showed that poliovirus RNA that contained a deletion of the entire 2C-coding sequence (i.e. DJB2 RNA) was a functional template for negative-strand synthesis. This indicates that the 2C CRE was not required in cis for negative-strand synthesis in preinitiation replication complexes in vitro. Although this finding appears to be in conflict with the published results referenced above, it should be noted that the helper RNA used in our in vitro assays contained the 2C coding-sequence including the 2C CRE. Therefore, it is possible that the 2C CRE was able to function in trans in the in vitro assays. Although additional work is required to investigate this possibility, there is a fundamental difference between the 5′ cloverleaf and the 2C CRE in terms of being required as a component of the template RNA during negative-strand synthesis in vitro.
Circular model for viral RNA replication
The results of this study indicate that sequences at the 5′ end of the viral genome are required for negative-strand synthesis. Previous studies have shown that sequences at the 3′ end including the poly(A) tail are also required for efficient negative-strand synthesis. Therefore, we propose a model for poliovirus replication that emphasizes a direct interaction between the seemingly distal RNA structures at the termini of the genomic RNA (Figure 5). This model is related to existing models of cellular translation that propose an interaction between the 5′ and 3′ terminal RNA elements of cellular mRNAs (Hentze, 1997; Sachs et al., 1997). Furthermore, poliovirus RNA may fold into structures that favor a proximal orientation between the 5′ and 3′ ends of the viral genome (Palmenberg and Sgro, 1997). A circular RNP complex, providing communication between the termini of viral RNA, could then coordinately regulate viral RNA stability, translation and negative-strand synthesis.
Since poliovirus genomic RNA is a linear molecule, the finding that the 5′ cloverleaf is required for the initiation of negative-strand synthesis at its 3′ end suggests that there is a direct interaction between the ends to form a circular complex. An RNP complex would form that is composed of the 5′ and 3′ ends as well as viral and cellular proteins (Figure 5). Interactions between the proteins bound to the two ends may form a protein bridge and help form a circular RNP complex. For example, 3CD is known to bind to the 5′ and 3′ UTRs (Andino et al., 1993; Harris et al., 1994; Xiang et al., 1995; Gamarnik and Andino, 1997). Therefore, a homodimer of 3CD bound to both ends might support the formation of a circular RNP complex. PCBP and PABP are known to bind to the 5′ and 3′ ends of poliovirus RNA, respectively. These two proteins are also known to bind to each other in an RNA-dependent fashion (Z.Wang et al., 1999; Wang and Kiledjian, 2000). PCBP is known to bind to the cytosine-rich element in the 3′ UTR of α-globin mRNA (Chkheidze et al., 1999) and to PABP bound to the poly(A) tail. PABP enhances the binding of PCBP to the α-globin 3′ UTR, and conversely, PCBP enhances the binding of PABP to the poly(A) tail (Wang and Kiledjian, 2000). In the case of poliovirus RNA, PCBP binds to a cytosine-rich element in the 5′ cloverleaf and PABP binds to the 3′ poly(A) tail. Therefore, an RNA-dependent PCBP–PABP interaction could also help stabilize the circular RNP complex depicted in Figure 5.
An attractive aspect of a circular RNP replication complex is that its formation could play a direct role in the switch from translation to replication, stabilize the RNA against nuclease digestion and facilitate the ability of the 5′ cloverleaf to play a direct role in the initiation of negative-strand synthesis. The formation of a circular RNP complex in the presence of the viral replication proteins may effectively block the continued binding of translation initiation factors and ribosomes to the 5′ terminal IRES. As a result, the viral RNA would be naturally cleared of translating ribosomes over a period of time via their continued elongation and termination at the 3′ boundary of the viral open reading frame (Figure 5, top). The cleared genomic RNA could then serve as a template for negative-strand synthesis. To avoid the dilemma of polymerase/ribosome collisions, it is likely that the synthesis of a VPg-pUpU primer (Figure 5, middle) and negative-strand initiation (Figure 5, bottom) does not occur until the viral RNA is cleared of translating ribosomes. One possibility is that the conserved stem–loop structure in the 2C CRE sequence, which is required for VPg uridylylation in vitro (Paul et al., 2000; Rieder et al., 2000), only folds into a functional stem–loop structure after this region of the viral RNA is cleared of translating ribosomes. This would act as a RNA molecular switch in the template that detects the end of translation and initiates VPg uridylylation and negative-strand RNA synthesis. The synthesis of negative-strand RNA would be expected to disrupt the circular orientation of the genomic RNA and prevent additional rounds of negative-strand synthesis. This could be an important factor in limiting negative-strand synthesis in establishing an asymmetric mode of replication that favors positive-strand synthesis overall. Finally, positive-strand RNA synthesis would involve repeated rounds of initiation on the newly synthesized negative-strand template essentially as previously described (Andino et al., 1993; Harris et al., 1994). Therefore, the 5′ cloverleaf appears to be a multifunctional cis-active structure that is used to regulate RNA stability, translation and both steps in the RNA replication cycle. It also plays a key role in the recognition of viral RNAs by the viral replication proteins and increases the fidelity of replication by insuring that viral RNAs with defective 5′ ends are not copied during the replication cycle. This is another example of how viruses have evolved to be very efficient in using the limited amount of genetic information in their genomes.
Materials and methods
Poliovirus cDNA clones
A cDNA clone of the Mahoney strain of type I poliovirus, pT7D-polio (Sarnow, 1989), was originally obtained from Dr Peter Sarnow (Stanford University, Palo Alto, CA). The pT7D-polio clone [pT7-PV1(A)12] was modified by lengthening the 3′ terminal poly(A) sequence from 12 to 80 nucleotides. This modified full-length cDNA clone of poliovirus type 1 was designated as pT7-PV1(A)80 (Figure 1A) and was used as the parental clone for the following constructs. (i) pDJB2 [pT7-PV1(A)80 ΔC869-T6011] contained a 5143 nt out-of-frame deletion and was constructed by digesting pT7-PV1(A)80 with SfuI, filling in the two nucleotide overhang with Klenow polymerase, and ligating the blunt ends. This created a new NruI site at the junction site at nt 866. (ii) pDJB17 [pT7-PV1(A)80ΔC869-T6011 G66+(GTAC)] was created by digesting pDJB2 with Acc65I, filling in the 5′ overhang and ligating the blunt ends. (iii) pDJB18 [pT7-PV1(A)80ΔC869-T6011 ΔG67-C70] was constructed by digesting pDJB2 with KpnI, blunt-end polishing with bacteriophage T4 DNA polymerase and blunt-end ligating. (iv) pDJB19 [pT7-PV1(A)80 G66+(GTAC)] was constructed by ligating the MluI to BlpI fragments of pDJB17 with the BlpI to MluI fragments of pT7-PV1(A)80. This created a full-length poliovirus clone with a GTAC insertion at nucleotide G66. (v) pDJB20 [pT7-PV1(A)80 ΔG67-C70)] was engineered by ligating the MluI to BlpI fragments of pDJB18 with the BlpI to MluI fragments of pT7-PV1(A)80 to create a full-length clone with a GTAC deletion following nucleotide G66. All mutations were confirmed by DNA sequencing. A poliovirus mutant similar to pDJB19 was previously described as 5NC-11 or DNC-11 and had a small-plaque phenotype in transfected cells (Trono et al., 1988; Andino et al., 1990a,b). Plasmid DNA similar to pDJB20 encoding poliovirus RNA with a lethal GTAC deletion following nucleotide G66 was previously described as pPN-2 (Trono et al., 1988) or ΔK (Kuge and Nomoto, 1987). (vi) pRNA2(A)12ΔGUA3 [pT7-PV1(A)12 ΔC1175-C2956 ΔG7418-A7422], a GTA3 deletion following nucleotide G7417, was engineered in the 3′ UTR of pT7-PV1(A)12 by site-directed mutagenesis (V.Chow and J.B.Flanegan, unpublished results). A 1782 nt deletion was then constructed in the P1 coding region by digesting with NruI and SnaBI and blunt-end ligating. Transcription of this plasmid DNA generated a subgenomic-sized viral RNA transcript with an in-frame deletion in the capsid-coding region and a GUA3 deletion in the 3′ UTR. Translation of this RNA resulted in the synthesis of all of the viral replication proteins from the P2 and P3 regions of the viral genome. The presence of the GUA3 deletion in the 3′ UTR had no effect on translation but partially inhibited negative-strand RNA synthesis in the in vitro replication assays used in this study. These features made RNA2(A)12ΔGUA3 a good helper RNA to use in the in vitro replication assays.
Viral RNA preparation
Viral RNA was transcribed in vitro from MluI-linearized plasmid DNA with bacteriophage T7 RNA polymerase in reactions containing 1 mM of each NTP as previously described (Barton et al., 1996). To synthesize viral RNAs with a 7-methyl guanosine cap analog, the concentration of GTP in the transcription reactions was lowered to 200 µM, and 1 mM 7-methyl guanosine cap analog (Epicenter Technologies, Madison, WI) was added. Radiolabeled viral RNA was prepared by including [α-32P]CTP (Amersham) in the transcription reactions. For all experiments, the viral RNAs were purified by chromatography on a Sephadex G50 column and by ethanol precipitation. RNA was quantitated by measuring UV absorption at 260 nm.
HeLa S10 translation–RNA replication reactions
HeLa S10 extracts and HeLa cell translation initiation factors were prepared as previously described (Barton et al., 1996). HeLa S10 translation–RNA replication reactions contained 50% (by volume) HeLa S10 extract, 20% (by volume) translation initiation factors, 10% (by volume) 10× nucleotide reaction mix (10 mM ATP, 2.5 mM GTP, 2.5 mM UTP, 600 mM KCH3CO2, 300 mM creatine phosphate, 4 mg/ml creatine kinase, 155 mM HEPES–KOH pH 7.4), and the indicated viral RNA(s) at 50–100 µg/ml as indicated. [35S]methionine (Amersham; >1000 Ci/mmol) was included in reactions to measure viral protein synthesis as previously described (Barton et al., 1996). 32P-labeled viral RNA transcripts were added at the start of these reactions to assay for RNA stability as described in the figure legends.
Viral negative-strand RNA synthesis
Poliovirus negative-strand RNA synthesis was measured using preinitiation RNA replication complexes as previously described (Barton et al., 1996, method 4). Briefly, HeLa S10 translation–replication reactions containing poliovirus transcript RNA were incubated for 4 h at 34°C in the presence of 2 mM guanidine HCl. Preinitiation RNA replication complexes were isolated from these reactions by centrifugation and resuspended in 50 µl reaction mixes containing 50 µg/ml puromycin, 5 µM CTP and 20 µCi of [α-32P]CTP. The reactions were incubated at 37°C for the indicated times in the figure legends and were terminated by the addition of 350 µl of 0.5% SDS buffer. The reaction products were then phenol extracted, ethanol precipitated, and analyzed by CH3HgOH–agarose gel electrophoresis. Labeled product RNA was detected by autoradiography of the dried gel. We and others have previously shown that the presence of two non-viral G nucleotides at the 5′ terminus of poliovirus transcript RNAs (Figure 1A) inhibits positive-strand RNA synthesis below detectable levels in these reactions (Barton et al., 1996, 1999; Herold and Andino, 2000). Therefore, the preinitiation replication complexes formed with transcript RNAs were only capable of supporting the synthesis of labeled negative-strand RNA.
Acknowledgments
Acknowledgements
We thank Joan Morasco for excellent technical assistance. This work was supported by Public Health Service grants AI15539 and AI32123 from the National Institute of Allergy and Infectious Diseases. D.J.B. was supported by Public Health Service training grant AI 07110 from the National Institute of Allergy and Infectious Diseases.
References
- Ambros V. and Baltimore,D. (1980) Purification and properties of a HeLa cell enzyme able to remove the 5′-terminal protein from poliovirus RNA. J. Biol. Chem., 255, 6739–6744. [PubMed] [Google Scholar]
- Ambros V., Pettersson,R.F. and Baltimore,D. (1978) An enzymatic activity in uninfected cells that cleaves the linkage between poliovirion RNA and the 5′ terminal protein. Cell, 15, 1439–1446. [DOI] [PubMed] [Google Scholar]
- Andino R., Rieckhof,G.E. and Baltimore,D. (1990a) A functional ribonucleoprotein complex forms around the 5′ end of poliovirus RNA. Cell, 63, 369–380. [DOI] [PubMed] [Google Scholar]
- Andino R., Rieckhof,G.E., Trono,D. and Baltimore,D. (1990b) Substitutions in the protease (3Cpro) gene of poliovirus can suppress a mutation in the 5′ noncoding region. J. Virol., 64, 607–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andino R., Rieckhof,G.E., Achacoso,P.L. and Baltimore,D. (1993) Poliovirus RNA synthesis utilizes an RNP complex formed around the 5′-end of viral RNA. EMBO J., 12, 3587–3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton D.J., Morasco,B.J. and Flanegan,J.B. (1996) Assays for poliovirus polymerase, 3Dpol, and authentic RNA replication in HeLa S10 extracts. Methods Enzymol., 275, 35–57. [DOI] [PubMed] [Google Scholar]
- Barton D.J., Morasco,B.J. and Flanegan,J.B. (1999) Translating ribosomes inhibit poliovirus negative-strand RNA synthesis. J. Virol., 73, 10104–10112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienz K., Egger,D. and Pasamontes,L. (1987) Association of polioviral proteins of the P2 genomic region with the viral replication complex and virus-induced membrane synthesis as visualized by electron microscopic immunocytochemistry and autoradiography. Virology, 160, 220–226. [DOI] [PubMed] [Google Scholar]
- Bienz K., Egger,D., Pfister,T. and Troxler,M. (1992) Structural and functional characterization of the poliovirus replication complex. J. Virol., 66, 2740–2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienz K., Egger,D. and Pfister,T. (1994) Characteristics of the poliovirus replication complex. Arch. Virol. Suppl., 9, 147–157. [DOI] [PubMed] [Google Scholar]
- Blyn L.B., Swiderek,K.M., Richards,O., Stahl,D.C., Semler,B.L. and Ehrenfeld,E. (1996) Poly(rC) binding protein 2 binds to stem–loop IV of the poliovirus RNA 5′ noncoding region: identification by automated liquid chromatography–tandem mass spectrometry. Proc. Natl Acad. Sci. USA, 93, 11115–11120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blyn L.B., Towner,J.S., Semler,B.L. and Ehrenfeld,E. (1997) Requirement of poly(C) binding protein 2 for translation of poliovirus RNA. J. Virol., 71, 6243–6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caliguiri L.A. and Tamm,I. (1969) Membranous structures associated with translation and transcription of poliovirus RNA. Science, 166, 885–886. [DOI] [PubMed] [Google Scholar]
- Caliguiri L.A. and Tamm,I. (1970) Characterization of poliovirus-specific structures associated with cytoplasmic membranes. Virology, 42, 112–122. [DOI] [PubMed] [Google Scholar]
- Chkheidze A.N., Lyakhov,D.L., Makeyev,A.V., Morales,J., Kong,J. and Liebhaber,S.A. (1999) Assembly of the α-globin mRNA stability complex reflects binary interaction between the pyrimidine-rich 3′ untranslated region determinant and poly(C) binding protein αCP. Mol. Cell. Biol., 19, 4572–4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker C.J. and Parker,R. (1994) Mechanisms of mRNA degradation in eukaryotes. Trends Biochem. Sci., 19, 336–340. [DOI] [PubMed] [Google Scholar]
- Egger D., Teterina,N., Ehrenfeld,E. and Bienz,K. (2000) Formation of the poliovirus replication complex requires coupled viral translation, vesicle production, and viral RNA synthesis. J. Virol., 74, 6570–6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanegan J.B., Petterson,R.F., Ambros,V., Hewlett,M.J. and Baltimore,D. (1977) Covalent linkage of a protein to a defined nucleotide sequence at the 5′-terminus of virion and replicative intermediate RNAs of poliovirus. Proc. Natl Acad. Sci. USA, 74, 961–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamarnik A.V. and Andino,R. (1997) Two functional complexes formed by KH domain containing proteins with the 5′ noncoding region of poliovirus RNA. RNA, 3, 882–892. [PMC free article] [PubMed] [Google Scholar]
- Gamarnik A.V. and Andino,R. (1998) Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev., 12, 2293–2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodfellow I., Chaudhry,Y., Richardson,A., Meredith,J., Almond,J.W., Barclay,W. and Evans,D.J. (2000) Identification of a cis-acting replication element within the poliovirus coding region. J. Virol., 74, 4590–4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris K.S., Xiang,W., Alexander,L., Lane,W.S., Paul,A.V. and Wimmer,E. (1994) Interaction of poliovirus polypeptide 3CDpro with the 5′ and 3′ termini of the poliovirus genome: identification of viral and cellular cofactors needed for efficient binding. J. Biol. Chem., 269, 27004–27014. [PubMed] [Google Scholar]
- Hentze M.W. (1997) eIF4G: a multipurpose ribosome adaptor? Science, 275, 500–501. [DOI] [PubMed] [Google Scholar]
- Herold J. and Andino,R. (2000) Poliovirus requires a precise 5′ end for efficient positive-strand RNA synthesis. J. Virol., 74, 6394–6400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewlett M.J., Rose,J.K. and Baltimore,D. (1976) 5′-terminal structure of poliovirus polyribosomal RNA is pUp. Proc. Natl Acad. Sci. USA, 73, 327–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiledjian M., Wang,X. and Liebhaber,S.A. (1995) Identification of two KH domain proteins in the α-globin mRNP stability complex. EMBO J., 14, 4357–4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolakofsky D. and Weissmann,C. (1971) Q-β replicase as repressor of Q-β RNA-directed protein synthesis. Biochim. Biophys. Acta, 246, 596–599. [DOI] [PubMed] [Google Scholar]
- Kuge S. and Nomoto,A. (1987) Construction of viable deletion and insertion mutants of the Sabin strain of type 1 poliovirus: function of the 5′ noncoding sequence in viral replication. J. Virol., 61, 1478–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobert P.E., Escriou,N., Ruelle,J. and Michiels,T. (1999) A coding RNA sequence acts as a replication signal in cardioviruses. Proc. Natl Acad. Sci. USA, 96, 11560–11565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight K.L. and Lemon,S.M. (1998) The rhinovirus type 14 genome contains an internally located RNA structure that is required for viral replication. RNA, 4, 1569–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchers W.J.G., Hoenderop,J.G.J., Bruins Slot,H.J., Pleij,C.W.A., Pilipenko,E.V., Agol,V.I. and Galama,J.M.D. (1997) Kissing of the two predominant hairpin loops in the Coxsackie B virus 3′ untranslated region is the essential structural feature of the origin of replication required for negative-strand RNA synthesis. J. Virol., 71, 686–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirmomeni M.H., Hughes,P.J. and Stanway,G. (1997) An RNA tertiary structure in the 3′ untranslated region of enteroviruses is necessary for efficient replication. J. Virol., 71, 2363–2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomoto A., Lee,Y.F. and Wimmer,E. (1976) The 5′ end of poliovirus mRNA is not capped with m7G(5′)ppp(5′)Np. Proc. Natl Acad. Sci. USA, 73, 375–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomoto A., Kitamura,N., Golini,F. and Wimmer,E. (1977) The 5′-terminal structures of poliovirion RNA and poliovirus mRNA differ only in the genome-linked protein VPg. Proc. Natl Acad. Sci. USA, 74, 5345–5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmenberg A.C. and Sgro,J.-Y. (1997) Topological organization of picornaviral genomes: statistical prediction of RNA structural signals. Semin. Virol., 8, 231–241. [Google Scholar]
- Parsley T.B., Towner,J.S., Blyn,L.B., Ehrenfeld,E. and Semler,B.L. (1997) Poly (rC) binding protein 2 forms a ternary complex with the 5′-terminal sequences of poliovirus RNA and the viral 3CD proteinase. RNA, 3, 1124–1134. [PMC free article] [PubMed] [Google Scholar]
- Paul A.V., Rieder,E., Kim,D.W., van Boom,J.H. and Wimmer,E. (2000) Identification of an RNA hairpin in poliovirus RNA that serves as the primary template in the in vitro uridylylation of VPg. J. Virol., 74, 10359–10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilipenko E.V., Poperechny,K., Maslova,S.V., Melchers,W.J.G., Bruins Slot,H.J. and Agol,V.I. (1996) Cis-element, oriR, involved in the initiation of (–) strand poliovirus RNA: a quasi-globular multi-domain RNA structure maintained by tertiary (‘kissing’) interactions. EMBO J., 15, 5428–5436. [PMC free article] [PubMed] [Google Scholar]
- Progue G.P. and Hall,T.C. (1992) The requirement for a 5′ stem–loop structure in brome mosaic virus replication supports a new model for viral positive-strand RNA initiation. J. Virol., 66, 674–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder E., Paul,A.V., Kim,D.W., van Boom,J.H. and Wimmer,E. (2000) Genetic and biochemical studies of poliovirus cis-acting replication element cre in relation to VPg uridylylation. J. Virol., 74, 10371–10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohll J.B., Moon,D.H., Evans,D.J. and Almond,J.W. (1995) The 3′ untranslated region of picornavirus RNA: features required for efficient genome replication. J. Virol., 69, 7835–7844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs A.B., Sarnow,P. and Hentze,M.W. (1997) Starting at the beginning, middle and end: translation initiation in eukaryotes. Cell, 89, 831–838. [DOI] [PubMed] [Google Scholar]
- Sarnow P. (1989) Role of 3′-end sequences in infectivity of poliovirus transcripts made in vitro. J. Virol., 63, 467–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarnow P., Bernstein,H.D. and Baltimore,D. (1986) A poliovirus temperature-sensitive RNA synthesis mutant in a noncoding region of the genome. Proc. Natl Acad. Sci. USA, 83, 571–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvera D., Gamarnik,A.V. and Andino,R. (1999) The N-terminal K homology domain of the poly(rC)-binding protein is a major determinant for binding to the poliovirus 5′-untranslated region and acts as an inhibitor of viral translation. J. Biol. Chem., 274, 38163–38170. [DOI] [PubMed] [Google Scholar]
- Simoes E.A. and Sarnow,P. (1991) An RNA hairpin at the extreme 5′ end of the poliovirus RNA genome modulates viral translation in human cells. J. Virol., 65, 913–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector D.H. and Baltimore,D. (1974) Requirement of 3′-terminal poly(A) for the infectivity of poliovirus RNA. Proc. Natl Acad. Sci. USA, 71, 2983–2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trono D., Andino,R. and Baltimore,D. (1988) An RNA sequence of hundreds of nucleotides at the 5′ end of poliovirus RNA is involved in allowing viral protein synthesis. J. Virol., 62, 2291–2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter B.L., Nguyen,J.H., Ehrenfeld,E. and Semler,B.L. (1999) Differential utilization of poly(rC) binding protein 2 in translation directed by picornavirus IRES elements. RNA, 5, 1570–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Bakkers,J.M., Galama,J.M., Bruins Slot,H.J., Pilipenko,E.V., Agol,V.I. and Melchers,W.J. (1999) Structural requirements of the higher order RNA kissing element in the enteroviral 3′ UTR. Nucleic Acids Res., 27, 485–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z. and Kiledjian,M. (2000) The poly(A)-binding protein and an mRNA stability protein jointly regulate an endoribonuclease activity. Mol. Cell. Biol., 20, 6334–6341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Day,N., Trifillis,P. and Kiledjian,M. (1999) An mRNA stability complex functions with poly(A)-binding protein to stabilize mRNA in vitro. Mol. Cell. Biol., 19, 4552–4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber H., Billeter,M.A., Kahane,S., Weissmann,C., Hindley,J. and Porter,A. (1972) Molecular basis for repressor activity of Q-β replicase. Nature New Biol., 237, 166–170. [DOI] [PubMed] [Google Scholar]
- Wimmer E., Hellen,C.U.T. and Cao,X. (1993) Genetics of poliovirus. Annu. Rev. Genet., 27, 353–436. [DOI] [PubMed] [Google Scholar]
- Xiang W., Harris,K.S., Alexander,L. and Wimmer,E. (1995) Interaction between the 5′-terminal cloverleaf and 3AB/3CDpro of poliovirus is essential for RNA replication. J. Virol., 69, 3658–3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zell R. and Stelzner,A. (1997) Application of genome sequence information to the classification of bovine enteroviruses: the importance of 5′- and 3′-nontranslated regions. Virus Res., 51, 213–229. [DOI] [PubMed] [Google Scholar]
- Zhao W.D., Lahser,F.C. and Wimmer,E. (2000) Genetic analysis of a poliovirus/hepatitis C virus (HCV) chimera: interaction between the poliovirus cloverleaf and a sequence in the HCV 5′ nontranslated region results in a replication phenotype. J. Virol., 74, 6223–6226. [DOI] [PMC free article] [PubMed] [Google Scholar]