Abstract
Oxa1p is a member of the conserved Oxa1/YidC/Alb3 protein family involved in the membrane insertion of proteins. Oxa1p has been shown previously to directly facilitate the export of the N-terminal domains of membrane proteins across the inner membrane to the intermembrane space of mitochondria. Here we report on a general role of Oxa1p in the membrane insertion of proteins. (i) The function of Oxa1p is not limited to the insertion of membrane proteins that undergo N-terminal tail export; rather, it also extends to the insertion of other polytopic proteins such as the mitochondrially encoded Cox1p and Cox3p proteins. These are proteins whose N-termini are retained in the mitochondrial matrix. (ii) Oxa1p interacts directly with these substrates prior to completion of their synthesis. (iii) The interaction of Oxa1p with its substrates is particularly strong when nascent polypeptide chains are inserted into the inner membrane, suggesting a direct function of Oxa1p in co-translational insertion from the matrix. Taken together, we conclude that the Oxa1 complex represents a general membrane protein insertion machinery in the inner membrane of mitochondria.
Keywords: membrane insertion/mitochondria/Oxa1
Introduction
The mitochondrial inner membrane harbors many proteins that display a wide variation in membrane topologies. A small percentage of these proteins are encoded by the mitochondrial genome, are synthesized in the matrix and become inserted into the inner membrane. The majority of inner membrane proteins, however, are encoded by the nuclear genome and are imported into the mitochondria following their synthesis in the cytoplasm. A subset of inner membrane proteins (both nuclear and mitochondrially encoded) whose N-termini reside in the intermembrane space (i.e. in the compartment between the outer and inner membranes) are sorted by way of insertion from the mitochondrial matrix into the inner membrane (Stuart and Neupert, 1996). This sorting event involves the insertion of hydrophobic transmembrane segments into the inner membrane, coupled to the translocation of hydrophilic segments across the membrane to the intermembrane space. These hydrophilic segments may not be limited to the N-terminal tail regions of proteins, but in the case of polytopic membrane proteins, may also include looped segments between neighboring transmembrane segments and, in some instances, C-terminal segments of polypeptides.
The process of N-tail protein export in mitochondria was found to resemble the N-terminal tail export event in prokaryotes, in both its signal sequence and energetic requirements (Herrmann et al., 1995; Rojo et al., 1995, 1999). The insertion of these proteins into the inner membrane and the coupled export of their N-termini into the intermembrane space require the action of an integral inner membrane protein, termed Oxa1p (He and Fox, 1997; Hell et al., 1997, 1998). Oxa1p is a member of the highly conserved Oxa1p/YidC/Alb3 protein family found throughout prokaryotes and eukaryotes. In eukaryotes, both mitochondrial and chloroplast homologs have been reported (Bauer et al., 1994; Bonnefoy et al., 1994; Sundberg et al., 1997). The prokaryotic homolog in Escherichia coli, YidC, has recently been shown to mediate protein insertion into the plasma membrane from the bacterial cytosol (Houben et al., 2000; Samuelson et al., 2000; Scotti et al., 2000). Furthermore, the chloroplast homolog of Oxa1p, Alb3, has been demonstrated to play a crucial role in the insertion of at least one protein (the light harvesting chlorophyll-binding protein, LHCP) into the thylakoid membrane of chloro plasts (Moore et al., 2000). Thus, the Oxa1p/YidC/Alb3 protein family represents a novel evolutionarily conserved membrane protein insertion machinery (Hell et al., 1998; Samuelson et al., 2000; Stuart and Neupert, 2000).
In mitochondria, Oxa1p is involved in the insertion of a variety of membrane proteins. Examples of proteins that use Oxa1p for their membrane insertion include the mitochondrially encoded subunit 2 of the cytochrome oxidase complex, Cox2p, which spans the inner membrane twice (Nout–Cout), and also Oxa1p itself, a nuclear-encoded polytopic protein, which spans the inner membrane five times in an Nout–Cin orientation. The function of Oxa1p is directly required for the export of at least the N-terminal tail region of these proteins (Hell et al., 1997, 1998). Export of other regions to the intermembrane space, such as hydrophilic loops between neighboring transmembrane domains and also C-terminal hydrophilic segments, was also observed to be inhibited in Oxa1p-deficient mitochondria. It is not known, however, whether the export of domains other than the N-terminal regions directly involves Oxa1p function (Hell et al., 1998). Export of the N-terminal tails across the inner membrane was kinetically found to precede, and even be a prerequisite for, the export step of internal or C-terminal hydrophilic regions of the protein (Herrmann et al., 1995, 1997). So far in our studies we have investigated the sorting of membrane proteins whose N-termini undergo export and reside in the intermembrane space, such as Cox2p.
Here we address the question of whether other mitochondrially encoded proteins require Oxa1p for their membrane insertion. Subunit 1 (Cox1p) and subunit 3 (Cox3p) of the cytochrome oxidase complex, and cytochrome b of the cytochrome bc1 complex, span the inner membrane several times (12, seven and eight times, respectively), in a manner whereby their N-termini reside in the mitochondrial matrix (Tsukihara et al., 1996; Hunte et al., 2000). Attainment of this topology involves the translocation of hydrophilic loops between neighboring transmembrane segments across the inner membrane to their final destination in the intermembrane space.
Using a yeast strain harboring a temperature-sensitive mutation in the OXA1 gene, oxa1-ts, we analyzed the membrane insertion of newly synthesized Cox1p, Cox3p and cytochrome b. Our results demonstrate that these mitochondrial gene products interact with Oxa1p during their synthesis and display a dependency on Oxa1p function for their efficient integration into the inner membrane. Furthermore, we demonstrate that the interaction of these nascent polypeptide chains with Oxa1p is stabilized by the presence of the associated ribosomes. We conclude that the function of Oxa1p is not limited to the export of N-terminal tails of membrane proteins, but rather represents a general membrane insertion machinery for a class of mitochondrial inner membrane proteins.
Results
Oxa1p is required for the efficient membrane insertion of Cox1p, Cox3p and cytochrome b
Isolated wild-type yeast mitoplasts were incubated in the presence of [35S]methionine under conditions that support translation of mitochondrial gene products. Following the translation incubation, mitoplasts were treated with proteinase K to probe for the accessibility of intermembrane space-exposed loops. The newly synthesized proteins Cox1p, Cox2p, Cox3p and cytochrome b were degraded by the added protease (Figure 1A). The inner membrane of the mitoplasts had remained intact under this procedure, as indicated by the inaccessibility of the radiolabeled Var1p protein, a matrix-soluble marker protein to the added proteinase K. This indicated that the hydrophilic loops had indeed become efficiently translocated across the inner membrane to the intermembrane space.
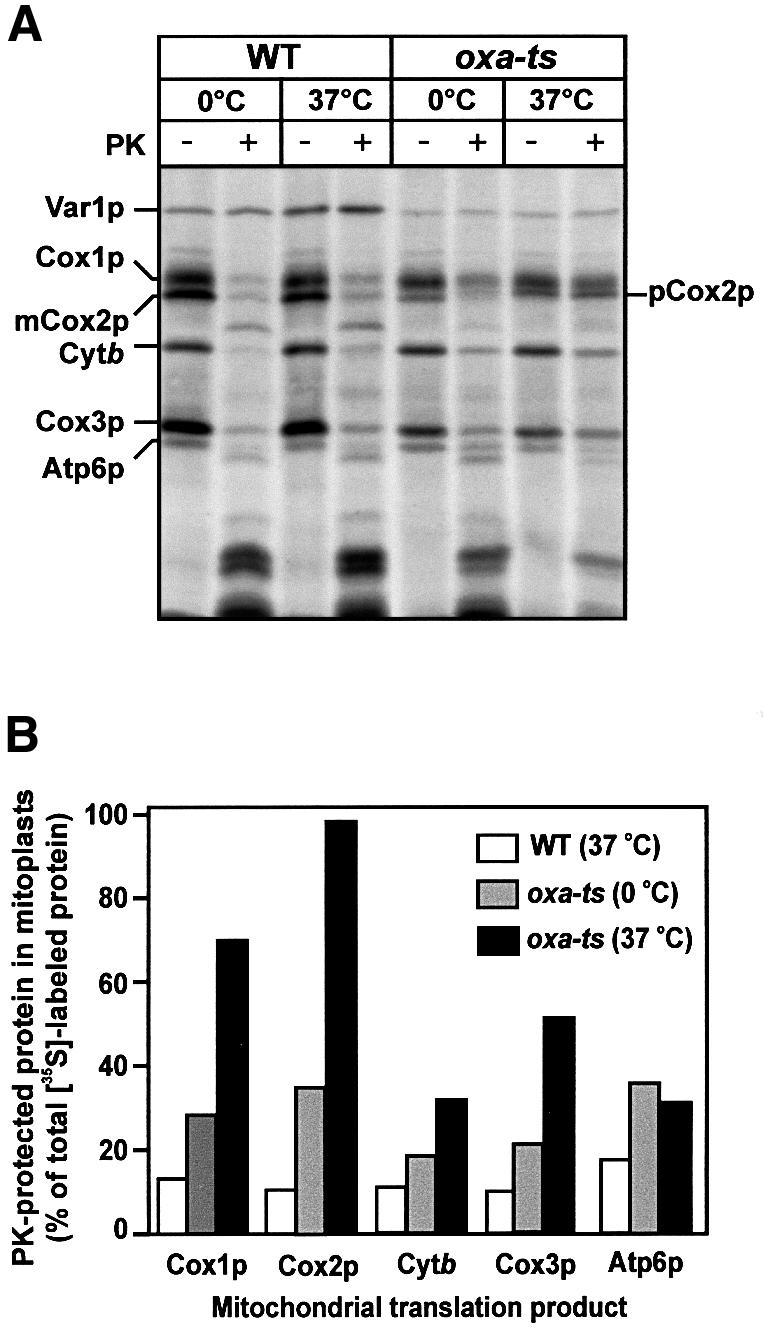
Fig. 1. Membrane insertion of newly synthesized mitochondrial translation products is inhibited in oxa-ts mitoplasts. Mitoplasts prepared from wild-type W303-1A (WT) and oxa-ts mitochondria were pre-incubated in translation buffer for 10 min at either 0 or 37°C, as indicated. In organello translation was performed in these mitoplasts for 20 min at 25°C in the presence of [35S]methionine. Following translation, samples were divided and either mock-treated or treated with proteinase K (PK), as indicated. Mitoplasts were re-isolated and analyzed by SDS–PAGE and autoradiography (A). The levels of radiolabeled proteins after pre-incubation of wild-type and oxa-ts mitoplasts were quantified using a phosphorimager (B). The accessibility of the newly synthesized mitochondrial gene products in both the 0 or 37°C pretreated wild-type mitoplasts was similar [see (A)], hence only the quantification of the 37°C pretreated wild-type sample is given. The levels of pCox2p and mCox2p in the oxa1-ts mitoplasts at 0°C were quantified and summed together, and a total Cox2p signal is given. Abbreviations used are as follows: cytochrome oxidase subunit 1, Cox1p; precursor of subunit 2, pCox2p; mature subunit 2, Cox2p; subunit 3, Cox3p; cytochrome b, Cytb; subunit 6 of ATPase, Atp6p; and the ribosomal protein, Var1p.
The role of Oxa1p in this insertion/export process was then investigated using mitoplasts generated from mitochondria isolated from a yeast strain harboring a temperature-sensitive mutation in the OXA1 gene (oxa1-ts), which had been grown at the permissive temperature (Figure 1A). As demonstrated previously, the Oxa1p function may be inhibited by a brief exposure of mitoplasts isolated from this strain to their non-permissive temperature, prior to the translation event (Hell et al., 1997, 1998). When mitochondrial protein synthesis was performed in the oxa1-ts mitoplasts, following their exposure to 37°C in this manner, Cox2p accumulated almost exclusively as its precursor form (pCox2p) in the mitochondrial matrix, where it was inaccessible to added protease. Furthermore, the membrane insertion of newly synthesized Cox1p, Cox3p and cytochrome b was also adversely affected in the oxa1-ts mitoplasts, as they were inaccessible to added protease, albeit to varying extents (Figure 1A). Exposure to this higher temperature did not adversely affect the export of these proteins in the control wild-type mitochondria. A partial inhibition of insertion of Cox1p, Cox2p, Cox3p and cytochrome b was also observed in the oxa1-ts mitoplasts that had not been exposed to the non-permissive temperature (Figure 1A and B). The function of Oxa1p was apparently already partly compromised in the oxa1-ts mitoplasts at lower temperatures (Figure 1A).
The insertion of another mitochondrially encoded polytopic protein, Atp6p, appeared to be affected in the oxa1-ts mutant. Atp6p, like cytochrome b, displayed a partial dependency on Oxa1p for its efficient insertion into the inner membrane. The inhibition of Atp6p insertion, however, appeared not to be dependent on the prior exposure of the oxa1-ts mitoplasts to the non-permissive temperature of 37°C.
The degree of dependency of the insertion process on the function of Oxa1p varied among the different mitochondrial proteins studied (Figure 1B). As reported previously, the export of pCox2p displayed a very tight requirement for Oxa1p for its membrane insertion (Hell et al., 1997): almost all of the newly synthesized pCox2p remained in the matrix in the oxa1-ts mutant mitoplasts following their exposure to the non-permissive temperature. Cox1p, like pCox2p, showed a strong dependency on Oxa1p function for its insertion, as ∼70% of the newly synthesized Cox1p remained inaccessible to added proteinase K in the oxa1-ts mitoplasts. The insertion of Cox3p and cytochrome b was reduced in the oxa1-ts mitoplasts: ∼50 and 30% of their newly synthesized forms, respectively, remained protease inaccessible in the oxa1-ts mitoplasts (Figure 1B).
Measurement of the turn-over rates of newly synthesized Cox1p, Cox2p and Cox3p indicated similar rates of degradation in both the oxa1-ts and wild-type mitochondria (M.Käser and T.Langer, personal communication). These observations would be inconsistent with the possibility that the newly synthesized Cox1p and Cox3p proteins are correctly inserted into the oxa1-ts mitoplasts, but undergo rapid degradation in the absence of exported Cox2p. Rather, our findings would be consistent with a direct involvement of Oxa1p in the membrane insertion of Cox1p and Cox3p.
In summary, membrane insertion of newly synthesized Cox1p, Cox3p and cytochrome b can be monitored in isolated wild-type mitoplasts. In the absence of Oxa1p function, newly synthesized Cox1p, Cox3p and cytochrome b failed to undergo efficient insertion into the inner membrane. The insertion of Cox1p, like that of pCox2p, was dependent on the presence of a functional Oxa1p, whereas the insertion of Cox3p and cytochrome b, in particular, was dependent to a lesser degree. In summary, the function of Oxa1p is not limited to mediating the membrane insertion and export of N-terminal regions of membrane proteins. Rather, Oxa1p also appears to play a role in the insertion of mitochondrially synthesized proteins whose N-termini are retained on the matrix side of the inner membrane.
Newly synthesized Cox1p, Cox2p and cytochrome b interact directly with Oxa1p
Using a chemical cross-linking analysis, we addressed whether the newly synthesized Cox1p, Cox3p and cytochrome b polypeptides interact directly with Oxa1p.
The reductant-cleavable cross-linking reagent N- succinimidyl-3-[2-pyridyldithio]propionate (SPDP) was added to isolated wild-type mitochondria in which protein synthesis was ongoing (Figure 2A). The proteins being synthesized were monitored by the incorporation of [35S]methionine. The cross-linking of radiolabeled nascent polypeptide chains to Oxa1p was subsequently assessed by immunoprecipitation using an Oxa1p-specific antiserum. Radiolabeled polypeptides cross-linked to Oxa1p were cleaved by the addition of the reductant βmercapto ethanol prior to the analysis of the Oxa1p immunoprecipitate by SDS gel electrophoresis. In addition to Cox2p, radiolabeled Cox3p, cytochrome b and some nascent chains were associated with Oxa1p in a cross-linking-dependent manner. Newly synthesized Cox1p was also found in association with Oxa1p following cross-linking, albeit to a lesser extent than Cox2p, Cox3p and cytochrome b. This relatively low efficiency of cross-linking using SPDP, a cysteine-lysine-specific hetero-bifunctional reagent, may be due to the low abundance of Cys residues in Cox1p polypeptide (one residue in total). In comparison, Cox2p, Cox3p and cytochrome b polypeptides contain more Cys residues (four, two and three residues, respectively). In fact, when cross-linking was performed using the lysine-specific homo-bifunctional cross-linking reagent dithiobis(succinimidyl propionate) (DSP) the efficiency of cross-linking of radiolabeled Cox1p to Oxa1p was significantly higher (Figure 2B). This most likely reflects the abundance of Lys residues in the Cox1p polypeptide (10 residues in total).
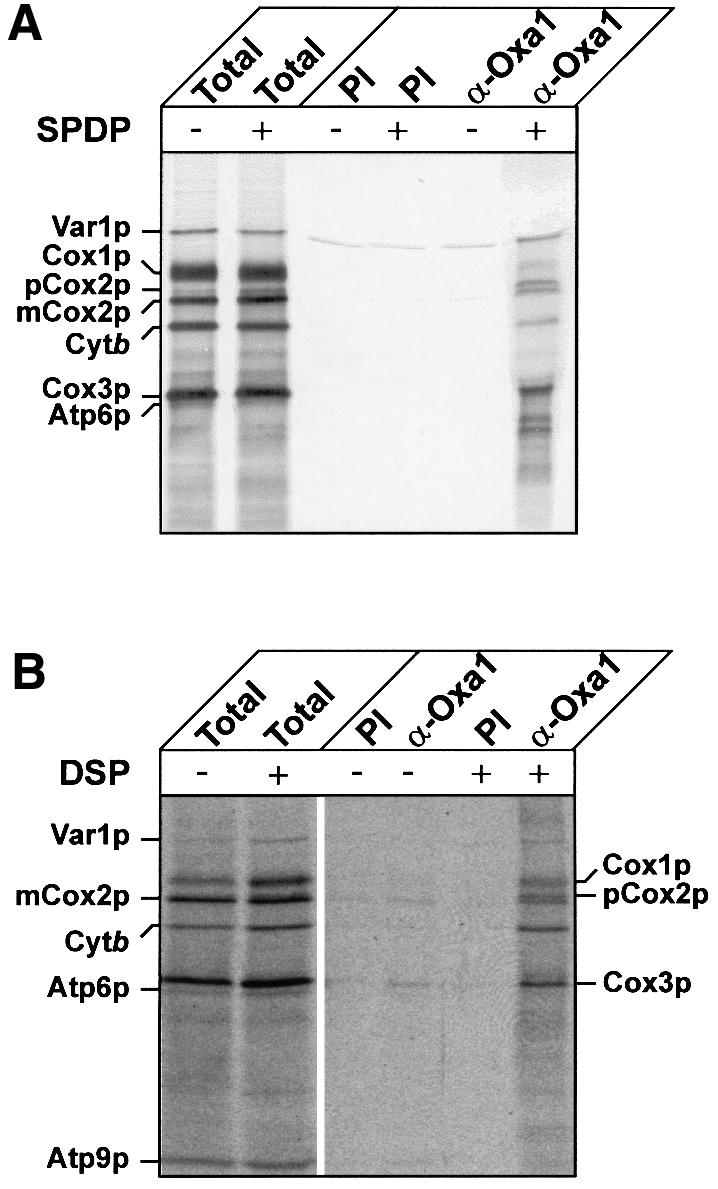
Fig. 2. Oxa1p interacts with proteins encoded by mitochondrial DNA. In vitro translation was performed for 15 min at 25°C in wild-type mitochondria in the presence of [35S]methionine, then the chemical cross-linker SPDP (0.5 mM) (A) or DSP (0.2 mM) (B) was added. The control samples (–) were mock-treated in parallel and received the solvent, dimethyl sulfoxide (DMSO), alone. Translation was allowed to proceed for another 15 min at 25°C. Cross-linking and translation were stopped by the addition of 100 mM glycine and 20 mM unlabeled methionine pH 8.0. Mitochondria were re-isolated, washed and lysed. After a clarifying spin, solubilized cross-linked products were immunoprecipitated with either pre-immune serum (PI) or antiserum specific for Oxa1p (α-Oxa1). Precipitated cross-linking products were eluted from the PAS-beads with sample buffer containing β-mercaptoethanol, which cleaves the cross-linker. Translation products were analyzed by SDS–PAGE and autoradiography. Total, 2% (A) or 1% (B) of cross-linking reaction that was used for immunoprecipitation. Note, in (B) less Oxa1p and pre-immune antisera was used for the immunoprecipitation reaction; thus, a reduction in the Var1p background signal was observed in this experiment.
Interaction of newly synthesized proteins with Oxa1p is not dependent on the presence of a membrane potential across the inner membrane
Next, we tested whether the association of newly synthesized proteins with Oxa1p was dependent on the presence of a membrane potential across the inner membrane during their synthesis. The membrane potential was dissipated prior to translation and cross-linking by the addition of valinomycin. Oxa1p with associated radiolabeled adducts obtained after cross-linking with SPDP were recovered by immunoprecipitation with Oxa1p-specific antiserum (Figure 3). The nature of the radio labeled adducts associated with Oxa1p was analyzed by subsequent cleavage of the cross-linking reagent with the reductant β-mercaptoethanol. When translation was performed in the presence of a membrane potential, Cox2p was observed to be associated with Oxa1p (Figure 3). In the absence of a membrane potential, efficient cross-linking of the accumulated precursor form of Cox2p, pCox2p, to Oxa1p was also obtained. The presence of an energized inner membrane is apparently not required for the interaction of the newly synthesized pCox2p and Oxa1p. On the other hand, as previously shown, the Oxa1p-mediated insertion of pCox2p into the inner membrane to an Nout–Cout orientation is dependent on a membrane potential (Herrmann et al., 1995).
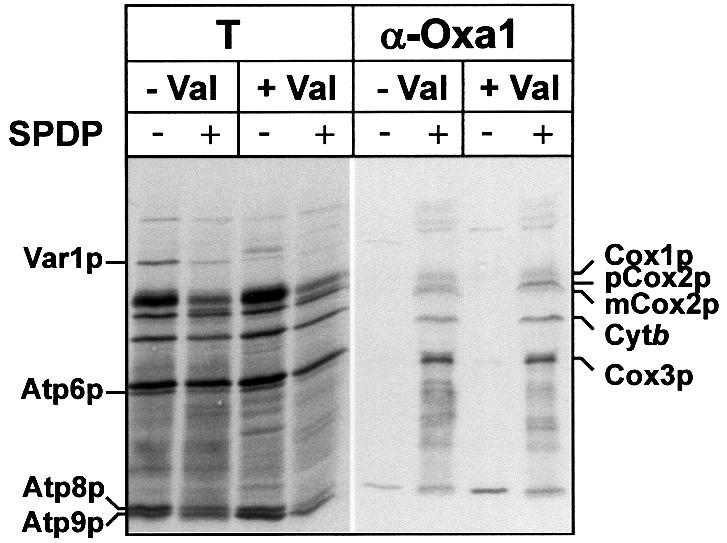
Fig. 3. Oxa1p can be cross-linked to mitochondrially encoded proteins in the absence of a membrane potential. Mitochondria were incubated in in vitro translation buffer in the absence (–Val) or presence (+Val) of 1 µM valinomycin for 5 min at 25°C. After the addition of [35S]methionine, the samples were incubated for a further 8 min at 25°C. Following the addition of the chemical cross-linker SPDP (0.5 mM) or DMSO (mock treatment), translation was allowed to proceed for another 30 min. Following cross-linking, Oxa1p and associated cross-linked adducts were immunoprecipitated, as described previously in Figure 2. Samples were analyzed by SDS–PAGE in the presence of β-mercaptoethanol and autoradiography. Abbreviations are as described in Figure 2. T, 2.5% of the cross-linking reaction that was used for immunoprecipitation. Note that the recovery of Atp9p in the Oxa1p immunoprecipitate was not dependent on the presence of cross-linker, and hence most likely represents a non-specific binding of this hydrophobic protein during the immunoprecipitation.
The efficiency of cross-linking of Cox1p, Cox3p and cytochrome b with Oxa1p was not adversely affected by the dissipation of the membrane potential prior to translation and cross-linking reactions. Thus, the interaction of these proteins with Oxa1p also occurs in a manner that is independent of a membrane potential. Furthermore, in contrast to Cox2p, the export of Cox1p, Cox3p and cytochrome b does not appear to display a requirement for the presence of a membrane potential (He and Fox, 1997; and our unpublished results).
In summary, the interaction of the newly synthesized proteins with Oxa1p occurs independently of a membrane potential. In addition, the accumulation of pCox2p with Oxa1p in the absence of a membrane potential indicates that the interaction of newly synthesized proteins with Oxa1p can occur even under conditions where subsequent export is blocked. The requirement of a membrane potential for the membrane insertion of pCox2p apparently does not reflect a need for energy for the initial interaction of this substrate with Oxa1p, but rather for a subsequent step in the biogenesis pathway of pCox2p, such as membrane insertion and/or translocation.
The association of nascent polypeptide chains with Oxa1p occurs in the presence of the associated ribosome
Mitochondrial ribosomes actively undergoing translation have been observed in close proximity to the mitochondrial inner membrane in vivo (Vignais et al., 1969; Watson, 1972). A close coupling of the synthesis of the membrane proteins encoded by mitochondrial DNA and their translocation into the membrane has been suggested to occur (Sanchirico et al., 1998). Furthermore, mitochondrially encoded proteins were observed to interact with Oxa1p early during their synthesis, as nascent chains, rather than after completion of synthesis (Hell et al., 1998). Consequently, the ribosome engaged in translation is likely to be in close proximity to the Oxa1p insertion machinery. We chose, therefore, to analyze the interaction of nascent chains with Oxa1p under conditions in which the completion of their synthesis to full-length polypeptide chains is inhibited. A block in protein synthesis was achieved through the addition of two well characterized protein synthesis inhibitors, chloramphenicol and puromycin.
Mitochondrial protein synthesis was performed in the presence of [35S]methionine for a short period, then elongation of the nascent chains was inhibited by the addition of either chloramphenicol or puromycin. In the control reaction, no inhibitor was added. The association of the radiolabeled polypeptides with Oxa1p was probed by chemical cross-linking using the non-cleavable reagent 1,5-difluoro-2,4-dinitrobenzene (DFDNB), and subsequent immunoprecipitation with Oxa1p-specific antisera (Figure 4A). When no inhibitor of translation was present, i.e. when protein synthesis was ongoing during cross-linking reaction, cross-linked adducts with apparent molecular weights between 36 and 80 kDa were found in association with Oxa1p. When, prior to cross-linking, further protein synthesis was prevented by the addition of chloramphenicol, cross-linked adducts of a similar size range were observed with Oxa1, albeit with lowered efficiency (Figure 4A and B). Chloramphenicol inhibits the peptidyl-transferase activity of the 50S ribosomal subunit, and hence serves to arrest further chain elongation. Thus, the stalled nascent chains, which remained with the ribosome, were found in association with the Oxa1p membrane protein insertion machinery. The lowered efficiency may be due to the fact that further initiation of nascent chain synthesis is also inhibited in the presence of chloramphenicol.
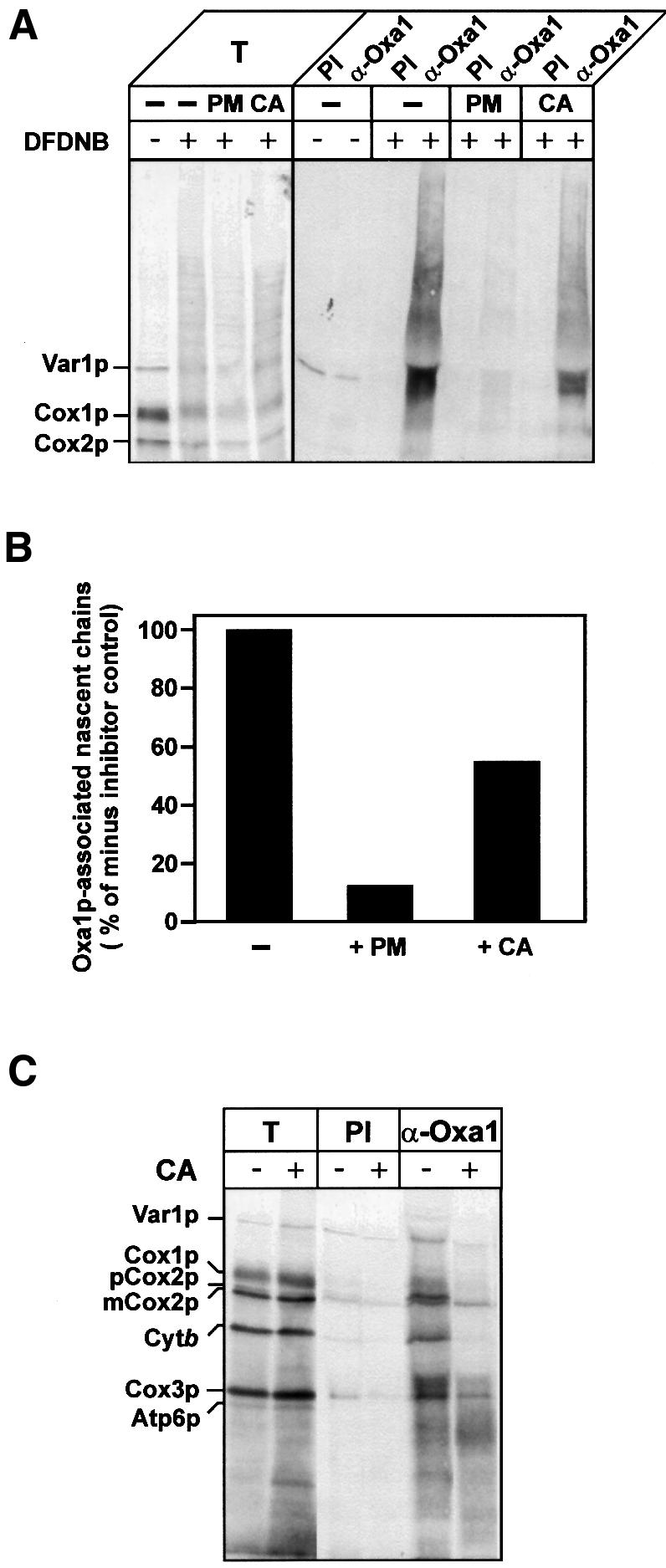
Fig. 4. Oxa1p interacts with mitochondrially encoded proteins predominantly during synthesis. (A and B) Translation in isolated wild-type mitochondria was performed in the presence of [35S]methionine, for 10 min at 25°C. Samples then received either puromycin (PM, 45 µg/ml) or chloramphenicol (CA, 3 mg/ml) or no addition (–), for 3 min at 25°C. The samples were incubated for a further 15 min, in the presence or absence of DFDNB (0.4 mM). Cross-linking and translation were stopped by the addition of 100 mM glycine pH 8.0 and 50 µg/ml puromycin. Re-isolated and washed mitochondria were lysed, and immunoprecipitation with either pre-immune serum (PI) or antiserum specific for Oxa1p (α-Oxa1) was performed. Immunoprecipitates were analyzed by SDS–PAGE and autoradiography (A). T, 0.5% of cross-linking reaction that was used for immunoprecipitation. (B) Quantification of the autoradiography. The total translation signals for each sample were initially quantified and were as follows (arbitrary units): no-inhibitor control, 156; PM-treated, 100; CA-treated, 150. The level of nascent chains associated with Oxa1p was then expressed as a percentage of the total translation signal for each of the treatments. The resulting value obtained in the absence of a translational inhibitor (control) was set to 100%, and the PM- and CA-treated values were then expressed as a percentage of the control. (C) Translation in isolated wild-type mitochondria was performed in the presence of [35S]methionine for 20 min at 25°C. Samples were placed on ice for 5 min. Aliquots were either treated with chloramphenicol (+CA, 3 mg/ml) or mock-treated (–CA). Following the addition of SPDP (0.3 mM), the samples were further incubated for 15 min at 25°C. The samples were processed and analyzed as described in Figure 2. T, 5% of cross-linking reaction that was used for immunoprecipitation.
Puromycin inhibits protein synthesis by causing premature release of the nascent polypeptide chain from the ribosome. Stopping protein synthesis in this manner resulted in a significant decrease (12% of no inhibitor control) of nascent chains that were found in association with Oxa1p (Figure 4A and B). The observed decrease in Oxa1p-associated nascent chains as compared with the control was much stronger than in the case of chloramphenicol treatment. Thus, efficient interaction of the nascent polypeptide chains with the Oxa1p insertion machinery appears to be achieved when these chains remain in association with the ribosome.
When cross-linking reaction was performed during ongoing protein synthesis, full-length radiolabeled proteins were found cross-linked to Oxa1p (Figure 4C). If, however, protein synthesis was arrested with chloramphenicol prior to the addition of the cross-linking reagent, a strong reduction in the level of full-length proteins cross-linked to Oxa1p was observed. A small percentage of the mature-size polypeptides was found in association with Oxa1p following chloramphenicol treatment. This minor fraction may represent polypeptide chains close to or at the completion of their synthesis, and that were still in the vicinity of Oxa1p at the time of cross-linking. Thus, upon cross-linking during ongoing protein synthesis, nascent chains appear to become linked to Oxa1p, but further synthesis may still proceed to their completion.
In summary, the interaction of Oxa1p with its substrates is initiated early during their synthesis as nascent polypeptide chains associated with ribosomes, rather than after completion of synthesis.
Discussion
The biogenesis of a group of proteins located in the mitochondrial inner membrane involves a sorting pathway that is evolutionarily conserved and reflects the prokaryotic ancestral nature of mitochondria. This pathway involves the insertion of proteins into the inner membrane from the level of the mitochondrial matrix and is facilitated by the Oxa1 complex. Oxa1p, a key component of this complex, is a member of the Oxa1/YidC/Alb3 family, proteins that are involved in analogous membrane protein insertion events in mitochondria, prokaryotes and chloroplasts, respectively. Our knowledge about the involvement of the mitochondrial homolog Oxa1p in membrane insertion, until now, has been limited to the sorting of membrane proteins whose N-termini undergo export from the matrix to the mitochondrial intermembrane space during their insertion event.
We report here on the topogenesis of polytopic proteins whose N-termini reside in the mitochondrial matrix and hence do not undergo N-terminal tail export. Our studies demonstrate an involvement of Oxa1p in the membrane insertion of Cox1p, Cox3p and cytochrome b, three mitochondrially encoded membrane proteins whose N-termini reside in the matrix. Newly synthesized Cox1p displayed a strong dependency on Oxa1p for its insertion. Furthermore, Oxa1p physically interacts with radiolabeled Cox1p early during its insertion process. We conclude that Oxa1p can directly facilitate the insertion of Cox1p into the inner membrane. Newly synthesized Cox3p and cytochrome b could also be cross-linked to Oxa1p, indicating a close physical relationship between Oxa1p and these proteins. The insertion of cytochrome b and Cox3p was clearly enhanced by the presence of a functional Oxa1p; however, a significant level of insertion of these proteins was observed under conditions where the insertion of Oxa1p-dependent Cox2p was almost completely blocked. We conclude, therefore, that Oxa1p can mediate the insertion of internal segments of polytopic proteins such as hairpin structures. Our observations are consistent with those made recently by Samuelson et al. (2000), where YidC, the prokaryotic homolog of Oxa1p, was found to facilitate the insertion of the M13 procoat protein into the periplasmic membrane. M13 procoat spans the membrane initially twice and inserts into the membrane as a hairpin loop structure.
In addition, our data indicate that there is variation in the degree of Oxa1p dependency displayed by different membrane proteins for their insertion into the mitochondrial inner membrane. What is the basis for this observed variation in Oxa1p dependency? These observations would indicate the existence of an Oxa1p-independent pathway that can facilitate the insertion of proteins into the inner membrane from the matrix. Indeed, a previous characterization of the oxa1 null mutant Δoxa1 is consistent with this possibility. Significant levels of the cytochrome bc1 complex were observed in an oxa1 deletion mutant (Meyer et al., 1997), indicating that the presence of Oxa1p is not essential for the insertion of cytochrome b (and possible other components of the cytochrome bc1 complex). Furthermore, extrachromosomal expression of the OXA1 gene rescued the growth phenotype of the Δoxa1 mutant (Bauer et al., 1994; Bonnefoy et al., 1994), despite the observation that the insertion of Oxa1p into the inner membrane is facilitated by Oxa1p itself (Hell et al., 1998). Such a complementation implies an alternative pathway assisting insertion of newly imported Oxa1p.
How does this Oxa1p-independent membrane insertion process occur? It can not be excluded that some proteins may undergo a limited spontaneous insertion into the inner membrane. Alternatively, the Oxa1p-independent insertion pathway may involve another protein-mediated insertion mechanism that displays partial overlapping specificity with the Oxa1 machinery. Candidate components for such an alternative mechanism may involve Pnt1p and/or Cox18p. Pnt1p, an integral inner membrane protein, was identified by Fox and colleagues in a genetic screen designed to identify proteins involved in the membrane insertion of Cox2p and Cox2p derivatives (He and Fox, 1999). Cox18p, an integral inner membrane protein required for the assembly of the cytochrome oxidase complex (Souza et al., 2000), shares some sequence similarity with Oxa1p. Finally, Dujardin and colleagues have reported that mutations in the C-terminal transmembrane segment of cytochrome c1, a subunit of the cytochrome bc1 complex, can cause partial restoration of growth of the Δoxa1 null mutant on non-fermentable carbon sources (Hamel et al., 1998). Although the molecular basis for this complementation is not yet fully understood, the observation illustrates that the inner membrane displays significant flexibility to facilitate insertion of its resident proteins.
In the present study we have used a cross-linking approach to map in more detail the nature of the association of proteins undergoing insertion with Oxa1p. The data presented here further our understanding of the membrane insertion of proteins in a number of ways. First, the interaction of the nascent polypeptide chain with Oxa1p is not dependent on the presence of a membrane potential across the inner membrane. The insertion of Cox2p into the inner membrane displays a strong dependency on both an energized inner membrane and on the function of Oxa1p. In the absence of a membrane potential, pCox2p was found in association with Oxa1p. The interaction of pCox2p with Oxa1p can occur early and independently of the insertion of Cox2p into the inner membrane.
Secondly, nascent polypeptide chains interact with Oxa1p early in their biogenesis. Cross-linking of full-length polypeptide chains to Oxa1p was obtained only when the cross-linking reaction was performed during ongoing protein synthesis. Upon block of protein synthesis, nascent polypeptide chains, rather than full-length proteins, were found in association with Oxa1p. These data indicate that: (i) the nascent polypeptide chain can be cross-linked to Oxa1p prior to, but independently of, completion of its synthesis; and (ii) further elongation of the nascent polypeptide chain can continue even if N-terminal regions of the chain are cross-linked to Oxa1p.
Thirdly, we observed that the association of Oxa1p with the nascent chain undergoing membrane insertion is enhanced by the presence of the associated ribosome. This might serve to ensure a close coupling of translation with the Oxa1p-mediated insertion into the membrane. The goal of such an intimate coupling of synthesis and translocation may be two-fold. First, coupling of these processes would ensure that translation was occurring at the site of insertion. Accumulation of these highly hydrophobic proteins in the matrix prior to their insertion, which could lead to their aggregation and loss of competence for subsequent membrane insertion, would thereby be prevented. Secondly, a co-translational insertion process could contribute energetically to the driving force required for the insertion and translocation events. A tight coupling of translation and insertion would imply the existence of a binding site for the translating ribosome on Oxa1p. A possible direct interaction of the ribosome and Oxa1p is currently being investigated.
Materials and methods
Yeast strains
Yeast strains used in this study were wild-type D273-10B (ATCC 24657), wild-type W303-1A (Mata, leu2, trp1, ura3, ade2, can1) and a temperature-sensitive mutant of OXA1, the oxa-ts strain. The oxa-ts strain was generated as follows: a PCR fragment containing the OXA1 gene with the temperature-sensitive mutation Leu-240 Ser was amplified using genomic DNA of the originally pet ts1402 strain (Bauer et al., 1994) as a template. This fragment was purified and transformed into an oxa1 null mutant, the Δoxa1 strain (W303-1A, OXA1::HIS3) (Hell et al., 1998). Via homologous recombination, the OXA1 gene containing the temperature-sensitive mutation was introduced in the OXA1 locus in the oxa1 null mutant, thereby replacing the disruption-marker HIS3 gene. Positive transformants were selected by growth on YP-glycerol at 24°C. The correct insertion of the mutated OXA1 gene was verified by PCR analysis of the OXA1 gene locus, sequence analysis of the OXA1 gene and by demonstration of the temperature-sensitive growth phenotype of the resulting oxa1-ts strain (results not shown).
In vitro labeling of mitochondrial translation products in mitoplasts
Isolated mitochondria were initially subjected to hypotonic swelling by diluting 10-fold in 20 mM HEPES pH 7.4 in the presence of 1 mM ATP for 30 min on ice. Resulting mitoplasts were re-isolated by centrifugation (Sigma, rotor 12154, 7 min, 10 000 r.p.m.) and resuspended to a protein concentration of 1.34 mg/ml in 230 µl of translation buffer containing 1% (v/v) pyruvate kinase (2 U/ml). After 5 min of incubation at 25°C, [35S]methionine was added and incubation was allowed to proceed for another 20 min. The translation was stopped by the addition of unlabeled methionine (30 mM) and puromycin (40 mg/ml). Following a 10-fold dilution with SH-buffer (0.6 M sorbitol, 20 mM HEPES pH 7.4), the samples were divided. One aliquot was treated with proteinase K (50 µg/ml), and the other aliquot was mock-treated in parallel. To inhibit the Oxa1p function in the oxa-ts mitoplasts the mitoplasts were pre-incubated in translation buffer at 0 or 37°C for 10 min, prior to the labeling reaction. Mitoplasts prepared from the isogenic wild-type strain were treated in parallel, as a control.
In vitro labeling of mitochondrial gene products and their cross-linking to Oxa1p
In vitro labeling of mitochondrial translation products was performed in the isolated mitochondria (3 mg mitochondrial protein/ml), as previously described (Hell et al., 2000), with the exceptions that the translation buffer contained no bovine serum albumin and the Tris–HCl buffer was substituted by HEPES–KOH. When indicated, the translation was performed in the absence of a membrane potential, followed by the addition of valinomycin (1 µM) to the mitoplasts. The concentration of valinomycin used was sufficient to dissipate the membrane potential, as judged by two independent criteria: (i) direct measurement of the membrane potential and (ii) measurement of preprotein import into the mitoplasts (a membrane potential-dependent process), where complete inhibition of import was observed in the presence of valinomycin (results not shown). Chemical cross-linking was performed using one of three different cross-linking reagents, DFDNB, SPDP and DSP, at the concentrations indicated in the figure legends. The cross-linking reagent was added to the translation reaction at the times indicated (see individual figure legends) and the translation was allowed to proceed at 25°C for the indicated time periods. The control samples (minus cross-linker) were mock-treated in parallel with buffer lacking the cross-linker. The cross-linker was quenched by the addition of 100 mM glycine pH 8.0 and translation was stopped by the addition of unlabeled methionine (20 mM) and/or puromycin (50 µg/ml), as indicated.
Immunoprecipitation of cross-linked in vitro translated proteins with Oxa1p-specific antisera was performed essentially as described previously (Hell et al., 1998).
Miscellaneous
Isolation of mitochondria (Herrmann et al., 1994), protein concentration determinations and SDS–PAGE were performed according to the published methods of Bradford (1976) and Laemmli (1970), respectively.
Acknowledgments
Acknowledgements
We thank Sandra Esser and Stefanie Glocker for excellent technical assistance. We are also grateful to Drs Hannes Herrmann, Michael Brunner and Thomas Langer for many helpful discussions. This work was supported in part by an NSF grant M.C.B. 0077961 to R.A.S. and a predoctoral fellowship from the Boehringer Ingelheim Fonds to K.H.
References
- Bauer M., Behrens,M., Esser,K., Michaelis,G. and Pratje,E. (1994) PET1402, a nuclear gene required for proteolytic processing of cytochrome oxidase subunit 2 in yeast. Mol. Gen. Genet., 245, 272–278. [DOI] [PubMed] [Google Scholar]
- Bonnefoy N., Chalvet,F., Hamel,P., Slominski,P.P. and Dujardin,G. (1994) OXA1, a Saccharomyces cerevisiae nuclear gene whose sequence is conserved from prokaryotes to eukaryotes controls cytochrome oxidase biogenesis. J. Mol. Biol., 239, 201–212. [DOI] [PubMed] [Google Scholar]
- Bradford M.M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilising the principle of protein-dye binding assay. Anal. Biochem., 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Hamel P., Lemaire,C., Bonnefoy,N., Brivet-Chevillotte,P. and Dujardin,G. (1998) Mutations in the membrane anchor of yeast cytochrome c1 compensate for the absence of Oxa1p and generate carbonate-extractable forms of cytochrome c1. Genetics, 150, 601–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S. and Fox,T.D. (1997) Membrane translocation of mitochondrially coded Cox2p: distinct requirements for export of N- and C-termini and dependence on the conserved protein Oxa1p. Mol. Biol. Cell, 8, 1449–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S. and Fox,T.D. (1999) Mutations affecting a yeast mitochondrial inner membrane protein, Pnt1p, block export of a mitochondrially synthesized fusion protein from the matrix. Mol. Cell. Biol., 19, 6598–6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hell K., Herrmann,J., Pratje,E., Neupert,W. and Stuart,R.A. (1997) Oxa1p mediates the export of the N- and C-termini of pCoxII from the mitochondrial matrix to the intermembrane space. FEBS Lett., 418, 367–370. [DOI] [PubMed] [Google Scholar]
- Hell K., Herrmann,J.M., Pratje,E., Neupert,W. and Stuart,R.A. (1998) Oxa1p, an essential component of the N-tail protein export machinery in mitochondria. Proc. Natl Acad. Sci. USA, 95, 2250–2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hell K., Tzagoloff,A., Neupert,W. and Stuart,R.A. (2000) Identification of Cox20p, a novel protein involved in the maturation and assembly of cytochrome oxidase subunit 2. J. Biol. Chem., 275, 4571–4578. [DOI] [PubMed] [Google Scholar]
- Herrmann J.M., Fölsch,H., Neupert,W. and Stuart,R.A. (1994) Isolation of yeast mitochondria and study of mitochondrial protein translation. In Celis,J.E. (ed.), Cell Biology: A Laboratory Handbook. Academic Press, San Diego, CA, pp. 538–544.
- Herrmann J.M., Koll,H., Cook,R.A., Neupert,W. and Stuart,R.A. (1995) Topogenesis of cytochrome oxidase subunit II—mechanisms of protein export from the mitochondrial matrix. J. Biol. Chem., 270, 27079–27086. [DOI] [PubMed] [Google Scholar]
- Herrmann J.M., Neupert,W. and Stuart,R.A. (1997) Insertion into the mitochondrial inner membrane of a polytopic protein, the nuclear encoded Oxa1p. EMBO J., 16, 2217–2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houben E.N., Scotti,P.A., Valent,Q.A., Brunner,J., de Gier,J.L., Oudega,B. and Luirink,J. (2000) Nascent Lep inserts into the Escherichia coli inner membrane in the vicinity of YidC, SecY and SecA. FEBS Lett., 476, 229–233. [DOI] [PubMed] [Google Scholar]
- Hunte C., Koepke,J., Lange,C., Rossmanith,T. and Michel,H. (2000) Structure at 2.3 Å resolution of the cytochrome bc1 complex from the yeast Saccharomyces cerevisiae co-crystallized with an antibody Fv fragment. Struct. Fold. Des., 8, 669–684. [DOI] [PubMed] [Google Scholar]
- Laemmli U.K. (1970) Cleavage of structural proteins during the assembly of the head bacteriophage T4. Nature, 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Meyer W., Bomer,U. and Pratje,E. (1997) Mitochondrial inner membrane bound Pet1402 protein is rapidly imported into mitochondria and affects the integrity of the cytochrome oxidase and ubiquinol–cytochrome c oxidoreductase complexes. Biol. Chem., 378, 1373–1379. [DOI] [PubMed] [Google Scholar]
- Moore M., Harrison,M.S., Peterson,E.C. and Henry,R. (2000) Chloroplast Oxa1p homolog albino3 is required for post-translational integration of the light harvesting chlorophyll-binding protein into thylakoid membranes. J. Biol. Chem., 275, 1529–1532. [DOI] [PubMed] [Google Scholar]
- Rojo E.E., Stuart,R.A. and Neupert,W. (1995) Conservative sorting of F0-ATPase subunit 9: export from matrix requires ΔpH across inner membrane and matrix ATP. EMBO J., 14, 3445–3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo E.E., Guiard,B., Neupert,W. and Stuart,R.A. (1999) N-terminal tail export from the mitochondrial matrix. Adherence to the prokaryotic ‘positive-inside’ rule of membrane protein topology. J. Biol. Chem., 274, 19617–19622. [DOI] [PubMed] [Google Scholar]
- Samuelson J.C., Chen,M., Jiang,F., Möller,I., Wiedmann,M., Kuhn,A., Phillips,G.J. and Dalbey,R.E. (2000) YidC mediates membrane protein insertion in bacteria. Nature, 406, 637–641. [DOI] [PubMed] [Google Scholar]
- Sanchirico M.E., Fox,T.D. and Mason,T.L. (1998) Accumulation of mitochondrially synthesized Saccharomyces cerevisiae Cox2p and Cox3p depends on targeting information in untranslated portions of their mRNAs. EMBO J., 17, 5796–5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotti P.A., Urbanus,M.L., Brunner,J., de Gier,J.L., von Heijne,G., van der Does,C., Driessen,A.J.M., Oudega,B. and Luirink,L. (2000) YidC, the Escherichia coli homologue of mitochondrial Oxa1p, is a component of the Sec translocase. EMBO J., 19, 542–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza R.L., Green-Willms,N.S., Fox,T.D., Tzagoloff,A. and Nobrega,F.G. (2000) Cloning and characterization of COX18, a Saccharomyces cerevisiae PET gene required for the assembly of cytochrome oxidase. J. Biol. Chem., 275, 14898–14902. [DOI] [PubMed] [Google Scholar]
- Stuart R.A. and Neupert,W. (1996) Topogenesis of inner membrane proteins of mitochondria. Trends Biochem. Sci., 21, 261–267. [PubMed] [Google Scholar]
- Stuart R.A. and Neupert,W. (2000) Making membranes in bacteria. Nature, 406, 575–577. [DOI] [PubMed] [Google Scholar]
- Sundberg E., Slagter,J.G., Fridborg,I., Cleary,S.P., Robinson,C. and Coupland,G. (1997) ALBINO3, an Arabidopsis nuclear gene essential for chloroplast differentiation, encodes a chloroplast protein that shows homology to proteins present in bacterial membranes and yeast mitochondria. Plant Cell, 9, 717–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukihara T., Aoyama,H., Yamashita,E., Tomizaki,T., Yamaguchi,H., Shinzawa-Itoh,K., Nakashima,R., Yaono,R. and Yosikawa,S. (1994) The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 Å. Science, 272, 1136–1144. [DOI] [PubMed] [Google Scholar]
- Vignais P.V., Huet,J. and André,J. (1969) Isolation and characterization of ribosomes from yeast mitochondria. FEBS Lett., 3, 177–181. [DOI] [PubMed] [Google Scholar]
- Watson K., (1972) The organization of ribosomal granules within mitochondrial structures of aerobic and anaerobic cells of Saccharomyces cerevisiae. J. Cell Biol., 55, 721–726. [DOI] [PMC free article] [PubMed] [Google Scholar]


