Abstract
GCN2 stimulates translation of GCN4 mRNA in amino acid-starved cells by phosphorylating translation initiation factor 2. GCN2 is activated by binding of uncharged tRNA to a domain related to histidyl-tRNA synthetase (HisRS). The HisRS-like region contains two dimerization domains (HisRS-N and HisRS-C) required for GCN2 function in vivo but dispensable for dimerization by full-length GCN2. Residues corresponding to amino acids at the dimer interface of Escherichia coli HisRS were required for dimerization of recombinant HisRS-N and for tRNA binding by full-length GCN2, suggesting that HisRS-N dimerization promotes tRNA binding and kinase activation. HisRS-N also interacted with the protein kinase (PK) domain, and a deletion impairing this interaction destroyed GCN2 function without reducing tRNA binding; thus, HisRS-N–PK interaction appears to stimulate PK function. The C-terminal domain of GCN2 (C-term) interacted with the PK domain in a manner disrupted by an activating PK mutation (E803V). These results suggest that the C-term is an autoinhibitory domain, counteracted by tRNA binding. We conclude that multiple domain interactions, positive and negative, mediate the activation of GCN2 by uncharged tRNA.
Keywords: dimerization/eIF2α kinase/GCN2/regulation/translation
Introduction
Yeast cells respond to amino acid or purine starvation by activating GCN2, a protein kinase (PK) that specifically phosphorylates the α subunit of translation initiation factor 2 (eIF2α) on serine-51. Phosphorylation of eIF2 converts it from substrate to inhibitor of its guanine nucleotide exchange factor eIF2B, decreasing formation of the ternary complex (eIF2-GTP–Met-tRNAiMet), which transfers Met-tRNAiMet to the 40S ribosome. A decrease in ternary complex levels stimulates translation of GCN4 mRNA, encoding a transcriptional activator of multiple amino acid biosynthetic enzymes. Four short open reading frames (uORFs) in the GCN4 mRNA leader function in a specialized re-initiation mechanism that derepresses GCN4 translation in response to reductions in ternary complex levels that are too small to impede general protein synthesis. Dominant activating mutations in GCN2 have been isolated that derepress GCN4 under nonstarvation conditions, and the strongest of these GCN2c alleles also impairs general translation and cell growth due to a severe depletion of the ternary complex.
GCN2 is activated in amino acid-starved cells through binding of uncharged tRNA to a region homologous to histidyl-tRNA synthetase (HisRS), located C-terminal to the kinase domain (Wek et al., 1989) (Figure 1). Mutations in the HisRS-like domain in conserved residues required for tRNA binding by class II aminoacyl-tRNA synthetases (the m2 motif) inactivated GCN2 kinase function, and destroyed tRNA binding by the isolated HisRS domain (Wek et al., 1995; Zhu et al., 1996) and by full-length GCN2 (Dong et al., 2000). It appears that starvation of yeast for any amino acid activates GCN2 and elicits derepression of GCN4 translation (Hinnebusch, 1992; Wek et al., 1995), implying that GCN2 can discriminate only between charged and uncharged forms of tRNA. Indeed, purified GCN2 bound various uncharged tRNAs with similar affinities, but interacted with charged tRNAPhe less strongly than with uncharged tRNAPhe (Dong et al., 2000). GCN2 homologs have been identified in Neurospora crassa (Sattlegger et al., 1998), Drosophila melanogaster (Santoyo et al., 1997; Olsen et al., 1998) and mouse (Berlanga et al., 1998; Sood et al., 2000), and the HisRS domain is conserved in all of these proteins; thus, activation by uncharged tRNA seems to be an evolutionarily conserved feature of the GCN2 family of PKs.
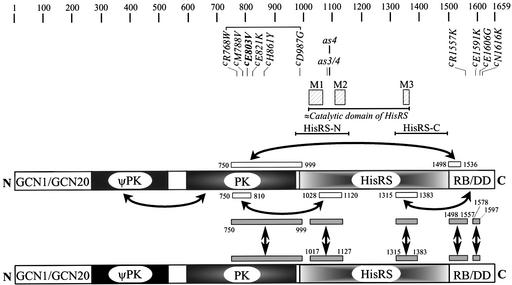
Fig. 1. Summary of GCN2 dimerization and domain interactions. Two GCN2 polypeptides of 1659 residues [numbered at the very top from amino (N) to carboxyl (C) terminus] are depicted schematically as rectangular boxes subdivided into the following functional domains: the conserved N-terminal region required for interaction with GCN1 and GCN20 (GCN1/GCN20) (Garcia-Barrio et al., 2000); a degenerate protein kinase domain (ψPK); the protein kinase domain (PK); a histidyl-tRNA synthetase-related region (HisRS); and a C-terminal region required for ribosome binding and dimerization by GCN2 (RB/DD). The rectangular gray boxes located between the two GCN2 schematics that are linked by two-headed arrows depict the segments involved in self-interactions, with the amino acid positions of their end points indicated. The open rectangular boxes above or below the top GCN2 schematic indicate the segments required for the interactions between different functional domains indicated by two-headed arrows. The interactions involving the HisRS domain are based on results presented below (see text). The locations of the N-terminal and C-terminal segments of the HisRS region designated in the text as HisRS-N and HisRS-C, respectively, are indicated above the top schematic. The region that is similar in sequence to the catalytic domain of authentic HisRS is indicated immediately below the hatched boxes labeled M1–3, the latter indicating the positions of three sequence motifs conserved among class II aminoacyl-tRNA synthetases. Immediately below the sequence numbering are the locations and nature of various GCN2c mutations (designated with ‘c’) and the as4 and as3/4 mutations.
The extreme C-terminal segment of yeast GCN2 (C-term) is also essential for GCN2 function in vivo (Wek et al., 1990). Both the C-term and PK domains have dimerization activities, but only the C-term was required for dimerization by full-length GCN2 in vivo (Qiu et al., 1998). The isolated C-term can bind double-stranded RNA in vitro and interact with ribosomes in cell extracts, suggesting that it mediates ribosome binding by GCN2 through interactions with double-stranded segments of rRNA (Zhu and Wek, 1998). It was shown recently that the C-term additionally promotes tRNA binding by the HisRS domain independently of its role in GCN2 dimerization (Dong et al., 2000). Moreover, the C-term can physically interact with both the HisRS and PK domains, and the HisRS region can interact with the PK domain (Qiu et al., 1998). Interestingly, tRNA antagonized interaction in vitro between the PK domain and a fragment containing the HisRS and C-term domains. Accordingly, we proposed that tRNA would activate GCN2 by impeding an inhibitory association between the PK and C-term domains (Dong et al., 2000).
Given the central role of the HisRS region in activation of GCN2 by uncharged tRNA, it was imperative to map precisely the interactions between this region and the PK and C-term domains. In particular, contacts between the HisRS and PK domains may influence the ability of tRNA to promote an activated conformation of the PK domain. Considering that Escherichia coli HisRS is a dimer, it was also important to determine whether the HisRS-like domain in GCN2 dimerizes, and the consequences of dimerization in this region for tRNA binding and kinase activation. We show here that the GCN2 HisRS region has two dimerization domains, of which the N-terminal moiety most likely resembles the dimerization surface in the catalytic domain of authentic HisRS. Mutational analysis suggests that this region makes a crucial contribution to GCN2 function by promoting tRNA binding and through a stimulatory physical interaction with the kinase domain. We also obtained evidence for physical interaction between the C-term and PK domains that negatively regulates kinase activity in a manner overcome by uncharged tRNA in amino acid-starved cells.
Results
The HisRS-like region of GCN2 contains two independent dimerization domains
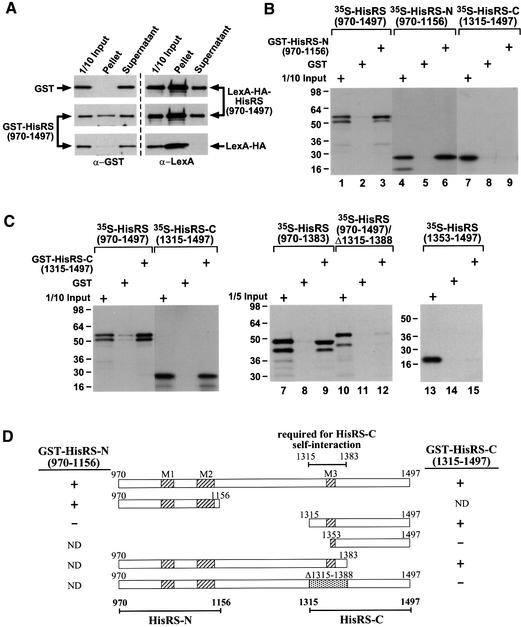
Because the HisRS-related region of GCN2 contains sequence similarities to residues that comprise the dimer interface in authentic E.coli HisRS (Arnez et al., 1995), we investigated whether the GCN2 HisRS-like domain can dimerize. In a first approach, we asked whether gluta thione S-transferase (GST) and LexA-HA (hemagglutinin) fusions to the HisRS-like segment (residues 970–1497), both expressed in yeast from the ADH1 promoter, could be co-immunoprecipitated from cell extracts. As shown in Figure 2A, a fraction of GST–HisRS(970–1497) was co-immunoprecipitated by anti-HA antibodies along with LexA-HA–HisRS(970–1497), but not with LexA-HA alone; additionally, GST alone did not co-immunoprecipitate with LexA-HA–HisRS(970–1497). Western analysis using antibodies against GCN2 showed that GST– HisRS(970–1497) and LexA-HA–HisRS(970–1497) were expressed at similar amounts (data not shown). Based on the recovery of GST–HisRS(970–1497) in immune complexes, we estimated that ∼1/4 of the total GST– HisRS(970–1497) was complexed with LexA-HA– HisRS(970–1497) in the extract. Given that the co-immunoprecipitation assay detects the heterodimers containing LexA- and GST-tagged HisRS fusions, but not the corresponding homodimers, our data suggest that a substantial proportion of the HisRS fusion proteins were dimerized in vivo.
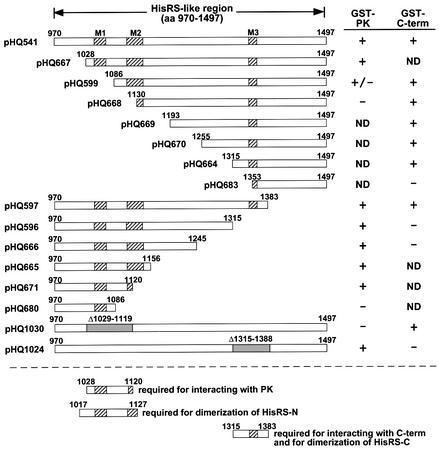
Fig. 2. The HisRS-like region contains two independent dimerization domains. (A) Co-immunoprecipitation of GST–HisRS with LexA-HA–HisRS from yeast cell extracts. Transformants of gcn2Δ strain HQY132 (Qiu et al., 1998) bearing HIS3 plasmids encoding LexA-HA–HisRS(970–1497) (pHQ588) or LexA-HA (p2247), and TRP1 plasmids encoding GST (pHQ242) or GST–HisRS(970–1497) (pHQ601), were grown in SC-His-Trp medium and whole-cell extracts were prepared. Aliquots of extracts containing 50 µg of protein were immunoprecipitated with anti-HA antibodies, and the immune complexes were resolved by SDS–PAGE and subjected to western blot analysis using anti-GST antibodies (left panels) or anti-LexA antibodies (right panels). The top panels show results from the transformant expressing GST and the LexA-HA–HisRS fusion; the middle panels derive from the transformant expressing the GST–HisRS and LexA-HA–HisRS fusions; the bottom two panels derive from the transformant expressing the GST–HisRS fusion and LexA-HA. (B and C) The HisRS-like region dimerizes in vitro through the N- and C-terminal subregions. The full-length HisRS-like region (aa 970–1497) or truncated segments indicated across the top of each panel were translated in vitro in the presence of [35S]methionine and incubated with either GST, GST–HisRS(970–1156) (B) or GST–HisRS(1315–1497) (C) proteins, which were expressed in E.coli and immobilized on glutathione–Sepharose beads. After extensive washing of the beads, the bound proteins were resolved by SDS–PAGE and visualized by fluorography. Numbers to the left of each gel give the positions of size standards of the indicated molecular weight in kilodaltons. (D) Summary of results from the binding assays shown in (B and C). Rectangular boxes with the locations of conserved motifs M1–3 (indicated with diagonal hatching) represent the in vitro translated proteins with GCN2 residue numbers indicated at their N- and C-termini. The binding (+) or failure to bind (–) of these segments to GST–HisRS(970–1156) or GST–HisRS(1315–1497) is summarized to the right and left of the schematics, respectively. ND, not determined. The results of these assays indicate that dimerization by the C-terminal segment of the HisRS-like region is dependent on residues 1315–1383, as indicated by the bar above the boxes. Bars below the boxes represent HisRS-N and HisRS-C subregions, respectively.
To map the interacting domain(s) within the HisRS segment, we carried out in vitro binding experiments using GST fusions expressed in E.coli containing the N- or C-terminus of the GCN2 HisRS-like region [residues 970– 1156 (GST–HisRS-N) or 1315–1497 (GST–HisRS-C), respectively] and 35S-labeled polypeptides synthesized in vitro containing the same GCN2 residues. As shown in Figure 2B (and summarized in Figure 2D), full-length and N-terminal [35S]HisRS fragments, but not the C-terminal [35S]HisRS fragment, bound to GST–HisRS-N but not to GST alone. Thus, the GCN2 HisRS-N region can dimerize in the absence of other yeast proteins. The GST–HisRS-C fusion interacted with full-length (970–1497) and C-terminal (1315–1497) segments of the HisRS-like domain (Figure 2C, lanes 1–6), indicating that an additional dimerization domain resides in the HisRS-C region. GST–HisRS-C also interacted with [35S]His RS(970–1383), but not with [35S]HisRS(1353–1497) (Figure 2C, lanes 7–9 and 13–15, respectively), suggesting that the 1315–1383 interval is required for dimerization by HisRS-C (summarized in Figure 2D). This conclusion was supported by the finding that deletion of the 1315–1388 interval from full-length [35S]HisRS abolished its binding to GST–HisRS-C (Figure 2C, lanes 10–11; see Figure 2D).
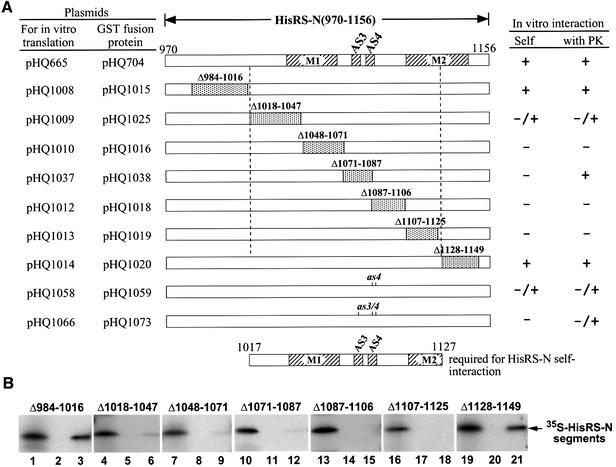
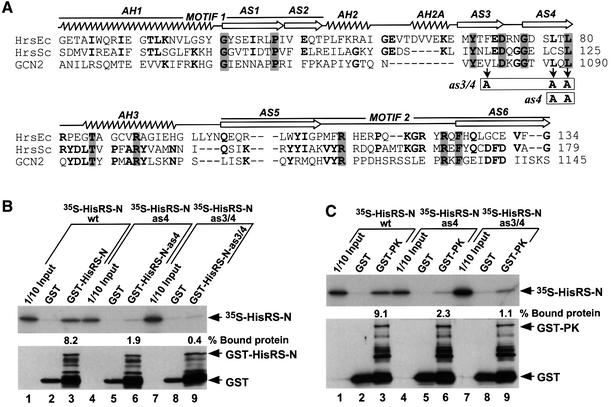
To localize the dimerization determinants in the HisRS-N domain, we carried out binding assays with GST–HisRS-N and derivatives of [35S]HisRS-N bearing successive internal deletions of 16–32 residues. Deleting blocks of residues in the 108 amino acid interval between 1018–1125 severely impaired or eliminated dimerization by these proteins in vitro (Figure 3, in vitro interaction, self). Interestingly, this segment of GCN2 HisRS-N corresponds to the dimeric interface between core catalytic domains in the crystal structure of E.coli HisRS (Arnez et al., 1995), including conserved residues in motif 1, β-strands AS3 and AS4, and the N-terminal half of motif 2 (Figure 3). Accordingly, we made substitution mutations in the GCN2 HisRS-N region at positions corresponding to amino acids that make symmetric hydrophobic contacts between the catalytic domains of the E.coli HisRS dimer (Arnez et al., 1995). Two residues in the predicted β-strand AS4, Leu1088 and Leu1090, were substituted simultaneously with alanines (as4 mutation), and these mutations were also combined with an Ala substitution of Val1080 in the predicted β-strand AS3 (as3/4 mutation) (see Figure 4A). Introduction of the as4 or as3/4 mutation into GST–HisRS-N and [35S]HisRS-N weakened or abolished, respectively, complex formation between these proteins in vitro (Figure 4B; summarized in Figure 3A). These findings suggest that GCN2 contains a dimerization interface resembling that of authentic HisRS.
Fig. 3. The minimal segment required for dimerization of the HisRS-N region also mediates interaction with the PK domain in vitro. (A) Summary of in vitro protein binding assays measuring dimerization or binding to the PK domain by wild-type or mutant versions of the HisRS-N segment. Wild-type HisRS-N is shown schematically at the top with the locations of motifs M1, M2 and predicted β-strands AS3 and AS4 indicated with hatching. Beneath the wild-type HisRS-N segment are shown mutant derivatives containing the indicated internal deletions (shown by shading) or the as4 or as3/4 point mutations. Each was synthesized in vitro and labeled with [35S]methionine, and also expressed as a GST fusion in E.coli, using the corresponding plasmids listed on the left. Each GST fusion protein was immobilized on glutathionine–Sepharose beads and incubated with the 35S-labeled HisRS-N protein containing the same deletion or mutation to assay for self-interactions. The results of these assays shown in (B) are summarized to the right of the schematics in (A) under the column headed ‘Self’. The GST–PK(568–998) fusion immobilized on glutathionine– Sepharose beads was also incubated with the same panel of 35S-labeled HisRS-N proteins and the results of these binding assays (data not shown) are summarized in (A) under the column headed ‘with PK’. The rectangular box at the bottom represents the minimal dimerization domain in the HisRS-N region deduced from these experiments. (B) The 35S-labeled HisRS-N segments containing the different deletions indicated above the gels were incubated with immobilized GST (lanes 2, 5, 8, 11, 14, 17 and 20) or GST–HisRS-N fusion proteins containing the same deletions present in the 35S-labeled HisRS-N segments (lanes 3, 6, 9, 12, 15, 18 and 21). After extensive washing, the bound proteins were resolved by SDS–PAGE and visualized by fluorography. Lanes 1, 4, 7, 10, 13, 16 and 19 contain 1/5 of the input radiolabeled proteins used in the binding experiments.
Fig. 4. Point mutations in the HisRS-N region abolish self-interaction and impair interaction with the PK domain in vitro. (A) Alignment of the HisRS-N region of GCN2 (amino acids 1036–1145) with segments from authentic histidyl-tRNA synthetases of E.coli (HrsEc) and Saccharomyces cerevisiae (HrsSc) encompassing motifs M1 and M2, the α-helices (AH1–AH3) and β-strands (AS1–AS6) present in the crystal structure of E.coli HisRS (Arnez et al., 1995), all indicated above the structure. The single or double mutations in β-strands AS3 (V1080A) and AS4 (L1088A, L1090A), designated as4 and as3/4, are indicated with boxes beneath the GCN2 sequence. (B and C) In vitro binding of [35S]HisRS-N segments (aa 970–1156) synthesized in vitro containing mutations as4 or as3/4 to GST fusion proteins containing the same HisRS-N segments (B), or the PK domain (C). The upper panels display the fluorograms of 35S-labeled proteins added to the reactions (1/10 Input) or bound to the GST or GST–HisRS-N proteins, as indicated. The percentages of the total 35S-labeled proteins bound in each reaction were quantified by phosphoimaging analysis and are indicated below the relevant lanes. The lower panels show the results of immunoblot analysis using GST antibodies of the GST proteins bound to glutathionine–Sepharose beads in each reaction. The positions of GST, GST–HisRS-N and GST–PK fusion proteins are indicated on the right.
Both dimerization domains in the HisRS-like region are required for GCN2 function in vivo
To determine whether dimerization of the HisRS-N region is required for GCN2 function in vivo, each of the internal deletions and point mutations shown in Figure 3A was generated in a plasmid-borne copy of GCN2, and the resulting gcn2 alleles were tested for complementation of the 3-aminotriazole-sensitive phenotype (3ATS) of a gcn2Δ mutant. GCN2-mediated derepression of GCN4 translation, with attendant derepression of histidine biosynthetic genes, is required for growth in the presence of 3AT, an inhibitor of the HIS3 product. None of the gcn2 alleles complemented the 3ATS phenotype of the gcn2Δ mutant. Western analysis of cell extracts with GCN2 antibodies indicated that all but three of the gcn2 alleles were expressed at levels similar to that of wild-type GCN2 (Table I). Although gcn2-Δ1018–1047, gcn2-as4 and gcn2-as3/4 products were expressed at only 60–70% of the wild-type level, these alleles still failed to complement the gcn2Δ mutant when overexpressed from high copy-number plasmids (data not shown). Deletion of the C-terminal dimerization domain in the HisRS-like region in gcn2-Δ1315–1383 also destroyed GCN2 function in vivo (Table I). We conclude that the dimerization domains in the N-terminal and C-terminal halves of the HisRS-like region are both required for GCN2 function in vivo.
Table I. Phenotypes and expression of plasmid-borne gcn2 alleles with mutations in the HisRS-like domaina.
| Plasmid | gcn2 allele | Growth on 3-AT | Relative GCN2 protein level |
|---|---|---|---|
| Vector | none | – | NA |
| pHQ644 | GCN2 | + | 100 |
| pHQ1041 | gcn2-Δ984–1016 | – | 90 |
| pHQ1039 | gcn2-Δ1018–1047 | – | 60 |
| pHQ1042 | gcn2-Δ1048–1071 | – | 90 |
| pHQ1047 | gcn2-Δ1071–1087 | – | 120 |
| pHQ1043 | gcn2-Δ1087–1106 | – | 110 |
| pHQ1040 | gcn2-Δ1107–1125 | – | 130 |
| pHQ1051 | gcn2-Δ1128–1149 | – | 130 |
| pHQ1092 | gcn2-Δ1315–1383 | – | 110 |
| pHQ1076 | gcn2-as4 | – | 70 |
| pHQ1077 | gcn2-as3/4 | – | 60 |
aWild-type GCN2 and the indicated gcn2 alleles were introduced on single-copy plasmids into gcn2Δ strain H1149 and the resulting transformants were tested for growth on synthetic minimal medium containing 30 mM 3AT. +, growth (Hinnebusch and Fink, 1983); –, no growth after 3 days at 30°C. The relative amount of GCN2 protein expressed from each allele was determined by subjecting whole-cell extracts from the same transformants to SDS–PAGE and immunoblot analysis using antibodies against GCN2.
Effects of mutations in the HisRS-N domain on tRNA binding and dimerization by full-length GCN2
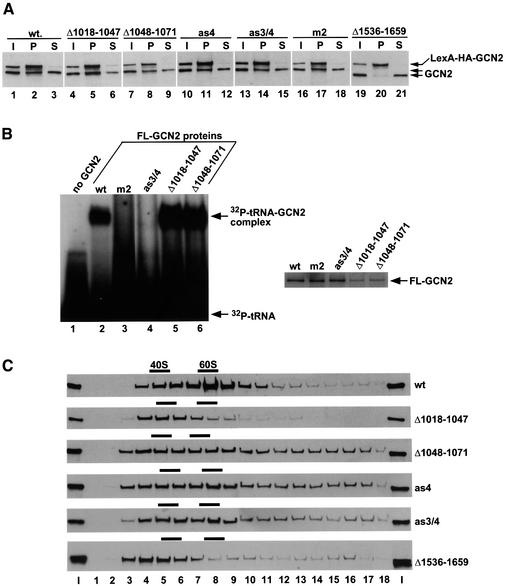
We showed previously that an internal deletion of all HisRS domain residues C-terminal to the HisRS-N domain had only a small effect on the ability of full-length GCN2 to dimerize with a full-length LexA-tagged GCN2 protein expressed in yeast (Qiu et al., 1998). In that study, the gcn2-Δ1161– 1570 product formed heterodimers with LexA–GCN2 at ∼75% of the level seen for wild-type GCN2. Similar results were obtained for a deletion of the PK domain, whereas deletion of the C-term domain reduced dimerization to only 8% of the wild-type level. Thus, dimerization by full-length GCN2 in vivo is more critically dependent on the C-term than on dimerization surfaces in the PK or HisRS-C domains. To evaluate the importance of the HisRS-N region for dimer formation by full-length GCN2, we asked whether the products of gcn2-Δ1018–1047, gcn2-Δ1048–1071, gcn2-as4 and gcn2-as3/4 could be co-immunoprecipitated with LexA–GCN2 from cell extracts. As shown above, all of these gcn2 mutations weakened or abolished dimerization of the isolated HisRS-N segment in vitro (Figure 3). In contrast, none of them reduced co-immunoprecipitation of full-length GCN2 with LexA– GCN2 from cell extracts (Figure 5A). The latter result was also obtained for the gcn2-m2 product, which lacks two key residues in motif 2 of the HisRS-like region required for tRNA binding in vitro (Wek et al., 1995). In agreement with previous findings (Qiu et al., 1998), only the gcn2-Δ1536–1659 product (lacking the C-term domain) failed to interact with LexA–GCN2 in these assays. We conclude that the HisRS-N and HisRS-C dimerization surfaces, and also the tRNA-binding activity of GCN2, are dispensable for dimerization by the full-length protein.
Fig. 5. Effects of mutations in the HisRS-N region on dimerization, tRNA binding and ribosome association by full-length GCN2 in vivo. (A) Co-immunoprecipitation assay of dimerization by wild-type or mutant GCN2 proteins with LexA-HA–GCN2. Whole-cell extracts were prepared from transformants of gcn2Δ strain HQY132 bearing high copy-number plasmid pHQ400 encoding LexA-HA–GCN2 and plasmids bearing wild-type GCN2 (p630), gcn2-Δ1018–1047 (pHQ1044), gcn2-Δ1048–1071 (pHQ1045), gcn2-as4 (pHQ1081), gcn2-as3/4 (pHQ1082), gcn2-m2 (p332) or gcn2-Δ1536–1659 (p2461). Aliquots of extracts containing 50 µg of protein were immunoprecipitated with anti-HA antibodies, and immune complexes were resolved by SDS–PAGE and subjected to immunoblot analysis with GCN2 antibodies, all as described previously (Qiu et al., 1998). Lanes 1, 4, 7, 10, 13, 16 and 19: 1/10 of the input (I) amounts of extract used for immunoprecipitations; lanes 2, 5, 8, 11, 14, 17 and 20: 1/2 of the pellet (P) fractions from the immunoprecipitations; lanes 3, 6, 9, 12, 15, 18 and 21: 1/10 of the supernatant (S) fractions from the immunoprecipitations. (B) GMSA of GCN2–tRNA complexes. The following affinity-purified FL-tagged GCN2 proteins were incubated with 32P-labeled total yeast tRNA (0.5 pmol) in 20 µl of RNA-binding buffer (20 mM HEPES pH 7.4, 150 mM NaCl, 7.5 mM MgCl2, 10% glycerol) at 30°C for 25 min: FL-GCN2 (2 pmol), FL-gcn2-m2 (2 pmol), FL-gcn2-as3/4 (2 pmol), FL-gcn2-Δ1018–1047 (∼0.4 pmol) and FL-gcn2-Δ1048–1071 (∼0.4 pmol). The [32P]tRNA–GCN2 complexes were resolved by electrophoresis in a 5% polyacrylamide gel in 0.5× Tris-borate-EDTA buffer (3 h, 130 V), dried under vacuum and visualized by autoradiography, with the results shown in the upper panel. The same amounts of each protein used in the binding reactions were resolved by SDS–PAGE and stained with Coomassie Brilliant Blue, with the results shown in the right panel. (C) Ribosome binding analysis. Cell extracts were prepared in a buffer lacking Mg2+ and cycloheximide (to dissociate polysomes and 80S ribosomes into 40S and 60S subunits) from transformants of yeast strain H1149 (Wek et al., 1990) harboring high-copy plasmids containing wild-type GCN2 (p630), gcn2-Δ1018–1047 (pHQ1044), gcn2-Δ1048–1071 (pHQ1045), gcn2-as4 (pHQ1081), gcn2-as3/4 (pHQ1082) or gcn2-Δ1536–1659 (p2461). Extracts were resolved by velocity sedimentation in sucrose density gradients and fractions were collected while scanning for A254, all as previously described (Ramirez et al., 1991), and subjected to immunoblot analysis using antibodies against the N-terminus of GCN2 (Romano et al., 1998). Fraction 1 is from the top of the gradient. The first and last lanes contain 1% of the input (I) amounts of extracts separated on the gradients. Positions of free 40S and 60S ribosomal subunits are indicated by bars above the gels.
Although the HisRS domain is dispensable for GCN2 dimerization, the binding of uncharged tRNA could depend on dimerization of the HisRS domain within GCN2. To address this possibility, we purified full-length GCN2 proteins [tagged with Flag (FL) epitope] bearing mutations in the HisRS-N domain that abolish dimerization of this moiety, and assayed them for tRNA binding using a gel mobility shift assay (GMSA). In agreement with recent findings (Dong et al., 2000), wild-type FL-GCN2 formed a complex with 32P-labeled total yeast tRNA ([32P]tRNA), whereas the gcn2-FL-m2 product (bearing motif 2 substitutions) did not (Figure 5B). The product of gcn2-FL-as3/4 was also impaired for tRNA binding (Figure 5B), suggesting that the symmetric contacts in the dimeric interface of the HisRS-N region are required for tRNA binding by GCN2. Surprisingly, the gcn2-FL-Δ1018–1047 and gcn2-FL-Δ1048–1071 products lacking predicted structural elements in motif 1 (most of AH1; C-terminus of AH1, AS1, AS2 and AH2, respectively) reproducibly displayed a higher affinity than the wild-type protein for tRNA (Figure 5B). The fact that these deletions destroy dimerization of the isolated HisRS-N domain in vitro, but do not impair tRNA binding by full-length GCN2, seems at odds with the idea that dimerization of the HisRS-N region is required for tRNA binding. This apparent contradiction is addressed below.
Having found that the Δ1018–1047 and Δ1048–1071 mutations in the HisRS-N domain did not impair dimerization or tRNA binding by full-length GCN2, we considered the possibility that they confer a Gcn– phenotype by disrupting ribosome binding by GCN2. We showed previously that removal of the C-term nearly abolished ribosome binding by GCN2, as judged by its failure to co-sediment with free 60S subunits in cell extracts resolved on sucrose gradients. In contrast, internal deletions in the HisRS-C region (residues 1161–1245 or 1401–1535) led to moderate reductions in ribosome binding (Ramirez et al., 1991). The results in Figure 5C confirm our previous observations by showing that a large proportion of wild-type GCN2, but little or no gcn2-Δ1536–1659 product (lacking the C-term), co-sedimented with 60S subunits (top and bottom panels). The as4 and as3/4 mutations had little or no effect on 60S subunit association, indicating that dimerization of the HisRS-N domain and tRNA binding are both dispensable for high-level ribosome association by GCN2. The Δ1048–1071 mutation also seemed to have little effect on ribosome binding, whereas the Δ1018–1047 deletion led to a strong reduction in 60S binding, comparable to that seen for deletion of the C-term (Δ1536–1659) (Figure 5C). Accordingly, the Gcn– phenotype of gcn2-Δ1018–1047 may result from diminished ribosome association by the mutant product. This seems unlikely for the gcn2-Δ1048–1071 product, however, implying that the HisRS-N region plays a role in stimulating PK function independently of tRNA binding or ribosome association by GCN2. As described below, this novel positive function may involve physical contact between the PK and HisRS-N regions.
The N- and C-terminal subdomains of the HisRS region interact with the PK and C-term domains, respectively
We showed previously that the isolated HisRS domain can interact strongly with the PK and C-term domains in vitro (Qiu et al., 1998). To determine whether these interactions require HisRS residues involved in dimerization, we mapped the binding determinants for the PK and C-term domains in the HisRS segment. A panel of 35S-labeled polypeptides containing the HisRS-like domain progressively truncated from the N- or C-terminus was tested for binding to GST fusions bearing PK residues 568–998 or C-term residues 1498–1659, using the same binding assay described in Figures 2–4. As summarized in Figure 6, residues 1028–1120 in the HisRS-N region were required for interaction with GST–PK(568–998), whereas residues 1315–1383 in the HisRS-C segment were needed for interaction with GST–C-term(1498–1659). It is interesting that the PK binding determinants are located in the PK-proximal portion of the HisRS-like domain, whereas the C-term binding determinants occur in the C-term-proximal section of the HisRS-like region.
Fig. 6. Mapping of amino acids in the HisRS-N and HisRS-C regions required for interaction with the PK and C-term domains in vitro. The rectangular boxes depict different segments of the HisRS-like region that were synthesized in vitro in the presence of [35S]methionine, using the plasmids listed to the left of the boxes. The gray segments in the pHQ1030 and pHQ1024 constructs depict deleted residues. 35S-labeled proteins were incubated with GST, GST–PK(568–998) or GST–C-term(1498–1659) immobilized on glutathione–Sepharose beads. After extensive washing, the proteins bound to the beads were resolved by SDS–PAGE and visualized by fluorography, all as described above. None of these radiolabeled proteins bound to GST alone (data not shown). Binding (+) or failure to bind (–) to GST–PK or GST–C-term is indicated on the right under the columns headed ‘GST–PK’ or ‘GST–C-term’, respectively. ND, not determined. The rectangular boxes under the dashed line depict the GCN2 residues required for binding of HisRS-N to PK, for dimerization of HisRS-C and for its binding to the C-term, and for dimerization of HisRS-N, deduced from results in this figure and in Figures 2 and 3. The hatched boxes indicate the locations of conserved motifs M1–3.
It was also intriguing that the segments in the HisRS region required for interaction with the PK or C-term were essentially co-extensive with the dimerization domains in the HisRS-N and HisRS-C regions, respectively (Figure 6, bottom). To explore whether PK binding and dimerization by the HisRS-N domain have identical sequence requirements, we carried out additional binding experiments using GST–PK(568–998) and the panel of [35S]HisRS-N polypeptides bearing internal deletions or substitutions described above. The results summarized in Figure 3A (column headed ‘with PK’) indicate that, with the exception of Δ1071–1087, all of the mutations that impaired self-interaction of HisRS-N similarly impaired its binding to GST–PK(568–998). Thus, there are nearly identical sequence requirements in the HisRS-N region for dimerization and binding to the PK domain.
Mapping residues in the PK domain required for interaction with the HisRS domain
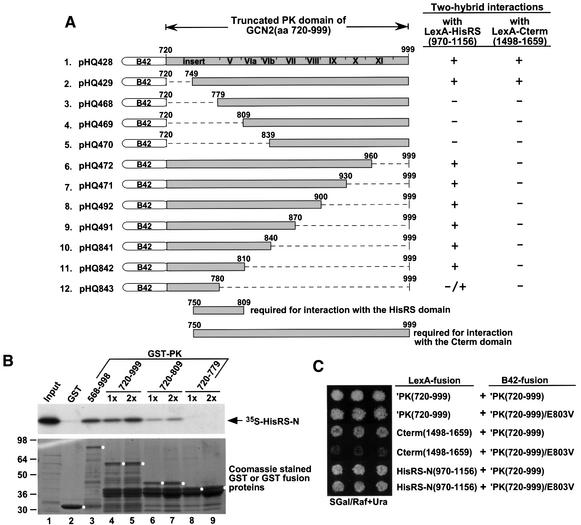
Given the role of the HisRS domain in binding tRNA and activating the kinase function of GCN2, it was important to identify the binding surface in the PK domain for the HisRS-N domain. Accordingly, we carried out two-hybrid assays using a LexA–HisRS-N construct and the panel of B42 activation domain fusions shown in Figure 7A containing different segments of the PK domain. These data showed that residues 750–809 of the PK domain are required for two-hybrid interaction with the HisRS-N segment. The results of GST pull-down experiments using [35S]HisRS-N and GST–PK fusions containing different sections of the PK domain confirmed that PK residues 720–809 are sufficient for the PK–HisRS-N interaction, although the GST–PK(720–809) segment bound [35S]His RS-N less tightly than the larger PK segment in GST– PK(720–999) (Figure 7B, lanes 4–7). Previously, we showed that a much larger segment of the PK domain, spanning residues 750–999, was required for interaction with the C-term segment of GCN2 in the two-hybrid assay (Qiu et al., 1998). These last results are summarized in Figure 7A for comparison with the current findings on PK–HisRS-N interactions.
Fig. 7. Mapping the binding determinants in the PK domain for the HisRS-N region and evidence that the GCN2c-E803V mutation impairs interaction between the PK and C-term domains in vivo. (A) Fusion proteins containing the truncated GCN2 PK domain (residues 720–999) or its deletion derivatives (depicted by gray rectangular boxes) and the B42 transcription activation domain were expressed from the plasmids listed on the left in transformants of yeast strain EGY48 coexpressing LexA–HisRS-N(970–1156) (from pHQ689) or LexA–C-term(1498–1659) (from pHQ311). Dashed lines represent the amino acids deleted from the PK domain, and the GCN2 residue numbers above the lines indicate the first and last residues deleted in each construct. The resulting transformants were tested for growth on SGal/Raf + Ura medium, indicative of a two-hybrid interaction that activates expression of the lexAop-LEU2 reporter resident in the strains, as previously described (Qiu et al., 1998). +, –/+ and – indicate strong, weak and no growth on the medium lacking leucine, respectively. (B) In vitro binding of [35S]HisRS-N (aa 970–1156) synthesized in vitro to GST fusion proteins containing different segments of the PK domain. The upper panel displays the amount of [35S]HisRS-N added to the reactions (1/10 Input) or bound to the GST or GST–PK fusion proteins as indicated above the panel. 1× and 2× represent two different relative concentrations of GST–PK fusion proteins used in the binding reactions.The lower panel shows the GST and GST–PK fusion proteins bound to the glutathione–Sepharose beads in the binding reactions stained with Coomassie Brilliant Blue. The full-length GST–PK fusion proteins are indicated by white dots next to the appropriate bands. (C) The GCN2c-E803V mutation impairs two-hybrid interaction between the PK and C-term domains in vivo. LexA fusion proteins containing truncated GCN2 PK(720–999) (encoded by pHQ433), C-term(1498–1659) (encoded by pHQ311) or HisRS-N(970–1156) (encoded by pHQ689) were expressed in transformants of EGY48 expressing B42 fusion proteins containing PK(720–999) (from pHQ428) or PK(720–999)/E803V (from pHQ823). The resulting transformants were tested for growth on SGal/Raf + Ura medium (lacking leucine), which is indicative of a two-hybrid interaction that activates expression of the lexAop-LEU2 reporter.
Evidence that PK–C-term interaction downregulates GCN2 kinase activity
GCN2c mutations conferring constitutive activation of GCN2 function in vivo map in the PK, HisRS and C-term domains, and the most potent of these mutations map in the C-term (Wek et al., 1990; Ramirez et al., 1992). Based on these observations, we speculated that the C-term might function as an autoinhibitory domain, and that binding of tRNA to the HisRS region would overcome this inhibition. We reasoned that if this model is correct, one or more GCN2c mutations might weaken the interaction between the isolated PK and C-term domains. To explore this possibility, we introduced each of five GCN2c mutations mapping in the PK domain (M788V, E803V, E821K, H861Y and R768W/D987G) into the LexA–′PK(720–999) and B42–′PK(720–999) constructs used for two-hybrid analysis, and tested them for interaction with LexA and B42 fusions bearing the C-term. The PK fusions were also tested for two-hybrid interactions with the appropriate constructs containing the wild-type ′PK(720–999) or HisRS-N segments to examine possible effects on PK dimerization or PK–HisRS interactions. Similarly, four GCN2c mutations mapping in the C-term (R1557K, E1591K, E1606G and N1616K) were introduced into the LexA and B42 fusions containing the C-term segment, and tested for interaction with the fusions bearing wild-type ′PK(720–999) or wild-type C-term. Interestingly, the E803V mutation in the kinase domain abolished interaction of the PK segment with the C-term, but had no effect on PK dimerization or PK interaction with the HisRS fragment (Figure 7C), and thus specifically impaired the PK–C-term interaction. None of the other GCN2c mutations affected any of the two-hybrid interactions (data not shown). These data suggest that the constitutive activation of GCN2 conferred by the E803V mutation involves the loss of an inhibitory physical interaction between the PK and C-term domains.
Discussion
The HisRS-N domain functions in dimerization of the HisRS region, tRNA binding and stimulatory interactions with the PK domain of GCN2
We have identified two dimerization domains in the N- and C-terminal halves of the HisRS-like segment of GCN2, located between residues 1017–1127 and 1315–1383, respectively (Figure 1). The HisRS-N dimerization domain represents a stretch of strong sequence similarity between GCN2 and authentic HisRSs (Ramirez et al., 1992; Arnez et al., 1995) that coincides with the dimerization interface in the catalytic domain of the E.coli enzyme (Arnez et al., 1995). Conserved amino acids in β-strands AS3 and AS4 make symmetric hydrophobic contacts that stabilize a four-stranded β-sheet forming an arch over the dimeric interface in the catalytic domain of E.coli HisRS (Arnez et al., 1995). Substituting the corresponding residues in GCN2 with alanines (the as3/4 mutations) abolished dimerization of the GCN2 HisRS-N segment in vitro, suggesting that the dimerization interface of this segment is structurally similar to that present in authentic HisRS. The fact that these residues are required for GCN2 function in vivo strongly suggests that dimerization of HisRS-N is important for activation of GCN2 by uncharged tRNA. The HisRS-C dimerization domain coincides with the C-terminal end of the catalytic domain of E.coli HisRS, encompassing βAS9, αH6 and motif 3 (Arnez et al., 1995). This region is not present at the dimer interface of E.coli HisRS, indicating a structural divergence between the GCN2 HisRS-C region and authentic HisRS. Although the HisRS-C domain is required for GCN2 function in vivo, it is unclear whether the dimerization activity of this segment is important for GCN2 activation.
Internal deletions in the HisRS-N region that abolished dimerization of this isolated segment in vitro did not reduce the ability of full-length GCN2 to form heterodimers in vivo with a LexA–GCN2 fusion. The same result was obtained previously for a deletion that included the HisRS-C dimerization domain (Qiu et al., 1998). These results can be explained by noting that GCN2 contains multiple dimerization domains (Figure 1) and that only the C-term appears to be critically required for dimerization by the full-length protein (Qiu et al., 1998). The m2 mutation that abolished tRNA binding did not impair dimerization between GCN2 and LexA–GCN2 (Figure 5A). This last finding implies that tRNA binding is not required for dimerization by GCN2. Accordingly, we propose that GCN2 occurs as a dimer under both non-starvation and starvation conditions. Rather than promoting dimerization, tRNA binding would trigger a conformational change in GCN2 that activates the kinase domain.
The as3/4 point mutations that impaired dimerization of the isolated HisRS-N domain (Figure 4) also destroyed tRNA binding by full-length GCN2 in vitro (Figure 5B). These results suggest that dimerization of the HisRS-N domain is required for tRNA binding by GCN2. In accordance with this conclusion, there is evidence that dimerization of class II aminoacyl-tRNA synthetases is required for proper folding of the catalytic domain, which encompasses the binding site for the acceptor stem of tRNA (Eriani et al., 1993; Agou et al., 1996). In contrast, a monomeric form of E.coli HisRS lacking the accessory C-terminal domain required for dimerization was active for tRNA binding, although its catalytic function was sharply impaired (Augustine and Francklyn, 1997). Given its considerable sequence divergence from authentic HisRS (Wek et al., 1989), and the negative influence of the adjacent PK domain on tRNA binding (see below), it seems plausible that dimerization of the HisRS-N domain in GCN2 could be required for tRNA binding, as suggested by the effects of the as3/4 mutation.
The gcn2-Δ1018–1047 mutation eliminates most of the predicted long helix of motif 1 (AH1) that forms part of the dimeric interface with its symmetrically related counterpart in the E.coli HisRS dimer (Arnez et al., 1995). The adjacent Δ1048–1071 mutation eliminates the remainder of motif 1 and the adjacent residues corresponding to Ile45 and Val46 in E.coli HisRS. The latter contribute hydrophobic contacts at the dimeric interface of the E.coli enzyme, although the fact that Ile45 is replaced by Arg1063 in wild-type GCN2 may signify a diminished role for motif 1 in dimerization of the GCN2 HisRS-N domain. The fact that these two deletions destroyed dimerization of the isolated HisRS-N segment in vitro supports the idea that motif 1 contributes to dimerization of HisRS-N in GCN2. If dimerization of HisRS-N is required for tRNA binding, how can we explain the fact that Δ1018–1047 and Δ1048–1071 did not impair tRNA binding by GCN2 (Figure 5B)? It is possible that dimerization by the PK domain upstream and the HisRS-C region downstream can compensate for the Δ1018–1047 and Δ1048–1071 mutations and maintain dimerization of the HisRS-N region in the absence of intact motif 1. In contrast, eliminating the critical dimerization contacts in β-strands AS3/AS4 by the as3/4 mutations would not be suppressed by dimerization of the adjacent PK and HisRS-C domains, resulting in loss of HisRS-N dimerization and tRNA binding.
Given that the Δ1048–1071 deletion did not impair tRNA binding or ribosome association by full-length GCN2, why is the gcn2-Δ1048–1071 product inactive in vivo? We found that HisRS-N residues 1028–1120 interact with the PK domain and that this interaction is impaired by the Δ1048–1071 mutation (Figure 3). Hence, we suggest that the PK–HisRS-N interaction has a stimulatory effect on kinase activity that is disrupted by Δ1048–1071. The portion of the HisRS-N region that interacts with the PK domain corresponds to the ‘top’ of the catalytic domain of E.coli HisRS, which forms the dimeric interface, and also includes structures AH3 and AS5, which immediately precede motif 2 (Figure 4A) (Arnez et al., 1995). By analogy with studies on Asp-RS (Eiler et al., 1999), tRNA binding to GCN2 may produce a conformational change in the HisRS-N domain that is transmitted to the PK domain and stimulates kinase activity (Figure 4). The segment of the PK domain that interacts with the HisRS-N region includes the C-terminal section of the large insert between kinase subdomains III and IV, portions of the ATP-binding and catalytic lobes, and the hinge region between the two lobes (Hanks and Quinn, 1991; Knighton et al., 1991; Ramirez et al., 1992). Thus, association of the HisRS-N region with this segment of the PK domain could enhance the correct orientation of the N- and C-terminal lobes of the PK domain (Johnson et al., 1996). This part of the PK domain also contains three of the five GCN2c mutations described previously (including E803V), providing independent evidence that it plays an important role in kinase activation by uncharged tRNA.
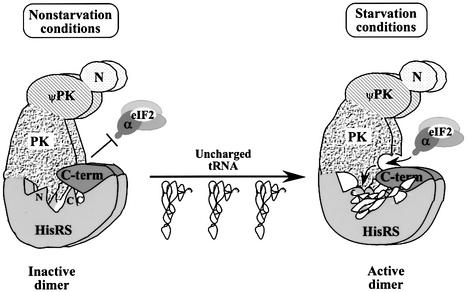
It was striking that the Δ1048–1071 mutation increased the affinity of GCN2 for tRNA. As this deletion should abolish association between the PK and HisRS-N domains (Figure 3), it appears that binding of the PK domain to the HisRS-N region interferes with tRNA binding. This conclusion may seem at odds with our suggestion that PK–HisRS-N interactions transmit the stimulatory effect of tRNA binding to the kinase domain; however, such opposing functions could be accommodated by proposing two different modes of PK–HisRS-N interaction in GCN2. One configuration would be incompatible with tRNA binding and would not stimulate PK function, whereas the other would allow tRNA binding and promote kinase activation (Figure 8). According to this hypothesis, the Δ1048–1071 mutation eliminated the inhibitory effect of PK–HisRS-N interaction on tRNA binding, but since it also destroyed the crucial positive effect of HisRS-N on the kinase domain, there was a net loss of GCN2 function. This model envisions a less extreme structural reorganization of GCN2 elicited by tRNA binding than we suggested previously, wherein all contacts between the PK and HisRS domains were abolished in the tRNA-bound state (Dong et al., 2000).
Fig. 8. A model depicting allosteric alterations in domain interactions in GCN2 elicited by bound tRNA that result in kinase activation. The domains of GCN2 are labeled as follows: binding domain for GCN1–GCN20 complex at the N-terminus, N; pseudokinase domain, ψPK; protein kinase domain, PK; HisRS-like region, HisRS; and C-term. The N- and C-terminal portions of the HisRS domain (labeled N and C in the HisRS region of the schematic on the left) interact with the PK and C-term segments, respectively. In the inactive form of GCN2 present in non-starvation conditions (left), productive association with the substrate eIF2 is prevented by binding of the C-term to the PK domain, and possibly also by HisRS-N–PK interactions (both inhibitory interactions are depicted by overlapped domains). The HisRS-N–PK interactions in this state are not compatible with tRNA binding. Under starvation conditions (right), uncharged tRNA binds to GCN2, making contacts with both the HisRS-N and C-term domains. The bound tRNA produces conformational changes in both domains that are transmitted to the PK domain. This opens up the substrate binding cleft in the PK domain and allows eIF2 binding and phosphorylation (the allosteric alteration in the PK domain is signified by the curved arrow in this domain.) The HisRS-N region remains engaged with the PK domain and contributes to its active conformation. The C-term also remains associated with the PK domain, but inhibitory C-term–PK contacts are lost upon tRNA binding. These inhibitory contacts are also weakened by the constitutively activating E803V mutation in the PK domain. Both inactive and active forms of GCN2 are depicted as dimers because GCN2 can dimerize in the absence of an intact HisRS domain and its tRNA binding activity. This model does not depict the potential ‘criss-cross’ interactions that could occur if the C-term of one subunit interacts with the HisRS-C region of the other subunit, as found in the crystal structure of authentic E.coli HisRS (see text for further details).
Evidence that the C-term–PK interaction contributes to the latency of GCN2 kinase activity
The isolated C-term domain of GCN2 can interact directly with the PK domain (Qiu et al., 1998), and residues 1498–1535 in the C-term are required for this interaction (Qiu et al., 1998). Previously, we showed that a large PK segment including residues 750–999 was required for binding to the C-term (Qiu et al., 1998) (summarized in Figure 7A and Figure 1), suggesting that the C-term makes multiple contacts across the PK domain. Interestingly, we discovered here that the GCN2c-E803V mutation weakened binding between the C-term and PK segments. This mutation maps in the small segment of the PK domain (residues 750–810) required for the PK–HisRS-N interaction, and is encompassed by the larger PK domain required for C-term binding (see Figures 1 and 7). The E803V mutation did not affect the PK–HisRS-N interaction, indicating that it specifically disrupts PK–C-term association. These observations suggest that PK–C-term interaction is required for inhibition of kinase activity, and that its alteration by E803V leads to inappropriate activation of GCN2 under non-starvation conditions. In the wild-type protein, the autoinhibitory function of the C-term would be reversed by tRNA binding to the composite HisRS–C-term domain.
Recently, we found that tRNA binding to a recombinant HisRS–C-term segment weakened its association with the isolated PK domain (Dong et al., 2000). This finding is consistent with the notion that tRNA binding disrupts an autoinhibitory interaction between the C-term and PK domains. Furthermore, the E803V mutation led to a large increase in the tRNA binding affinity of full-length GCN2 (Dong et al., 2000). This last result, combined with our observation that PK–C-term interaction is impaired by the E803V mutation, supports the idea that tRNA binding stabilizes a conformation in which the C-term dissociates from the PK domain and becomes more accessible to tRNA. The E803V mutation would shift the equilibrium towards a conformation of GCN2 that can bind tRNA and become activated at low concentrations of uncharged tRNA present in non-starved cells.
The fact that other GCN2c mutations in the PK or C-term domains did not impair the PK–C-term two-hybrid interaction could be explained by proposing that these mutations have more subtle effects than E803V on PK–C-term association, and overcome the inhibitory function of the C-term without disrupting the strongest contacts between these two domains. In this view, the C-term is not completely dissociated from the PK domain upon tRNA binding, and only the subset of interactions required for PK inhibition are replaced by C-term–tRNA interactions. This hypothesis is akin to the suggestion above that tRNA binding switches the PK–HisRS-N interaction between inhibitory and stimulatory modes rather than completely dissociating the two domains (Figure 8). Thus, several results obtained here imply that tRNA binding produces a more subtle rearrangement of domain interactions than previously imagined (Dong et al., 2000).
Finally, what could be the role of interactions between the C-term domain and the HisRS-C region of GCN2? The authentic HisRS contains an accessory domain at its extreme C-terminus that interacts with the catalytic domain of the opposing subunit in the dimer (Arnez et al., 1995). It is tempting to propose a similar ‘criss-cross’ interaction for GCN2 in which the C-term of one subunit interacts with the HisRS-C region of the other subunit in the dimer. In this way, dimerization at the C-terminus of GCN2 would be stabilized by HisRS-C– HisRS-C and HisRS-C–C-term interactions in addition to the C-term–C-term interactions described previously (Qiu et al., 1998). This speculation is also in accordance with the proposed role of the accessory C-terminal domain of E.coli HisRS in tRNA binding (Arnez et al., 1995) and our finding that tRNA binding by GCN2 requires both the C-term and HisRS domains (Dong et al., 2000).
Materials and methods
Plasmids
Plasmids used in this study are listed in Table II. Details of their construction are available as Supplementary data at The EMBO Journal Online.
Table II. Plasmids used in this study.
| Plasmid | Allele | Reference |
|---|---|---|
| Plasmids encoding GST fusion proteins | ||
| pHQ242 | GST under ADH1 promoter in 2µ TRP1 plasmid pPM237 (L.Prakash) | this study |
| pHQ601 | GST-GCN2(970–1497) in pHQ242 backbone | this study |
| pHQ529 | GST-GCN2(720–999) in pGEX-5X-1 (Pharmacia Biotech.) | this study |
| pHQ531 | GST-GCN2(1498–1659) in pGEX-5X-1 | Qiu et al. (1998) |
| pHQ551 | GST-GCN2(568–998) in pGEX-5X-1 | Qiu et al. (1998) |
| pHQ701 | GST-GCN2(1315–1497) in pGEX-5X-1 | this study |
| pHQ704 | GST-GCN2(970–1156) in pGEX-5X-1 | this study |
| pHQ993 | GST-GCN2(720–809) in pGEX-5X-1 | this study |
| pHQ994 | GST-GCN2(720–779) in pGEX-5X-1 | this study |
| pHQ1015 | GST-GCN2(970–1156)/Δ984–1016 in pGEX-5X-1 | this study |
| pHQ1016 | GST-GCN2(970–1156)/Δ1048–1071 in pGEX-5X-1 | this study |
| pHQ1018 | GST-GCN2(970–1156)/Δ1087–1106 in pGEX-5X-1 | this study |
| pHQ1019 | GST-GCN2(970–1156)/Δ1107–1125 in pGEX-5X-1 | this study |
| pHQ1020 | GST-GCN2(970–1156)/Δ1128–1149 in pGEX-5X-1 | this study |
| pHQ1025 | GST-GCN2(970–1156)/Δ1018–1047 in pGEX-5X-1 | this study |
| pHQ1038 | GST-GCN2(970–1156)/Δ1071–1087 in pGEX-5X-1 | this study |
| pHQ1059 | GST-GCN2(970–1156)/as4 in pGEX-5X-1 | this study |
| pHQ1073 | GST-GCN2(970–1156)/as3/4 in pGEX-5X-1 | this study |
| Plasmids for in vitro translation of GCN2 segments | ||
| pHQ541 | GCN2(970–1497) under T7 promoter in pGEM-3Z (Promega) | Qiu et al. (1998) |
| pHQ596 | GCN2(970–1315) in pGEM-3Z | this study |
| pHQ597 | GCN2(970–1383) in pGEM-3Z | this study |
| pHQ599 | GCN2(1086–1497) in pGEM-3Z | this study |
| pHQ664 | GCN2(1315–1497) in pGEM-3Z | this study |
| pHQ665 | GCN2(970–1156) in pGEM-3Z | this study |
| pHQ666 | GCN2(970–1245) in pGEM-3Z | this study |
| pHQ667 | GCN2(1028–1497) in pGEM-3Z | this study |
| pHQ668 | GCN2(1130–1497) in pGEM-3Z | this study |
| pHQ669 | GCN2(1193–1497) in pGEM-3Z | this study |
| pHQ670 | GCN2(1255–1497) in pGEM-3Z | this study |
| pHQ671 | GCN2(970–1120) in pGEM-3Z | this study |
| pHQ680 | GCN2(970–1086) in pGEM-3Z | this study |
| pHQ683 | GCN2(1353–1497) in pGEM-3Z | this study |
| pHQ1008 | GCN2(970–1156)/Δ984–1016 in pGEM-3Z | this study |
| pHQ1009 | GCN2(970–1156)/Δ1018–1047 in pGEM-3Z | this study |
| pHQ1010 | GCN2(970–1156)/Δ1048–1071 in pGEM-3Z | this study |
| pHQ1012 | GCN2(970–1156)/Δ1087–1106 in pGEM-3Z | this study |
| pHQ1013 | GCN2(970–1156)/Δ1107–1125 in pGEM-3Z | this study |
| pHQ1014 | GCN2(970–1156)/Δ1128–1149 in pGEM-3Z | this study |
| pHQ1024 | GCN2(970–1497)/Δ1315–1388 in pGEM-3Z | this study |
| pHQ1030 | GCN2(970–1497)/Δ1029–1119 in pGEM-3Z | this study |
| pHQ1037 | GCN2(970–1156)/Δ1071–1087 in pGEM-3Z | this study |
| pHQ1058 | GCN2(970–1156)/as4 in pGEM-3Z | this study |
| pHQ1066 | GCN2(970–1156)/as3/4 in pGEM-3Z | this study |
| Plasmids encoding GCN2 and derivatives | ||
| p332 | gcn2-m2 in p630 backbone | Wek et al. (1995) |
| p630 | wild-type GCN2 in in YEp24 (Botstein et al., 1979) 2µ, URA3 | Wek et al. (1990) |
| p2461 | gcn2-Δ1536–1659 in p630 backbone | Qiu et al. (1998) |
| pHQ644 | GCN2, CEN6, ARSH4, URA3, a derivative of p722 (Wek et al., 1990) | this study |
| pHQ1039 | gcn2-Δ1018–1047 in pHQ644 backbone | this study |
| pHQ1040 | gcn2-Δ1107–1125 in pHQ644 backbone | this study |
| pHQ1041 | gcn2-Δ984–1016 in pHQ644 backbone | this study |
| pHQ1042 | gcn2-Δ1048–1071 in pHQ644 backbone | this study |
| pHQ1043 | gcn2-Δ1087–1106 in pHQ644 backbone | this study |
| pHQ1044 | gcn2-Δ1018–1047 in p630 backbone | this study |
| pHQ1045 | gcn2-Δ1048–1071 in p630 backbone | this study |
| pHQ1047 | gcn2-Δ1071–1087 in pHQ644 backbone | this study |
| pHQ1051 | gcn2-Δ1128–1149 in pHQ644 backbone | this study |
| pHQ1076 | gcn2-as4 in pHQ644 backbone | this study |
| pHQ1077 | gcn2-as3/4 in pHQ644 backbone | this study |
| pHQ1081 | gcn2-as4 in p630 backbone | this study |
| pHQ1082 | gcn2-as3/4 in p630 backbone | this study |
| pHQ1092 | gcn2-Δ1315–1383 in pHQ644 backbone | this study |
| pDH103 | GCN2-FL in pEMBLyex4 (Cesareni and Murray, 1987) backbone | Dong et al. (2000) |
| pDH104 | gcn2-FL-m2 in pEMBLyex4 backbone | Dong et al. (2000) |
| pHQ1048 | gcn2-FL-Δ1018–1047 in pEMBLyex4 backbone | this study |
| pHQ1049 | gcn2-FL-Δ1048–1071 in pEMBLyex4 backbone | this study |
| pHQ1084 | gcn2-FL-as3/4 in pEMBLyex4 backbone | this study |
| Plasmids for two-hybrid analysis of GCN2 segments | ||
| p2247 | LexA-HA in pEG202 (Golemis et al., 1996) backbone | Zhang et al. (1997) |
| pHQ311 | LexA-GCN2(1498–1659) in pEG202 backbone | Qiu et al. (1998) |
| pHQ400 | LexA-HA-GCN2(27–1659) in p2247 backbone | Qiu et al. (1998) |
| pHQ433 | LexA-GCN2(720–999) in pEG202 backbone | Qiu et al. (1998) |
| pHQ588 | LexA-HA-GCN2(970–1497) in p2247 backbone | Qiu et al. (1998) |
| pHQ689 | LexA-GCN2(970–1156) in pEG202 backbone | this study |
| pHQ428 | B42-GCN2(720–999) in pJG4-5 (Golemis et al., 1996) backbone | Qiu et al. (1998) |
| pHQ429 | B42-GCN2(750–999) in pJG4-5 backbone | Qiu et al. (1998) |
| pHQ468 | B42-GCN2(780–999) in pJG4-5 backbone | this study |
| pHQ469 | B42-GCN2(810–999) in pJG4-5 backbone | this study |
| pHQ470 | B42-GCN2(740–999) in pJG4-5 backbone | this study |
| pHQ471 | B42-GCN2(720–929) in pJG4-5 backbone | this study |
| pHQ472 | B42-GCN2(720–959) in pJG4-5 backbone | this study |
| pHQ491 | B42-GCN2(720–869) in pJG4-5 backbone | this study |
| pHQ492 | B42-GCN2(720–899) in pJG4-5 backbone | this study |
| pHQ823 | B42-GCN2(720–999)/E803V in pJG4-5 backbone | this study |
| pHQ841 | B42-GCN2(720–839) in pJG4-5 backbone | this study |
| pHQ842 | B42-GCN2(720–809) in pJG4-5 backbone | this study |
| pHQ843 | B42-GCN2(720–779) in pJG4-5 backbone | this study |
Biochemical methods
Co-immunoprecipitation and immunoblotting were conducted as described previously (Qiu et al., 1998) using GCN2 antibodies described previously (Romano et al., 1998) and GST antibodies purchased from Santa Cruz (1:2000 dilution). Preparation of GST and GST fusion proteins, in vitro translation of GCN2 fragments and GST pull-down assays were carried out as described previously (Qiu et al., 1998). Preparation of FL-His6-tagged GCN2 proteins, 32P-labeling of total yeast tRNA and GMSAs of GCN2 binding to 32P-tRNA were all conducted as described previously (Dong et al., 2000). Ribosome binding of GCN2 was analyzed as described previously (Ramirez et al., 1991).
Supplementary data
Supplementary data for this paper are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We are indebted to Ron Wek for plasmid p332. We thank Tom Dever and Evelyn Sattlegger for critical reading of the manuscript and colleagues in the LEGR for suggestions and discussions. J.D. was supported as a Research Associate by the National Research Council. C.S.F. was supported by NIH grant GM54899.
References
- Agou F., Waller,J.P. and Mirande,M. (1996) Expression of rat aspartyl-tRNA synthetase in Saccharomyces cerevisiae. J. Biol. Chem., 271, 29295–29303. [DOI] [PubMed] [Google Scholar]
- Arnez J.G., Harris,D.C., Mitschler,A., Rees,B., Francklyn,C.S. and Moras,D. (1995) Crystal structure of histidyl-tRNA synthetase from Escherichia coli complexed with histidyl-adenylate. EMBO J., 14, 4143–4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustine J. and Francklyn,C. (1997) Design of an active fragment of a class II aminoacyl-tRNA synthetase and its significance for synthetase evolution. Biochemistry, 36, 3473–3482. [DOI] [PubMed] [Google Scholar]
- Berlanga J.J., Herrero,S. and de Haro,C. (1998) Characterization of the hemin-sensitive eukaryotic initiation factor 2α kinase from mouse nonerythroid cells. J. Biol. Chem., 273, 32340–32346. [DOI] [PubMed] [Google Scholar]
- Botstein D., Falco,S.C., Stewart,S.E., Brennan,M., Scherer,S., Stinchcomb,D.T., Struhl,K. and Davis,R.W. (1979) Sterile host yeasts (SHY): a eukaryotic system of biological containment for recombinant DNA experiments. Gene, 8, 17–24. [DOI] [PubMed] [Google Scholar]
- Cesareni G. and Murray,J.A.H. (1987) Plasmid vectors carrying the replication origin of filamentous single-stranded phages. In Setlow,J.K. and Hollaender,A. (eds), Genetic Engineering: Principles and Methods. Vol. 9. Plenum Press, New York, NY, pp. 135–154.
- Dong J., Qiu,H., Garcia-Barrio,M., Anderson,J. and Hinnebusch,A.G. (2000) Uncharged tRNA activates GCN2 by displacing the protein kinase moiety from a bipartite tRNA-binding domain. Mol. Cell, 6, 269–279. [DOI] [PubMed] [Google Scholar]
- Eiler S., Dock-Bregeon,A., Moulinier,L., Thierry,J.C. and Moras,D. (1999) Synthesis of aspartyl-tRNAAsp in Escherichia coli—a snapshot of the second step. EMBO J., 18, 6532–6541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriani G., Cavarelli,J., Martin,F., Dirheimer,G., Moras,D. and Gangloff,J. (1993) Role of dimerization in yeast aspartyl-tRNA synthetase and importance of the class II invariant proline. Proc. Natl Acad. Sci. USA, 90, 10816–10820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Barrio M., Dong,J., Ufano,S. and Hinnebusch,A.G. (2000) Association of GCN1–GCN20 regulatory complex with the conserved N-terminal domain of eIF2α kinase GCN2 is required for GCN2 activation in vivo. EMBO J., 19, 1887–1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golemis E.A., Gyuris,J. and Brent,R. (1996) Interaction trap/two-hybrid system to identify interacting proteins. In Ausubel,F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl,K. (eds), Current Protocols in Molecular Biology. John Wiley, New York, NY, pp. 20.1.1–20.1.28.
- Hanks S.K. and Quinn,A.M. (1991) Protein kinase catalytic domain sequence database: identification of conserved features of primary structure and classification of family members. Methods Enzymol., 200, 38–62. [DOI] [PubMed] [Google Scholar]
- Hinnebusch A.G. (1992) General and pathway-specific regulatory mechanisms controlling the synthesis of amino acid biosynthetic enzymes in Saccharomyces cerevisiae. In Broach,J.R., Jones,E.W. and Pringle,J.R. (eds), The Molecular and Cellular Biology of the Yeast Saccharomyces: Gene Expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 319–414.
- Hinnebusch A.G. and Fink,G.R. (1983) Positive regulation in the general amino acid control of Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 80, 5374–5378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson L.N., Noble,M.E.M. and Owen,D.J. (1996) Active and inactive protein kinases: structural basis for regulation. Cell, 85, 149–158. [DOI] [PubMed] [Google Scholar]
- Knighton D.R., Zheng,J., Ten Eyck,L.F., Xuong,N.H., Taylor,S.S. and Sowadski,J.M. (1991) Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science, 253, 407–414. [DOI] [PubMed] [Google Scholar]
- Olsen D.S., Jordan,B., Chen,D., Wek,R.C. and Cavener,D.R. (1998) Isolation of the gene encoding the Drosophila melanogaster homolog of the Saccharomyces cerevisiae GCN2 eIF-2α kinase. Genetics, 149, 1495–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu H., Garcia-Barrio,M.T. and Hinnebusch,A.G. (1998) Dimerization by translation initiation factor 2 kinase GCN2 is mediated by interactions in the C-terminal ribosome-binding region and the protein kinase domain. Mol. Cell. Biol., 18, 2697–2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez M., Wek,R.C. and Hinnebusch,A.G. (1991) Ribosome-association of GCN2 protein kinase, a translational activator of the GCN4 gene of Saccharomyces cerevisae. Mol. Cell. Biol., 11, 3027–3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez M., Wek,R.C., Vazquez de Aldana,C.R., Jackson,B.M., Freeman,B. and Hinnebusch,A.G. (1992) Mutations activating the yeast eIF-2α kinase GCN2: isolation of alleles altering the domain related to histidyl-tRNA synthetases. Mol. Cell. Biol., 12, 5801–5815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano P.R. et al. (1998) Autophosphorylation in the activation loop is required for full kinase activity in vivo of human and yeast eukaryotic initiation factor 2α kinases PKR and GCN2. Mol. Cell. Biol., 18, 2282–2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoyo J., Alcalde,J., Mendez,R., Pulido,D. and de Haro,C. (1997) Cloning and characterization of a cDNA encoding a protein synthesis initiation factor-2α (eIF-2α) kinase from Drosophila melanogaster. J. Biol. Chem., 272, 12544–12550. [DOI] [PubMed] [Google Scholar]
- Sattlegger E., Hinnebusch,A.G. and Barthelmess,I.B. (1998) cpc-3, the Neurospora crassa homologue of yeast GCN2, encodes a polypeptide with juxtaposed eIF2α kinase and histidyl-tRNA synthetase-related domains required for general amino acid control. J. Biol. Chem., 273, 20404–20416. [DOI] [PubMed] [Google Scholar]
- Sood R., Porter,A.C., Olsen,D., Cavener,D.R. and Wek,R.C. (2000) A mammalian homologue of GCN2 protein kinase important for translational control by phosphorylation of eukaryotic initiation factor-2α. Genetics, 154, 787–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wek R.C., Jackson,B.M. and Hinnebusch,A.G. (1989) Juxtaposition of domains homologous to protein kinases and histidyl-tRNA synthetases in GCN2 protein suggests a mechanism for coupling GCN4 expression to amino acid availability. Proc. Natl Acad. Sci. USA, 86, 4579–4583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wek R.C., Ramirez,M., Jackson,B.M. and Hinnebusch,A.G. (1990) Identification of positive-acting domains in GCN2 protein kinase required for translational activation of GCN4 expression. Mol. Cell. Biol., 10, 2820–2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wek S.A., Zhu,S. and Wek,R.C. (1995) The histidyl-tRNA synthetase-related sequence in the eIF-2α protein kinase GCN2 interacts with tRNA and is required for activation in response to starvation for different amino acids. Mol. Cell. Biol., 15, 4497–4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Kirouac,M., Zhu,N., Hinnebusch,A.G. and Rolfes,R.J. (1997) Evidence that complex formation by Bas1p and Bas2p (Pho2p) unmasks the activation function of Bas1p in an adenine-repressible step of ADE gene transcription. Mol. Cell. Biol., 17, 3272–3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S. and Wek,R.C. (1998) Ribosome-binding domain of eukaryotic initiation factor-2 kinase GCN2 facilitates translation control. J. Biol. Chem., 273, 1808–1814. [DOI] [PubMed] [Google Scholar]
- Zhu S., Sobolev,A.Y. and Wek,R.C. (1996) Histidyl-tRNA synthetase-related sequences in GCN2 protein kinase regulate in vitro phosphorylation of eIF-2. J. Biol. Chem., 271, 24989–24994. [DOI] [PubMed] [Google Scholar]