Abstract
Human pluripotent stem cells (hPSCs) have been derived from the inner cell mass cells of blastocysts (embryonic stem cells) and primordial germ cells of the developing gonadal ridge (embryonic germ cells). Like their mouse counterparts, hPSCs can be maintained in culture in an undifferentiated state and, upon differentiation, generate a wide variety of cell types. Embryoid body (EB) formation is a requisite step in the process of in vitro differentiation of these stem cells and has been used to derive neurons and glia, vascular endothelium, hematopoietic cells, cardiomyocytes, and glucose-responsive insulin-producing cells from mouse PSCs. EBs generated from human embryonic germ cell cultures have also been found to contain a wide variety of cell types, including neural cells, vascular endothelium, muscle cells, and endodermal derivatives. Here, we report the isolation and culture of cells from human EBs as well as a characterization of their gene expression during growth in several different culture environments. These heterogeneous cell cultures are capable of robust and long-term [>70 population doublings (PD)] proliferation in culture, have normal karyotypes, and can be cryopreserved, clonally isolated, and stably transfected. Cell cultures and clonal lines retain a broad pattern of gene expression including simultaneous expression of markers normally associated with cells of neural, vascular/hematopoietic, muscle, and endoderm lineages. The growth and expression characteristics of these EB-derived cells suggest that they are relatively uncommitted precursor or progenitor cells. EB-derived cells may be suited to studies of human cell differentiation and may play a role in future transplantation therapies.
Mouse pluripotent stem cells (mPSCs) have been derived from the inner cell mass cells of blastocysts and from primordial germ cells colonizing the developing gonadal ridge and are referred to as embryonic stem (ES) cells and embryonic germ (EG) cells, respectively. When mPSCs differentiate in vitro, they form complex three-dimensional cell aggregates termed embryoid bodies (EBs). Some early developmental processes are recapitulated within the environment of an EB, resulting in a haphazard collection of precursor and more fully differentiated cells from a wide variety of lineages. Through this intermediate step, mPSCs can generate cells of the hematopoietic lineage (1, 2), cardiomyocytes (3, 4), neurons (5) and glial precursors (6), skeletal muscle (7), vascular endothelial cells (8), visceral endoderm (9, 10), and glucose-responsive insulin-producing cells (11).
When human EG cells differentiate, they also form EBs composed of endodermal, ectodermal, and mesodermal derivatives (12). Although a compelling demonstration of the potential of human EG cells, the limited growth characteristics of differentiated cells within EBs and difficulties associated with their isolation would make extensive experimental manipulation difficult and limit their use in future cellular transplantation therapies. We reasoned that precursor or progenitor cells of the observed constituent cell types must have been present at some point in EB formation, and if they could be isolated, these cells may have desirable proliferation and expression characteristics.
Here, we describe cells isolated from human EBs that are capable of long-term and robust proliferation in culture. Mixed cell EB-derived (EBD) cultures and clonally isolated EBD cell lines simultaneously express a wide array of mRNA and protein markers that are normally associated with distinct developmental lineages. The proliferation and expression characteristics of these cells suggest they may be useful in the study of human cell differentiation and as a resource for cellular transplantation therapies.
Materials and Methods
Pluripotent Stem Cell Culture, EB Formation, and Establishment of EBD Cell Cultures.
Human EG cultures LV (XX), SL (XY), LU2 (XY), and SD (XX) were derived and cultured from 5-, 6-, 7-, and 11-week postfertilization primordial germ cells, respectively, as described (12). EBs were formed in the presence of leukemia inhibitory factor (1000 U/ml), basic fibroblast growth factor (2 ng/ml), forskolin (10 μM), and 15% FCS (HyClone). Approximately 10 cystic EBs from each culture were dissociated by digestion in 1 mg/ml Collagenase/Dispase (Roche Molecular Biochemicals) for 30 min to 1 h at 37°C. Cells were then spun at 1,000 rpm for 5 min and then resuspended in various growth media/matrix environments. RPMI growth media included RPMI 1640 (LTI)/15% FCS/0.1 mM nonessential amino acids/2 mM L-glutamine/100 U/ml penicillin/100 μg/ml streptomycin. EGM2MV media (Clonetics, San Diego) included 5% FCS, hydrocortisone, human basic fibroblast growth factor, human vascular epidermal growth factor, R(3)-insulin-like growth factor I, ascorbic acid, human epidermal growth factor, heparin, gentamycin, and amphotericin. Matrices were bovine collagen I (Collaborative Biomedical Products, Bedford, MA; 10 μg/cm2), human extracellular matrix (Collaborative Biomedical Products, 5 μg/cm2), and tissue culture plastic. Cells were cultured at 37°C, 5% CO2, 95% humidity and routinely passaged 1:10 to 1:40 by using 0.025% Trypsin, 0.01% EDTA (Clonetics) for 5 min at 37°C. Low serum cultures were treated with trypsin inhibitor (Clonetics) and then spun down and resuspended in growth media. Cell were cryopreserved in the presence of 50% FCS, 10% DMSO in a controlled rate freezing vessel, and stored in liquid nitrogen. Cells prepared for cytogenetic analysis were incubated in growth media with 0.1 μg/ml Colcemid/2.5 μg/ml ethidium bromide for 3 h, trypsinized, resuspended in 0.075 M KCl, incubated for 35 min at 37°C, and then fixed in 3:1 methanol/acetic acid. Cell proliferation assays were carried out by plating 1 × 104 cells of EBD culture SD into 35-mm dishes containing the growth environment in which they were derived. Cells from three wells were grown until subconfluent and then trypsinized, diluted 1:10, replated, grown, and counted. Student's t tests (n = 3) were performed to assign significance. Clonal lines were generated from EBD culture LV by low-density plating in EGM2MV media on collagen I (LVEC) followed by cloning cylinder isolation and expansion to >1 × 106 cells. Cloning efficiency was determined by low density plating of a total of 600 LVEC cells. Methylene Blue staining to identify colonies was performed 10 days after plating. PD levels were calculated as 3.32 (log cellsharvested − log cellsplated) and do not include cell division during the initial phase of culture derivation.
Gene Transfer into EBD Cell Cultures.
Stable transfection of human EBD cultures was carried out by lipofection. Briefly, 1–5 μg of a construct containing the neomycin phosphotransferase gene flanked by the mouse phosphoglycerate kinase-1 was used to transfect ≈2 × 105 cells by using Lipofectamine plus lipid (LTI). Stably transfected cells were selected by growth on collagen I in EGM2MV media supplemented with 200–400 μg/ml G418 and isolated by using cloning cylinders.
Retroviral transduction of culture LVEC was carried out by using the MGIN vector and amphotropic viral envelope as reported (13). This vector uses the retroviral long terminal repeat to drive transcription of enhanced green fluorescence protein (EGFP) and neomycin phosphotransferase coding regions. Lentiviral transduction was carried out by using the EF.GFP vector in which the human elongation factor 1α promoter drives the transcription of EGFP in an HIV-1 based self-inactivating lentiviral backbone. EF.GFP virus was produced by cotransfection of 293T cells with a packaging plasmid expressing HIV-1 gag/pol, REV and TAT proteins, and a plasmid expressing the VSV-G envelope. Viral titers were determined by the number of GFP expressing 293T cells after infection. For the EBD cell transduction, 1 × 105 LVEC cells were infected with 1 × 106 transducing units of either retrovirus or lentivirus in the presence of 8 μg/ml polybrene overnight for two successive days. Transduced cells were analyzed for GFP expression by using fluorescence-activated cell sorting analysis 6 days after plating and proliferation in the absence of drug selection. Mock-infected cells were used to establish the level of background fluorescence. Retrovirally transduced LVEC cells were selected by growth in the presence of 400 μg/ml G418.
Immunocytochemistry and Telomerase Activity Assay.
Approximately 1 × 105 cells were plated in each well of an 8-well glass bottom chamber slide. Cells were fixed in either 4% paraformaldehyde in PBS or a 1:1 mixture of methanol/acetone for 10 min as recommended by the antibody manufacturer. Cells were permeabilized in 0.1% Triton X-100, 1 × PBS for 10 min if required, then blocked in Powerblock (BioGenex), 5% FBS, or 1–5% goat serum supplemented with 0.5% BSA for 10–60 min as recommended by the antibody manufacturer. Primary antibodies and dilutions were as follows: neurofilament 68 kDa (Roche, 1:4), neuron-specific enolase (PharMingen, 1:100), tau (PharMingen, 5 μg/ml), vimentin (Roche, 1:10), human nestin (gift from Ron McKay, National Institutes of Health, 1:250), galactocerebroside (Sigma, 1:500), 2′,3′-cyclic nucleotide 3′-phosphodiesterase (Sigma, 1:500), O4 (Roche, 10 μg/ml), and SMI32 (Sternberger monoclonal, 1:5000). Antibodies reactive to the astrocyte marker glial fibrillary acidic protein (GFAP) and neuronal marker β tubulin type III were not included, as conditions for their specific reactivity could not be established. Detection was carried out by secondary antibodies conjugated to biotin, streptavidin-conjugated horseradish peroxidase, and 3-amino-9-ethylcarbazole chromagen (BioGenex). Telomerase assays were performed by using a telomeric repeat amplification protocol followed by ELISA detection of amplified products (TeloTAGGG PCR ELISA PLUS, Roche).
mRNA Expression Profiles.
RNA was prepared from cells growing on 60-mm tissue culture plates by using the Qiagen miniprep kit. RNA preparations were digested with RNase-free DNase (Roche) for 30 min at 37°C, and then the digest was inactivated at 75°C for 5 min. Synthesis of cDNA was performed on 5 μg of RNA by using oligo (dT) primers and a standard Moloney murine leukemia virus (LTI) reaction carried out at 42°C. Thirty cycles of PCR were carried out in the presence of 1.5 mM MgCl2 with an annealing temperature of 55°C and incubation times of 30 s. PCR reactions were resolved on a 1.8% agarose gel. The efficacy of all PCRs was established by using appropriate commercially available human tissue RNA (CLONTECH). Some gels were subject to Southern blot analysis by using oligonucleotide probes end-labeled with [32P]ATP, hybridized in 6 × SSC/5 × Denhardt's solution/0.1% SDS/0.05% sodium pyrophosphate/100 μg/ml sheared and denatured salmon sperm DNA at 45°C. cDNA synthesis and genomic DNA contamination were monitored by primers specific to human phosphoglycerate kinase-1, which give products of ≈250 bp and ≈500 bp when amplifying cDNA and genomic DNA, respectively. Ethidium bromide fluorescence of agarose gel resolved PCR amplimers, and immunocytochemical reactivities were subjectively assigned to one of four intensity categories: very strong, strong, detected, and not detected. PCR primer and probe sequences appear in Table 1, which is published as supplemental data on the PNAS web site, www.pnas.org.
Tumor Formation.
Three female 6-week-old SCID-NOD mice were injected with 3 × 106 EBD cells (LVEC) or 2.5 × 105 to 1 × 106 mouse ES cells (ES D3) in the left and right calf muscles, respectively. After 1 month, animals were killed and visually examined for tumors. Injected calf muscles were dissected intact, fixed in 4% phosphate-buffered paraformaldehyde overnight, then processed and imbedded in paraffin. Sections stained with hematoxylin and eosin were examined.
Results
Human pluripotent stem cell cultures derived from primordial germ cells were isolated and cultured as described (12). During routine growth, 1–5% of the multicellular EG colonies formed large fluid-filled cystic EBs that were loosely attached to a remaining EG colony or to the fibroblast feeder layer. Four genetically distinct EG cultures (LV, SL, LU2, and SD) were selected to represent the range of developmental stages at which human EG cultures can be initiated (5–11 weeks postfertilization). Karyotypically, two cultures were 46, XX (LV, SD), and two were 46, XY (SL, LU2). Approximately 10 EBs were removed from each EG culture and were dissociated in a mixture of collagenase I and dispase I (Roche). EB constituent cells were then replated in three (LV) or six (SL, LU2, SD) growth media and biomatrix combinations in an effort to identify environments that promoted vigorous cell proliferation with the possibility of differential enrichment of outgrowth populations.
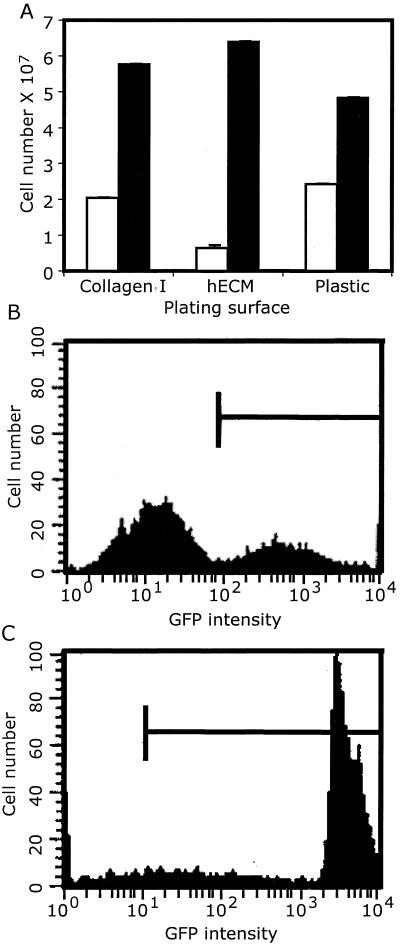
Two growth media were selected to investigate the effects of serum and specific mitogens on the proliferation of these human cells. RPMI 1640 supplemented with 15% FCS is a simple base media that relies on serum to support cell proliferation. EGM2MV(Clonetics) has a reduced serum content (5%) and contains basic fibroblast growth factor, epidermal growth factor, vascular epidermal growth factor, and insulin-like growth factor I. Three cell attachment surfaces were used: tissue culture-treated plastic, bovine collagen I, and human extracellular matrix extract. All six growth environments supported cell proliferation, and the resultant cells were termed EBD cell cultures. Cell proliferation studies carried out on several genetically distinct EBD cultures indicated that EGM2MV medium was superior to RPMI 1640 medium (P < 0.001) and that extracellular matrix and collagen I were superior to tissue culture plastic (P < 0.001). The results from a proliferation study on culture SD is shown in Fig. 1A. Karyotypic analysis performed on each culture at approximately 20 PD indicated that the cells had a normal diploid chromosomal complement.
Figure 1.
EBD cell proliferation rate and flow cytometric (fluorescence-activated cell sorting) analyses. (A) 1 × 104 cells from EBD culture SL grown 6 days in RPMI/15% serum (unshaded bars) or EGM2MV media (shaded bars) on three plating surfaces then trypsinized and counted. Fluorescence-activated cell sorting analyses of retroviral (B) and lentiviral (C) transduction efficiency and EGFP expression level. Bars indicate levels above background.
The ability of EBD cells to be stably transfected in the EGM2MV/collagen I environment was examined by lipofection of a neomycin resistance gene driven by the mouse phosphoglucokinase I promoter. Stable integration efficiencies of ≈1 × 10−5 were routinely obtained, and neomycin-resistant clonal lines could be expanded to >1 × 106 cells. In an effort to improve integration efficiency, retroviral and lentiviral transduction were investigated. When cultures of LVEC were infected with equal titers of either retrovirus or lentivirus carrying EGFP expression vectors, the efficiencies were ≈30% and ≈98%, respectively, and remained constant for >2 weeks (Fig. 1 B and C). The retrovirally transduced LVEC culture was essentially 100% EGFP positive after 2 weeks of drug selection and has remained so for >30 PD.
In an effort to classify EBD cells by their expression characteristics, we initially chose to look for the presence of neural progenitor and neuronal and glial markers. Neural progenitors are capable of generating both neurons and glia and are known to express the intermediate filament proteins nestin (14) and vimentin (15–17). Additionally, we used several neuronal and glial markers in our survey, including neuronal markers neurofilament light isoform, tau, neurofilament heavy isoform (SMI32), and neuron-specific enolase; and glial markers 2′,3′-cyclic nucleotide 3′-phosphodiesterase (CNPase), galactocerebroside, and O4 antigen.
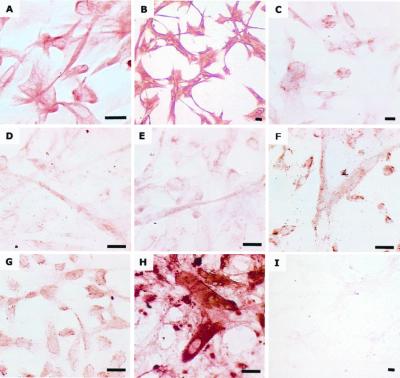
As evident in Fig. 2, cells of the EBD culture LVEC were strongly immunoreactive to the nestin- and vimentin-specific antibodies (>95% cells positive). Cells were less strongly and/or less consistently immunoreactive (10–50% cells positive) to neurofilament light isoform-, tau-, neuron-specific enolase-, SMI32-, CNPase-, and galactocerebroside-specific antibodies. No cells immunoreactive for the O4 antigen were detected.
Figure 2.
Immunocytochemical analysis of EBD culture LVEC. Antibody epitopes are as follows: (A) nestin, (B) vimentin, (C) neurofilament light isoform, (D) tau, (E) neuron-specific enolase, (F) neurofilament heavy isoform, (G) CNPase, (H) galactocerebroside, and (I) O4 antigen, no immunoreactivity. (Bars represent 10 μm)
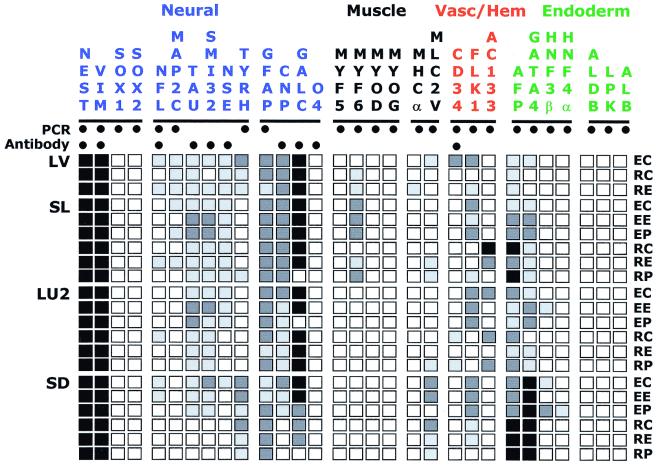
To confirm some of the antibody staining results, and to expand the range of markers examined, a 24-gene reverse transcriptase-PCR expression profile was carried out on the LVEC culture and other EBD cultures. Markers were chosen to indicate expression profiles of four cell lineages known to be present in human EBs: neural, muscle, vascular/hematopoietic, and endoderm (12). In this study, vascular and hematopoietic cells were grouped because they share the markers used. Additionally, the nine antibodies used in Fig. 2 were used, confirming the expression status of four PCR reactions, and extending the analysis to antigens not readily amenable to PCR. Expression of markers not verified by antibody staining was confirmed by Southern blot hybridization of PCR products to specific internal oligonucleotide probes (data not shown). When possible, several markers of a lineage or cell type were used. As seen in Fig. 3, the neural lineage markers were the most strongly and consistently expressed. Neural progenitor markers nestin and vimentin and astrocyte marker glial fibrillary acidic protein (GFAP) were expressed in all cultures. The neuronal markers neurofilament light isoform, microtubule associated protein 2C, tau, nonphosphorylated neurofilament heavy isoform (SMI32), neuron-specific enolase, and tyrosine hydroxylase were weakly expressed in many of the cultures, with occasionally stronger expression of tau and SMI32 when cultures SL and LU2 were grown in EGM2MV media on human extracellular matrix or on tissue culture plastic. CNPase and galactocerebroside are specifically expressed in oligodendrocytes and Schwann cells (18, 19). Both markers were strongly expressed in most of the EBD cultures. No expression of SOX1, SOX2, or O4 was detected.
Figure 3.
Expression profiles of four EBD cultures. Markers and lineages are
listed above and are grouped by lineage affiliation. Assay type is
indicated by ● for PCR, antibody, or both.
Culture identity is indicated at left, growth environment on right, as
follows: E, EGM2MV media; R, RPMI/15% serum media; C, collagen
I-coated surface; E, human extracellular matrix-coated surface; P,
tissue culture-treated plastic surface. Expression levels are indicated
as follows: ■, very strong;
 , strong;
, strong;
 , detected;
□, not detected.
, detected;
□, not detected.
In general, the muscle markers were most weakly and sporadically expressed. Expression of the muscle-specific developmental genes myf5, myogenin, and myoD or myosin heavy chain α was not detected. However, expression of myf6 was detected in cultures SL and LV, and expression of myosin light-chain 2 ventricular isoform was detected in some cultures.
The vascular/hematopoietic stem cell marker CD34 was expressed most strongly by culture LVEC but was detectable in culture LU2. Flk1 (vascular epidermal growth factor receptor-2) was expressed by all four cultures, with strongest expression by cells growing in EGM2MV media. AC133 (CD133) is a cell surface marker of vascular/hematopoietic stem and progenitor cells (20, 21) that is also expressed in some human epithelial cells (22). Expression of AC133 in EBD cell cultures was restricted to SL and LU2, with the strongest expression in SL growing in RPMI media on collagen I (SLRC).
The endoderm marker α-1-fetoprotein was expressed in all instances. GATA4, which is expressed in endoderm and heart (23), was also expressed in most of the cultures. Hepatic nuclear factor 3β is expressed in many endodermal derivatives such as liver (24) and is an essential early factor in pancreatic development (25). Hepatic nuclear factor 3β expression was only detected when EGM2MV media was used, and then only in two cultures. Hepatic nuclear factor 4α expression is regulated by hepatic nuclear factor 3β (26) and was only detected in one instance, coincident with the strongest expression of hepatic nuclear factor 3β. No expression of the liver-specific markers aldolase B, liver pyruvate kinase, or albumin was detected in any of the cultures.
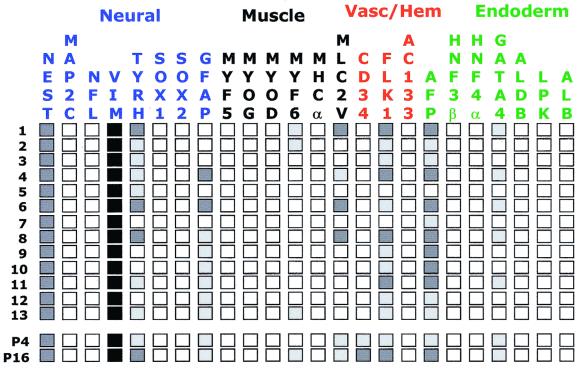
Simultaneous expression of markers from several different lineages, as they have been commonly defined, can be due to multiple cell types within the culture or result from cells capable of multilineage expression. To resolve this issue in these human cell cultures, mRNA expression profiles were carried out on 13 clonal lines isolated from the LVEC culture. As seen in Fig. 4, nestin and vimentin expression was retained in all of the LVEC clonal lines, consistent with the uniform immunocytochemical staining of the LVEC culture for these markers. Expression of the other markers varied substantially. In 11 of 13 lines, mRNA expression of markers from all four lineages was detected. Although the mixed culture expression should be the sum representative of the individual clones, variation during clone isolation and enrichment for certain mRNAs allow for clonal lines that express mRNA not detected in the mixed culture. For example, the muscle marker myf6 is detected in LV-1, -2, and -13 but not in the low passage LVEC mixed culture. In general, the cloning efficiency of LVEC cells was ≈29% (173 colonies/600 cells plated). In the isolation of these LV clones, 69% (27 of 39) of the isolated colonies were capable of expansion to >106 cells.
Figure 4.
mRNA expression profiles of 13 LVEC clonal cell lines and 2 LVEC
cultures. Cells were grown in EGM2MV media on collagen I. Markers and
lineages are listed above and are grouped by lineage affiliation. Lines
(1–13) and LVEC culture at passage 4 (P4) and passage 16 (P16) are
indicated at left. Expression levels are indicated as follows:
■, very strong;
 , strong;
, strong;
 , detected;
□, not detected.
, detected;
□, not detected.
The possibilities of culture expression drift and variability due to assay artifacts was further examined by comparison of the mRNA expression of low-passage LVEC culture and one that had undergone ≈70 PD in 16 passages. Little difference was observed in the marker expression levels except for the apparent accentuation of some markers, including myf6, in the later passage cells (Fig. 4). In this case, when signal levels were normalized against the expression of nestin or a phosphoglycerate kinase-1 control, no obvious differences were apparent (data not shown). The change in expression levels of markers not expressed in the LVEC culture were not determined; however, decreased immunoreactivity to AC133 has been noted following continuous passage of culture SLRC.
To determine the proliferative capacity of EBD cultures, LVEC, SLEC, LU2EC, and SDEC were continuously passaged. After approximately 70–80 PD, these cultures failed to divide. Continuous passage of cultures in environments less favorable to proliferation has not been carried out; however, most EBD cultures are capable of at least 40 PD. To determine whether the proliferation of EBD cultures may be limited by the absence of telomerase, telomeric repeat amplification protocol assays were performed on LVEC and SDEC cultures that had undergone approximately 20 PD after EBD cell establishment. No telomerase activity was detected in either culture (data not shown), consistent with the hypothesis that in cells that have not been transformed, cell division in the absence of telomerase activity leads to cellular senescence.
The ability of EBD cells to proliferate in vivo was examined by injection of 3 × 106 LVEC cells into the calf muscle of 3 immunocompromised mice. No tumors were detected 2 months postinjection, whereas large teratocarcinomas formed when an equal number of, or as little as 2.5 × 105, mouse ES cells were injected similarly.
Discussion
The promise of hPSCs as a resource to study cellular aspects of developmental processes and as a source of cells for transplantation therapies lies in their ability to differentiate into a wide variety of cell types. The cell cultures and clonal cell lines described in this report were derived from EBs formed in human PSC cultures established from primordial germ cells. These cell cultures and lines are capable of robust and extensive proliferation and express a wide range of markers normally associated with a number of different cell lineages. Unlike mouse cells, many karyotypically normal human cell types are capable of long-term proliferation in vitro, possibly reflecting inherent species differences in senescence (27). The in vitro proliferative capacity of EBD cells allows for routine genetic and epigenetic manipulation as well as clonal isolation. Furthermore, extended proliferation in an environment nonpermissive for EG cells also reduces the possibility of stem cell contamination.
The formation of EBs and the proliferation characteristics of EBD cells from the EG cultures contrast with a reported effort to isolate differentiated cells from human ES cultures. In one report, differentiation of ES cultures did not result in recognizable EBs, and the constituent cells of the aggregates that did form, did not proliferate extensively (28). More recently, trypsin-dissociated EB cells formed from human ES cultures were plated onto gelatin-coated plastic dishes, but the proliferative capacity of the resultant cells was not evident, as they were assayed after 10 days, and no evidence of PD was presented (29). The significant difference in proliferative capacity between the EBD cells and reported differentiated cells derived from human ES cultures may be due to some aspect of EG growth or EBD isolation protocols, or to inherent differences between pluripotent stem cell types. Irrespective of cause, the capacity of the EBD cells to divide extensively in vitro has important implications for the study, manipulation, and therapeutic use of these cells.
The growth environments we studied had a significant effect on the proliferation of EBD cells but did not predictably influence their gene expression profiles. This was not unexpected, as EBs are heterogeneous with respect to cell type content and the environments were designed to be generally supportive rather than specifically tailored to a particular cell type. This was substantiated by differences in EBD expression profiles when multiple cultures were initiated in parallel or serially from one EG culture (data not shown). The strongest and most consistent antibody and PCR markers were markers associated with neural lineages. A neural identity of cells derived from hPSCs is not surprising, as neural cells can be routinely obtained from mouse ES, EG, and embryonal carcinoma cultures as well as human embryonal carcinoma cultures (30). However, EBD cell cultures and clonal lines cannot be viewed simply as neural progenitors, as they simultaneously express markers from multiple, distinct cell lineages. Multilineage gene expression has been reported in other precursor or progenitor cell populations but not with such a broad range. Neural progenitors simultaneously express neuronal and glial markers (16, 17), and multipotent hematopoietic progenitors simultaneously express a variety of lineage-affiliated transcription factors and cytokine receptors (31). The breadth of expression exhibited by EBD cultures and clonal lines may be unique to these cells or an outcome of the experimental culture design. The multilineage expression exhibited by EBD cells may represent the basis for the developmental plasticity observed after the differentiation of bone marrow (32) and central nervous system stem cells (33). In this model, multilineage gene expression by precursor or progenitor cells defines a ground state from which cell-extrinsic and cell-intrinsic signals work to continuously define a differentiated expression pattern and phenotype (34). In this regard, it would be useful to similarly assay the gene expression profiles of other stem, precursor, or progenitor cell populations.
The human cells described here have several properties that suggest their usefulness as models of human cell differentiation and in transplantation therapies. These include robust and long-term proliferation with a normal karyotype; ability to be cryopreserved, cloned, and genetically manipulated; and a developmentally broad multilineage expression profile. In vitro and in vivo animal model experiments are currently underway to evaluate the efficacy of EBD cells, with or without further differentiation, in the treatment of Parkinson's disease, amyotrophic lateral sclerosis (ALS), stroke, spinal cord injury, and diabetes. For PSCs to be of practical use, methods to generate large numbers of homogeneous cell types must be developed.
Supplementary Material
Acknowledgments
Geron, Inc. provided funding for the study described in this article. Under a licensing agreement between Geron and Johns Hopkins University, the University and J.D.G. and M.J.S. are entitled to a share of sales royalty from Geron. The University and J.D.G. and M.J.S. own stock in Geron, the sale of which is subject to certain restrictions under University policy. The terms of this arrangement are being managed by the University in accordance with its conflict-of-interest policies.
Abbreviations
- PD
population doublings
- ES
embryonic stem
- EG
embryonic germ
- EBD
embryoid body-derived
- PSC
pluripotent stem cell
- EB
embryoid body
- EGFP
enhanced green fluorescence protein
- CNPase
2′,3′-cyclic nucleotide 3′-phosphodiesterase
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.021537998.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.021537998
References
- 1.Wiles M V, Keller G. Development (Cambridge, UK) 1991;111:259–267. doi: 10.1242/dev.111.2.259. [DOI] [PubMed] [Google Scholar]
- 2.Keller G, Kennedy M, Papayannopoulou T, Wiles M V. Mol Cell Biol. 1993;13:473–486. doi: 10.1128/mcb.13.1.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klug M, Soonpaa M, Field L. Am J Physiol. 1995;269:H1913–H1921. doi: 10.1152/ajpheart.1995.269.6.H1913. [DOI] [PubMed] [Google Scholar]
- 4.Rohwedel J, Sehlmeyer U, Shan J, Meister A, Wobus A. Cell Biol Int. 1996;20:579–587. doi: 10.1006/cbir.1996.0076. [DOI] [PubMed] [Google Scholar]
- 5.Bain G, Kitchens D, Yao M, Huettner J E, Gottlieb D I. Dev Biol. 1995;168:342–357. doi: 10.1006/dbio.1995.1085. [DOI] [PubMed] [Google Scholar]
- 6.Brustle O, Jones K N, Learish R D, Karram K, Choudhary K, Wiestler O D, Duncan I D, McKay R D. Science. 1999;285:754–756. doi: 10.1126/science.285.5428.754. [DOI] [PubMed] [Google Scholar]
- 7.Rohwedel J, Maltsev V, Bober E, Arnold H-H, Hescheler J, Wobus A. Dev Biol. 1994;164:87–101. doi: 10.1006/dbio.1994.1182. [DOI] [PubMed] [Google Scholar]
- 8.Wang R, Clark R, Bautch V. Development (Cambridge, UK) 1992;114:303–316. doi: 10.1242/dev.114.2.303. [DOI] [PubMed] [Google Scholar]
- 9.Abe K, Niwa H, Iwase K, Takiguchi M, Mori M, Abe S-I, Abe K, Yamura K-I. Exp Cell Res. 1996;229:27–34. doi: 10.1006/excr.1996.0340. [DOI] [PubMed] [Google Scholar]
- 10.Doetschman T C, Eistetter H, Katz M, Schmidt W, Kemler R. J Embryol Exp Morphol. 1985;87:27–45. [PubMed] [Google Scholar]
- 11.Soria B, Roche E, Berna G, Leon-Quinto T, Reig J A, Martin F. Diabetes. 2000;49:157–162. doi: 10.2337/diabetes.49.2.157. [DOI] [PubMed] [Google Scholar]
- 12.Shamblott M J, Axelman J, Wang S, Bugg E M, Littlefield J W, Donovan P J, Blumenthal P D, Huggins G R, Gearhart J D. Proc Natl Acad Sci USA. 1998;95:13726–13731. doi: 10.1073/pnas.95.23.13726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng L, Du C, Murray D, Tong X, Zhang Y A, Chen B P, Hawley R G. Gene Ther. 1997;4:1013–1022. doi: 10.1038/sj.gt.3300507. [DOI] [PubMed] [Google Scholar]
- 14.Lendahl U, Zimmerman L B, McKay R D. Cell. 1990;60:585–595. doi: 10.1016/0092-8674(90)90662-x. [DOI] [PubMed] [Google Scholar]
- 15.Pleasure S J, Lee V M. J Neurosci Res. 1993;35:585–602. doi: 10.1002/jnr.490350603. [DOI] [PubMed] [Google Scholar]
- 16.Colucci-D'Amato G L, Tino A, Pernas-Alonso R, ffrench-Mullen J M, di Porzio U. Exp Cell Res. 1999;252:383–391. doi: 10.1006/excr.1999.4636. [DOI] [PubMed] [Google Scholar]
- 17.Piper D R, Mujtaba T, Rao M S, Lucero M T. J Neurophysiol. 2000;84:534–548. doi: 10.1152/jn.2000.84.1.534. [DOI] [PubMed] [Google Scholar]
- 18.Sprinkle T J, Agee J F, Tippins R B, Chamberlain C R, Faguet G B, De Vries G H. Brain Res. 1987;426:349–357. doi: 10.1016/0006-8993(87)90888-2. [DOI] [PubMed] [Google Scholar]
- 19.Raff M C, Mirsky R, Fields K L, Lisak R P, Dorfman S H, Silberberg D H, Gregson N A, Leibowitz S, Kennedy M C. Nature (London) 1978;274:813–816. [PubMed] [Google Scholar]
- 20.Yin A H, Miraglia S, Zanjani E D, Almeida-Porada G, Ogawa M, Leary A G, Olweus J, Kearney J, Buck D W. Blood. 1997;90:5002–5012. [PubMed] [Google Scholar]
- 21.Miraglia S, Godfrey W, Buck D. Blood. 1998;91:4390–4391. [PubMed] [Google Scholar]
- 22.Corbeil D, Roper K, Hellwig A, Tavian M, Miraglia S, Watt S M, Simmons P J, Peault B, Buck D W, Huttner W B. J Biol Chem. 2000;275:5512–5520. doi: 10.1074/jbc.275.8.5512. [DOI] [PubMed] [Google Scholar]
- 23.Arceci R J, King A A, Simon M C, Orkin S H, Wilson D B. Mol Cell Biol. 1993;13:2235–2246. doi: 10.1128/mcb.13.4.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ang S L, Wierda A, Wong D, Stevens K A, Cascio S, Rossant J, Zaret K S. Development (Cambridge, UK) 1993;119:1301–1315. doi: 10.1242/dev.119.4.1301. [DOI] [PubMed] [Google Scholar]
- 25.Wu K L, Gannon M, Peshavaria M, Offield M F, Henderson E, Ray M, Marks A, Gamer L W, Wright C V, Stein R. Mol Cell Biol. 1997;17:6002–6013. doi: 10.1128/mcb.17.10.6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duncan S A, Navas M A, Dufort D, Rossant J, Stoffel M. Science. 1998;281:692–695. doi: 10.1126/science.281.5377.692. [DOI] [PubMed] [Google Scholar]
- 27.Littlefield J W. Exp Gerontol. 1996;31:29–32. doi: 10.1016/0531-5565(95)00018-6. [DOI] [PubMed] [Google Scholar]
- 28.Reubinoff B E, Pera M F, Fong C Y, Trounson A, Bongso A. Nat Biotechnol. 2000;18:399–404. doi: 10.1038/74447. [DOI] [PubMed] [Google Scholar]
- 29.Schuldiner M, Yanuka O, Itskovitz-Eldor J, Melton D, Benvenisty N. Proc Natl Acad Sci USA. 2000;97:11307–11312. doi: 10.1073/pnas.97.21.11307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andrews P W. Dev Biol. 1984;103:285–293. doi: 10.1016/0012-1606(84)90316-6. [DOI] [PubMed] [Google Scholar]
- 31.Hu M, Krause D, Greaves M, Sharkis S, Dexter M, Heyworth C, Enver T. Genes Dev. 1997;11:774–785. doi: 10.1101/gad.11.6.774. [DOI] [PubMed] [Google Scholar]
- 32.Petersen B E, Bowen W C, Patrene K D, Mars W M, Sullivan A K, Murase N, Boggs S S, Greenberger J S, Goff J P. Science. 1999;284:1168–1170. doi: 10.1126/science.284.5417.1168. [DOI] [PubMed] [Google Scholar]
- 33.Bjornson C R, Rietze R L, Reynolds B A, Magli M C, Vescovi A L. Science. 1999;283:534–537. doi: 10.1126/science.283.5401.534. [DOI] [PubMed] [Google Scholar]
- 34.Enver T, Heyworth C M, Dexter T M. Blood. 1998;92:348–352. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.