Abstract
The initiator protein DnaA of Escherichia coli binds to a 9mer consensus sequence, the DnaA box (5′-TTA/TTNCACA). If complexed with ATP it adopts a new binding specificity for a 6mer consensus sequence, the ATP-DnaA box (5′-AGatct). Using DNase footprinting and surface plasmon resonance we show that binding to ATP-DnaA boxes in the AT-rich region of oriC of E.coli requires binding to the 9mer DnaA box R1. Cooperative binding of ATP-DnaA to the AT-rich region results in its unwinding. ATP-DnaA subsequently binds to the single-stranded region, thereby stabilizing it. This demonstrates an additional binding specificity of DnaA protein to single-stranded ATP-DnaA boxes. Binding affinities, as judged by the DnaA concentrations required for site protection in footprinting, were ∼1 nM for DnaA box R1, 400 nM for double-stranded ATP-DnaA boxes and 40 nM for single-stranded ATP-DnaA boxes, respectively. We propose that sequential recognition of high- and low-affinity sites, and binding to single-stranded origin DNA may be general properties of initiator proteins in initiation complexes.
Keywords: DNA unwinding element/oriC/protein–DNA interaction/replication initiation/surface plasmon resonance
Introduction
Bacterial initiation of replication is characterized by sequential steps (Kornberg and Baker, 1992). The initiator protein DnaA binds to 9mer DnaA boxes, five in the case of Escherichia coli and 15 for Bacillus subtilis. In the presence of DNA-structuring proteins HU or IHF, and provided that DnaA protein is complexed with ATP, this leads to a local unwinding of the AT-rich region in the vicinity of the DnaA boxes (Bramhill and Kornberg, 1988; Gille and Messer, 1991; Hwang and Kornberg, 1992; Krause et al., 1997). The unwound region provides the substrate for the loading of the replicative helicase, DnaB in the case of E.coli, and the landing zone for all other proteins that form the replication fork (Fang et al., 1999). Initially 28 bp are unwound, both in E.coli and in B.subtilis oriC, followed by 16–25 additional base pairs upon addition of single-stranded DNA-binding (SSB) protein (Krause and Messer, 1999). So far, it is not clear why ATP-DnaA is required for the unwinding reaction, and the biochemical mechanism for this basic step in the initiation of replication is unknown.
ADP- and ATP-DnaA bind similarly to 9mer DnaA boxes (Schaper and Messer, 1995). We have shown recently that ATP-DnaA, in addition, has a binding specificity for a 6mer consensus sequence, 5′-AGatct, not seen with ADP-DnaA (Speck et al., 1999), which we called an ATP-DnaA box. ATP-DnaA boxes are low-affinity binding sites, and binding to them requires the presence of a 9mer high-affinity binding site close by. In the dnaA promoter region, cooperative binding of ATP-DnaA to 9mer DnaA boxes and 6mer ATP-DnaA boxes is required for efficient repression (Speck et al., 1999). In this paper we analyze the interaction of ATP-DnaA protein with the AT-rich region of oriC. We show that ATP-DnaA binds sequentially to the 9mer DnaA box R1, to the AT-rich region, and to single-stranded ATP-DnaA boxes in this sequence. We incorporate the results in a model for the unwinding reaction.
Results
ATP- and ADP-DnaA bind differently to the AT-rich region in oriC
The AT-rich region in the replication origin oriC of E.coli contains three 13mer repeats and the so-called AT cluster adjacent to DnaA box R1 (Bramhill and Kornberg, 1988; Asai et al., 1990). Within this fragment are four sequences that conform to the consensus sequence for ATP-DnaA boxes. Each of the 13mer repeats starts with a GATC Dam recognition site that is also part of a potential ATP-DnaA box. Binding of ATP- and ADP-DnaA to this fragment was analyzed by DNase I footprinting.
A 428 bp PCR fragment containing DnaA box R1 and the AT-rich region of oriC was generated using primer Pr1, 5′-end-labeled with 32P, and primer Pr2, respectively. The labeled fragment was incubated with increasing concentrations of ATP- or ADP-DnaA, probed with DNase I, and subjected to electrophoresis on a sequencing gel (see Materials and methods). As shown in Figure 1, both ADP- and ATP-DnaA already protected the DnaA box R1 at the lowest DnaA concentration (150 nM). ADP-DnaA did not show any additional protection, even at the highest concentration (600 nM). ATP-DnaA, on the contrary, bound to the AT-rich region, starting at 300 nM, and showed partial protection of the whole segment from the AT cluster to the rightmost 13mer starting at 400 nM. This suggests a cooperative binding of ATP-DnaA to the AT-rich region. A concentration of 400 nM corresponds approximately to the in vivo ATP-DnaA concentration at the time of initiation (Hansen et al., 1991; Kurokawa et al., 1999).
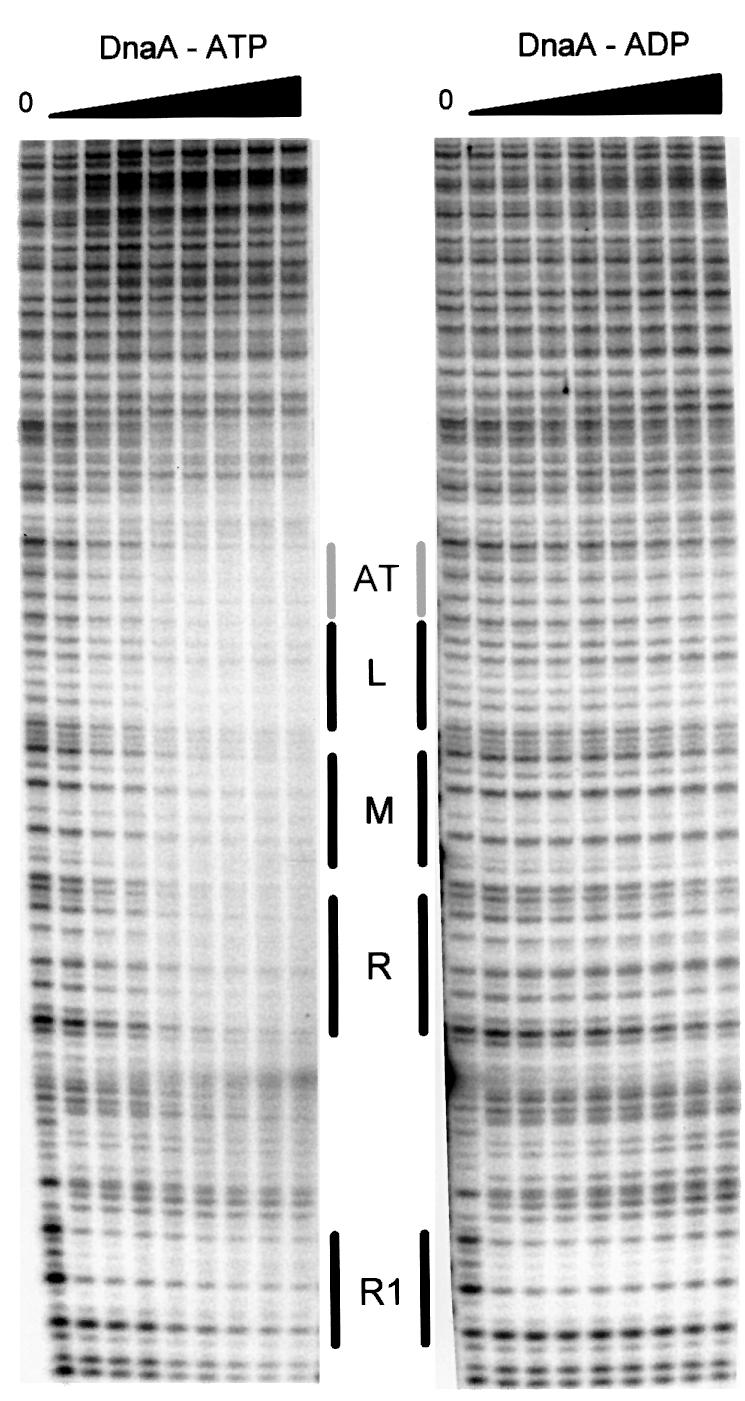
Fig. 1. DNase I footprint of the AT-rich region. DNase I footprinting was performed using a 5′-end-labeled 428 bp PCR DNA fragment. Increasing amounts of ATP- or ADP-DnaA were added, giving final concentrations of 150, 300, 350, 400, 450, 500, 550 and 600 nM. Incubation was for 10 min at 37°C. DnaA box R1, the AT cluster and the three 13mers are indicated by bars; the lower strands are shown.
Binding of ATP-DnaA to the AT-rich region requires the presence of DnaA box R1
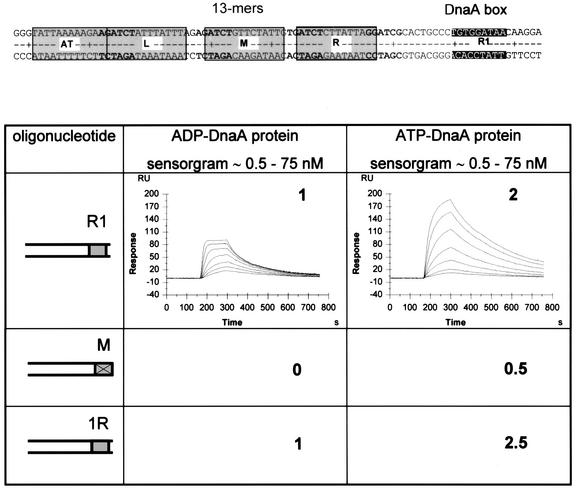
For a more detailed analysis of complex formation at the AT-rich region of oriC, we determined the kinetics and the stoichiometry of the complexes using surface plasmon resonance (SPR) with the BIAcore instrument. SPR measures the change in mass at the surface of a chip. Details of the technique as used for DnaA–DNA interaction have been described recently (Speck et al., 1999). For SPR analysis we used a DNA fragment from oriC containing the AT-rich region, the AT cluster, all three 13mers and DnaA box R1 (oriC coordinates 8–94; all oriC coordinates refer to Buhk and Messer, 1983; Figure 2). Oligonucleotides were chemically synthesized with a biotin label at one 5′-end, and were bound to a streptavidin-coated chip. A flow of ATP- or ADP-DnaA was applied past the chip. The major advantage of this technique is that protein–DNA interactions can be monitored in real time, very accurately and very sensitively. A response of 1 RU (resonance unit) corresponds to 1 pg/mm2 protein or 0.73 pg/mm2 DNA, respectively (Speck et al., 1999). Since the BIAcore measures mass differences, one can calculate the mean stoichiometry of the complexes at any given DnaA protein concentration. Limitations of the BIAcore technique are as follows. (i) Kinetic constants can only be determined for 1:1 interactions, using the Langmuir binding model, since correct assignment of a binding model is too error prone for more complex reactions. It is, however, possible to recognize a complex, potentially cooperative interaction from the shape of the sensorgrams. (ii) Escherichia coli DnaA tends to bind non-specifically to the carboxymethyl dextran matrix of the sensor chips, since it is a very basic protein. Therefore, we can not probe concentrations >100 nM. In contrast, we used 600 nM as the highest concentration for the DNase I footprints.
Fig. 2. Binding reactions of ATP- and ADP-DnaA protein to the AT-rich region of oriC as measured by SPR. The fragment used in these measurements is shown at the top. DnaA box R1 is indicated in light gray, ATP-DnaA boxes are in bold. The three 13mer repeats are boxed. The stoichiometry of the complexes obtained at a protein concentration of 75 nM is indicated in the figure as x:1 (protein:DNA). ATP- and ADP-DnaA protein binding is shown for the wild type (R1), a DnaA box R1 mutant (M) and a fragment that contains an inverted DnaA box R1 (indicated as 1R). One resonance unit corresponds to a change in mass of 1 pg/mm2. Within each sensorgram, individual curves were obtained with protein concentrations (bottom to top) of 1.2, 2.3, 4.7, 9.4, 18.8, 37.5 and 75 nM.
Oligonucleotides with variants of DnaA box R1 were analyzed for their binding kinetics and stoichiometry with ATP- and ADP-DnaA protein. Representative binding experiments are shown in Figure 2. ADP-DnaA bound to a fragment with a wild-type DnaA box R1 with 1:1 kinetics and a stoichiometry of 1. The equilibrium dissociation constant was 3 nM, which is similar to results found by band-shift experiments (Schaper and Messer, 1995). ATP-DnaA showed complex binding kinetics, compatible with cooperative binding, and a stoichiometry ≥2, although saturation of the binding reaction was not reached. When saturation of a reaction is approached, the injection of higher protein concentrations does not result in the binding of proportionally more protein to the DNA. This is visible as compression of the curves in the sensorgrams, which is seen with ADP-DnaA, but clearly not in the sensorgrams of ATP-DnaA shown in Figure 2. If DnaA box R1 was scrambled, ADP-DnaA could not bind and ATP-DnaA bound very inefficiently. Surprisingly, inversion of DnaA box R1 gave similar results to the wild-type configuration. The results show that ATP-DnaA, but not ADP-DnaA, can bind to the AT-rich region in a presumably cooperative manner. A wild-type DnaA box R1 is required for the reaction as an anchor point, in agreement with genetic experiments (Langer et al., 1996).
The 13mer region contains several GATC recognition sites for Dam methyltransferase, and methylation of these sites is required for efficient replication initiation (Messer et al., 1985). In order to analyze whether methylated ATP-DnaA boxes are a better substrate for ATP-DnaA, we methylated the fragment with Dam methyltransferase in vitro, and subjected it to the same SPR analysis as shown in Figure 2. ATP-DnaA bound to the methylated fragment with an approximately three times higher stoichiometry than to the unmethylated one (7:1 versus 2:1 at 75 nM DnaA).
ATP-DnaA also binds to the AT-rich region when it is single stranded
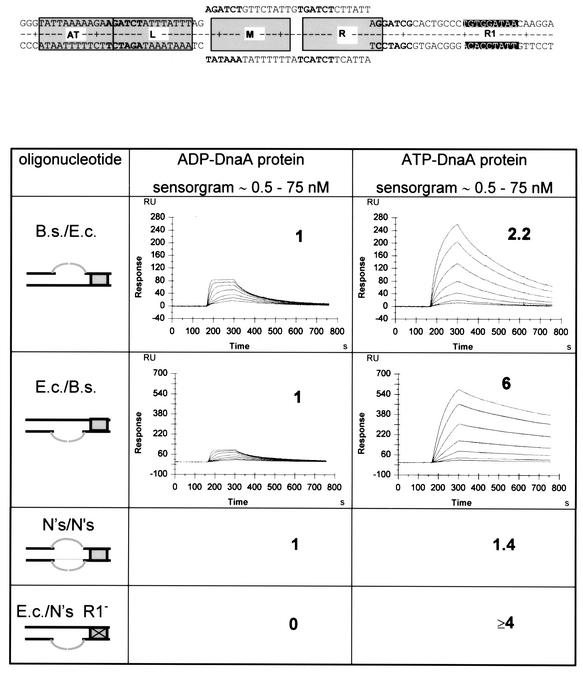
DnaA-mediated unwinding is only observed with supercoiled template. In order to be able to measure DnaA binding to an unwound AT-rich region, we constructed artificial substrates. In one strand, these contained the E.coli sequence of the whole AT-rich region and DnaA box R1. In the other strand, the M and R 13mers were replaced by the corresponding region from B.subtilis oriC (Krause and Messer, 1999), thereby avoiding hybridization in this segment (‘bubble’ substrate; Figure 3).
Fig. 3. Binding reactions of ATP- and ADP-DnaA protein to an artificial bubble as measured by SPR. At the top, a version of the artificial bubble DNA is indicated. The DnaA box R1 is indicated in gray, the three 13mers are boxed and the ATP-DnaA boxes are shown in bold. The upper strand corresponds to the E.coli sequence. In the lower strand, the M and R 13mers were replaced by the corresponding region from B.subtilis oriC (Krause and Messer, 1999), thereby avoiding hybridization in this segment. The stoichiometry of the complexes obtained at a protein concentration of 75 nM is indicated in the figure as x:1 (protein:DNA). Fragments that contain scrambled sequences in the lower or upper strand of the bubble or in the flanking sequence are indicated by N′s. One resonance unit corresponds to a change in mass of 1 pg/mm2. Within each sensorgram, individual curves were obtained with protein concentrations (bottom to top) of 1.2, 2.3, 4.7, 9.4, 18.8, 37.5 and 75 nM.
ADP-DnaA bound to all fragments that contained a wild-type DnaA box R1 with a stoichiometry of 1, demonstrating that it binds to the double-stranded DnaA box but to no other part of the substrate. For ATP-DnaA the binding kinetics and the stoichiometry were very similar to a double-stranded fragment if the E.coli sequence was in the ‘lower’ strand, indicating that ATP-DnaA can bind to single-stranded ATP-DnaA boxes. If the E.coli sequence was in the ‘upper’ strand, cooperative binding seemed to be extreme. The stoichiometry was ≥6 at a DnaA concentration far from saturation. The dissociation rate was very slow, as seen from the slow decrease of the curves after the protein flow was replaced by buffer. A similar fragment but with a scrambled sequence flanking the bubble, including a scrambled DnaA box R1, had similar binding characteristics. The ‘lower’ strand also contained a scrambled sequence in this fragment at the M and R 13mers (Figure 3, E.c./N′s R1–). This means that once the region is unwound, i.e. for DnaA binding to single-stranded 13mers, the anchor point at R1 is no longer required. If both the flanking region and the bubble contained a scrambled sequence, and no E.coli-specific 13mers but an intact DnaA box R1 (Figure 3, N′s/N′s), both forms of DnaA bound with a stoichiometry of only 1, i.e. to the DnaA box R1. This means that the specific E.coli or B.subtilis sequence in the single-stranded region is clearly required for ATP-DnaA binding. The results, summarized in Figure 3, show that ATP-DnaA binds cooperatively and with sequence specificity to single-stranded DNA of the AT-rich region of oriC.
DnaA domain 1 is not required for binding to ATP-DnaA boxes
It has been suggested that a truncated form of DnaA protein without domain 1 is able to perform the unwinding reaction (Sutton et al., 1998). Domain 1 has been shown to mediate oligomerization of DnaA (Weigel et al., 1999), and it is involved in loading of DnaB helicase (Sutton et al., 1998; Seitz et al., 2000). DnaA without the oligomerization domain 1 can, however, bind cooperatively to adjacent DnaA boxes (Messer et al., 1999; Jakimowicz et al., 2000; Majka et al., 2001). For an analysis of the binding characteristics of DnaA protein lacking domain 1, DnaA[87–467] with an N-terminal His6 tag, we determined the stoichiometry of binding using the double-stranded oriC substrate used in the experiment shown in Figure 2. Table I shows that this DnaA deletion derivative bound identically to wild type, with a higher stoichiometry for the ATP form. A derivative containing the DNA binding domain 4 only bound with a stoichiometry of 1:1. This is not surprising since this protein lacks the ATP-binding site.
Table I. Stoichiometry of DnaA derivatives with the AT-rich region.
| Protein | ADP form | ATP form |
|---|---|---|
| Wild-type DnaA | 1:1 | 2.2:1 |
| DnaA[87–467]-His6a | 1:1 | 2.2:1 |
| DnaA[335–467]a | 1:1 |
aNumbers in square brackets indicate expressed amino acid residues.
DnaA protein protects the single-stranded AT-rich region against cleavage by nuclease P1
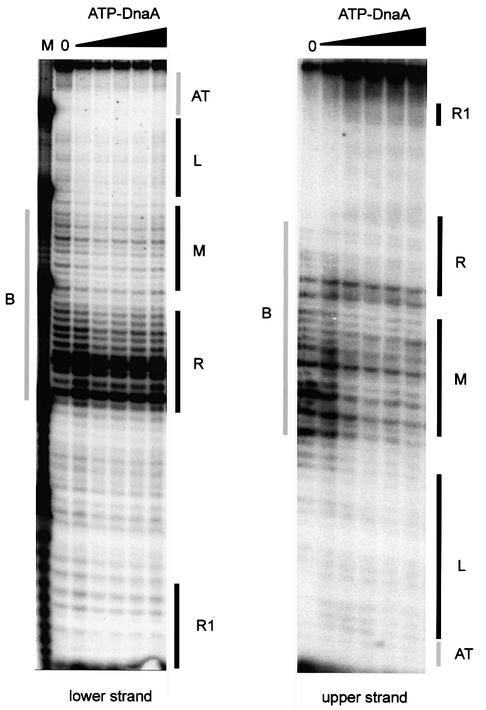
The non-complementary ‘bubble’ region of the E.coli– B.subtilis hybrid substrate is susceptible to cleavage by the single-strand specific nuclease P1 (Figure 4, tracks without ATP-DnaA). In order to determine DnaA protein binding to single-stranded DNA by an independent method, we developed a nuclease P1 footprinting technique. ATP-DnaA already protected the ‘bubble’ at a concentration of 40 nM, both in the ‘upper’ and in the ‘lower’ strand (Figure 4). Protection of the ‘upper’ strand extended over the middle and right 13mer, covering nearly the whole single-stranded region. On the ‘lower’ strand, only a part of the right 13mer and the middle 13mer were protected, and in only ∼50% of the DNA strands. In both strands a few single nucleotides were especially sensitive to the nuclease with and without DnaA. Binding of ATP-DnaA also protected the single-stranded ‘upper’ strand of the AT-rich region against the action of DNase I (H.Seitz and W.Messer, unpublished).
Fig. 4. Nuclease P1 footprint of the artificial bubble substrate and ATP-DnaA. Nuclease PI footprinting was performed using the DNA fragment shown in Figure 3 (top). The DNA was 5′-end-labeled. Increasing amounts of ATP-DnaA were added, giving final concentrations of 10, 40, 75, 100 and 150 nM. Incubation was for 10 min at 37°C. DnaA box R1, the AT cluster, the three 13mers (L, M and R) and the single-stranded region (B) are indicated by bars.
Discussion
The AT-rich region of E.coli oriC contains three 13mer repeats, each of them starting with a GATC site that is part of a potential ATP-DnaA box. In total, there are six potential ATP-DnaA boxes in this region, four within the AT-rich part and two just left of it (Figure 5). Also, B.subtilis oriC has several ATP-DnaA boxes in its AT-rich region with about the same spacing but a slightly different sequence. We have probed this chromosome segment by DNase I footprinting, SPR and nuclease P1 footprinting. DNase I footprinting shows that ATP-DnaA, but not ADP-DnaA, binds to the AT-rich region of E.coli oriC with high cooperativity. Protection against digestion was obtained with 300–400 nM ATP-DnaA. This is about the in vivo concentration of ATP-DnaA at the time of initiation (Hansen et al., 1991; Kurokawa et al., 1999). This is compatible with the notion that the concentration of ATP-DnaA regulates initiation.
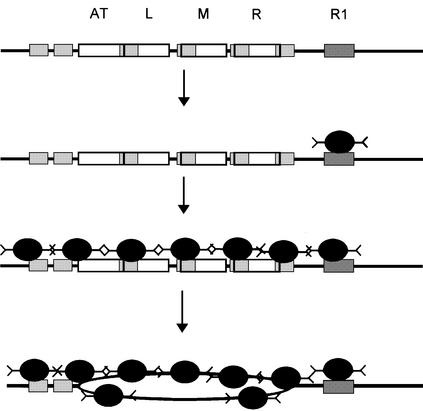
Fig. 5. A model describing the DnaA-dependent unwinding reaction at oriC. The DnaA box R1 and the three 13mers are labeled. The six ATP-DnaA boxes are indicated by small gray boxes. Initially, DnaA protein binds with a high affinity to DnaA box R1. This complex serves as an anchor for cooperative binding of ATP-DnaA to the ATP-DnaA boxes, positioning the protein to the region of unwinding. The formation of this complex needs high concentrations of ATP-DnaA protein and can be considered, therefore, as the rate-limiting reaction in unwinding. Single-stranded DNA resulting from unwinding will then be stabilized by cooperative binding of ATP-DnaA. The affinity of ATP-DnaA is higher to single-stranded ATP-DnaA boxes than to double-stranded ATP-DnaA boxes.
SPR analysis allowed a definition of the quantitative aspects of complex formation between the oriC AT-rich region and DnaA. ADP-DnaA bound to the single 9mer DnaA box on the fragment used for the analysis with simple 1:1 kinetics and a 1:1 stoichiometry, and to no other part of the region. ATP-DnaA bound with a stoichiometry >2 and exhibited complex binding kinetics suggesting cooperative binding. Although for ATP-DnaA we could not reach the saturation level of the binding reaction, it is very clear that the stoichiometry was >2:1. A wild-type 9mer DnaA box R1 was required, supporting the model that DnaA bound to R1 is used as an anchor that directs cooperative binding of several additional ATP-DnaA molecules to the ATP-DnaA boxes in the vicinity. Until now there has been only an isolated and indirect observation suggesting binding of DnaA to this region. DnaA binding to oriC, as measured by filter binding, could be competed by a 13mer-containing double-stranded oligonucleotide (Yung and Kornberg, 1989).
In E.coli and other Enterobacteriaceae, Dam methylation of GATC sites regulates replication initiation in two ways. Shortly after replication, origins are exempt from re-initiation because hemimethylated oriC DNA is sequestered (Ogden et al., 1988; Campbell and Kleckner, 1990). In addition, fully methylated GATC sites are required for optimal oriC function (Messer et al., 1985). Our observation that the Dam-methylated AT-rich region binds ATP-DnaA with higher stoichiometry at non-saturating concentrations, i.e. with higher affinity, may be an explanation for the latter effect.
ATP-DnaA bound with especially high cooperativity to single-stranded DNA from the AT-rich region. The 9mer DnaA box R1 was no longer required. However, the specific sequence of the AT-rich region was essential. Substrates with the E.coli sequence in the top strand were more effective than the complementary substrates. ATP-DnaA could protect both strands of an artificial single-strand ‘bubble’ substrate against cleavage by single-strand-specific nuclease P1. Protection was detected at a concentration of 40 nM. In comparison, ATP-DnaA boxes in the double-stranded form were protected at much higher concentrations, 400 nM, indicating a higher affinity of ATP-DnaA for single-stranded DNA.
All these experiments, footprints and SPR analysis show unambiguously that ATP-DnaA binds to the AT-rich region in single- and double-stranded form.
From these results we propose a new model for the mechanism of origin unwinding (Figure 5). Initial binding is directed to the high-affinity 9mer DnaA boxes. One of these, DnaA box R1, is most important since it serves as an anchor for cooperative binding of ATP-DnaA to the 6mer ATP-DnaA boxes in the still double-stranded AT-rich region. Interaction with the ATP-DnaA boxes depends on a high ATP-DnaA concentration. DNA unwinding was detected in vitro at a concentration similar to that at which we found moderate protection of the AT-rich region. This suggests that binding to ATP-DnaA boxes reflects a molecular barrier for the following steps, and with that presumably regulates the time point of initiation. The AT-rich region overlaps with a sequence that is thermodynamically unstable, as seen in vitro by an opening of the DNA helix in the absence of Mg2+ ions. This sequence has been called a DNA unwinding element (DUE) (Kowalski and Eddy, 1989). The ATP-DnaA–13mer complex destabilizes the DNA helix such that it promotes unwinding under physiological salt conditions. This could be by a breathing of the region or by a sudden unwinding of the whole segment (Kowalski et al., 1988). The unwound region is then stabilized by binding of additional ATP-DnaA molecules to single-stranded ATP-DnaA boxes. The affinity of ATP-DnaA for single-stranded ATP-DnaA boxes is ∼10 times higher than for double-stranded ATP-DnaA boxes, as judged from the footprints. In addition, the ATP-DnaA–single-stranded ATP-DnaA box complex is characterized by a very slow dissociation rate (Figure 3). Therefore, the single-stranded nature of the complex is guaranteed. The model is illustrated in Figure 5.
The critical elements of a replication origin are, thus, the presence of the initial initiator binding site(s), secondary binding sites (e.g. ATP-DnaA boxes) that augment the thermodynamic instability of a DUE, and a tertiary binding site(s) within the single-stranded DNA to finally stabilize the unwound DNA. Factors that influence the stability of base pairing in the AT-rich region are the temperature and the degree of supercoiling of the origin domain, which is in part influenced by divergent transcription, i.e. transcriptional activation. The spatial arrangement of the different binding sites is also important, as is the ability of the initiator protein to bind sequentially to high- and low-affinity sites, as shown here. The importance of all these factors has been demonstrated for E.coli oriC and for other origins. The DUE is an essential cis-acting element of oriC (Kowalski and Eddy, 1989), the degree of supercoiling determines the initiation competence of oriC (for review see Messer and Weigel, 1996), and transcriptional activation is required in vivo (Messer, 1972) and under near-physiological conditions in vitro (Skarstad et al., 1990). DnaA box R1 is essential (Langer et al., 1996), as is its precise distance to the AT-rich region (Hsu et al., 1994). Finally, as shown here, the ability of DnaA protein to acquire additional binding specificities upon interaction with ATP, binding to double-stranded 6mer ATP-DnaA boxes and to single-stranded ones, ensures that the DnaA–oriC complex is assembled first at the five high-affinity DnaA boxes and then becomes activated at the correct time in the cell cycle. Once the correct number of ATP-DnaA monomers has assembled, low affinity of double-stranded and moderate affinity of single-stranded recognition sites follows. We suggest that such a sequential recognition and cooperative interaction may be a general method to accomplish specific positioning of complexes on sequences with low binding affinity, like the 13mer repeats.
The crucial role of the ATP form of DnaA allows a cyclic regulation. Upon synthesis, DnaA protein is likely to be in its ATP form since the cellular ATP concentration is high. In this form, it is initiation competent and active as an autorepressor (Speck et al., 1999). At the end of the initiation cycle, the ATPase activity of DnaA is induced by the loading of the β-subunit of DNA polymerase III, thereby inactivating DnaA protein for further initiation (Katayama et al., 1998).
We propose that the mechanism of origin unwinding outlined here is universal throughout all organisms. All eubacteria have DnaA proteins, and many bacteriophages (Dodson et al., 1986) and plasmids (Helinski et al., 1996) have comparable origins and initiator proteins. Eukaryotic viruses like SV40, polyoma or papilloma have origins with initiator binding sites, a DUE element and AT-rich regions (DePamphilis, 1996). Their initiator proteins are functional equivalents of DnaA protein in the sense that they recognize the origin, unwind DNA and initially stabilize the single-stranded DNA before further stabilization by replication protein A (RPA) (Wessel et al., 1992). Yeast is the paradigm for origins active in the initiation of eukaryotic genome replication. Here also, AT-rich regions and a DUE are associated with an initiator binding site that accepts a six-protein origin recognition complex (ORC). In addition, ORC binds like DnaA to single-stranded DNA, and a change in the double- and single-strand-specific conformation of ORC is associated with an ATP/ADP switch (Lee and Bell, 2000; Lee et al., 2000). ORCs and corresponding origins, which may (Abdurashidova et al., 2000) or may not be AT rich (Delgado et al., 1998), have been found in all metazoa that have been studied, e.g. Drosophila, Xenopus and mammals (for review see DePamphilis, 1999; Ritzi and Knippers, 2000). It seems to be common to all initiator proteins that they recognize a double-stranded binding site, build a larger complex that promotes helical instability, and finally stabilize the unwound region by binding to the single-stranded DNA (e.g. DnaA, SV40 and probably ORC). It is not yet clear whether all initiator proteins have a weaker affinity for the double-stranded AT-rich region, as shown here for the DnaA protein, but it seems to be favorable to bring the initiator into close proximity to the unwound region in order to quickly stabilize the unwound DNA for subsequent helicase loading.
Materials and methods
Chemicals, proteins and oligonucleotides
Standard chemicals were obtained from Sigma (St Louis, MO) or Merck (Darmstadt, Germany), and radioactive [γ-32P]ATP (3000 Ci/mmol) from Amersham (Little Chalfont, UK). Restriction enzymes were supplied by Roche Molecular Biochemicals (Mannheim, Germany) or New England Biolabs (Beverly, MA). Oligonucleotides used for PCR and BIAcore were chemically synthesized (Metabion, Martinsried, Germany). Nuclease P1 was from Roche Molecular Biochemicals (Mannheim, Germany). DnaA[87–467]-His6 protein was constructed by inserting into the SalI and PstI sites of pdnaA116DnaA[87–467] (Weigel et al., 1999) a double-stranded oligonucleotide (upper, 5′-TCGAGCCATCATCATCATCATCATCATGCA; lower, 5′-GATGATGATGATGATGATGGC) encoding a His6 tag. Wild-type DnaA and DnaA[87–467]-His6 protein were overexpressed from plasmid pdnaA116 and purified as described (Krause et al., 1997). DnaA[335–467]-His6 was a gift from S.Schaper. Standard methods were used for DNA manipulations (Sambrook et al., 1989).
DNase I protection assays
DNase I protection assays were performed as described (Galas and Schmitz, 1978). The DNA fragment used in the footprints was amplified using the primers Pr1 (5′-GCCTTTGAGTGAGCTGATACCGCTTCCTTGTTATCCACAGGGCAGTGCG) and Pr2 (5′-CGAGCGCAGCGAGTCAGTGAG). Primer Pr1 was 5′-end-labeled with 32P; nucleotides 1–25 represent a sequence not present in oriC to provide a neutral spacer between the 5′-end label and DnaA box R1.
Surface plasmon resonance
SPR was determined using a BIAcore 2000 instrument (BIAcore AB, Uppsala, Sweden) as described, with minor modifications (Speck et al., 1999). Samples in a concentration range of 1.2–75 nM DnaA were injected using the Kinject command defining an association time of 140 s and a dissociation time of 450 s. Reactions were at 22°C. For data analysis we used the BIAevaluation 3.0 program (BIAcore AB, Sweden, Uppsala). The following oligonucleotides were used (for oligonucleotides that base pair completely only the biotinylated strand is shown): wt up, 5′-GGGTATTAAAAAGAAGATCTATTTATTTAGAGATCTGTTCTATTGTGATCTCTTATTAGGATCGCACTGCCCTGTGGATAACAAGGA; R1 Mut up, 5′-GGGTATTAAAAAGAAGATCTATTTATTTAG AGATCTGTTCTATTGTGATCTCTTATTAGGATCGCACTGCCCGCAAGGAAGCAAGGA; R1 inv up, 5′-GGGTATTAAAAAGAA GATCTATTTATTTAGAGATCTGTTCTATTGTGATCTCTTATTA GGATCGCACTGCCCTTATCCACACAAGGA; B.s. in bubble up, 5′-GGGTATTAAAAAGAAGATCTATTTATTTAGATATTTATAAAAAATAGTAGAAGTAATAGGATCGCACTGCCCTGTGGATAACAAGGA; B.s. in bubble lo, 5′-TCCTTGTTATCCACAGGGCAGTGCGATCCTATTACTTCTACTATTTTTTATAAATATCTAAATAAATAGATCTTCTTTTTAATACCC.
In order to scramble sequences flanking the bubble and/or within the bubble but keep the AT ratio, we used the lower strand 3′-sequence as the upper strand 5′-sequence: Mut all bubble, R1 wt up, 5′-CCCATA ATTTTTCTTCTAGATAAATAAATCACGGTTACGCTAGCGTAGGTGACTGACTCCTAGCGTCTGCCCTGTGGATAACAAGGA; Mut all bubble, R1 wt lo, 5′-TCCTTGTTATCCACAGGGCAGACGCTAGGACATTCGACGAGCGTACGAGCGTCAGCGGATTTATTTATCTAGAAGAAAAATTATGGG; Mut all, bubble wt up, 5′-CCCATA ATTTTTCTTCTAGATAAATAAATCAGATCTGTTCTATTGTGATCTCTTATTTCCTAGCGTGACGGGACACCTATTGTTCCT; Mut all, bubble wt lo, 5′-AGGAACAATAGGTGTCCCGTCACGCTAGGAATTACTTCTACTATTTTTTATAAATATGATTTATTTATCTAGAAGAAAAATTATGGG.
Nuclease P1 protection assays
Nuclease P1 footprints were performed similarly to the DNase I protection assays. DNA fragments were generated by annealing two complementary oligonucleotides, including an artificial single-stranded region as shown in Figure 3. Before annealing, oligonucleotides were 5′-end-labeled with 32P and T4 polynucleotide kinase. Binding reactions were carried out with 0.2 ng of labeled double-stranded DNA in 20 µl of the binding buffer (25 mM HEPES pH 7.6, 100 mM potassium acetate, 5 mM magnesium acetate, 4 mM dithiothreitol, 0.2% Triton X-100, 0.5 mg/ml bovine serum albumin, 100 µM ATP or ADP) and the indicated amounts of ATP- or ADP-DnaA. Mixtures were incubated at 37°C for 10 min, nuclease P1 (0.00005 U) was added, and samples were incubated at 37°C for 4 min. After addition of an equal volume of stop buffer (1% SDS, 200 mM NaCl, 20 mM EDTA pH 8.0, 1 mg/ml glycogen), the samples were purified by phenol/chloroform extraction, DNA was precipitated with ethanol and resuspended in 1 µl of TE buffer (10 mM Tris–HCl pH 8.0, 0.1 mM EDTA). Following the addition of 5 µl of sequencing gel loading buffer (98% formamide, 0.025% bromophenol blue, 0.025% xylene cyanol), samples were incubated at 96°C for 5 min and loaded onto 8% sequencing gels. After electrophoresis, gels were dried and analyzed in a PhosphorImager (Molecular Dynamics).
Acknowledgments
Acknowledgements
We thank F.Blaesing, J.Majka, J.Nardmann, H.Seitz and C.Weigel for many stimulating discussions. This work was supported by grant Me 659/6-1 of the Deutsche Forschungsgemeinschaft.
References
- Abdurashidova G., Deganuto,M., Klima,R., Riva,S., Biamonti,G., Giacca,M. and Falaschi,A. (2000) Start sites of bidirectional DNA synthesis at the human lamin B2 origin. Science, 287, 2023–2026. [DOI] [PubMed] [Google Scholar]
- Asai T., Takanami,M. and Imai,M. (1990) The AT richness and gid transcription determine the left border of the replication origin of the E.coli chromosome. EMBO J., 9, 4065–4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramhill D. and Kornberg,A. (1988) Duplex opening by dnaA protein at novel sequences in initiation of replication at the origin of the E.coli chromosome. Cell, 52, 743–755. [DOI] [PubMed] [Google Scholar]
- Buhk H.J. and Messer,W. (1983) Replication origin region of Escherichia coli: nucleotide sequence and functional units. Gene, 24, 265–279. [DOI] [PubMed] [Google Scholar]
- Campbell J.L. and Kleckner,N. (1990) E.coli oriC and the dnaA gene promoter are sequestered from dam methyltransferase following the passage of the chromosomal replication fork. Cell, 62, 967–979. [DOI] [PubMed] [Google Scholar]
- Delgado S., Gomez,M., Bird,A. and Antequera,F. (1998) Initiation of DNA replication at CpG islands in mammalian chromosomes. EMBO J., 17, 2426–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePamphilis M.L. (1996) Origins of DNA replication. In DePamphilis,M.L. (ed.), DNA Replication in Eukaryotic Cells. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 45–86.
- DePamphilis M.L. (1999) Replication origins in metazoan chromosomes: fact or fiction? BioEssays, 21, 5–16. [DOI] [PubMed] [Google Scholar]
- Dodson M., Echols,H., Wickner,S., Alfano,C., Mensa-Wilmot,K., Gomes,B., LeBowitz,J., Roberts,J.D. and McMacken,R. (1986) Specialized nucleoprotein structures at the origin of replication of bacteriophage λ: localized unwinding of duplex DNA by a six-protein reaction. Proc. Natl Acad. Sci. USA, 83, 7638–7642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang L.H., Davey,M.J. and O’Donnell,M. (1999) Replisome assembly at oriC, the replication origin of E.coli, reveals an explanation for initiation sites outside an origin. Mol. Cell, 4, 541–553. [DOI] [PubMed] [Google Scholar]
- Galas D. and Schmitz,A. (1978) Dnase footprinting: a simple method for determining protein–DNA binding specificity. Nucleic Acids Res., 5, 3157–3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gille H. and Messer,W. (1991) Localized unwinding and structural perturbations in the origin of replication, oriC, of Escherichia coli in vitro and in vivo. EMBO J., 10, 1579–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen F.G., Atlung,T., Braun,R.E., Wright,A., Hughes,P. and Kohiyama,M. (1991) Initiator (DnaA) protein concentration as a function of growth rate in Escherichia coli and Salmonella typhimurium. J. Bacteriol., 173, 5194–5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helinski D.R., Toukdarian,A.E. and Novick,R.P. (1996) Replication control and other stable maintenance mechanisms of plasmids. In Neidhardt,F.C. et al. (eds), Escherichia coli and Salmonella: Cellular and Molecular Biology. ASM Press, Washington, DC, pp. 2295–2324.
- Hsu J., Bramhill,D. and Thompson,C.M. (1994) Open complex formation by DnaA initiation protein at the E.coli chromosomal origin requires the 13-mers precisely spaced relative to the 9-mers. Mol. Microbiol., 11, 903–911. [DOI] [PubMed] [Google Scholar]
- Hwang D.S. and Kornberg,A. (1992) Opening of the replication origin of Escherichia coli by DnaA protein with protein HU or IHF. J. Biol. Chem., 267, 23083–23086. [PubMed] [Google Scholar]
- Jakimowicz D., Majka,J., Konopa,G., Wegrzyn,G., Messer,W., Schrempf,H. and Zakrzewska-Czerwinska,J. (2000) Architecture of the Streptomyces lividans DnaA protein–replication origin complexes. J. Mol. Biol., 298, 351–364. [DOI] [PubMed] [Google Scholar]
- Katayama T., Kubota,T., Kurokawa,K., Crooke,E. and Sekimizu,K. (1998) The initiator function of DnaA protein is negatively regulated by the sliding clamp of the E.coli chromosomal replicase. Cell, 94, 61–71. [DOI] [PubMed] [Google Scholar]
- Kornberg A. and Baker,T.A. (1992) DNA Replication. W.H.Freeman and Co., New York, NY.
- Kowalski D. and Eddy,M.J. (1989) The DNA unwinding element: a novel, cis-acting component that facilitates opening of the Escherichia coli replication origin. EMBO J., 8, 4335–4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalski D., Natale,D.A. and Eddy,M.J. (1988) Stable DNA unwinding, not ‘breathing’ accounts for single-strand-specific nuclease hypersensitivity of specific A+T-rich sequences. Proc. Natl Acad. Sci. USA, 85, 9464–9468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause M. and Messer,W. (1999) DnaA proteins of Escherichia coli and Bacillus subtilis: coordinate actions with single-stranded DNA-binding protein and interspecies inhibition during open complex formation at the replication origins. Gene, 228, 123–132. [DOI] [PubMed] [Google Scholar]
- Krause M., Lurz,R., Rückert,B. and Messer,W. (1997) Complexes at the replication origin of Bacillus subtilis with homologous and heterologous DnaA protein. J. Mol. Biol., 274, 365–380. [DOI] [PubMed] [Google Scholar]
- Kurokawa K., Nishida,S., Emoto,A., Sekimizu,K. and Katayama,T. (1999) Replication cycle-coordinated change of the adenine nucleotide-bound forms of DnaA protein in Escherichia coli. EMBO J., 18, 6642–6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer U., Richter,S., Roth,A., Weigel,C. and Messer,W. (1996) A comprehensive set of DnaA box mutations in the replication origin, oriC, of Escherichia coli. Mol. Microbiol., 21, 301–311. [DOI] [PubMed] [Google Scholar]
- Lee D.G. and Bell,S.P. (2000) ATPase switches controlling DNA replication initiation. Curr. Opin. Cell Biol., 12, 280–285. [DOI] [PubMed] [Google Scholar]
- Lee D.G., Makhov,A.M., Klemm,R.D., Griffith,J.D. and Bell,S.P. (2000) Regulation of origin recognition complex conformation and ATPase activity: differential effects of single-stranded and double-stranded DNA binding. EMBO J., 19, 4774–4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majka J., Zakrzewska-Czerwinska,J. and Messer,W. (2001) Sequence recognition, cooperative interaction and dimerization of the initiator protein DnaA of Streptomyces. J. Biol. Chem., in press. [DOI] [PubMed] [Google Scholar]
- Messer W. (1972) Initiation of DNA replication in E.coli B/r. Chronology of events and transcriptional control of initiation. J. Bacteriol., 112, 7–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messer W. and Weigel,C. (1996) Initation of chromosome replication. In Neidhardt,F.C. et al. (eds), Escherichia coli and Salmonella: Cellular and Molecular Biology. ASM Press, Washington, DC, pp. 1579–1601.
- Messer W., Bellekes,U. and Lother,H. (1985) Effect of dam-methylation on the activity of the replication origin, oriC. EMBO J., 4, 1327–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messer W. et al. (1999) Functional domains of DnaA proteins. Biochimie, 81, 819–825. [DOI] [PubMed] [Google Scholar]
- Ogden G.B., Pratt,M.J. and Schaechter,M. (1988) The replicative origin of the E.coli chromosome binds to cell membranes only when hemimethylated. Cell, 54, 127–135. [DOI] [PubMed] [Google Scholar]
- Ritzi M. and Knippers,R. (2000) Initiation of genome replication: assembly and disassembly of replication-competent chromatin. Gene, 245, 13–20. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Schaper S. and Messer,W. (1995) Interaction of the initiator protein DnaA of Escherichia coli with its DNA target. J. Biol. Chem., 270, 17622–17626. [DOI] [PubMed] [Google Scholar]
- Seitz H., Weigel,C. and Messer,W. (2000) The interaction domains of the DnaA and DnaB replication proteins of Escherichia coli. Mol. Microbiol., 37, 1270–1279. [DOI] [PubMed] [Google Scholar]
- Skarstad K., Baker,T.A. and Kornberg,A. (1990) Strand separation required for initiation of replication at the chromosomal origin of E.coli is facilitated by a distant RNA–DNA hybrid. EMBO J., 9, 2341–2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speck C., Weigel,C. and Messer,W. (1999) ATP- and ADP-DnaA protein, a molecular switch in gene regulation. EMBO J., 18, 6169–6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton M.D., Carr,K.M., Vicente,M. and Kaguni,J.M. (1998) Escherichia coli DnaA protein. The N-terminal domain and loading of DnaB helicase at the E.coli chromosomal origin. J. Biol. Chem., 273, 34255–34262. [DOI] [PubMed] [Google Scholar]
- Weigel C., Schmidt,A., Seitz,H., Tuengler,D., Welzeck,M. and Messer,W. (1999) The N-terminus promotes oligomerisation of the Escherichia coli initiator protein DnaA. Mol. Microbiol., 34, 53–66. [DOI] [PubMed] [Google Scholar]
- Wessel R., Schweizer,J. and Stahl,H. (1992) Simian virus T-antigen DNA helicase is a hexamer which forms a binary complex during bidirectional unwinding from the viral origin of DNA replication. J. Virol., 66, 804–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yung B.Y.M. and Kornberg,A. (1989) The dnaA initiator protein binds separate domains in the replication origin of Escherichia coli. J. Biol. Chem., 264, 6146–6150. [PubMed] [Google Scholar]