Abstract
We have determined the 3.0 Å resolution structure of wild-type HIV-1 reverse transcriptase in complex with an RNA:DNA oligonucleotide whose sequence includes a purine-rich segment from the HIV-1 genome called the polypurine tract (PPT). The PPT is resistant to ribonuclease H (RNase H) cleavage and is used as a primer for second DNA strand synthesis. The ‘RNase H primer grip’, consisting of amino acids that interact with the DNA primer strand, may contribute to RNase H catalysis and cleavage specificity. Cleavage specificity is also controlled by the width of the minor groove and the trajectory of the RNA:DNA, both of which are sequence dependent. An unusual ‘unzipping’ of 7 bp occurs in the adenine stretch of the PPT: an unpaired base on the template strand takes the base pairing out of register and then, following two offset base pairs, an unpaired base on the primer strand re-establishes the normal register. The structural aberration extends to the RNase H active site and may play a role in the resistance of PPT to RNase H cleavage.
Keywords: HIV-1/polypurine tract/reverse transcriptase/RNase H/RNA:DNA
Introduction
HIV-1 reverse transcriptase (RT) is a multifunctional enzyme that is responsible for copying the single-stranded viral RNA genome into double-stranded DNA (Telesnitsky and Goff, 1997). RT contains a DNA polymerase that can copy either an RNA or DNA template and a ribonuclease H (RNase H) activity that cleaves the RNA strand in RNA:DNA hybrids. In addition to degrading the RNA genome after it has been copied to DNA, the RNase H cleavages define the ends of the double-stranded genome that are the substrates for integration into the host genome. In vivo studies demonstrate that inactivation of RNase H results in non-infectious virus particles (Tanese and Goff, 1988; Schatz et al., 1989).
HIV-1 RT is a heterodimer consisting of p66 and p51 subunits. Both subunits are derived from a gag-pol polyprotein, which is cleaved by the viral protease. The two subunits have a common N-terminus; p51 lacks the C-terminal RNase H domain present in p66. Crystal structures of HIV-1 RT complexes with DNA:DNA have been determined in the presence (Huang et al., 1998) and absence (Jacobo-Molina et al., 1993; Ding et al., 1998) of a bound dNTP. While both subunits contain fingers, palm, thumb and connection subdomains, the arrangement of the subdomains in the two subunits is very different (Kohlstaedt et al., 1992; Jacobo-Molina et al., 1993). The crystal structure of the binary complex of HIV-1 RT and a DNA:DNA substrate showed that the DNA:DNA substrate is bent by ∼40° (Jacobo-Molina et al., 1993; Ding et al., 1998). Near the polymerase active site the duplex adopts A-form geometry; near the RNase H active site the duplex adopts B-form geometry (Ding et al., 1997). The structure of the isolated RNase H domain of HIV-1 RT was solved (Davies et al., 1991) before the structure of the intact HIV-1 RT was known. The HIV-1 RNase H domain has a structure that is very similar to the RNase HI of Escherichia coli (Katayanagi et al., 1990; Yang et al., 1990) and of Thermus thermophilus (Ishikawa et al., 1993), but none of the RNases H has been cocrystallized with RNA:DNA, their natural substrate, and there are no published structures of HIV-1 RT in complex with an RNA:DNA duplex.
Polypurine tract in retroviral replication
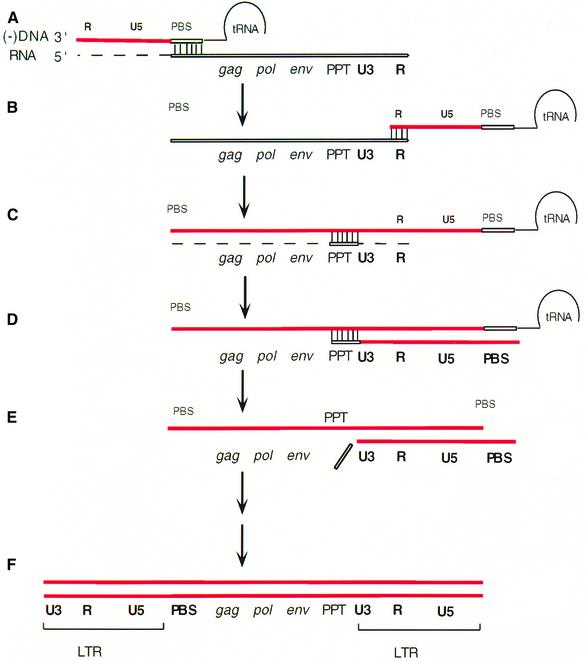
Viral DNA synthesis (see Figure 1) is initiated from a cellular tRNA base paired to the genome of HIV-1 at the primer binding site (PBS). As the minus (–) DNA strand is synthesized, the RNA strand is digested by RNase H. The PBS is near the 5′ end of the genome. Degradation of the RNA by RNase H allows DNA synthesis to be transferred to the 3′ end of the RNA. After strand transfer, (–) strand synthesis can continue, accompanied by RNase H degradation of the RNA genome, but the degradation is not complete. The purine-rich polypurine tract (PPT) is resistant to RNase H cleavage and serves as the primer for plus (+) strand synthesis (reviewed in Telesnitsky and Goff, 1997). The PPT sequence is just 5′ of U3 (Figure 1). Removal of the PPT primer by RNase H defines the left end of the upstream long terminal repeat (LTR) (Figure 1E), which, together with the downstream LTR, is the substrate for the viral integrase enzyme that inserts the linear viral DNA in the host genome. Unlike many retroviruses that have only one PPT sequence, HIV-1 has a second copy of the PPT (central PPT) located near the center of the genome (Charneau et al., 1992). Mutations replacing purines by pyrimidines in the HIV-1 and TY1 central PPTs, which do not modify amino acid sequence, slow down viral growth (Charneau et al., 1992; Hungnes et al., 1992; Heyman et al., 1995), suggesting that the central PPT is important, but not necessary for replication of retroviruses. While the sequence of multiple copies of PPTs is identical, it is likely that the relative importance of a PPT is determined by the neighboring sequences that flank different copies of PPT.
Fig. 1. Process of reverse transcription of the HIV-1 genome. (A) Minus strand DNA synthesis (DNA strand in red) is initiated using a cellular tRNA annealed to the PBS. The RNA strand of the RNA:DNA duplex is degraded by RNase H of HIV-1 RT. (B) First strand transfer allows annealing of the newly formed DNA to the 3′ end of the viral genome. Transfer is mediated by identical repeated (R) sequences. (C) Minus strand DNA synthesis resumes, accompanied by RNase H digestion of all template RNA except PPT. (D) PPT is used as a primer for second strand DNA synthesis. (E) RNase H removes the tRNA and the PPT. In HIV-1, a single RNA nucleotide (from tRNA) is left by RNase H at the RNA/DNA PBS junction. (F) During second strand transfer (not shown) the newly formed PBS DNA (second strand) anneals to the PBS DNA from the first strand. Completion of second strand synthesis results in a linear DNA duplex with LTRs at both ends.
There are at least three requirements for the end of the viral genome to be synthesized correctly. First, the PPT RNA must be resistant to cleavage by RNase H during (–) strand DNA synthesis (Figure 1C). Secondly, RNase H cleavage must occur precisely at the end of the PPT to generate the correct primer for the proper initiation of (+) strand DNA synthesis (Figure 1C). Thirdly, after the PPT primer has been used to initiate DNA synthesis, it must be precisely removed from the end of the viral DNA (Figure 1E).
These three requirements have been extensively studied using biochemical methods in the HIV-1, avian sarcoma leukosis virus (ASLV) and Moloney murine leukemia virus (MuLV) systems (reviewed in Telesnitsky and Goff, 1997). The PPT is relatively resistant to RNase H degradation in vitro, although cleavages within the PPT can occur (Champoux et al., 1984; Resnick et al., 1984; Rattray and Champoux, 1989; Wöhrl and Moelling, 1990; Fuentes et al., 1995; Gao et al., 1998, 1999). There is evidence to suggest that both RT and the PPT are important for determining the specificity of cleavage and for controlling plus strand priming: (i) mutations at the primer grip or thumb subdomain of RT dramatically affect the ability of the enzyme to cleave specifically at the 3′ end of the PPT (Ghosh et al., 1997; Palaniappan et al., 1997; Powell et al., 1997, 1999; Gao et al., 1998); (ii) the isolated RNase H domain of MuLV RT exhibits different cleavage specificity of PPT than the intact MuLV RT (Zhan and Crouch, 1997); (iii) with the exception of a few changes at the G-rich 3′ end of PPT, most single base mutations of the PPT sequence can be tolerated without altering the specificity of cleavage and plus strand priming by HIV-1 RT (Huber et al., 1989; Rattray and Champoux, 1989; Luo et al., 1990; Wöhrl and Moelling, 1990; Pullen et al., 1993; Powell and Levin, 1996); and finally, (iv) the NMR structure of an 8 bp RNA:DNA oligonucleotide containing the last four residues of PPT (with a mutation of one G to A) and the first 4 bp of U3, shows that the width and shape of the major groove is unusual (Fedoroff et al., 1997). Despite the considerable body of experimental data, however, the molecular details of PPT recognition remain unclear both in terms of the generation of the primer and its removal.
Mechanism and specificity of RNase H cleavage
NMR structural studies (Fedoroff et al., 1993; Lane et al., 1993) have suggested that RNA:DNA duplexes are neither A- nor B-form structures in solution. This led to the hypothesis that RNase H distinguishes DNA:RNA and RNA:RNA duplexes by recognizing differences in the width of the minor groove, and suggested that a minor groove width of ∼9–10 Å should be optimal for efficient recognition by RNase H (Fedoroff et al., 1993, 1997; Salazar et al., 1994; Zhu et al., 1995; Horton and Finzel, 1996; Han et al., 1997; Bachelin et al., 1998; Szyperski et al., 1999). In some cases, however, RNase H can cleave single-stranded RNA adjacent to the RNA:DNA duplex region, albeit with low efficiency (Gao et al., 1997; Lima and Crooke, 1997). Furthermore, under certain conditions the RNases H of both MuLV RT and HIV-1 RT can cleave an RNA:RNA substrate (Ben-Artzi et al., 1992; Blain and Goff, 1993; Smith and Roth, 1993; Hostomsky et al., 1994; Götte et al., 1995). Finally, HIV-1 RT can cleave chimeric hybrid duplexes (RNA-DNA annealed to DNA) at the RNA/DNA junction where the minor grooves tend to be very narrow [as small as 4.5–5.5 Å (Szyperski et al., 1999)] and bent (Salazar et al., 1994; Szyperski et al., 1999). These data suggest that the specificity of RNase H cleavage does not depend solely on the width of the minor groove (Szyperski et al., 1999).
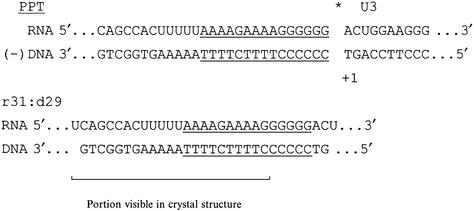
In an effort to study the interactions of HIV-1 RT with RNA:DNA, to discern the molecular details of PPT recognition and to understand better the mechanism and specificity of RNase H cleavage, we determined the 3.0 Å crystal structure of HIV-1 RT in complex with a PPT-containing RNA:DNA substrate and the Fab fragment of a monoclonal antibody. The PPT-containing RNA:DNA oligonucleotide (r31:d29) was bound to HIV-1 RT with the 3′ end of DNA at the polymerase active site and the middle part of the PPT (defined as a stretch of rAs) near the RNase H active site (Figure 2). This binding mode was designed to investigate the inefficient cleavage of the PPT by HIV RNase H. This complex has extensive protein– nucleic acid interactions, and the nucleic acid has unusual structural features that provide a basis for understanding why the PPT is resistant to degradation by HIV-1 RNase H.
Fig. 2. Top: HIV genome sequence at the PPT (underlined) and U3 region. The minus strand synthesis initiation site is marked with an asterisk; +1 is the first nucleotide of U3. Bottom: sequence of the RNA:DNA oligonucleotide in our RT–RNA:DNA complex.
Results and discussion
Overall structure of the HIV-1 RT–RNA:DNA complex
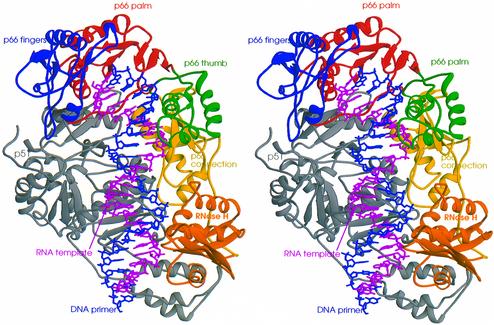
The overall conformation of the protein in the HIV-1 RT–RNA:DNA complex (Figure 3) is similar to that in the HIV-1 RT–DNA:DNA complex. There are extensive interactions between the nucleic acid and amino acids of all subdomains of the p66 subunit; there are also interactions between the nucleic acid and the fingers and connection subdomains of the p51 subunit (Figures 3 and 4). The nucleic acid binding cleft is ∼60 Å in length, extending from the polymerase active site to the RNase H active site. The distance in nucleotides between the polymerase and RNase H catalytic sites in this structure is 18 bp, which agrees with past biochemical studies with HIV-1 RT (Schatz et al., 1990; Wöhrl and Moelling, 1990; Gopalakrishnan et al., 1992; Ghosh et al., 1995; Götte et al., 1995; Gao et al., 1998, 1999). In HIV-1 RT complexes containing double-stranded DNA template-primers, the distance between the polymerase and RNase H active sites is 17 bp (Jacobo-Molina et al., 1993; Huang et al., 1998). This nucleic acid-dependent difference in the distance between the polymerase and RNase H active sites agrees with previous biochemical studies (Götte et al., 1998). The final model of RNA:DNA template-primer includes 45 nucleotides (23 template, 22 primer) encompassing more than two full helical turns of RNA:DNA. The major groove is exposed to the solvent, as might be expected for a non-specific DNA binding protein. The electron density of the nucleic acid is stronger in regions where there are extensive interactions with the enzyme, including the polymerase and RNase H active sites. Simulated annealing omit maps for nucleotides in key locations are shown in Figure 5.
Fig. 3. Stereo view of a ribbon representation of the structure of HIV-1 RT in complex with the polypurine RNA:DNA. The fingers, palm, thumb, connection and RNase H subdomains of p66 are colored blue, red, green, yellow and orange, respectively. The p51 subunit is colored gray. The RNA template and DNA primer strands are shown in magenta and blue, respectively.
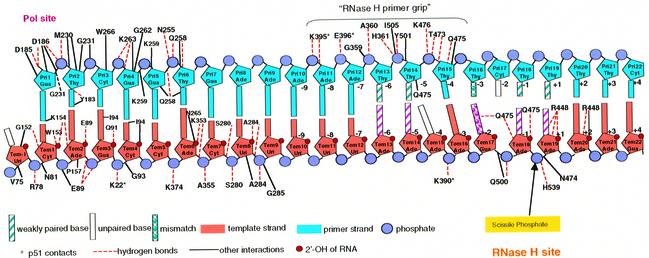
Fig. 4. The sequence and numbering scheme of the RNA:DNA PPT and the interactions between the nucleic acid and amino acid residues of HIV-1 RT (≤3.8 Å). The RNA (orange) and DNA (cyan) strands are designated Tem and Pri, respectively. The nucleotide site positions are labeled with ascending numbers from the polymerase domain toward the RNase H domain. Amino acids of the p51 subunit are designated by an asterisk following the residue number; all others are in p66. RNase H nucleotide site positions are designated positive (+1 to +4) for positions 3′ to, and negative (–1 to –9) for positions 5′ to, the scissile phosphate, where the 3′ and 5′ orientations are for the RNA strand. Hydrogen bonds are shown in red dashed lines and other types of interaction are shown in solid black lines. 2′-OH groups of RNA and phosphate groups are shown in red and gray spheres. Weakly paired (distance ≥3.6 Å), mismatched and unpaired bases are shown filled with stripes, spheres and empty, respectively. Residues Gly359 and Ala360 of the RNase H primer grip interact with the nucleic acid through their main-chain atoms. Arg284 was modeled as Ala because of weak density for the side chain. N474 interacts with Pri15-Thy through a water molecule (not shown).
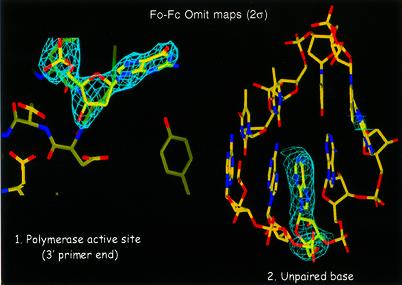
Fig. 5. Simulated annealing (Fo – Fc) omit electron density maps contoured at the 2σ level at the polymerase active site (1) (omitting nucleic acid) and of the unpaired residue of template (2) (omitting unpaired residue Tem-15-Ade).
RT has similar interactions with the DNA primer strand in RT–RNA:DNA and RT–DNA:DNA structures
While the contacts between RT and the DNA primer strands are very similar in the RT complexes with RNA:DNA and DNA:DNA template-primers, the contacts with the RNA template (Figures 3 and 4) are different from those with the DNA template. However, there are relatively modest changes in the protein structure.
As seen in the RT–DNA:DNA structure, many of the RT contacts with the nucleic acid in the RT–RNA:DNA complex involve the sugar–phosphate backbone, consistent with the fact that RT can copy a wide variety of different templates. RT has numerous interactions with 2′-OH groups of the RNA template in the RT–RNA:DNA complex. Such interactions (indicated in magenta in Figure 6) include residues 280 and 284 of helix I of the p66 thumb, residues of the template grip including 89 and 91 of the p66 palm and residues of the RNase H domain (Figures 4 and 6). The more extensive contacts between RT and RNA:DNA versus DNA:DNA may account for the increased polymerization activity and processivity of the enzyme with RNA templates.
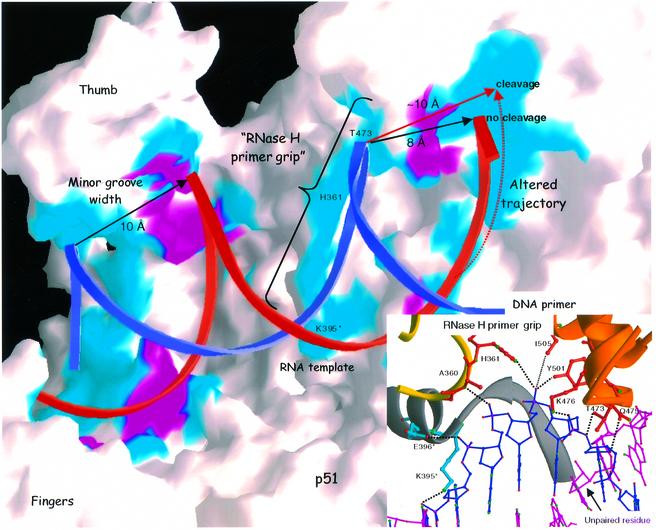
Fig. 6. Molecular surface representation of HIV-1 RT showing the nucleic acid binding cleft and the RNase H primer grip. Residues colored in cyan or magenta are amino acids within 3.8 Å of the 2′-OH of RNA template nucleotides (magenta) or any other part of nucleic acid (cyan). The RNA template is shown as red ribbon and the DNA primer in blue ribbon. Minor groove widths proximal to the thumb area or at the RNase H active site are indicated (∼10 and ∼8 Å, respectively). The trajectory and minor groove width of a hypothetical RNA strand that can be cleaved efficiently by RNase H are shown in red. The RNase H primer grip region is shown in ball and stick representation in the figure inset.
There is also an increased involvement of the p51 subunit in binding the RNA:DNA relative to the DNA:DNA template-primer. Although residues Lys395 and Glu396 of the p51 subunit interact with both the DNA:DNA and RNA:DNA duplexes (with Pri10-Ade and Pri11-Ade, Figure 4), residues Lys22 and Lys390 of p51 interact only with the RNA:DNA duplex (with the phosphates of Tem4-Gua and Tem16-Ade).
Nucleic acid geometry
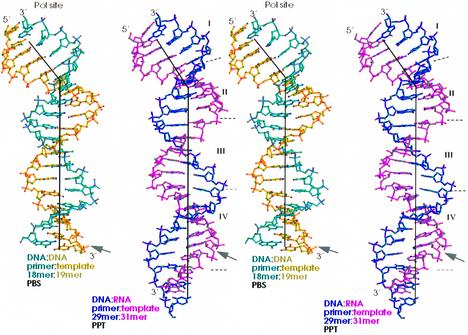
The nucleic acid geometry was analyzed using CURVES (Lavery and Sklenar, 1988) and SCHNAP (Lu et al., 1997). Both programs yielded similar results. Despite the difference in the nature of the duplex (RNA:DNA versus DNA:DNA), the sequence (PPT versus PBS) and the length (31:29 versus 19:18), the overall conformations of these two nucleic acids are remarkably similar. Both have a bend of ∼40° with the helical curvature occurring smoothly over bp 5–9 from the polymerase active site (Jacobo-Molina et al., 1993; Ding et al., 1998) (Figure 7). This bend is a hallmark of nucleic acids bound to a variety of polymerases and is associated with a transition from A- to B-form geometry.
Fig. 7. Stereo view of structures of the nucleic acid template-primers in the RT–DNA:DNA (Ding et al., 1998) and RT–RNA:DNA complexes. The 19mer DNA and 31mer RNA templates are shown in yellow and magenta, respectively. The 18mer DNA and 29mer DNA primers are cyan and blue. Region I contains the 4 bp near the polymerase active site. Region II consists of the next 4 bp at the bend of the nucleic acid. The next 5 bp compose region III, followed by region IV that contains residues of the ‘unzipped’ part of PPT.
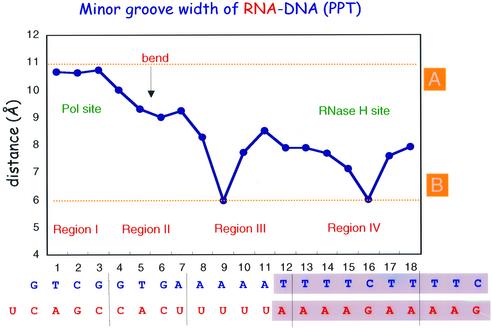
The helical parameters of the RNA:DNA duplex bound to HIV-1 RT were compared with values for canonical A- and B-form DNA:DNA and A-form RNA:RNA duplexes and with other RNA:DNA hybrids (Table I). The values obtained for the RNA:DNA in complex with RT were averaged within four separate regions of the nucleic acid. Region I contains the 4 bp near the polymerase active site. Region II consists of the next 4 bp at the bend of the nucleic acid. Region III includes the next 5 bp, followed by region IV, which contains residues of the ‘unzipped’ portion of the PPT (see below). The parameters that define nucleic acid conformation (Table I) suggest that none of the four regions has canonical A- or B-type geometry. However, the geometry of region I is significantly closer to that of A-form than the other regions. Regions II–IV have a conformation closer to an intermediate between A- and B-form, consistent with the suggestion of Arnott et al. (1986) that RNA:DNA duplexes adopt ‘H-form’ geometry. The H-form conformation is characterized by values for the inclination of the base pairs with respect to the helical axis, the dislocation of the base pairs from the helix axis (Xdisp) and the helical rise, which are intermediate between those of canonical A- and B-form helices (Table I). NMR studies of RNA:DNA duplexes in solution showed that unliganded RNA:DNA duplexes adopt H-form geometry (Fedoroff et al., 1993; Lane et al., 1993). The width of the minor groove of the RNA:DNA (PPT) varies between canonical A- and B-forms (Figure 8), however, and it is closer to B-form in the stretch of rA:dTs (region IV, average minor groove width 7 Å). This is similar to the minor groove width in B-DNA (6 Å) determined by X-ray diffraction of DNA fibers (Chandrasekaran et al., 1989), and markedly lower than the average minor groove width in the RNA:DNA duplexes whose structures have been solved by NMR [∼9 Å, (Arnott et al., 1986; Fedoroff et al., 1993; Lane et al., 1993; Han et al., 1997)] or crystallography (8.7–10.5 Å) (Horton and Finzel, 1996), as shown in Figure 8. A purine-rich sequence with only two consecutive As (RNA:DNA 5′-GAAGAAGAA:CTTCTTCTT) had a considerably wider minor groove (9.4–10.1 Å) (Xiong and Sundaralingam, 1998), suggesting that the narrowing of the minor groove in 5′-oligo(rA): 3′-oligo(dT) tracts of RNA:DNA follows the same rules as in DNA:DNA [5′-oligo(dA):3′-oligo(dT) tracts], i.e. maximal narrowing of the minor groove requires at least four consecutive As. The narrow minor groove we report here for the rA:dT tract of the PPT is remarkably close to the minor groove narrowing predicted by Dickerson and coworkers (Han et al., 1997).
Table I. Nucleic acid parameters.
| Xdisp (Å) | Inclination (°) | Rise (Å) | Twist (ω) | Slide (Å) | |
|---|---|---|---|---|---|
| Region I (bp 1–4) | –2.8 | 0.1 | 3.1 | 31.5 | –0.1 |
| Region II (bp 5–8) | –2.5 | –1.5 | 3.2 | 33.8 | –0.1 |
| Region III (bp 9–13) | –2.3 | –3.7 | 3.3 | 33.2 | –0.2 |
| Region IV (‘unzipped’ 14–19) | –2.0 | –3.9 | 3.2 | 32.2 | 0.1 |
| RNA:DNAa | –4.7 | 12.1 | 2.9 | 30.4 | –1.6 |
| RNA:DNAb | –3.7 | 5.5 | 3.1 | 32.0 | –1.9 |
| RNA:DNAc | –3.3 | 13.9 | 2.9 | 33.0 | –1.2 |
| RNA:DNAd | 19 | 2.5 | 33.1 | ||
| A-RNAe | 13 | 2.8 | 32.7 | –1.1 | |
| A-DNAf | –4.1 | 12 | 2.9 | 31.1 | –1.6 |
| B-DNAf | –0.14 | –6 | 3.4 | 36.0 | 0.4 |
| r(R10)d(Y10)g | –3.3 | 6 | 2.9 | 33.7 | |
| r(R10)r(Y10)h | –5.2 | 8.1 | 2.6 | 31 |
Fig. 8. Variation in minor groove width of the RNA:DNA template-primer. The four regions of nucleic acid are defined in the legend of Figure 8. The minor groove width values for canonical A- and B-type DNA are 11 and 6 Å, respectively.
The geometry of the RNA:DNA hybrid in complex with RT is considerably less uniform than the geometry of unliganded RNA:DNA duplexes, either in solution or in crystals. The variation in the width of the minor groove of the protein-bound RNA:DNA complex (up to 4.5 Å) (Figure 8) is considerably larger than that of free RNA:DNA hybrids (typically 0.5–2 Å), suggesting that the structure of the RNA:DNA duplex is significantly affected by the contacts with the enzyme. The effects of RT are manifested in several ways: (i) the RNA:DNA template-primer has a bend near the RT polymerase active site that is similar to the bend in complexes of RT and a DNA:DNA duplex; (ii) both the RNA:DNA (PPT) and DNA:DNA (PBS) duplexes have similar A-form geometry near the RT polymerase active site, where the majority of the protein–nucleic acid contacts occur; and (iii) there are structural irregularities (discussed below in more detail) in the RNA:DNA (PPT) that may be either caused or stabilized by specific contacts with the enzyme (Figure 4).
Role of A-tracts in the structure of PPT
A-tracts [in this case, stretches of four or more consecu tive 5′-oligo(dA):3′-oligo(dT) (dA:dT) or 5′-oligo(rA): 3′-oligo(dT) (rA:dT)] have long been known to display several unusual features: they are straight (Han et al., 1997) and have a narrow minor groove and a large propeller twist, presumably because there are only two rather than three hydrogen bonds between the bases. A-tracts (dA:dT) are highly resistant to reconstitution around nucleosome cores (Rhodes, 1979) and interact poorly with TATA binding protein, which requires an ∼80° bend at the binding site (Kim et al., 1993). Integration host factor (IHF), a small protein that specifically recognizes an A-tract (dA:dT) by binding the phosphates of the narrow minor groove, appears to recognize an A-rich (dA:dT) DNA duplex by structure rather than by base-specific contacts (Rice et al., 1996).
It is possible that special structural features of A-tracts (rA:dT) are recognized by HIV RT and affect the specificity of RNase H cleavage. While A-tracts in DNA (dA:dT) are unbent, bends are commonplace at junctions between A-tracts (dA:dT) and a dG:dC base pair (Dickerson et al., 1996). A dG:dC/dA:dT junction has been reported to be a flexible hinge, capable of adopting either a straight or a bent conformation under the influence of local forces (Dickerson et al., 1994). While this information is based primarily on crystallographic studies with DNA:DNA oligonucleotides, the same factors, i.e. large propeller twist and narrow minor groove, exist in an rA:dT RNA:DNA duplex (Table I). Therefore, the PPT sequence may have a natural propensity for (i) bending at the rG:dC/rA:dT junction and for (ii) ‘stiffness’ in the stretches that contain multiple As. These two properties, combined with the extensive protein–nucleic acid interactions in this region (Figure 4; Table II), may contribute to an unusual structural deformation that we term ‘unzipping of the PPT’, which is centered at the rG:dC/rA:dT junction and is discussed below. Furthermore, they are likely to affect the trajectory of the template-primer and its positioning at the RNase H active site.
Table II. Template-primer contacts affecting RNase H activity.
| RNase H primer grip contacts with the DNA primer strand in HIV-1 RT and functional equivalents in other enzymes | ||||||
|---|---|---|---|---|---|---|
| HIV-1 RT | HIV-2 RT | MuLV RT | E.coli RNase H | Nucleotide site positiona | Contacting nucleotide | Comments |
| G359 | N359 | –7 | Pri12-Ade | main-chain contactb | ||
| A360 | T360 | –7 | Pri12-Ade | main-chain contactb | ||
| H361 | H361 | –6 | Pri13-Thy | conservedb | ||
| K395c | R395c | –9/–8 | Pri10-Ade/Pri11-Ade | homologousb | ||
| E396c | E396c | –8 | Pri11-Ade | conservedb | ||
| T473 | T473 | S557 | T43 | –5/–4 | Pri14-Thy/Pri15-Thy | homologous |
| N474 | N474 | A558 | N44 | –4 | Pri15-Thy | mostly conservedd |
| Q475 | Q475 | Q559 | N45 | –4/–5 | Pri15-Thy | mostly conserved |
| K476 | K476 | R560 | R46 | –5 | Pri14-Thy | homologous |
| Y501 | Y501 | Y586 | Y73 | –5/–6 | Pri14-Thy | conserved |
| I505 | I505 | T590 | G77 | –6 | Pri13-Thy | poorly conserved |
| RNase H contacts with the RNA template strand | ||||||
| K390c | H390c | –3 | Tem16-Ade | homologousb | ||
| R448 | R448 | L529 | G15 | +1/+2 | Tem19-Ade/Tem20-Ade | poorly conserved |
| N474 | N474 | A558 | N44 | +1 | Tem19-Ade | mostly conserved |
| Q475 | Q475 | Q559 | N45 | –1/–2 | Tem18-Ade/Tem17-Gua | mostly conserved |
| Q500 | Q500 | R585 | Q72 | –1/–2 | Tem18-Ade/Tem17-Gua | homologous |
| H539 | H539 | H638 | H124 | +1 | Tem19-Ade | conserved |
aNucleotide site positions are defined in Figure 4.
bEscherichia coli RNase HI lacks residues equivalent to these residues in the p66 and p51 connection subdomains; instead, the basic loop and helix present both in E.coli RNase HI and MuLV RT RNase H may provide functionally equivalent residues.
cResidue in p51 subunit.
dInteraction mediated by a water molecule.
‘Unzipping’ in the PPT
By far the most striking structural feature of the PPT structure is a departure from Watson–Crick base pairing involving several nucleotide pairs. We refer to this as ‘unzipping of the PPT’. The structural aberration starts at the 5′ end of the PPT with the ‘melting’ of the first 2 bp of the PPT (bp 13 and 14, Figure 4), leaving an unpaired template base (Tem15-Ade, Figures 4 and 5). The unpaired template nucleotide is followed by a frame-shifted A-T base pair (Tem16-Ade:Pri15-Thy) and a G-T mismatch (Tem17-Gua:Pri16-Thy). The next primer nucleotide (Pri17-Cyt) is unpaired, compensating for the unpaired residue of the template (Tem15-Ade), bringing the nucleotide pairs back into frame. The next 4 bp are located adjacent to the RNase H active site and linked by what would normally be the scissile phosphate. For these 4 bp, although the nucleic acid duplex is realigned, the separations between the bases are relatively large. This means that the bases of the RNA template positioned at the RNase H active site are loosely bound to the corresponding bases of the DNA primer and held in place mostly by stacking interactions. Finally, the nucleic acid duplex appears to ‘heal’ and restore normal register in the remaining base pairs immediately following the RNase H active site.
Throughout the length of this structural aberration there are numerous contacts between amino acids of RT and phosphate groups of the ‘stretched’ or ‘slipped’ base pairs (Figures 4 and 6). The protein–nucleic acid interactions may contribute to the fact that the unusual nucleic acid structure in this region is well defined. It is not clear whether the unzipped PPT structure results from the protein–RNA:DNA contacts or is an intrinsic property of the PPT sequence. However, it seems plausible that the unusual structure depends on three factors: (i) the stiffness of A-tracts; (ii) the propensity to bend at the end of A-tracts (in particular at an AT/GC junction); and (iii) the large number of contacts with the amino acids of the RNase H domain.
Contacts of RNase H with template-primer: the RNase H primer grip
In the RT–RNA:DNA complex, as well in RT complexes with DNA:DNA, a network of amino acids interacts with the DNA primer strand near the RNase H active site (Table II). We propose that these amino acids form an element of the RT structure (the ‘RNase H primer grip’) that serves to position the DNA primer strand near the RNase H active site and helps to determines the trajectory of the template strand in relation to the RNase H active site. The RNase H primer grip makes contacts with nucleic acid site positions –4 to –9 relative to the scissile phosphate base pair (see Figure 4 for a definition of nucleic acid site position). The RNase H primer grip, by interacting with the DNA primer strand, may also control the interactions of the RNA template substrate and the RNase H catalytic site in a nucleic acid sequence-dependent and structure-dependent manner. The interactions between the PPT and the RNase H primer grip of HIV-1 RT include residues Lys395 and Glu396 of the p51 subunit, Gly359 (main chain), Ala360 (main chain) and His361 of the p66 connection subdomain, and Thr473, Asn474 (mediated by a water molecule), Gln475, Lys476, Tyr501 and Ile505 of the RNase H domain of HIV-1 RT (Table II). HIV-1 RT residues that contact the RNA template strand (positions –2 to +2, centered around the scissile phosphate) include the side chain of Lys390 of p51, and the side chains of Arg448, Asn474, Gln475, Gln500 and His539 of p66.
Sequence alignment of the RNase H domain of HIV-1 RT and the RNase H domains of HIV-2 RT, MuLV RT and E.coli RNase HI suggests a significant conservation of the residues of the RNase H primer grip in all these enzymes. Many of the residues of HIV-1 RT RNase H that contact the RNA:DNA template-primer have conserved functionally important counterparts in E.coli RNase HI. Specifically, mutations Thr43Cys, Asn44Ala, Asn45Ala, Gln72Ala, Tyr73Leu and Gly77Ala at residues of E.coli RNase HI, which are the functional equivalents of Thr473, Asn474, Gln475, Gln500, Tyr501 and Ile505, respectively, of HIV-1 RT, resulted in decreased efficiency of RNA:DNA cleavage (as judged by changes in the efficiency ratio kcat/Km in the wild type compared with the mutant enzymes) by 140-, 8-, 10-, 3-, 3- and 5-fold, respectively (Kanaya, 1997 and references therein). Similarly, mutations of the Gln72 and His124 of E.coli RNase HI, which are equivalent to Gln500 and His539 of HIV-1 RT, which contact the RNA template at the RNase H active site of HIV-1 RT, substantially decrease the catalytic efficiency of the enzymes (3- and 70-fold for Gln72Ala and His124Ala, respectively). Finally, the Gln475Glu, His539Phe and His539Asp HIV-1 RT mutants have decreased RNase H activity and defective PPT cleavage (Tisdale et al., 1991; Volkmann et al., 1993).
The RNase H primer grip together with minor groove width and trajectory determines retroviral RNase H cleavage specificity
It has been suggested that minor groove width determines susceptibility of nucleic acids to RNase H cleavage (Fedoroff et al., 1997; Han et al., 1997; Xiong and Sundaralingam, 1998). However, the ability of RNase H to cleave nucleic acids with diverse minor grooves (Ben-Artzi et al., 1992; Blain and Goff, 1993; Smith and Roth, 1993; Hostomsky et al., 1994; Salazar et al., 1994; Götte et al., 1995; Gao et al., 1997; Lima and Crooke, 1997; Szyperski et al., 1999) suggests that additional factors are likely to determine the specificity of RNase H.
We have identified the RNase H primer grip as a structural element of HIV RT that is involved in the control of RNase H cleavage specificity (Figure 6). Since the RNase H primer grip makes numerous contacts with the DNA primer strand, appropriate access to the RNase H active site can only occur with substrates that have the correct minor groove width. The RNase H primer grip also places additional constraints on the structure of nucleic acids leading up to the RNase H active site: our model suggests that curvatures associated with particular RNA:DNA sequences could also affect cleavage specificity. This model allows for the possibility that some RNA:DNA substrates that have a minor groove width appropriate for cleavage may have an inappropriate trajectory and may not be able to interact properly with the RNase H active site.
Despite the fact that the T-rich primer interacts strongly with the RNase H primer grip, the rA-stretch of the PPT is a poor substrate for RNase H cleavage. There is an unusually narrow minor groove that might prevent the rA-rich RNA template strand from reaching the catalytic residues of RNase H. Interestingly, the approximate distance between the active site residues of RNase H and what would normally be the scissile phosphate of the A-tract (rA:dT) in our structure is ∼3 Å, similar to the reduction in minor groove width, presumably caused by the A-tract (rA:dT) on the RNA. It is possible that RNase H of HIV-1 RT has evolved to interact unproductively with A-tracts to prevent cleavage within the PPT.
The trajectory model may also account for the inability of HIV-1 RT to cleave within the (dC:rG)6 sequence at the end of PPT. Precise trajectory might also be a determining factor in certain rare cleavage events such as cutting of RNA:RNA or RNA:RNA-DNA.
Effect of PPT structure on the cleavage specificity of RNase H at the U3/PPT junction; a biological role for the nucleic acid bend near the polymerase active site
The specificity of cleavage at the 3′ end of the PPT (after the G-stretch) is critical since it helps to determine one end of the viral DNA. If the PPT/U3 junction is modeled at the RNase H active site using our structure, the A-tract (rA:dT), including the unzipped region, would be in position to contact residues of the H and I helices of the p66 thumb. Biochemical studies have shown that mutation of several residues of this region of the p66 thumb dramatically affects cleavage at the PPT/U3 junction (Powell et al., 1997, 1999; Gao et al., 1998). Hence our structural and modeling data suggest that the p66 thumb may contribute to precise cleavage at the end of PPT by interacting with the A-tract (rA:dT) region at the opposite end of the PPT. The stiffness of the A-tract (rA:dT) sequences may cause pausing of polymerization at the point where the A-tract (rA:dT) at the 5′ end of PPT must be bent to create the 40–45° bend, normally found 5–9 bp from the polymerase active site, in the vicinity of the p66 thumb. Pausing at this site could promote RNase H cleavage at the PPT/U3 junction, which is concomitantly aligned at the RNase H active site. Thus, the bend may have a biological role in ensuring precise cleavage at the PPT/U3 junction, which is required for retroviral replication.
The sequence features important for positioning RNase H for the cleavage reaction that generates the plus strand primer of HIV-1 and MuLV have been studied extensively (Rattray and Champoux, 1989; Pullen et al., 1993; Powell and Levin, 1996). For HIV-1, it has been reported that the –2G at the 3′ end of HIV-1 PPT is crucial for specific cleavage at the PPT/U3 junction (between –1G and +1A in Figure 1). In our complex, a G residue is also positioned two registers 3′ of the scissile phosphate. This G makes a specific interaction with Gln475 (hydrogen bond between 2-amino of G and Oε1 of Gln475) that would be lost if there were another base present at this position. The contact of Gln475 with the –2G base at the PPT 3′ end may also contribute to precise cleavage at the PPT/U3 junction.
The HIV PPT as a target for drug design
The unusual structural features of HIV-1 PPT bound to HIV-1 RT may provide attractive targets for drug design. Specific inhibitors designed and/or selected to recognize preferentially the structural characteristics of PPT could become anti-AIDS drugs. Several studies have shown that the minor groove of DNA:DNA and RNA:DNA can be targeted by minor groove binding drugs. Furthermore, substantial progress has been made in making such inhibitors sequence specific (White et al., 1998). Binding of drugs that target the minor groove of DNA results in expansion of the minor groove and shortening of the major groove width; it is possible that such changes could render PPT susceptible to HIV-1 RT cleavage. Alternatively, the PPT can be targeted to give a PPT–drug complex with poor ability to be used as a primer for second strand synthesis. Dickerson and colleagues have specifically suggested (Han et al., 1997) that inhibitors such as a bis-linked distamycin–anthramycin target the HIV-1 RT–RNA:DNA PPT; our results support that strategy. The present study has revealed a new potential target and highlights the importance of structure in drug development efforts.
Materials and methods
Crystallization and data collection
Wild-type HIV-1 RT was purified as described previously (Clark et al., 1995). Nucleic acids were purified and annealed as described before, using diethyl pyrocarbonate-treated aqueous solutions (Sarafianos et al., 1999). The crystallization protocols for the RT–RNA:DNA–Fab complex were similar to those used to prepare the corresponding crystals of HIV-1 RT complexed with DNA:DNA (Ding et al., 1998; Sarafianos et al., 1999). Specifically, purified RT was mixed with Fab at a 1:0.8 mass ratio and with template-primer to final concentrations of 25 mg/ml protein and 0.2 mM nucleic acid. Hanging drops were prepared by mixing equal volumes of the complex and crystallization solutions [100 mM cacodylate pH 5.6, 29–31% saturated ammonium sulfate (SAS)] at 4°C. Before freezing, crystals were transferred into soaking solutions containing 36% SAS, 18% glucose (w/v) and 18% glycerol (v/v), reaching the final concentration in three steps of 20–30 min each (Ding et al., 1998). Soaking solutions contained 0.2 mM RNA:DNA. Several diffraction datasets were collected at the Brookhaven National Laboratory Synchrotron Light Source X25 beamline, and at the Cornell High-Energy Synchrotron Source (CHESS) F1 beamline. The main dataset was collected from one crystal frozen at –165°C and recorded on image plates (Table III). Diffraction data were processed and scaled with DENZO and SCALEPACK, respectively (Otwinowski and Minor, 1996). The diffraction data statistics are summarized in Table III.
Table III. Summary of data collection and refinement statistics.
| Temperature (°C) | –165 |
| Resolution range (Å) | 40–3.0 |
| Space group | P3212 |
| Unit cell (a, b, c; Å) | 166.2, 166.2, 218.8 |
| Rsym (%) | 12.8 |
| Reflections (total/unique) | 611 137/58 390 |
| Completeness (total/3.05–3.0 Å, %) | 85.0/68.7 |
| Radiation source | NSLS-BNL (X25) |
| Refinement statistics | |
| resolution range (Å) | 8.0–3.0 Å |
| number of atoms | 11 930 |
| Rcryst/Rfree | 27.4/31.6 |
| r.m.s. deviation bonds (Å) | 0.012 |
| r.m.s. deviation angles (°) | 1.72 |
| Luzzati coordinate error (Å) | 0.46 |
Structure determination
The structure was solved by molecular replacement [X-PLOR (Brunger, 1993)] using the wild-type HIV RT–DNA:DNA–Fab structure (Ding et al., 1998) as a starting model. To avoid bias no DNA:DNA or RNA:DNA model was included in the initial stages of structure determination. In order to improve phasing quality at the early stages of structure refinement, we used the program RAVE to average the electron density maps from several frozen and unfrozen datasets at 3.5 Å resolution. Rounds of model building were guided by both normal Fourier maps and averaged maps. In the early steps of refinement simulated annealing omit maps and averaged electron density maps computed for the RT–RNA:DNA–Fab complex clearly indicated the position and conformation of nucleic acid (Figure 4). These maps showed that there was departure from canonical base pairing for several nucleotides of PPT. Even when the nucleic acid was included in the phasing and was restrained to a canonical Watson–Crick base-paired form the density clearly showed the unusual structure. The parameters used for refinement of nucleic acid were from Parkinson et al. (1996). The aberrant region of nucleic acid (region IV) was refined without any base-pairing or sugar-puckering restraints. The structure was refined using the torsion-restrained slow-cooling protocol in program X-PLOR (Rice and Brunger, 1994), followed by positional and individual isotropic thermal parameter refinement. Later stages of refinement incorporated resolution-dependent normalization and bulk-solvent correction of structure factors (Brunger, 1993). The final cycle of refinement using data between 8.0 and 3.0 Å resolution converged with a conventional crystallographic R factor of 27.4% and a free R factor of 31.6%. Coordinates and structure factors are available from the Protein Data Bank (entry 1HYS).
Acknowledgments
Acknowledgements
We thank Michael Gait and colleagues at the MRC Laboratory of Molecular Biology, Cambridge, for instruction in synthesizing RNA oligonucleotides, and the staff personnel at BNLS and CHESS and other members of E.A.’s group for help with data collection. S.G.S. was supported by an NIH-NIAID NRSA fellowship (AI 09578). The research in E.A.’s laboratory has been supported by NIH MERIT award AI 27690. S.H.H.’s laboratory is sponsored in part by the National Cancer Institute, DHHS, under contract with ABL, and by NIGMS.
References
- Arnott S., Chandrasekaran,R., Millane,R.P. and Park,H.S. (1986) DNA-RNA hybrid secondary structures. J. Mol. Biol., 188, 631–640. [DOI] [PubMed] [Google Scholar]
- Bachelin M., Hessler,G., Kurz,G., Hacia,J.G., Dervan,P.B. and Kessler,H. (1998) Structure of a stereoregular phosphorothioate DNA/RNA duplex. Nature Struct. Biol., 5, 271–276. [DOI] [PubMed] [Google Scholar]
- Ben-Artzi H., Zeelon,E., Le Grice,S.F.J., Gorecki,M. and Panet,A. (1992) Characterization of the double stranded RNA dependent RNase activity associated with recombinant retroviruses. Nucleic Acids Res., 20, 5115–5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blain S.W. and Goff,S.P. (1993) Nuclease activities of Moloney murine leukemia virus reverse transcriptase. Mutants with altered substrate specificities. J. Biol. Chem., 268, 23585–23592. [PubMed] [Google Scholar]
- Brünger A.T. (1993) X-PLOR Manual Version 3.1: A System for X-ray Crystallography and NMR. Yale University Press, New Haven, CT.
- Champoux J.J., Gilboa,E. and Baltimore,D. (1984) Mechanism of RNA primer removal by the RNase H activity of avian myeloblastosis virus reverse transcriptase. J. Virol., 49, 686–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekaran R., Wang,M., He,R.G., Puigjaner,L.C., Byler,M.A., Millane,R.P. and Arnott,S. (1989) A re-examination of the crystal structure of A-DNA using fiber diffraction data. J. Biomol. Struct. Dyn., 6, 1189–1202. [DOI] [PubMed] [Google Scholar]
- Charneau P., Alizon,M. and Clavel,F. (1992) A second origin of DNA plus-strand synthesis is required for optimal HIV replication. J. Virol., 66, 2814–2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark A.D. Jr, Jacobo-Molina,A., Clark,P., Hughes,S.H. and Arnold,E. (1995) Crystallization of HIV-1 RT with and without nucleic acid substrates, inhibitors and an antibody Fab fragment. Methods Enzymol., 262, 171–185. [DOI] [PubMed] [Google Scholar]
- Conn G.L., Brown,T. and Leonard,G.A. (1999) The crystal structure of the RNA/DNA hybrid r(GAAGAGAAGC). d(GCTTCTCTTC) shows significant differences to that found in solution. Nucleic Acids Res., 27, 555–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies J.F., Hostomska,Z., Hostomsky,Z., Jordan,S.R. and Matthews,D.A. (1991) Crystal structure of the RNase H domain of HIV-1 RT. Science, 252, 88–95. [DOI] [PubMed] [Google Scholar]
- Dickerson R.E. (1992) DNA structure from A to Z. Methods Enzymol., 211, 67–111. [DOI] [PubMed] [Google Scholar]
- Dickerson R.E., Goodsell,D.S. and Neidle,S. (1994) ‘…the tyranny of the lattice …’ Proc. Natl Acad. Sci. USA, 91, 3579–3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickerson R.E., Goodsell,D. and Kopka,M.L. (1996) MPD and DNA bending in crystals and in solution. J. Mol. Biol., 256, 108–125. [DOI] [PubMed] [Google Scholar]
- Ding J., Hughes,S.H. and Arnold,E. (1997) Protein–nucleic acid interactions and DNA conformation in a complex of HIV-1 RT with a double-stranded DNA template-primer. Biopolymers, 44, 125–138. [DOI] [PubMed] [Google Scholar]
- Ding J., Das,K., Hsiou,Y., Sarafianos,S.G., Clark,A.D.,Jr, Jacobo-Molina,A., Tantillo,C., Hughes,S.H. and Arnold,E. (1998) Structure and functional implications of the polymerase active site region in a complex of HIV-1 RT with double-stranded DNA and an antibody Fab fragment at 2.8 Å resolution. J. Mol. Biol., 284, 1095–1111. [DOI] [PubMed] [Google Scholar]
- Fedoroff O.Y., Salazar,M. and Reid,B.R. (1993) Structure of a DNA:RNA hybrid duplex. Why RNase H does not cleave pure RNA. J. Mol. Biol., 233, 509–523. [DOI] [PubMed] [Google Scholar]
- Fedoroff O.Y., Ge,Y. and Reid,B.R. (1997) Solution structure of r(gaggacug):d(CAGTCCTC) hybrid: implications for the initiation of HIV-1 (+)-strand synthesis. J. Mol. Biol., 269, 225–239. [DOI] [PubMed] [Google Scholar]
- Fuentes G.M., Rodriguez-Rodriguez,L., Fay,P.J. and Bambara,R.A. (1995) Use of an oligoribonucleotide containing the polypurine tract sequence as a primer by HIV RT. J. Biol. Chem., 270, 28169–28176. [DOI] [PubMed] [Google Scholar]
- Gao G., Orlova,M., Georgiadis,M.M., Hendrickson,W.A. and Goff,S.P. (1997) Conferring RNA polymerase activity to a DNA polymerase: a single residue controls substrate selection. Proc. Natl Acad. Sci. USA, 94, 407–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H.Q., Boyer,P.L., Arnold,E. and Hughes,S.H. (1998) Effects of mutations in the polymerase domain on the polymerase, RNase H and strand transfer activities of HIV-1 RT. J. Mol. Biol., 277, 559–572. [DOI] [PubMed] [Google Scholar]
- Gao H.Q., Sarafianos,S.G., Arnold,E. and Hughes,S.H. (1999) Similarities and differences in the RNase H activities of HIV-1 RT and Moloney murine leukemia virus RT. J. Mol. Biol., 294, 1097–1113. [DOI] [PubMed] [Google Scholar]
- Ghosh M., Howard,K.J., Cameron,C.E., Benkovic,S.J., Hughes,S.H. and Le Grice,S.F. (1995) Truncating α-helix E′ of p66 HIV RT modulates RNase H function and impairs DNA strand transfer. J. Biol. Chem., 270, 7068–7076. [DOI] [PubMed] [Google Scholar]
- Ghosh M., Williams,J., Powell,M.D., Levin,J.G. and Le Grice,S.F. (1997) Mutating a conserved motif of the HIV-1 RT palm subdomain alters primer utilization. Biochemistry, 36, 5758–5768. [DOI] [PubMed] [Google Scholar]
- Gopalakrishnan V., Peliska,J.A. and Benkovic,S.J. (1992) HIV-1 RT: Spatial and temporal relationship between the polymerase and RNase H activities. Proc. Natl Acad. Sci. USA, 89, 10763–10767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Götte M., Fackler,S., Hermann,T., Perola,E., Cellai,L., Gross,H.J., Le Grice,S.F. and Heumann,H. (1995) HIV-1 RT-associated RNase H cleaves RNA/RNA in arrested complexes: implications for the mechanism by which RNase H discriminates between RNA/RNA and RNA/DNA. EMBO J., 14, 833–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Götte M., Maier,G., Gross,H.J. and Heumann,H. (1998) Localization of the active site of HIV-1 RT-associated RNase H domain on a DNA template using site-specific generated hydroxyl radicals. J. Biol. Chem., 273, 10139–10146. [DOI] [PubMed] [Google Scholar]
- Gyi J.I., Conn,G.L., Lane,A.N. and Brown,T. (1996) Comparison of the thermodynamic stabilities and solution conformations of DNA.RNA hybrids containing purine-rich and pyrimidine-rich strands with DNA and RNA duplexes. Biochemistry, 35, 12538–12548. [DOI] [PubMed] [Google Scholar]
- Gyi J.I., Lane,A.N., Conn,G.L. and Brown,T. (1998) Solution structures of DNA.RNA hybrids with purine-rich and pyrimidine-rich strands: comparison with the homologous DNA and RNA duplexes. Biochemistry, 37, 73–80. [DOI] [PubMed] [Google Scholar]
- Han G.W., Kopka,M.L., Cascio,D., Grzeskowiak,K. and Dickerson,R.E. (1997) Structure of a DNA analog of the primer for HIV-1 RT second strand synthesis. J. Mol. Biol., 269, 811–826. [DOI] [PubMed] [Google Scholar]
- Heyman T., Agoutin,B., Friant,S., Wilhelm,F.X. and Wilhelm,M.L. (1995) Plus-strand DNA synthesis of the yeast retrotransposon Ty1 is initiated at two sites, PPT1 next to the 3′ LTR and PPT2 within the pol gene. PPT1 is sufficient for Ty1 transposition. J. Mol. Biol., 253, 291–303. [DOI] [PubMed] [Google Scholar]
- Horton N.C. and Finzel,B.C. (1996) The structure of an RNA/DNA hybrid: a substrate of the RNase activity of HIV-1 RT. J. Mol. Biol., 264, 521–533. [DOI] [PubMed] [Google Scholar]
- Hostomsky Z., Hughes,S.H., Goff,S.P. and Le Grice,S.F. (1994) Redesignation of the RNase D activity associated with retroviral RT as RNase H*. J. Virol., 68, 1970–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Chopra,R., Verdine,G.L. and Harrison,S.C. (1998) Structure of a covalently trapped catalytic complex of HIV-1 RT: implications for drug resistance. Science, 282, 1669–1675. [DOI] [PubMed] [Google Scholar]
- Huber H.E., McCoy,J.M., Seehra,J.S. and Richardson,C.C. (1989) HIV-1 RT. Template binding, processivity, strand displacement synthesis and template switching. J. Biol. Chem., 264, 4669–4678. [PubMed] [Google Scholar]
- Hungnes O., Tjotta,E. and Grinde,B. (1992) Mutations in the central polypurine tract of HIV-1 result in delayed replication. Virology, 190, 440–442. [DOI] [PubMed] [Google Scholar]
- Ishikawa K., Okumura,M., Katayanagi,K., Kimura,S., Kanaya,S., Nakamura,H. and Morikawa,K. (1993) Crystal structure of RNase H from Thermus thermophilus HB8 refined at 2.8 Å resolution. J. Mol. Biol., 230, 529–542. [DOI] [PubMed] [Google Scholar]
- Jacobo-Molina A. et al. (1993) Crystal structure of HIV-1 RT complexed with double-stranded DNA at 3.0 Å resolution shows bent DNA. Proc. Natl Acad. Sci. USA, 90, 6320–6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaya S. (1997) Enzymatic activity and protein stability of E.coli RNase HI. In Crouch,R.J. and Toulme,J.J. (eds), Ribonucleases H. Les editions INSERM, Paris, France.
- Katayanagi K., Miyagawa,M., Matsushima,M., Ishikawa,M., Kanaya,S., Ikehara,M., Matsuzaki,T. and Morikawa,K. (1990) Three-dimensional structure of RNase H from E. coli. Nature, 347, 306–309. [DOI] [PubMed] [Google Scholar]
- Kim J.L., Nikolov,D.B. and Burley,S.K. (1993) Co-crystal structure of TBP recognizing the minor groove of a TATA element. Nature, 365, 520–527. [DOI] [PubMed] [Google Scholar]
- Kohlstaedt L.A., Wang,J., Friedman,J.M., Rice,P.A. and Steitz,T.A. (1992) Crystal structure at 3.5 Å resolution of HIV-1 RT complexed with an inhibitor. Science, 256, 1783–1790. [DOI] [PubMed] [Google Scholar]
- Lane A.N., Ebel,S. and Brown,T. (1993) NMR assignments and solution conformation of the DNA.RNA hybrid duplex d(GTGAACTT).r(AAGUUCAC). Eur. J. Biochem., 215, 297–306. [DOI] [PubMed] [Google Scholar]
- Lavery R. and Sklenar,H. (1988) The definition of generalized helicoidal parameters and of axis curvature for irregular nucleic acids. J. Biomol. Struct. Dyn., 6, 63–91. [DOI] [PubMed] [Google Scholar]
- Leonard G.A., McAuley-Hecht,K.E., Ebel,S., Lough,D.M., Brown,T. and Hunter,W.N. (1994) Crystal and molecular structure of r(CGCGAAUUAGCG): an RNA duplex containing two G(anti).A(anti) base pairs. Structure, 2, 483–494. [DOI] [PubMed] [Google Scholar]
- Lima W.F. and Crooke,S.T. (1997) Cleavage of single strand RNA adjacent to RNA-DNA duplex regions by Escherichia coli RNase H1. J. Biol. Chem., 272, 27513–27516. [DOI] [PubMed] [Google Scholar]
- Lu X.J., El Hassan,M.A. and Hunter,C.A. (1997) Structure and conformation of helical nucleic acids: analysis program (SCHNAaP). J. Mol. Biol., 273, 668–680. [DOI] [PubMed] [Google Scholar]
- Luo G.X., Sharmeen,L. and Taylor,J. (1990) Specificities involved in the initiation of retroviral plus-strand DNA. J. Virol., 64, 592–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor,W. (1996) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 55–62. [DOI] [PubMed] [Google Scholar]
- Palaniappan C., Wisniewski,M., Jacques,P.S., Le Grice,S.F., Fay,P.J. and Bambara,R.A. (1997) Mutations within the primer grip region of HIV-1 RT result in loss of RNase H function. J. Biol. Chem., 272, 11157–11164. [DOI] [PubMed] [Google Scholar]
- Parkinson G., Vojtechovsky,J., Clowney,L., Brunger,A.T. and Berman,H.M. (1996) New parameters for the refinement of nucleic acid containing structures. Acta Crystallogr. D., 52, 57–64. [DOI] [PubMed] [Google Scholar]
- Powell M.D. and Levin,J.G. (1996) Sequence and structural determinants required for priming of plus-strand DNA synthesis by the HIV-1 polypurine tract. J. Virol., 70, 5288–5296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell M.D., Ghosh,M., Jacques,P.S., Howard,K.J., Le Grice,S.F. and Levin,J.G. (1997) Alanine-scanning mutations in the ‘primer grip’ of p66 HIV-1 RT result in selective loss of RNA priming activity. J. Biol. Chem., 272, 13262–13269. [DOI] [PubMed] [Google Scholar]
- Powell M.D., Beard,W.A., Bebenek,K., Howard,K.J., Le Grice,S.F., Darden,T.A., Kunkel,T.A., Wilson,S.H. and Levin,J.G. (1999) Residues in the αH and αI helices of the HIV-1 RT thumb subdomain required for the specificity of RNase H-catalyzed removal of the polypurine tract primer. J. Biol. Chem., 274, 19885–19893. [DOI] [PubMed] [Google Scholar]
- Pullen K.A., Rattray,A.J. and Champoux,J.J. (1993) The sequence features important for plus strand priming by HIV-1 RT. J. Biol. Chem., 268, 6221–6227. [PubMed] [Google Scholar]
- Rattray A.J. and Champoux,J.J. (1989) Plus-strand priming by Moloney murine leukemia virus. The sequence features important for cleavage by RNase H. J. Mol. Biol., 208, 445–456. [DOI] [PubMed] [Google Scholar]
- Resnick R., Omer,C.A. and Faras,A.J. (1984) Involvement of retrovirus RT-associated RNase H in the initiation of strong-stop (+) DNA synthesis and the generation of the long terminal repeat. J. Virol., 51, 813–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes D. (1979) Nucleosome cores reconstituted from poly(dA-dT) and the octamer of histones. Nucleic Acids Res., 6, 1805–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice L.M. and Brunger,A.T. (1994) Torsion angle dynamics: reduced variable conformational sampling enhances crystallographic structure refinement. Proteins, 19, 277–290. [DOI] [PubMed] [Google Scholar]
- Rice P.A., Yang,S., Mizuuchi,K. and Nash,H.A. (1996) Crystal structure of an IHF–DNA complex: a protein-induced DNA U-turn. Cell, 87, 1295–1306. [DOI] [PubMed] [Google Scholar]
- Salazar M., Fedoroff,O.Y., Zhu,L. and Reid,B.R. (1994) The solution structure of the r(gcg)d(TATACCC):d(GGGTATACGC) Okazaki fragment contains two distinct duplex morphologies connected by a junction. J. Mol. Biol., 241, 440–455. [DOI] [PubMed] [Google Scholar]
- Sarafianos S.G., Das,K., Clark,A.D.,Jr, Ding,J., Boyer,P.L., Hughes,S.H. and Arnold,E. (1999) Lamivudine (3TC) resistance in HIV-1 RT involves steric hindrance with β-branched amino acids. Proc. Natl Acad. Sci. USA, 96, 10027–10032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz O., Cromme,F.V., Gruninger-Leitch,F. and Le Grice,S.F.J. (1989) Point mutations in conserved amino acid residues within the C-terminal domain of HIV-1 reverse transcriptase specifically repress RNase H function. FEBS Lett., 257, 311–314. [DOI] [PubMed] [Google Scholar]
- Schatz O., Mous,J. and Le Grice,S.F.J. (1990) HIV-1 RT-associated ribonuclease H displays both endonuclease and 3′→5′ exonuclease activity. EMBO J., 9, 1171–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J.S. and Roth,M.J. (1993) Purification and characterization of an active HIV-1 RNase H domain. J. Virol., 67, 4037–4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyperski T., Götte,M., Billeter,M., Perola,E., Cellai,L., Heumann,H. and Wuthrich,K. (1999) NMR structure of the chimeric hybrid duplex r(gcaguggc).r(gcca)d(CTGC) comprising the tRNA-DNA junction formed during initiation of HIV-1 reverse transcription. J. Biomol. NMR, 13, 343–355. [DOI] [PubMed] [Google Scholar]
- Tanese N. and Goff,S.P. (1988) Domain structure of the Moloney murine leukemia virus RT: mutational analysis and separate expression of the DNA polymerase and RNase H activities. Proc. Natl Acad. Sci. USA, 85, 1777–1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telesnitsky A. and Goff,S.P. (1997) Reverse transcriptase and the generation of retroviral DNA. In Coffin,J.M., Hughes,S.H. and Varmus,H.E. (eds), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed]
- Tisdale M., Schulze,T., Larder,B.A. and Moelling,K. (1991) Mutations within the RNase H domain of HIV-1 RT abolish virus infectivity. J. Gen. Virol., 72, 59–66. [DOI] [PubMed] [Google Scholar]
- Volkmann S., Wöhrl,B.M., Tisdale,M. and Moelling,K. (1993) Enzymatic analysis of two HIV-1 RT mutants with mutations in carboxyl-terminal amino acid residues conserved among retroviral RNase H. J. Biol. Chem., 268, 2674–2683. [PubMed] [Google Scholar]
- Wang A.H.J., Fuji,S., van Boom,J.H., van der Marel,G.A., van Boekel,S.A.A. and Rich,A. (1982) Molecular structure of r(GCG)d(TATACGC): a DNA-RNA hybrid helix joined to double helical DNA. Nature, 299, 601–604. [DOI] [PubMed] [Google Scholar]
- White S., Szewczyk,J.W., Turner,J.M., Baird,E.E. and Dervan,P.B. (1998) Recognition of the four Watson–Crick base pairs in the DNA minor groove by synthetic ligands. Nature, 391, 468–471. [DOI] [PubMed] [Google Scholar]
- Wöhrl B.M. and Moelling,K. (1990) Interaction of HIV-1 RNase H with polypurine tract containing RNA-DNA hybrids. Biochemistry, 29, 10141–10147. [DOI] [PubMed] [Google Scholar]
- Xiong Y. and Sundaralingam,M. (1998) Crystal structure and conformation of a DNA-RNA hybrid duplex with a polypurine RNA strand: d(TTCTTBr5CTTC)-r(GAAGAAGAA). Structure, 6, 1493–1501. [DOI] [PubMed] [Google Scholar]
- Yang W., Hendrickson,W.A., Crouch,R.J. and Satow,Y. (1990) Structure of RNase H phased at 2 Å resolution by MAD analysis of the selenomethionyl protein. Science, 249, 1398–1405. [DOI] [PubMed] [Google Scholar]
- Zhan X. and Crouch,R.J. (1997) The isolated RNase H domain of murine leukemia virus RT. Retention of activity with concomitant loss of specificity. J. Biol. Chem., 272, 22023–22029. [DOI] [PubMed] [Google Scholar]
- Zhu L., Salazar,M. and Reid,B.R. (1995) DNA duplexes flanked by hybrid duplexes: the solution structure of chimeric junctions in [r(cgcg)d(TATACGCG)]2. Biochemistry, 34, 2372–2380. [DOI] [PubMed] [Google Scholar]