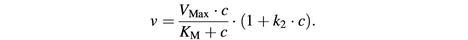Abstract
The mechanism of facilitated translocation through nuclear pore complexes (NPCs) is only poorly understood. Here, we present a kinetic analysis of the process using various model substrates. We find that the translocation capacity of NPCs is unexpectedly high, with a single NPC allowing a mass flow of nearly 100 MDa/s and rates in the order of 103 translocation events per second. Our data further indicate that high affinity interactions between the translocation substrate and NPC components are dispensable for translocation. We propose a ‘selective phase model’ that could explain how NPCs function as a permeability barrier for inert molecules and yet become selectively permeable for nuclear transport receptors and receptor–cargo complexes.
Keywords: importin/kinetics/nuclear pore complex/nuclear transport/nuclear transport factor 2
Introduction
The nuclear envelope (NE) separates the nuclear from the cytoplasmic compartment and thereby necessitates nucleocytoplasmic transport. All nuclear proteins, for example, need to be imported from the cytoplasm, while the major cellular RNAs, mRNA, tRNA and rRNA, are synthesized by transcription in the nucleus and need to be exported to the cytoplasm, where they function in translation.
Nucleocytoplasmic exchange of small molecules and macromolecules proceeds through nuclear pore complexes (NPCs), which are embedded in the NE (Feldherr, 1962). NPCs have a mass of ∼125 MDa in higher eukaryotes (Reichelt et al., 1990; Akey and Radermacher, 1993) or 66 MDa in Saccharomyces cerevisiae (Rout and Blobel, 1993; Yang et al., 1998), and allow passage of material in essentially two modes: passive diffusion and facilitated translocation. Passive diffusion does not require any specific interactions between the diffusing species and components of the NPC; it is fast for small molecules, but becomes restricted and inefficient as the diffusing objects approach a size limit of 20–40 kDa (Paine et al., 1975; Bonner, 1978).
In contrast, facilitated translocation allows the passage of objects as large as several megadaltons. It is often coupled to an input of metabolic energy, which in turn permits transport against a gradient of chemical activity (reviewed in Mattaj and Englmeier, 1998; Görlich and Kutay, 1999). Facilitated translocation requires specific interactions between the translocating species and constituents of the NPC and is therefore a highly selective process. Only some proteins, such as nuclear transport receptors, possess ‘translocation-promoting properties’ that allow a direct interaction with NPCs and facilitated translocation without the aid of trans-acting factors. Objects that lack such translocation-promoting properties will from now on be referred to as ‘inert molecules’. Inert molecules above the passive diffusion limit remain excluded from nucleocytoplasmic exchange, unless they are recognized and bound by appropriate nuclear transport receptors, which can mediate translocation-relevant interactions with the NPC also in trans.
The superfamily of importin β-related factors constitutes the best characterized class of nuclear transport receptors so far (Mattaj and Englmeier, 1998; Weis, 1998; Görlich and Kutay, 1999; Nakielny and Dreyfuss, 1999). These receptors bind cargo molecules on one side of the NE, translocate through the NPC to the other side, release their cargo and finally return to the original compartment to mediate another round of transport. According to the direction in which they carry a cargo, they can be classified as importins or exportins. The cargo–receptor interactions are controlled by a RanGTP gradient across the NE. Importins and exportins bind RanGTP, which allows them to respond to the gradient by cargo loading and release in the appropriate compartment. Importins bind cargoes at low RanGTP levels in the cytoplasm and release their cargo at high RanGTP concentrations into the nucleus. They are recycled as RanGTP complexes back to the cytoplasm, where Ran is removed from the receptor and GTP is hydrolysed, allowing the importin to bind and import another cargo molecule. Cargo binding to exportin is regulated in an exactly converse manner to importins.
Importins and exportins constantly export RanGTP from the nucleus, which necessitates the nuclear RanGTP pool being efficiently replenished. This is accomplished by nuclear transport factor 2 (NTF2) (Ribbeck et al., 1998; Smith et al., 1998) and the exclusively nuclear localized nucleotide exchange factor RanGEF (Bischoff and Ponstingl, 1991). NTF2 mediates nuclear import of RanGDP, while RanGEF recharges Ran with GTP in the nucleus.
Importin- and exportin-mediated transport cycles can accumulate cargoes against a gradient of chemical activity, which is an energy-consuming task. This energy originates from the chemical potential of the RanGTP gradient, which in turn is maintained by RanGEF-mediated nucleotide exchange on Ran and the subsequent cytoplasmic hydrolysis of Ran-bound GTP. The facilitated translocation process per se is, however, not directly coupled to nucleotide hydrolysis or any other irreversible event (Kose et al., 1997; Nakielny and Dreyfuss, 1998; Ribbeck et al., 1998, 1999; Schwoebel et al., 1998; Englmeier et al., 1999), and thus constitutes a fully reversible process. The molecular mechanism of this facilitated translocation process is a central, but as yet largely unresolved problem in the field of nucleocytoplasmic transport. The same applies to the question of the nature of the permeability barrier for inert objects.
Vertebrate NPCs are estimated to be composed of 30–50 different proteins (nucleoporins), many of which contain numerous phenylalanine-rich (Phe-rich) repeats (Fabre and Hurt, 1997; Ryan and Wente, 2000). These repeats interact specifically with transport receptors of the importin β superfamily (see for example Radu et al., 1995), with NTF2 (Paschal and Gerace, 1995; Clarkson et al., 1996) and factors involved in mRNA export (Bachi et al., 2000; Strasser et al., 2000), and are therefore believed to play a crucial role in the translocation process. This is directly supported by the observation that point mutations in NTF2 or importin β, which impair their interaction with repeat domains, also compromise translocation (Bayliss et al., 1999, 2000). How the interaction between the translocating species and the repeats can facilitate NPC passage has so far remained unclear.
Here, we study the dynamics of the translocation process. We found that NPCs can mediate translocation at a much higher rate (∼103 s–1) and accommodate a much higher mass flow (nearly 100 MDa/s) than previously anticipated. Furthermore, we can estimate that translocating material can cross the central channel of the NPC at speeds of at least 0.5 µm/s. We propose a model for NPC function in which barrier function and the phenomenon of facilitated translocation are intimately linked to each other. In this model, the permeability barrier within the central channel is built by a mutual attraction between the hydrophobic Phe-rich clusters of the nucleoporin repeats. This should result in a meshwork that restricts the flow of inert molecules. Translocating material, however, can be incorporated into the meshwork, because it is able to interact with the Phe-rich clusters and thus locally compete the mutual attraction between the repeats. The translocating species could thus selectively partition into the central plug and use this ‘selective solvation’ to cross this permeability barrier at a high rate.
Results
In recent years, much progress has been made in identifying nuclear transport factors that mediate translocation through NPCs and also a considerable body of data has accumulated describing interactions between such transport factors and individual NPC components (see for example Damelin and Silver, 2000). However, the fundamental question as to how a translocating species is transferred from one side of the NPC to the other has remained largely unanswered. To shed some light on this process, we wanted to ask the apparently simple question regarding the rate(s) at which intact NPCs can mediate translocation. It obviously matters for possible models of the translocation process whether 1, 10, 100 or even more translocation events per second need to be explained. In fact, experimentally determined rate constants probably constitute the most crucial boundary condition that a faithful translocation model must satisfy. The availability of such data would finally permit testing of translocation models by computer simulation.
One way to determine rate constants for NPC passage is to measure the influx of a given fluorescent substrate into nuclei as the time-dependent increase in nuclear fluorescence. Considering the nuclear volume and the number of NPCs per nucleus, the rate can then be expressed as translocations per NPC per second. As numerous nuclei, each containing ∼2800 NPCs (see Materials and methods), can be viewed by fluorescence microscopy at a time, this procedure would average the translocation activity of a great number of NPCs. In order to come to meaningful numbers, however, a number of problems need first to be considered.
First, it is well established that the various transport pathways compete with each other for binding sites at the NPC. This complicates the kinetic analysis when transport processes are studied in vivo or in the presence of cytosolic extracts. This problem can be avoided by using permeabilized cells, which are largely depleted of endogenous nuclear transport receptors, and re-addition of a defined concentration of exogenous recombinant nuclear transport factors and transport substrates. A second complication is that translocation through NPCs is not necessarily the rate-limiting step of the overall transport process. Importin β-dependent import is a good example to illustrate this problem. In this case, translocation into the nucleus needs to be terminated by direct binding of RanGTP to importin β, which releases the substrate from importin β and importin β from the NPC (Moore and Blobel, 1993; Rexach and Blobel, 1995; Görlich et al., 1996). The rate of nuclear substrate accumulation will thus be a result of at least four individual rate constants: those for translocation of the importin β–substrate complex into the nucleus, import of RanGDP, nucleotide exchange to RanGTP and, finally, RanGTP binding to importin β. The translocation rate of the substrate–importin β complex cannot therefore be derived from the rate of substrate accumulation alone (although the latter sets a lower limit for the translocation rate). For an initial characterization, we therefore wanted to study translocation events that are not coupled to any other rate-limiting step.
Translocation of ‘empty transportin’ is exceedingly fast and mediated by weak interactions with NPCs
Such a minimalistic translocation system is represented by NPC passage of cargo-free (‘empty’) transport receptors. Empty exportins, for example, easily enter nuclei (Kutay et al., 1997a) to pick up export substrates; but also, importins can cross the NE independently of cargo loading (Kose et al., 1997; Nakielny and Dreyfuss, 1998). Transportin appeared to be particularly suitable for this study, because it has a surprisingly high concentration (>100 µM) of nuclear binding sites, from which it is normally released by RanGTP (not shown). In the absence of Ran, however, these binding sites trap transportin upon nuclear entry and make the otherwise fully reversible NPC passage pseudo-unidirectional.
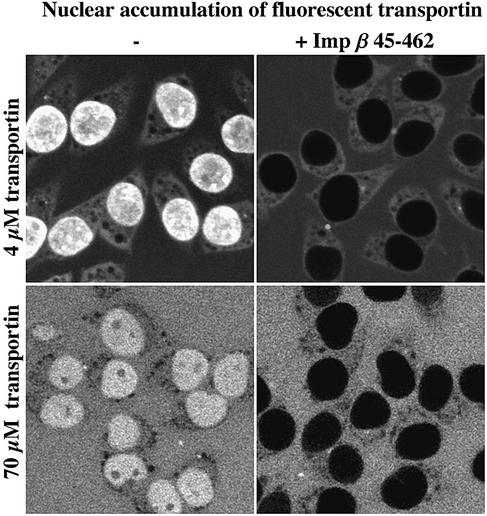
To allow detection by confocal fluorescence microscopy, we fluorescently labelled transportin (see Materials and methods). When added to permeabilized cells (Adam et al., 1990), the fluorescent protein accumulated brightly in the nuclei within 1 min (see Figure 1). At this point, it was crucial to test whether the nuclear influx of transportin represents a bona fide translocation process. To distinguish translocation from passive diffusion, we made use of the dominant-negative importin β 45–462 mutant, which binds virtually irreversibly to NPCs (Kutay et al., 1997b). This mutant has only a very mild effect on NPC passage by passive diffusion, reducing the influx of lysozyme, small dextrans or the green fluorescent protein (GFP) by no more than ∼30% (K.Ribbeck and D.Görlich, unpublished data). However, it imposes a very tight block (>100-fold inhibition) on facilitated translocation events, such as the classical nuclear localization signal (NLS) import pathway (Kutay et al., 1997b). Figure 1 shows that this mutant completely blocked nuclear influx of empty transportin (>200-fold reduction). The experiment therefore establishes that this process also depends on specific interactions with constituents of the NPC and thus represents, just as NLS import or mRNA export, a facilitated translocation event. For reasons detailed below, it is important to note that the mutant is effective at low (4 µM) as well as at high (70 µM) transportin concentrations.
Fig. 1. Transportin—a simple model substrate for facilitated translocation through NPCs. Alexa-labelled transportin (4 or 70 µM) was added to permeabilized cells and its distribution subsequently recorded by scanning with a confocal microscope through the unfixed samples. One minute was sufficient for a clear nuclear accumulation of transportin (left). The dominant-negative importin β 45–462 mutant (3 µM) blocked the transportin influx completely (right).
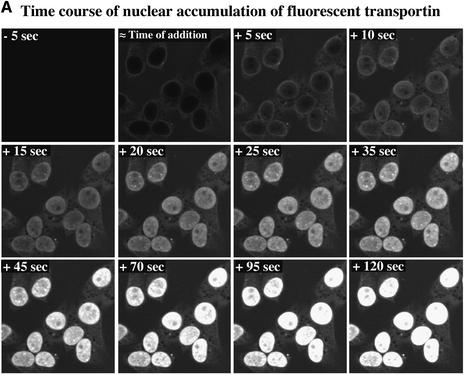
Nuclear accumulation of transportin turned out to be very fast and we therefore recorded the import reaction from the first few seconds onwards. Figure 2 shows that transportin equilibrated between cytoplasm and nucleus within ∼5 s, and accumulated rapidly in the nuclei, reaching an endpoint after ∼2 min.
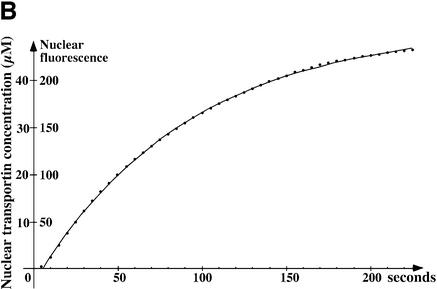
Fig. 2. Time course of transportin influx into nuclei. Fluorescent transportin (4 µM) was added to the permeabilized cells and its influx into nuclei recorded in real time by confocal microscopy. (A) Frames taken at indicated time points. (B) Integrated nuclear fluorescence (excluding the NPC signal) is plotted against time. The time course fits an exponential curve of the form f[t] = fMax(1 – e–kt), where t is time, f[t] is the nuclear fluorescence, fMax is the endpoint of the reaction and k is the first-order rate constant. For conversion of fluorescence intensities into absolute substrate concentration see Materials and methods. The initial rate of nuclear accumulation (given by ri = kfMax) for 4 µM transportin was found to be 0.6 µM/s, corresponding to ∼150 translocations·NPC–1·s–1 (average from 10 independent measurements).
To obtain quantitative values, we plotted the integrated intranuclear fluorescence against time (Figure 2B). The data points fit well to an exponential curve of the form f[t] = fMax(1 – e–kt), suggesting that the substrate enters nuclei according to first order kinetics. A calibration of the system permitted a conversion of the arbitrary fluorescence units into absolute substrate concentrations, while the nuclear volume and NPC density of HeLa nuclei imply that a change in nuclear concentration of 1 µM/s requires a flux of 250 molecules·NPC–1·s–1 (detailed in Materials and methods). At a transportin concentration of 4 µM, the nuclear concentration increased at an initial rate of 0.6 µM/s, corresponding to 150 translocations·NPC–1·s–1.
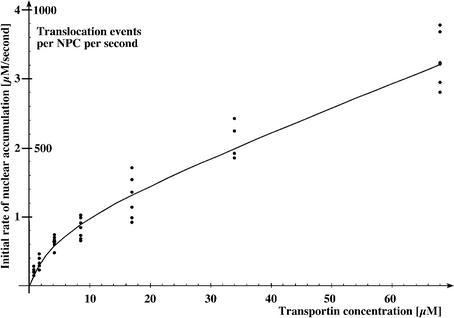
The net flux of transportin through NPCs should depend on its concentration difference between nucleus and cytoplasm. On the other hand, translocation proceeds through NPC-bound intermediates, and as the number of binding sites at the NPC is limited, an increasing substrate concentration should gradually saturate the translocation process. The characteristics of the saturation kinetics should, in turn, allow conclusions about affinities of the translocation-relevant interactions and the translocation capacity of an NPC. With this rationale in mind, we determined initial translocation rates for a wide range (0.85–68 µM) of transportin concentrations. A first conclusion from this series of experiments is that the translocation capacity of NPCs is indeed enormous; the data from Figure 3 suggest that a single NPC is able to translocate 800 transportin molecules per second, which is 30 times higher than the previously reported maximum translocation rate (Nemergut and Macara, 2000).
Fig. 3. Concentration dependence of transportin flux through NPCs. The initial rates of influx of fluorescent transportin were determined for concentrations of 0.85–68 µM (42 data points). A saturation of influx by increasing transportin concentrations is clearly evident. However, the dose dependence deviates from an ideal Michaelis–Menten curve of the form v = VMaxc/(KM + c), apparently because cooperativity in NPC passage becomes significant at higher transportin concentration. The fit was improved by introducing a correction factor (1 + k2c), which takes cooperativity into account and gives a modified Michaelis–Menten equation of the form
The numerically obtained best-fit parameters are: VMax = 300 events·NPC–1·s–1; KM = 4 µM; k2 = 0.03 µM–1.
For low substrate concentrations, the translocation rate increased initially linearly with transportin concentration; however, saturation became clearly evident from concentrations of ∼5–10 µM and above. The Michaelis–Menten equation (Michaelis and Menten, 1913) describes the simplest case of saturation kinetics, and applies provided two conditions are fulfilled: (i) the rate-limiting intermediates all form with identical dissociation constants; and (ii) no cooperativity between substrate molecules or other allosteric effects occur. For substrate concentrations up to 10–20 µM, the data points are consistent with a KM of ∼4 µM (see Figure 3). The KM approximates the dissociation constants for the translocation-relevant transportin–NPC interactions and the value of 4 µM implies that translocation is brought about by very weak and thus transient interactions. For comparison, the transportin–RanGTP interaction has a KD of ∼1 nM and is thus ∼4000-fold stronger (Bischoff and Görlich, 1997).
At substrate concentrations of ≥20 µM, the relationship between the translocation rate and substrate concentration deviated unexpectedly from an ideal Michaelis–Menten curve. With increasing substrate concentrations, a Michaelis–Menten curve approaches a fixed maximum rate, with its slope becoming zero. In contrast, the best-fitting curve to the data points approaches a line whose slope is clearly greater than zero (Figure 3). This could indicate that transportin can also use alternative paths through the NPC that become saturated only at extremely high substrate concentrations. It is important to note, however, that the enormous fluxes observed at high transportin concentrations can still be completely blocked by the importin β dominant-negative mutant (see Figure 1). This crucial control strongly suggests that these very high fluxes are not just caused by some non-selective damage to NPCs or an impaired integrity of the NE, but instead represent bona fide translocation events.
NPC passage of transportin–cargo complexes
A crucial question at this point was whether the characteristics we observed for NPC passage of empty transport receptors apply only to this type of translocating species or also to larger cargo–receptor complexes. To address this question, we studied import of a large fusion between the nucleoplasmin core domain and the M9 signal, which confers transportin-dependent nuclear import (Pollard et al., 1996). The fusion protein forms pentamers of 120 kDa and initial experiments showed that its import rate depends on the stoichiometry with respect to transportin. Import was comparably slow for one and fastest for five transportin molecules per pentamer (not shown).
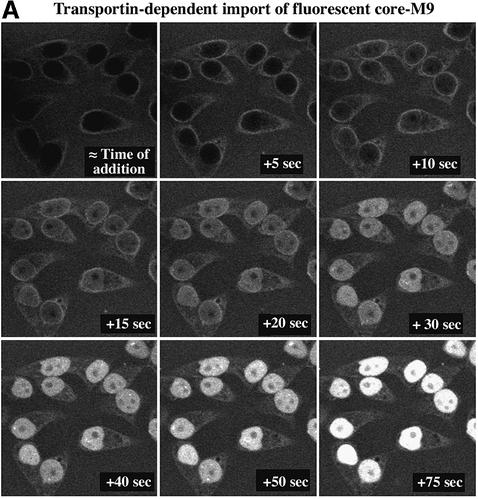
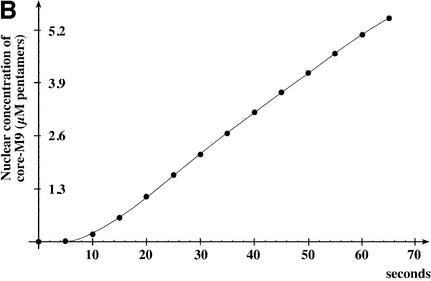
When 1 µM core–M9 pentamers were imported in the presence of Ran, an energy-regenerating system and 5 µM transportin, they accumulated in the nucleus at a rate of 0.11 µM/s, i.e. ∼28 pentameric core–M9–transportin complexes were translocated per NPC per second (Figure 4). This pentameric complex has a mol. wt of ∼620 kDa and therefore accounts for a mass flow of ∼17 MDa·NPC–1·s–1 (28 NPC–1 s–1 × 620 kDa). For comparison, 5 µM cargo-free transportin (100 kDa) results in a flux of ∼170 translocations·NPC–1·s–1 or a mass flow of ∼17 MDa·NPC–1·s–1. We can thus conclude that the mass flow of optimal substrate–transport receptor complexes through NPCs can be as high as that of the corresponding cargo-free receptors. This conclusion, however, only applies to optimal substrates; cargo domains that are not ‘streamlined’ for NPC passage can slow down the translocation process considerably (S.Jäkel, K.Ribbeck and D.Görlich, unpublished observation).
Fig. 4. Rate of core–M9 import into nuclei. The pentameric core–M9 fusion (Alexa labelled) was complexed with transportin at a 1:5 molar ratio. Import into nuclei was with 1 µM of this 630 kDa complex in the presence of Ran and an energy-regenerating system. (A) Time lapse images of M9 import. (B) Plot of integrated nuclear fluorescence (nuclear core–M9 concentration) versus time. The initial rate of nuclear accumulation was 0.11 µM/s, corresponding to 28 translocations·NPC–1·s–1 or a mass flow of ∼17 MDa·NPC–1·s–1.
NPC passage of NTF2
The concept of facilitated translocation through NPCs is clear and well established for objects that are larger than the assumed passive diffusion limit (40–60 kDa). Can facilitated translocation then also apply to smaller objects and, if so, how would it relate to ‘passive’ diffusion? NTF2 is an ideal object to investigate this question. It is a homodimer of two 15 kDa subunits (Moore and Blobel, 1994; Paschal and Gerace, 1995; Stewart et al., 1998), it mediates import of RanGDP (Ribbeck et al., 1998; Smith et al., 1998) and needs to return to the cytoplasm in an empty state to pick up another Ran molecule.
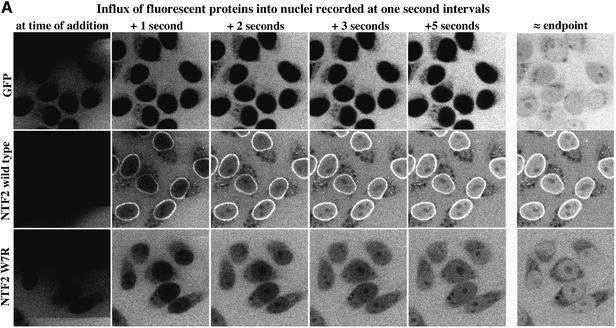
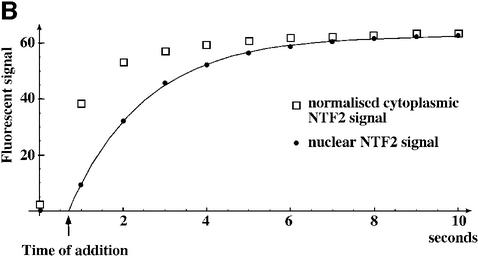
To study NPC passage of empty NTF2, we fluorescently labelled the protein to allow detection by fluorescence microscopy. NTF2 lacks prominent intranuclear binding sites and so the endpoint of the import reaction is a fairly even distribution between nucleus and cytoplasm (see the corresponding panels in Figure 5). Upon addition to the permeabilized cells, NTF2 instantaneously bound to NPCs and equilibrated between nucleus and cytoplasm with a half-time of 0.7 s (Figure 5A and B).
Fig. 5. Comparison of NTF2 flux through NPCs with that of GFP. (A) Comparison of influx of GFP (5 µM), wild-type NTF2 (2.5 µM dimers) and the NTF2 W7R mutant into nuclei. Note that NTF2 nearly instantaneously equilibrated between nucleus and cytoplasm, while GFP remained excluded from the nuclei when tested on the same time scale. Endpoints were 2 min for GFP, 12 s for wild-type NTF2 and 60 s for the NTF2 W7R mutant. (B) Quantitation of NTF2 (wild type) influx into nuclei. Equilibration between the nuclear and cytoplasmic NTF2 pools occurs with a first-order rate constant of k = 1 s–1 or a half-time of ∼0.7 s. This implies that a nucleocytoplasmic concentration difference of 2.5 µM causes a change in nuclear NTF2 concentration at an initial rate of 2.5 µM/s, which corresponds to a flux of ∼620 NTF2 dimers·NPC–1·s–1 (for details see Supplementary data).
NTF2 might pass through NPCs so fast because it is subject to a similar translocation mechanism to transportin or because any molecule of this size can diffuse passively through NPCs at this extremely high rate. One can distinguish between these two possibilities by comparing the flux of NTF2 through NPCs with that of other molecules of similar size and comparable diffusion coefficient. We chose GFP as a reference molecule, because it has a similar molecular weight (28 kDa) to the NTF2 dimer (29 kDa). Gel filtration experiments (see Materials and methods; Table I) indicated that GFP has a slightly smaller Stokes’ radius (2.35 nm) than NTF2 (2.50 nm), suggesting that GFP diffuses in aqueous medium somewhat faster than NTF2. If the flux of small macromolecules through NPCs was solely determined by their dimensions and diffusion coefficients, then GFP should enter nuclei at a similar rate to or even faster than NTF2. This assumption is contradicted by the data from Figure 5 and Table I: GFP traverses NPCs ∼120 times slower than NTF2. Similar results to those for GFP were also obtained using dextrans as inert reference molecules (data not shown). This strongly suggests that NTF2 crosses the NPC in a facilitated manner, which is also consistent with the observation that NPC passage of NTF2 is very sensitive towards specific inhibitors of facilitated translocation (see Supplementary data, available at The EMBO Journal Online).
Table I.
| Transport species | Mol. wt (kDa) | Radius (nm) | Translocations through NPCs at Δc = 1 µM (NPC–1 s–1) | Diffusion rate through a hypothetical ‘plugless’ NPC at Δc = 1 µM (pore–1 s–1) |
|---|---|---|---|---|
| Complex of core–M9 pentamers + five transportins | 630 | 8.17 | 28 | 120 |
| Transportin | 100 | 4.13 | 65 | 430 |
| BSA | 68 | 3.55 | <0.1 | 540 |
| GFP | 29 | 2.36 | 2 | 920 |
| NTF2 | 29.5 | 2.51 | 250 | 850 |
| NTF2 W7R | 29.5 | 2.51 | 60 | 850 |
Tryptophan 7 in NTF2 has previously been shown to be crucial for interaction with Phe-rich repeats and for translocation of the NTF2–RanGDP complex (Bayliss et al., 1999). Mutation of this residue to arginine (W7R) not only abrogates the prominent NPC staining of NTF2, but also significantly delays passage through NPCs (Figure 5). It is, however, interesting to note that W7R-NTF2 still passes NPCs ∼30 times faster than GFP, suggesting that the exposed tryptophan 7 is not the only residue in NTF2 with translocation-promoting properties and that the remaining extremely low affinity for NPC components is sufficient to facilitate NPC passage. We are currently performing systematic mutagenesis to test which surface properties of NTF2 and GFP account for their different translocation behaviours.
Facilitated translocation of optimal substrates occurs at rates expected for a purely diffusion-controlled reaction
The quantitation of NTF2 influx into nuclei (see Figure 5B; Table I) shows that a nucleocytoplasmic concentration difference of 2.5 µM NTF2 dimers results in a flux of ∼620 dimers·NPC–1·s–1, which is approximately four times faster than the transportin flux at the same concentration (see Figures 3 and 4; Table I). Higher NTF2 concentrations also resulted in higher fluxes, and the dose dependence of NPC passage of NTF2 appears analogous to that of transportin (data not shown). At 100 µM NTF2 dimers, for example, ∼2500 translocations·NPC–1·s–1 were observed.
It is, however, most remarkable that very high translocation rates occur even at comparably low NTF2 concentrations. Under subsaturating conditions, NPCs catalyse nucleocytoplasmic exchange of NTF2 with a specificity constant (kcat/KM) of 2.5 × 108 M–1 s–1 (see Materials and methods), which is close to the number expected for purely diffusion-controlled reactions. This prompted us to compare the rates for facilitated translocation with free diffusion through a hypothetical NPC that lacks its permeability barrier and is instead filled only by aqueous medium.
The rate of free diffusion depends on the dimension of the channel. The functional diameter of NPCs has been determined using nucleoplasmin-coated gold and the largest particles that could still pass NPCs had a diameter of 25 nm, including the nucleoplasmin coat (Feldherr et al., 1984). Since nucleoplasmin has to bind importin α/β heterodimers before NPC passage, which probably adds an ∼8 nm layer of importins, we can conclude that the actual translocating species (and thus the channel) probably had a total diameter of ∼40 nm.
Assuming the central channel has a diameter of 40 nm and a length of 40 nm, we calculated the rates of free diffusion as shown in Table I (for details see Materials and methods). The numbers show that the permeability barrier of the NPC restricts the flux of GFP and bovine serum albumin (BSA) by factors of 500 and 5000, respectively, as compared with the diffusion through a hypothetical plug-free channel. The passage of NTF2 is, however, <4-fold restricted (see Table I). In this context, it is interesting to note that the high concentration of macromolecules in nucleus and cytoplasm makes the medium there so viscous that diffusion of a 30 kDa protein is also slowed down 3- to 5-fold compared with a protein-free solution (Wachsmuth et al., 2000). This in turn implies that NTF2 moves through the permeability barrier of the NPC not significantly slower than it diffuses through the nucleus or cytoplasm. The same even applies to the 620 kDa core–M9–transportin complex (Table I).
Discussion
Translocation through NPCs is a massive process
The mechanism of facilitated translocation through NPCs is one of the central problems in nucleocytoplasmic transport. Here we present a detailed study on the kinetics of the process. We observed that the maximum translocation rates and the translocation capacity of NPCs are much higher than previously anticipated. For example, a single NPC is capable of translocating 2500 NTF2 homodimers or ∼800 transportin molecules per second. Considering transportin’s mol. wt of 100 kDa, this corresponds to a mass flow of 80 MDa/s. In other words, it takes only a little more than 1 s to shuffle the mass of an NPC (125 MDa) through an NPC.
Given the dimensions of an NPC, translocation must occur over a significant distance and at a remarkable velocity. The observed rate of M9 import, for example, indicates that M9–transportin complexes move at at least 0.5 µm/s from the cytoplasmic to the nuclear side of the central channel (see Supplementary data).
A mass flow of 80 MDa·NPC–1·s–1, as in the case of cargo-free transportin, appears to be a very large value. It is therefore important to note that import of M9 substrates (Figure 4), importin β-dependent import or NTF2- mediated import of Ran (K.Ribbeck and D.Görlich, unpublished) can occur in vitro with total mass flows of ∼15–40 MDa·NPC–1·s–1. As detailed in the Supplementary data, we estimate that proliferating HeLa cells in vivo use a nuclear transport capacity of 10–20 MDa·NPC–1·s–1, indicating that the available capacity is probably not a limiting factor for cellular processes.
Others have estimated an NPC capacity of only ∼4 translocations·NPC–1·s–1 (Keminer et al., 1999). They used importin α/β or CRM1-dependent transport substrates with crude cellular extracts as a source for nuclear transport receptors. It therefore needs to be considered that their system assayed complex transport cycles and not actual translocation rates, simply because other steps (such as binding of the substrate to the limiting receptors) were probably rate limiting. In fact, our preliminary data indicate that, for example, importin β-dependent substrates can be imported at rates of ≥100 translocations·NPC–1·s–1.
Translocation is mediated by very low affinity interactions
For sufficiently low concentrations (≤10 µM), the saturation of transportin’s NPC passage by an increasing transportin concentration can be approximated by a Michaelis–Menten equation with a KM of ∼4 µM. This suggests that no obligatory translocation intermediates form with a KD lower than this KM for the overall process (otherwise, the process would become saturated at a lower transportin concentration). This in turn implies that all interactions relevant to transportin translocation must be of very low affinity (KD ≥4 µM). Considering the translocation process, this low affinity for NPC components and the associated high off-rates make perfect sense and are probably crucial for the high translocation rates observed for transportin, simply because the overall translocation process cannot be faster than the terminal dissociation from the NPC.
The KM of ∼4 µM is similar to the dissociation constants reported for binding of NTF2 to an individual Phe-rich nucleoporin repeat (KD ≈ 20 µM) or to an 18-repeat domain (KD ≈ 1.4 µM) (Bayliss et al., 1999; Chaillan-Huntington et al., 2000); it is therefore consistent with the assumption that such interactions with Phe-rich repeats represent the translocation-relevant ones. However, even much lower affinities for NPC components can be sufficient to promote facilitated translocation. This is illustrated by the W7R NTF2 mutant, where the principal binding site for Phe-rich repeats has been eliminated (Bayliss et al., 1999). In contrast to wild-type NTF2, this mutant does not become concentrated at NPCs (Figure 5A) and thus binds NPC components with a KD of probably >100 µM (see also Bayliss et al., 1999). Yet, it permeates through the pores 30 times faster than GFP (Figure 5A; Table I).
Rout et al. (2000) proposed that NPC passage becomes facilitated when the residence time of transport substrates at the entrance of the translocation channel is increased by a binding of these substrates to nucleoporins. This might contribute to the translocation of some transport species; however, our data make it unlikely that this pinpoints the general principle of facilitated translocation. According to Rout’s model, facilitation of NPC passage should be directly proportional to the factor by which the translocating species becomes concentrated at NPCs. The W7R NTF2 mutant, however, demonstrates that concentrating the translocating species at NPCs is not a prerequisite for a facilitated NPC passage.
A model for the facilitated translocation process: the selective phase hypothesis
NTF2 is of similar size to GFP, but passes through NPCs 120 times faster than GFP (Figure 5; Table I). This is, however, still ∼4-fold slower than an unrestricted, free diffusion through a hypothetical NPC whose central channel is filled only with aqueous medium. This is an important point and implies that no mechanism needs to be postulated by which the translocating species is ‘propelled’ from one side of the NPC to the other. Instead, the phenomena of ‘translocation’ and selectivity of NPC can be more simply explained by NPCs restricting the free flow of some molecules more strongly than that of others. We consider it unlikely that NPCs contain differently sized channels for passive diffusion and facilitated translocation. Instead, we propose that translocation proceeds through a rather homogeneous medium that restricts movement of inert molecules, but is permeable for objects with certain ‘translocation-promoting’ properties that in turn correlate with an affinity for certain NPC components.
How could such a mechanism work? Before we attempt an answer for NPCs, we would like to draw a parallel to lipid bilayers, another type of cellular permeability barrier. Lipid bilayers also show a size-selective permeability and are, for example, more permeable for water (mol. wt 18) than for sucrose (mol. wt 342). However, size is not the only criterion. Lipid bilayers are also more permeable for lipophilic molecules than for charged ones, simply because the transfer of ions from water into the hydrophobic lipid phase is energetically very unfavourable. In contrast, a lipophilic molecule can easily partition into the hydrophobic core, subsequently exit the bilayer on the other side and thus cross the lipid bilayer at a high rate.
The only candidate for the morphological equivalent of the permeability barrier within the NPC is the central plug, a 12 MDa structure with low electron density that fills the central channel (Reichelt et al., 1990). The central plug might work analogously to a lipid bilayer, although on a much larger scale. It might constitute a semi-liquid phase, into which transport receptors can easily partition, but inert molecules above a certain size cannot. Transport receptors would then promote the translocation of cargo molecules simply by increasing their ‘solubility’ in the central plug. How then could the central plug form a discrete, semi-liquid phase with such properties?
Several lines of evidence support the assumption that Phe-rich repeats are major and functionally relevant constituents of the central plug. First, monoclonal antibodies recognizing such repeats, or wheat germ agglutinin, which binds sugars within the repeat regions, also stain the central channel (Akey and Goldfarb, 1989). Secondly, facilitated passage of empty NTF2 (this study), NTF2–Ran (Bayliss et al., 1999) or importin β–cargo complexes (Bayliss et al., 2000) through the central plug requires a certain affinity between the translocating species and Phe-rich repeats. Finally, the repeats are estimated to be present in >1000 copies/NPC (Bayliss et al., 1999) and would thus be sufficiently abundant to constitute the principal structural element of the central plug.
The properties of liquids are largely determined by the interactions between the solvent molecules. Accordingly, a key element of our model is a mutual attraction between repeats, possibly through hydrophobic interactions between the Phe-rich clusters. This attraction would ensure the structural integrity of the plug, while the presence of the hydrophilic spacers between the Phe-rich clusters would prevent a collapse of the structure. It is easy to imagine how these interactions could create a sieve-like structure that allows the passage of small molecules but restricts the flow of larger ones (see Figure 6). Passage of objects larger than the mesh size would be required to break the meshes locally. This could occur when the translocating species (such as a transport receptor) locally competes the repeat–repeat interaction by a direct binding to the repeats (Figure 6). The translocating species would then become part of the meshwork. In other words, the plug would seal around the translocating species and remain a barrier for inert molecules even when large objects pass.
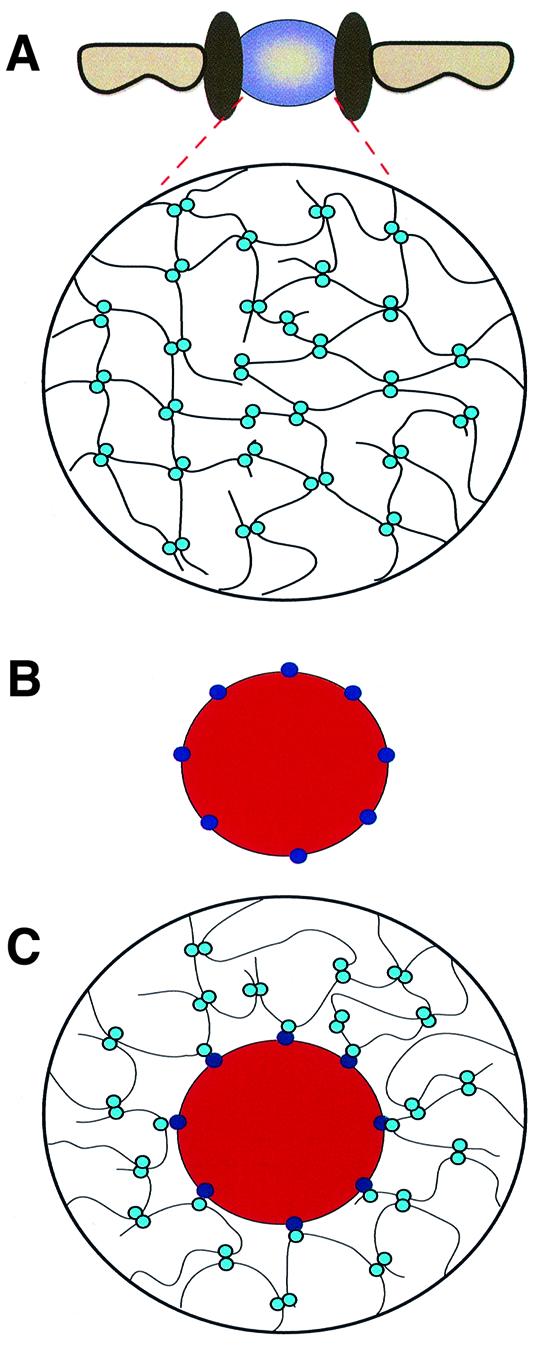
Fig. 6. The selective phase model for facilitated translocation through NPCs. (A) The permeability barrier of the central plug ‘at rest’. Phe-rich nucleoporin repeats (light blue circles) attract each other and form a meshwork that restricts passage of inert objects. (B) An object capable of facilitated translocation, distinguished from inert objects by binding sites (dark blue circles) for the Phe-rich repeats. (C) The translocating species interacts with the Phe-rich repeats and thus becomes part of the meshwork. It dissolves in the central plug, which allows crossing of the permeability barrier.
The local concentration of Phe-rich repeats inside the central plug has been estimated to be 50 mM (Bayliss et al., 1999). The permeability barrier could therefore be maintained by millimolar dissociation constants for the repeat–repeat interactions. This predicted very weak nature of these interactions has two consequences that are relevant to our model. First, even weak receptor–repeat interactions could efficiently compete the attraction between the repeats. Secondly, a rearrangement of all these interactions during translocation would require only very low activation energies. This would make the meshwork sufficiently dynamic to accommodate the tremendous fluxes that occur through NPCs.
The barrier created by the mechanisms described above cannot be an absolute one. Inert objects above the passive diffusion limit could also break into the meshes. However, this would be energetically unfavourable because the free energy required to disrupt the inter-repeat attractions would not be sufficiently compensated by the energy released from newly formed interactions between the translocating species and the repeats.
One can also see the contribution of the Phe-rich repeats to the selectivity of NPCs from a slightly different perspective. Exposed hydrophobic groups cause a reorganization of the normal hydrogen bonding network in water and force the nearby water molecules into a more ordered state. The central plug with ∼50 mM hydrophobic clusters should contain very little ‘undisturbed’ water and therefore constitute an energetically unfavourable environment for highly hydrated macromolecules. Such an entropic effect should also restrict passage of (hydrophilic) inert molecules but not that of transport receptors with a sufficiently hydrophobic surface.
The question remains as to how transport receptors could promote NPC passage of cargoes that cannot cross the NPC on their own. First, the receptor can cover those regions of the cargo whose interaction with the Phe-rich repeat phase is energetically unfavourable. This scenario probably applies to histone H1, whose import receptor, the importin β–importin 7 heterodimer, appears to cover most of its highly charged surface (Jäkel et al., 1999). In cases where a small signal such as an SV-40 NLS is recognized, the receptor will probably not cover much more than the signal. However, the receptor–repeat interaction can compensate for the free energy required to transfer the cargo into an environment where it is normally poorly soluble, and thereby promote translocation.
Materials and methods
Fluorescent proteins
Transportin, Ran, NTF2 and the core–M9 fusion protein were expressed and purified essentially as described (Ribbeck et al., 1999). The W7R NTF2 mutant was generated by PCR and verified by DNA sequencing. Expression was from pQE60. Gel filtration confirmed that the mutation did not affect homodimerization. GFP was expressed with a C-terminal His tag and purified on Ni-NTA–agarose followed by chromatography on Superdex 75.
Labelling with Alexa 488 maleimide was performed in Tris buffer pH 7.5 on ice, using 0.8 Alexa molecules per transportin or per NTF2 homodimer, or two Alexa molecules per core–M9 pentamer.
Import assays
HeLa cells (‘HeLa B’; European Cell Culture Collection, ECACC code b85060701) were grown on coverslips to ∼30–50% confluence, washed in cold permeabilization buffer (20 mM HEPES–KOH pH 7.4, 120 mM potassium acetate, 5 mM magnesium acetate, 0.5 mM EGTA, 250 mM sucrose), permeabilized for 15 min with 60 µg/ml digitonin, washed three times in digitonin-free buffer and kept on ice until use.
The coverslip with permeabilized cells was then fixed to the stage of an inverted confocal microscope (Leica TCS NT, used with a 63× water-immersion objective). Texas red dextran 70 000 (which remains excluded from the nuclei) was added, scans in the fluorescein and Texas channels were started in time-lapse mode and the focal plane was adjusted to the equator of the nuclei. The import reaction was then started by replacing the dextran solution by the import mixture containing Alexa 488-, fluorescein- or GFP-labelled proteins. Import was performed at 20°C in permeabilization buffer, except for influx of transportin (Figures 1–3). Transportin without substrates appears unstable in a HEPES–potassium acetate buffer. Therefore, HEPES was replaced by Tris–HCl, and 120 mM potassium acetate by 30 mM NaCl + 90 mM potassium acetate.
Quantitation of import
The signals over nuclei (excluding the signal at NPCs) were integrated using an automated routine and the histogram function of Adobe Photoshop 5.0™. Data points were then interpolated using the ‘Fit’, ‘NonlinearFit’ or ‘PolynomialFit’ functions from Mathematica 4.0.
Factors to convert fluorescence intensities into absolute substrate concentrations were obtained for each scan from the fluorescence signal outside the cells, which corresponds to substrate concentration added to the import reaction. Under the microscope settings used, the recorded signal was found to be linear with the concentration of added fluorescently labelled protein.
These calculations gave the concentration of the fluorescent protein as a function of time (f[t]). The derivative of this function (df[t]/dt) gives the flux at given time points (as µM/s) and was used to calculate initial influx rates.
Number of NPCs and volume in HeLa nuclei and calculation of fluxes
The factor for converting µM/s into translocations·NPC–1·s–1 depends on the nuclear volume and the number of NPCs per nucleus. To determine the density of NPC in NEs, we decorated NPCs in permeabilized HeLa cells with a fluroescent 45–462 importin β fragment and visualized them by confocal microscopy. Counting their number in given areas gave 5.1 ± 0.2 NPCs/µm2, which agrees well with the number reported by Kubitscheck et al. (1996) for permeabilized 3T3 cells.
The nuclei we used have an ellipsoid shape with average radii of 8.0 ± 1.0, 6.4 ± 0.8 and 5.2 ± 0.7 µm in the three axes, a volume of ∼1130 µm3 and a surface area of ∼540 µm2. They contain ∼2770 NPCs, which agrees well with 2000 and 4000 NPCs for G1 and G2 cells, respectively, as determined by electron microscopy (Maul et al., 1972). Our nuclei have a larger volume than those in the Maul study, which might be due to shrinking during fixation for electron microscopy and nuclear decondensation upon cell permeabilization. For our study, it was crucial, however, to consider the nuclear dimensions under import conditions.
The total flux of a given species between nucleus and cytoplasm can be described by
where dc/dt is the change in concentration with time, VNuc is the nuclear volume, NA is Avogadro’s number and NNPC is the number of NPCs per nucleus. A change in nuclear concentration by 1 µM/s therefore equals in HeLa cells a flux (FluxNPC) of ∼250 molecules·NPC–1·s–1.
Determination of Stokes’ radii
A Superdex 200 column was calibrated using standards with known Stokes’ radii (Cabré and Canela, 1989), namely thyroglobulin (8.6 nm), ferritin (6.06 nm), aldolase (4.60 nm), BSA (3.55 nm), ovalbumin (2.73 nm), chymotrypsinogen (2.24 nm), myglobin (2.08 nm) and cytochrome c (1.65 nm). Plasmid DNA and GDP were used to determine the void and total volumes, respectively. This calibration allowed us to estimate the Stokes radii of GFP (2.36 nm), NTF2 (2.51 nm), transportin (4.13 nm) and the transportin–core–M9 complex (8.17 nm) from their retention times.
Calculation of diffusion coefficients
The diffusion coefficients (D) for GFP and NTF2 were calculated according to D = kT/6πηRS, where k is the Boltzmann constant, T is the temperature (293K), RS is the Stokes radius and η is the viscosity of the medium (assuming 0.0145 g·cm–1·s–1 at 20°C for our import buffer that contains 250 mM sucrose). The following values were obtained: 0.627 × 10–6 cm2/s for GFP, 0.59 × 10–6 cm2/s for NTF2, 0.417 × 10–6 cm2/s for BSA, 0.358 × 10–6 cm2/s for transportin and 0.181 × 10–6 cm2/s for the pentameric transportin–core–M9 complex.
Estimation of fluxes through a hypothetical NPC that lacks the central plug
The flux through a plugless NPC (Table I) can be described by Fick’s first law:
where D is the diffusion coefficient, R is the radius of the channel (20 nm), Δc is the nucleocytoplasmic concentration difference, NA is Avogadro’s number and L is the channel length (40 nm). The correction term Kcorr = (1 – r/R)2 extends the validity of Fick’s law from point-like objects to objects with the radius r.
Supplementary data
Supplementary data to this paper are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank Drs M.Pool, T.A.Rapoport, I.Vetter and D.Oesterhelt, the members of our laboratory, I.W.Mattaj and the anonymous reviewers for stimulating discussions/constructive comments on the manuscript. This work was supported by grants from the DFG and the HFSPO (RG0198/1998M).
References
- Adam S.A., Marr,R.S. and Gerace,L. (1990) Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J. Cell Biol., 111, 807–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akey C.W. and Goldfarb,D.S. (1989) Protein import through the nuclear pore complex is a multistep process. J. Cell Biol., 109, 971–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akey C.W. and Radermacher,M. (1993) Architecture of the Xenopus nuclear pore complex revealed by three-dimensional cryo-electron microscopy. J. Cell Biol., 122, 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachi A. et al. (2000) The C-terminal domain of TAP interacts with the nuclear pore complex and promotes export of specific CTE-bearing RNA substrates. RNA, 6, 136–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayliss R., Ribbeck,K., Akin,D., Kent,H.M., Feldherr,C.M., Görlich,D. and Stewart,M. (1999) Interaction between NTF2 and xFxFG-containing nucleoporins is required to mediate nuclear import of RanGDP. J. Mol. Biol., 293, 579–593. [DOI] [PubMed] [Google Scholar]
- Bayliss R., Littlewood,T. and Stewart,M. (2000) Structural basis for the interaction between FxFG nucleoporin repeats and importin-β in nuclear trafficking. Cell, 102, 99–108. [DOI] [PubMed] [Google Scholar]
- Bischoff F.R. and Görlich,D. (1997) RanBP1 is crucial for the release of RanGTP from importin β-related nuclear transport factors. FEBS Lett., 419, 249–254. [DOI] [PubMed] [Google Scholar]
- Bischoff F.R. and Ponstingl,H. (1991) Catalysis of guanine nucleotide exchange on Ran by the mitotic regulator RCC1. Nature, 354, 80–82. [DOI] [PubMed] [Google Scholar]
- Bonner W.M. (1978) Protein migration and accumulation in nuclei. In Busch,H. (ed.), The Cell Nucleus. Vol. 6, Part C. Academic Press, New York, NY, pp. 97–148.
- Cabré F. and Canela,E. (1989) Accuracy and precision in the determination of Stokes radii and molecular masses of proteins by gel filtration chromatography. J. Chromatogr., 472, 347–356. [DOI] [PubMed] [Google Scholar]
- Chaillan-Huntington C., Braslavsky,C.V., Kuhlmann,J. and Stewart,M. (2000) Dissecting the interactions between NTF2, RanGDP and the nucleoporin XFXFG repeats. J. Biol. Chem., 275, 5874–5879. [DOI] [PubMed] [Google Scholar]
- Clarkson W.D., Kent,H.M. and Stewart,M. (1996) Separate binding sites on nuclear transport factor 2 (NTF2) for GDP-Ran and the phenylalanine-rich repeat regions of nucleoporins p62 and Nsp1p. J. Mol. Biol., 263, 517–524. [DOI] [PubMed] [Google Scholar]
- Damelin M. and Silver,P.A. (2000) Mapping interactions between nuclear transport factors in living cells reveals pathways through the nuclear pore complex. Mol. Cell, 5, 133–140. [DOI] [PubMed] [Google Scholar]
- Englmeier L., Olivo,J.C. and Mattaj,I.W. (1999) Receptor-mediated substrate translocation through the nuclear pore complex without nucleotide triphosphate hydrolysis. Curr. Biol., 9, 30–41. [DOI] [PubMed] [Google Scholar]
- Fabre E. and Hurt,E. (1997) Yeast genetics to dissect the nuclear pore complex and nucleocytoplasmic trafficking. Annu. Rev. Genet., 31, 277–313. [DOI] [PubMed] [Google Scholar]
- Feldherr C.M. (1962) The nuclear annuli as pathways for nucleocytoplasmic exchanges. J. Cell Biol., 14, 65–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldherr C.M., Kallenbach,E. and Schultz,N. (1984) Movement of karyophilic protein through the nuclear pores of oocytes. J. Cell Biol., 99, 2216–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich D. and Kutay,U. (1999) Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol., 15, 607–660. [DOI] [PubMed] [Google Scholar]
- Görlich D., Pante,N., Kutay,U., Aebi,U. and Bischoff,F.R. (1996) Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO J., 15, 5584–5594. [PMC free article] [PubMed] [Google Scholar]
- Jäkel S., Albig,W., Kutay,U., Bischoff,F.R., Schwamborn,K., Doenecke,D. and Görlich,D. (1999) The importin β/importin 7 heterodimer is a functional nuclear import receptor for histone H1. EMBO J., 18, 2411–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keminer O., Siebrasse,J.P., Zerf,K. and Peters,R. (1999) Optical recording of signal-mediated protein transport through single nuclear pore complexes. Proc. Natl Acad. Sci. USA, 96, 11842–11847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kose S., Imamoto,N., Tachibana,T., Shimamoto,T. and Yoneda,Y. (1997) Ran-unassisted nuclear migration of a 97-kD component of nuclear pore-targeting complex. J. Cell Biol., 139, 841–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubitscheck U., Wedekind,P., Zeidler,O., Grote,M. and Peters,R. (1996) Single nuclear pores visualized by confocal microscopy and image processing. Biophys. J., 70, 2067–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutay U., Bischoff,F.R., Kostka,S., Kraft,R. and Görlich,D. (1997a) Export of importin α from the nucleus is mediated by a specific nuclear transport factor. Cell, 90, 1061–1071. [DOI] [PubMed] [Google Scholar]
- Kutay U., Izaurralde,E., Bischoff,F.R., Mattaj,I.W. and Görlich,D. (1997b) Dominant-negative mutants of importin-β block multiple pathways of import and export through the nuclear pore complex. EMBO J., 16, 1153–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattaj I.W. and Englmeier,L. (1998) Nucleocytoplasmic transport: the soluble phase. Annu. Rev. Biochem., 67, 265–306. [DOI] [PubMed] [Google Scholar]
- Maul G.G., Maul,H.M., Scogna,J.E., Lieberman,M.W., Stein,G.S., Hsu,B.Y. and Borun,T.W. (1972) Time sequence of nuclear pore formation in phytohemagglutinin-stimulated lymphocytes and in HeLa cells during the cell cycle. J. Cell Biol., 55, 433–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis L. and Menten,M. (1913) Biochem. Z., 49, 333. [Google Scholar]
- Moore M.S. and Blobel,G. (1993) The GTP-binding protein Ran/TC4 is required for protein import into the nucleus. Nature, 365, 661–663. [DOI] [PubMed] [Google Scholar]
- Moore M.S. and Blobel,G. (1994) Purification of a Ran-interacting protein that is required for protein import into the nucleus. Proc. Natl Acad. Sci. USA, 91, 10212–10216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakielny S. and Dreyfuss,G. (1998) Import and export of the nuclear protein import receptor transportin by a mechanism independent of GTP hydrolysis. Curr. Biol., 8, 89–95. [DOI] [PubMed] [Google Scholar]
- Nakielny S. and Dreyfuss,G. (1999) Transport of proteins and RNAs in and out of the nucleus. Cell, 99, 677–690. [DOI] [PubMed] [Google Scholar]
- Nemergut M.E. and Macara,I.G. (2000) Nuclear import of the Ran exchange factor, RCC1, is mediated by at least two distinct mechanisms. J. Cell Biol., 149, 835–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paine P.L., Moore,L.C. and Horowitz,S.B. (1975) Nuclear envelope permeability. Nature, 254, 109–114. [DOI] [PubMed] [Google Scholar]
- Paschal B.M. and Gerace,L. (1995) Identification of NTF2, a cytosolic factor for nuclear import that interacts with nuclear pore protein p62. J. Cell Biol., 129, 925–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard V.W., Michael,W.M., Nakielny,S., Siomi,M.C., Wang,F. and Dreyfuss,G. (1996) A novel receptor-mediated nuclear protein import pathway. Cell, 86, 985–994. [DOI] [PubMed] [Google Scholar]
- Radu A., Moore,M.S. and Blobel,G. (1995) The peptide repeat domain of nucleoporin Nup98 functions as a docking site in transport across the nuclear pore complex. Cell, 81, 215–222. [DOI] [PubMed] [Google Scholar]
- Reichelt R., Holzenburg,A., Buhle,E.L.,Jr, Jarnik,M., Engel,A. and Aebi,U. (1990) Correlation between structure and mass distribution of the nuclear pore complex and of distinct pore complex components. J. Cell Biol., 110, 883–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rexach M. and Blobel,G. (1995) Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors and nucleoporins. Cell, 83, 683–692. [DOI] [PubMed] [Google Scholar]
- Ribbeck K., Lipowsky,G., Kent,H.M., Stewart,M. and Görlich,D. (1998) NTF2 mediates nuclear import of Ran. EMBO J., 17, 6587–6598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribbeck K., Kutay,U., Paraskeva,E. and Görlich,D. (1999) The translocation of transportin–cargo complexes through nuclear pores is independent of both Ran and energy. Curr. Biol., 9, 47–50. [DOI] [PubMed] [Google Scholar]
- Rout M.P. and Blobel,G. (1993) Isolation of the yeast nuclear pore complex. J. Cell Biol., 123, 771–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout M.P., Aitchison,J.D., Suprapto,A., Hjertaas,K., Zhao,Y. and Chait,B.T. (2000) The yeast nuclear pore complex: composition, architecture and transport mechanism. J. Cell Biol., 148, 635–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan K.J. and Wente,S.R. (2000) The nuclear pore complex: a protein machine bridging the nucleus and cytoplasm. Curr. Opin. Cell Biol., 12, 361–371. [DOI] [PubMed] [Google Scholar]
- Schwoebel E.D., Talcott,B., Cushman,I. and Moore,M.S. (1998) Ran-dependent signal-mediated nuclear import does not require GTP hydrolysis by Ran. J. Biol. Chem., 273, 35170–35175. [DOI] [PubMed] [Google Scholar]
- Smith A., Brownawell,A. and Macara,I.G. (1998) Nuclear import of Ran is mediated by the transport factor NTF2. Curr. Biol., 8, 1403–1406. [DOI] [PubMed] [Google Scholar]
- Stewart M., Kent,H.M. and McCoy,A.J. (1998) Structural basis for molecular recognition between nuclear transport factor 2 (NTF2) and the GDP-bound form of the Ras-family GTPase Ran. J. Mol. Biol., 277, 635–646. [DOI] [PubMed] [Google Scholar]
- Strasser K., Bassler,J. and Hurt,E. (2000) Binding of the Mex67p/Mtr2p heterodimer to FXFG, GLFG and FG repeat nucleoporins is essential for nuclear mRNA export. J. Cell Biol., 150, 695–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachsmuth M., Waldeck,W. and Langowski,J. (2000) Anomalous diffusion of fluorescent probes inside living cell nuclei investigated by spatially-resolved fluorescence correlation spectroscopy. J. Mol. Biol., 298, 677–689. [DOI] [PubMed] [Google Scholar]
- Weis K. (1998) Importins and exportins: how to get in and out of the nucleus. Trends Biochem. Sci., 23, 185–189. [DOI] [PubMed] [Google Scholar]
- Yang Q., Rout,M.P. and Akey,C.W. (1998) Three-dimensional architecture of the isolated yeast nuclear pore complex: functional and evolutionary implications. Mol. Cell, 1, 223–234. [DOI] [PubMed] [Google Scholar]