Abstract
Simple base damages are repaired through a short-patch base excision pathway where a single damaged nucleotide is removed and replaced. DNA polymerase β (Pol β) is responsible for the repair synthesis in this pathway and also removes a 5′-sugar phosphate residue by catalyzing a β-elimination reaction. How ever, some DNA lesions that render deoxyribose resistant to β-elimination are removed through a long-patch repair pathway that involves strand displacement synthesis and removal of the generated flap by specific endonuclease. Three human DNA polymerases (Pol β, Pol δ and Pol ε) have been proposed to play a role in this pathway, however the identity of the polymerase involved and the polymerase selection mechanism are not clear. In repair reactions catalyzed by cell extracts we have used a substrate containing a reduced apurinic/apyrimidinic (AP) site resistant to β-elimination and inhibitors that selectively affect different DNA polymerases. Using this approach we find that in human cell extracts Pol β is the major DNA polymerase incorporating the first nucleotide during repair of reduced AP sites, thus initiating long-patch base excision repair synthesis.
Keywords: AP sites/base excision repair/DNA polymerase β/human cell extracts
Introduction
Abasic (apurinic/apyrimidinic, AP) sites can arise in DNA as a result of spontaneous hydrolysis of the N-glycosylic bond or the removal of altered bases by DNA glycosylases (Lindahl, 1993). It has been estimated that ∼10 000 AP sites are formed in each mammalian cell per day under normal physiological conditions (Lindahl and Nyberg, 1972). An AP site is a non-coding lesion that can lead to misincorporation during replication and transcription (Friedberg et al., 1995), and thus it should be promptly removed from DNA. Repair of AP sites is initiated by apurinic/apyrimidinic endonuclease (AP endonuclease), which binds to the AP site and hydrolyzes the phosphodiester bond 5′ to the abasic site, generating a 5′-terminal sugar phosphate (dRP) (Lindahl, 1990; Lindahl and Wood, 1999). In mammalian cells the dRP is removed by an AP lyase activity associated with DNA polymerase β (Pol β) (Matsumoto and Kim, 1995; Sobol et al., 2000). Pol β removes the dRP and simultaneously adds one nucleotide to the 3′-end of the nick. The nick is finally sealed by DNA ligase and the entire repair reaction results in the removal of the abasic sugar and its replacement with the normal nucleotide (single-nucleotide or short-patch base excision repair, BER) (Dianov et al., 1992; Dianov and Lindahl, 1994).
Pol β exerts its AP lyase activity through the β-elimination mechanism, which is very sensitive to chemical modifications of a dRP. For example, oxidized or reduced AP sites are resistant to β-elimination. Repair of modified AP sites resistant to β-elimination is accomplished by long-patch BER (Matsumoto et al., 1994; Klungland and Lindahl, 1997). It is assumed that during this mode of repair, DNA polymerase adds a few more nucleotides into the repair gap and displaces the dRP residue as part of a ‘flap’ oligonucleotide. This flap is removed by the flap endonuclease (FEN1) and DNA ligase finally seals the ends (Klungland and Lindahl, 1997; Kim et al., 1998). The role of different DNA polymerases in long-patch BER is uncertain. Both Pol β and Pol δ/ε are able to support long-patch BER in the repair reaction reconstituted with purified proteins (Klungland and Lindahl, 1997; Stucki et al., 1998; Pascucci et al., 1999); however, it is still unclear which polymerase is the polymerase of choice. In other words, it is not clear which polymerase is involved in long-patch BER catalyzed by cell-free extracts under conditions where all three polymerases are present at their physiological levels, or how the particular polymerase is selected. In this study we characterized the repair of normal and reduced AP sites by human cell extracts under conditions that allow us to discriminate between repair synthesis performed by different polymerases. We find that, independently of the nature of the AP site, BER is always initiated by Pol β, which adds the first nucleotide into the repair gap. These data suggest that both short-patch and long-patch BER pathways are initiated by this polymerase.
Results
Characterization of the substrate
In this study we used an oligonucleotide duplex substrate bearing a single uracil/adenine base pair at the defined position (Figure 1A). The uracil-containing strand of the substrate was labeled at the 5′-end. Prior to reactions, uracil was removed by incubation with human uracil-DNA glycosylase (UDG) to generate an abasic site, which was immediately reduced by incubation with sodium borohydride. The identity of the substrate was verified by an in vitro repair reaction reconstituted with purified proteins. Incubation of the substrate containing the reduced AP site with an AP endonuclease (APE1) and DNA ligase resulted in a 16mer product generated by incision of the oligonucleotide at the AP site (Figure 1B, lane 2). However, no full-length product was observed because ligation was blocked by dRP. Pol β, when added to the reaction, was not able to remove the reduced dRP residue, but stimulated extension of the 16mer incision product by addition of 4–6 nt, thus displacing the dRP residue as part of a flap oligonucleotide (Figure 1B, lane 3). The complete repair and restoration of the full-length 33mer product was observed only when all four enzymes (APE1, Pol β, FEN1 and DNA ligase) were present (Figure 1B, lane 4). These data demonstrated that our substrate indeed contained a reduced AP site and was repaired exclusively through the long-patch repair mechanism.
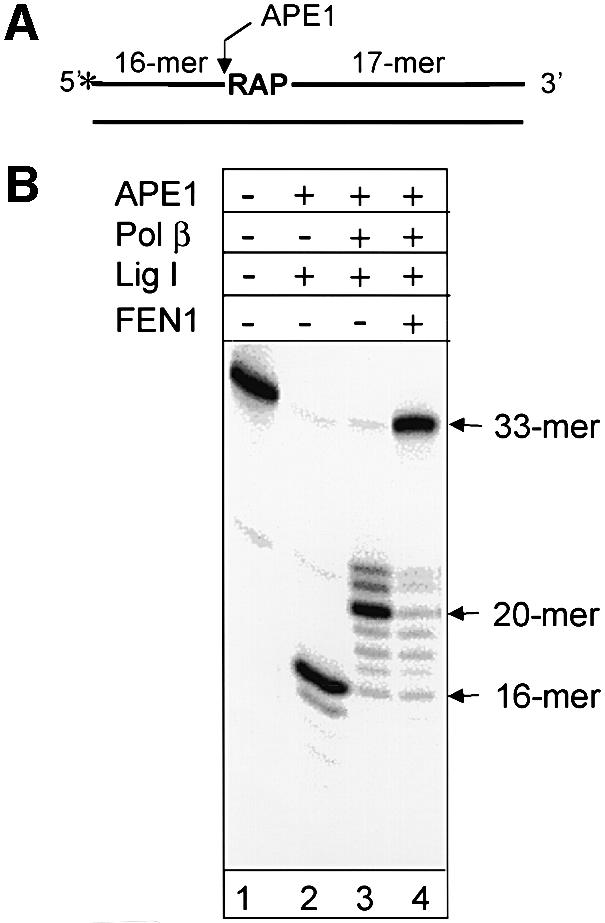
Fig. 1. Repair of reduced AP site reconstituted with purified enzymes. (A) Schematic presentation of the 32P-5′-labeled (*) oligonucleotide substrate containing a reduced AP site (RAP). The arrow indicates the incision site for APE1. (B) A 33 bp duplex oligonucleotide (10 ng) containing a uracil residue at position 17 was pretreated with UDG to generate an AP site, which was then reduced by incubation with sodium borohydride as described in Materials and methods. The buffer containing magnesium, dNTPs and indicated proteins (10 ng of APE1, 2 ng of Pol β, 10 ng of FEN1 and 35 ng of DNA ligase I) was then added and incubation was carried out for 20 min at 37°C. Reaction products were analyzed by electrophoresis in 20% denaturing polyacrylamide gel.
Pol δ is not able to incorporate ddNTPs during repair synthesis
Mammalian Pol β and Pol δ have different abilities to incorporate dideoxyribonucleotide monophosphates (ddNMPs) during DNA synthesis (Weiser et al., 1991). Indeed, in repair reactions reconstituted with purified proteins, Pol β efficiently used substrate with a pre-incised reduced AP site and added 4 nt to the 3′-end of the nick, thus generating a strand displacement flap at the 5′-end (Figure 2, lane 2). When normal deoxyribonucleotide triphosphates (dNTPs) were gradually substituted with ddNTPs, Pol β could efficiently incorporate ddNMPs, and DNA synthesis was terminated after addition of the first nucleotide (Figure 2, lanes 3 and 4). In contrast, Pol δ could incorporate the first normal dNMP and then slowly catalyze further strand displacement (Figure 2, lane 5), but was unable to incorporate ddNMPs into DNA. Addition of increasing amounts of ddNTPs gradually reduced incorporation of the first nucleotide into the repair gap by Pol δ (Figure 2, lanes 6 and 7). We used this differential ability of the two DNA polymerases to incorporate ddNMPs to address the question of which of them would be initiating DNA repair synthesis at the reduced AP site in mammalian cell extracts.
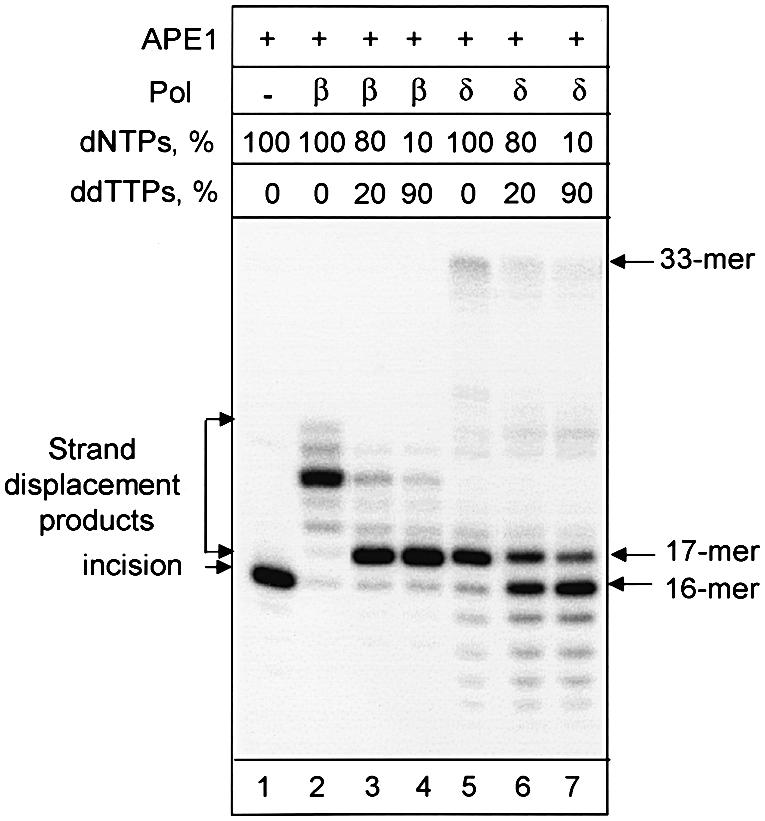
Fig. 2. Pol δ is not able to incorporate ddNTPs during DNA repair synthesis. A 33 bp duplex oligonucleotide (10 ng) containing a reduced AP site was incubated for 20 min at 37°C in the buffer containing magnesium, NTPs and indicated proteins. The reaction mixtures contain 10 ng of APE1 and 2 ng of Pol β (lanes 2–4) or 10 ng of APE1, 1 U of Pol δ and 50 ng of PCNA (lanes 5–7). Normal dNTPs were used in reactions shown in lanes 1, 2 and 5. Reactions shown in lanes 3, 4, 6 and 7 contained a mixture of dNTPs and ddNTPs at the indicated ratio. Reaction products were analyzed by electrophoresis in 20% denaturing polyacrylamide gel.
Initiation of DNA repair synthesis at reduced AP sites is not affected by ddNTPs
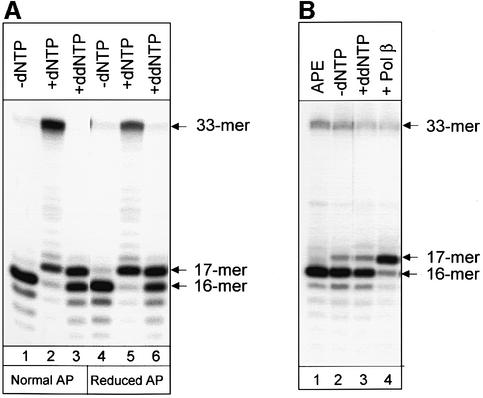
In this set of experiments we measured repair reactions catalyzed by human and mouse cell extracts. The extracts, prepared by the method of Manley et al. (1980), were mainly depleted of dNTPs and the repair reactions were dependent on the addition of exogenous dNTPs. Incu bation of the substrate containing a normal AP site in cell extracts, without dNTPs added, led to generation of the 16mer incision product, but further repair was blocked (Figure 3A, lane 1). An addition of dNTPs restored the repair reaction, which was detected by the appearance of the full-length 33mer repair product (Figure 3A, lane 2). It is well established that Pol β is responsible for the initiation of repair synthesis at normal AP sites. As expected, during processing of the normal AP site, human cell extracts efficiently catalyzed the addition of ddNMPs into the repair gap, thus terminating the repair reaction and generating the 17mer product (Figure 3A, lane 3). The repair efficiency of the reduced AP site was approximately half of that for the normal AP site (compare Figure 3A, lanes 2 and 5, 33mer band), but the incorporation of the first nucleotide (dNMP and ddNMP) into the repair gap was equally efficient for the normal and reduced AP sites (Figure 3A, lanes 2 and 3, and 5 and 6). These data suggest that a DNA polymerase that is able to incorporate ddNMPs is involved in the initiation of repair synthesis at the reduced AP site. Pol β is an obvious candidate for such a function. We thus examined the ability of Pol β-deficient mouse cell extracts to incorporate ddNMPs into the repair gap and found normal incision activity, but significantly reduced incorporation of ddNMPs, in these extracts (Figure 3B, lane 3, 17mer product). To confirm that the observed reduction was due to the absence of Pol β, purified enzyme was added to the Pol β-deficient cell extracts. Addition of 5 ng of Pol β completely restored the ability of these extracts to incorporate ddNMPs (Figure 3, lane 4).
Fig. 3. DNA repair incorporation of ddNTPs catalyzed by whole-cell extracts. (A) Reaction mixtures (10 µl) containing 10 ng of substrate oligonucleotide duplex with normal (lanes 1–3) or reduced (lanes 4–6) AP sites were incubated with 10 µg of human cell extracts for 20 min at 37°C without dNTPs (lanes 1 and 4) in the presence of normal dNTPs (lanes 2 and 5) or with ddNTPs added (lanes 3 and 6). (B) Reaction mixtures (10 µl) containing 10 ng of substrate oligo nucleotide duplex with reduced AP sites were incubated with 10 ng of APE1 (lane 1) or 10 µg of Pol β-deficient cell extracts (lanes 2–4) for 20 min at 37°C without dNTPs (lane 2) or in the presence of ddNTPs (lanes 3 and 4). The reaction shown in lane 4 also contains 5 ng of purified Pol β. Reaction products were analyzed by electrophoresis in 20% denaturing polyacrylamide gel.
Incorporation of the first nucleotide into the repair gap is insensitive to aphidicolin
Three major mammalian polymerases that are involved in DNA replication, α, δ and ε, are extremely sensitive to aphidicolin. At 100 µg/ml concentration of aphidicolin, DNA synthesis by these polymerases is completely blocked. In contrast, Pol β is not sensitive to aphidicolin (Wang, 1996). In our system we find that addition of 1% dimethylsulfoxide (DMSO) (aphidicolin solvent) only slightly inhibited (3–5%) an addition of the first nucleotide for both normal and reduced AP site-containing substrates, but do not find any inhibitory effect of aphidicolin at the concentration tested (100 µg/ml) (Figure 4). However, aphidicolin completely inhibited minor strand displacement, probably catalyzed by polymerases other than Pol β (Figure 4, compare lanes 2 and 5, and 3 and 6). Taken together these data provide clear evidence for the role of Pol β in initiating long-patch repair synthesis.
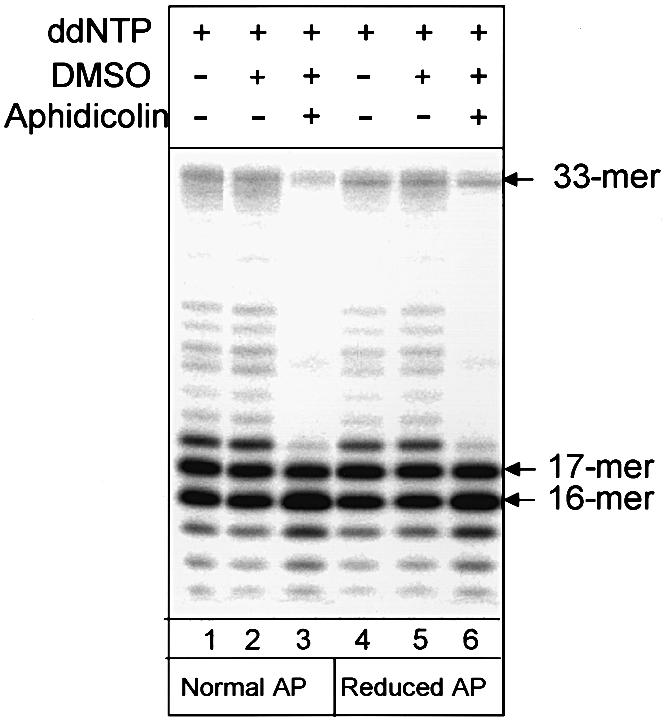
Fig. 4. Polymerase incorporating the first nucleotide into the repair gap is not sensitive to aphidicolin. Reaction mixtures (10 µl) containing 10 ng of substrate oligonucleotide duplex with normal (1–3) or reduced (4–6) AP sites were incubated with 10 µg of human cell extract supplemented with ddNTPs at 37°C in a reaction buffer described in Materials and methods. Reactions 2 and 5 contained 1% DMSO. Reactions 3 and 6 contained 1% DMSO and 100 μg/ml aphidicolin. Reaction products were analyzed by electrophoresis in 20% denaturing polyacrylamide gel.
Strand displacement is a limiting factor in repair of reduced AP sites
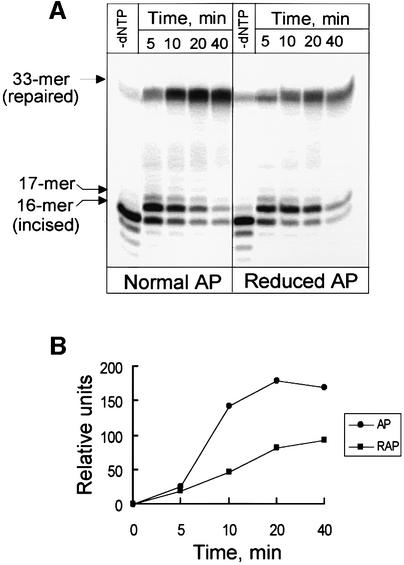
In human cell extracts we find that the rate of repair of reduced AP sites is ∼2-fold slower than the repair of normal AP sites (Figure 5). There are at least two possible explanations for this observation: multiple rounds of initiation of DNA polymerase(s) are required for strand displacement, or strand displacement is less efficient at reduced AP sites than at normal AP sites. To distinguish between those possibilities we measured the efficiency of strand displacement catalyzed by purified Pol β at normal and reduced AP sites, and found that strand displacement is equal or even slightly more efficient on reduced AP sites when compared with normal AP sites (Figure 6). However, addition of a limited amount of Pol β to the cell extracts significantly increases their ability to repair reduced AP sites (Figure 7, compare lanes 3 and 4), indicating that DNA polymerase activity in these cell extracts is a limiting factor for repair of reduced AP sites.
Fig. 5. Repair of normal and reduced AP sites by human cell extracts. (A) Reaction mixtures (10 µl) containing 10 ng of substrate oligo nucleotide duplex with normal or reduced AP sites were incubated with 10 µg of human cell extract supplemented with dNTPs at 37°C in a reaction buffer described in Materials and methods. After the indicated time, reaction products were analyzed by electrophoresis in 20% denaturing polyacrylamide gel. (B) Phosphoimager quantification of the gel shown in Figure 5A.
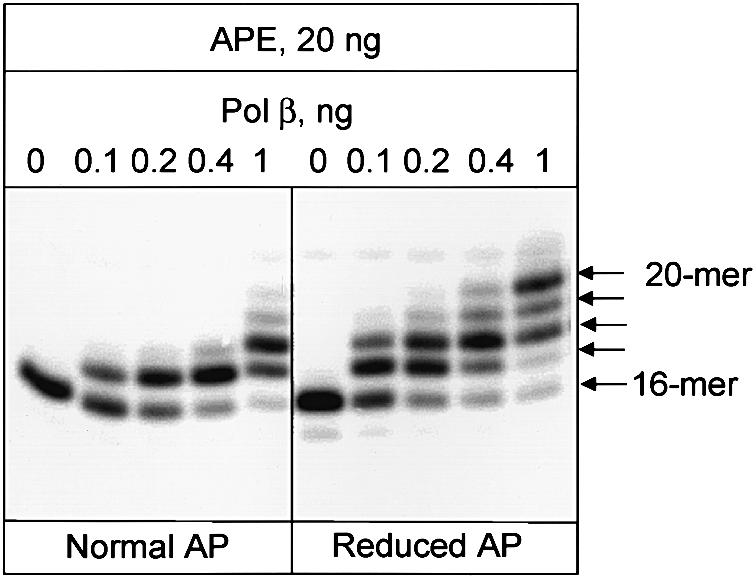
Fig. 6. Strand displacement reaction catalyzed by Pol β at the incised AP site. 5′-labeled oligonucleotide duplex (10 ng) containing a normal or reduced AP site at position 17 was incubated under conditions described in Materials and methods with 20 ng of APE1 and the indicated amount of Pol β. Reaction products were analyzed by electrophoresis in 20% denaturing polyacrylamide gel.
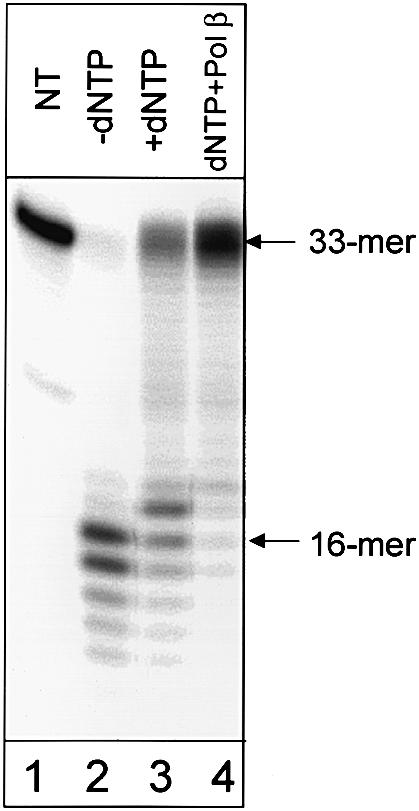
Fig. 7. Stimulation of repair of reduced AP sites in human cell extracts by Pol β. 5′-labeled oligonucleotide duplex (10 ng) containing reduced AP site at position 17 was incubated under conditions described in Materials and methods without cell extract (lane 1, NT), with 10 µg of human cell extract, but without dNTPS (lane 2), in a cell extract supplemented with dNTPs (lane 3) and in a cell extract supplemented with dNTPs and 2 ng of Pol β (lane 4). Reaction products were analyzed by electrophoresis in 20% denaturing polyacrylamide gel.
Discussion
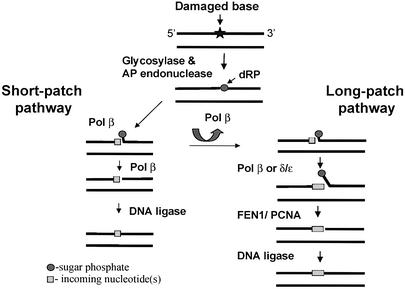
The identity of the DNA polymerases involved in different repair pathways is very important for understanding the mechanism coordinating DNA repair in mammalian cells. There are two major BER pathways, short-patch and long-patch repair, involving replacement of only one or a few nucleotides, respectively (Dianov and Lindahl, 1994; Lindahl et al., 1995, 1997). Pol β is the major DNA polymerase participating in the short-patch pathway that performs the majority of BER (Wiebauer and Jiricny, 1990; Dianov et al., 1992; Sobol et al., 1996, 2000). Long-patch repair, which is believed to be the major pathway when the 5′-sugar phosphate repair intermediate is resistant to β-elimination, was found to be proliferating cell nuclear antigen (PCNA) dependent (Matsumoto et al., 1994; Frosina et al., 1996). Based on these data it was suggested that PCNA-dependent Pol δ was the major DNA polymerase involved in long-patch BER (Matsumoto et al., 1994; Frosina et al., 1996). The potential problem with this model is the mechanism for coordination of the two repair pathways. If two different polymerases are involved in BER, how does the cell decide which pathway to use in the early stages of BER before the 5′-sugar phosphate intermediate is reached? Previous results have suggested a role for Pol β in the long-patch repair pathway. Klungland and Lindahl (1997) demonstrated that neutralizing antibodies to Pol β inhibited repair of reduced AP sites in human cell extracts. Dianov et al. (1999) also observed Pol β-dependent release of the flap oligonucleotide during long-patch repair of uracil residues in DNA by human cell extracts. Based on these data we hypothesized that a single DNA polymerase (Pol β) is involved in initiation of BER and that the decision of which repair pathway to use is based on the nature of the AP site (Figure 8). At normal AP sites Pol β would add the first nucleotide during repair synthesis, remove the dRP residue, and the repair would be accomplished by the short-patch repair mechanism (Figure 8, left). However, we proposed that Pol β would dissociate from the repair complex if AP sites were resistant to β-elimination. Then, either Pol β or Pol δ/ε would bind to the complex and continue strand displacement. Finally, FEN1 and DNA ligase would accomplish repair through the long-patch pathway (Figure 8, right). In this study we tested this model experimentally. As shown before, although the rates were different, both linear and closed circular DNA containing AP sites resistant to β-elimination are repaired by mammalian cell extracts (Biade et al., 1998). This suggests that the oligonucleotide substrates used in our study are reasonably well processed by mammalian cell extracts via both short-patch and long-patch repair pathways. The repair assay employed in our study allowed us to identify the polymerase that is responsible for incorporation of the first nucleotide during repair synthesis. Using this assay we now demonstrate that, independently of the nature of the AP site, repair synthesis is initiated by a polymerase with characteristics similar to those of Pol β, which is able to incorporate ddNMPs and is insensitive to aphidicolin. We demonstrate further that incorporation of the ddNMPs is significantly reduced in extracts derived from Pol β-deficient cells and that repair incorporation was complemented by addition of purified Pol β. These data clearly demonstrate that Pol β can be responsible for incorporation of the first nucleotide into the repair gap. An interesting unanswered question is which polymerase continues strand displacement? However, our experimental design does not allow us to answer this question because in our conditions repair synthesis is terminated after incorporation of the first ddNMP. Inclusion of the first normal dNTP and then second ddNTPs in the repair reaction would bring some uncertainties to the data interpretation, as we would not be able to say which polymerase initiated repair synthesis. However, the data from other laboratories suggest that Pol δ/ε may contribute to the repair synthesis following Pol β dissociation. Frosina et al. (1996) previously showed that while PCNA antibodies did not inhibit incorporation of the first nucleotide into the repair gap, further incorporation was dependent on PCNA (Frosina et al., 1996). However, in the light of the more recent findings that PCNA stimulates flap removal by FEN1 protein (Klungland and Lindahl, 1997; Gary et al., 1999), the effect of the PCNA antibodies may also be interpreted as an indication of the role of PCNA in the flap removal during long-patch repair. Why is repair of reduced AP sites slower than repair of normal AP sites? Incorporation of the first nucleotide into the repair gap was equally efficient for both normal and reduced AP sites (Figure 5). These data indicate that this first step is not the rate-limiting reaction during repair of reduced AP sites. We also demonstrate that addition of purified Pol β to the cell extracts significantly improved repair of reduced AP sites (Figure 7). Based on these data we suggest that a distributive DNA polymerase like Pol β, which requires multiple rounds of initiation, is involved in the strand displacement reaction, and further binding to the substrate may be the step that determines the rate of repair of reduced AP sites.
Fig. 8. Model for repair of reduced AP sites. For details see the text.
Materials and methods
Materials
Synthetic oligodeoxyribonucleotides purified by high-pressure liquid chromatography were obtained from Midland. [γ-32P]ATP (3000 Ci/mmol) was purchased from Amersham Pharmacia Biotech. Recombinant human Pol β was a gift from S.Wilson. Human UDG was purified as described (Slupphaug et al., 1995). Calf thymus Pol δ was purified as previously described (Weiser et al., 1991). DNA ligase I was a gift from A.Tomkinson. Histidine-tagged human PCNA, APE1 and FEN1 proteins were purified on Ni2+-charged His-Bind Resin (Novagen, Cambridge, MA) as recommended by the manufacturer.
Oligonucleotide DNA substrates
The 33mer oligonucleotide (5′-TACCGCGGCCGGCCGAUCAAGCTTATTGGGTAC-3′) containing a single uracil residue at position 17 was 5′-end labeled and annealed to the complementary strand as previously described (Dianov and Lindahl, 1991). Prior to assembly of the excision reaction, the oligonucleotide substrate (100 ng) was pretreated with UDG (100 ng) in 10 mM HEPES pH 7.9, 1 mM EDTA and 70 mM KCl. The reaction mixture was incubated at 37°C for 30 min. Due to the instability of the AP-containing DNA, the substrates were prepared just before performing the BER reactions. To prepare the reduced AP site, AP-containing oligonucleotide was treated with 0.1 M sodium borohydride for 30 min on ice and then filtered through a Sephadex G-25 spin column.
BER reactions
The BER reactions were carried out in a reaction mixture (10 µl) that contained 45 mM HEPES–KOH pH 7.8, 70 mM KCl, 7.5 mM MgCl2, 0.5 mM EDTA, 1 mM dithiothreitol (DTT), 2 mM ATP, 0.4 mg/ml bovine serum albumin, 20 µM each of the indicated dNTPs or ddNTPs, and 32P-labeled oligonucleotide substrate (10 ng). When aphidicolin (Sigma) was added to a final concentration of 100 µg/ml, the buffer contained 1% DMSO. The reactions were initiated by adding whole-cell extracts (10 µg) or purified APE1, FEN1, Pol β and DNA ligase I at the amount indicated in the figure legends. After incubation for 20 min at 37°C the reactions were stopped by addition of 10 µl of 0.2 M EDTA, proteins were removed by extraction with an equal volume of chloroform–isoamyl alcohol (24:1), and mixed with an equal volume of gel loading buffer (95% formamide, 20 mM EDTA, 0.02% bromophenol blue and 0.02% xylene cyanol). Following incubation at 90°C for 2–5 min, the reaction products were separated by electrophoresis in a 20% polyacrylamide gel containing 7 M urea in 89 mM Tris–HCl, 89 mM boric acid and 2 mM EDTA pH 8.0.
Cells and extracts
Normal human lymphoid cells AG9387 were obtained from the Human Genetic Mutant Cell Repository (Coriell Institute, Camden, NJ). Cells were grown in medium recommended by the Repository. The DNA Pol β-knockout mouse fibroblast cell line (MB19tsA) was grown as described (Sobol et al., 1996). Whole-cell extracts were prepared by the method of Manley et al. (1980) and dialyzed overnight against buffer containing 25 mM HEPES–KOH pH 7.9, 2 mM DTT, 12 mM MgCl2, 0.1 mM EDTA, 17% glycerol and 0.1 M KCl. Extracts were aliquoted and stored at –80°C. All experiments were repeated at least three to five times and representative gels are shown.
Acknowledgments
Acknowledgements
We thank Sam Wilson and Alan Tomkinson for providing reagent proteins and expression constructs. John Thacker, Tim Humphrey and Helen Budworth are thanked for critical reading of the manuscript. V.N.P. was supported by the NIH grant GM52948 to Ellen Fanning.
REFERENCES
- Biade S., Sobol,R.W., Wilson,S.H. and Matsumoto,Y. (1998) Impair ment of proliferating cell nuclear antigen-dependent apurinic/apyrimidinic site repair on linear DNA. J. Biol. Chem., 273, 898–902. [DOI] [PubMed] [Google Scholar]
- Dianov G. and Lindahl,T. (1991) Preferential recognition of I–T base-pairs in the initiation of excision-repair by hypoxanthine-DNA glycosylase. Nucleic Acids Res., 19, 3829–3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dianov G. and Lindahl,T. (1994) Reconstitution of the DNA base excision-repair pathway. Curr. Biol., 4, 1069–1076. [DOI] [PubMed] [Google Scholar]
- Dianov G., Price,A. and Lindahl,T. (1992) Generation of single-nucleotide repair patches following excision of uracil residues from DNA. Mol. Cell. Biol., 12, 1605–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dianov G.L., Prasad,R., Wilson,S.H. and Bohr,V.A. (1999) Role of DNA polymerase β in the excision step of long patch mammalian base excision repair. J. Biol. Chem., 274, 13741–13743. [DOI] [PubMed] [Google Scholar]
- Friedberg E.C., Walker,G.C. and Siede,W. (1995) DNA Repair and Mutagenesis. American Society of Microbiology Press, Washington, DC.
- Frosina G., Fortini,P., Rossi,O., Carrozzino,F., Raspaglio,G., Cox,L.S., Lane,D.P., Abbondandolo,A. and Dogliotti,E. (1996) Two pathways for base excision repair in mammalian cells. J. Biol. Chem., 271, 9573–9578. [DOI] [PubMed] [Google Scholar]
- Gary R., Kim,K., Cornelius,H.L., Park,M.S. and Matsumoto,Y. (1999) Proliferating cell nuclear antigen facilitates excision in long-patch base excision repair. J. Biol. Chem., 274, 4354–4363. [DOI] [PubMed] [Google Scholar]
- Kim K., Biade,S. and Matsumoto,Y. (1998) Involvement of flap endonuclease 1 in base excision DNA repair. J. Biol. Chem., 273, 8842–8848. [DOI] [PubMed] [Google Scholar]
- Klungland A. and Lindahl,T. (1997) Second pathway for completion of human DNA base excision-repair: reconstitution with purified proteins and requirement for DNase IV (FEN1). EMBO J., 16, 3341–3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T. (1990) Repair of intrinsic DNA lesions. Mutat. Res., 238, 305–311. [DOI] [PubMed] [Google Scholar]
- Lindahl T. (1993) Instability and decay of the primary structure of DNA. Nature, 362, 709–715. [DOI] [PubMed] [Google Scholar]
- Lindahl T. and Nyberg,B. (1972) Rate of depurination of native deoxyribonucleic acid. Biochemistry, 11, 3610–3618. [DOI] [PubMed] [Google Scholar]
- Lindahl T. and Wood,R.D. (1999) Quality control by DNA repair. Science, 286, 1897–1905. [DOI] [PubMed] [Google Scholar]
- Lindahl T., Satoh,M.S. and Dianov,G. (1995) Enzymes acting at strand interruptions in DNA. Philos. Trans. R. Soc. Lond. B Biol. Sci., 347, 57–62. [DOI] [PubMed] [Google Scholar]
- Lindahl T., Karran,P. and Wood,R.D. (1997) DNA excision repair pathways. Curr. Opin. Genet. Dev., 7, 158–169. [DOI] [PubMed] [Google Scholar]
- Manley J.L., Fire,A., Cano,A., Sharp,P.A. and Gefter,M.L. (1980) DNA-dependent transcription of adenovirus genes in a soluble whole-cell extract. Proc. Natl Acad. Sci. USA, 77, 3855–3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto Y. and Kim,K. (1995) Excision of deoxyribose phosphate residues by DNA polymerase β during DNA repair. Science, 269, 699–702. [DOI] [PubMed] [Google Scholar]
- Matsumoto Y., Kim,K. and Bogenhagen,D.F. (1994) Proliferating cell nuclear antigen-dependent abasic site repair in Xenopus laevis oocytes: an alternative pathway of base excision DNA repair. Mol. Cell. Biol., 14, 6187–6197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascucci B., Stucki,M., Jonsson,Z.O., Dogliotti,E. and Hubsher,U. (1999) Long patch base excision repair with purified proteins. J. Biol. Chem., 274, 33696–33702. [DOI] [PubMed] [Google Scholar]
- Slupphaug G., Eftedal,I., Kavli,B., Bharati,S., Helle,N.M., Haug,T., Levine,D.W. and Krokan,H.E. (1995) Properties of a recombinant human uracil-DNA glycosylase from the UNG gene and evidence that UNG encodes the major uracil-DNA glycosylase. Biochemistry, 34, 128–138. [DOI] [PubMed] [Google Scholar]
- Sobol R.W., Horton,J.K., Kuhn,R., Gu,H., Singhal,R.K., Prasad,R., Rajewsky,K. and Wilson,S.H. (1996) Requirement of mammalian DNA polymerase-β in base-excision repair. Nature, 379, 183–186. [DOI] [PubMed] [Google Scholar]
- Sobol R.W., Prasad,R., Evenski,A., Baker,A., Yang,X.-P., Horton,J. and Wilson,S.H. (2000) The lyase activity of the DNA repair protein β-polymerase protects from DNA-damage-induced cytotoxicity. Nature, 405, 807–809. [DOI] [PubMed] [Google Scholar]
- Stucki M., Pascucci,B., Parlanti,E., Fortini,P., Wilson,S.H., Hubscher,U. and Dogliotti,E. (1998) Mammalian base excision repair by DNA polymerases δ and ε. Oncogene, 17, 835–843. [DOI] [PubMed] [Google Scholar]
- Wang T.S.-F. (1996) Cellular DNA polymerases. In DePamphilis,M.L. (ed.), DNA Replication in Eukaryotic Cells. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY, pp. 461–463.
- Weiser T., Gassmann,M., Thommes,P., Ferraru,E., Spadari,S. and Hubsher,U. (1991) Biochemical and functional comparison of DNA polymerases α, δ and ε from calf thymus. J. Biol. Chem., 266, 10420–10428. [PubMed] [Google Scholar]
- Wiebauer K. and Jiricny,J. (1990) Mismatch-specific thymine DNA glycosylase and DNA polymerase β mediate the correction of G–T mispairs in nuclear extracts from human cells. Proc. Natl Acad. Sci. USA, 87, 5842–5845. [DOI] [PMC free article] [PubMed] [Google Scholar]