Abstract
The role of PKN, a fatty acid- and Rho small GTPase-activated protein kinase, in cell-cycle regulation was analyzed. Microinjection of the active form of PKN into a Xenopus embryo caused cleavage arrest, whereas normal cell division proceeded in the control embryo microinjected with buffer or the inactive form of PKN. Exogenous addition of the active form of PKN delayed mitotic timing in Xenopus egg cycling extracts judging by morphology of sperm nuclei and Cdc2/cyclin B histone H1 kinase activity. The kinase-negative form of PKN did not affect the timing, suggesting that delayed mitotic timing depends on the kinase activity of PKN. The dephosphorylation of Tyr-15 of Cdc2 was also delayed in correlation with Cdc2/cyclin B histone H1 kinase activation in extracts containing active PKN. The Cdc25C activity for the dephosphorylation of Tyr-15 in Cdc2 was suppressed by pretreatment with the active form of PKN. Furthermore, PKN efficiently phosphorylated Cdc25C in vitro, indicating that PKN directly inhibits Cdc25C activity by phosphorylation. These results suggest that PKN plays a significant role in the control of mitotic timing by inhibition of Cdc25C.
Protein phosphorylation/dephosphorylation reaction is a key event in the regulation of cell division. In dividing eukaryotic cells, entry into mitosis is governed by the M phase-promoting factor. This factor consists of the Cdc2 protein kinase and cyclin B and acts by phosphorylating substrates that are essential for the execution of mitotic processes (1). Before mitosis, the activity of Cdc2/cyclin B histone H1 kinase is suppressed through inhibitory phosphorylation of Tyr-15 and Thr-14 residues of Cdc2 by the Wee1 and Myt1 kinases (2–5). At onset of mitosis, the phosphatase Cdc25C removes these inhibitory phosphate groups from Cdc2 and thereby activates Cdc2/cyclin B histone H1 kinase. The activity of Cdc25C is strictly regulated, probably by phosphorylation, and is low during interphase and high at mitosis (6, 7). Thus, entry into mitosis is under the control of a tightly regulated network of protein kinases and phosphatases. Although upstream players such as Chk1 and Cds1 (8–10) in the tyrosine phosphorylation/dephosphorylation of Cdc2 have been identified, it remains unclear how these enzymes are controlled during the cell cycle. There is a possibility that the unidentified protein kinase or kinases might participate in upstream regulation.
PKN is a serine/threonine protein kinase that has a catalytic domain highly homologous to protein kinase C (PKC) in the carboxyl-terminal region and a unique regulatory domain in the amino-terminal region (11–13). The amino-terminal region of PKN contains three repeats of a leucine zipper-like motif, and its kinase activity is stimulated by fatty acids such as arachidonic acid (13). We reported that PKN translocates from the cytosol to the nucleus of fibroblasts on exposure to stress such as heat shock and serum starvation (14) and that PKN is cleaved during apoptosis, presumably by caspase-3, which generates a constitutively active kinase fragment (15). Furthermore, we found that PKN is one of the downstream targets of small GTPase, Rho (16–18). Rho regulates actin-based cytoskeletal structures, including focal adhesions, stress fibers, and the contractile ring, and works as a switch in stimulus-evoked cell adhesion and cytokinesis (19, 20). Expression of the dominant-negative form of Rho kinase, another protein kinase regulated by Rho, inhibits the cytokinesis of the Xenopus embryo and mammalian cells (21). The overexpression of citron (another downstream target of Rho) mutant results in the production of multinucleate cells, and its kinase-active mutant causes abnormal contraction during cytokinesis (22). These findings suggest that PKN also may participate in cell-cycle control.
The present study analyzed the role of PKN in the regulation of the cell cycle. We found that exogenous addition of the active form of PKN delays the mitosis of Xenopus egg cycling extracts. In vitro experiments indicated that PKN phosphorylates and inhibits Cdc25C, revealing that delayed mitotic timing in the egg extracts is caused by inhibition of Cdc25C.
Materials and Methods
Antibodies.
Antiphospho-Tyr-15 of Cdc2 polyclonal antibody and anti-Cdc2 monoclonal antibody were obtained from New England Biolabs and Santa Cruz Biotechnology, respectively.
Recombinant Baculoviruses and Protein Production.
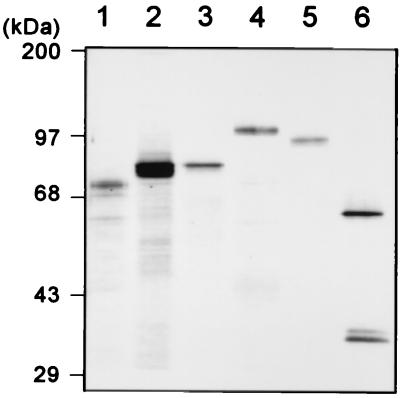
Recombinant viruses encoding glutathione S-transferase (GST)/PKN () (the constitutively active form of human PKN) and GST/PKN ()-K644E (the kinase-negative form of PKN) have been described (23). The recombinant viruses for GST/Wee1 (full-length Xenopus Wee1), GST/cyclin B1 (the indestructible form of Xenopus cyclin B1), and histidine (His)6/Cdc2 (N133A) (the kinase-negative form of Xenopus Cdc2) were provided by M. Iwabuchi (Tokyo Institute of Technology, Yokohama, Japan). Recombinant virus encoding GST/PKCɛ () (the constitutively active form of rat PKCɛ) was generated by using the baculovirus transfer vector of pBlueBacHis/GST (23) and the cDNA fragment encoding the catalytic domain of rat PKCɛ (24). Recombinant virus encoding GST/Cdc25C was generated by using the baculovirus transfer vector of pBlueBacHis/GST and the full-length Xenopus Cdc25C cDNA (25) provided by N. Nakajo (Kyushu University, Fukuoka, Japan). All GST-fused proteins were expressed in Sf9 cells and purified by glutathione Sepharose 4B (Amersham Pharmacia) chromatography as described (23). Cdc2 (N133A)/cyclin B complex was purified by using glutathione Sepharose 4B from Sf9 cells that had been simultaneously infected with recombinant baculoviruses encoding (His)6/Cdc2 (N133A) and GST/cyclin B. Fig. 1 shows the protein preparations used in this study.
Figure 1.
Protein preparations. An aliquot of each recombinant protein was resolved by SDS/PAGE and silver stained. Lane 1, GST/PKCɛ (); lane 2, GST/PKN ()-K644E; lane 3, GST/PKN (); lane 4, GST/Wee1; lane 5, GST/Cdc25; and lane 6, the complex of GST/cyclin B and (His)6/Cdc2 (N133A).
Microinjection into Xenopus Embryo.
Proteins were concentrated, and solvents were replaced by microinjection buffer (20 mM Tris⋅HCl, pH 7.5/88 mM NaCl) with Centricon-10 (Amicon). Sexually mature females of Xenopus laevis were induced to ovulate by injection of human chorionic gonadotropin (500 units per female). Eggs were artificially inseminated, and embryos were cultured in 20% MMR (1 mM Hepes-KOH, pH 7.8/20 mM NaCl/0.4 mM KCl/0.2 mM MgCl2/0.4 mM CaCl2/0.02 mM EDTA) containing 5% Ficoll (Amersham Pharmacia). Embryos were selected for injection at the beginning of the first cleavage furrow formation, and 10 nl of protein sample was injected into one blastomere. Cleavage arrest of the injected blastomere was observed 5 h after fertilization.
Cycling Xenopus Egg Extracts.
Xenopus egg extracts that reproduce the progression of embryonic mitotic cycles were prepared according to a modification of the procedure described by Murray (26). Briefly, eggs were dejellied with 2.5% thioglycolic acid (pH 8.2) and activated by treatment with 0.2 μg/ml of calcium ionophore A23187 for 3 min. Activated eggs were incubated in 20% MMR for 25 min, then washed with extraction buffer (20 mM Hepes-KOH, pH 7.4/100 mM KCl/5 mM MgCl2/0.1 mM CaCl2) containing 50 μg/ml of cytochalasin B. After being transferred to a test tube, eggs were chilled on ice for 5 min, and cytoplasmic extract was obtained by centrifugation at 15,000 × g for 10 min. The extracts were recentrifuged to remove contaminating lipid and mixed with 1:50 vol/vol of ATP solution (500 mM phosphocreatine/50 mM MgCl2/50 mM ATP) for final cycling extracts. To monitor the cell-cycle phase of egg extracts, sperm that had been demembranated with 0.05% lysolecithin were added to the extracts at the start of incubation, and the morphology of sperm chromatin was visualized by fluorescence with the DNA-binding dye Hoechst 33342. The purified protein was added to the extracts (1 ng protein per 1 μl extracts) at the same time as sperm chromatin.
Assay of Histone H1 Kinase.
For histone H1 kinase assay (27), extracts were quickly frozen in liquid nitrogen and stored at −80°C. Frozen extracts were thawed by adding 9 volumes of an ice-cold dilution buffer (20 mM Hepes-KOH, pH 7.5/80 mM β-glycerophosphate/20 mM EGTA/5 mM MgCl2). A total of 10 μl of diluted extract was mixed with 20 μl of reaction buffer (80 mM β-glycerophosphate/20 mM MgCl2/0.6 mM ATP/30 μg/ml leupeptin/30 μg/ml aprotinin/0.6 mg/ml histone H1/37 kBq [γ-32P] ATP) and incubated for 30 min at 25°C. Reactions were stopped by addition of SDS sample buffer and boiled for 2 min. Histone H1 was separated by SDS/PAGE and stained with Coomassie blue, and the band was excised. The amount of 32P incorporation into the gel slice was quantified by Cerenkov counting.
Effect of PKN on Wee1 Activity.
Wee1 activity was assayed by using the Cdc2 (N133A)/cyclin B complex as a substrate. Affinity-purified GST/Wee1 (100 ng) was incubated in the reaction mixture [20 mM Tris⋅HCl, pH 7.5/5 mM MgCl2/1 mM ATP/100 ng GST/PKN () or GST/PKN ()-K644E] for 20 min at 30°C. After incubation, the Cdc2 (N133A)/cyclin B complex (1 μg) was added to the reaction mixture, and samples were incubated for 15 min at 25°C. Reactions were stopped by boiling the mixture in SDS-sample buffer. Phosphorylation of Tyr-15 of Cdc2 was analyzed by Western blotting with the antiphospho-Tyr-15 of Cdc2 antibody. Blots were visualized by the enhanced chemiluminescence method.
Effect of PKN on Cdc25 Tyrosine Phosphatase Activity.
Cdc25C tyrosine phosphatase activity was assayed by using the Cdc2 (N133A)/cyclin B complex purified from Sf9 cells as a phosphorylated substrate. Affinity-purified GST/Cdc25C (100 ng) was incubated in the reaction mixture [20 mM Tris⋅HCl, pH 7.5/5 mM MgCl2/1 mM ATP/100 ng GST/PKN () or GST/PKN ()-K644E] for 20 min at 30°C. Dephosphorylation of Cdc2 was initiated by adding Cdc2 (N133A)/cyclin B complex (1 μg), EDTA (10 mM), and DTT (10 mM). Samples were incubated for 30 min at 25°C, and reactions were stopped by boiling the mixture in SDS sample. Dephosphorylation of Tyr-15 of Cdc2 was analyzed by Western blotting with the antiphospho-Tyr-15 of Cdc2 antibody.
In Vitro Phosphorylation of Cdc25C.
The in vitro phosphorylation reaction proceeded at 30°C for 5 min in the reaction mixture containing 20 mM Tris⋅HCl, pH 7.5, 5 mM MgCl2, 40 μM ATP, 370 kBq of [γ-32P] ATP, 200 ng GST/Cdc25C, and 100 ng recombinant enzyme. The reaction was terminated by boiling the mixture in SDS-sample buffer. The samples were resolved by SDS/PAGE, and the gel was dried under vacuum. The phosphorylation of the proteins was analyzed by an image analyzer (packard instant imager).
Results and Discussion
Microinjection of the Active Form of PKN Inhibits Cell Division of Xenopus Embryos.
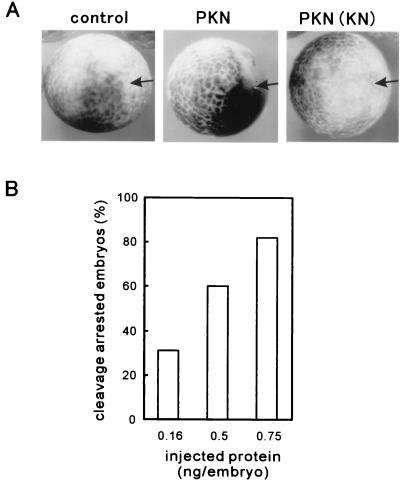
To analyze the effect of PKN on cell division, we microinjected the active form of PKN into one blastomere of Xenopus embryos at the two-cell stage. The active form of PKN was affinity purified from Sf9 cells as a GST-fusion protein containing the catalytic domain of PKN (Fig. 1). This protein is constitutively active as reported (23). Fig. 2A shows that microinjection of the active form of PKN clearly caused cleavage arrest, whereas normal cell division proceeded in the control embryos microinjected with buffer or the inactive form of PKN. Fig. 2B shows that the arrest depended on the amounts of the active PKN. Nuclear staining of the embryos revealed normal daughter nuclei in the uninjected area but one nucleus in the cleavage-arrested area (data not shown). These results suggest that PKN affects cell-cycle progression at the mitotic phase rather than at cytokinesis.
Figure 2.
Microinjection of the active form of PKN inhibits cell division of the Xenopus embryo. (A) Effects of microinjection of the active form of PKN on cell division of Xenopus embryos. Control buffer, 0.75 ng per embryo of GST/PKN () or 4 ng per embryo of GST/PKN ()-K644E, was injected into one blastomere at the two-cell stage. Embryos were photographed 5 h after fertilization. Arrows indicate the position of injection. GST/PKN () and GST/PKN ()-K644E are indicated as PKN and PKN(KN), respectively. (B) Dose dependency of the active form of PKN for cleavage arrest of Xenopus embryos. The indicated amounts of GST/PKN () were microinjected as in A. A typical result of three independent experiments is shown.
Exogenous Addition of the Active Form of PKN Delays Mitosis in Egg Extracts.
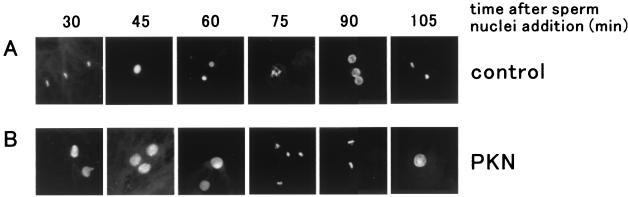
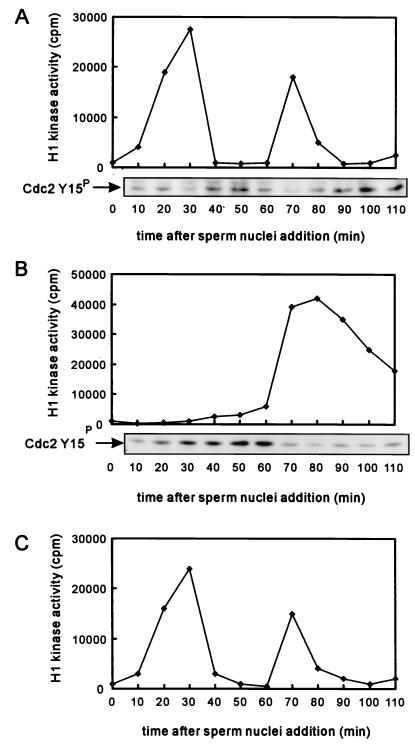
Xenopus egg cycling extracts have been widely used to investigate the regulation of the cell cycle and faithfully mimic many aspects of the cell cycle in vivo, including nuclear envelope breakdown and reformation (26). Thus, to assess the role of PKN in mitotic timing, we added the active form of PKN exogenously to cycling Xenopus egg extracts. Nuclear envelope breakdown and chromatin condensation were observed in the control egg extracts 30 min after the addition of sperm nuclei, and the chromatin condensation that followed was at 75 min (Fig. 3A). In contrast, Fig. 3B shows that the active form of PKN elicited a substantial delay of mitosis 75–90 min after the addition of sperm nuclei. The kinase-negative form of PKN and the kinase-active form of PKCɛ, which has a catalytic domain related to PKN, did not affect the timing of nuclear envelope breakdown (data not shown), suggesting that this process depends on the kinase activity of PKN. Early stages of the Xenopus cell cycle are controlled by activation and inactivation of the Cdc2/cyclin B complex, the activity of which is measured as histone H1 phosphorylation. Fig. 4A shows the oscillation of histone H1 kinase activity in the Xenopus egg-cycling extracts. The histone H1 kinase activity oscillated in the control extracts in correlation with the cell cycling shown in Fig. 3A. Addition of the kinase-active form of PKCɛ to the egg-cycling extracts did not change the oscillation of Cdc2/cyclin B histone H1 kinase activity (Fig. 4C). However, when the active form of PKN was added to the cycling extracts, histone H1 kinase activation was delayed (Fig. 4B). These results suggest that the delay of mitotic timing in the egg extracts containing the active form of PKN is caused by the suppression of Cdc2 activity. Thus, we analyzed the phosphorylation of Tyr-15 of Cdc2 by immunoblot with the antibody specific for the phospho-Tyr-15 peptide of Cdc2. Cdc2 is retained in an inactive state by Thr-14 and Tyr-15 phosphorylation, and the dephosphorylation of Thr-14 and Tyr-15 by Cdc25C leads to the activation of the Cdc2/cyclin B complex. Fig. 4A Lower shows that the oscillation of dephosphorylation of Tyr-15 of Cdc2 correlated with Cdc2/cyclin B histone H1 kinase activity in the control extracts. The dephosphorylation of Tyr-15 was delayed in extracts containing active PKN (Fig. 4B Lower). These observations indicate that the suppression of Cdc2 activity is derived from the prolonged inhibitory phosphorylation of Tyr-15 of Cdc2.
Figure 3.
Exogenous addition of PKN delays mitosis in egg-cycling extracts. The morphology of sperm nuclei is shown at indicated times after adding sperm nuclei and control buffer (A) or GST/PKN () (B) to a cycling extract. GST/PKN () is indicated as PKN. An input sperm nucleus was visualized by fluorescence with Hoechst 33342.
Figure 4.
Exogenous addition of PKN delays activation of Cdc2/cyclin B histone H1 kinase in egg-cycling extracts. Samples withdrawn from extracts at each time point were assayed for Cdc2/cyclin B histone H1 kinase activity and phosphorylation of Tyr-15 of Cdc2. Oscillation of histone H1 kinase activity in the cycling extracts containing buffer (A), GST/PKN () (B), and GST/PKCɛ (386−737) (C) are shown. A Lower and B Lower show phosphorylation of Tyr-15 of Cdc2 in extracts containing buffer or GST/PKN () by Western blotting with antiphospho-Tyr-15 antibody.
PKN Phosphorylates and Inhibits Cdc25C in Vitro.
We addressed the question of how PKN causes the prolonged inhibitory phosphorylation of Tyr-15 of Cdc2. Three possible mechanisms can be suggested as follows: (i) PKN directly phosphorylates Tyr-15 of Cdc2; (ii) PKN activates Wee1, the upstream protein tyrosine kinase that phosphorylates Tyr-15 of Cdc2; or (iii) PKN inhibits the activity of Cdc25C, the protein phosphatase that removes the inhibitory phosphate of Cdc2.
First, we examined whether PKN can phosphorylate Cdc2 in vitro by using the full-length GST fusion protein expressed in Escherichia coli. PKN phosphorylated Cdc2 protein but not Tyr-15 (data not shown). Furthermore, PKN did not change the activity of Cdc2 in vitro (data not shown). These results suggest that PKN does not regulate directly Cdc2/cyclin B histone H1 kinase activity.
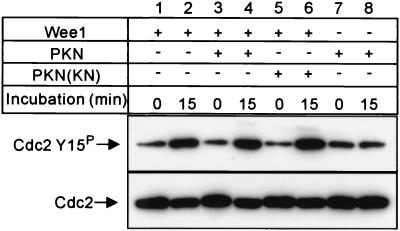
We then analyzed the effect of PKN on Wee1 activity in vitro by using the Cdc2/cyclin B complex affinity purified from Sf9 cells as substrates for Wee1. Fig. 5 (lanes 1 and 2) shows that GST-fused Wee1 affinity purified from Sf9 cells substantially phosphorylated Tyr-15 of Cdc2 in the complex. Incubating Wee1 with the active form of PKN did not enhance the phosphorylation level of Tyr-15 of Cdc2 in the complex (Fig. 5, lanes 3 and 4), indicating that PKN does not directly activate Wee1.
Figure 5.
PKN does not enhance Wee1 activity in vitro. Wee1 activity was assayed by using the substrate Cdc2 (N133A)/cyclin B complex. Affinity-purified GST/Wee1 was incubated with GST/PKN () or GST/PKN ()-K644E. The Cdc2 (N133A)/cyclin B complex was then added, and samples were incubated for 15 min at 25°C. Phosphorylation of Tyr-15 of Cdc2 was analyzed by Western blotting with antiphospho-Tyr-15 antibody (Upper). The same blot was reprobed with anti-Cdc2 antibody (Lower). GST/Wee1, GST/PKN (), and GST/PKN ()-K644E are indicated as Wee1, PKN, and PKN(KN), respectively.
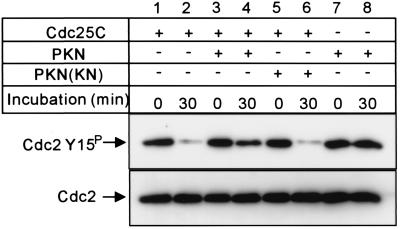
We therefore investigated whether PKN inhibits the dephosphorylation of Tyr-15 of Cdc2 by Cdc25C. We prepared GST/Cdc25C from Sf9 cells and assayed the activity for dephosphorylation of Tyr-15 of Cdc2. The complex of the kinase-negative form of Cdc2 and cyclin B constituted the phosphorylated substrate because active Cdc2 can itself phosphorylate and activate Cdc25C (28). Fig. 6 (lanes 1 and 2) shows that GST/Cdc25C clearly dephosphorylated Tyr-15 of Cdc2. Tyr-15 dephosphorylation was suppressed after Cdc25C was incubated with the active (Fig. 6, lanes 3 and 4) but not the inactive (Fig. 6, lanes 5 and 6) form of PKN.
Figure 6.
PKN inhibits Cdc25C activity in vitro. Cdc25C tyrosine phosphatase activity was assayed by using the Cdc2 (N133A)/cyclin B complex purified from Sf9 cells as the phosphorylated substrate. Affinity-purified GST/Cdc25C was incubated with GST/PKN () or GST/PKN ()-K644E; then Cdc2 dephosphorylation was initiated by addition of the Cdc2 (N133A)/cyclin B complex. Dephosphorylation of Tyr-15 of Cdc2 was analyzed by Western blotting with antiphospho-Tyr-15 antibody (Upper). The same blot was reprobed with anti-Cdc2 antibody (Lower). GST/Cdc25C, GST/PKN (), and GST/PKN ()-K644E are indicated as Cdc25C, PKN, and PKN(KN), respectively.
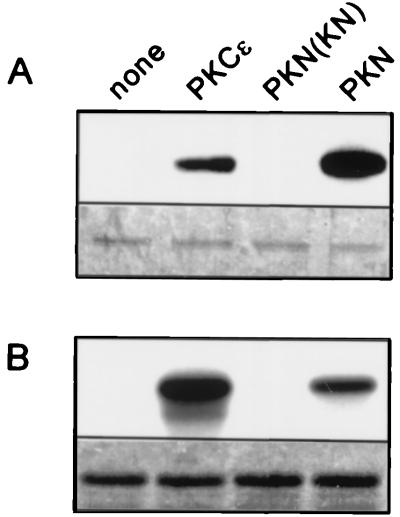
We examined whether PKN can phosphorylate Cdc25C in vitro. Fig. 7 shows that PKN efficiently phosphorylated Cdc25C in vitro. These results indicate that PKN directly inhibits Cdc25C activity by phosphorylation. PKCɛ also phosphorylated Cdc25C. However, compared with myelin basic protein, the efficiency of PKCɛ is lower than that of PKN. Because PKCɛ does not delay mitotic timing, as indicated in Fig. 4, PKN and PKCɛ may recognize the distinct sites of Cdc25C.
Figure 7.
PKN phosphorylates Cdc25C in vitro. (A) Affinity-purified GST/Cdc25C from Sf9 cells was phosphorylated in vitro by GST/PKCɛ (), GST/PKN ()-K644E, or GST/PKN (). (B) Myelin basic protein was phosphorylated by using the same amount of each enzyme as in A. GST/PKCɛ (), GST/PKN ()-K644E, and GST/PKN () are indicated as PKCɛ, PKN(KN), and PKN, respectively. A Lower and B Lower show Coomassie blue staining of each input substrate.
The present study analyzed the biochemical properties of PKN and its role in the regulation of the cell cycle and found that PKN directly phosphorylates and inhibits mitotic regulatory phosphatase of Cdc25C. These results suggest that PKN plays a significant role in the control of mitotic timing by inhibition of Cdc25C. Although the catalytic domain of PKN is highly homologous with the PKC family, and the substrate specificity of PKN is similar to that of the PKC family (13), PKCɛ did not delay mitosis in the cycling extracts, and it phosphorylated Cdc25C less efficiently than PKN. These results suggest that PKN has a specific function for the control of mitotic timing among the PKC-related super family.
PKN has been identified as one of the downstream targets of Rho. Rho has been considered to participate in cytokinesis. It is reported that inhibition of Rho function by botulinum ADP-ribosyltransferase C3 or the dominant-negative form of Rho-kinase blocked cytokinesis in Xenopus embryos and in sand dollars (29, 30). However, our present results suggest that PKN activation delays mitotic timing. Although whether PKN participates in cytokinesis remains unclear, PKN may have a different function in cell-cycle regulation from other downstream targets of Rho.
We reported that PKN translocates to the nucleus on exposure to stress such as heat shock and that the constitutively active form of PKN is generated by apoptotic signaling (14, 15). Such changes of PKN in vivo might affect cell-cycle regulation. Taken together, we speculate that PKN functions as a cell-cycle regulator.
Further analysis with a mammalian system may provide important insights into cell-cycle control of PKN.
Acknowledgments
We thank Y. Nishizuka for encouragement. We also thank N. Nakajo for providing Xenopus Cdc25C cDNA. This work was supported in part by research grants from the Ministry of Education, Science, Sports and Culture, Japan and by the Research for the Future program, the Japan Society for the Promotion of Science.
Abbreviations
- GST
glutathione S-transferase
- His
histidine
- PKC
protein kinase C
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.021541498.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.021541498
References
- 1.King R W, Jackson P K, Kirschner M W. Cell. 1994;79:563–571. doi: 10.1016/0092-8674(94)90542-8. [DOI] [PubMed] [Google Scholar]
- 2.Parker L L, Atherton-Fessler S, Piwnica-Worms H. Proc Natl Acad Sci USA. 1992;89:2917–2921. doi: 10.1073/pnas.89.7.2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mueller P R, Coleman T R, Dunphy W G. Mol Cell Biol. 1995;6:119–134. doi: 10.1091/mbc.6.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mueller P R, Coleman T R, Kumagai A, Dunphy W G. Science. 1995;270:86–90. doi: 10.1126/science.270.5233.86. [DOI] [PubMed] [Google Scholar]
- 5.Watanabe N, Broome M, Hunter T. EMBO J. 1995;14:1878–1891. doi: 10.1002/j.1460-2075.1995.tb07180.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Izumi T, Walker D H, Maller J L. Mol Biol Cell. 1992;3:927–939. doi: 10.1091/mbc.3.8.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kumagai A, Dunphy M W. Cell. 1992;70:139–151. doi: 10.1016/0092-8674(92)90540-s. [DOI] [PubMed] [Google Scholar]
- 8.Blasina A, Van de Weyer I, Laus M C, Luyten W H M L, Parker A E, McGowan C H. Curr Biol. 1999;9:1–10. doi: 10.1016/s0960-9822(99)80041-4. [DOI] [PubMed] [Google Scholar]
- 9.Furnari B, Blasina A, Boddy M N, Mcgowan C H, Russell P. Mol Biol Cell. 1999;10:833–845. doi: 10.1091/mbc.10.4.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zeng Y, Forbes K C, Wu Z, Moreno S, Piwnica-Worms H, Enoch T. Nature (London) 1998;395:507–510. doi: 10.1038/26766. [DOI] [PubMed] [Google Scholar]
- 11.Mukai H, Ono Y. Biochem Biophys Res Commun. 1994;199:897–904. doi: 10.1006/bbrc.1994.1313. [DOI] [PubMed] [Google Scholar]
- 12.Mukai H, Kitagawa M, Shibata H, Takanaga H, Mori K, Shimakawa M, Miyahara M, Hirao K, Ono Y. Biochem Biophys Res Commun. 1994;204:348–356. doi: 10.1006/bbrc.1994.2466. [DOI] [PubMed] [Google Scholar]
- 13.Kitagawa M, Mukai H, Shibata H, Ono Y. Biochem J. 1995;310:657–664. doi: 10.1042/bj3100657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mukai H, Miyahara M, Sunakawa H, Shibata H, Toshimori M, Kitagawa M, Shimakawa M, Takanaga H, Ono Y. Proc Natl Acad Sci USA. 1996;93:10195–10199. doi: 10.1073/pnas.93.19.10195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takahashi M, Mukai H, Toshimori M, Miyamoto M, Ono Y. Proc Natl Acad Sci USA. 1998;95:11566–11571. doi: 10.1073/pnas.95.20.11566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amano M, Mukai H, Ono Y, Chihara K, Matsui T, Hamajima Y, Okawa K, Iwamatsu A, Kaibuchi K. Science. 1996;271:648–650. doi: 10.1126/science.271.5249.648. [DOI] [PubMed] [Google Scholar]
- 17.Watanabe G, Saito Y, Madaule P, Ishizaki T, Fujisawa K, Mori N, Mukai H, Ono Y, Kakizuka A, Narumiya S. Science. 1996;271:645–648. doi: 10.1126/science.271.5249.645. [DOI] [PubMed] [Google Scholar]
- 18.Shibata H, Mukai H, Inagaki Y, Homma Y, Kimura K, Kaibuchi K, Narumiya S, Ono Y. FEBS Lett. 1996;385:221–224. doi: 10.1016/0014-5793(96)00385-7. [DOI] [PubMed] [Google Scholar]
- 19.Machesky L M, Hall A. Trends Cell Biol. 1996;6:304–310. doi: 10.1016/0962-8924(96)10026-x. [DOI] [PubMed] [Google Scholar]
- 20.Hall A. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 21.Yasui Y, Amano M, Nagata K, Inagaki N, Nakamura H, Saya H, Kaibuchi K, Inagaki M. J Cell Biol. 1998;143:1249–1258. doi: 10.1083/jcb.143.5.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Madaule P, Eda M, Watanabe N, Fujisawa K, Matsuoka T, Bito H, Ishizaki T, Narumiya S. Nature (London) 1998;394:491–494. doi: 10.1038/28873. [DOI] [PubMed] [Google Scholar]
- 23.Yoshinaga C, Mukai H, Toshimori M, Miyamoto M, Ono Y. J Biochem (Tokyo) 1999;126:475–484. doi: 10.1093/oxfordjournals.jbchem.a022476. [DOI] [PubMed] [Google Scholar]
- 24.Ono Y, Fujii T, Ogita K, Kikkawa U, Igarashi K, Nishizuka Y. J Biol Chem. 1988;263:6927–6932. [PubMed] [Google Scholar]
- 25.Nakajo N, Yoshitome S, Iwashita J, Iida M, Uto K, Ueno S, Okamoto K, Sagata N. Genes Dev. 2000;14:328–338. [PMC free article] [PubMed] [Google Scholar]
- 26.Murray A W. Methods Cell Biol. 1991;36:581–605. [PubMed] [Google Scholar]
- 27.Ohsumi K, Sawada W, Kishimoto T. J Cell Sci. 1994;107:3005–3013. doi: 10.1242/jcs.107.11.3005. [DOI] [PubMed] [Google Scholar]
- 28.Hoffmann I, Clarke P R, Jessus Marcote M, Karsenti E, Draetta G. EMBO J. 1993;12:53–63. doi: 10.1002/j.1460-2075.1993.tb05631.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kishi K, Sasaki T, Kuroda S, Itoh T, Takai Y. J Cell Biol. 1993;120:1187–1195. doi: 10.1083/jcb.120.5.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mabuchi I, Hamaguchi Y, Fujimoto H, Morii N, Mishima M, Narumiya S. Zygote. 1993;1:325–331. doi: 10.1017/s0967199400001659. [DOI] [PubMed] [Google Scholar]