Abstract
The crystal structure of biotin synthase from Escherichia coli in complex with S-adenosyl-l-methionine and dethiobiotin has been determined to 3.4 angstrom resolution. This structure addresses how “AdoMet radical” or “radical SAM” enzymes use Fe4S4 clusters and S-adenosyl-l-methionine to generate organic radicals. Biotin synthase catalyzes the radical-mediated insertion of sulfur into dethiobiotin to form biotin. The structure places the substrates between the Fe4S4 cluster, essential for radical generation, and the Fe2S2 cluster, postulated to be the source of sulfur, with both clusters in unprecedented coordination environments.
Biotin synthase (BioB) catalyzes the final step in the biotin biosynthetic pathway, the conversion of dethiobiotin (DTB) to biotin. This remarkable reaction uses organic radical chemistry for the insertion of a sulfur atom between nonactivated carbons C6 and C9 of DTB (Scheme 1). BioB is a member of the “AdoMet radical” or “radical SAM” superfamily, which is characterized by the presence of a conserved CxxxCxxC sequence motif (C, Cys; x, any amino acid) that coordinates an essential Fe4S4 cluster, as well as by the use of S-adenosyl-l-methionine (AdoMet or SAM) for radical generation (1-3). AdoMet radical enzymes act on a wide variety of biomolecules. For example, BioB and lipoyl-acyl carrier protein synthase (LipA) are involved in vitamin biosynthesis; lysine 2,3-aminomutase (LAM) facilitates the fermentation of lysine; class III ribonucleotide reductase (RNR) activase and pyruvate formate-lyase (PFL) activase catalyze the formation of glycyl radicals in their respective target proteins; and spore photoproduct lyase repairs ultraviolet light-induced DNA damage.
Scheme 1.
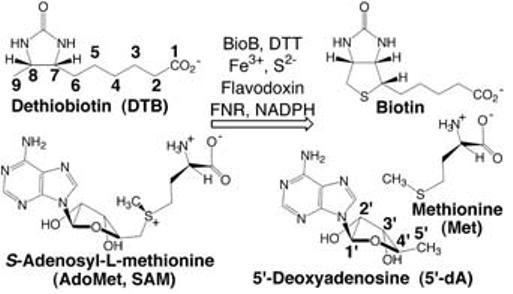
Overall reaction catalyzed by BioB.
AdoMet has been referred to as the “poor man's adenosylcobalamin” (4) because of the ability of both cofactors to generate a highly reactive 5′-deoxyadenosyl radical (5′-dA·), formed through homolytic cleavage of a C-Co bond in the case of adenosylcobalamin (AdoCbl) and through reductive cleavage of a C-S bond in the case of AdoMet (5). In AdoMet radical enzymes, the formation of 5′-dA· requires the addition of one electron, provided in E. coli by reduced flavodoxin and transferred first into an Fe4S4 cluster and then into AdoMet (3). In the reaction catalyzed by BioB, there is general agreement that 5′-dA· generated from AdoMet oxidizes DTB (6), but the number and types of FeS clusters and of other cofactors involved in the reaction have been a subject of controversy (7-14). Protein preparation-dependent cofactor differences have led to two mechanistic proposals for the method of S insertion in BioB. One proposal involves the use of an Fe2S2 cluster as the sulfur source for biotin, and is consistent with 34S isotopic labeling studies (15) and with the observed destruction of an Fe2S2 cluster that accompanies BioB turnover (16, 17). Another proposal is based on the activity of BioB from which the Fe2S2 cluster has been removed, and suggests that S is provided from an enzyme-bound persulfide generated via an intrinsic pyridoxal-5′-phosphate (PLP)-dependent cysteine desulfurase activity (10, 18). Irrespective of whether BioB preparations contain an Fe2S2 cluster or PLP, assay mixtures produce one or fewer turnovers per monomer over the course of several minutes to hours (10, 16, 19). Thus, it has been suggested that BioB is either subject to strong product inhibition (18) or may be a “suicide enzyme” (i.e., a stoichiometric reactant rather than a true catalyst).
The crystal structure of BioB complexed with AdoMet and DTB reveals the general architecture of the AdoMet radical superfamily, for which there are an estimated 600 unique protein members (20). Here, we describe the fold of an AdoMet radical protein, the structural basis of AdoMet binding, and the novel FeS clusters of BioB. This work addresses the mechanism of radical generation and sulfur insertion, and sheds light on the possible evolutionary relationships of radical enzymes.
The structure of BioB has been determined to 3.4 Å resolution by iron multiwavelength anomalous dispersion (MAD) techniques (21). The fold of each subunit of the BioB dimer is a triosephosphate isomerase (TIM) type (α/β)8 barrel, with two additional helices at the N terminus and a disordered region at the C terminus (Fig. 1). BioB joins at least 25 enzyme superfamilies incorporating TIM barrel architecture and three AdoCbl-dependent enzymes that use this fold for catalysis of radical-based chemistry (22). However, it is unusual to find FeS clusters bound within and on top of a TIM barrel.
Fig.1.
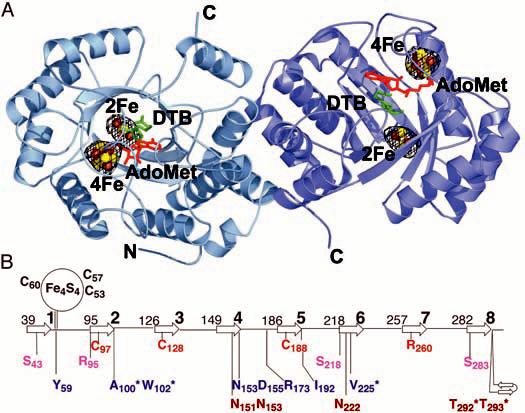
(A) Structure of BioB, FeS clusters, and bound AdoMet and DTB. BioB exists as a homodimer in solution (11) and we find two possible dimeric relationships between BioB monomers in the crystal. The dimer shown here buries 17.6% of the monomer surface area (13,249 Å2) and is likely to be physiologically relevant. An anomalous Fourier electron density map, calculated with data collected at the Fe absorption peak wavelength (1.73827 Å) and phases from the polypeptide portion of the model, is contoured at 3σ in black mesh. These electron density peaks represent the positions of the four FeS clusters in this dimeric structure. There are no other features of a similar size in the electron density map. The FeS clusters are shown as ball-and-stick representations, with brown Fe atoms and yellow S atoms. In addition, we find one AdoMet (red) and one DTB (green) per subunit. Figures 1A and 2 were prepared with PyMOL (42). (B) Topology diagram of the BioB TIM barrel showing the location of important residues with respect to the β strands (arrows, numbered 1 to 8). The numbers to the left of each β strand correspond to the N-terminal residue of that secondary structure element. Ligands to the Fe4S4 cluster are in black, ligands to the Fe2S2 cluster are in red, and residues that contact the Fe2S2 cluster ligand Arg260 are in pink. AdoMet contacts (blue) include Ala100, Trp102, and Arg173, which form hydrogen bonds to the amino acid moiety; Asp155 and Asn153, which form hydrogen bonds to the ribose hydroxyl groups (Fig. 2A); Tyr59 and Ile192, which stack against the adenine ring (Fig. 2A); and Val225, which forms backbone hydrogen bonds to the adenine ring. Residues in position to form hydrogen bonds to DTB (brown) include Asn151, Asn153, and Asn222, which contact the DTB ureido ring (Fig. 2C), and Thr292 and Thr293, which contact the carboxylate tail. Asterisks denote main-chain interactions.
An Fe4S4 cluster is located at the C-terminal end of the TIM barrel, far (∼30 Å) from the dimer interface, and an Fe2S2 cluster is located deep inside the barrel, ∼25 Å from the C-terminal end (Fig. 1A). The active site, containing AdoMet and DTB, is situated between these two clusters (Fig. 1A). This cluster arrangement suggests an explanation for the reported differences in FeS cluster content: A surface FeS cluster may be more readily lost and reconstituted than a deeply buried cluster. The Fe2S2 cluster is observed in recombinant BioB in vivo by Mössbauer spectroscopy (7, 23) and is always retained in the initial purified protein (8, 11, 12, 14, 24). Treatment of BioB with strong chemical reductants and metal chelators, followed by FeS reconstitution, yields an Fe2S2-depleted form of BioB with an intact Fe4S4 cluster (8, 9, 13, 25, 26). Reconstituted protein not subjected to a reduction or chelation step (such as used herein) contains both an Fe4S4 and an Fe2S2 cluster (7, 13, 14).
With respect to the proposal that PLP is a cofactor in BioB (10, 18), we did not add PLP during protein purification or crystallization and do not find it in our structure. Additionally, the structure does not reveal an obvious binding site for PLP. Indeed, cysteine desulfurases typically bind PLP via an imine linkage to a Lys residue, and all Lys residues, including the one highly conserved Lys (Lys49 in BioB), are found at the surface of the protein, far from the putative site of a cysteine persulfide (10) and the DTB binding site. Therefore, this x-ray structure analysis favors a mechanism that invokes a role for an Fe2S2 cluster over one that requires PLP.
The Fe2S2 cluster of BioB is unique in terms of its coordination by β-strand residues in the core of a barrel, as well as in terms of the identity of the amino acid ligands: Cys97, Cys128, Cys188, and Arg260 (Fig. 1B and Fig. 2B). All four of these residues are absolutely conserved, and the assignment of the fourth ligand as Arg is unambiguous even at the moderate resolution of this structure; the amino acid sequence of the TIM barrel has been fully assigned with no breaks or gaps, and the shape of the omit electron density at position 260 is completely consistent with an Arg side chain (Fig. 2B). The assignment of an Arg ligand to a metal is unprecedented in biology, although guanidine species have been observed to ligate CoIII, OsIII, PtII, NiII, and ZnII in small molecules in aqueous solution at a pH range of ∼3.5 to 10 (27-29). It has been predicted that Arg could serve as a metal ligand in a protein environment where the Arg side chain was uncharged (29), and in BioB, Arg260 has an unusual environment that should indeed alter its pKa. It is buried in the center of a TIM barrel and has a high number of potential hydrogen-bonding partners (Ser43, Ser218, Ser283, and Arg95) (Fig. 2B). The conservation of Arg as an FeS cluster ligand in BioB, rather than the more common Cys or His residues, suggests an important role for this residue in modulating the properties of the cluster or facilitating catalysis. The choice of metal ligands may be important for preserving charge neutrality within the core of the (α/β)8 barrel, because one common mechanism for FeS cluster redox modulation by backbone NH-to-S hydrogen bonds is not possible with this structural motif. All surrounding backbone NHs are involved in hydrogen bonding between the β strands of the barrel and cannot interact with the cluster. Thus, the ligands and neighboring side chains are likely to play an important role in redox modulation, and an Arg (or His) ligand would be a better choice than Cys in terms of reducing the overall net negative charge of this buried cluster. The use of Arg rather than His is intriguing. Arg260 could play a structural role, in that the long length of the Arg side chain allows for a barrel that is less compact and able to fit both substrates, or this residue could play a catalytic role in the S insertion reaction. During turnover of BioB, the Fe2S2 cluster is destroyed as one S is transferred into biotin (16). In our structure, the Arg260 side chain could rearrange to bridge the two Fe atoms and facilitate this proposed S transfer. Our knowledge of how FeS clusters are assembled in biological systems is still in its infancy, and it will be interesting to see whether any of the features observed in the BioB structure are mimicked in proteins involved in cluster assembly.
Fig.2.
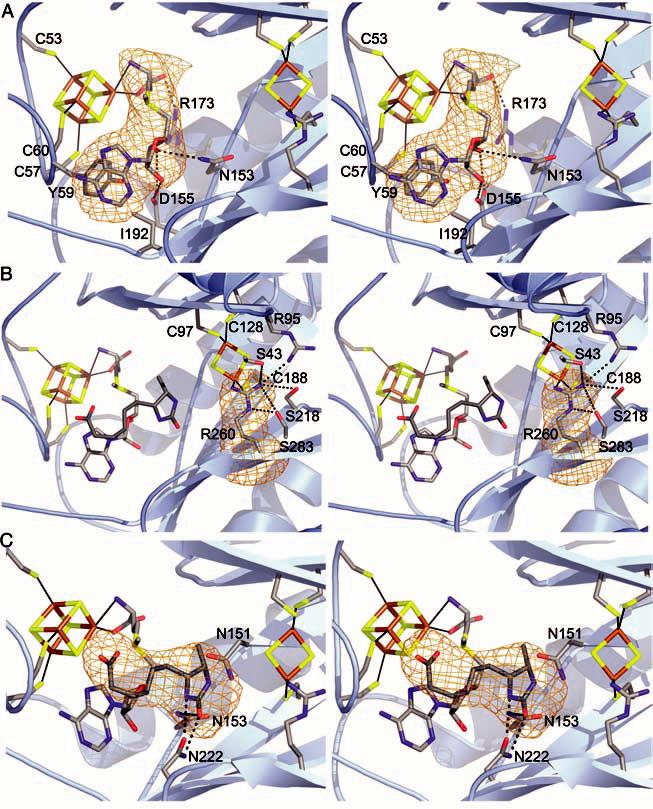
(A) Stereo view of the Fe4S4 cluster with AdoMet bound. Conserved side-chain contacts between BioB and AdoMet are indicated, and AdoMet is shown in a simulated annealing omit map contoured at 4.5σ (orange). DTB is omitted for clarity. Color code: C, gray; O, red; N, blue; S, yellow; Fe, brown. (B) Stereo view of the active site, focusing on the Fe2S2 cluster and its ligands. The unusual Arg260 ligand is shown in a simulated annealing omit map contoured at 4.5σ. In addition to the Fe2S2 cluster, Arg260 interacts with Ser43, Ser218, Ser283, and Arg95. Also shown are the positions of the Fe4S4 cluster, AdoMet, and DTB with respect to the Fe2S2 cluster. (C) Stereo view of DTB interacting with AdoMet and conserved residues Asn222, Asn151, and Asn153 in the active site. Potential hydrogen bonds between Asn222 and DTB are drawn as dashed lines. The stacking of the carboxylate tail of DTB and the adenine ring of AdoMet is visible in the orientation, although contacts with Thr292 and Thr293 are not. DTB is shown in a simulated annealing omit map contoured at 4.0σ.
Proteins are classified as members of the AdoMet radical superfamily on the basis of the presence of the sequence motif CxxxCxxC (Cys53, Cys57, and Cys60 for BioB) (20). This motif is contained in a 28-residue loop extending from β strand 1 to helix 1 of the TIM barrel, and the three cysteines coordinate three of the four irons of the Fe4S4 cluster, as expected from mutagenesis studies (24, 30). AdoMet is the fourth ligand to the cluster and binds as an N/O chelate to a unique Fe position through its amino-group nitrogen and carboxyl-group oxygen (Fig. 2A). This binding mode is consistent with spectroscopic results for PFL activase (31, 32) and is likely to be common to AdoMet radical proteins, because spectral changes associated with AdoMet binding have been reported for several family members (33-35). Also consistent with spectroscopic data on BioB and PFL activase (36, 37), the sulfonium of AdoMet does not appear to be a ligand to Fe and is ∼4.0 Å away from the nearest cluster Fe. In addition to its interaction with the Fe4S4 cluster, the AdoMet binding site comprises residues from disparate parts of the sequence and of the three-dimensional structure (Fig. 1B). AdoMet is bound in an extended conformation (Fig. 2A), stretching across the top of the barrel, such that contacts are made to AdoMet by residues in or following β strands 1, 2, and 4 to 6. The net result of these interactions is that AdoMet is completely buried from solvent and appears to be ideally positioned for electron transfer from the Fe4S4 cluster and hydrogen atom abstraction from DTB.
The enzyme preparation used for crystallization has DTB bound (38), and we observe electron density in the active site that fits this substrate (Fig. 2C). DTB binds in the core of the TIM barrel, between the Fe2S2 cluster and AdoMet (Fig. 1A). In contrast to the reported stoichiometry of bound substrates (38), we find one DTB and one AdoMet in each BioB subunit. AdoMet binding to BioB is greatly enhanced (by a factor of >20) when DTB is also bound (38), and we find that DTB makes substantial van der Waals contacts with AdoMet, covering 50% of its surface. These interactions include the stacking of the carboxylate tail against the AdoMet adenine and the stacking of the DTB ureido ring with the AdoMet ribose (Fig. 2, B and C). DTB makes a series of contacts with protein as well, the most important of which is the bidentate interaction with Asn222 (Fig. 2C) that may play a role in orienting the substrate for hydrogen atom abstraction. Finally, the DTB carboxylate interacts with the backbone amides of Thr292 and Thr293 as well as the side chain of Thr292 (Fig. 1B). Interactions with these residues could serve to close a loop over the top of the barrel upon DTB binding, sealing the barrel for the radical-mediated reaction that follows.
Although the elucidation of a detailed enzyme mechanism awaits further biochemical studies, the structure does provide insight into key steps of the BioB reaction. The observed cooperativity of substrate binding, for example, can be explained by the extensive interaction between AdoMet and DTB. With respect to radical generation, the electron transfer from flavodoxin to the Fe4S4 cluster is made possible by the binding of the cluster close to the protein surface (∼6 to 7 Å). The subsequent electron transfer from the Fe4S4 cluster to the AdoMet sulfonium is facilitated by direct O/N coordination of the AdoMet to Fe, thereby restraining the position of the sulfonium to the proximity of the Fe4S4 cluster. The resulting formation of the 5′-dA· species is likely coupled to hydrogen atom abstraction from C9 of DTB, and we find that DTB is positioned accordingly, with C9 ∼3.9 Å away from the 5′ carbon of AdoMet. Completion of the biotin thiophane ring requires the abstraction of a second hydrogen atom from C6 of DTB. Deuterium transfer from of the C6 position of (2H)DTB has shown that a second 5′-dA· is likely also responsible for this second hydrogen atom abstraction (6), although the requirement for two equivalents of AdoMet has recently been disputed (18). Here, we find that C6 of DTB is positioned ∼4.1 Å from the 5′ carbon position of AdoMet, which suggests that 5′-dA· also accomplishes the second hydrogen atom abstraction.
The sulfur insertion step is the most controversial part of the mechanism of BioB. The structure analysis shows that the Fe2S2 cluster is ideally positioned to play a role in sulfur insertion (Fig. 2B). The closest bridging S of the Fe2S2 cluster is 4.6 Å away from C9 of DTB, a position consistent with transfer of this sulfur to biotin, as suggested by results from 34S-labeling experiments (15) and the observation of Fe2S2 cluster degradation during turnover (16, 17). If the in vivo sulfur source is the Fe2S2 cluster, then is BioB a “suicide enzyme” capable of performing only a single turnover because of the destruction of the Fe2S2 cluster? Only two biotin-requiring proteins are present in E. coli: acetyl-CoA carboxylase and the bifunctional biotin operon repressor/holocarboxylase synthetase (BirA). Therefore, a single turnover of multiple BioB proteins may provide enough biotin for the life cycle of E. coli. Alternatively, the Fe2S2 cluster may be rebuilt after each turnover by cysteine desulfurases (IcsS or SufS) and FeS cluster assembly proteins (IscU/A or SufA), conferring true catalytic capability to BioB. Although the Fe2S2 cluster is buried, the movement of Arg95 and/or Tyr149 could open access to the Fe2S2 cluster from the bottom of the barrel. Thus, the cluster may be destroyed from the top and rebuilt from the bottom.
An open question is whether all AdoMet radical enzymes will share the TIM barrel fold. The consensus motif CxxxCxxC, used to identify AdoMet radical proteins, represents a single loop that could be inserted into a series of different structural scaffolds. In contrast, the AdoMet binding residues are spread throughout the primary sequence (Fig. 1B). Unfortunately, the sequence motif for AdoMet binding is not as obvious as that for the Fe4S4 cluster, because several contacts to AdoMet are from the protein backbone (Fig. 1B), making it difficult to say with confidence that AdoMet radical enzymes will bind AdoMet with identical protein structural motifs. However, there are a few short sequence motifs, such as the one surrounding Ile192 (Fig. 2A), that have homologs in other AdoMet radical proteins: IxGxxE in BioB, MxGxxE in LipA, VxGxxD in PFL activase, and LxGxxD in LAM, for example (D, Asp; E, Glu; G, Gly; I, Ile; L, Leu; M, Met; V, Val). The primary sequences of the known and predicted AdoMet radical proteins are also of reasonable length for TIM barrel folds, with the exception of the class III RNR activase sequence, which is likely too short to contain a full (α/β)8 barrel. If the protein fold does turn out to be the same for most or all AdoMet radical proteins, this would raise an interesting question as to how these enzymes generate radicals on such structurally divergent substrates. A barrel fold makes sense for small substrates like DTB that can fit into the active site and be sealed away from solvent for catalytic steps involving radical intermediates. The choice of this fold has less obvious value for the members of the superfamily whose substrates are proteins or DNA.
From an evolutionary perspective, it is interesting to compare the AdoMet-dependent radical enzyme BioB with the structures of AdoCbl-dependent radical enzymes. With the exception of the class II ribonucleotide reductases, AdoCbl-dependent radical enzymes use the TIM barrel architecture for substrate binding (39-41). AdoCbl is bound to a separate domain that docks with the TIM barrel at its C-terminal end. Superposition of the TIM barrel of BioB with the TIM barrel of AdoCbl-dependent diol dehydratase (40) shows that the corrin ring occupies the same position as the BioB Fe4S4 cluster. Evolutionary conversion of an AdoMet radical enzyme to an AdoCbl radical enzyme may be as simple as deleting the CxxxCxxC loop to make room for the docking of AdoCbl bound to an additional protein or domain. The surprising similarity between the structures of BioB and AdoCbl radical enzymes indicates that the TIM barrel fold may be a preferred scaffold for 5′-dA· chemistry and suggests an evolutionary relationship between these two enzyme superfamilies.
Supplementary Material
Footnotes
Supporting Online Material www.sciencemag.org/cgi/content/full/303/5654/76/DC1 Materials and Methods References Table S1
References and Notes
- 1.Frey PA, Booker SJ. Adv. Protein Chem. 2001;58:1. doi: 10.1016/s0065-3233(01)58001-8. [DOI] [PubMed] [Google Scholar]
- 2.Cheek J, Broderick JB. J. Biol. Inorg. Chem. 2001;6:209. doi: 10.1007/s007750100210. [DOI] [PubMed] [Google Scholar]
- 3.Jarrett JT. Curr. Opin. Chem. Biol. 2003;7:174. doi: 10.1016/s1367-5931(03)00022-x. [DOI] [PubMed] [Google Scholar]
- 4.Frey PA. FASEB J. 1993;7:662. doi: 10.1096/fasebj.7.8.8500691. [DOI] [PubMed] [Google Scholar]
- 5.Frey PA, Reed GH. Arch. Biochem. Biophys. 2000;382:6. doi: 10.1006/abbi.2000.2010. [DOI] [PubMed] [Google Scholar]
- 6.Escalettes F, Florentin D, Tse Sum Bui B, Lesage D, Marquet A. J. Am. Chem. Soc. 1999;121:3571. [Google Scholar]
- 7.Benda R, et al. Biochemistry. 2002;41:15000. doi: 10.1021/bi026590q. [DOI] [PubMed] [Google Scholar]
- 8.Duin EC, et al. Biochemistry. 1997;36:11811. doi: 10.1021/bi9706430. [DOI] [PubMed] [Google Scholar]
- 9.Ollagnier-de Choudens S, et al. Biochemistry. 2000;39:4165. doi: 10.1021/bi992090u. [DOI] [PubMed] [Google Scholar]
- 10.Ollagnier-de Choudens S, Mulliez E, Hewiston KS, Fontecave M. Biochemistry. 2002;41:9145. doi: 10.1021/bi0122011. [DOI] [PubMed] [Google Scholar]
- 11.Sanyal I, Cohen G, Flint DH. Biochemistry. 1994;33:3625. doi: 10.1021/bi00178a020. [DOI] [PubMed] [Google Scholar]
- 12.Tse Sum Bui B, Florentin D, Marquet A, Benda R, Trautwein AX. FEBS Lett. 1999;459:411. doi: 10.1016/s0014-5793(99)01300-9. [DOI] [PubMed] [Google Scholar]
- 13.Ugulava NB, Gibney BR, Jarrett JT. Biochemistry. 2001;40:8343. doi: 10.1021/bi0104625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ugulava NB, Surerus KK, Jarrett JT. J. Am. Chem. Soc. 2002;124:9050. doi: 10.1021/ja027004j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tse Sum Bui B, et al. FEBS Lett. 1998;440:226. [Google Scholar]
- 16.Ugulava NB, Sacanell CJ, Jarrett JT. Biochemistry. 2001;40:8352. doi: 10.1021/bi010463x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tse Sum Bui B, et al. Biochemistry. 2003;42:8791. doi: 10.1021/bi034426c. [DOI] [PubMed] [Google Scholar]
- 18.Ollagnier-de Choudens S, Mulliez E, Fontecave M. FEBS Lett. 2002;532:465. doi: 10.1016/s0014-5793(02)03733-x. [DOI] [PubMed] [Google Scholar]
- 19.Sanyal I, Gibson KJ, Flint DH. Arch. Biochem. Biophys. 1996;326:48. doi: 10.1006/abbi.1996.0045. [DOI] [PubMed] [Google Scholar]
- 20.Sofia HJ, Chen G, Hetzler BG, Reyes-Spindola JF, Miller NE. Nucleic Acids Res. 2001;29:1097. doi: 10.1093/nar/29.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.The crystallographic R factor is 25.6% and Rfree is 30.0%. For a description of the structure elucidation and a table of data and refinement statistics, see the supporting online material [Google Scholar]
- 22.Murzin AG, et al. J. Mol. Biol. 1995;247:536. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 23.Cosper MM, Jameson GNL, Eidsness MK, Huynh BH, Johnson MK. FEBS Lett. 2002;529:332. doi: 10.1016/s0014-5793(02)03390-2. [DOI] [PubMed] [Google Scholar]
- 24.Hewitson KS, Baldwin JE, Shaw NM, Roach PL. FEBS Lett. 2000;466:372. doi: 10.1016/s0014-5793(00)01101-7. [DOI] [PubMed] [Google Scholar]
- 25.Ollagnier-de Choudens S, Sanakis Y, Hewiston KS, Roach P, Munck E. J. Biol. Chem. 2002;277:13449. doi: 10.1074/jbc.M111324200. [DOI] [PubMed] [Google Scholar]
- 26.Ugulava NB, Gibney BR, Jarrett JT. Biochemistry. 2000;39:5206. doi: 10.1021/bi9926227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baranska M, Gumienna-Kontecka E, Kozlowski H, Proniewicz LM. J. Inorg. Biochem. 2002;92:112. doi: 10.1016/s0162-0134(02)00485-3. [DOI] [PubMed] [Google Scholar]
- 28.Aoki S, Iwaida K, Hanamoto N, Shiro M, Kimura E. J. Am. Chem. Soc. 2002;124:5256. doi: 10.1021/ja020029y. [DOI] [PubMed] [Google Scholar]
- 29.Fairlie DP, et al. Inorg. Chem. 1997;36:1020. doi: 10.1021/ic961138e. [DOI] [PubMed] [Google Scholar]
- 30.Hewitson KS, et al. J. Biol. Inorg. Chem. 2002;7:83. doi: 10.1007/s007750100268. [DOI] [PubMed] [Google Scholar]
- 31.Krebs C, Broderick WE, Henshaw TF, Broderick JB, Huynh BH. J. Am. Chem. Soc. 2002;124:912. doi: 10.1021/ja017562i. [DOI] [PubMed] [Google Scholar]
- 32.Walsby CJ, Ortillo D, Broderick WE, Broderick JB, Hoffman BM. J. Am. Chem. Soc. 2002;124:11270. doi: 10.1021/ja027078v. [DOI] [PubMed] [Google Scholar]
- 33.Cosper MM, et al. J. Am. Chem. Soc. 2002;124:14006. doi: 10.1021/ja0283044. [DOI] [PubMed] [Google Scholar]
- 34.Liu A, Graslund A. J. Biol. Chem. 2000;275:12367. doi: 10.1074/jbc.275.17.12367. [DOI] [PubMed] [Google Scholar]
- 35.Padovani D, Thomas F, Trautwein AX, Mulliez E, Fontecave M. Biochemistry. 2001;40:6713. doi: 10.1021/bi002936q. [DOI] [PubMed] [Google Scholar]
- 36.Walsby CJ, et al. J. Am. Chem. Soc. 2002;124:3143. doi: 10.1021/ja012034s. [DOI] [PubMed] [Google Scholar]
- 37.Cosper MM, et al. Protein Sci. 2003;12:1573. doi: 10.1110/ps.0302203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ugulava NB, Frederick KK, Jarrett JT. Biochemistry. 2003;42:2708. doi: 10.1021/bi0261084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mancia F, et al. Structure. 1996;4:339. doi: 10.1016/s0969-2126(96)00037-8. [DOI] [PubMed] [Google Scholar]
- 40.Masuda J, Shibata N, Morimoto Y, Toraya T, Yasuoka N. Struct. Fold. Des. 2000;8:775. doi: 10.1016/s0969-2126(00)00164-7. [DOI] [PubMed] [Google Scholar]
- 41.Reitzer R, et al. Struct. Fold. Des. 1999;7:891. [Google Scholar]
- 42.DeLano WL. http://www.pymol.org The PyMOL Molecular Graphics System. 2002 [Google Scholar]
- 43.Supported by NIH grants R01-GM59175 (J.T.J.) and R01-GM65337 (C.L.D.), the Searle Scholars Program (C.L.D.), the Cecil and Ida Green Career Development Fund (C.L.D.), a Lester Wolfe Predoctoral Fellowship (F.B.), and a Cellular, Biochemical, and Molecular Sciences NIH training grant T32-GM07229 (J.T.W.). Data were collected at the National Synchrotron Light Source (NSLS), Advanced Light Source (ALS), and Stanford Synchrotron Radiation Laboratory (SSRL). Synchrotron facilities are funded by the U.S. Department of Energy (ALS 5.0.2, NSLS X25, SSRL), NIH National Center of Research Resources (NSLS X25), and National Institute of General Medical Sciences (NSLS X25). Coordinates and structure factors have been deposited in the Protein Data Bank with accession number 1R30 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


