Abstract
eIF5A is a highly conserved putative eukaryotic translation initiation factor that has been implicated in translation initiation, nucleocytoplasmic transport, mRNA decay, and cell proliferation, but with no precise function assigned so far. We have previously shown that high-copy PKC1 suppresses the phenotype of tif51A-1, a temperature-sensitive mutant of eIF5A in S. cerevisiae. Here, in an attempt to further understand how Pkc1 functionally interacts with eIF-5A, it was determined that PKC1 suppression of tif51A-1 is independent of the cell integrity MAP kinase cascade. Furthermore, two new suppressor genes, ZDS1 and GIC1, were identified. We demonstrated that ZDS1 and ZDS2 are necessary for PKC1, but not for GIC1 suppression. Moreover, high-copy GIC1 also suppresses the growth defect of a PKC1 mutant (stt1), suggesting the existence of a Pkc1-Zds1-Gic1 pathway. Consistent with the function of Gic1 in actin organization, the tif51A-1 strain shows an actin polarity defect that is partially recovered by overexpression of Pkc1 and Zds1 as well as Gic1. Additionally, PCL1 and BNI1, important regulators of yeast cell polarity, also suppress tif51A-1 temperature sensitivity. Taken together, these data strongly support the correlated involvement of Pkc1 and eIF5A in establishing actin polarity, which is essential for bud formation and G1/S transition in S. cerevisiae.
THE putative translation initiation factor 5A (eIF5A) is a highly conserved and essential protein present in all organisms from archebacteria to mammals but not in eubacteria (Schnier et al. 1991; Chen and Liu 1997). Despite its highly conserved essential function, the critical cellular role of eIF5A is not known. A number of studies implicate eIF5A in a variety of cellular processes.
The involvement of eIF5A with translation initiation was proposed due to its purification from ribosomes of reticulocyte lysates and to its stimulatory effect in the methionyl-puromycin assay used to implicate factors in the first peptide bond formation (Benne and Hershey 1978). However, depletion of eIF5A in yeast caused only a slight decrease in the protein synthesis rate, arguing against a role as a general translation initiation factor. Therefore, it was hypothesized that eIF5A may function in the translation of a specific subset of mRNAs. Since depletion of eIF5A in yeast also causes an increase of G1-arrested cells, judged by cell morphology, it was proposed that eIF5A may be important for translating mRNAs encoding proteins required for cell cycle progression (Kang and Hershey 1994). This connection between eIF5A and cell cycle progression is further supported by the observation that blocking any step of hypusination, its essential post-translational modification, in mammalian cells inhibits cell proliferation (Park et al. 1997), placing eIF5A among the potential targets for cancer therapy (Caraglia et al. 2001).
eIF5A has also been implicated in nucleocytoplasmic export of Rev-dependent HIV-1 transcripts and mRNA decay (Ruhl et al. 1993; Bevec et al. 1996; Bevec and Hauber 1997; Zuk and Jacobson 1998). However, subsequent studies have not confirmed the involvement of eIF5A with Rev-dependent nuclear export in either mammalian or yeast systems (Shi et al. 1996, 1997; Henderson and Percipalle 1997; Lipowsky et al. 2000; Li-En Jao and Chen 2002; Valentini et al. 2002). Moreover, the effect of eIF5A on mRNA decay seems to be secondary, as arrest of cell growth of eIF5A temperature-sensitive mutants does not directly correlate with mRNA accumulation (Valentini et al. 2002).
Thus, although eIF5A has been associated with different cellular events, the role played by this essential factor remains unclear. In an attempt to identify cellular partners for eIF5A and understand its critical cellular function, a temperature-sensitive mutant of TIF51A (tif51A-1), one of the genes encoding eIF5A in Saccharomyces cerevisiae, was used in a high-copy suppressor screen. PKC1, encoding for the only yeast protein kinase C, was one of the suppressors isolated together with three members of the cell integrity pathway, WSC1, WSC2, and WSC3 (Valentini et al. 2002).
Pkc1 in S. cerevisiae controls a variety of cellular processes such as cell cycle progression, mating, nutrient sensing, and the structural organization of the cytoskeleton (Heinisch et al. 1999). Pkc1 is activated by a small GTPase of the Rho family, Rho1, which receives upstream signals from the Slg1 (Wsc1) and Mid2 transmembrane sensors (Philip and Levin 2001). The downstream PKC1-mitogen-activated protein kinase (MAPK) cascade, consisting of Bck1, Mkk1/Mkk2, and Mpk1 kinases, phosphorylates transcription factors that regulate cell wall remodeling and cytoskeleton organization in polarized cell growth (Heinisch et al. 1999). Although less well defined, several studies report evidence for other biochemical pathways branching out from Pkc1 (Ketela et al. 1999; Andrews and Stark 2000; Li et al. 2000; Nanduri and Tartakoff 2001; Chai et al. 2002; Valdivia and Schekman 2003; Vilella et al. 2005).
In this work, we further analyzed the functional interaction between eIF5A and the Pkc1-cell integrity pathway. Our results demonstrate that PKC1 suppression of tif51A-1 is MAP kinase independent. Furthermore, we present data that suggest the existence of a novel pathway independent of the MAP kinases. This pathway links Pkc1 to the Cdc42 effector, Gic1, in a Zds1/Zds2-dependent manner. The results herein favor a more direct involvement of Pkc1 in actin polarization, which is necessary for bud formation during G1/S transition. Finally, we discuss a possible role for eIF5A in this process.
MATERIALS AND METHODS
Yeast strains manipulation and plasmids:
S. cerevisiae strains used in this work are listed in Table 1. Procedures for cell growth and genetic manipulations were carried out according to standard protocols (Guthrie and Fink 1991). Plasmids used in this work are listed in Table 2. The functionality of all plasmids showing negative results in the high-copy suppression analysis (Table 3) was confirmed by complementation or phenotypic suppression of known mutants (supplemental material at http://www.genetics.org/supplemental/). Cloning by PCR was performed with Pfx DNA polymerase (Invitrogen, San Diego) or with Vent DNA polymerase (New England Biolabs, Beverly, MA) following standard molecular biology procedures (Ausubel et al. 2005).
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| SVL14 | MATaade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 ssd1-d tif51A-1 | Valentini et al. (2002) |
| SVL26 | MATaade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 ssd1-d tif51A-2 | Valentini et al. (2002) |
| SVL32 | MATaade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 ssd1-d tif51A-3 | Valentini et al. (2002) |
| SVL248 | MATaade2 his3 leu2 trp1 ura3 tif51A(ts1159) | This study |
| SVL82 (W303) | MATaade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 ssd1-d | Pamela Silver |
| SVL170 (DLY486) | MATα ade8 his3 leu2 met3 ura3 stt1 (pkc1) | David Levin |
| SVL230 (YEF877) | MATα his3 leu2 lys2 trp1 ura3 zds1ΔHIS3 | Bi and Pringle (1996) |
| SVL321 (CY832) | MATaade2-1 his3-11,15 leu2-3,112 ura3-1 can1-100 trp1-1 ssd1ΔLEU2 | Rosenwald et al. (2002) |
| SVL327 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 zds2ΔkanMX4 | Anita Corbett |
| SVL411 | MATaade2 his3 leu2 ura3 trp1 zds1ΔHIS3 zds2ΔkanMX4 ssd1-d tif51A-1 | This study |
| SVL412 | MATaade2 his3 leu2 ura3 trp1 ssd1ΔLEU2 tif51A-1 | This study |
| SVL531 | MATα ade2 his3 leu2 ura3 trp1 zds1ΔHIS3 ssd1ΔLEU2 | This study |
| SVL532 | MATα ura3 leu2 his3 trp1 zds1ΔHIS3 zds2ΔkanMX4 ssd1ΔLEU2 | This study |
TABLE 2.
Plasmids used in this study
| Plasmid | Feature | Source |
|---|---|---|
| pSV58 (pRS314) | TRP1, CEN | Sikorski and Hieter (1989) |
| pSV64 (pRS425) | LEU2, 2μ | Christianson et al. (1992) |
| pSV65 (pRS426) | URA3, 2μ | Christianson et al. (1992) |
| pSV107 | TIF51A, URA3, 2μ | Valentini et al. (2002) |
| pSV108 | TIF51A, LEU2, 2μ | Valentini et al. (2002) |
| pSV115 | WSC1, URA3, 2μ | Valentini et al. (2002) |
| pSV116 | WSC2, URA3, 2μ | Valentini et al. (2002) |
| pSV117 | WSC3, URA3, 2μ | Valentini et al. (2002) |
| pSV181 | PKC1, URA3, 2μ | Valentini et al. (2002) |
| pSV292 (pROM2) | ROM2, TRP1, 2μ | Helliwell et al. (1998) |
| pSV293 (pRHO2) | RHO2, TRP1, 2μ | Helliwell et al. (1998) |
| pSV294 (pBCK1-20) | BCK1-20, URA3, CEN | Helliwell et al. (1998) |
| pSV295 (pMKK1) | MKK1, URA3, 2μ | Helliwell et al. (1998) |
| pSV296 (pMPK1) | MPK1 (SLT2), URA3, 2μ | Helliwell et al. (1998) |
| pSV304 | GIC1, URA3, 2μ | This study |
| pSV352 (YEp24-BNI1) | BNI1, URA3, 2μ | Helliwell et al. (1998) |
| pSV376 (YEp351-CDC24) | CDC24, LEU2, 2μ | Richman et al. (1999) |
| pSV377 (YEp351-CDC42) | CDC42, LEU2, 2μ | Richman et al. (1999) |
| pSV381 (YEp13-BEM3) | BEM3, LEU2, 2μ | Richman et al. (1999) |
| pSV383 (YEp13-RGA1) | RGA1, LEU2, 2μ | Richman et al. (1999) |
| pSV384 (pFK2CU) | SSD1, URA3, CEN | Uesono et al. (1994) |
| pSV387 (YEp13-MID2) | MID2, LEU2, 2μ | de Bettignies et al. (2001) |
| pSV388 (YEp13-MTL1) | MTL1, LEU2, 2μ | de Bettignies et al. (2001) |
| pSV396 (EBO459) | CLN2, URA3, 2μ | Lenburg and O'Shea (2001) |
| pSV401 (pDLB678) | BEM1, URA3, 2μ | Gladfelter et al. (2002) |
| pSV403 (pDLB722) | CLA4, URA3, 2μ | Gladfelter et al. (2002) |
| pSV407 (YEp352-STE20) | STE20, URA3, 2μ | Gladfelter et al. (2002) |
| pSV411 (MPO120) | PCL1, URA3, 2μ | Lenburg and O'Shea (2001) |
| pSV412 (MPO121) | PCL2, URA3, 2μ | Lenburg and O'Shea (2001) |
| pSV454 (pKT1057) | BEM2, LEU2, 2μ | Kawasaki et al. (2003) |
| pSV455 (pKT1130) | BUD1, LEU2, 2μ | Kawasaki et al. (2003) |
| pSV501 | TIF51A, TRP1, CEN | This study |
| pSV531 (pTH123) | GIC1, LEU2, 2μ | Höfken and Schiebel (2004) |
| pSV532 (pTH124) | GIC2, LEU2, 2μ | Höfken and Schiebel (2004) |
| pSV534 (pTH179) | GIC1CRIB−, LEU2, 2μ | Höfken and Schiebel (2004) |
| pSV535 (pTH185) | GIC1-pr, LEU2, 2μ | Höfken and Schiebel (2004) |
| pSV588 (YEp-ZDS2) | ZDS2, URA3, 2μ | Yu et al. (1996) |
| pSV589 | ZDS1, URA3, 2μ | This study |
TABLE 3.
Genes analyzed for their ability to suppress the temperature-sensitive phenotype of the tif51A-1 mutant
| Gene | Functional information | Suppression |
|---|---|---|
| TIF51A | Putative translation initiation factor eIF5A | + |
| WSC1 | Sensor for cell wall integrity signaling, plays a role in activation of the Pkc1-MAPK pathway; Wsc family | + |
| WSC2 | Putative sensor for cell wall integrity signaling; Wsc family | + |
| WSC3 | Putative sensor for cell wall integrity signaling; Wsc family | + |
| MID2 | Sensor for cell wall integrity signaling, plays a role in activation of the Pkc1-MAPK pathway; Mtl1 homolog | + |
| MTL1 | Protein that acts in signal transduction of cell wall stress, plays a role in regulation of cell integrity pathway; Mid2 homolog | + |
| ROM2 | GDP/GTP exchange factor (GEF) for Rho1 and Rho2 | + |
| RHO2 | GTP-binding protein, member of the rho subfamily of ras-like proteins | + |
| PKC1 | Protein kinase C, regulates MAP kinase cascade involved in regulating cell wall metabolism (cell integrity pathway) | + |
| BCK1-20a | MAP kinase kinase kinase component of cell integrity pathway | − |
| MKK1 | MAP kinase kinase component of cell integrity pathway | − |
| MPK1 | MAP kinase component of cell integrity pathway | − |
| ZDS1 | Protein with effects on cell polarity and transcriptional silencing, homolog of Zds2 | + |
| GIC1 | Effector of Cdc42, important for bud emergence; Gic2 homolog | + |
| PCL1 | G1/S-specific Pho85 cyclin | + |
| PCL2 | G1/S-specific Pho85 cyclin | − |
| CLN2 | G1/S-specific Cdc28 cyclin | − |
| GIC2 | Effector of Cdc42, important for bud emergence; Gic1 homolog | − |
| CDC42 | Rho-type GTPase involved in bud site assembly and cell polarity | − |
| CLA4 | PAK kinase required for cytokinesis, effector of Cdc42 | − |
| STE20 | PAK kinase of the pheromone pathway; also regulates polarized growth, effector of Cdc42 | − |
| CDC24 | GEF for Cdc42, involved in bud emergence, bud site selection, and growth of mating projection | − |
| BUD1 | GTP-binding protein of the ras superfamily involved in bud site selection | − |
| BEM1 | Scaffold protein for complexes involving cell polarity establishment and morphogenesis factors such as Cdc24 and Bud1 | + |
| BEM2 | Rho-type GTPase-activating protein (GAP) for Rho1 | − |
| BNI1 | Formin protein involved in cytoskeletal polarization and cytokinesis | + |
| ZDS2 | Protein with effects on cell polarity and transcriptional silencing, homolog of Zds1 | + |
| RGA1 | Rho-type GAP for Cdc42 | − |
| BEM3 | Rho-type GAP for Cdc42 | − |
| SSD1 | mRNA binding protein; plays a role in maintenance of cellular integrity | + |
a Encodes a constitutively activated version of Bck1 (Lee and Levin 1992).
High-copy suppressor screen:
A URA3/2μ genomic yeast library (Connelly and Hieter 1996) was transformed into a tif51A-1 strain (SVL14). Approximately 40,000 transformants were selected by plating on uracil dropout plates, incubating at 25° overnight, and shifting plates to 36° for 3–4 days. Plasmids were rescued from temperature-resistant clones and retransformed into SVL14 to test for plasmid linkage. The genomic segment present in each of the selected clones was determined by sequencing the ends with T3 and T7 primers and using these sequences to search the S. cerevisiae genome database. High-copy suppressor genes were characterized by subcloning different segments of the original clone into pSV65 (pRS426) and testing them in SVL14. GIC1 and ZDS1 were also cloned into pSV65 (pRS426) using the following primers: GIC1-A, 5′-CGG GGT ACC AAT ACG TAC CCG GGT AGT AG-3′; GIC1-B, 5′-CGG GGT ACC GTC TGA GCA GGA ATA AAG AG-3′; and ZDS1-A, 5′-CGC GGA TCC TGG AAT TCT ATC GAG CGA CC-3′; ZDS1-B, 5′-CGC GGA TCC CTC TGT TCT TAT ACG GTT CC-5′. The plasmid pSV501 was constructed by subcloning the BamHI fragment containing TIF51A from pSV107 into pSV58 (pRS314).
Phalloidin staining:
Staining of actin filaments was carried out essentially as described (Amberg 1998). Cells from exponential phase cultures were fixed at room temperature for 1 hr with 3.7% formaldehyde in the culture medium plus 1 hr incubation in PBS with 3.7% formaldehyde. Cells were subsequently washed twice with PBS and resuspended in 500 μl of PBS. Staining was performed by adding 10 μl of rhodamine-phalloidin (Molecular Probes, Eugene, OR) to 100 μl of cell suspension. Cells were incubated in the dark for 1 hr, washed five times with 1 ml of PBS, and finally suspended in 100 μl of mount solution [90% glycerol, 0.1× PBS, 92.5 mm p-phenylenediamine (Sigma, St. Louis), pH adjusted to 0.8 with 0.5 m sodium carbonate, pH 9.0]. Stained cells were stored at −20° until microscopic analysis. Rhodamine-phalloidin-stained cells (2.5 μl) were visualized by fluorescence microscopy using a rhodamine filter and a Nikon TE300 inverted microscope. Images were captured with a MicroMax 5-MHz CCD (Princeton Instruments, Princeton, NJ) and the software Image-Pro Plus (Media Cybernetics).
RESULTS
PKC1 suppression of a temperature-sensitive mutant of eIF5A occurs in a MAP kinase-independent pathway:
In a previous study, three temperature-sensitive alleles of TIF51A, tif51A-1, tif51A-2, and tif51A-3 were characterized and used to further investigate eIF5A function in yeast (Valentini et al. 2002). As PKC1 and WSC1-3 were identified as high-copy suppressors of the tif51A-1 mutant (Valentini et al. 2002), we decided to analyze the mechanism governing this suppression. Pkc1, the yeast protein kinase C counterpart, with its putative upstream regulators Wsc1 to -3, plays an important role in cell integrity maintenance. This function is performed by Pkc1 signaling through different downstream effectors to achieve cell wall remodeling and actin reorganization in response to several stimuli, including heat shock, pheromone, low osmolarity, nutrient starvation, and cell cycle progression (Heinisch et al. 1999). A well-established effector cascade downstream of Pkc1 is the MAP kinase pathway, which is composed of the kinases Bck1, Mkk1/Mkk2, and Mpk1. Activation of Mpk1 upregulates transcription of a series of genes involved in cell cycle progression and cell wall synthesis (Gustin et al. 1998). While transcription of a great number of genes is mediated by the MAP kinases cascade in response to different cell wall impairments, little is known about other Pkc1 downstream effectors (Ketela et al. 1999; Andrews and Stark 2000; Li et al. 2000; Nanduri and Tartakoff 2001; Chai et al. 2002; Valdivia and Schekman 2003).
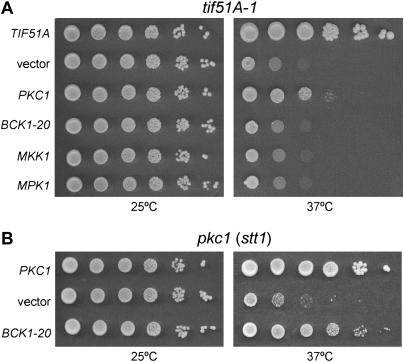
To determine whether the suppression of tif51A-1 promoted by high-copy PKC1 occurs via the MAP kinases, we examined whether high-copy MKK1, MPK1, or an activated allele of BCK1, BCK1-20, could suppress the temperature-sensitive phenotype of tif51A-1. Unlike PKC1, none of the MAP kinase genes was able to suppress the tif51A-1 growth defect at the nonpermissive temperature of 37° (Figure 1A). However, as a control, the activated allele BCK1-20 successfully overcame the defect of a PKC1 temperature-sensitive mutant, stt1 (Yoshida et al. 1994; Figure 1B). This result suggests that, although BCK1-20 can suppress the temperature-sensitive phenotype of the pkc1 mutant (stt1), activation of the cell integrity MAP kinase pathway is not sufficient to overcome the tif51A-1 growth phenotype. In contrast to the results with genes that function downstream of Pkc1, high-copy plasmids containing genes for known and putative upstream activators of Pkc1 (including WSC1-3, MID2, MTL1, ROM2, and RHO2) do suppress the tif51A-1 mutant phenotype (Hohmann 2002; Table 3). Taken together, these results strongly suggest that Pkc1 does not act through its downstream MAP kinase cascade to promote suppression of the tif51A-1 temperature-sensitive phenotype.
Figure 1.
PKC1 suppression of the tif51A-1 mutant is independent of its downstream MAP kinase effectors. (A) Tenfold serial dilutions of early saturated tif51A-1 cells (SVL14) harboring pSV107 (TIF51A), pRS426 (vector), pSV181 (PKC1), pSV294 (BCK1-20), pSV295 (MKK1), or pSV296 (MPK1) were plated onto SC plates to determine growth at permissive (25°) or nonpermissive (37°) temperatures. (B) PKC1 temperature-sensitive mutant cells (SVL170) harboring pSV181 (PKC1), pRS426 (vector), or pSV294 (BCK1-20) were assayed as described in A. The plates were photographed after 3–4 days of growth.
New suppressors of a temperature-sensitive mutant of eIF5A suggest a novel pathway linking Pkc1 to cell polarity:
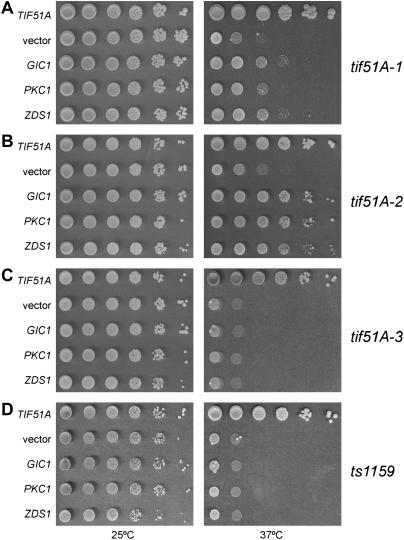
Since enhanced activity of cell integrity MAP kinase cascade members failed to suppress the temperature-sensitive phenotype of the tif51A-1 mutant, we screened for new suppressors that could help us to understand the functional relationship between Pkc1 and eIF5A. Two additional high-copy suppressors were identified: GIC1 and ZDS1 (Figure 2A).
Figure 2.
High-copy suppressors of eIF5A mutants. Dilutions of the tif51A-1 (A, SVL14), tif51A-2 (B, SVL26), tif51A-3 (C, SVL32), and ts1159 strains (D, SVL248) bearing the plasmids pSV107 (TIF51A), pRS426 (vector), pSV304 (GIC1), pSV181 (PKC1), and pSV589 (ZDS1) were grown for 3–4 days on SC plates at 25° and 37°.
The mutant used in the screen above is one of the four yeast eIF5A mutants described so far: ts1159 (Zuk and Jacobson 1998), tif51A-1, tif51A-2, and tif51A-3 (Valentini et al. 2002). To analyze whether PKC1, ZDS1, and GIC1 suppression is allele specific, we tested the temperature sensitivity of eIF5A mutants other than tif51A-1 in the presence of the suppressors (Figure 2, B–D). As observed, only tif51A-2 is also suppressed by PKC1, ZDS1, and GIC1, while no growth improvement is conferred to the alleles ts1159 and tif51A-3. Interestingly, tif51A-1 and tif51A-2 mutants contain amino acid substitutions in the same residue (P83S and P83L, respectively). On the other hand, the mutants that were not suppressed harbor amino acid changes in different points of eIF5A: ts1159 (S149P) and tif51A-3 (C39Y and G118D). Therefore, this allele-specific suppression may reflect the presence of similar defects in tif51A-1 and tif51A-2 mutants that can be bypassed by PKC1, ZDS1, and GIC1. However, as mutants ts1159 and tif51A-3 are much more sick than the others (data not shown; Valentini et al. 2002), the lack of suppression by these genes may not reflect the occurrence of completely different phenotypes between ts1159/tif51A-3 and tif51A-1/tif51A-2, but rather may be due to a broader range of defects in the former mutants. In agreement with this hypothesis is the fact that PKC1, ZDS1, and GIC1 are not able to suppress tif51AΔ, or tif51A-1 and tif51A-2 at higher temperatures (data not shown), demonstrating that these suppressors can only partially correct the defects of the mutants analyzed.
Gic1 is an effector of Cdc42 that is important for bud emergence and contains a Cdc42/Rac-interactive-binding (CRIB) domain, which mediates interaction with GTP-bound Cdc42 (Brown et al. 1997). Gic1, together with its homolog Gic2, seems to exert its function during the G1/S cell cycle transition by linking the major polarization organizer Cdc42 and the formin Bni1, which is responsible for induction of actin polymerization (Pruyne et al. 2004). Zds1 has also been implicated in cell cycle progression but with a less well-defined function (Ma et al. 1996). ZDS1 and ZDS2, its functionally redundant homolog, have been isolated in a series of high-copy suppressor screens (Schwer and Shuman 1996; Tsuchiya et al. 1996; Bourbonnais et al. 2001; Sekiya-Kawasaki et al. 2002). Interestingly, high levels of Zds1 can decrease Cdc42 activity (Bi and Pringle 1996) and high-throughput two-hybrid data have shown physical interactions among Pkc1, Zds2, Zds1, and Gic1 (Drees et al. 2001).
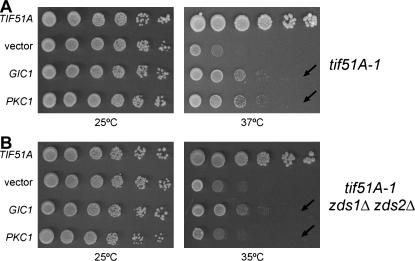
To further investigate the relationship among Pkc1, Zds1, and Gic1, we tested whether Zds function is important for the suppression mediated by high-copy PKC1 or GIC1. To address this question, a tif51A-1 strain lacking both ZDS1 and ZDS2 was generated (SVL411), and the ability of PKC1 or GIC1 to suppress the temperature-sensitive growth phenotype of this mutant was examined (Figure 3). To avoid possible influences of the genetic background on PKC1 suppression, this assay was performed in parallel with the strain SVL412, obtained from the same tetrad as SVL411, but containing only the tif51A-1 mutation. Interestingly, PKC1 could not suppress tif51A-1 in the absence of ZDS1 and ZDS2, while GIC1 could (Figure 3, A and B, compare rows indicated by arrows). Also, the tif51A-1 zds1Δ zds2Δ strain demonstrates enhanced temperature sensitivity, lowering from 37° to 35° the restrictive temperatures of the triple mutant (Figure 3, A and B, right). This synthetic sickness between these genes strengthens their functional connection. These data, together with the previous physical interactions described (Drees et al. 2001), strongly support a model in which Pkc1 acts through a downstream pathway different from the MAP kinase cascade. Furthermore, these data suggest that both Zds1 and Gic1 participate in this signaling pathway. Thus, as all these factors act in the same pathway to promote eIF5A mutant suppression, it is possible that eIF5A plays a role in cell polarity.
Figure 3.
Suppression of the tif51A-1 mutant by PKC1 is dependent on the ZDS1 and ZDS2 genes. Dilutions of the tif51A-1 (A, SVL412) and tif51A-1 zds1Δ zds2Δ (B, SVL411) strains harboring plasmids pSV107 (TIF51A), pRS426 (vector), pSV304 (GIC1), or pSV181 (PKC1) were grown for 3–4 days on SC plates at 25°, 35°, or 37°.
The eIF5A mutant tif51A-1 genetically interacts with G1/S transition factors that are involved with cell polarity and important for proper actin organization:
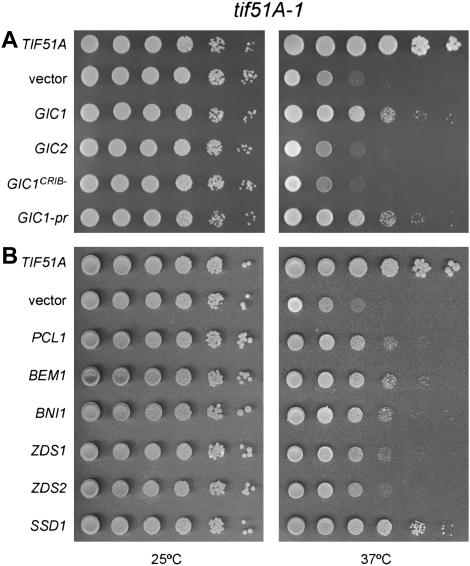
To better understand the function of eIF5A, other genes functionally related to those suppressors identified in the screen were tested to determine whether they could also suppress the growth defect of the tif51A-1 mutant (Table 3). As mentioned before, for those genes related to the cell integrity pathway, only members proposed to act upstream of Pkc1 (Wsc1-3, Mid2, Mtl1, Rom2, and Rho2) (Hohmann 2002) were able to suppress the temperature-sensitive phenotype of the tif51A-1 mutant. In addition to the role of Pkc1 in the cell integrity pathway, this kinase is also genetically linked to G1 cyclins via an interaction network (Lenburg and O'shea 2001). Thus, we tested whether overexpression of the G1 cyclins PCL1, PCL2, and CLN2 would suppress the tif51A-1 mutant. Notably, only Pcl1 could suppress the tif51A-1 mutant (Figure 4B; Table 3).
Figure 4.
Suppression of the tif51A-1 mutant requires different factors related to cell cycle progression. Serial dilutions of tif51A-1 cells (SVL14) carrying the indicated genes on LEU2 (A) or URA3 (B) plasmids were spotted on SC plates and incubated for 3–5 days at 25° and 37°.
Then, genes functionally linked to Gic1 were tested for suppression of the tif51A-1 mutant. Initially, the GIC1 homolog, GIC2, and the gene encoding the Gic1 recruiter, CDC42, were tested. Unexpectedly, neither one was able to suppress the tif51A-1 mutant phenotype, suggesting that the suppression is dependent on a Gic1-specific function (Figure 4A; Table 3). Genes encoding other effectors of Cdc42, CLA4 and STE20, and the Cdc42 activator, CDC24, also could not suppress the tif51A-1 phenotype (Table 3). These findings reinforce the hypothesis that a Gic1-specific function is required for tif51A-1 suppression.
To determine whether the interaction between Gic1 and Cdc42 is required for GIC1-mediated suppression of tif51A-1, it was tested if GIC1CRIB−, an allele known to disrupt protein-protein interactions with Cdc42 (Brown et al. 1997), can suppress the tif51A-1 mutant. As shown in Figure 4A, GIC1 lacking the CRIB domain could not suppress tif51A-1, suggesting that although CDC42 does not suppress by itself, a Gic1-Cdc42 physical interaction may be necessary for high-copy GIC1 suppression of the tif51A-1 temperature-sensitive phenotype.
In addition to a Gic1 function in actin polarization, a high-copy suppressor screen with let1Δ, a mitotic exit network component mutant, has recently implicated Gic1 in this process. This new role for Gic1 is separable from that in the G1/S transition, since the function in mitotic exit requires a pool of Gic1 not associated with the cell cortex, as a prenylated form of Gic1 (Gic1-pr) cannot work in the mitotic exit (Höfken and Schiebel 2004). To determine which Gic1 function is necessary to suppress the tif51A-1 mutant phenotype, we tested the cortex-restricted Gic1 for suppression of tif51A-1. As shown in Figure 4A, this form of Gic1 promotes growth of the tif51A-1 mutant at the nonpermissive temperature. This result supports the model that the G1/S transition function of Gic1 is responsible for the tif51A-1 mutant suppression.
The last set of genes functionally linked to GIC1 includes BEM2, BEM1, BNI1, and BUD1. These genes are involved with polarized cell growth and show genetic defects with gic1Δ gic2Δ (Chen et al. 1997; Jaquenoud and Peter 2000; Kawasaki et al. 2003). We tested whether any of these genes could suppress the phenotype of the tif51A-1 mutant. This analysis revealed that both BEM1 and BNI1 are high-copy suppressors of tif51A-1 (Figure 4B; Table 3). These results show that other factors important for G1/S transition are also able to suppress the tif51A-1 mutant.
Finally, we tested whether the following ZDS1-correlated genes could suppress tif51A-1: ZDS2, its homolog; RGA1 and BEM3, encoding negative regulators of Cdc42; and SSD1, a gene of unknown function that suppresses defects in the cell integrity pathway and has its null mutant suppressed by ZDS1 (Tsuchiya et al. 1996; Kaeberlein and Guarente 2002). Among these genes, only ZDS2 and SSD1 suppress the tif51A-1 phenotype (Figure 4B; Table 3). These results show that ZDS2 shares the ZDS1 function necessary for tif51A-1 mutant suppression. The fact that presence of SSD1 can greatly compensate for loss of eIF5A function is considered later (see discussion). The observation that neither RGA1 nor BEM3 was identified as suppressors suggests that the Zds1 function important for suppression is, most probably, not related to its role in negative regulation of Cdc42 (Bi and Pringle 1996).
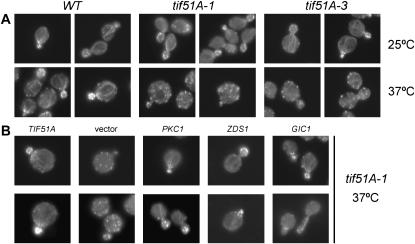
Taken together, the results point to a G1/S defect in the tif51A-1 mutant as most of the suppressors described above act in cell polarity during cell cycle progression. An important event in G1/S transition in S. cerevisiae is the establishment of an axis of polarity. Immediately after the positioning of the Cdc42-related factors at the bud site, the assembly of a polarized actin cytoskeleton is crucial for progression of the cell cycle (Pruyne et al. 2004). Therefore, we tested whether two mutants of TIF51A, tif51A-1 and tif51A-3, exibit any actin polarization defect. Interestingly, both mutants showed marked defects in actin cytoskeleton organization in budding cells at the nonpermissive temperature. In contrast, actin cables and patches appeared normal during growth at the permissive temperature (Figure 5A).
Figure 5.
The actin cytoskeleton defect of the tif51A-1 mutant is also suppressed by the temperature-sensitive phenotype suppressors PKC1, ZDS1, and GIC1. (A) Wild-type strain SVL82 (W303) and tif51A-1 and tif51A-3 mutant (SVL14 and SVL32) cells were grown at 25° and then shifted to 37° for 4 hr. Actin was stained with rhodamin-conjugated phalloidin and visualized by fluorescence microscopy. (B) tif51A-1 cells (SVL14) bearing pSV107 (TIF51A), pRS426 (vector), pSV181 (PKC1), pSV589 (ZDS1), and pSV304 (GIC1) were analyzed as described in A.
To test if PKC1 and the newly identified suppressors, GIC1 and ZDS1, can suppress the actin organization defect as well as the growth phenotype of the tif51A-1 mutant at the nonpermissive temperature, the actin cytoskeleton organization of this tif51A-1 mutant in the presence of high-copy PKC1, GIC1, and ZDS1 was analyzed (Figure 5B). The results demonstrate that suppression of the temperature-sensitive growth phenotype is correlated with a suppression of the actin organization defect at the restrictive temperature and suggest that eIF5A plays a role in the establishment of cell polarity.
GIC1 overexpression can cause cell hyperpolarization:
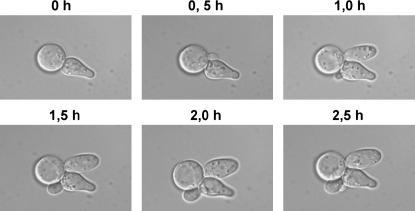
While analyzing the effect of overexpression of GIC1 on the actin cytoskeleton defect of the tif51A-1 mutant, it was noted that ∼10–20% of cells exhibited elongated buds with a hyperpolarized actin cytoskeleton (Figure 5B, GIC1, bottom). To investigate if this hyperpolarization was correlated with the tif51A-1 mutant suppression by GIC1, the morphology of cells growing on a plate at the restrictive temperature was inspected. We observed that both elongated and normal morphologies were present, but elongated ones occurred at a much lower number, apparently at the same frequency as that seen during actin cytoskeleton observation (data not shown). Furthermore, to check if cells acquiring elongated morphology are able to successfully progress through the cell cycle and thus are not sick or dead, time-lapse microscopy was performed. This analysis revealed no apparent defect in growth and, moreover, the generation of new buds was also detected (Figure 6). These data imply that actin hyperpolarization is not necessary for GIC1 suppression and that the resulting elongated cells can properly progress through the cell cycle.
Figure 6.
Expression of GIC1 from a high-copy plasmid generates elongated viable cells at low frequency. Time-lapse images of tif51A-1 cells (SVL14) containing pSV304 (GIC1) are shown. An aliquot of midlog phase cells growing in SC liquid medium was collected and monitored by time-lapse microscopy after immobilizing cells on SC pads. Cells were photographed at 30-min intervals for 150 min at room temperature.
GIC1 and BNI1 can bypass the growth defect of a PKC1 temperature-sensitive mutant:
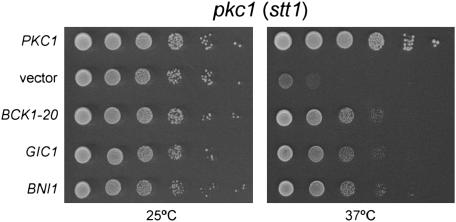
The results presented above suggest a pathway branching from Pkc1 to Gic1. Thus, we hypothesized that increased levels of Gic1 would suppress a PKC1 mutant. To test this hypothesis, the ability of high-copy GIC1, as well as of the downstream acting formin BNI1, to suppress the temperature-sensitive phenotype of the PKC1 mutant stt1 was evaluated (Figure 7). In agreement with our hypothesis, high-copy GIC1 and BNI1 could improve the PKC1 mutant (stt1) growth at the restrictive temperature. This finding further supports the existence of a novel Gic1 pathway downstream of Pkc1.
Figure 7.
High-copy GIC1 and BNI1 suppress the temperature-sensitive phenotype of a pkc1 (stt1) mutant. Tenfold dilutions of the pkc1 (stt1) strain (SVL170) harboring plasmids pSV181 (PKC1), pRS426 (vector), pSV294 (BCK1-20), or pSV304 (GIC1) were grown for 3–4 days on SC plates at 25° and 37°.
DISCUSSION
PKC1 suppression of the eIF5A mutant is MAP kinase independent:
In this work, we have analyzed the mechanism that underlies the high-copy PKC1 suppression of the temperature-sensitive eIF5A mutant tif51A-1. Although PKC1 and some of its putative upstream activators show genetic interactions with the tif51A-1 mutant, our genetic analysis indicates that the downstream MAP kinase cascade members of the cell integrity pathway are not able to suppress the tif51A-1 mutant, suggesting that a less well-characterized different branch of Pkc1 signaling is involved in this suppression. It has been proposed that Pkc1 has other downstream effectors, as the pkc1 null mutant phenotype is more severe than those associated with the lack of the genes encoding the downstream components of the MAP kinase cascade (Heinisch et al. 1999). In fact, several cellular mechanisms in which Pkc1 plays a role are MAP kinase independent (Ketela et al. 1999; Andrews and Stark 2000; Li et al. 2000; Nanduri and Tartakoff 2001; Chai et al. 2002; Valdivia and Schekman 2003; Vilella et al. 2005).
Moreover, consistent with the suppression data, although eIF5A mutants show enhanced growth on media containing 1 m sorbitol (Valentini et al. 2002), they do not demonstrate other phenotypes associated with defects in cell integrity of MAP kinase mutants such as cell lysis at the restrictive temperature and sensitivity to caffeine, staurosporine (a specific inhibitor of PKC isozymes), and calcofluor white (Heinisch et al. 1999) (data not shown). Thus, considering the fact that PKC1 suppression is MAP kinase independent, we searched for new suppressors of the eIF5A mutant tif51A-1 that could unveil the effectors downstream of Pkc1 that are rescuing the growth impairment of this eIF5A mutant.
A novel pathway links Pkc1 to the Cdc42 effector Gic1:
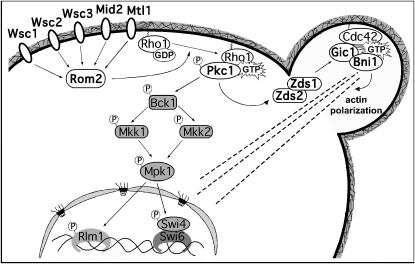
In addition to the well-known tif51A-1 mutant suppressor PKC1, we show here that ZDS1 and GIC1 are also suppressors of tif51A-1. As the proteins encoded by these three genes participate in a network of physical interactions (Drees et al. 2001), we tested the hypothesis that Pkc1-Zds1-Gic1 constitutes a new signaling pathway, illustrated in the model in Figure 8. In agreement with this model, PKC1 suppression is abolished in tif51A-1 cells lacking the redundant genes ZDS1 and ZDS2. In contrast, high-copy GIC1 still suppresses the tif51A-1 zds1Δ zds2Δ triple mutant. Considering that Pkc1 acts in response to Rho1 (Heinisch et al. 1999), this is the first study demonstrating functional data linking Rho1- and Cdc42-regulated pathways through Zds1 and Zds2.
Figure 8.
Schematic of pathways acting downstream of Pkc1. In addition to the well-known MAP kinase cascade, we propose a link between Pkc1 and Gic1 via Zds1 and Zds2. This novel pathway connects Pkc1 directly to cell polarity factors and appears to be responsible for high-copy suppression of the eIF5A mutant tif51A-1.
In addition to the fact that the Zds proteins physically interact with factors important for polarized growth, septin organization, and cytokinesis (Drees et al. 2001), ZDS1 and ZDS2 genetically interact with CDC42 and SWE1, important factors regulating cell cycle progression and morphogenesis (Bi and Pringle 1996; Ma et al. 1996). Thus, Zds proteins may act as an integration point for distinct signaling pathways that helps maintain a balance among different signals. This model could explain why Zds1 downregulates both Pkc1-MAP kinase and Cdc42 activity (Bi and Pringle 1996; Griffioen et al. 2003). In fact, high-copy ZDS1 does not suppress the temperature-sensitive phenotype of the PKC1 mutant stt1 (data not shown), as high-copy GIC1 does (discussed below). However, although overexpression of ZDS1 causes a cell integrity defect at elevated temperatures (Griffioen et al. 2003), high-copy PKC1 is not toxic to zds1Δ zds2Δ cells at any temperature, and it does not exacerbate even the cold sensitivity of this double mutant (data not shown). Therefore, the inability of PKC1 to act as a suppressor in the strain tif51A-1 zds1Δ zds2Δ is not likely the result of Pkc1 overactivation and toxicity. Thus, Zds1 may exert Pkc1-MAP kinase negative feedback and also act as a downstream member of a pathway leading to the Cdc42 effector Gic1.
Furthermore, the influence of Zds1 and Zds2 over Cdc42 may not occur only as negative regulation. Two pieces of data support this idea: (1) high-copy ZDS1 is able to induce actin polarization in a tif51A-1 mutant at the restrictive temperature; and (2) decreasing Cdc42 activity through overexpression of its known GTPase-activating proteins (GAPs) Rga1 and Bem3 does not suppress the tif51A-1 mutant, suggesting that Zds1 is acting to promote suppression through a mechanism that does not involve Cdc42 inhibition.
It is important to note that high-copy CDC42 does not suppress the tif51A-1 mutant, but its function may be necessary for GIC1 suppression, since abolishing the Cdc42-Gic1 interaction, via CRIB domain mutations, also abolishes GIC1 suppression. This indication of the involvement of a specific Gic1 function in tif51A-1 suppression is reinforced by the fact that no other Cdc42 effectors were identified as suppressors of this eIF5A mutant. Also consistent with this observation, overexpression of Bni1, a formin proposed to act after and in a manner dependent on Gic1 (Jaquenoud and Peter 2000), also suppresses the tif51A-1 temperature-sensitive phenotype.
Finally, we demonstrated that high levels of Gic1 and Bni1 can bypass the temperature-sensitive growth defect of the PKC1 mutant stt1, indicating that these factors may function downstream of Pkc1. On the other hand, ZDS1 does not suppress the stt1 mutant (data not shown), but this fact could be due to its proposed role in maintaining the balance of different signaling pathways, as mentioned above. Taken together, these data strongly support the existence of the proposed pathway linking Pkc1 to Gic1 through Zds1 and Zds2 (Figure 8) and connecting Pkc1 signaling to polarized growth.
Interestingly, another protein, Tos2, also connects Pkc1 to Cdc42, but via Cdc24 (Drees et al. 2001), and, recently, this protein has been shown to have a possible role in anchoring Cdc24 to the plasma membrane (Toenjes et al. 2004). Like Zds1 and Zds2, Tos2 contains multiple protein kinase C consensus phosphorylation sites and physically interacts with Pkc1 (Drees et al. 2001; Toenjes et al. 2004). Moreover, Skg6, which also interacts with both Zds1 and Zds2, shows interesting homology (35% identity, 48% similarity) to Tos2 (Drees et al. 2001; Toenjes et al. 2004). These protein interactions suggest a connection between Pkc1 and members of cell polarity determination in a common protein-protein network and further support the link between Pkc1 and Gic1 mediated by Zds1 and Zds2 as a novel pathway.
Curiously, here we also characterized SSD1 as a tif51A-1 suppressor in a low-copy plasmid. SSD1 is a polymorphic gene that encodes a protein that may or may not be functional, depending on the allele present in the genetic background of the yeast strain considered (Stettler et al. 1993). Although its specific role has not been determined, a functional allele of SSD1 can suppress different mutants related to cell integrity and also the gic1Δ gic2Δ double mutant. Futhermore, ssd1Δ phenotypes can be suppressed by ZDS1 (Tsuchiya et al. 1996; Chen et al. 1997; Kaeberlein and Guarente 2002). Therefore, the isolation of SSD1 as a tif51A-1 suppressor strengthens the functional interaction between eIF5A and the Pkc1-Zds1-Gic1 pathway.
eIF5A function is important for actin cytoskeleton organization:
The identity of the eIF5A mutant suppressors described herein raised the hypothesis that a defect in establishment of cell polarity occurs in the tif51A-1 mutant. Subsequent analysis of the tif51A-1 strain actin cytoskeleton confirmed that this mutant shows defects in actin organization at the restrictive temperature. These data agree with a previous study, in which eIF5A was proposed to be important for translation of a subset of mRNAs involved in the G1/S transition, since depletion of this factor in yeast causes only a minor defect of total translation rate and an increase of enlarged cells with G1 morphology (Kang and Hershey 1994). The question of how eIF5A acts to assure correct polarized growth in S. cerevisiae is being investigated currently.
eIF5A is highly conserved throughout evolution, from archeabacteria to mammals, and this may reflect at some level a conservation of function. Therefore, as budding is not a mechanism ubiquitously used for eukaryotes to progress in the cell cycle, it would not be appropriate to propose a direct function for eIF5A in establishment of cell polarity in S. cerevisiae. Moreover, as mentioned before, overexpression of Pkc1, Zds1, and Gic1 cannot completely rescue eIF5A mutant defects, demonstrating that actin polarization is not the only function of this essential protein. Thus, it is unwise to assume that these proteins, including eIF5A, act directly in the same pathway. Conversely, eIF5A could control the expression of some factors important for G1/S transition such as the suppressors of the eIF5A mutant. Therefore, future studies involving the factors revealed herein may contribute to the elucidation of the role played by eIF5A toward specific gene expression.
Acknowledgments
We are grateful to Anita Corbett, Daniel Lew, David Levin, Douglas Johnson, Elmar Schiebel, Enrique Herrero, Erin O'Shea, John Pringle, Kazuma Tanaka, Keith Kozminski, Marc Crouzet, Michael Hall, Pamela Silver, and Yoshiko Kikuchi for strains and plasmids. We also thank Anita Corbett, Anne McBride, and Michelle Harreman for the critical reading of this manuscript. This work was supported by grants to S.R.V. and a Ph.D. fellowship to C.F.Z., both from Fundação de Amparo à Pesquisa do Estado de São Paulo.
References
- Amberg, D. C., 1998. Three-dimensional imaging of the yeast actin cytoskeleton through the budding cell cycle. Mol. Biol. Cell 9: 3259–3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews, P. D., and M. J. Stark, 2000. Type 1 protein phosphatase is required for maintenance of cell wall integrity, morphogenesis and cell cycle progression in Saccharomyces cerevisiae. J. Cell Sci. 113: 507–520. [DOI] [PubMed] [Google Scholar]
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman et al., 2005. Current Protocols in Molecular Biology. John Wiley & Sons, New York.
- Benne, R., and J. W. Hershey, 1978. The mechanism of action of protein synthesis initiation factors from rabbit reticulocytes. J. Biol. Chem. 253: 3078–3087. [PubMed] [Google Scholar]
- Bevec, D., H. Jaksche, M. Oft, T. Wohl, M. Himmelspach et al., 1996. Inhibition of HIV-1 replication in lymphocytes by mutants of the Rev cofactor eIF-5A. Science 271: 1858–1860. [DOI] [PubMed] [Google Scholar]
- Bevec, D., and J. Hauber, 1997. Eukaryotic initiation factor 5A activity and HIV-1 Rev function. Biol. Signals 6: 124–133. [DOI] [PubMed] [Google Scholar]
- Bi, E., and J. R. Pringle, 1996. ZDS1 and ZDS2, genes whose products may regulate Cdc42 in Saccharomyces cerevisiae. Mol. Cell. Biol. 16: 5264–5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourbonnais, Y., N. Faucher, D. Pallotta and C. Larouche, 2001. Multiple cellular processes affected by the absence of the Rpb4 subunit of RNA polymerase II contribute to the deficiency in the stress response of the yeast rpb4delta mutant. Mol. Gen. Genet. 264: 763–772. [DOI] [PubMed] [Google Scholar]
- Brown, J. L., M. Jaquenoud, M. P. Gulli, J. Chant and M. Peter, 1997. Novel Cdc42-binding proteins Gic1 and Gic2 control cell polarity in yeast. Genes Dev. 11: 2972–2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caraglia, M., M. Marra, G. Giuberti, A. M. D'Alessandro, A. Budillon et al., 2001. The role of eukaryotic initiation factor 5A in the control of cell proliferation and apoptosis. Amino Acids 20: 91–104. [DOI] [PubMed] [Google Scholar]
- Chai, B., J. M. Hsu, J. Du and B. C. Laurent, 2002. Yeast RSC function is required for organization of the cellular cytoskeleton via an alternative PKC1 pathway. Genetics 161: 575–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, G. C, Y. J. Kim and C. S. Chan, 1997. The Cdc42 GTPase-associated proteins Gic1 and Gic2 are required for polarized cell growth in Saccharomyces cerevisiae. Genes Dev. 11: 2958–2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, K. Y., and A. Y. Liu, 1997. Biochemistry and function of hypusine formation on eukaryotic initiation factor 5A. Biol. Signals 6: 105–109. [DOI] [PubMed] [Google Scholar]
- Christianson, T. W., R. S. Sikorski, M. Dante, J. H. Shero and P. Hieter, 1992. Multifunctional yeast high-copy-number shuttle vectors. Gene 110: 119–122. [DOI] [PubMed] [Google Scholar]
- Connelly, C., and P. Hieter, 1996. Budding yeast SKP1 encodes an evolutionarily conserved kinetochore protein required for cell cycle progression. Cell 86: 275–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bettignies, G., D. Thoraval, C. Morel, M. F. Peypouquet and M. Crouzet, 2001. Overactivation of the protein kinase C-signaling pathway suppresses the defects of cells lacking the Rho3/Rho4-GAP Rgd1p in Saccharomyces cerevisiae. Genetics 159: 1435–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drees, B. L., B. Sundin, E. Brazeau, J. P. Caviston, G. C. Chen et al., 2001. A protein interaction map for cell polarity development. J. Cell Biol. 154: 549–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladfelter, A. S., I. Bose, T. R. Zyla, E. S. Bardes and D. J. Lew, 2002. Septin ring assembly involves cycles of GTP loading and hydrolysis by Cdc42p. J. Cell Biol. 156: 315–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffioen, G., S. Swinnen and J. M. Thevelein, 2003. Feedback inhibition on cell wall integrity signaling by Zds1 involves Gsk3 phosphorylation of a cAMP-dependent protein kinase regulatory subunit. J. Biol. Chem. 278: 23460–23471. [DOI] [PubMed] [Google Scholar]
- Gustin, M. C., J. Albertyn, M. Alexander and K. Davenport, 1998. MAP kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 62: 1264–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie, C., and G. R. Fink, 1991. Guide to Yeast Genetics. Academic Press, New York.
- Heinisch, J. J., A. Lorberg, H. P. Schmitz and J. J. Jacoby, 1999. The protein kinase C-mediated MAP kinase pathway involved in the maintenance of cellular integrity in Saccharomyces cerevisiae. Mol. Microbiol. 32: 671–680. [DOI] [PubMed] [Google Scholar]
- Helliwell, S. B., A. Schmidt, Y. Ohya and M. N. Hall, 1998. The Rho1 effector Pkc1, but not Bni1, mediates signalling from Tor2 to the actin cytoskeleton. Curr. Biol. 8: 1211–1214. [DOI] [PubMed] [Google Scholar]
- Henderson, B. R., and P. Percipalle, 1997. Interactions between HIV Rev and nuclear import and export factors: the Rev nuclear localisation signal mediates specific binding to human importin-beta. J. Mol. Biol. 274: 693–707. [DOI] [PubMed] [Google Scholar]
- Höfken, T., and E. Schiebel, 2004. Novel regulation of mitotic exit by the Cdc42 effectors Gic1 and Gic2. J. Cell Biol. 164: 219–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohmann, S., 2002. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol. Mol. Biol. Rev. 66: 300–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaquenoud, M., and M. Peter, 2000. Gic2p may link activated Cdc42p to components involved in actin polarization, including Bni1p and Bud6p (Aip3p). Mol. Cell. Biol. 20: 6244–6258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein, M., and L. Guarente, 2002. Saccharomyces cerevisiae MPT5 and SSD1 function in parallel pathways to promote cell wall integrity. Genetics 60: 83–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, H. A., and J. W. Hershey, 1994. Effect of initiation factor eIF-5A depletion on protein synthesis and proliferation of Saccharomyces cerevisiae. J. Biol. Chem. 269: 3934–3940. [PubMed] [Google Scholar]
- Kawasaki, R., K. Fujimura-Kamada, H. Toi, H. Kato and K. Tanaka, 2003. The upstream regulator, Rsr1p, and downstream effectors, Gic1p and Gic2p, of the Cdc42p small GTPase coordinately regulate initiation of budding in Saccharomyces cerevisiae. Genes Cells 8: 235–250. [DOI] [PubMed] [Google Scholar]
- Ketela, T., R. Green and H. Bussey, 1999. Saccharomyces cerevisiae Mid2p is a potential cell wall stress sensor and upstream activator of the PKC1–MPK1 cell integrity pathway. J. Bacteriol. 181: 3330–3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, K. S., and D. E. Levin, 1992. Dominant mutations in a gene encoding a putative protein kinase (BCK1) bypass the requirement for a Saccharomyces cerevisiae protein kinase C homolog. Mol. Cell. Biol. 12: 172–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenburg, M. E, and E. K. O'Shea, 2001. Genetic evidence for a morphogenetic function of the Saccharomyces cerevisiae Pho85 cyclin-dependent kinase. Genetics 157: 39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y., R. D. Moir, I. K. Sethy-Coraci, J. R. Warner and I. M. Willis, 2000. Repression of ribosome and tRNA synthesis in secretion-defective cells is signaled by a novel branch of the cell integrity pathway. Mol. Cell. Biol. 20: 3843–3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-En Jao, D., and K. Yu Chen, 2002. Subcellular localization of the hypusine-containing eukaryotic initiation factor 5A by immunofluorescent staining and green fluorescent protein tagging. J. Cell Biochem. 86: 590–600. [DOI] [PubMed] [Google Scholar]
- Lipowsky, G., F. R. Bischoff, P. Schwarzmaier, R. Kraft, S. Kostka et al., 2000. Exportin 4: a mediator of a novel nuclear export pathway in higher eukaryotes. EMBO J. 19: 4362–4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, X. J., Q. Lu and M. Grunstein, 1996. A search for proteins that interact genetically with histone H3 and H4 amino termini uncovers novel regulators of the Swe1 kinase in Saccharomyces cerevisiae. Genes Dev. 10: 1327–1340. [DOI] [PubMed] [Google Scholar]
- Nanduri, J., and A. M. Tartakoff, 2001. Perturbation of the nucleus: a novel Hog1p-independent, Pkc1p-dependent consequence of hypertonic shock in yeast. Mol. Biol. Cell 12: 1835–1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, M. H., Y. B. Lee and A. Joe, 1997. Hypusine is essential for eukaryotic cell proliferation. Biol. Signals 6: 15–23. [DOI] [PubMed] [Google Scholar]
- Philip, B., and D. E. Levin, 2001. Wsc1 and Mid2 are cell surface sensors for cell wall integrity signaling that act through Rom2, a guanine nucleotide exchange factor for Rho1. Mol. Cell. Biol. 21: 271–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruyne, D., A. Legesse-Miller, L. Gao, Y. Dong and A. Bretscher, 2004. Mechanisms of polarized growth and organelle segregation in yeast. Annu. Rev. Cell Dev. Biol. 20: 559–591. [DOI] [PubMed] [Google Scholar]
- Richman, T. J., M. M. Sawyer and D. I. Johnson, 1999. The Cdc42p GTPase is involved in a G2/M morphogenetic checkpoint regulating the apical-isotropic switch and nuclear division in yeast. J. Biol. Chem. 274: 16861–16870. [DOI] [PubMed] [Google Scholar]
- Rosenwald, A. G., M. A. Rhodes, H. Van Valkenburgh, V. Palanivel, G. Chapman et al., 2002. ARL1 and membrane traffic in Saccharomyces cerevisiae. Yeast 19: 1039–1056. [DOI] [PubMed] [Google Scholar]
- Ruhl, M., M. Himmelspach, G. M. Bahr, F. Hammerschmid, H. Jaksche et al., 1993. Eukaryotic initiation factor 5A is a cellular target of the human immunodeficiency virus type 1 Rev activation domain mediating trans-activation. J. Cell Biol. 123: 1309–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnier, J., H. G. Schwelberger, Z. Smit-McBride, H. A. Kang and J. W. Hershey, 1991. Translation initiation factor 5A and its hypusine modification are essential for cell viability in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 11: 3105–3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer, B., and S. Shuman, 1996. Multicopy suppressors of temperature-sensitive mutations of yeast mRNA capping enzyme. Gene Exp. 5: 331–344. [PMC free article] [PubMed] [Google Scholar]
- Sekiya-Kawasaki, M., M. Abe, A. Saka, D. Watanabe, K. Kono et al., 2002. Dissection of upstream regulatory components of the Rho1p effector, 1,3-beta-glucan synthase, in Saccharomyces cerevisiae. Genetics 162: 663–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, X. P., K. C. Yin, Z. A. Zimolo, A. M. Stern and L. Waxman, 1996. The subcellular distribution of eukaryotic translation initiation factor, eIF-5A, in cultured cells. Exp. Cell Res. 225: 348–356. [DOI] [PubMed] [Google Scholar]
- Shi, X. P., K. C. Yin and L. Waxman, 1997. Effects of inhibitors of RNA and protein synthesis on the subcellular distribution of the eukaryotic translation initiation factor, eIF-5A, and the HIV-1 Rev protein. Biol. Signals 6: 143–149. [DOI] [PubMed] [Google Scholar]
- Sikorski, R. S., and P. Hieter, 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stettler, S., N. Chiannilkulchai, S. Hermann-Le Denmat, D. Lalo, F. Lacroute et al., 1993. A general suppressor of RNA polymerase I, II and III mutations in Saccharomyces cerevisiae. Mol. Gen. Genet. 239: 169–176. [DOI] [PubMed] [Google Scholar]
- Toenjes, K. A., D. Simpson and D. I. Johnson, 2004. Separate membrane targeting and anchoring domains function in the localization of the S. cerevisiae Cdc24p guanine nucleotide exchange factor. Curr. Genet. 45: 257–264. [DOI] [PubMed] [Google Scholar]
- Tsuchiya, E., G. Matsuzaki, K. Kurano, T. Fukuchi, A. Tsukao et al., 1996. The Saccharomyces cerevisiae SSD1 gene is involved in the tolerance to high concentration of Ca2+ with the participation of HST1/NRC1/BFR1. Gene 176: 35–38. [DOI] [PubMed] [Google Scholar]
- Uesono, Y., A. Fujita, A. Toh-E and Y. Kikuchi, 1994. The MCS1/SSD1/SRK1/SSL1 gene is involved in stable maintenance of the chromosome in yeast. Gene 143: 135–138. [DOI] [PubMed] [Google Scholar]
- Valdivia, R. H, and R. Schekman, 2003. The yeasts Rho1p and Pkc1p regulate the transport of chitin synthase III (Chs3p) from internal stores to the plasma membrane. Proc. Natl. Acad. Sci. USA 100: 10287–10292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentini, S. R., J. M. Casolari, C. C. Oliveira, P. A. Silver and A. McBride, 2002. Genetic interactions of yeast eukaryotic translation initiation factor 5A (eIF5A) reveal connections to poly(A)-binding protein and protein kinase C signaling. Genetics 160: 393–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilella, F., E. Herrero, J. Torres and M. A. de la Torre-Ruiz, 2005. Pkc1 and the upstream elements of the cell integrity pathway in Saccharomyces cerevisiae, Rom2 and Mtl1, are required for cellular responses to oxidative stress. J. Biol. Chem. 280: 9149–9159. [DOI] [PubMed] [Google Scholar]
- Yoshida, S., Y. Ohya, A. Nakano and Y. Anraku, 1994. Genetic interactions among genes involved in the STT4–PKC1 pathway of Saccharomyces cerevisiae. Mol. Gen. Genet. 242: 631–640. [DOI] [PubMed] [Google Scholar]
- Yu, Y., Y. W. Jiang, R. J. Wellinger, K. Carlson, J. M. Roberts et al., 1996. Mutations in the homologous ZDS1 and ZDS2 genes affect cell cycle progression. Mol. Cell. Biol. 16: 5254–5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuk, D., and A. Jacobson, 1998. A single amino acid substitution in yeast eIF-5A results in mRNA stabilization. EMBO J. 17: 2914–2925. [DOI] [PMC free article] [PubMed] [Google Scholar]