Abstract
In the growth plate of endochondral bones, parathyroid hormone (PTH)-related peptide (PTHrP) regulates the rate of chondrocyte maturation from prehypertrophic chondrocytes to hypertrophic chondrocytes. Using an antibody specific for Sox9 phosphorylated at serine 181 (S181), one of the two consensus protein kinase A phosphorylation sites of Sox9, we showed that the addition of PTHrP strongly increased the phosphorylation of SOX9 in COS7 cells transfected with both SOX9- and PTH/PTHrP receptor-expressing vectors. PTHrP also increased the SOX9-dependent activity of chondrocyte-specific enhancers in the gene for type II collagen (Col2a1) in transient transfection experiments. This increased enhancer activity did not occur with a Sox9 mutant harboring serine-to-alanine substitutions in its two consensus protein kinase A phosphorylation sites. Consistent with these results, PTHrP also increased Col2a1 mRNA levels in rat chondrosarcoma cells as well as 10T1/2 mesenchymal cells transfected with a PTH/PTHrP receptor expressing plasmid. No phosphorylation of Sox9 at S181 was detected in prehypertrophic chondrocytes of the growth plate or any chondrocytes of PTH/PTHrP receptor null mutants. In contrast in wild-type mouse embryos, previous immunohistochemistry experiments indicated that Sox9 phosphorylated at S181 was detected almost exclusively in chondrocytes of the prehypertrophic zone. Sox9, regardless of the phosphorylation state, was present in all chondrocytes of both genotypes except hypertrophic chondrocytes. Our results indicated that Sox9 is a target of PTHrP signaling in prehypertrophic chondrocytes in the growth plate. We hypothesize that Sox9 mediates at least some effects of PTHrP in the growth plate and that the PTHrP-dependent increased transcriptional activity of Sox9 helps maintain the chondrocyte phenotype of cells in the prehypertrophic zone and inhibits their maturation to hypertrophic chondrocytes.
The differentiation of mesenchymal cells into chondrocytes occurs along a multistep pathway (1). Mesenchymal progenitor cells first undergo condensation, which is followed by their overt differentiation into chondrocytes. In the growth plate of endochondral skeletal elements, chondrocytes then flatten and proliferate unidirectionally. After these cells stop proliferating, their genetic program changes and they become hypertrophic. The most distal hypertrophic chondrocytes acquire the ability to mineralize their extracellular matrix before they undergo apoptosis and are replaced by bone cells. Several cytokines, including bone morphogenetic proteins, insulin-like growth factor 1, fibroblast growth factors, parathyroid hormone (PTH)-related peptide (PTHrP), Indian hedgehog (Ihh), transforming growth factor-β, and others are known to influence discrete steps in this pathway (2, 3).
PTHrP, which was first identified as a factor involved in humoral hypercalcemia of malignancy (4, 5), plays a key role in regulating the rate of differentiation of prehypertrophic chondrocytes into hypertrophic chondrocytes (6). PTHrP and PTH share a highly homologous N-terminal region, and both peptides activate a common PTH/PTHrP receptor (7, 8). In PTHrP null mice, skeletal abnormalities (9) consist of shorter epiphyseal cartilages, mostly because of a smaller zone of proliferating chondrocytes and accelerated mineralization and ossification. The finding that the same phenotype was observed in PTH/PTHrP receptor homozygous mutant mice (10) was evidence that the PTH/PTHrP receptor mediated the actions of PTHrP during endochondral bone formation. The mRNA of PTH/PTHrP receptor has a low level but widespread expression in growth plate cartilages, but it is expressed at much higher levels in chondrocytes that are at the border between the proliferation and hypertrophy zones (11, 12). In contrast to PTHrP and PTH/PTHrP receptor knockout mice, overexpression of PTHrP in chondrocytes in transgenic mice has been shown to cause short-limbed dwarfism and a delay in endochondral ossification, probably due to a slower rate of chondrocyte maturation into hypertrophic chondrocytes (13). Similarly, in humans, a constitutively active mutation in the PTH/PTHrP receptor causes Jansen-type metaphyseal chondrodysplasia, which is also characterized by a delay in endochondral maturation (14, 15). Furthermore, transgenic mice having expression of this mutant PTH/PTHrP receptor have demonstrated slower conversion of proliferating chondrocytes into hypertrophic chondrocytes (16).
Ihh, a member of the hedgehog family of secreted polypeptides, is expressed predominantly in the prehypertrophic chondrocytes of the growth plate in a pattern that is slightly distal to and overlaps that of the PTH/PTHrP receptor (17). In the developing embryo, Ihh stimulates expression of PTHrP. In contrast, PTHrP delays the conversion of chondrocytes into prehypertrophic chondrocytes that express Ihh. Thus, these two factors form a negative feedback loop that regulates the rate of differentiation of proliferating chondrocytes into hypertrophic chondrocytes (9, 18). Another cytokine, transforming growth factor-β, was also shown to stimulate the expression of PTHrP mRNA and regulate the rate of hypertrophic differentiation (19).
The PTH/PTHrP receptor is a member of the G protein-coupled receptor family having seven transmembrane segments (20). Ligand binding to the receptor stimulates two second-messenger pathways, one controlled by adenylate cyclase and the other by phospholipase C (21). PTHrP or its N-terminal peptide (1–34) has been shown to increase intracellular cAMP in chondrocytes (22).
SOX9 is a high-mobility group box-containing transcription factor that harbors a strong transactivation domain. The Sox9 gene is expressed in all chondrocyte progenitors and chondrocytes, but its expression is completely turned off in hypertrophic chondrocytes (23–25). This expression parallels that of the gene for type II collagen (Col2a1), a specific marker of chondrocyte differentiation. Sox9 binds to and activates Col2a1 chondrocyte-specific enhancers and activates the Col2a1 gene (26, 27), strongly suggesting that this gene is a direct target of Sox9. In addition, the finding that Sox9 also binds and activates similar chondrocyte-specific enhancers in Col11a2 (28) and CDRAP (29), two other chondrocyte-specific genes, suggested that Sox9 controls a large set of chondrocyte-specific genes. This hypothesis was strongly supported by our recent genetic experiments, which established that Sox9 is completely required for chondrocyte differentiation and cartilage formation (30). Indeed, Sox9 null mutant cells are unable to differentiate into chondrocytes and express a series of chondrocyte marker genes, such as Col2a1, and genes for type IX (Col9a2) and type XI (Col11a2) collagen as well as the aggrecan gene. This block in differentiation occurs at the stage of mesenchymal condensation. The properties of Sox9− null mutant cells in vivo thus indicate that Sox9 is a master factor that has an essential role during chondrogenesis. Additionally, heterozygous mutations in and around the SOX9 gene in humans cause campomelic dysplasia, a lethal perinatal disease involving abnormalities in most skeletal structures derived from cartilage (31–35). This disease is usually associated with sex reversal, implying that SOX9 also functions in sex determination (36, 37). Two other members of the Sox family of transcription factors, L-Sox5 and Sox6, are also likely to have major roles in chondrogenesis (38).
Given the central role of Sox9 in chondrocyte differentiation, it is not unreasonable to think that this transcription factor may be a target of some of the signaling molecules that are known to affect discrete steps in the chondrocyte differentiation pathway. We have shown recently that SOX9 contains two consensus phosphorylation sites for cAMP-dependent protein kinase A (PKA) and that PKA phosphorylation of SOX9 increases its DNA binding and transcriptional activities. Moreover, using a phosphospecific antibody that specifically recognized Sox9 phosphorylated at serine 181, one of its two consensus PKA phosphorylation sites, we demonstrated that cAMP increased the phosphorylation of Sox9 in chondrocytes. More importantly, we demonstrated that phosphorylated Sox9 was mainly localized in prehypertrophic chondrocytes in E16.5 mouse embryos, overlapping the major expression site of the PTH/PTHrP receptor (39). Because PTHrP signals through cAMP, we therefore examined whether Sox9 was a target of PTHrP signaling.
Materials and Methods
Northern Hybridization.
Rat chondrosarcoma (RCS) cells are stable differentiated chondrocytes that express type II, IX, and XI collagens but not type I and type X collagens (40). RCS cells express PTH/PTHrP receptor RNA in a Northern hybridization experiment (data not shown). 10T1/2 mesenchymal cells have a low-level expression of Sox9 and Col2a1 but do not express PTH/PTHrP receptor (41). RCS or 10T1/2 cells transfected with an expression plasmid of the PTH/PTHrP receptor were cultured for 24 h as described previously (39). The medium was then changed to DMEM with or without increasing concentrations of PTHrP, and the cells were cultured over different intervals before they were collected as indicated in Fig. 1. Total RNA was extracted by using the RNeasy Mini Kit (Qiagen, Chatsworth, CA). Ten micrograms of each RNA sample was loaded, and the blot was hybridized with a Col2a1-specific probe as described (39). Hybridization membranes were stripped and rehybridized by using a Sox9-specific probe or glyceraldehyde-3-phosphate dehydrogenase probe as an RNA loading control.
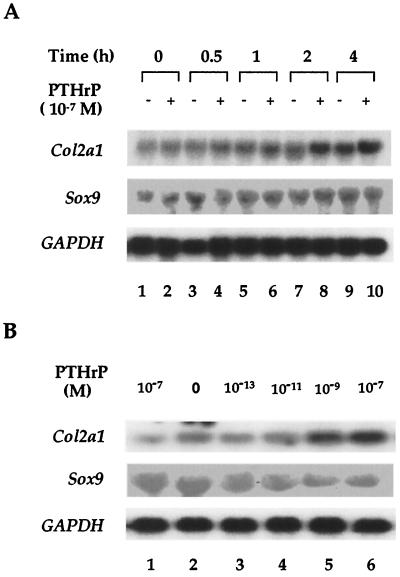
Figure 1.
PTHrP-increased expression of Col2a1 RNA. (A) RCS cells were cultured for 24 h before 10−7 M PTHrP was added; the cells were cultured further for different intervals as indicated. Cells were then collected, and total RNA was extracted and hybridized with a Col2a1 probe. The hybridization membrane was rehybridized with Sox9 and glyceraldehyde-3-phosphate dehydrogenase probes. (B) 10T1/2 cells were transfected with an empty vector (lane 1) or a PTH/PTHrP receptor expression plasmid (lanes 2–6) for 24 h. Different concentrations of PTHrP were then added as indicated, and the cells were cultured for another 4 h. Northern hybridization was performed as in A.
Cell Culture and Transfection Experiments.
RCS and COS7 cells were cultured as described (39). Cells were then transfected with luciferase reporter plasmids containing an 89-bp Col2a1 promoter without (p89Luc) or with four copies of a 48-bp chondrocyte-specific Col2a1 enhancer element (4 × 48-p89Luc) or two copies of a 100-bp chondrocyte-specific enhancer element including the 48-bp DNA segment (2 × 100-p89Luc) and the pSV2-β-galactosidase (βgal) plasmid in a ratio of 3:1 as described (39). All transfections were done by using FuGene6 (Roche Molecular Biochemicals) according to the manufacturer's instructions. Expression plasmids (100 ng) for wild-type (wt) or m12 mutant SOX9 containing serine-to-alanine mutations in PKA phosphorylation sites and the PTH/PTHrP receptor (200 ng) were transfected as indicated in Fig. 2D. Luciferase and βgal activity was assayed in cell lysates prepared as described (39). Different concentrations of PTHrP and 10 or 30 μM H89 (a PKA-specific inhibitor) or okadaic acid (OA; a serine–threonine phosphatase inhibitor) were added 24 h after transfection, and the cells were incubated for 8 h before being collected. Luciferase reporter activity was represented as the average of triplicate cultures in one of several representative experiments as described (39).
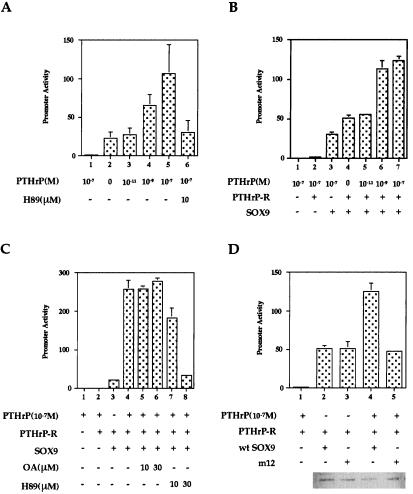
Figure 2.
PTHrP-increased activity of a Col2a1 chondrocyte-specific enhancer. (A) RCS cells were transfected with p89Luc (lane 1) or 4 × 48-p89Luc (lanes 2–6) and pSV2-βgal at a ratio of 3:1. Eight hours later, increasing concentrations of PTHrP with or without 10 μM H89 were added as indicated, and the cells were cultured for an additional 8 h. Luciferase activity was normalized by using the level of βgal activity. (B and C) COS7 cells were cotransfected with expression plasmids of SOX9 (100 ng) and PTH/PTHrP receptor (PTHrP-R) (200 ng) together with the 4 × 48-p89Luc reporter (B) or 2 × 100-p89Luc reporter (C) and pSV2-βgal. In both experiments, cells were transfected with an empty vector (lane 1), a PTH/PTHrP receptor expression plasmid (lanes 2 and 4–7 in B; lanes 2–8 in C), and a SOX9 expression plasmid (lanes 3–7 in B, lanes 3–8 in C). H89 or OA was added 8 h later as indicated. Cells were cultured for an additional 8 h and, after lysis, used to measure luciferase and βgal activity. (D) An empty vector (lane 1), 100 ng of wt SOX9 (lanes 2 and 4), m12 (lanes 3 and 5), and 200 ng of PTH/PTHrP receptor expression plasmids (lanes 1–5) were cotransfected into COS7 cells together with 4 × 48-p89Luc and pSV2-βgal as described (39). Cells were treated (lanes 1, 4, and 5) with 10−7 M PTHrP for 8 h. A Western blot using a SOX9 antibody that recognizes the carboxyl terminus of SOX9 was performed to verify that similar levels of wt SOX9 and m12 were present.
Western Blotting.
COS7 cell lysates were prepared in an extraction buffer (50 mM Tris·HCl, pH 8.0/150 mM NaCl/1% Nonidet P-40/1 mM DTT/1 mM PMSF). Western blotting was done by using the enhanced chemiluminescence kit (Amersham Pharmacia). The SOX9.P antibody was used at a 1:1,000 dilution. To reprobe the membrane with the SOX9 antibody, the SOX9.P antibody was stripped off, and the blots were reprobed with the SOX9 antibody diluted at 1:1,000. The SOX9.P and Sox9 antibodies have been described (39).
Immunofluorescence.
COS7 cells were cotransfected with expression plasmids of SOX9 (100 ng) and the PTH/PTHrP receptor (200 ng) for 24 h. The medium was then changed to new DMEM with or without 10−7 M PTHrP. Eight hours later, the cell monolayers were fixed with methanol for 15 min and incubated with blocking buffer (PBS with 5% goat serum and 3% BSA) for 1 h. Next, SOX9 or SOX9.P antibodies (diluted at 1:500 with blocking buffer) were added and incubated for 1 h at room temperature. After washing, a secondary antibody consisting of biotin-conjugated anti-rabbit IgG (Jackson ImmunoResearch) diluted at 1:200 in blocking buffer was added, and the mixture was incubated at room temperature for 1 h. The cells were then incubated with extrAvidin-fluorescein isothiocyanate conjugate (Sigma) diluted 1:250 in blocking buffer for 1 h. Cells were then washed three times with cold PBS; the last wash was supplemented with 5 μg/ml of the DNA dye 4′,6-diamidino-2 phenylindole (Sigma).
Immunohistochemical Analysis.
In this analysis, 16.5 days postcoitum, wt or PTH/PTHrP receptor null mutant mouse littermates were generated in a mixed background of C57BL/6 and 129/SVJ mice (10). The embryonic hind legs were fixed in 4% paraformaldehyde in PBS for 12 h. After dehydration, tissues were embedded in paraffin and sectioned. Immunohistochemical staining was performed by using the Dako EnVision+ System (Dako) as described (39). SOX9 and SOX9.P antibodies were both used at a dilution of 1:30 with 10% goat serum in PBS.
Results
Increased Col2a1 Gene Expression by PTHrP.
PTHrP modulates the maturation of chondrocytes by inhibiting the rate of their differentiation into hypertrophic chondrocytes (6). Also, expression of the Col2a1 gene is increased in prehypertrophic chondrocytes, the area of the growth plate where the PTH/PTHrP receptor is expressed at high level (12). To determine whether PTHrP increased the expression of Col2a1 in RCS cells in culture, we treated these cells with PTHrP and measured Col2a1 RNA. Treatment with 10−7 M PTHrP peptide 1–34 (designed hereafter as PTHrP) produced a detectable increase in Col2a1 RNA after 2 h and a 2- to 3-fold increase after 4 h. In addition, when 10T1/2 cells were transfected with a PTH/PTHrP receptor-expressing plasmid, PTHrP treatment increased the level of Col2a1 RNA in a dose-dependent manner (Fig. 1B). However, no significant change of Sox9 RNA was observed after PTHrP treatment in both cell types. These results suggested that activation of the signaling pathway downstream of the PTH/PTHrP receptor results in an increase in Col2a1 expression without changes in the level of Sox9 RNA.
Increased Activity of Sox9-Dependent Col2a1 Chondrocyte-Specific Enhancers by PTHrP.
We previously showed that cAMP increased the activity of a Sox9-dependent 48-bp Col2a1 chondrocyte-specific enhancer in RCS cells (39). Because cAMP is a major mediator of PTHrP signaling, we tested whether PTHrP had a similar effect. Fig. 2A shows that PTHrP increased the activity of this 4 × 48-bp Col2a1 enhancer and that H89, a PKA-specific inhibitor, blocked this enhanced activity. These results suggested that PKA mediated the effects of PTHrP. Because the activity of the 48-bp Col2a1 enhancer was previously shown to depend on Sox9, we then tested whether SOX9 was a target of PTHrP signaling. Reporter vectors driven by either a 4 × 48-bp or 2 × 100-bp enhancer, which includes the 48-bp Col2a1 chondrocyte-specific enhancer, were used to test for the effect of PTHrP. Cotransfection of SOX9 and the PTH/PTHrP receptor in COS7 cells (with addition of 10−7M PTHrP) increased the activity of the 4 × 48-bp Col2a1 enhancer about 2.5-fold over that of the control without addition of PTHrP and 4-fold compared with transfection of SOX9 alone (Fig. 2B). This enhanced activity was blocked by H89 but was not affected by OA, an inhibitor of phosphatase. Thus, the inhibition of phosphatase activity had no effect on the Sox9-dependent increase of the Col2a1 enhancer in response to PTHrP. A higher level of enhancer activation in response to PTHrP was seen in the 2 × 100-bp Col2a1 enhancer, which may have been due to the PTHrP-induced recruitment of other transcription factors to this larger enhancer and their cooperation with Sox9 (Fig. 2C). Because PTHrP did not increase the activity of the Col2a1 enhancers in the absence of SOX9, these results suggested that SOX9 was required to mediate the response of the Col2a1 enhancer to PTHrP and that PTHrP acted upstream of the cAMP signaling pathway.
Abolishment of PTHrP Activation of the Col2a1 Enhancer by Mutations in Two Consensus PKA Phosphorylation Sites of SOX9.
Previously, we showed that SOX9 contains two consensus PKA phosphorylation sites and that the phosphorylation of SOX9 by PKA increased its transcriptional activity as assayed by the increased activity of a Sox9-dependent Col2a1 chondrocyte-specific enhancer. To provide evidence that PTHrP acted through PKA phosphorylation to regulate the activity of SOX9, we compared the effect of PTHrP on the activity of wt SOX9 with that of m12, a mutant SOX9 harboring serine-to-alanine substitutions in the two consensus PKA phosphorylation sites (Fig. 2D). In transfection experiments using mutant Sox9, PTHrP did not increase the Sox9-dependent activity of the 4 × 48-bp Col2a1 enhancer, which contrasts with the increase observed using wt Sox9. A control Western blot indicated equal levels of expression of wt SOX9 and m12. We concluded that one or both of the PKA phosphorylation sites of SOX9 were required to mediate the PTHrP-dependent increase in Col2a1 enhancer activity.
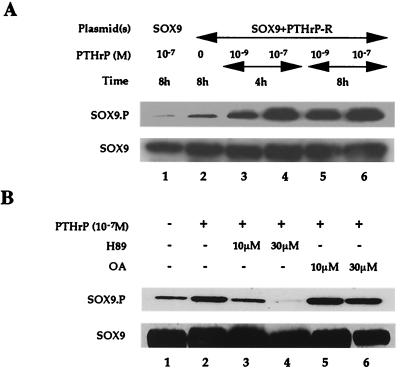
PTHrP-Dependent Increase in Phosphorylation of SOX9 at S181.
We next used a phosphospecific antibody (SOX9.P) that specifically recognizes SOX9 phosphorylated at S181, one of the two consensus PKA phosphorylation sites (39), to directly test whether PTHrP treatment resulted in phosphorylation of Sox9. In previous experiments, we showed that a Sox9 mutant having a serine-to-alanine substitution at S181 had greatly reduced transcriptional activation in response to PKA compared with that of wt Sox9. Thus, phosphorylation of S181 has functional consequences with respect to the transcriptional activity of SOX9. After cotransfection with SOX9 and PTH/PTHrP receptor expression plasmids, COS7 cells were treated with different concentrations of PTHrP for 4 or 8 h. PTHrP increased the phosphorylation of SOX9 at S181 in a dose- and time-dependent manner (Fig. 3A). This phosphorylation was blocked by H89, a PKA-specific inhibitor, but not OA, a phosphatase inhibitor (Fig. 3B), suggesting that PTHrP targets Sox9 through PKA phosphorylation. These results are also in agreement with the inhibition of the PTHrP-dependent increase in Col2a1 enhancer activity by H89 but not OA.
Figure 3.
PTHrP-increased phosphorylation of SOX9 at S181. (A) COS7 cells were transfected with a SOX9 expression plasmid (100 ng) (lane 1) or cotransfected with SOX9 (100 ng) and PTH/PTHrP receptor (200 ng) expression plasmids (lanes 2–6). PTHrP was (lanes 1 and 3–6) or was not (lane 2) added as indicated. Cell lysates were fractionated by using SDS/PAGE and blotted, and the blots were incubated with Sox9.P and Sox9 antibodies. (B) COS7 cells were cotransfected with SOX9 (100 ng) and PTH/PTHrP receptor (200 ng) expression plasmids. No PTHrP (lane 1) or 10−7 M PTHrP (lanes 2–6) with H89 (lanes 3 and 4) or OA (lanes 5 and 6) was added. Cells were then cultured for an additional 4 h before lysis.
PTHrP Treatment Induced Phosphorylation of SOX9 in Intact Cells.
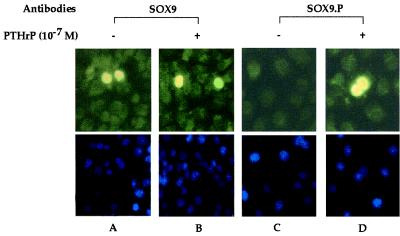
We next examined the effects of PTHrP on the cellular localization of SOX9 by using the SOX9.P and SOX9 antibodies. COS7 cells were cotransfected with SOX9 and PTH/PTHrP receptor expression plasmids and then treated with 10−7 M PTHrP. In these experiments, SOX9 was present in the cell nucleus whether PTHrP was present or not (Fig. 4 A and B). Also, only cells treated with PTHrP reacted with the SOX9.P antibody, indicating that PTHrP was responsible for SOX9 phosphorylation (Fig. 4A). In addition, the phosphorylated SOX9 remained in the cell nucleus. This experiment also demonstrated that phosphorylation of SOX9 observed in Western blots was not due to cross-activation of kinases after cell lysis but was a direct consequence of PTHrP treatment.
Figure 4.
Lack of effect of PTHrP-dependent phosphorylation of SOX9 on nuclear localization in intact cells. COS7 cells were cotransfected with expression plasmids for SOX9 (100 ng) and PTH/PTHrP receptor (200 ng) without (A and C) or with (B and D) addition of 10−7 M PTHrP for 8 h. Cells were fixed and incubated with SOX9 or SOX9.P antibodies as described in Materials and Methods. (Upper) The nuclear localization of SOX9 (A and B) and SOX9 phosphorylated at S181 (D). A phosphorylated SOX9 signal was not detected without PTHrP treatment (C). (Lower) The same fields showing cell nuclei stained with 4′,6-diamidino-2 phenylindole.
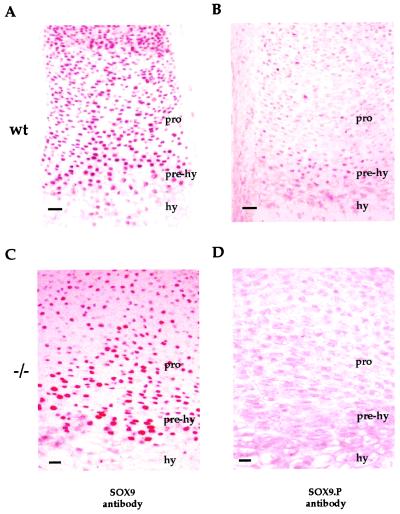
Absence of Sox9 Phosphorylated at S181 in the Prehypertrophic Zone of the Growth Plate in PTH/PTHrP Receptor Mutant Mice.
Prehypertrophic chondrocytes, which express high levels of the PTH/PTHrP receptor, are very likely to be the major site of action of PTHrP in the growth plate (4). Our previous experiments showed that Sox9 phosphorylated at S181 was detected mainly in the prehypertrophic zone of the mouse growth plate (39), overlapping the major site of expression of the PTH/PTHrP receptor. To further explore the relationship between PKA phosphorylation of Sox9 and PTHrP signaling in vivo, we performed an immunohistochemical comparison of the growth plates of E16.5 PTH/PTHrP receptor null mutant and wild-type mice by using the SOX9.P antibody. As previously reported, Sox9 was evenly distributed in all except hypertrophic chondrocytes of wt mice (39). In addition, Sox9 phosphorylated at S181 was essentially detected only in the prehypertrophic zone of the growth plate (Fig. 5 A and B). In contrast, although the distribution of Sox9 in the growth plate in PTH/PTHrP receptor null mice was the same as that in wt controls (Fig. 5C), phosphorylated Sox9 was not detected in the prehypertrophic zone (Fig. 5D). This result indicated that in the absence of the PTH/PTHrP receptor, chondrocytes were unable to phosphorylate Sox9 at S181 in the prehypertrophic zone of the growth plate.
Figure 5.
Abolishment of phosphorylation of Sox9 at S181 in the prehypertrophic zone of the growth plate in PTH/PTHrP receptor null mutant mice. Immunohistochemistry of a hindleg section of E16.5 wt and PTH/PTHrP receptor null mutant (−/−) mouse embryos with SOX9 or SOX9.P antibodies. Sox9 was distributed evenly in resting, proliferating (pro), and prehypertrophic (prehy) chondrocytes but was absent in hypertrophic (hy) chondrocytes (A), whereas phosphorylated Sox9 was seen only in prehypertrophic chondrocytes (B) of wt embryos. In E16.5 PTH/PTHrP receptor null mutant mouse hindlegs, Sox9 showed the same distribution as in wt embryos (C), but the phosphorylated Sox9 signal was absent in the prehypertrophic chondrocytes (D). (Bars represent 50 μm.)
Discussion
PTHrP and PTH/PTHrP receptor knockout mice have very similar skeletal abnormalities and show an accelerated differentiation of chondrocytes into hypertrophic chondrocytes. In addition, Ihh, which is secreted by newly formed hypertrophic chondrocytes, stimulates PTHrP expression, whereas PTHrP retards formation of these cells. This negative feedback loop presumably serves to accurately regulate the transition of chondrocytes into hypertrophic chondrocytes, a process that is critical for the longitudinal growth of endochondral bones (18).
We present here evidence that the master chondrogenic factor Sox9 is a downstream target of PTHrP signaling in vivo, that PTHrP produces an increase in SOX9 transcriptional activity, and that this increase is mediated through phosphorylation by PKA, a known mediator of PTHrP signaling in the growth plate. First, in DNA transfection experiments, the activity of SOX9-dependent Col2a1 enhancers was increased by PTHrP. In this assay, we have used the activity of the Col2a1 enhancer as a functional measurement of the transcriptional activity of SOX9 (41). Consistent with the response of the Col2a1 enhancer to PTHrP, we found that PTHrP increased the levels of Col2a1 but not Sox9 in RNA in both RCS cells and 10T1/2 cells transfected with a PTH/PTHrP receptor-expressing vector. Second, SOX9 mutants containing serine-to-alanine substitutions in the two PKA consensus phosphorylation sites were unable to mediate the increased activity of the Col2a1 enhancer in response to PTHrP, although the activity of mutant SOX9 was the same as that of wt SOX9 in the absence of PTHrP. Earlier experiments had shown that both of these sites were phosphorylated by PKA in vitro but that the enzyme was unable to phosphorylate the mutant SOX9 containing serine-to-alanine substitutions in the two consensus PKA phosphorylation sites. We thus conclude that the PTHrP-dependent increase in Col2a1 enhancer activity was due to an increase in SOX9 transcriptional activity and that this increased activity was very likely a consequence of PKA phosphorylation. This increased transcriptional activity of SOX9 may be accounted for by the increased DNA binding activity produced by PKA phosphorylation of SOX9 (39). Moreover, phosphorylated SOX9 may interact more efficiently with components of the transcriptional machinery. In addition, increases in the intracellular concentration of cAMP should also increase the activity of other cAMP-responsive transcription factors such as CREB, which may cooperate with SOX9 to activate downstream genes.
Next, we showed that the addition of increasing concentrations of PTHrP to cells cotransfected with both SOX9 and PTH/PTHrP receptor-expressing vectors resulted in a time- and dose-dependent phosphorylation of SOX9 as measured by the phosphorylation of S181, one of the two consensus PKA phosphorylation sites. Evidence that this PTHrP-dependent phosphorylation of SOX9 was mediated by PKA came from the inhibition of SOX9 phosphorylation by H89, a specific inhibitor of PKA. This inhibition of PTHrP-dependent Sox9 phosphorylation by H89 paralleled the inhibition of the PTHrP-dependent increase in Col2a1 enhancer activity by H89. Thus, our results strongly suggest that both the PTHrP-dependent phosphorylation of SOX9 and its increased transcriptional activity in response to PTHrP were mediated by PKA. Other earlier experiments showed that Sox9 phosphorylated at S181, one of the two consensus PKA phosphorylation sites, was detected mainly in the prehypertrophic zone of the growth plate, which is the area where the PTHrP receptor is highly expressed. Evidence that phosphorylation of Sox9 at S181 in the prehypertrophic zone of the growth plate in vivo occurred as a consequence of PTHrP signaling was demonstrated by the absence of Sox9 phosphorylation in the growth plate of PTH/PTHrP receptor null mutant mice.
We conclude from these experiments that Sox9 is a target of PTHrP signaling. Based on this result, we propose that the increase in Sox9 activity, brought about by its PKA phosphorylation in prehypertrophic chondrocytes, mediates, at least in part, the function of PTHrP in the growth plate. According to this model, the PTHrP-dependent increase in the transcriptional activity of the master transcription factor Sox9 would help maintain the chondrocytic phenotype of cells in the prehypertrophic zone of the growth plate and prevent their further differentiation into hypertrophic chondrocytes.
Independent evidence that Sox9 has a role in the transition of prehypertrophic chondrocytes to hypertrophic chondrocytes came from the observation that the hypertrophic zone in heterozygous Sox9 mutant mice was larger than that in wt mice and that premature mineralization occurred in the growth plate (W. Bi, J. Deng, Z. Zhang, W.H., R. R. Behringer, and B.d.C., unpublished data). Because expression of Sox9 is completely shut off in the hypertrophic zone and mineralized areas of the growth plate, both the enlarged hypertrophic zone and premature mineralization of endochondral bones in Sox9 heterozygous mice were very likely consequences of an increased transition rate from prehypertrophic chondrocytes to hypertrophic chondrocytes. We have hypothesized that this was caused by Sox9 haploinsufficiency in cells that express Sox9, including those in the prehypertrophic zone. Since our previous studies had established that Sox9 is required for the condensation of mesenchymal chondrocyte progenitor cells, the premature mineralization of the growth plate in heterozygous Sox9 mutants and the absence of phosphorylation of Sox9 at S181 in PTHrP receptor null mice favor the view that Sox9 has a role in chondrogenesis beyond mesenchymal condensation.
In summary, our results establish a rational link between signaling by PTHrP, which is known to control a discrete step in the chondrogenesis pathway, and the transcription factor Sox9, a master factor for chondrocyte differentiation.
Acknowledgments
We thank Heidi Eberspaecher for help in immunohistochemistry and Patricia Arubaleze and Janie Finch for editorial assistance. DNA sequencing was performed by The University of Texas M. D. Anderson Cancer Center core sequencing facility, which is supported by National Cancer Institute Grant CA 16672. This work was funded by National Institutes of Health Grants R01 AR42909 and P01 AR 42919-02 (to B.d.C.) and National Institutes of Health Grant DK47038 (to H.M.K.).
Abbreviations
- PTH
parathyroid hormone
- PTHrP
PTH-related peptide
- PKA
cAMP-dependent protein kinase A
- Col2a1
proα1 (II) collagen gene
- Ihh
Indian hedgehog
- RCS
rat chondrosarcoma
- OA
okadaic acid
- wt
wild type
- βgal
β-galactosidase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.011393998.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.011393998
References
- 1.Erlebacher A, Filvarof E H, Gitelman S E, Derynck R. Cell. 1995;80:271–278. doi: 10.1016/0092-8674(95)90487-5. [DOI] [PubMed] [Google Scholar]
- 2.Vortkamp A, Lee K, Lanske B, Segre G V, Kronenberg H M, Tabin C J. Science. 1996;273:613–622. doi: 10.1126/science.273.5275.613. [DOI] [PubMed] [Google Scholar]
- 3.Pateder D B, Rosier R N, Schwarz E M, Reynolds P R, Puzas J E, D'Souza M, O'Keefe R J. Exp Cell Res. 2000;256:555–562. doi: 10.1006/excr.2000.4860. [DOI] [PubMed] [Google Scholar]
- 4.Moseley J M, Kubota M, Diefenbach-Jagger H, Wettenhall R E H, Kemp B E, Suva L J, Rodda C P, Ebeling P R, Hudson P J, Zajac J D, et al. Proc Natl Acad Sci USA. 1987;84:5048–5052. doi: 10.1073/pnas.84.14.5048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suva L J, Winslow G A, Wettenhall R E H, Kemp B E, Hudson P J, Diefenbach-Jagger H, Moseley J M, Rodda C P, Martin T J, Wood W I. Science. 1987;237:893–896. doi: 10.1126/science.3616618. [DOI] [PubMed] [Google Scholar]
- 6.Lanske B, Kronenberg H M. Crit Rev Eukaryotic Gene Expression. 1998;8:297–320. doi: 10.1615/critreveukargeneexpr.v8.i3-4.40. [DOI] [PubMed] [Google Scholar]
- 7.Juppner H, Abou-Samra A-B, Uneno S, Gu W X, Potts J T, Jr, Segre G V. J Biol Chem. 1988;263:8557–8560. [PubMed] [Google Scholar]
- 8.Fraher L J, Hodsman A B, Jonas K, Saunders D, Rose C I, Henderson J E, Hendy G N, Goltzman D. J Clin Endocrinol Metab. 1992;75:417–423. doi: 10.1210/jcem.75.2.1322424. [DOI] [PubMed] [Google Scholar]
- 9.Kapaplis A C, Luz A, Glowacki J, Bronson R T, Tybulewicz V L J, Kronenberg H M, Mulligan R C. Genes Dev. 1994;8:277–289. doi: 10.1101/gad.8.3.277. [DOI] [PubMed] [Google Scholar]
- 10.Lanske B, Karaplis A C, Lee K, Luz A, Vortkamp A, Pirro A, Karperien M, Defice L H K, Ho C, Mulligan R C, et al. Science. 1996;273:663–666. doi: 10.1126/science.273.5275.663. [DOI] [PubMed] [Google Scholar]
- 11.Iwamoto M, Jikko A, Murakami H, Shimazu A, Nakashima K, Takigawa M, Baba H, Suzuki F, Kato Y. J Biol Chem. 1994;269:17245–17251. [PubMed] [Google Scholar]
- 12.Lee K, Lanske B, Karaplis A C, Deeds J D, Kohno H, Nissenson R A, Kronenberg H M, Segre G V. Endocrinology. 1996;137:5109–5118. doi: 10.1210/endo.137.11.8895385. [DOI] [PubMed] [Google Scholar]
- 13.Weir E C, Philbrick W M, Amling M, Neff L A, Baron R, Broadus A E. Proc Natl Acad Sci USA. 1996;93:10240–10245. doi: 10.1073/pnas.93.19.10240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schipani E, Kruse K, Juppner H. Science. 1995;268:98–100. doi: 10.1126/science.7701349. [DOI] [PubMed] [Google Scholar]
- 15.Schipani E, Langman C B, Parfitt A M, Jensen G S, Kikuchi S, Kooh S W, Cole W G, Juppner H. N Engl J Med. 1996;335:708–714. doi: 10.1056/NEJM199609053351004. [DOI] [PubMed] [Google Scholar]
- 16.Bitgood M J, Mcmahon A P. Dev Biol. 1995;172:126–138. doi: 10.1006/dbio.1995.0010. [DOI] [PubMed] [Google Scholar]
- 17.Schipani E, Lanske B, Hunzelman J, Luz A, Kovacs C S, Lee K, Pirro A, Kronenberg H M. J Bone Miner Res. 1997;12:30. doi: 10.1073/pnas.94.25.13689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vortkamp A, Lee K, Lanske B, Segre G V, Kronenberg H M, Tabin C J. Science. 1996;273:613–621. doi: 10.1126/science.273.5275.613. [DOI] [PubMed] [Google Scholar]
- 19.Serra R, Karaplis A, Sohn P. J Cell Biol. 1999;4:783–794. doi: 10.1083/jcb.145.4.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jüppner H, Abou-Samra A, Freeman M, Kong X, Schipani E, Richards J, Kolakowski L F, Hock J, Potts J T, Kronenberg H M, et al. Science. 1991;254:1024–1026. doi: 10.1126/science.1658941. [DOI] [PubMed] [Google Scholar]
- 21.Abou-Samra A-B, Jüppner H, Force T, Freeman M, Kong X-F, Schipani E, Urena P, Richards J, Bonventre J V, Potts J T, Jr, et al. Proc Natl Acad Sci USA. 1992;89:2732–2736. doi: 10.1073/pnas.89.7.2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsukazaki T, Ohtsuru A, Namba H, Oda J, Motomura K, Osaki M, Kiriyama T, Iwasaki K, Yamashita S. J Endocrinol. 1996;150:359–368. doi: 10.1677/joe.0.1500359. [DOI] [PubMed] [Google Scholar]
- 23.Ng L-J, Wheatley S, Muscat G E O, Conway-Campbell J, Bowles J, Wright E, Bell D M, Tam P P L, Cheah K S E, Koopman P. Dev Biol. 1997;183:108–121. doi: 10.1006/dbio.1996.8487. [DOI] [PubMed] [Google Scholar]
- 24.Wright E, Hargrave M R, Christiansen J, Cooper L, Kun J, Evans T, Gangadharan U, Greenfield A, Koopman P. Nat Genet. 1995;9:15–20. doi: 10.1038/ng0195-15. [DOI] [PubMed] [Google Scholar]
- 25.Zhao Q, Eberspaecher H, Lefebvre V, de Crombrugghe B. Dev Dyn. 1997;209:377–386. doi: 10.1002/(SICI)1097-0177(199708)209:4<377::AID-AJA5>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 26.Bell D M, Leung K K H, Whearley S C, Ng L J, Zhou S, Ling K W, Sham M H, Koopman P, Tam P P L, Cheah K S E. Nat Genet. 1997;16:174–178. doi: 10.1038/ng0697-174. [DOI] [PubMed] [Google Scholar]
- 27.Lefebvre V, Huang W, Harley V R, Goodfellow P N, de Crombrugghe B. Mol Cell Biol. 1997;17:2336–2346. doi: 10.1128/mcb.17.4.2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bridgewater L C, Lefebvre V, de Crombrugghe B. J Biol Chem. 1998;273:14998–15006. doi: 10.1074/jbc.273.24.14998. [DOI] [PubMed] [Google Scholar]
- 29.Xie W F, Zhang X, Sakano S, Lefebvre V, Sandell L J. J Bone Miner Res. 1999;14:757–763. doi: 10.1359/jbmr.1999.14.5.757. [DOI] [PubMed] [Google Scholar]
- 30.Bi W, Deng J, Zhang Z, Behringer R R, de Crombrugghe B. Nat Genet. 1999;22:85–89. doi: 10.1038/8792. [DOI] [PubMed] [Google Scholar]
- 31.Foster J W, Dominguez-Steglich M A, Guioli S, Kwok C, Weller P A, Stevanovic M, Weissenbach J, Mansour S, Young I D, Goodfellow P N, et al. Nature (London) 1994;372:525–530. doi: 10.1038/372525a0. [DOI] [PubMed] [Google Scholar]
- 32.Houston C S, Opitz J M, Spranger J W, Macpherson R I, Reed M H, Gilbert E F, Herrman J, Schinzel A. Am J Med Genet. 1993;15:2–28. doi: 10.1002/ajmg.1320150103. [DOI] [PubMed] [Google Scholar]
- 33.Kwok C, Weller P A, Guioli S, Foster J W, Mansour S, Zuffardi O, Punett H H, Dominguez-Steglich M A, Brook J D, Young I D, et al. Am J Hum Genet. 1995;57:1028–1036. [PMC free article] [PubMed] [Google Scholar]
- 34.Meyer J, Südbeck P, Held M, Wagner T, Schmitz M L, Bricarelli F D, Eggermont E, Friedrich U, Haas O A, Kobelt A, et al. Hum Mol Genet. 1997;6:91–98. doi: 10.1093/hmg/6.1.91. [DOI] [PubMed] [Google Scholar]
- 35.Wagner T, Wirth J, Meyer J, Zabel B, Held M, Zimmer J, Pasantes J, Dagna Bricarelli F, Keutel J, Hustert E, et al. Cell. 1994;79:1111–1120. doi: 10.1016/0092-8674(94)90041-8. [DOI] [PubMed] [Google Scholar]
- 36.Mansour S, Hall C M, Pembrey M E, Young I D. J Med Genet. 1995;32:415–420. doi: 10.1136/jmg.32.6.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wirth J, Wagner T, Meyer J, Pfeiffer R A, Tietze H-U, Schempp W, Scherer G. Hum Genet. 1996;97:186–193. doi: 10.1007/BF02265263. [DOI] [PubMed] [Google Scholar]
- 38.Lefebvre V, Li P, de Crombrugghe B. EMBO J. 1998;17:5718–5733. doi: 10.1093/emboj/17.19.5718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang W, Zhou X, Lefebvre V, de Crombrugghe B. Mol Cell Biol. 2000;20:4149–4158. doi: 10.1128/mcb.20.11.4149-4158.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mukhopadhyay K, Lefebvre V, Zhou G, Garofalo G, Kimora J H, de Crombrugghe B. J Biol Chem. 1995;270:27711–27719. doi: 10.1074/jbc.270.46.27711. [DOI] [PubMed] [Google Scholar]
- 41.Hollnagel A, Ahrens M, Gross G. J Bone Miner Res. 1997;12:1993–2004. doi: 10.1359/jbmr.1997.12.12.1993. [DOI] [PubMed] [Google Scholar]