Abstract
We describe a developmental, genetic, and molecular analysis of the sole Drosophila member of the BAG family of genes, which is implicated in stress response and survival in mammalian cells. We show that the gene, termed starvin (stv), is expressed in a highly tissue-specific manner, accumulating primarily in tendon cells following germ-band retraction and later in somatic muscles and the esophagus during embryonic stage 15. We show that stv expression falls within known tendon and muscle cell transcriptional regulatory cascades, being downstream of stripe, but not of another tendon transcriptional regulator, delilah, and downstream of the muscle regulator, mef-2. We generated a series of stv alleles and, surprisingly, given the muscle and tendon-specific embryonic expression of stv, found that the gross morphology and function of somatic muscles is normal in stv mutants. Nonetheless, stv mutant larvae exhibit a striking and fully penetrant mutant phenotype of failure to grow after hatching and a severely impaired ability to take up food. Our study provides the first report of an essential, developmentally regulated BAG-family gene.
MAMMALIAN Bcl1-associated athanogene (BAG)-domain proteins, named after the founding member BAG-1 (Takayama et al. 1995), are Hsp70-family cochaperones implicated in cell survival, intracellular signaling, gene expression, and human disorders such as Parkinson's disease (Takayama et al. 1995, 1997, 1999; Gebauer et al. 1997; Höhfeld and Jentsch 1997; Zeiner et al. 1997; Liu et al. 1998; Antoku et al. 2001; Briknarova et al. 2002; Kalia et al. 2004; reviewed in Doong et al. 2002; Alberti et al. 2003; Gehring 2004; Townsend et al. 2005). The ∼80-amino-acid C-terminal BAG domain comprises an antiparallel, amphipathic, three-helix bundle structure (Briknarova et al. 2001; Sondermann et al. 2001; Brockmann et al. 2004; Symersky et al. 2004), which interacts with the ATPase domain of Hsc70 and Hsp70 (Höhfeld and Jentsch 1997; Takayama et al. 1997). BAG-domain proteins can act as nucleotide exchange factors that influence cycling between ADP-bound and ATP-bound forms, directly regulating the activity of HSP70 and HSC70 (Höhfeld and Jentsch 1997; Höhfeld 1998; Gassler et al. 2001). BAG-domain proteins also form complexes with other proteins, such as Bcl-2 (Takayama et al. 1995; Antoku et al. 2001), Raf kinase (Wang et al. 1996; Song et al. 2001), steroid receptors (Froesch et al. 1998; Kullmann et al. 1998; Shatkina et al. 2003), tyrosine kinase receptors (Bardelli et al. 1996), the cellular stress response protein GADD34 (Hung et al. 2003), and Siah (Matsuzawa et al. 1998). Some of these interactions have been shown to be independent of Hsp70-family chaperones, potentially expanding the functions of this family of proteins beyond those of the cellular stress response, although Hsc70 and Hsp70 are the main binding partners of BAG-1 in cellular extracts (Höhfeld and Jentsch 1997; Takayama et al. 1997).
Six mammalian BAG-family members have been identified: BAG 1, BAG-2, BAG-3/CAIR-1, BAG-4/SODD, BAG-5, and HLA-B/BAT3, the human ortholog of Scythe (Takayama et al. 1999; Manchen and Hubberstey 2001).
Despite the broad significance of this family of proteins, BAG-family members are yet to be analyzed from a developmental or genetic perspective. Here we present a developmental and genetic analysis of a Drosophila melanogaster BAG-family gene that we have named starvin (stv). Surprisingly, we found stv expression to be regulated in a highly developmentally specific fashion, being expressed primarily in developing larval somatic muscles and their epidermal tendon (muscle attachment) cells and less strongly in the esophagus. This tissue-specific expression appears not to be required for gross muscle morphology, which showed no apparent disruption in stv mutant embryos. However, we show that stv is essential for viability, specifically for the ability of newly hatched larvae to ingest food and grow, indicating that subtle or nonmorphological muscle or esophageal-specific defects required for feeding are disrupted in stv mutant embryos.
MATERIALS AND METHODS
Drosophila stocks:
UAS∷dei was obtained from A. Michelson (Howard Hughes Medical Institute, Brigham and Women's Hospital, Boston); UAS∷sr138 (third chromosome) was obtained from T. Volk (Weizmann Institute of Science, Rehovot, Israel); 69B Gal4/CyO was obtained from M. Muskavitch (Indiana University, Bloomington, IN); β3Tubulin∷lacZ was obtained from D. Buttgereit (Phillips-University Marburg, Marburg, Germany); UAS∷mef2 was obtained from B. Bour (Pennsylvania State University, University Park, PA); en∷Gal4 was obtained from A. Brand (University of Cambridge, Cambridge, UK). Other Drosophila stocks were obtained from the Bloomington (Indiana) stock center.
Antibodies:
Rabbit anti-Alien antibody (Goubeaud et al. 1996) was obtained from A. Paululat (Phillips-University Marburg, Marburg, Germany). Rabbit anti-GFP was purchased from CLONTECH (Pala Alto, CA). Rabbit antimuscle myosin (Kiehart and Feghali 1986) was obtained from D. Kiehart (Duke University Medical School, Durham, NC). Other monoclonal antibodies used in this study were obtained from the Developmental Studies Hybridoma Bank under the auspices of the National Institute of Child Health and Human Development and maintained at the University of Iowa. Rabbit antibodies were raised against a Starvin (STV)-maltose binding protein (MBP) fusion protein and affinity purified first by passing the serum over a MBP column to deplete MBP-specific antibodies and then by passing the eluate over a STV∷MBP column and eluting the bound fraction. Rat anti-STV antibodies were raised against a STV∷GST fusion protein and affinity purified using a MBP∷STV column. The secondary antibodies goat anti-rabbit biotin, donkey anti-rabbit HRP, donkey anti-rat biotin, goat anti-rat HRP, and donkey anti-mouse biotin were affinity purified polyclonals purchased from Jackson ImmunoResearch (West Grove, PA). The Vectastain ABC kit (Vector Laboratories, Burlingame, CA) was used to visualize the antibody stains.
Drosophila embryo whole-mount antibody staining:
Embryos were collected from egg-laying chambers on grape juice agar plates partially spread with yeast paste. Eggs were collected, dechorionated by soaking in 50% bleach, then washed thoroughly in water or 7.5 mm Na2HPO4, 2.5 mm NaH2PO4, 145 mm NaCl, 1% Tween 20 (PBT). Embryos were fixed in 3.7% formaldehyde. Blocking (with either 5% Blotto or 2% goat serum) was performed for a minimum of 30 min. Embryos were incubated with primary antibody overnight at 4° with gentle agitation. After washing, embryos were incubated in secondary antibody for at least 1 hr at 25°, or overnight at 4°, and washed repeatedly before the color reaction was carried out using diaminobenzidine (DAB). If a blue/black staining was desired, rather than the default brown, the final DAB solution also contained a 0.08% NiCl2. Embryos were rinsed four to eight times in PBT, followed by one 5-min wash before being placed under 80% glycerol in PBT for mounting.
P-element and EMS mutagenesis:
The P-element insertion line l(3)00543 (FlyBase 1999) was obtained from the Bloomington Drosophila Stock Center. Imprecise excisions were achieved by crossing in a source of Δ2-3 transposase and selecting for loss of the ry+ marker and for decreased viability when trans-heterozygous with l(3)00543. Chemical mutagenesis was performed on an isogenized ru st e ca strain. Male flies were fed ethyl methanesulfonate in a sucrose solution as described in Grigliatti (1986) and progeny were screened for lethality when trans-heterozygous with the excision deletion Df(3L)stv3c. Three lethal lines (stv1, stv2, and stv3) and one semilethal line (stv4) were obtained from 5000 mutagenized third chromosomes. To aid sequencing of alleles, interspecific hybrids were obtained by crossing stv−/TM3rySb D. melanogaster females to D. simulans males (obtained from the Bloomington stock center) and selecting for non-Sb progeny as the source of DNA to be amplified using melanogaster-specific primers. All stv mutant phenotypic analyses were performed on maternal− zygotic− (m−z−) germline clone mutants produced following the method of Chou and Perrimon (1996).
Feeding ability assay:
Larvae were separated into m−z− and m−z+ classes on the basis of a GFP-marked balancer chromosome and placed onto a fresh grape agar plate, which contained yeast paste mixed with bromophenol blue, an indigestible dye that is visible in the alimentary system of the larva. Larvae were scored for feeding status every 12–24 hr, at which point they were transferred to a fresh indicator plate.
RESULTS
starvin encodes a BAG-domain protein:
Drosophila stv was isolated as a λ-phage expression plaque that bound radiolabeled Polycomblike (Pcl) protein using a previously described filet-based technique (Robert and Saint 1998). Subsequent extensive genetic interaction analysis (data not shown) and developmental and subcellular localization data (see below) indicated that this interaction was almost certainly artifactual. Nevertheless, we pursued the analysis of this gene because we found it to be a member of a family of genes that are postulated to play important roles in mammalian cells (see below), yet no member of this gene family had been subjected to genetic analysis. Sequence analysis of the cDNA revealed a 79-aa region in the carboxy terminal region of STV that shares sequence similarity with three known human proteins, BAG-3/CAIR-1, BAG-4, and BAG-5 (Figure 1A). The region of similarity between these BAG-family members and BAG-1, BAG-2, and HLA-B/BAT3/Scythe defines the BAG domain (Takayama et al. 1999; Thress et al. 2001). The region of recognizable similarity between STV and BAG-3, BAG-4, and the two BAG domains of BAG-5 extends beyond the BAG domain defined by Takayama et al. (1999), enlarging it to 79 aa (Figure 1A). STV has the highest sequence similarity over this region with BAG-4, a lower similarity with BAG-3 and BAG-5 (Figure 1A), and much less similarity to the BAG-1, BAG-2, and Scythe BAG domains (data not shown). BAG-1, BAG-2, and Scythe also have relatively low levels of similarity with all other BAG-family proteins. All BAG-family proteins identified so far, including STV, have a BAG domain close to the carboxyl terminus of the protein, although BAG-5 also has an amino-located BAG domain (Takayama et al. 1999). BLAST analysis of the amino acid sequence of BAG-domain proteins, including STV, failed to reveal any other members of this family in the D. melanogaster genome (results not shown).
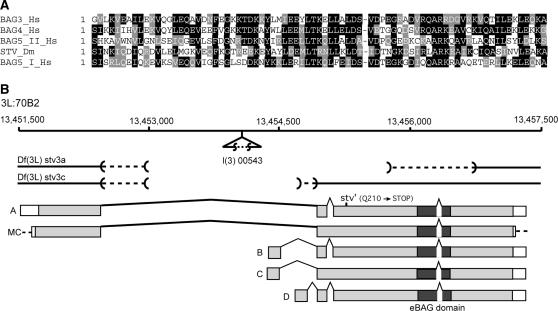
Figure 1.
(A) Alignments of selected BAG domains, showing identical (solid background) and similar (shaded background) residues. STV is most similar to the human BAG-4 protein, while BAG-1 and BAG-2 are the least similar of the five human BAG-family members. Dm, D. melanogaster; Hs, Homo sapiens. Accession nos: BAG-1 (H. sapiens) gpAAC34258; BAG-2 (H. sapiens) gpAAD16121; BAG-3 (H. sapiens) gpAAD16122; BAG-4 (H. sapiens) gpAAD16123; BAG-5 (H. sapiens) gpAAD16124. (B) The genomic region at 70B2 covering the stv locus and showing the intron-exon structure of the longest stv cDNA (labeled A) and the open reading frames (shaded) of this cDNA and three other open reading frames listed in FlyBase (labeled B, C, and D, respectively). In addition, a cDNA clone that we analyzed and that has a structure different from the FlyBase cDNA sequences is also represented (labeled MC). The location of the l(3)00543 P element is marked. The extent of two of the P-element-induced lethal deficiencies is shown, as is the position of the mutated codon in the stv1 allele. The dashed lines indicate the region within which the breakpoints lie. The numbers indicate the genome sequence position along the third chromosome.
Sequence analysis revealed that stv corresponds to CG32130, a computer-annotated gene located at 70B2 on the left arm of the D. melanogaster third chromosome. Conceptual translation of a full-length cDNA (Brown and Kafatos 1988) revealed a 635-amino-acid, 69-kDa protein with an exon structure different from those derived from the Drosophila genome project analyses (Figure 1B). In addition to the BAG domain, another prominent feature of STV is the relative abundance of glutamine, proline, and alanine residues, which make up 38% of the amino acid content (data not shown), although they are particularly sparse over the BAG-domain region.
starvin is expressed specifically in embryonic somatic muscle and tendon cells:
To determine the subcellular localization and tissue distribution of STV, rabbit and rat antibodies were raised against bacterially expressed STV fusion proteins (see materials and methods). Staining of whole-mount embryos showed that STV is not expressed prior to the completion of germ-band retraction (data not shown) but is first detected in a small number of cells during the early stages of dorsal closure (Figure 2A). STV becomes more widely distributed as embryogenesis proceeds (Figure 2, B and C), with faint stripes of expression appearing and increasing in intensity as dorsal closure progresses. These stripes correspond to tendon (muscle attachment) cells at the segmental boundaries. During dorsal closure, in addition to the stripes, various small clusters of cells expressing STV between the stripes were observed, and a somewhat more general staining developed laterally (Figure 2, D and E). The nature of these clusters has not been determined. It is likely that they represent cells that will become the attachment points for the lateral transverse muscles and other muscles that do not attach at the segment border. The tendon-specific transcription factor, Stripe, localizes during stage 13 in similar intrasegmental clusters, which later refine to become the attachment sites of muscles LT1–3 (Lee et al. 1995; Frommer et al. 1996). The bulk of the remaining intrasegmental staining is due to the presence of STV in the somatic muscles (see below). By stage 15, the embryonic pattern of STV localization is essentially mature (Figure 2F).
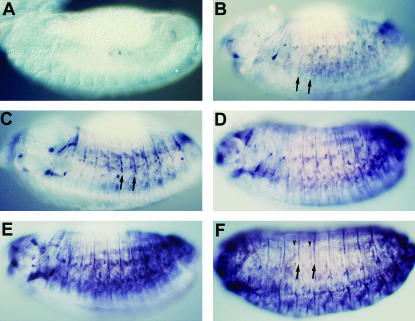
Figure 2.
Developmental profile of stv embryonic expression revealed by whole-mount anti-STV staining. All views are lateral. (A) A stage 13 embryo showing the earliest detectable STV expression, after germ-band retraction has been completed. (B) A mid-stage 13 embryo. STV localization is much more extensive, although very faint compared with older embryos. Intersegmental stripes (arrows) can be seen, in addition to spots of localization. (C) A late stage 13 embryo. Staining is darker, especially in the intersegmental stripes (arrows), and the level of intrasegmental staining is increasing. (D and E) Early and late stage 14 embryos. (F) The mature embryonic STV expression pattern at stage 15. This corresponds to the stage at which most muscles have formed their attachments with the epidermis. STV is present in intersegmental (arrows) and intrasegmental (arrowheads) attachment cells and in muscles.
starvin is expressed in tendon cells that attach body wall and head muscles:
Similarities between the pattern of expression of STV and the published expression patterns of other genes suggested that STV is present in epidermal tendon cells. To confirm this, we directly compared the dorsal, lateral, and ventral aspects of STV localization with genes known to be expressed in epidermal tendon cells. STV has a very distinctive dorsal expression pattern of stripes at the segment boundary (Figure 3A). In segments A2–A6 these stripes are disrupted at the dorsal midline, and there is additional localization in clusters of cells anterior to the main stripe (Figure 3, A–C). Double immunostainings using an enhancer trap that expresses in the dorsal vessel confirmed that both aspects of dorsal STV localization are epidermal (Figure 3, D and E). Each displaced cell cluster at the dorsal midline consists of two cells, one per hemi-segment, which are shifted one cell to the anterior of the main stripe. These cells function as attachment cells for a fragment of muscle DA1, which splits into two at its posterior end and forms two attachments at the dorsal midline (Armand et al. 1994). The dorsal aspects of STV localization are identical to those of Alien (Figure 3F), a known marker of epidermal tendon cells (Goubeaud et al. 1996), confirming that STV is present in these cells.
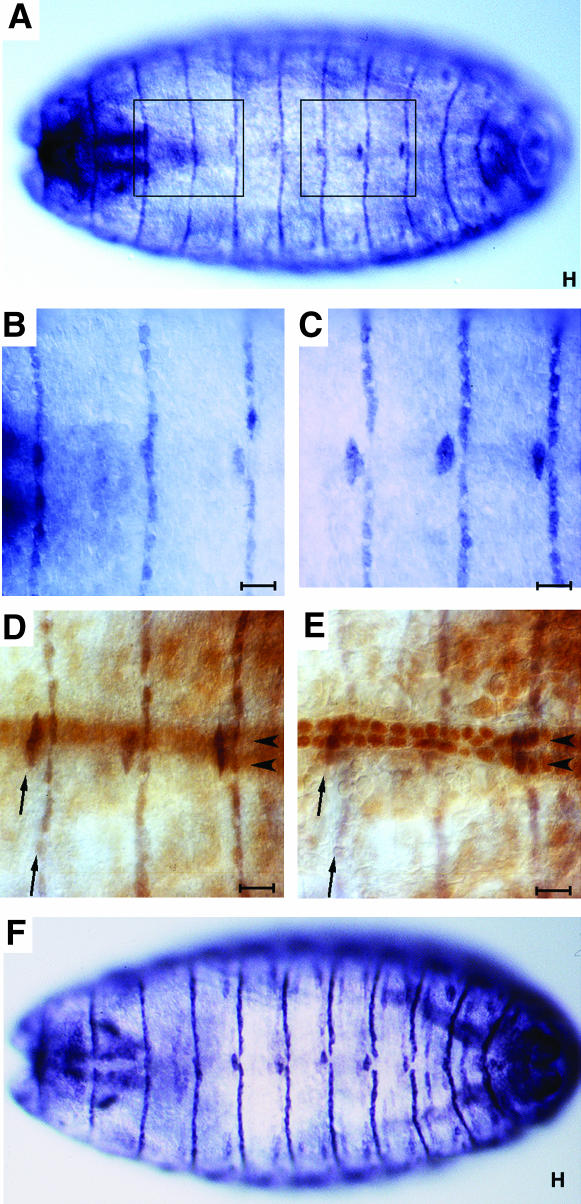
Figure 3.
STV is present in all dorsal epidermal tendon cells. All panels are dorsal views of stage 16 embryos. (A) Anti-STV staining reveals one stripe of STV per segment, extending across the dorsal epidermis of the embryo. Five of these stripes, those at the anterior of segments A2–A6, are broken at the dorsal midline. (B) Higher magnification of the anterior segments boxed in A showing stripes anterior to segments T3, A1, and A2. Only that of A2 is broken at the dorsal midline. (C) Higher magnification of the posterior segments boxed in A showing three stripes of STV localization which are all broken at the dorsal midline (anterior of segments A4–A6). (D) A higher-magnification view of a howE7-3-4 lacZ embryo, an enhancer trap that expresses strongly in nuclei of dorsal vessel cells that flank the dorsal midline. The darker staining in focus shows STV expression (arrows) while the lighter brown color is anti-βGal staining of the dorsal vessel nuclei (arrowheads, out of focal plane). (E) The same embryo with the dorsal vessel nuclei staining in focus and STV staining out of focus. The STV stripes have a width of one cell and the broken portion of the stripe represents two cells, one on either side of the dorsal midline, which are displaced one cell from the main stripe. (F) Anti-Alien staining, showing the same pattern as STV localization. Alien is present in epidermal tendon cells (Goubeaud et al. 1996). All bars, 10 μm.
The lateral localization of STV was found to be more complex than its dorsal localization (Figure 4), a reflection of the diversity of muscles in the lateral region. The three major regions of attachment cells were all found to express stv (groups I, II, and III of Armand et al. 1994; Figure 4A). These regions represent cells that form attachments with multiple muscles. STV was also detected in the attachment cells for the lateral transverse muscles, but staining was not clearly seen in most other cells that form attachments with single muscles (Figure 4A). However, in this regard STV behaves in the same manner as Kakapo, Alien, and β1Tubulin (Alien and β1Tubulin shown in Figure 4, C and E, respectively; Buttgereit 1996; Goubeaud et al. 1996; Strumpf and Volk 1998).
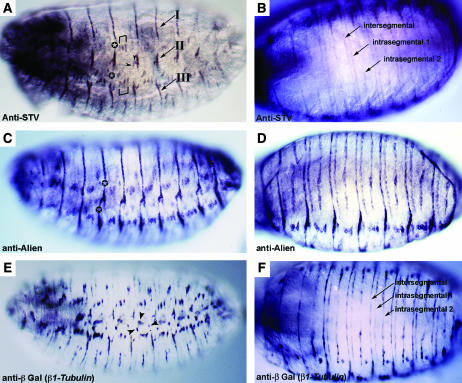
Figure 4.
STV is present in lateral epidermal and ventral epidermal tendon cells. All embryos are at stage 16. (A, C, and E) Lateral views. (B, D, and F) Ventral views. (A and B) Anti-STV staining. (A) The stripes of STV localization that are observed dorsally (Figure 3) are disrupted laterally. The dorsal stripes form attachment group I. STV is also present in the short lateral stripe representing group II and the more ventral group III. Brackets mark the dorsal and ventral attachments of muscles LT1 (21), LT2 (22), and LT3 (23). Arrowheads indicate the ventral attachment of muscle DT1 (18) and the posterior attachment of muscle DO5 (20). Other single attachment points cannot be readily identified. (B) Three stripes of STV localization per segment are observed ventrally. Morphologically, these stripes will generate three furrows, or apodemes: one intersegmental and two intrasegmental (Campos-Ortega and Hartenstein 1997). (C and D) Anti-Alien staining. (C) Alien expression differs from STV expression by continuing across the lateral regions among the attachment groups I, II, and III (indicated by a star). (D) A ventral view shows a pattern identical to that of STV. (E and F) Anti-βGal staining of an embryo carrying a β1Tubulin∷lacZ construct. (E) lacZ is expressed in all tendon cells plus the chordotonal organs (arrowheads). Like STV, β1Tubulin expression is absent among the attachment groups. (F) β1Tubulin is expressed in the same three stripes per segment as STV and Alien.
The pattern of STV localization in the ventral region of the embryo is also representative of genes known to be expressed in epidermal tendon cells (Figure 4, B, D, and F). Three stripes of STV localization were observed in each abdominal segment (Figure 4B), the same pattern seen for β1Tubulin and Alien (Figure 4, D and F; Buttgereit 1996; Goubeaud et al. 1996). The three stripes represent the intersegmental and two intrasegmental apodemes (the morphological indentation produced by tendon cell differentiation and muscle attachment), the intrasegmental apodemes being present only at the ventral surface of the embryo (Campos-Ortega and Hartenstein 1997). The presence of three apodemes is a consequence of muscles attaching at three different points per segment. Localization of all markers of epidermal tendon cells continues along the portion of the apodemes where muscles are not attached, in contrast to the lateral gaps in staining (see above).
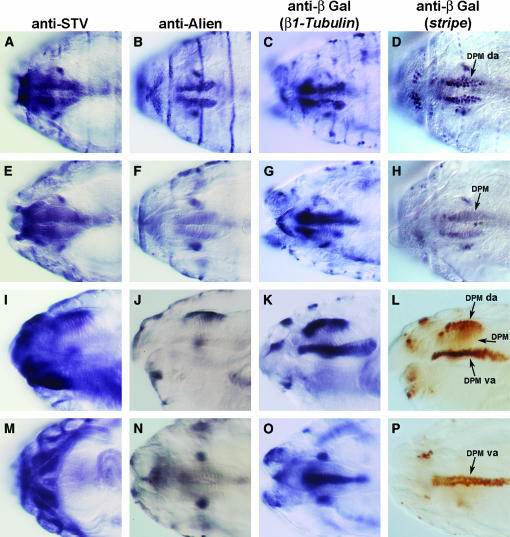
We also observed strong STV expression in the head (Figure 5). The most prominent muscle group of the head is the dorsal pharyngeal musculature (DPM), which is formed from numerous fibers that run dorsoventrally. Dorsally, the DPM attaches at the ventral plane of the dorsal pouch. The ventral attachments are at the dorsal pharyngeal epithelium (Campos-Ortega and Hartenstein 1997). STV was observed in two broad rows of cells in a manner identical to Alien, Stripe, and β1Tubulin, which identifies these cells as the dorsal attachment cells of the DPM (Figure 5, A–D). Visualizing the distinctive morphology of the DPM showed that the attachment cells, rather than the musculature itself, were stained (Figure 5, E–L). STV and Alien are present in the dorsal attachments of the DPM only, while Stripe and β1Tubulin are localized to both dorsal and ventral attachments (Figure 5, I–L). Goubeaud et al. (1996) showed the presence of Alien in the DPM ventral attachments, but we saw no such localization (compare Figure 5J with K and L and Figure 5N with O and P). It is possible that Alien is present in the ventral attachments at a lower concentration than in the dorsal attachment, although the clear lateral view of both sets of attachments shows strong staining dorsally, but no detectable staining ventrally. From the ventral side of the embryo the ventral attachments can be discerned as an alignment of two rows of cells, as observed for Stripe and β1Tubulin, but not for STV and Alien (Figure 5, M–P).
Figure 5.
The pattern of STV expression in the stage 16 embryonic head. A, E, I, and M are stained with anti-STV. B, F, J, and N are stained with anti-Alien. (C, G, K, and O) Embryos carrying a β1Tubulin∷lacZ promoter construct stained with anti-β̃galactosidase. (D, H, L, and P) Embryos carrying an enhancer trap inserted in stripe and stained with anti-β-galactosidase. (A–H) Dorsal views. (A) STV is present in the dorsal attachment of the DPM. STV is expressed in essentially the same pattern as Alien, β1Tubulin, and Stripe markers (B–D), confirming that STV is expressed in tendon cells. (E–H) The same embryos shown in A–E viewed in a different focal plane, focused on the DPM, showing staining for STV to be out of focus and confirming that STV is present in the attachment cells of the DPM. (I–L) Lateral views. (I) STV is localized to the dorsal attachments of the DPM, but not to the DPM itself. The ventral attachment cells of the DPM did not stain. (J–L) Alien, β1Tubulin, and Stripe expression, respectively. Dorsal attachment but not ventral attachment localization is evident for Alien, while β1Tubulin and Stripe are present in both dorsal and ventral attachments of the DPM. (M–P) Ventral views. STV is expressed strongly in the large ventral intersegmental muscles (VIS), but Alien (N), β1Tubulin (O), and Stripe (P) are not. The stripe enhancer trap clearly shows the two rows of cells that make up the ventral attachments of the DPM. DPM da, dorsal attachment of DPM; DPM va, ventral attachment of DPM.
In addition to attachment cells of the DPM, STV is localized in the head to a cluster of cells either side of the DPM, as is Alien, Stripe, and β1Tubulin (Figure 5, A–D). These cells are tendon cells that attach to other cephalic muscles (Goubeaud et al. 1996). Although the identities of the muscles involved have not been determined, it is most likely that the cells attach to the dorsal mouthpart muscles, since this pair of muscles extends dorsolaterally from the wall of the pharynx to the epidermis (Campos-Ortega and Hartenstein 1997). The level of resolution of STV localization was insufficient to reveal whether or not stv is expressed in tendon cells that attach to other cephalic muscles, such as the ventral mouthparts muscle.
Other Starvin-expressing tissues:
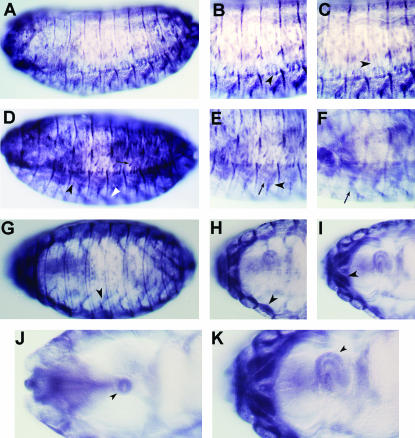
In addition to the distinctive pattern of expression in epidermal tendon cells, stv is also expressed in muscles (Figures 5 and 6). A ventral view of the head region revealed the presence of staining for STV in large muscles, a striking difference between the localization of STV and that of Alien, Stripe, and β1Tubulin (Figure 5M). stv is not expressed in all muscles, as STV staining was not observed in the dorsal pharyngeal musculature (Figure 5I). However, stv does appear to be expressed in all major somatic muscle groups (Figure 6). Specifically, STV was observed in the ventral oblique muscles, the dense ventral lateral clusters, the lateral transverse muscles, and the ventral intersegmental muscles. Significantly, given the mutant phenotype described below, STV localization was observed to be associated with the esophagus and related junctions, notably the proventriculus (Figure 6, J and K).
Figure 6.
STV is present in somatic muscles and the esophagus. (A) Lateral view of a stage 15 embryo, showing staining in epidermal tendon cells as well as in muscles. B and C are higher magnification views of A in different focal planes. Arrowheads indicate ventral attachments of the lateral transverse muscles (LT1–3). The comparison between B and C suggests that STV is present in both LT muscles and their attachment cells. (D) Lateral view of a stage 16 embryo showing lateral transverse muscles (arrow), ventral oblique muscles (white arrowhead), and a cluster of ventral longitudinal, ventral oblique, and ventral acute muscles (black arrowhead). (E and F) Higher magnification views of D in different focal planes showing ventral olique muscles (arrow) and ventral acute muscles (arrowhead). (G) Ventral view of a stage 16 embryo, showing strong staining for STV in the ventral oblique muscles (VO4–6; arrowhead). (H and I) Higher magnification views of G in different focal planes. The short bars of staining perpendicular to the cuticle represent epidermal tendon cells, while the extended parallel staining is in the muscles. The arrowhead in H indicates very strong staining in longitudinal muscles of T3, which most likely represents ventral intersegmental muscle 5 (VIS5) in combination with the ventral longitudinal muscles. There is also very strong staining in muscles of the first and second thoracic segments. The arrowhead in I indicates muscles in the VIS muscles of the first thoracic segment and possibly the ventral pharyngeal muscles. (J) Dorsal view of the head region of a stage 16 embryo. Staining is observed in a ring structure that is part of the proventriculus (arrowhead). (K) Higher magnification of I showing expression in the tubular esophagus (arrowhead).
stv expression is induced by the stripe and mef2 transcriptional regulatory pathways:
stripe function is necessary and sufficient for the epidermal tendon cell fate (Lee et al. 1995; Frommer et al. 1996; Becker et al. 1997). Two stripe transcripts are known: stripe a, which encodes a 1180-amino-acid protein, and stripe b, which encodes a 906-amino-acid protein, consisting of the carboxyl 906 residues encoded by stripe a. Both isoforms include the early growth response 1 and 2 (EGR)-like triple zinc-finger domain (Frommer et al. 1996). stripe b is expressed earlier and more generally, but the final expression pattern is a combination of both transcripts (Frommer et al. 1996).
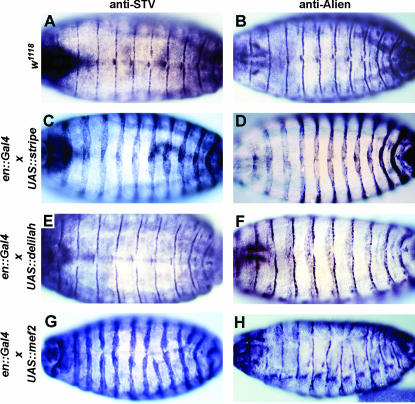
Ectopic expression of stripe b can induce ectopic expression of kakapo (kak), delilah (dei), alien, and β1Tubulin (Becker et al. 1997; Vorbrüggen and Jäckle 1997). As stv is also expressed in the tendon cells, we tested whether ectopic expression of stv is also induced by stripe. Embryos expressing stripe b in the pattern of engrailed, utilizing an en∷GAL4 UAS∷str ectopic expression system (Brand and Perrimon 1993; Becker et al. 1997), showed ectopic stv expression identical to that seen for alien (Figure 7, A–D). Using the 24B GAL4 mesoderm-specific driver to express stripe in the muscles (Brand and Perrimon 1993) did not result in ectopic expression of stv or alien (data not shown), consistent with the previous demonstration that ectopic stripe expression cannot induce expression of kak, dei, alien, or β1Tubulin in the mesoderm (Vorbrüggen and Jäckle 1997).
Figure 7.
Ectopic expression of stripe and mef2, but not delilah, induces STV. All embryos shown are at stage 16 and viewed dorsally or dorso-laterally. A, C, E, and G are stained with anti-STV and B, D, F, H are stained with anti-Alien. (A and B) w1118 control embryos. (C and D) enGAL4; UASstripe embryos. (E and F) enGAL4; UASdelilah embryos. (G and H) enGAL4; UASmef2 embryos. STV is clearly induced by ectopic stripe (C), but not by ectopic delilah (E). Ectopic mef2 is also sufficient to induce ectopic STV in the ectoderm (G). The irregularity of engrailed domains in the enGAL4; UASmef2 embryos is due to ectopic mef2 affecting the patterning of the epidermis and disrupting dorsal closure. Ectopic alien expression is not induced in an enGAL4; UASmef2 embryo (H), showing that the ectopic induction of stv by mef2 is not a consequence of induction of ectopic tendon cells.
dei, which also encodes a transcription factor, is known to be downstream of stripe function (Becker et al. 1997; Vorbrüggen and Jäckle 1997). DEI is therefore a candidate activator of genes that are known to be downstream of stripe, but not known to be direct targets. Ectopic expression of dei in the epidermis, driven by the en∷Gal4 driver, induced ectopic alien but not stv expression (Figure 7, E and F). Thus, alien is induced by both stripe and dei, while stv is induced by stripe but not by dei. It is not known whether stv is a direct or an indirect target of stripe.
The transcription factor MEF2 is known to be required for the expression of numerous somatic muscle genes. mef2 expression in the epidermis is known to induce expression of β3Tubulin, nautilus (nau), and myospheroid (mys; Lin et al. 1997). Ectopic STV was induced when UAS∷mef2 (Bour et al. 1995) was expressed in the epidermis using the en∷Gal4 or 69B Gal4 drivers (Figure 7G and data not shown). In contrast, Alien expression is only mildly disrupted by en∷Gal4 and 69B Gal4-induced expression of mef2 (Figure 7H and data not shown), indicating that the induction of stv by mef2 is not a secondary consequence of an ectopic induction of tendon cell fates.
Generation and analysis of starvin mutations:
l(3)00543 is a semilethal allele caused by the insertion of a P element within the stv transcription unit, although not within the protein-coding sequences. This allele was considered likely to be hypomorphic, so we took two approaches, imprecise P-element excision and EMS mutagenesis, to generate amorphic stv alleles. Multiple P-element reversion strains that exhibited complete lethality were generated and shown by Southern blot analysis to be deletions of regions of stv. Two examples are shown diagrammatically in Figure 1B. Imprecise P-element excision is not a reliable method of inducing specific null mutations, as deletions may not result in null alleles or may remove functionally unrelated flanking loci. For this reason a screen for EMS-induced mutations that are lethal when trans-heterozygous with one of the P reversion stocks, Df(3L)stv3c, was carried out.
Four alleles, stv1, stv2, stv3, and stv4, were generated. stv1, stv2, and stv3 are homozygous and trans-heterozygous lethal, while stv4 is homozygous semilethal. stv4 is lethal in combination with stv1 and stv3, but 20% viable when trans-heterozygous with stv2. The alleles do not form a simple severity series, as stv1/stv3 individuals exhibit a milder larval phenotype than stv1/stv2 individuals do (see below). The allelic strengths may best be represented as stv1 > stv3 ≈ stv2 > stv4. Sequence analysis revealed that stv1 carries a nonsense mutation, which would result in a truncated protein of 209 aa (23 kDa), one-third the size of wild-type STV (Figure 1) and lacking the BAG domain. Although the molecular lesions in the other alleles were not determined, anti-STV staining could not be detected in stv1, stv2, and stv3 trans-heterozygous mutant embryos (data not shown). We conclude that these EMS-induced alleles correspond to mutations in the CG32130/starvin gene.
Although we did not observe any STV protein that was deposited maternally, we avoided complications from any possible maternal contribution by generating germline clone maternal mutant embryos using the FLP/FRT OvoD method developed by Chou and Perrimon (1996). The germline clone females were crossed to males heterozygous for stv1. stv mutant homozygotes were found to die during the first larval instar. While the number of newly hatched first instar larvae was the expected ratio of 1:1 homozygote:heterozygote, very few homozygous mutant second instar larvae were observed (Table 1). The time of lethality was further examined by separating newly hatched germline clone homozygous and heterozygous larvae using the GFP fluorescence of a marker on the balancer chromosome of the heterozygote. Heterozygotes followed wild-type developmental timing, with a first molt occurring ∼24 hr after hatching and a second at ∼48 hr (Table 1). In contrast, stv1/stv2 germline clone mutants could remain as first instars for ≥3 days (Table 1). Only a minority of stv1/stv2 m−z− mutants reached the second larval instar, and these took at least twice as long to do so after hatching, relative to their heterozygous siblings.
TABLE 1.
stv mutant larvae display delayed or arrested growth
| stv1/stv2
|
stv2/stv3
|
stv1/stv3
|
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| m−z+
|
m−z−
|
m−z+
|
m−z−
|
m−z+
|
m−z−
|
|||||||||||||
| Days | L1 | L2 | L3 | L1 | L2 | L3 | L1 | L2 | L3 | L1 | L2 | L3 | L1 | L2 | L3 | L1 | L2 | L3 |
| 1 | 8 | 24 | 0 | 38 | 0 | 0 | 4 | 26 | 0 | 25 | 0 | 0 | 5 | 45 | 0 | 13 | 26 | 0 |
| 2 | 0 | 2 | 22 | 14 | 6 | 0 | 0 | 7 | 19 | 9 | 2 | 0 | 0 | 11 | 37 | 9 | 24 | 5 |
| 3 | 0 | 0 | 21 | 5 | 3 | 1 | 0 | 0 | 25 | 1 | 3 | 1 | 0 | 2 | 42 | 4 | 16 | 18 |
Larvae were separated into m−z+ and m−z− classes by the presence or absence, respectively, of GFP fluorescence and allowed to continue to develop on yeast paste. The numbers of live first, second, and third instar larvae were counted at ∼1, 2, and 3 days after hatching. Larval instars were distinguished by the size and structure of the mouth-hook apparatus and the structure of the posterior spiracles.
stv mutant larvae exhibit a severe feeding disability:
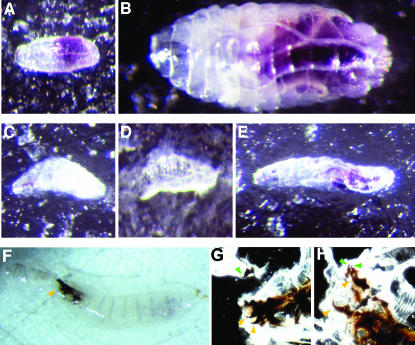
One explanation for the failure of the stv larvae to grow, or to do so only slowly, is that they may not have been able to feed (see, for example, Zinke et al. 1999). Uptake of food can be tested by feeding larvae yeast paste containing bromophenol blue, which acts as a nondigestible dye that is visible in the foregut and midgut (Dubreuil et al. 1998). Newly hatched progeny of germline clone crosses were sorted into m−z+ and m−z− classes by the presence or absence, respectively, of GFP fluorescence and then allowed to develop on grape agar plates supplemented with yeast paste containing bromophenol blue. stv1/stv2 and stv2/stv3 larvae demonstrated severely reduced or no detectable food uptake, as observed by a minimal amount or absence of bromophenol blue visible in the gut (Figure 8, A–E). Larvae that showed no blue food in the gut failed to grow above hatching size (Figure 8, C and D), although some were observed to live for at least 4 days after hatching. Larvae that showed some food uptake grew from hatching size, but the amount of growth was minimal compared to heterozygous siblings (Figure 8, B and E). The majority of heterozygous larvae increased their weight many times over and molted to second instars ∼1 day after hatching. In contrast, all m−z− larvae were still in the first instar (Table 1). Two days after hatching, the majority of heterozygous larvae had undergone the second molt, with no first instars observed, and by 3 days all heterozygous larvae had entered the third larval instar. A minority of m−z− larvae underwent the first molt after 2 days, as determined by mouth-hook size and structure of posterior spiracles, but these larvae were smaller than normal at the second molt (Table 1). By 3 days, rare m−z− mutant third instar larvae were observed (Table 1). These failed to pupate, and in some instances were observed alive 10 days after hatching, grown since molting, but still far smaller than wild-type wandering third instars. However, the majority of stv mutant larvae did not develop beyond first instar stage and died well short of reaching wild-type size. The commonly observed size difference between m−z− and m−z+ larvae is shown in Figure 8. We also examined the phenotype of zygotic trans-heterozygous mutant combinations and found the same slow-growth phenotype and an inability to ingest bromophenol-stained yeast paste, consistent with the absence of observable maternal stv product.
Figure 8.
stv mutants have feeding and molting disabilities. (A and B) stv1/TM6B, Tb germline clone-derived zygotically rescued (m−z+) larvae. (C–E) stv1/stv2 maternal zygotic mutant (m−z−) larvae. (F and G) stv1/stv3 m−z− larvae. All larvae were placed on bromophenol-blue-containing yeast paste <2 hr after hatching. (A and C) Approximately 8 hr after hatching, mutant and zygotically rescued larvae are of a comparable size, although blue food can be seen in the gut of the latter but not the former. (B) By ∼48 hr after hatching, this zygotically rescued larva has grown substantially and completed the first molt, while others progress through the second molt (not shown). (D) In contrast, a stv1/stv2 mutant larva, shown at the same magnification as the heterozygous larva in B, has not grown at all. (E) In contrast to D, this larva has doubled in length since hatching, and some blue dye is visible. However, the size increase is minor compared with the zygotically rescued larva of the same age (B), and no molts have occurred. All larvae are shown at the same magnification. (F–H) A hypomorphic allelic combination does not result in feeding difficulties, but a significant proportion of larvae become stuck at the second molt. (F) An example of a stv1/stv3 larva (m−z−) that has failed to molt correctly between the second and third larval instar. This larva has managed to free itself of its old cuticle, but the cuticle remains attached to the mouth-hook apparatus (arrowhead). (G) Larva shown in F squashed between two microscope slides to visualize the mouth hooks. Both the smaller second instar mouth hooks (green arrowheads) and the third instar mouth hooks (red arrowheads) can be seen, indicating that the cuticle remained attached because the mouth hooks have not resolved. (H) Squash of a second larva, which failed to separate the new and old cuticles. Again, both second and third instar mouth hooks can be seen (green and yellow arrowheads, respectively).
In contrast to the severely compromised feeding phenotype and associated stunted growth rate of larvae of the allele combinations stv1/stv2 and stv2/stv3, the development of stv1/stv3 larvae was only slightly retarded. Three days after hatching, when virtually all heterozygous larvae were in the third instar, only a small proportion of m−z− larvae remained arrested in the first larval instar, and the remainder were approximately equally divided between the second and third larval instars (Table 1). However, a striking phenotype seen in a proportion of stv1/stv3 larvae was the failure of second instar and third instar mouth hooks to separate, preventing completion of the second molt (Figure 8, F–H, and data not shown).
Muscle and head involution defects are not evident in stv mutant embryos:
There are numerous possible behavioral, morphological, and physiological reasons for reduced or absent feeding. Given the expression of stv in muscle and tendon cells, the most likely explanation was some type of muscle defect. The most common phenotypes for mutants of genes expressed in epidermal tendon cells are misguidance of muscles (and thus misattachment) and the failure of maintenance of attachment. For example, stripe mutant embryos exhibit elongated muscle processes and occasional rounding up of muscles that have broken their attachments (Frommer et al. 1996). Rounding up of muscles is the prominent phenotype of mys and irregular facets (if) mutant embryos (Leptin et al. 1989; Brabant and Brower 1993; Brown 1994). Muscles of kak mutant embryos detach from the epidermis during stage 17 because of a lack of structural integrity of the epidermal cells, which tear in two under the stress of muscle contraction (Gregory and Brown 1998; Prokop et al. 1998b). A severe somatic muscle attachment phenotype should also result in embryonic lethality or, at least, larval movement defects. Neither of these was observed in stv mutant homozygotes. The tracks left by stv mutants did not obviously differ in length or complexity from those of wild-type larvae. Like normal larvae, stv mutant larvae were found to converge on yeast paste and mouth-hook movements were indistinguishable from those of normal larvae while feeding (data not shown). The gross feeding behavior of stv mutant larvae therefore appeared to be normal.
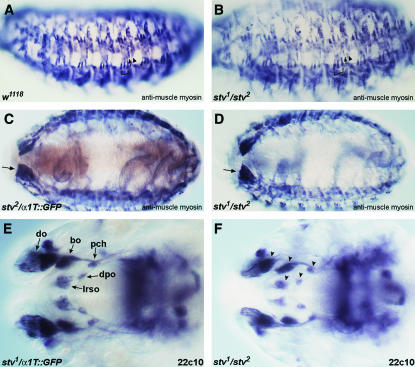
To further test whether muscle structure was affected, we used antimuscle myosin to visualize somatic muscles in stv mutant embryos (Figure 9, A–D). Germline clone mutant embryos, lacking both maternal and zygotic stv products, exhibited no muscle abnormalities. All major muscle groups were observed to be present and showed no sign of disorganization.
Figure 9.
Muscle defects are not evident in stv mutants, and head involution proceeds normally. (A–D) Muscle myosin was detected with antimuscle myosin (blue/black). Light brown staining in A, C, and E is anti-GFP to distinguish zygotically rescued embryos. (A) Lateral view of a stage 16 w1118 embryo (shown instead of a m−z+ embryo to show the wild-type pattern of lateral muscles in the absence of anti-GFP staining). Arrowhead indicates muscle DT1, arrow indicates LT4, and LT1–3 are bracketed. (B) Lateral view of a S16 stv1/ stv2 m−z− embryo. Symbols as in A. The muscle pattern appears normal. (C) Ventral view of a S16 stv2/TM6B, α1Tubulin∷GFP m−z+ embryo. Arrow indicates the large VIS muscles. (D) Ventral view of a S16 stv1/stv2 m−z− embryo. Arrow indicates the large VIS muscles. The muscle pattern appears normal. (E and F) Mab 22C10 staining of stage 16 embryos. (E) Dorsal view of the head region of a S16 stv1/TM6B, α1Tubulin∷GFP m−z+ embryo. do, dorsal organ; bo, Bolwig's organ; pch, dorsal pharyngeal chordotonal organ; dpo, dorsal pharyngeal organ; lrso, labral sensory complex. Before head involution, lrso and dpo extend anteriorly to do. (F) Dorsal view of the head region of a S16 stv1/stv2 m−z−. The relative positions of the sense organs (arrowheads) indicate that head involution has proceeded normally.
One known cause of larval feeding failure is incomplete head involution during embryogenesis. For example, mutants specific for the cytoskeletal isoform of tropomyosin die as first instar larvae because they cannot feed (Tetzlaff et al. 1996). These mutants are normal until stage 16 of embryogenesis, but head involution fails to occur, and the hatching larvae have a protruding pharyngeal mass (Tetzlaff et al. 1996). Progression of head involution can be followed by the relative positions of the head sensory organs. Before head involution occurs, the labral sensory complex (lrso) and dorsal pharyngeal organ (dpo) are positioned the farthest anteriorly, such that the lrso is anterior to the dorsal organ (do) (Campos-Ortega and Hartenstein 1997). Staining with monoclonal antibody 22C10 showed that the relative positions of the sense organs in the head are the same in late mutant and heterozygous larvae (Figure 9, E and F), indicating that head involution occurred normally in stv mutant larvae.
We conclude, therefore, that feeding is compromised in stv mutant larvae not as a consequence of a more general defect, but because of something that specifically affects the ingestion of food.
DISCUSSION
We report here a developmental and genetic characterization of the Drosophila gene stv. The derived amino acid sequence of STV revealed significant similarity to the BAG family of genes. The region of similarity in STV extends almost 80 aa and includes the 47-aa BAG domain originally defined by Takayama et al. (1999). The BAG family is an important gene family implicated in a variety of biological roles, most often through their role as Hsp70 and Hsc70 cochaperones.
stv is expressed in a highly tissue-specific manner in epidermal muscle attachment cells and in somatic muscles, as well as in parts of the esophagus or associated tissues. It should be noted that the Berkeley Drosophila Genome Project in situ data (http://www.fruitfly.org/cgi-bin/ex/insitu.pl) show midgut expression that we did not observe. We may have missed this expression because it is transient or because of translational regulation or because of limited antibody penetration.
Very few genes are expressed specifically in both epidermal muscle attachment cells and associated somatic muscles. myospheroid (encoding the βPS integrin subunit) is expressed in both tissues, and βPS integrin localizes to the membrane, concentrated at the sites of muscle-epidermal attachment (Brown et al. 1993). The second gene known to be expressed in both muscles and their attachments is held out wings (how), which encodes an RNA-binding domain protein that regulates tendon cell differentiation (Nabel-Rosen et al. 2002 and references therein). In contrast to the localization of STV, expression of how begins early, zygotic message being observed in the mesoderm from the beginning of gastrulation. how remains expressed in cells that will form the somatic, visceral, and cardiac muscles. During dorsal closure, expression also becomes apparent in the epidermal muscle attachment cells. Levels of HOW remain highest in the cardiac muscles (dorsal vessel) and epidermal muscle attachment cells (Baehrecke 1997; Fyrberg et al. 1997; Lo and Frasch 1997; Zaffran et al. 1997). The latter pattern of HOW localization is similar to that of STV, although no appreciable levels of STV are detected in the dorsal vessel. Thus, STV has a unique distribution, being present in fewer tissues than HOW and more generally localized within the cell than βPS.
The accumulation of STV in epidermal muscle attachment cells is most similar to the accumulation of β1Tubulin and Delilah, which are first observed in weak intersegmental stripes during stage 13, which increase in intensity during stages 15 and 16 (Armand et al. 1994; Buttgereit 1996; Yarnitzky et al. 1997). In contrast, kak, alien, and stripe begin to be expressed earlier, at stage 11, and always at their final intensity (Lee et al. 1995; Frommer et al. 1996; Goubeaud et al. 1996; Strumpf and Volk 1998). stv regulation falls within previously characterized muscle and tendon transcriptional regulatory pathways. stv is downstream of stripe function in the epidermis, but not of delilah. In contrast, expression of alien was found to be downstream of both stripe and delilah. In somatic muscle, stv appears to be under the regulation of the transcription factor mef2.
stv mutant individuals lack the ability to take up food and die most often as first or, occasionally, as second or third instar larvae. The survival for days after hatching, even of larvae that apparently fail to eat at all, is consistent with previously observed rates of survival under starvation conditions. Larvae hatched onto PBS solution live for 2–3 days, and on 20% sucrose in PBS for ∼8 days (Britton and Edgar 1998; B. Edgar, personal communication). In both instances, larvae remain small and arrested in the first larval instar. The delayed development and reduced growth of stv mutant larvae is therefore most likely due to a severely reduced uptake of nutrients necessary for growth.
We were unable to determine the precise cause of the feeding impairment. The localization of STV to epidermal tendon cells and somatic muscles suggests a role in muscle development, attachment, or function, but morphological defects in muscle structures were not observed. Consistent with this, stv mutant larvae hatch, crawl around apparently normally, and exhibit normal mouth-hook movements. It is possible that a specific muscle involved in control of the esophagus is particularly sensitive to disruption by the absence of stv function, manifesting as a phenotype that is more specific than would be predicted from the broad tendon and muscle cell expression of stv. Alternatively, defects in the foregut, proventriculus, or gastric cecum can result in feeding defects. However, again, no obvious defects were observed in the morphology of these structures in stv mutant larvae. The gross anatomy of the muscles and gut of late stv mutant embryos appeared normal, but subtle changes may have been missed in our analysis. Alternatively, stv may not be required for tissue structure, but may be required for proper physiological function of muscle or esophageal structures. Consistent with this, STV appears in muscles only after attachment, so it is likely to have a role in the later aspects of muscle maturation or function. Subtle changes in muscle function may also explain the stv1/stv3 mutant larvae that die at the molt between the second and third larval instars from failure of the old and new sets of mouth hooks to separate, as the muscles responsible for moving the mouth hooks are thought to detach from the set to be discarded and to establish connections to the new mouth hooks (Crossley 1978).
The functions of mammalian BAG-domain proteins shed little light on the stv mutant phenotype. The BAG domain of STV is most similar to those of human BAG-4, which is also referred to as SODD (suppressor of the death domain), human BAG-3, and one of the human BAG-5 BAG domains. BAG-4 is a 457-aa binding partner of tumor necrosis factor receptor 1 (TNF-R1) and death receptor 3 (Jiang et al. 1999; Miki and Eddy 2002) and, like other human BAG-domain proteins, also interacts with Bcl2. BAG-4/SODD appears to have roles similar to other human BAG-domain proteins in acting in an antipapoptotic manner, for example, suppressing apoptosis when overexpressed in pancreated cancer cells (Liao et al. 2001) and suppressing TNF-induced apoptosis (Miki and Eddy 2002). BAG-3 forms a ternary complex with Hsc70 and phospholipase Cγ (PLCγ) in response to EGF. PLCγ is contacted by BAG-3 through a domain comprising multiple PXXP motifs leaving the BAG domain free to contact Hsp70 (Doong et al. 2000). BAG-5 also exhibits Hsp70-binding ability, along with an ability to interact directly with parkin, an E3 ubiquitin ligase frequently mutated in early-onset Parkinson's disease (Kalia et al. 2004). In this case, BAG-5 inhibits parkin E3 ubiquitin ligase activity and Hsp70-mediated refolding of misfolded proteins and, in contrast to other BAG-domain proteins, acts as an antisurvival factor in dopaminergic neurons (Kalia et al. 2004). Consequently, although we can be confident that STV will act as an Hsp70-family chaperone and, most likely, bind to other proteins to modify their function, the potential targets are numerous, making their cellular and developmental roles impossible to predict. Curiously, some mutant alleles of Actin88F induce heat-shock proteins at normal temperatures. In these mutants, heat-shock proteins are bound to the myofiber, suggesting that they stabilize the mutant Actin forms (reviewed by Bernstein et al. (1993). However, the predominantly normal function of the muscles in stv mutant embryos suggests that if such a role exists for STV, it must be relatively minor.
Irrespective of the difficulty in establishing a developmental or physiological explanation for the stv mutant feeding phenotype, our analysis has yielded several important observations that shed new light onto the function of the BAG-family genes. The developmental specificity, larval lethality, and feeding and molting phenotypes raise the possibility that one or more of the BAG-family genes plays developmentally specific roles during mammalian development.
Acknowledgments
We thank B. Bour, D. Buttgereit, A. Michelson, A. Paululat, and T. Volk for the gifts of fly strains and antibodies. This work was funded by the Australian Research Council and the National Health and Medical Research Council of Australia. M. Coulson was supported by an Australian Postgraduate Research Award and S. Robert was supported by a University of Adelaide Faculty of Science Scholarship.
References
- Alberti, S., C. Esser and J. Höhfeld, 2003. BAG-1—a nucleotide exchange factor of Hsc70 with multiple cellular functions. Cell Stress Chaperones 8: 225–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoku, K., R. S. Maser, W. J. Scully, Jr., S. M. Delach and D. E. Jonhson, 2001. Isolation of Bcl-2 binding proteins that exhibit homology with BAG-1 and suppressor of death domains protein. Biochem. Biophys. Res. Commun. 286: 1003–1010. [DOI] [PubMed] [Google Scholar]
- Armand, P., A. C. Knapp, A. J. Hirsch, E. F. Wiechaus and M. D. Cole, 1994. A novel basic helix-loop-helix protein is expressed in muscle attachment sites of the Drosophila epidermis. Mol. Cell. Biol. 14: 4145–4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baehrecke, E. H., 1997. who encodes a KH RNA binding protein that functions in muscle development. Development 124: 1323–1332. [DOI] [PubMed] [Google Scholar]
- Bardelli, A., P. Longati, D. Albero, S. Goruppi, C. Schneider et al., 1996. HGF receptor associates with the anti-apoptotic protein BAG-1 and prevents cell death. EMBO J. 15: 6205–6212. [PMC free article] [PubMed] [Google Scholar]
- Becker, S., G. Pasca, D. Strumpf, L. Min and T. Volk, 1997. Reciprocal signaling between Drosophila epidermal muscle attachment cells and their corresponding muscles. Development 124: 2615–2622. [DOI] [PubMed] [Google Scholar]
- Bernstein, S. I., P. T. O'Donnell and R. M. Cripps, 1993. Molecular genetic analysis of muscle development, structure, and function in Drosophila. Int. Rev. Cytol. 143: 63–152. [DOI] [PubMed] [Google Scholar]
- Bour, B. A., M. A. O'Brien, W. L. Lockwood, E. S. Goldstein, R. Bodmer et al., 1995. Drosophila MEF2, a transcription factor that is essential for myogenesis. Genes Dev. 9: 730–741. [DOI] [PubMed] [Google Scholar]
- Brabant, M. C., and D. L. Brower, 1993. PS2 integrin requirements in Drosophila embryo and wing morphogenesis. Dev. Biol. 157: 49–59. [DOI] [PubMed] [Google Scholar]
- Brand, A. H., and N. Perrimon, 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415. [DOI] [PubMed] [Google Scholar]
- Briknarova, K., S. Takayama, L. Brive, M. L. Havert, D. A. Knee et al., 2001. Structural analysis of BAG1 cochaperone and its interactions with Hsc70 heat shock protein. Nat. Struct. Biol. 8: 349–352. [DOI] [PubMed] [Google Scholar]
- Briknarova, K., S. Takayama, S. Homma, K. Baker, E. Cabezas et al., 2002. BAG4/SODD protein contains a short BAG domain. J. Biol. Chem. 277: 31172–31178. [DOI] [PubMed] [Google Scholar]
- Britton, J. S., and B. A. Edgar, 1998. Environmental control of the cell cycle in Drosophila: nutrition activates mitotic and endoreplicative cells by distinct mechanisms. Development 125: 2149–2158. [DOI] [PubMed] [Google Scholar]
- Brockmann, C., D. Leitner, D. Labudde, A. Diehl, V. Sievert et al., 2004. The solution structure of the SODD BAG domain reveals additional electrostatic interactions in the HSP70 complexes of SODD subfamily BAG domains. FEBS Lett. 558: 101–106. [DOI] [PubMed] [Google Scholar]
- Brown, N. H., 1994. Null mutations in the αPS2 and βPS integrin subunit genes have distinct phenotypes. Development 120: 1221–1231. [DOI] [PubMed] [Google Scholar]
- Brown, N. H., and T. C. Kafatos, 1988. Functional cDNA libraries from Drosophila embryos. J. Mol. Biol. 203: 425–437. [DOI] [PubMed] [Google Scholar]
- Brown, N. H., J. W. Bloor, O. Dunin-Borkowski and M. D. Martin-Bermudo, 1993. Integrins and morphogenesis. Development 1993(Suppl.): 177–183. [PubMed] [Google Scholar]
- Buttgereit, D., 1996. Transcription of the β1 tubulin (βTub56D) gene in apodemes is strictly dependent on muscle insertion during embryogenesis in Drosophila melanogaster. Eur. J. Cell Biol. 71: 183–191. [PubMed] [Google Scholar]
- Campos-Ortega, J. A., and V. Hartenstein, 1997. The Embryonic Development of Drosophila melanogaster. Springer-Verlag, Berlin.
- Chou, T.-B., and N. Perrimon, 1996. The autosomal FLP-DFS technique for generating germline mosaics in Drosophila melanogaster. Genetics 144: 1673–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossley, A. C., 1978. The morphology and development of the Drosophila musculature system, pp. 499–560 in The Genetics and Biology of Drosophila, edited by M. Ashburner and T. R. F. Wright. Academic Press, London.
- Doong, H., J. Price, Y. S. Kim, C. Gasbarre, J. Probst et al., 2000. CAIR-1/BAG-3 forms an EGF-regulated ternary complex with phospholipase C-gamma and Hsp70/Hsc70. Oncogene 19: 4385–4395. [DOI] [PubMed] [Google Scholar]
- Doong, H., A. Vrailas and E. C. Kohn, 2002. What's in the ‘BAG’?—a functional domain analysis of the BAG family proteins. Cancer Lett. 188: 25–32. [DOI] [PubMed] [Google Scholar]
- Dubreuil, R. R., J. Frankel, P. Wang, J. Howrylak, M. Kappil et al., 1998. Mutations of α spectrin and labial block cuprophilic cell differentiation and acid secretion in the middle midgut of Drosophila larvae. Dev. Biol. 194: 1–11. [DOI] [PubMed] [Google Scholar]
- FlyBase, 1999. The FlyBase database of the Drosophila genome projects and community literature. Nucleic Acids Res. 27: 85–88 (http://flybase.bio.indiana.edu). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froesch, B. A., S. Takayama and J. C. Reed, 1998. BAG-1L protein enhances androgen receptor function. J. Biol. Chem. 273: 11660–11666. [DOI] [PubMed] [Google Scholar]
- Frommer, G., G. Vorbrüggen, G. Pasca, H. Jäckle and T. Volk, 1996. Epidermal egr-like zinc finger protein of Drosophila participates in myotube guidance. EMBO J. 15: 1642–1649. [PMC free article] [PubMed] [Google Scholar]
- Fyrberg, C., J. Becker, P. Barthmaier, J. Mahaffey and E. Fyrberg, 1997. A Drosophila muscle-specific gene related to the mouse quaking locus. Gene 197: 315–323. [DOI] [PubMed] [Google Scholar]
- Gassler, C. S., T. Wiederkehr, D. Brehmer, B. Bukau and M. P. Mayer, 2001. Bag-1M accelerates nucleotide release for human Hsc70 and Hsp70 and can act concentration-dependent as positive and negative cofactor. J. Biol. Chem. 276: 32538–32544. [DOI] [PubMed] [Google Scholar]
- Gebauer, M., M. Zeiner and U. Gehring, 1997. Proteins interacting with the molecular chaperone hsp70/hsc70: physical associations and effects on refolding activity. FEBS Lett. 417: 109–113. [DOI] [PubMed] [Google Scholar]
- Gehring, U., 2004. Biological activities of HAP46/BAG-1. EMBO Rep. 5: 148–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goubeaud, A., S. Knirr, R. Renkawitz-Pohl and A. Paululat, 1996. The Drosophila gene alien is expressed in the muscle attachment sites during embryogenesis and encodes a protein highly conserved between plants, Drosophila and vertebrates. Mech. Dev. 57: 59–68. [DOI] [PubMed] [Google Scholar]
- Gregory, S. L., and N. H. Brown, 1998. kakapo, a gene required for adhesion between and within cell layers in Drosophila, encodes a large cytoskeletal linker protein related to plectin and dystrophin. J. Cell Biol. 143: 1271–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigliatti, T., 1986. Mutagenesis, pp. 55–83 in Drosophila: A Practical Approach, edited by D. B. Roberts. IRL Press, Oxford.
- Höhfeld, J., 1998. Regulation of the heat shock conjugate Hsc70 in the mammalian cell: the characterization of the anti-apoptotic protein BAG-1 provides novel insights. Biol. Chem. 379: 269–274. [PubMed] [Google Scholar]
- Höhfeld, J., and S. Jentsch, 1997. GrpE-like regulation of the Hsc70 chaperone by the anti-apoptotic protein BAG-1. EMBO J. 16: 6209–6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung, W. J., R. S. Roberson, J. Taft and D. Y. Wu, 2003. Human BAG-1 proteins bind to the cellular stress response protein GADD34 and interfere with GADD34 functions. Mol. Cell. Biol. 23: 3477–3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, Y., J. D. Woronicz, W. Liu and D. V. Goeddel, 1999. Prevention of constitutive TNF receptor 1 signaling by silencer of death domains. Science 283: 543–546. [DOI] [PubMed] [Google Scholar]
- Kalia, S. K., S. Lee, P. D. Smith, L. Liu, S. J. Crocker et al., 2004. BAG5 inhibits Parkin and enhances dopaminergic neuron degeneration. Neuron 44: 931–945. [DOI] [PubMed] [Google Scholar]
- Kiehart, D. P., and R. Feghali, 1986. Cytoplasmic myosin from Drosophila melanogaster. J. Cell Biol. 103: 1517–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kullmann, M., J. Schneikert, J. Moll, S. Heck, M. Zeiner et al., 1998. RAP46 is a negative regulator of glucocorticoid receptor action and hormone-induced apoptosis. J. Biol. Chem. 273: 14620–14625. [DOI] [PubMed] [Google Scholar]
- Lee, J. C., K. VijayRaghavan, S. E. Celniker and M. A. Tanouye, 1995. Identification of a Drosophila muscle development gene with structural homology to mammalian early growth response transcription factors. Proc. Natl. Acad. Sci. USA 92: 10344–10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leptin, M., T. Bogaert, R. Lehmann and M. Wilcox, 1989. The function of PS integrins during Drosophila embryogenesis. Cell 56: 401–408. [DOI] [PubMed] [Google Scholar]
- Liao, Q., F. Ozawa, H. Friess, A. Zimmermann, S. Takayama et al., 2001. The anti-apoptotic protein BAG-3 is overexpressed in pancreatic cancer and induced by heat stress in pancreatic cancer cell lines. FEBS Lett. 503: 151–157. [DOI] [PubMed] [Google Scholar]
- Lin, M.-H., B. A. Bour, S. M. Abmayr and R. V. Storti, 1997. Ectopic expression of MEF2 in the epidermis induces epidermal expression of muscle genes and abnormal muscle development in Drosophila. Dev. Biol. 182: 240–255. [DOI] [PubMed] [Google Scholar]
- Liu, R., S. Takayama, Y. Zheng, B. Froesch, G. Q. Chen et al., 1998. Interaction of BAG-1 with retinoic acid receptor and its inhibition of retinoic acid-induced apoptosis in cancer cells. J. Biol. Chem. 273: 16985–16992. [DOI] [PubMed] [Google Scholar]
- Lo, P. C. H., and M. Frasch, 1997. A novel KH-domain protein mediates cell adhesion processes in Drosophila. Dev. Biol. 190: 241–256. [DOI] [PubMed] [Google Scholar]
- Manchen, S. T., and A. V. Hubberstey, 2001. Human Scythe contains a functional nuclear localization sequence and remains in the nucleus during staurosporine-induced apoptosis. Biochem. Biophys. Res. Commun. 287: 1075–1082. [DOI] [PubMed] [Google Scholar]
- Matsuzawa, S., S. Takayama, B. A. Froesch, J. M. Zapata and J. C. Reed, 1998. p53-inducible human homologue of Drosophila seven in absentia (Siah) inhibits cell growth: suppression by BAG-1. EMBO J. 17: 2736–2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki, K., and E. M. Eddy, 2002. Tumor necrosis factor receptor 1 is an ATPase regulated by silencer of death domain. Mol. Cell. Biol. 22: 2536–2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabel-Rosen, H., G. Volohonsky, A. Reuveny, R. Zaidel-Bar and T. Volk, 2002. Two isoforms of the Drosophila RNA binding protein, how, act in opposing directions to regulate tendon cell differentiation. Dev. Cell 2: 183–193. [DOI] [PubMed] [Google Scholar]
- Prokop, A., J. Uhler, J. Roote and M. Bate, 1998. b The kakapo mutation affects terminal arborization and central dendritic sprouting of Drosophila motorneurons. J. Cell Biol. 143: 1283–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert, S., and R. Saint, 1998. Rapid screening for protein interactors using in vitro translated protein and an expression library. Technical Tips Online (http://www.elsevier.com/locate/tto).
- Shatkina, L., S. Mink, H. Rogatsch, H. Klocker, G. Langer et al., 2003. The cochaperone Bag-1L enhances androgen receptor action via interaction with the NH2-terminal region of the receptor. Mol. Cell. Biol. 23: 7189–7197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sondermann, H., C. Scheufler, C. Schneider, J. Hohfeld, F. U. Hartl et al., 2001. Structure of a Bag/Hsc70 complex: convergent functional evolution of Hsp70 nucleotide exchange factors. Science 291: 1553–1557. [DOI] [PubMed] [Google Scholar]
- Song, J., M. Takeda and R. I. Morimoto, 2001. Bag1-Hsp70 mediates a physiological stress signalling pathway that regulates Raf-1/ERK and cell growth. Nat. Cell Biol. 3: 276–282. [DOI] [PubMed] [Google Scholar]
- Strumpf, D., and T. Volk, 1998. Kakapo, a novel cytoskeletal-associated protein is essential for the restricted localization of the neuregulin-like factor, Vein, at the muscle-tendon junction site. J. Cell Biol. 143: 1259–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symersky, J., Y. Zhang, N. Schormann, S. Li, R. Bunzel et al., 2004. Structural genomics of Caenorhabditis elegans: structure of the BAG domain. Acta Crystallogr. D Biol. Crystallogr. 60: 1606–1610. [DOI] [PubMed] [Google Scholar]
- Takayama, S., T. Sato, S. Krajewski, K. Kochel, S. Irie et al., 1995. Cloning and functional analysis of BAG-1: a novel Bcl-2-binding protein with anti-cell death activity. Cell 80: 279–284. [DOI] [PubMed] [Google Scholar]
- Takayama, S., D. N. Bimston, S. Matsuzawa, B. C. Freeman, C. Aime-Sempe et al., 1997. BAG-1 modulates the chaperone activity of Hsp70/Hsc70. EMBO J. 16: 4887–4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama, S., Z. Xie and J. C. Reed, 1999. An evolutionarily conserved family of Hsp70/Hsc70 molecular chaperone regulators. J. Biol. Chem. 274: 781–786. [DOI] [PubMed] [Google Scholar]
- Tetzlaff, M. T., H. Jäckle and M. J. Pankratz, 1996. Lack of Drosophila cytoskeletal tropomyosin affects head morphogenesis and the accumulation of oskar mRNA required for germ cell formation. EMBO J. 15: 1247–1254. [PMC free article] [PubMed] [Google Scholar]
- Thress, K., J. Song, R. I. Morimoto and S. Kornbluth, 2001. Reversible inhibition of Hsp70 chaperone function by Scythe and Reaper. EMBO J. 20: 1033–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend, P. A., A. Stephanou, G. Packham and D. S. Latchman, 2005. BAG-1: a multi-functional pro-survival molecule. Int. J. Biochem. Cell Biol. 37: 251–259. [DOI] [PubMed] [Google Scholar]
- Vorbrüggen, G., and H. Jäckle, 1997. Epidermal muscle attachment site-specific target gene expression and interference with myotube guidance in response to ectopic stripe expression in the developing Drosophila epidermis. Proc. Natl. Acad. Sci. USA 94: 8608–8611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H. G., S. Takayama, U. R. Rapp and J. C. Reed, 1996. Bcl-2 interacting protein, BAG-1, binds to and activates the kinase Raf-1. Proc. Natl. Acad. Sci. USA 93: 7063–7068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarnitzky, T., L. Min and T. Volk, 1997. The Drosophila neuregulin homolog Vein mediates inductive interactions between myotubes and their epidermal attachment cells. Genes Dev. 11: 2691–2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaffran, S., M. Astier, D. Gratecos and M. Sémériva, 1997. The held out wings (how) Drosophila gene encodes a putative RNA-binding protein involved in the control of muscular and cardiac activity. Development 124: 2087–2098. [DOI] [PubMed] [Google Scholar]
- Zeiner, M., M. Gebauer and U. Gehring, 1997. Mammalian protein RAP46: an interaction partner and modulator of 70kDa heat shock proteins. EMBO J. 16: 5483–5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinke, I., C. Kirchner, L. C. Chao, M. T. Tetzlaff and M. J. Pankratz, 1999. Suppression of food intake and growth by amino acids in Drosophila: the role of pumpless, a fat body expressed gene with homology to vertebrate glycine cleavage system. Development 126: 5275–5284. [DOI] [PubMed] [Google Scholar]