Abstract
Non-LTR retrotransposons R1 and R2 have persisted in rRNA gene loci (rDNA) since the origin of arthropods despite their continued elimination by the recombinational mechanisms of concerted evolution. This study evaluated the short-term evolutionary dynamics of the rDNA locus by measuring the divergence among replicate Drosophila melanogaster lines after 400 generations. The total number of rDNA units on the X chromosome of each line varied from 140 to 310, while the fraction of units inserted with R1 and R2 retrotransposons ranged from 37 to 65%. This level of variation is comparable to that found in natural population surveys. Variation in locus size and retrotransposon load was correlated with large changes in the number of uninserted and R1-inserted units, yet the numbers of R2-inserted units were relatively unchanged. Intergenic spacer (IGS) region length variants were also used to evaluate changes in the rDNA loci. All IGS length variants present in the lines showed significant increases and decreases of copy number. These studies, combined with previous data following specific R1 and R2 insertions in these lines, help to define the type and distribution, both within the locus and within the individual units, of recombinational events that give rise to the concerted evolution of the rDNA locus.
TANDEMLY repeated multigene families frequently undergo concerted evolution, a phenomenon in which genes in a gene family show more sequence homogeneity within a species than between species. It has been suggested that homogenization occurs most rapidly within a chromosome (Schlötterer and Tautz 1994) by recombinational mechanisms such as gene conversion, intrachromosomal loop deletions, and unequal crossovers between sister chromatids (Dover 1994; Elder and Turner 1995; Liao 1999). New sequence variants within an array can also increase in frequency and spread through a population by segregation and recombination between homologs and can eventually become fixed in the species by natural selection, molecular drive, or drift.
The ribosomal RNA gene locus (rDNA) is of particular interest for the study of concerted evolution. In eukaryotes, the rDNA locus is composed of hundreds to thousands of tandemly repeated rRNA genes interspersed with noncoding, intergenic spacer (IGS) regions (Long and Dawid 1980). High redundancy of rRNA genes is critical for fitness because the ribosomal translational machinery of the cell is necessary in large quantities for growth and the RNA components of the ribosome structure do not benefit from translational amplification. Homogenization of the repeats within species is thought to be beneficial to the organism by ensuring that all ribosomal subunits are equally compatible with other components of the translational machinery. On the basis of population genetic studies, various recombinational models for explaining the homogeneity of the tandemly repeated rRNA genes have been proposed (Coen et al. 1982; Lyckegaard and Clark 1991; Schlötterer and Tautz 1994; Polanco et al. 1998, 2000).
An added complexity to understanding the evolution of the rDNA loci is that in many animal phyla these loci are home to specialized transposable elements (Eickbush 2002; Burke et al. 2003; Kojima and Fujiwara 2004; Penton and Crease 2004). Best studied are the R1 and R2 non-LTR retrotransposable elements of arthropods. These elements insert into the 28S gene and render the inserted genes nonfunctional (Long and Dawid 1979; Kidd and Glover 1981; Eickbush and Eickbush 2003). R1 and R2 have persisted via vertical descent in arthropods since the origin of the phylum, suggesting that occasional retrotransposition has been an effective strategy to evade elimination from the rDNA locus by the recombinational mechanisms of concerted evolution (Burke et al. 1998; Malik et al. 1999; Gentile et al. 2001).
To derive a comprehensive population genetics model for the evolution of the rDNA locus and its R1 and R2 inhabitants, one must measure changes in multiple properties of the locus over time. These properties include the number of rDNA units, the sequence of the units, IGS length variation, the fraction of the units inserted with R1 and R2, the frequency of R1 and R2 retrotransposition, and, finally, the distribution of this variation across the locus. To acquire these data, we are conducting a long-term study of the rDNA loci in the Harwich mutation-accumulation lines of Drosophila melanogaster. The Harwich lines are replicate stocks derived from a highly inbred line, separated over 400 generations ago (Mackay et al. 1992). Using the highly variable 5′ ends of R1 and R2 that are generated during insertion, the rates of R1 and R2 retrotransposition and elimination in these lines have been previously estimated (Pérez-González and Eickbush 2002; Pérez-González et al. 2003). This report continues our characterization of the evolutionary dynamics of the Harwich rDNA loci, this time with an emphasis on locus structure. We quantitated differences in the X-chromosome-linked rDNA loci, including the number of units in each locus, the load of transposable element insertions, and variation in the IGS. Our results show a remarkably dynamic locus with significant changes in its size and composition in only 400 generations. The combined analysis of R1, R2, and IGS markers in these lines provides insights into the properties of the recombinational mechanisms that drive the concerted evolution of the rDNA locus.
MATERIALS AND METHODS
Fly stocks and DNA isolation:
The Harwich stocks were a gift from T. F. C. Mackay. Line number designations were consistent with line numbers in Mackay et al. (1992), and flies were collected at various times from the 395th to the 415th generation. Genomic DNA was isolated from ∼50 females and 75 males per line as described in Eickbush and Eickbush (1995).
Southern genomic blots:
For the genomic blots, ∼3 μg DNA was restriction digested and separated on 1.0% agarose gels. After transfer of the genomic DNA to nitrocellulose paper, the paper was hybridized in 2× SSC, 5× Denhardt's at 65° for 14 hr as described in Eickbush and Eickbush (1995). Final washing of the filters was in 0.5× SSC at 65°. Gene sequences used for the hybridization probes were amplified via PCR from genomic DNA. PCR primers 5′-TTAGTGGGAGATATTAGACCTC-3′ and 5′-TGAACACCGAGATCAAGTC-3′, which amplified a region extending from position 6100 to 6521 (Tautz et al. 1988), were used to generate the 28S probe, and 5′-GCCGACCTCGCATTGTTC-3′ and 5′-TTTGTATTATACCGTAACG-3′, which amplified a region extending from position 10881 to 11184 (Tautz et al. 1988), were used to generate the external transcribed spacer (ETS) probe. Primers were synthesized by Invitrogen Custom Primers. Final PCR products to be used as probes were gel purified and random primer labeled with [α-32P]dCTP as described for the Rediprime II Random Prime labeling system (Amersham Biosciences). Hybridized and washed filters were exposed to a PhosphorImager screen for 36–72 hr, and the images were scanned with a Molecular Dynamics PhosphorImager scanner. The image was analyzed with ImageQuant software to produce signal traces and to quantitate band intensities.
Quantitation of full-length R2 elements:
The number of full-length R2 elements was determined by PCR amplification using the 5′ end-labeled primer 5′-GTTTAGCATTACCGGGACCAC-3′, which anneals to bases 145–165 of full-length R2 elements, and the primer 5′-TGCCCAGTGCTCTGAATGTC-3′, which anneals to the 28S gene sequence 60–80 bp upstream of the R2 insertion site. End labeling of the R2 primer, PCR amplification of both male and female genomic DNA, and separation of the PCR products on 8% high-voltage denaturing polyacrylamide gels are described in Pérez-González and Eickbush (2002). PCR amplifications of 5′ ends of full-length R2 elements produced multiple distinct product lengths (see Pérez-González and Eickbush 2002, Figure 3A). The relative intensity of each band was quantitated from the PhosphorImager scan. To adjust for the increased PCR amplification efficiency of shorter DNA fragments, the expected intensity per copy was calculated with a regression analysis using single copy variants as reference markers. PCR amplification of the full-length R2 elements in males produced 11–16 different PCR fragments, representing variants in both the X- and the Y-linked rDNA locus. Within each amplification reaction, bands were defined as representing single copy or multicopy variants. The copy numbers of more intense, multicopy bands were then determined. Female DNA in all stocks produced either four or five fragments (X-linked locus only). Comparison of the bands generated from male and female DNA from each line was used to confirm which of the X-linked bands represented single copies and to provide data for the linear regression.
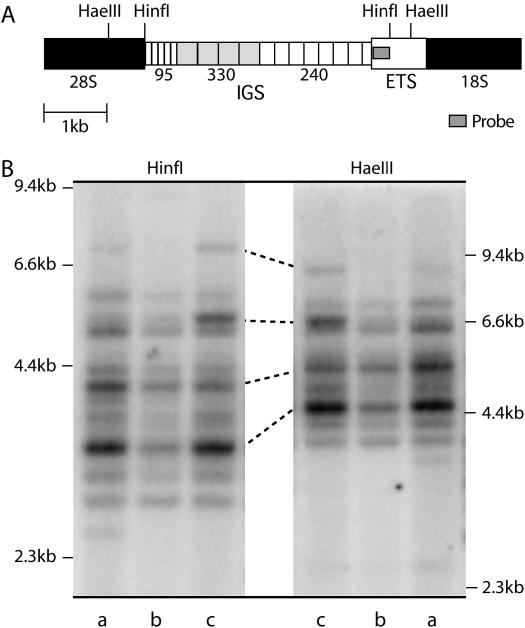
Figure 3.
IGS length profiles reproduced with two different restriction enzymes. (A) A diagram of the IGS region showing restriction sites and probe location. The IGS is mostly composed of tandem repeats of 95, 330, and 240 bp. (B) Genomic DNA was digested with HinfI or HaeIII, fractioned through a 1% agarose gel, and transferred to nitrocellulose. The resulting Southern blot was probed with a region of the ETS, marked with a shaded bar in A. The position of DNA standards in kilobases is indicated on the left and right. Lanes labeled a contain DNA from Harwich line 3, lanes labeled b contain DNA from line 20, and lanes labeled c contain DNA from line 2. Several corresponding bands in these last two lanes are connected with lines. The pattern of IGS bands is identical between the HinfI digests and HaeIII digests, with HinfI producing smaller fragments that are better separated on the gel.
RESULTS
Variation in the fraction of 28S genes containing R1 and R2:
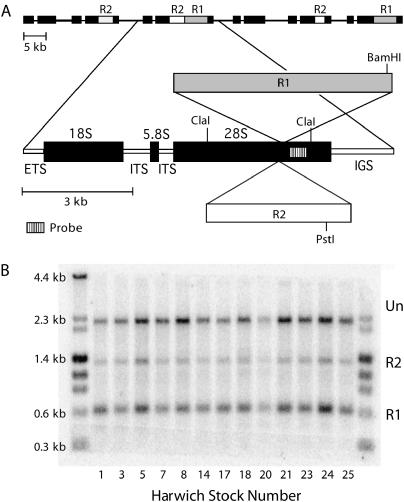
The fraction of X-chromosome-linked 28S rRNA genes inserted with R1 and R2 was determined for each Harwich line by quantitative Southern analysis of genomic DNA. This approach was possible because of the high level of sequence uniformity of the 28S rRNA gene, the R1 and R2 elements, and the 3′ junction of these elements with the 28S gene (Eickbush and Eickbush 1995; Lathe et al. 1995; Lathe and Eickbush 1997). To conduct the analysis, Southern blots of triple restriction digested (ClaI, BamHI, and PstI) female DNA from each stock was probed with a 28S gene fragment located downstream of the R1 and R2 insertion sites. A diagram of the 28S gene indicating the locations of restriction sites and the probe used in the analysis is shown in Figure 1A, and an example of a resulting genomic blot is shown in Figure 1B. The restriction digest produced three fragment types: uninserted 28S genes (2.3-kb ClaI-ClaI), 28S genes inserted with R2 (1.5-kb PstI-ClaI), and 28S genes inserted with R1 (0.7-kb BamHI-ClaI). The rare 28S genes with both R1 and R2 insertions (double insertions) were scored only as R1-inserted genes in this blot because the R1 insertion site is located on the 28S gene between the hybridization probe region and the R2 insertion site. The relative intensity of the hybridization to these three bands represented the relative proportions of the three types of rDNA units in the locus. This Southern analysis was repeated multiple times for each Harwich line, and the mean fractions of rDNA units that were uninserted, R1 inserted, and R2 inserted were calculated (Table 1).
Figure 1.
Retrotransposable element insertions in the rRNA gene (rDNA) loci of D. melanogaster. (A) The tandemly repeated rDNA units (top) are shown with no insertion, an R1 element, an R2 element, or both R1 and R2 elements in the 28S gene. An expanded diagram of an rDNA unit includes both full-length R1 and R2 insertions. Solid bars represent the 18S, 5.8S, and 28S rRNA genes, and thin open bars represent the ETS, internal transcribed spacers (ITS), and IGS. The locations of restriction enzyme cleavage sites and the probe used in B are indicated. (B) A sample Southern blot used to determine the fraction of inserted and uninserted rDNA units in the Harwich lines. Genomic DNA from females of each line was digested with BamHI, ClaI, and PstI, fractioned through a 1% agarose gel, and transferred to nitrocellulose. The resulting blot was probed with the 28S gene segment indicated in A. Uninserted rDNA units were represented by a 2.2-kb ClaI-ClaI fragment, R1-inserted and doubly inserted units by a 0.6-kb BamHI-ClaI fragment, and R2-inserted units by a 1.4-kb PstI-ClaI fragment. DNA markers were run in the first and last lanes with the fragment sizes indicated on the left.
TABLE 1.
Fraction of units in the rDNA locus inserted with transposable elements
| Harwich stock no. | Na | % uninserted | % R1 inserted | % R2 insertedb | Counted R2 elementsb |
|---|---|---|---|---|---|
| 1 | 5 | 34.8 ± 1.6 | 53.3 ± 1.8 | 12.0 ± 0.6 | 27 |
| 2 | 8 | 63.2 ± 2.1 | 24.6 ± 1.9 | 12.3 ± 1.1 | 26 |
| 3 | 5 | 61.9 ± 4.6 | 28.3 ± 3.9 | 9.9 ± 1.1 | 26 |
| 4 | 6 | 59.0 ± 1.7 | 28.7 ± 2.0 | 12.3 ± 0.6 | 25 |
| 5 | 5 | 50.7 ± 2.6 | 32.1 ± 2.9 | 17.2 ± 0.3 | 24 |
| 7 | 4 | 58.8 ± 3.2 | 29.3 ± 3.6 | 11.9 ± 0.7 | 26 |
| 8 | 5 | 62.2 ± 2.5 | 29.3 ± 2.3 | 8.4 ± 0.4 | 26 |
| 14 | 5 | 52.3 ± 2.2 | 37.9 ± 2.4 | 9.8 ± 0.4 | 25 |
| 15 | 6 | 60.6 ± 3.6 | 29.7 ± 3.7 | 9.7 ± 1.1 | 24 |
| 17 | 5 | 45.2 ± 2.7 | 41.3 ± 3.4 | 13.5 ± 1.0 | 25 |
| 18 | 5 | 50.4 ± 2.8 | 36.7 ± 3.0 | 13.0 ± 0.7 | 24 |
| 21 | 3 | 56.9 ± 6.1 | 34.0 ± 6.3 | 9.1 ± 0.8 | 22 |
| 22 | 5 | 38.5 ± 3.2 | 48.3 ± 3.4 | 13.0 ± 0.9 | 24 |
| 23 | 4 | 53.2 ± 4.1 | 32.2 ± 5.2 | 14.7 ± 1.1 | 25 |
| 24 | 6 | 54.5 ± 4.0 | 32.0 ± 4.1 | 13.5 ± 0.5 | 24 |
| 25 | 6 | 53.5 ± 2.5 | 32.8 ± 2.8 | 13.7 ± 0.8 | 22 |
| Mean | 53.5 | 34.4 | 12.1 | 24.7 | |
| SD | 8.3 | 7.6 | 2.3 | 1.4 |
Standard errors were calculated as standard deviation divided by the square root of N.
Number of gels quantitated for each Harwich stock.
Does not include the five R2 elements that are inserted in 28S genes also containing an R1 element.
The fraction of uninserted rDNA units among the Harwich lines ranged from 35 to 63%, the fraction of the units inserted with R1 elements ranged from 25 to 53%, and the fraction inserted with only an R2 element ranged from 8 to 17%. These twofold differences suggested that significant changes had occurred in the rDNA loci of the Harwich lines.
Variation in the size of the rDNA locus:
Unlike the highly uniform sequence found at the 3′ ends of the R1 and R2 elements, the 5′ ends are variable. This variation includes large deletions of the 5′ end of the element and small duplications and/or deletions of the 28S sequences upstream of the insertion site. Variation of R1 and R2 5′ junctions is similar to that found in other non-LTR retrotransposons and is the result of the retrotransposition mechanism used by these elements (Luan et al. 1993). PCR amplification has previously been used to score all 5′ variants of R1 and R2 in the Harwich lines (see Pérez-González and Eickbush 2002; Pérez-González et al. 2003 for a complete description of the R1 and R2 5′ variants found in these lines).
The size of the rDNA locus on the X chromosome in each of the Harwich lines was calculated by counting the total number of R2 elements and dividing that number by the fraction of the rDNA units represented by those elements. R2-inserted units provided better accuracy because the R2 elements were present in lower numbers, they showed less variation between lines, and their retrotransposition machinery is particularly prone to generating 5′-end variation. In contrast, R1 elements were two to four times more abundant and >70% of the copies were full-length elements with similar 5′ ends, making it more difficult to quantitate their absolute numbers. The number of 5′-truncated R2 elements identified on the X chromosome of each stock varied from 7 to 10 (Pérez-González et al. 2003, Figure 3B). To determine the number of full-length R2 elements on the X chromosome, PCR amplifications of the 5′ ends of full-length R2 elements were separated on high-voltage denaturing gels (see example in Pérez-González and Eickbush 2002, Figure 2A). One primer was end labeled with 32P, which allowed quantitation of bands from a phosphorimage scan of the gel. Separation on the gel resulted in a dominant band representing the canonical full-length R2 elements and a series of variant bands ranging from 20 bp longer to 43 bp shorter than the canonical band. Band intensities were quantitated, and the copy number for each band was determined using a regression analysis to compensate for differential PCR amplification of different length DNA fragments (see materials and methods for a description of this quantitation). The total number of full-length and 5′-truncated R2 elements scored in each of the Harwich stocks by these PCR approaches ranged from 27 to 32 elements.
Figure 2.
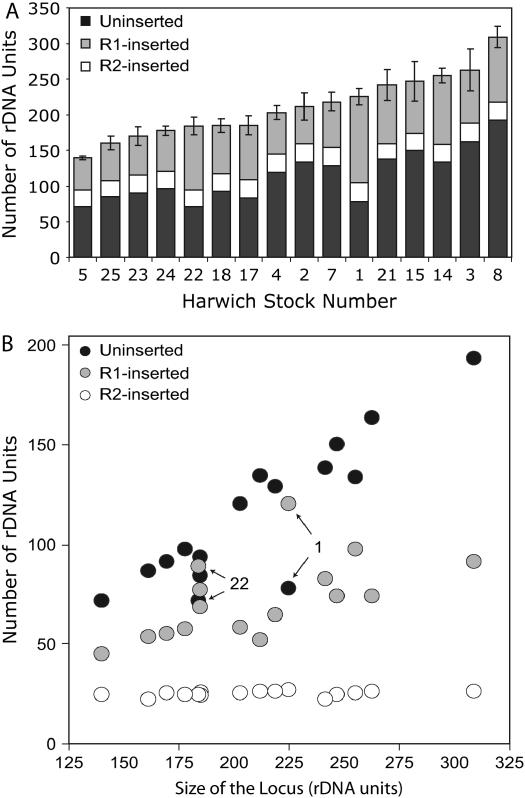
Variation in X chromosome rDNA locus size and load of R1 and R2 elements in 16 Harwich replicate lines. (A) The total number of rDNA units in each line is indicated by the total height of each bar. The numbers of uninserted, R1-inserted, and R2-inserted units are indicated by the shading. The total number of rDNA units in each line was determined by counting all R2 elements via a series of PCR reactions and dividing that number by the fraction of the total rDNA units containing R2 insertions (Table 1). Five R2 elements in each line were inserted upstream of an R1 and, thus, were not scored as R2 inserted by the Southern assay approach in Figure 1B. On the basis of the total number of units, the numbers of uninserted and R1-inserted units were also calculated. (B) The numbers of uninserted, R1-inserted, and R2-inserted units for each line are plotted relative to total locus size.
Occasionally, both R1 and R2 elements can insert in one rDNA unit. Such units were scored as R1-inserted units in the Southern blots and, subsequently, in the fractions presented in Table 1. To calculate locus size on the basis of the fraction of singly inserted R2 units, it was necessary to subtract those R2 elements that are part of double insertions from the total number of R2 elements determined above. The number of doubly inserted units was scored with a series of PCR amplifications in which one primer annealed to R2 sequences near the 3′ end of the element and a series of primers annealed to different regions within the R1 element (Pérez-González and Eickbush 2002). Five rDNA units on the X chromosome were found to be inserted with both R1 and R2 in each of the Harwich lines. Subtracting the five double-inserted units from the total number of R2 insertions, the number of R2-only inserted rDNA units was found to vary from 22 to 27 (Table 1). Dividing these numbers by the percentage of the locus containing R2 insertions, the total rDNA locus sizes of the Harwich lines were calculated to range from 140 to 310 units. These values are graphed in Figure 2A in order of increasing total locus size. The standard error for locus size was calculated from the error associated with the Southern blot determinations of the fraction of rDNA units with R2 insertions and ranged from 2 to 11% (mean 6%). On the basis of these determinations, the number of uninserted rDNA units in the Harwich lines was estimated to range from 70 to 195 and the number of R1-inserted units was estimated to range from 45 to 120.
In Figure 2B, the numbers of uninserted, R1-inserted, and R2-inserted units in each of the Harwich lines are plotted vs. the total size of the rDNA locus. The changes in total locus size were strongly correlated with changes in the number of uninserted and R1-inserted units, while the number of R2-inserted units remained relatively unchanged. A few lines, in particular lines 1 and 22, exhibited atypical increases in the number of R1-inserted units. The implications of these findings with respect to the likely changes that have occurred within the rDNA locus are discussed below.
Intergenic spacer length variants:
The IGS region of the D. melanogaster rDNA unit is highly variable in length with any chromosome containing an assortment of IGS length variants (Tautz et al. 1988; Williams et al. 1989; Polanco et al. 1998, 2000). This IGS length variation results from different multiples of tandemly repeated 95-, 330-, and 240-bp sequences (Figure 3A). As an independent means to monitor changes in the rDNA locus of the Harwich lines, the IGS length variants present in each line were cataloged and the copy numbers of each variant were determined.
IGS length profiles were generated by Southern analysis using genomic DNA restriction digested with HaeIII or HinfI, enzymes that cleave in the highly conserved regions flanking the IGS but not within the 95-, 240-, and 330-bp repeats (Figure 3A). Fragments generated with HaeIII included the IGS and 1.3 kb of flanking ETS and 28S gene, while HinfI included only 0.3 kb of flanking DNA. The Southern blots were probed with a DNA fragment from the ETS sequence. By using a probe outside of the repetitive IGS region, the hybridization signal would be independent of length and thus similar for all IGS fragment sizes. To show that the observed length variation was due to differences in the number of 95-, 330-, or 240-bp repeats and not restriction site polymorphisms within the IGS lengths, independent blots of three lines digested with HaeIII or HinfI are shown in Figure 3B. The relative intensity of all bands seen in the HinfI digest was reproduced with the HaeIII digest. HinfI was used in the final quantitation of IGS profiles from each Harwich line because the smaller fragment sizes gave better resolution of length variants.
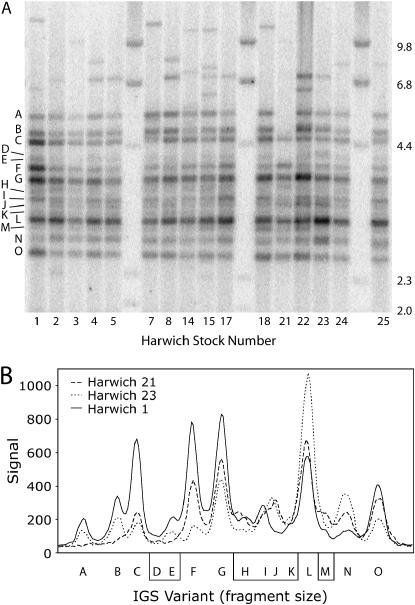
The HinfI-generated IGS profiles for all the Harwich stocks are shown in Figure 4A. In general, all stocks shared the same predominant set of IGS length variants in the size range of 2.5–5.3 kb. This profile is highly diagnostic of the Harwich lines when compared to other D. melanogaster stocks (Coen et al. 1982; Williams et al. 1989; Polanco et al. 1998). The Harwich IGS lengths were designated with letters A–O. Most of the hybridizing bands suggest a precise fragment length, indicating multiple copies of identical length variants. However, it was possible that some of these bands represented several variants of similar length. Indeed, the weaker bands (D, E, H–K) appeared less distinct in size, indicating that they may represent multiple variants. While IGS variants <2.5 kb were rare, longer variants up to 15 kb were found in many of the stocks. These longer IGS variants were not shared among the different Harwich lines.
Figure 4.
IGS profiles of all Harwich lines. (A) Genomic DNA was digested with HinfI, fractionated through 1% agarose gel, transferred to a nitrocellulose filter, and hybridized with the ETS probe (see Figure 3A). The blot reveals a similar set of length variants present in all Harwich lines but significant differences in the abundance of each variant. Predominant bands have been labeled with letters A–O. (B) Signal tracings of three Harwich lines from a phosphorimage of the probed filter show that the relative intensities of the variant bands are significantly different across the lines.
Changes in the relative abundance and copy number of IGS variants:
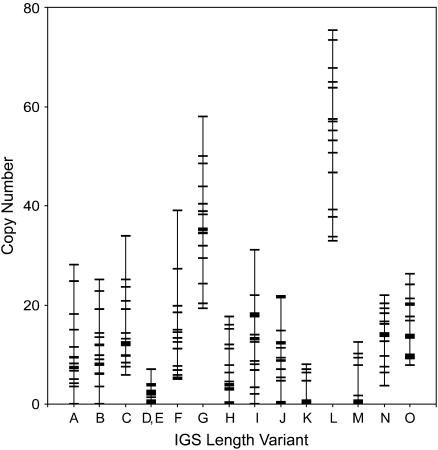
Although the Harwich stocks shared the same basic set of IGS length variants, the relative intensity of the bands corresponding to these lengths varied significantly among lines, indicating dramatic changes in copy number. This can be seen in the hybridization density tracings for three of the Harwich lines shown in Figure 4B. On the basis of these tracings, the IGS variants C, F, and G were most abundant in line 1, while variant L was clearly most abundant in line 23. In some instances, all copies of a particular variant were eliminated from a line (e.g., the loss of variants A and B in line 21).
The fraction of the total hybridization signal attributed to each variant was determined by quantitating band intensities from each IGS profile. The copy number for each IGS variant was then calculated by multiplying the fraction of hybridization signal per variant by the total number of units in the locus (Figure 2). The error associated with these estimates is larger because it includes both the error associated with the determination of locus size (mean 6%, Table 1) and the error of detecting the relative percentage of each IGS length variant. Standard errors for IGS blots were calculated for two Harwich lines on three blots and ranged from 0.1 to 5% (data not shown). In Figure 5, each IGS length variant is represented by a vertical bar with hatch marks indicating the copy number found in the Harwich lines. Variants G and L were generally the most abundant IGS type, ranging from an estimated 20 to 60 and 30 to 75 copies, respectively. All other variants also showed a wide distribution in copy number, many ranging from an estimated 0 to 20–30 copies.
Figure 5.
Range of copy number for each IGS length variant. Variants are labeled A–O and correspond to the bands shown in Figure 4. Copy number for each IGS variant was calculated for each Harwich line from the fraction of hybridization signal quantitated per band multiplied by the total number of units in the locus. For all IGS variants, a set of horizontal hatch marks indicates the copy numbers determined for the 16 Harwich lines. Lines that have the same copy number for a particular variant (e.g., 0 copies) are represented by a single hatch mark. Vertical lines represent the range of copy numbers observed.
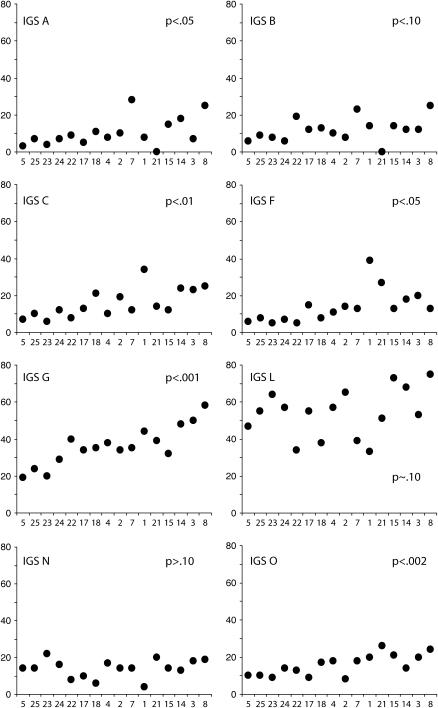
In Figure 6, the number of rDNA units containing the most abundant IGS variant types is shown arranged in order of increasing locus size. All IGS variant types exhibited a positive correlation between abundance and locus size, although this correlation was not always highly significant. In addition, many Harwich lines contained idiosyncratic expansions or reductions of particular variants that did not correlate with locus size. For example, IGS variant A was unusually abundant in Harwich 7, while all copies were lost in Harwich 21. IGS variants C and F were unusually abundant in Harwich 1. IGS variant L was reduced in copy number in Harwich 1, 7, 18, and 22.
Figure 6.
Copy number for eight major IGS variants arranged by rDNA locus size from smallest to largest. Across the lines, the copy number of variants is positively correlated with the size of the rDNA locus, but the strength of the correlation (P-values are shown) varies from very strong (G and O) to weak (L and N).
DISCUSSION
Population genetic approaches have frequently been used to infer from extant genetic variation the recombinational processes that drive concerted evolution of the D. melanogaster rDNA locus (Hillis et al. 1991; Lyckegaard and Clark 1991; Jakubczak et al. 1992; Schlötterer and Tautz 1994; Polanco et al. 1998, 2000; Pérez-González and Eickbush 2001). These studies have provided insights into the possible mechanisms responsible for concerted evolution but do not allow for the estimation of the rates of such events. In an attempt to directly measure rates of change, Coen et al. (1982) followed changes in the structure of the rDNA locus of D. melanogaster isofemale lines over time. They estimated that the rDNA loci could remain stable in the laboratory for >1000 generations. Previous studies in our laboratory using the Harwich mutation-accumulation lines have estimated the rate of insertion and deletion of marked R1 and R2 elements within the rDNA loci (Pérez-González and Eickbush 2002; Pérez-González et al. 2003). Although 300 new insertions and deletions were scored after 400 generations, most events (74%) occurred on the Y chromosomes. The limited number of events detected on the X chromosomes (0–9/line) suggested that significant structural changes had not occurred in the X-linked rDNA loci.
As described in this report, the X chromosome rDNA locus in these Harwich lines had in fact changed dramatically in the 400 generations since the lines were separated. The size of the locus varied from 140 rDNA units to 310 units, a range similar to the 110–280 units found in the X chromosomes from a natural population of D. melanogaster (Lyckegaard and Clark 1991). The range of uninserted units among the Harwich lines varied from 35 to 63% and was consistent with levels seen in a survey of 27 geographical stocks of D. melanogaster (23–68%) (Jakubczak et al. 1992). The average proportion of uninserted rDNA units among the Harwich lines was 54%, also similar to the 51% mean seen in the survey of geographical stocks (Jakubczak et al. 1992). It is remarkable that fluctuations in locus size and level of element insertion as a consequence of recombination events, retrotransposition, selection, and drift could so rapidly reproduce from a single inbred laboratory stock the wide variation seen in natural populations.
Recombination within the rDNA locus:
The dramatic differences in locus size among the Harwich lines suggest frequent expansion and contraction in the number of rDNA units as a result of unequal crossovers. As shown in Figure 2B, with only a few exceptions (e.g., lines 1 and 21, which will be discussed below) the copy number of R1-inserted and uninserted units changed uniformly relative to the overall size of the locus (Figure 2B). The slope of each correlation indicated the proportion of the changes in locus size that could be attributed to that unit type. Uninserted units, which averaged 54% of the units, accounted for 73% of the change in locus size (i.e., slope = 0.73); R1-inserted units, which averaged 34% of the locus, accounted for 26% of the change; and R2-inserted units, which averaged 12% of the locus, accounted for <1% of the change. Therefore, uninserted units were overrepresented in the recombinational activity of the locus, R1-inserted units were underrepresented, and R2-inserted units were essentially excluded from the recombinational expansions and contractions of the locus. This recombinational bias suggests that population genetic models to explain the concerted evolution of the rDNA locus should not assume that recombination events are evenly nor randomly distributed across the locus if insertions are present in that locus.
One model for explaining the relative participation of inserted and uninserted units in the recombinational events within the rDNA locus is that R2 insertions and many R1 insertions are clustered in the rDNA locus in regions of low recombination. For example, McAllister and Werren (1999) have shown that the edges of tandem arrays undergo less frequent recombination. A second model is that unequal crossovers are short (i.e., involve offsets of only a small number of units) and require multiple contiguous units to be similar in type. In this model, R1- and R2-inserted elements could be dispersed throughout the locus, but the low frequency of R2-inserted units and of 5′-truncated R1 elements in the locus would reduce the probability of successful offset alignments involving these units. In contrast, the high frequency of uninserted units or units with full-length R1 elements would provide many opportunities for offset alignment and subsequent recombination. Ultimate resolution of the patterns of unequal crossover across the rDNA locus in the Harwich lines will require knowledge of the distribution of the insertions across the locus.
The similar number of R2-inserted units in all lines was consistent with the low level of insertion and deletion of marked R2 elements previously detected in these lines (Pérez-González and Eickbush 2002). Surprisingly, the number of R1-inserted units characterized here varied from 45 to 120 units, a range far larger than expected given our previous detection of only limited numbers of marked R1 insertions and deletions on each X chromosome (Pérez-González et al. 2003). This difference is explained by the PCR approach used in our previous study, which scored the appearance of only new (unique) 5′ variants, not insertions or duplications of identical-in-length elements. Similarly, the previous PCR survey detected deletions only if all copies of that length variant were deleted. The large changes in R1 copy number that we observed are mostly associated with full-length copies, many of which are associated with identical-in-length 5′ ends (Pérez-González and Eickbush 2002).
Our analysis of IGS length variation among the replicate Harwich lines also provides insights into the distribution of the recombination events within the rDNA locus. All Harwich lines contained similar IGS length variants in the size range from 2.5 to 5.3 kb (Figure 4). Such long-term stability in IGS profiles in laboratory-maintained lines has also been observed by Coen et al. (1982). However, as shown in Figures 5 and 6, this apparent stability belies the many recombination events that have occurred in the locus. Each of the IGS length variants exhibited significant differences in copy number among the Harwich lines. Because all IGS length variants experienced changes in number of units, our results suggest that most of the IGS variant types are interspersed and that recombination events associated with the changes in locus size have occurred at many locations throughout the locus. Williams et al. (1989) have previously suggested that many IGS variants are widely distributed across the loci, on the basis of an analysis of the rare recombinations that can be scored between the X and Y chromosome loci. Presumably, those IGS variants we observed whose abundance best correlate with locus size (e.g., variants G, Figure 6) are widely distributed across the X chromosome locus, while variant types whose abundance only weakly correlates with locus size (e.g., variants L) are more localized in the locus, perhaps in regions with lower rates of recombination.
The IGS analysis of the Harwich lines can also be used to address the question of whether recombination events between rDNA units occur within the genic (transcribed) regions or intergenic (IGS) regions. Unequal crossovers within the genes would change the IGS copy number but not their lengths, while crossovers within the IGS could produce new length variants. Polanco et al. (2000) have shown that individual IGS variants in D. melanogaster contain variable numbers of the 95-, 330-, and 240-bp repeats. Repeated recombination between these repeats would give rise to a continuous spread of IGS length variants. However, the IGS profiles observed for the Harwich lines have a distinctive pattern that changed little among the lines. The dramatic changes in IGS variant copy number without dispersing the common profile of IGS variants support recombination models in which crossovers seldom occur in the intergenic region of the rDNA units.
Many Harwich lines contained longer, unique IGS variants (i.e., not shared between lines) from 6 to 15 kb in length. It is unlikely that these long variants were imported from the Y chromosome because the rate of recombination between the rDNA arrays on the X and Y chromosomes is extremely low (Williams et al. 1989) and our previous studies did not detect the movement of marked R1 or R2 elements from the Y to the X chromosome in the Harwich lines (Pérez-González et al. 2003). Thus, the long IGS variants detected on the X chromosome appeared to have been produced by recombination. Unlike the common IGS variants, these longer variants appeared to be sites of recombination, as most Harwich lines contained their own distinct pattern of long variants. Some of these long variants also became as abundant as the more common shorter types. Determining the distribution of these long variants in the locus would provide valuable insights into the recombination mechanisms that have occurred.
Possible effects of R1 retrotransposition on the rDNA locus:
While most of the changes observed on the X chromosome rDNA locus appear to have been the result of unequal crossovers, several lines have undergone dramatic shifts in the relative abundance of R1-inserted units compared to uninserted units. In the two most extreme examples, R1-inserted units represented 53% of the rDNA units in line 1 and 48% in line 22 compared with ∼30% in most other lines. It is possible that these increases in the fraction of R1-inserted units were a result of unequal crossover events, which preferentially duplicated R1-inserted units or eliminated uninserted units from the locus. However, no Harwich lines showed the opposite scenario—preferential duplication of uninserted units or deletion of R1-inserted units—even though such events would likely confer a selective advantage in these laboratory populations. Consistent with a model that active R1 retrotransposition increased the number of R1 elements at the expense of uninserted units, Harwich lines 1 and 22 were shown to contain X chromosomes with the greatest number of new 5′ R1 junctions (six in line 1, four in line 22) (Pérez-González et al. 2003). Only a small fraction of R1 retrotransposition events produce new 5′ junctions that were scored by our PCR assay (Pérez-González and Eickbush 2002), suggesting that many more retrotransposition events occurred in these lines.
In conclusion, this study revealed that one can follow changes in the size and composition of the rDNA locus in inbred lines maintained in the laboratory. While the focus of this study was the X-linked rDNA loci of the Harwich lines, we expect that even more extensive changes have occurred in the Y-linked rDNA loci, on the basis of the threefold higher level of new insertions detected on the Y chromosomes (Pérez-González et al. 2003) and greater variation in Y-linked IGS variants between lines (K. T. Averbeck, unpublished data). The differences in rDNA loci that we observed strongly support population genetic models of frequent unequal crossovers widely spread across the locus but suggest that these recombinations are not distributed evenly or randomly. New questions concerning the evolution of the rDNA locus have arisen out of this study, such as why R2 elements are preferentially excluded from recombination events, whether long IGS variants stimulate recombination, and whether R1 activity has contributed to changes in the locus in some lines. These questions can be adequately addressed only with a physical map of the distribution of R1 and R2 insertions and IGS variants across the rDNA locus of the Harwich lines. A BAC cloning project is currently under way, and significant portions of the X chromosome locus of Harwich line 21 have been recovered (W. D. Burke, K. T. Averbeck and T. H. Eickbush, unpublished results). With a map of the distribution of variation along large segments of the rDNA locus from this line, we will be able to use the variation scored in all 16 Harwich lines to provide new insights into the relentless forces that rapidly shape the landscape of the rDNA locus.
Acknowledgments
We thank D. G. Eickbush and W. D. Burke for helpful discussions and comments on the manuscript. We thank T. F. C. Mackay for originally supplying the Harwich lines. This research was supported by National Science Foundation grant MCB-9974606 to T.H.E.
References
- Burke, W. D., H. S. Malik, W. C. Lathe III and T. H. Eickbush, 1998. Are retrotransposons long-term hitchhikers? Nature 392: 141–142. [DOI] [PubMed] [Google Scholar]
- Burke, W. D., D. Singh and T. H. Eickbush, 2003. R5 retrotransposons insert into a family of infrequently transcribed 28S rRNA genes of Planaria. Mol. Biol. Evol. 20: 1260–1270. [DOI] [PubMed] [Google Scholar]
- Coen, E. S., J. M. Thoday and G. Dover, 1982. Rate of turnover of structural variants in the rDNA gene family of Drosophila melanogaster. Nature 295: 564–568. [DOI] [PubMed] [Google Scholar]
- Dover, G., 1994. Concerted evolution, molecular drive and natural selection. Curr. Biol. 4: 1165–1166. [DOI] [PubMed] [Google Scholar]
- Eickbush, T. H., 2002. R2 and related site-specific non-long terminal repeat retrotransposons, pp. 813–835 in Mobile DNA II, edited by N. L. Craig, R. Craigie, M. Gellart and A. M. Lambowitz. American Society for Microbiology, Washington, DC.
- Eickbush, D. G., and T. H. Eickbush, 1995. Vertical transmission of the retrotransposable elements R1 and R2 during the evolution of the Drosophila melanogaster species subgroup. Genetics 139: 671–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickbush, D. G., and T. H. Eickbush, 2003. Transcription of endogenous and exogenous R2 elements in the rRNA gene locus of Drosophila melanogaster. Mol. Cell Biol. 23: 3825–3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elder J. F., Jr., and B. J. Turner, 1995. Concerted evolution of repetitive DNA sequences in eukaryotes. Quart. Rev. Biol. 70: 297–320. [DOI] [PubMed] [Google Scholar]
- Gentile, K. L., W. D. Burke and T. H. Eickbush, 2001. Multiple lineages of R1 retrotransposable elements can coexist in the rDNA loci of Drosophila. Mol. Biol. Evol. 18: 235–245. [DOI] [PubMed] [Google Scholar]
- Hillis, D. M., C. Moritz, C. A. Porter and R. J. Baker, 1991. Evidence for biased gene conversion in concerted evolution of ribosomal DNA. Science 251: 308–310. [DOI] [PubMed] [Google Scholar]
- Jakubczak, J., M. Zenni, R. Woodruff and T. H. Eickbush, 1992. Turnover of R1 (Type I) and R2 (Type II) retrotransposable elements in the ribosomal DNA of Drosophila melanogaster. Genetics 131: 129–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd, S. J., and D. M. Glover, 1981. Drosophila melanogaster ribosomal DNA containing type II insertions is variably transcribed in different strains and tissues. J. Mol. Biol. 151: 645–662. [DOI] [PubMed] [Google Scholar]
- Kojima, K. K., and H. Fujiwara, 2004. Cross-genome screening of novel sequence-specific non-LTR retrotransposons: various multicopy RNA genes and microsatellites are selected as targets. Mol. Biol. Evol. 21: 207–217. [DOI] [PubMed] [Google Scholar]
- Lathe, W. C., III, and T. H. Eickbush, 1997. A single lineage of R2 retrotransposable element is an active, evolutionarily stable component of the Drosophila rDNA locus. Mol. Biol. Evol. 14: 1232–1241. [DOI] [PubMed] [Google Scholar]
- Lathe, W. C., III, W. D. Burke, D. G. Eickbush and T. H. Eickbush, 1995. Evolutionary stability of the R1 retrotransposable element in the genus Drosophila. Mol. Biol. Evol. 12: 1094–1105. [DOI] [PubMed] [Google Scholar]
- Liao, D. Q., 1999. Concerted evolution: molecular mechanism and biological implications. Am. J. Hum. Genet. 64: 24–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, E. O., and I. B. Dawid, 1979. Expression of ribosomal DNA insertions in Drosophila melanogaster. Cell 18: 1185–1196. [DOI] [PubMed] [Google Scholar]
- Long, E. O., and I. B. Dawid, 1980. Repeated genes in eukaryotes. Annu. Rev. Biochem. 49: 727–764. [DOI] [PubMed] [Google Scholar]
- Luan, D. D., M. H. Korman, J. L. Jakubczak and T. H. Eickbush, 1993. Reverse transcription of R2Bm is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell 72: 595–605. [DOI] [PubMed] [Google Scholar]
- Lyckegaard, E. M. S., and A. G. Clark, 1991. Evolution of ribosomal RNA gene copy number on the sex chromosomes of Drosophila melanogaster. Mol. Biol. Evol. 8: 458–474. [DOI] [PubMed] [Google Scholar]
- Mackay, T. F. C., R. F. Lyman, M. S. Jackson, C. Terzian and W. G. Hill, 1992. Polygenic mutation in Drosophila melanogaster: estimates from divergence among inbred strains. Evolution 46: 300–316. [DOI] [PubMed] [Google Scholar]
- Malik, H. S., W. D. Burke and T. H. Eickbush, 1999. The age and evolution of non-LTR retrotransposable elements. Mol. Biol. Evol. 16: 793–805. [DOI] [PubMed] [Google Scholar]
- McAllister, B. F, and J. H. Werren, 1999. Evolution of tandemly repeated sequences: What happens at the end of an array? J. Mol. Biol. 48: 469–481. [DOI] [PubMed] [Google Scholar]
- Penton, E. H., and T. J. Crease, 2004. Evolution of the transposable element Pokey in the ribosomal DNA of species in the subgenus Daphnia (Crustacea: Cladocera). Mol. Biol. Evol. 21: 1727–1739. [DOI] [PubMed] [Google Scholar]
- Pérez-González, C. E., and T. H. Eickbush, 2001. Dynamics of R1 and R2 elements in the rDNA locus of Drosophila simulans. Genetics 158: 1557–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-González, C. E., and T. H. Eickbush, 2002. Rates of R1 and R2 retrotransposition and elimination from the rDNA locus of Drosophila melanogaster. Genetics 162: 799–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-González, C. E., W. D. Burke and T. H. Eickbush, 2003. R1 and R2 retrotransposition and deletion in the rDNA loci on the X and Y chromosomes of Drosophila melanogaster. Genetics 165: 675–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polanco, C., A. I. González, Á. de la Fuente and G. A. Dover, 1998. Multigene family of ribosomal DNA in Drosophila melanogaster reveals contrasting patterns of homogenization for IGS and ITS spacer regions: a possible mechanism to resolve this paradox. Genetics 149: 243–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polanco, C., A. I. González and G. A. Dover, 2000. Patterns of variation in the intergenic spacers of ribosomal DNA in Drosophila melanogaster support a model for genetic exchanges during X-Y pairing. Genetics 155: 1221–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlötterer, C., and D. Tautz, 1994. Chromosomal homogeneity of Drosophila ribosomal DNA arrays suggests intrachromosomal exchanges drive concerted evolution. Curr. Biol. 4: 777–783. [DOI] [PubMed] [Google Scholar]
- Tautz, D., J. M. Hancock, D. A. Webb, C. Tautz and G. A. Dover, 1988. Complete sequences of the rRNA genes of Drosophila melanogaster. Mol. Biol. Evol. 5: 366–376. [DOI] [PubMed] [Google Scholar]
- Williams, S. M., J. A. Kennison, L. G. Robbins and C. Strobeck, 1989. Reciprocal recombination and the evolution of the ribosomal gene family of Drosophila melanogaster. Genetics 122: 617–624. [DOI] [PMC free article] [PubMed] [Google Scholar]