Abstract
Somatolactin (SL) in fish belongs to the growth hormone/prolactin family. Its ortholog in tetrapods has not been identified and its function(s) remains largely unknown. The SL-deficient mutant of medaka (color interfere, ci) and an SL receptor (SLR) recently identified in salmon provide a fascinating field for investigating SL's function(s) in vivo. Here we isolated a medaka ortholog of the salmon SLR. The mRNA is transcribed in variable organs. Triglycerides and cholesterol contents in the ci are significantly higher than those in the wild type, providing the first evidence of SL's function in suppressing lipid accumulation to organs. Interestingly, phylogenetic comparisons between the medaka SLR and growth hormone receptor (GHR), which is also isolated in this study, in relation to GHRs of other fish, suggested that all GHRs reported from nonsalmonid species are, at least phylogenetically, SLRs. An extra intron inserted in medaka and pufferfish SLRs and flounder and sea bream GHRs also supports their orthologous relationship, but not with tetrapod GHRs. These results may indicate lineage-specific diversification of SLR and GHR functions among fish or just an inappropriate naming of these receptors. Further functional and comparative reassessments are necessary to address this question.
SOMATOLACTIN (SL) belongs to the growth hormone (GH)/prolactin (PRL) family, but its biological activities are mostly unknown and remain debatable. Researchers have isolated SLs from many fish species and detected up- and downregulation of its expression during variable physiological processes (see references in Fukada et al. 2005). Experiments using recombinant proteins of SLs show certain biological activities in vitro and in vivo (Lu et al. 1995; Calduch-Giner et al. 1998; Vega-Rubin de Celis et al. 2003). However, none has been supported by clear and direct evidence explaining the mechanisms or actions of target cells, tissues, and organs of SLs.
Medaka (Oryzias latipes), a small freshwater fish, is a powerful model organism in genetic and developmental studies. We have recently identified an SL mutant of medaka, color interfere (ci), providing genetic evidence for SL's function (Fukamachi et al. 2004). This mutant exhibits a remarkably pale body color due to constitutively increased white pigment cells (leucophores) and reduced orange pigment cells (xanthophores) in the skin (Figure 1). Transcription of the medaka SL in the brain (pituitary) is dramatically enhanced or suppressed when the fish is kept in a black or white tank, respectively (i.e., due to morphological background adaptation of the body color), which is similar to that experienced in red drum and Atlantic croaker (Zhu and Thomas 1995). These results strongly suggest a conserved and major role of SLs in chromatophore regulation.
Figure 1.
Lateral views of the wild-type and ci medaka. Body color of the ci (top) is less yellow than that of the wild type (bottom) due to reduced xanthophores (orange pigment cells) on the skin. As shown, the ci fish are in good health accompanied by no apparent morphological or behavioral anomaly under normal breeding conditions.
More recently, however, an SL receptor (SLR) was identified in masu salmon, Oncorhynchus masou (Fukada et al. 2005). Its amino acid sequence is similar to the GH receptors (GHRs) of teleosts (38–58% identity), but it binds to SL rather than to GH. The mRNA was shown to be strongly expressed in the liver and fat, and these authors suggested SL's main function in lipid metabolism. SL's role in fat metabolism had actually been suggested earlier from studies of another salmonid species, the rainbow trout (O. mykiss), in which phenotypes of its cobalt variants were observed. The cobalt fish have a severe defect in SL expression and are obese, with lipid accumulation evident in the body cavity (Kaneko et al. 1993; Yada et al. 2002). However, the pituitary of the cobalt is separated from the hypothalamus and lacks most of the pars intermedial (in which somatolactin-producing cells are distributed). Therefore, it is unclear whether the SL itself or other hormones released from the pars intermedia of the pituitary (or both) is the cause of the lipid accumulation.
These results from two salmonid species are somewhat intriguing to us because the ci medaka does not exhibit any apparent obesity (see Figure 1) nor does it require special care for breeding, unlike the cobalt. These inconsistent observations led us to consider that the ci medaka may have additional physiological anomalies, although they are not apparent in situ. In this article, we describe cloning of a medaka ortholog of the salmon SLR, its broad expression in organs, and extraordinary phenotypes of the ci fish other than in pigmentation. During our series of experiments, we recognized that most of the genes reported as GH receptor (GHR) from other fish species are, at least phylogenetically, SLRs. We look at this more closely in the discussion.
MATERIALS AND METHODS
Medaka:
We used the HNI strain (northern inbred) as the wild type for SLR cloning and phenotypic comparison. Genomic sequences on the genome database are of Hd-rR (southern inbred) and we used this strain for gap filling of the SLR locus (see results). In terms of the GHR locus, however, we used the HNI strain because genomic PCR could not efficiently amplify the gap of the Hd-rR allele, probably because of the poly(C) repeat within the gap (it should correspond to AACCCCCCACCCCCCCAG of the HNI allele). The OLc08.02d clone is of Hd-rR. The ci are offspring of fish formerly transferred to our laboratory from Nagoya University (see Kelsh et al. 2004), which have northern alleles at least around the ci locus (S. Fukamachi, unpublished data). All fish were bred at 27° and under 14 hr light/10 hr dark conditions.
RT-PCR and 5′- and 3′-RACE:
The total RNA of embryo, larva, or adult organs was extracted using ISOGEN (Nippon Gene). After its quantification by electrophoresis and spectrometer, the first-strand cDNAs were synthesized using ReverTra Ace (Toyobo) with 5′- and 3′-adaptors and used as PCR templates. All PCRs were carried out using LA Taq polymerase (Takara, Berkeley, CA) following parameters of 94° for 1 min; 30 cycles of 98° for 20 sec, 60° for 1 min, 72° for 2 min and a final extension at 72° for 10 min. Products were separated on 1% agarose gel and observed on an ultraviolet transilluminator after ethidium bromide staining. We performed nested PCRs in RACEs. Strongest bands were cut, and DNAs were purified using the QIAquick gel extraction kit (QIAGEN, Chatsworth, CA) and directly sequenced.
Sequencing:
We used PCR product presequencing kit (USB) for template preparation from PCR products. BigDye Terminator (version 3, Applied Biosystems, Foster City, CA) was used for sequencing reaction and products were electrophoresed on an ABI PRISM 3100 Genetic Analyzer. We assembled the sequences by using SeqManII software (DNASTAR, Madison, WI).
Phylogenetic analyses:
cDNA sequences of SLR, GHR, and PRL receptor (PRLR) were obtained from the GenBank of NCBI (http://www.ncbi.nlm.nih.gov/). Exon-intron boundaries of human, mouse, and chicken were identified by comparing the cDNA and genomic sequences by BLAST (http://www.ncbi.nlm.nih.gov/BLAST/) (see results for those of medaka and fugu). Corresponding exons of other species were predicted from amino acid sequences aligned by using MegAlign software (DNASTAR).
Lipids measurement:
After anesthetizing with 0.02% 2-phenoxyethanol, body weight was measured and ∼2 μl of blood was collected directly from the caudal vessels using a fine-tipped capillary tube as described previously (Yada and Ito 1998). Blood plasma was obtained by centrifugation at 3000 × g for 5 min and stored at −80° until analyses. Muscle was taken from the dorsal side after removal of the skin. The muscle and liver were weighed and stored at −80°. The hepatosomatic index was calculated as liver weight × 100/body weight. Gill filaments were cut from ceratobranchials and placed in ice-cold sucrose-EDTA-imidazole solution (150 mm sucrose, 10 mm disodium ethylenediamine tetraacetate, 50 mm imidazole, pH 7.3) and stored at −80°.
Plasma cortisol concentrations were determined using a commercially available kit (enzyme immunoassay for cortisol, Oxford Biomedical Research, Oxford, MI). Gill Na+, K+-ATPase activities were measured as described previously (Yada and Ito 1998). Optical density was measured with a microplate reader (SpectraMax 190, Nihon Molecular Devices).
Lipids were extracted from tissue in 20 volumes of chloroform/methanol (2:1, v/v), using the method of Folch et al. (1957). Lipid classes were separated by thin-layer chromatography on chromatorod SIII (Iatron Laboratories) using the two-solvent system with chloroform/methanol/water (40:20:2.5, v/v/v) and hexane/ether/formic acid (54:6:0.08, v/v/v). After separation, lipid contents were quantified by Iatroscan MK 5 flame-ionization detector (Iatron Laboratories). Quantitative standards of tripalmitin and cholesterol were obtained from Tokyo Kasei Kogyo.
RESULTS
Cloning of a medaka SLR:
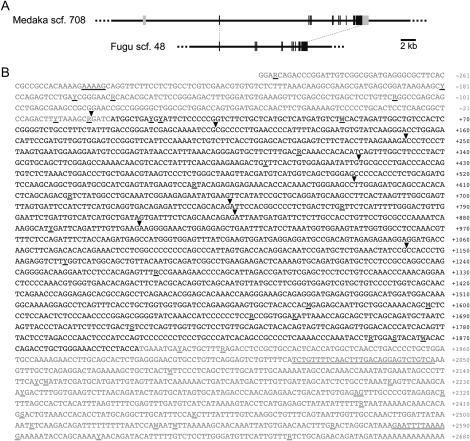
Whole-genome sequence is a very powerful tool for rapid gene cloning. Although still not complete in medaka, an ongoing sequencing project provides assembled sequences on the Web [http://medaka.utgenome.org/ (revision 200406)]. However, it allows only a blastn (nucleotide-nucleotide) search that is not suitable for finding an orthologous relationship between genes of divergent species such as medaka and salmonids. Therefore, we first performed a tblastn (protein-translated nucleotide) search on the JGI Takifugu rubripes (fugu) genome database (http://genome.jgi-psf.org/fugu6/fugu6.home.html, version 3.0) using an amino acid sequence of the SLR of masu salmon (GenBank accession no. BAD51998) and found an orthologous region on scaffold 48. Fugu is evolutionally much closer to medaka than to salmon (see Miya et al. 2003) and nucleotide sequences of its translated regions enabled us to identify corresponding regions on scaffold 708 of the medaka genome by the blastn search (∼67% identity). Although both the fugu and medaka databases provide statistically predicted exon-intron boundaries (and deduced amino acid sequences), some seem to be incorrect because their positions are not conserved between these species. Therefore, we manually repredicted their exons by a comparative-genomic approach using mVISTA as an alignment tool (http://genome.lbl.gov/vista/index.shtml) and, as a result, we could determine putative exon-intron boundaries that are perfectly conserved between medaka and fugu (Figure 2A).
Figure 2.
Cloning of the medaka SLR. (A) Comparative-genomic prediction of medaka and fugu SLR exons. Solid and shaded boxes indicate translated and untranslated regions, respectively. Corresponding exons of medaka and fugu retain identical exon-intron boundaries. Note that their SLR proteins are encoded by nine exons. (B) Nucleotide sequence of the medaka SLR cDNA (GenBank accession no. DQ002886). Arrowheads indicate position of introns inserted. Shaded residues indicate 5′- and 3′-UTRs. Underlining indicates polymorphic residues detected between the HNI (experimentally determined) and Hd-rR (on the genome sequence) alleles.
According to these newly predicted exons of medaka, we constructed pairs of primers that amplify overlapping regions of the translated region, performed RT-PCR using liver cDNA as a template, and directly sequenced the amplified products. We also determined medaka's 5′- and 3′-UTRs by RACEs (Figure 2B). By comparing this cDNA sequence with the corresponding genomic sequence on scaffold 708 (which had a gap of 484 bp that we filled experimentally), we confirmed that the medaka SLR locus consists of 10 exons, 9 of which encode the protein.
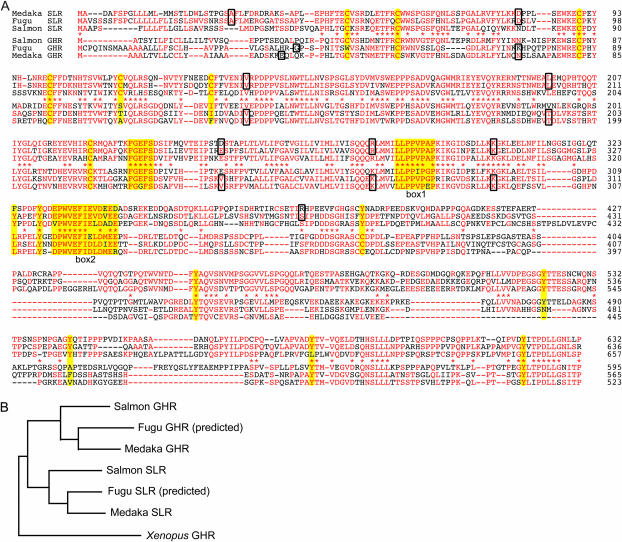
Deduced protein of the medaka SLR consists of 632 amino acids and shares 324 identical residues (∼48%) with the salmon SLR (Figure 3A). Fugu SLR protein, which should be more accurately predicted than the sequence on the database, shares ∼62% of identical amino acids with the medaka SLR (Figure 3A). Phylogenetic analysis, however, did not necessarily show a closer relationship of the medaka and fugu SLRs to the salmon SLR than to the GHRs of other fish (data not shown; see Figure 7). Because GHR of medaka or fugu has not been cloned and the alternative (i.e., the genes that we cloned/predicted are not SLRs but GHRs) seemed possible, we further attempted to isolate a medaka ortholog for salmon GHR as well.
Figure 3.
Amino acid sequences of the medaka SLR and GHR. (A) Amino acid alignment of the SLRs and GHRs of medaka, fugu, and masu salmon. Those of fugu are putative (see text). Major residues among each group are in red. Asterisks indicate that the major residues for SLRs and GHRs are identical. Boxes indicate residues that contain exon-intron boundaries in their codons or the first residues in each exon. The conserved cysteines (six disulfide linked and one unpaired), FGXFS motif, box 1, box 2, and tyrosines (see text) are highlighted in yellow. (B) A phylogenic tree drawn by the Neighbor program in the PHYLIP package (http://evolution.genetics.washington.edu/phylip.html) using entire sequences of the SLRs and GHRs. All nodes are supported by bootstrap values of 100, demonstrating an orthologous relationship among the SLRs and GHRs of these fish (see also Figure 7).
Figure 7.
Phylogenetic relationships among SLRs, GHRs, and PRLRs of vertebrates. (A) A phylogenic tree drawn by clustalW available at EMBL-EBI (http://www.ebi.ac.uk/clustalw/). Numbers indicate bootstrap values calculated using MEGA3 software (Kumar et al. 2004). To avoid disturbances from possible splicing variations reported in tetrapods (e.g., an extra exon in mouse, loss of an exon in opossums and avians), proteins encoded by the last seven exons of the medaka SLR (see Figure 2B) and corresponding regions of other SLRs, GHRs, and PRLRs were used for analysis (a similar tree was actually drawn by using the full length of these proteins). In this analysis, we added SLR and GHR of another pufferfish (Tetraodon nigroviridis) predicted from its genome database (http://www.genoscope.cns.fr/externe/tetraodon/), although several putative sequencing errors on the database were manually corrected (e.g., from TCCCCCT to TCCCCT and from GCCCA to GCCA). A yellow background indicates PRLRs, although the position of the sea bream PRLR (Santos et al. 2001) is arguable (white background). Blue and pink backgrounds indicate GHRs and SLRs, respectively. Considering positions of eel GHR1 and GHR2 (although their bootstrap values, indicated in gray, are relatively low), GHRs in a green background seem more likely to be SLRs. Similar trees were drawn by parsimony, neighbor-joining, or maximum-likelihood methods using the programs (Protpars, Neighbor, or Proml) in the PHYLIP package. (B) The positions of the extra intron inserted at the SLR (GHR) loci reported to date. Red indicates major amino acids at each site among the four species. Their coding sequences are indicated below. The intron is inserted at the positions indicated by arrowheads.
Cloning of a medaka GHR:
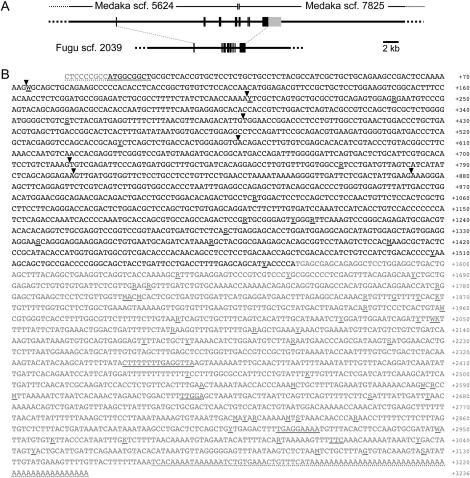
Starting from an amino acid sequence of masu salmon GHR (GenBank accession no. BAB64911), we adopted the same strategy to predict exons of medaka and fugu GHRs, except that an EST clone of medaka (OLc08.02d) was fortunately available from the Mbase database (http://mbase.bioweb.ne.jp/∼dclust/medaka_top.html). We found orthologous regions on scaffold 5624 and 7825 of medaka (i.e., they have not been assembled into one scaffold, and we experimentally determined the interscaffold sequence of 178 bp) and scaffold 2039 of fugu (Figure 4A). We determined the complete nucleotide sequence of the OLc08.02d by primer walking (Figure 4B) and revealed that the medaka GHR protein is encoded by eight exons, instead of the nine of the medaka SLR. The deduced protein consists of 523 amino acids and shares 275 identical residues (∼46%) with the salmon GHR (Figure 3A). The predicted fugu GHR is also encoded by eight exons and shares ∼59% of nucleotide and ∼49% of amino acid identities with the medaka GHR (Figure 3A).
Figure 4.
Cloning of the medaka GHR. (A) Comparative-genomic prediction of medaka and fugu GHR exons. Solid and shaded boxes indicate translated and untranslated regions, respectively. Corresponding exons of medaka and fugu retain identical exon-intron boundaries. Note that their GHR proteins are encoded by eight exons. (B) Nucleotide sequence of the medaka GHR cDNA contained in OLc08.02d (GenBank accession no. DQ010539). Arrowheads indicate position of introns inserted. Shaded residues indicate 5′- and 3′-UTRs. Another exon in the 5′ upstream region that does not encode protein as reported in tetrapod GHRs might be missing from this EST clone. Underlining indicates polymorphic residues detected between the HNI (experimentally determined by direct sequencing of RT-PCR products) and Hd-rR (contained in OLc08.02d) alleles. Polymorphisms in residues with dashed underlining were not examined.
There are no scaffolds other than scaffolds 48 and 2039 in the fugu genome that encode a protein more similar to the salmon SLR or GHR. Phylogenetic analysis using the SLRs and GHRs of medaka, fugu, and masu salmon clearly supported their orthologous relationships (Figure 3B). Although exon-intron boundaries in masu salmon have not been identified, existence of an extra intron in the SLRs but not GHRs of medaka (and fugu) also supports the idea that the latter are orthologs of tetrapod GHRs (which do not have the extra intron).
Expression of the medaka SLR and GHR genes:
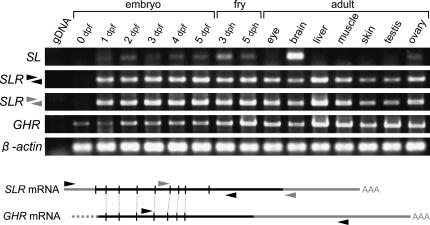
The above results strongly suggest that a medaka ortholog of the salmon SLR was successfully cloned. We then analyzed its expression by RT-PCR. As shown in Figure 5, expression of its ligand, SL, was detectable from day 1 post-fertilization (1 dpf) until hatching (5 dpf). Similarly, a weak expression was detected in larvae of day 3 and 5 post-hatching (dph). In adults, the strongest expression was detected in the brain (containing the pituitary), consistent with our former results (Fukamachi et al. 2004). SLR expression was also detectable at 1–5 dph. In adults, this was detected from all organs examined, with the strongest expression in the liver. The muscle and gonad (ovary) showed relatively strong expression similar to the result in coho salmon, O. kisutch (Fukada et al. 2005). Unexpectedly, the expression in the skin was as weak as that in the eye or brain.
Figure 5.
Expression of the medaka SLR and GHR. (Bottom) SLR and GHR mRNAs and the approximate positions of primers used in RT-PCR. Vertical solid lines indicate the position of introns.
We also analyzed expression of the medaka GHR and obtained similar results to that of the SLR, except that the GHR expression was detectable from 0 dpf (i.e., maternal storage) and seems enhanced after hatching (Figure 5). Thus, both the medaka SLR and GHR are expressed from very early stages of embryonic development and in variable organs in adults.
Accumulation of lipids in the ci mutants:
Given the broad expression of the medaka SLR in adults, anomalies of the SL-deficient mutant, the ci, would not necessarily be restricted to chromatophores in the skin, but would appear in other tissues and organs, most likely in the liver. From this standpoint, the phenotypes of the cobalt rainbow trout are suggestive, and we investigated whether or not the ci shares common defects, other than pigmentation, with the cobalt.
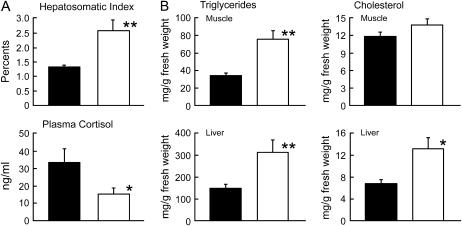
We compared 10 wild-type and 10 ci medaka that had been kept under identical breeding conditions for more than 3 months. Body weight of the ci (0.334 ± 0.032 g) was slightly but significantly (P < 0.05) lower than that of the wild type (0.458 ± 0.024 g). However, liver weight of the ci (0.009 ± 0.003 g) was 50% heavier than that of the wild type (0.006 ± 0.001 g), resulting in a significantly (P < 0.01) higher hepatosomatic index (Figure 6A). Plasma cortisol concentrations of the ci medaka were about half the levels observed in the wild type (Figure 6A). There was no significant difference in the gill Na+, K+-ATPase activities between the ci and wild types (0.83 ± 0.14 and 1.01 ± 0.04 μmol ADP/mg protein h, respectively).
Figure 6.
Physiological defects in the ci medaka. Results are shown as means ± SEM. Solid bar, wild type; open bar, ci. Significance of differences between two groups was analyzed by ANOVA followed by Student's t-test or Mann-Whitney U-test. (A) Hepatosomatic index and cortisol concentration in plasma were significantly higher and lower in the ci, respectively. *P < 0.05; **P < 0.01. (B) Triglycerides are significantly accumulated in both the muscle and the liver of the ci, while accumulation of cholesterol is apparent only in the liver.
The ci medaka showed significantly higher accumulations of triglycerides in the liver and muscle than the wild type did (Figure 6B). Cholesterol contents in the liver of the ci were also higher than those of the wild type, although there was no obvious difference in the muscle contents between the two groups. All of these defects in the ci are consistent with those of the cobalt (Yada et al. 2002; T. Yada, K. Muto, T. Azuma, S. Fukamachi and T. Hirano, unpublished data) and indicate that the anomalies in the cobalt are also caused by lack of SL itself, rather than by other hormones secreted from the pituitary (see discussion). Taken together, our results demonstrate that SLs have at least two functions conserved widely in teleosts: suppressing fat accumulation in organs and regulating chromatophore development in the skin.
DISCUSSION
Phylogenetic paradox among fish SLRs and GHRs:
Although we strongly suggest that the two medaka genes that we isolated are true orthologs of the salmon SLR and GHR (Figure 3B), even more careful gene definition might be necessary as discussed below.
The first definition of fish GHR was almost solely based on sequence similarity to the tetrapod GHRs, although interaction with human GH was shown (Calduch-Giner et al. 2001; Lee et al. 2001). According to these sequences, GHRs of other fish were isolated (Calduch-Giner et al. 2003; Nakao et al. 2004; and other unpublished sequences in the GenBank database). A few studies showed binding of the GHR to native GH (Tse et al. 2003; Kajimura et al. 2004), but they did not examine interaction between the GHR and SL. Importantly, the recent studies by Fukada et al. (2004, 2005) demonstrated that (1) salmonid GHR binds to GH but not to SL or PRL; (2) salmonid SLR binds more to SL than to GH or PRL; and (3) the SLR can significantly (cross-) react with GH but not with PRL. According to these authors' results, the salmonid GHR and SLR seem to have been correctly termed and we believe that the GHRs and SLRs of medaka and fugu, which were carefully designated by surveying whole-genome sequences, are their true orthologs (Figure 3B). However, GHRs of other nonsalmonid species and those of medaka, fugu, and salmonids seem to not be orthologous to each other.
As shown in Figure 3A (and other data not shown), the FGXFS motif (which is altered to YGXFS in placentals; see Goffin and Kelly 1997) and four proline residues in box 1 (Dinerstein et al. 1995) are perfectly conserved among all the fish receptors. Box 2, which is less conserved even in mammals, also seems conserved. Interestingly, the fifth and sixth bisulfide-linked cysteines in the extracellular domain are not conserved in the GHRs of medaka, fugu, and 4 species of salmonids, as they are among the six GHRs of tetrapods (18 species). In contrast, the SLRs and GHRs of other nonsalmonids (13 species) retain all six cysteines (with the exception of the first one in fugu, which must be experimentally confirmed). Furthermore, eight tyrosine residues in the cytoplasmic domain, at least some of which are important for GH-induced cellular responses (Lobie et al. 1995; Hansen et al. 1997) and are highly conserved among tetrapods [93% = 133 tyrosines/(8 sites × 18 species)], are more conserved in the SLRs (22/24) than in the GHRs (43/72) of medaka, fugu, and salmonids. Again, these tyrosines tend to be more conserved in the nonsalmonid GHRs (108/112) as SLRs.
Thus, the SLRs of medaka, fugu, and salmonids (and GHRs of other nonsalmonids) are more similar in structure to the tetrapod GHRs than are their own GHRs. This may be the cause of their significant (cross-) interaction with the mammalian or fish GH (Lee et al. 2001; Tse et al. 2003; Kajimura et al. 2004; Fukada et al. 2005). Therefore, even though the nonsalmonid GHRs had been shown to interact with GH, it remains unclear whether they are GHRs or SLRs (i.e., they may interact more with SL as the salmon SLR).
Indeed, phylogenetic analyses apparently classified the GHRs of sea breams, turbot, flounder, and tilapia as SLR (Figure 7A). An extra intron inserted at the last exon of the sea bream and flounder GHRs (Tse et al. 2003; Nakao et al. 2004), similar to the medaka and fugu SLRs (Figure 3A), also supports this conclusion, although we noted that its position reported in flounder is not identical to those of medaka, fugu, and sea bream (Figure 7B). GHRs of goldfish, carps, catra, rohu, hawk fish, and catfish are less apparent, but are more likely to be SLR than GHR (Figure 7A). One (or even both) of eel GHRs also seems to be SLR (both retain the six cysteines). Therefore, the medaka (and fugu) GHR may be the first successful isolation of nonsalmonid GHR, and these sequences might be of more help to clone “true” GHR orthologs of other nonsalmonid species.
The possibility of functional switching of fish SLRs and GHRs during evolution:
Needless to say, however, the above consideration is based on phylogenetic analyses, and we cannot exclude the possibility that these nonsalmonid GHRs (and even the SLRs of medaka and fugu) may be the “functional” equivalent of GHR, i.e., the functional switching of the family genes during evolution. From this standpoint, the structural dissimilarity between tetrapod and fish GHRs (see above) is interesting. Because SL seems to be missing from mammalian and avian genomes (S. Fukamachi, unpublished observation), but not from lungfish (Amemiya et al. 1999), ancestral existence and consequent loss of SL in the higher vertebrates seems likely.
Disruption of SL-SLR relationships might have triggered functional reorganization of, or compensation by, their closely related genes, such as GH and GHR. This probably occurred in the higher vertebrates instead, but such functional alteration(s) might have taken place in some fish lineage after the genome/gene duplication. While salmonids and sea bream retain the duplicated GHRs and SLs, respectively (see Figure 7A; Cavari et al. 2000), only one copy of them is found in the medaka or fugu genome (although it is still incomplete in medaka). In addition, a novel SL gene has been reported in some fish species (Cheng et al. 1997; Yang and Chen 2003; Zhu et al. 2004), while it seems to be missing in medaka or fugu. These inconsistent gene sets among some fish might reflect the putative lineage-specific functional diversification of SL, SLR, GH, and GHR (and a putative receptor for the novel SL). Further extensive phylogenetic and functional evaluations using variable species are necessary to resolve these complicated but intriguing questions.
Common defects in the ci and the cobalt caused by lack of SL:
Although we encountered the above difficulties in confirming that the gene that we cloned in Figure 2B is a true ortholog of the salmon SLR, the initial purpose of this study was, of course, to investigate the phenotypes of the medaka SL-deficient mutant, the ci, to assess SL's function other than in pigmentation. We detected the strongest expression of the medaka SLR (and GHR) and excessive accumulation of lipids in the liver (Figures 5 and 6B). In the cobalt rainbow trout, triglycerides and cholesterol content in the liver and muscle are several times higher than those in normal fish (Yada et al. 2002), whereas those in the ci medaka are only twice as high only in the liver (Figure 6). This qualitative and quantitative difference might reflect a difference in species, different defects in SL expression (i.e., lack of SL-producing cells in the cobalt and a mutation in the SL gene in the ci), or, most likely, lack of other hormones in the cobalt (as discussed in Yada et al. 2002). In coho salmon, SLR mRNA is similarly expressed with the highest levels in the liver and visceral fat among all tissues examined (Fukada et al. 2005). Regardless of whether SLR or GHR actually mediates SL signaling, these results strongly suggest that the liver and fat tissues are some of the target organs and tissues of SL and that SLs of both medaka and rainbow trout function to suppress accumulation of lipids in organs. Considering that even the putative SL-null mutant (the ci) can normally grow and reproduce, commercially valuable fish (e.g., fish with high- and low-fat meat or beautiful body color) may be able to be produced by controlling SL expression.
The other defect in the ci was a reduced circulating level of cortisol. A level of plasma cortisol lower than that of normal fish is also observed in the cobalt (Yada et al. 2002). The cobalt fish shows hypertrophy in interrenal tissue, which is equivalent to the adrenal of tetrapods, suggesting an interruption in the regulation of cortisol secretion without SL (Oguri 1992; Kaneko et al. 1993). Although this study does not examine morphological differences in interrenal tissue between the two types of medaka, the lower levels of plasma cortisol observed in the ci fish suggest an important involvement of SL in cortisol secretion. Since stress-responding elevation in the circulating SL level has been observed in salmonid species, mediation of stress response is thought to be a candidate in physiological roles of SLs (Rand-Weaver et al. 1993; Kakizawa et al. 1995). It would be interesting, in the future, to examine the stress response in SL levels in the ci medaka.
Further detailed investigation of the ci phenotype and its comparison to that of the cobalt will reveal common and disparate defects other than in body color, lipid metabolism, and cortisol secretion, which in turn will provide clear-cut evidence for the physiological functions of SLs.
Acknowledgments
This study could not have been conducted without the medaka and fugu genome databases (see text for their URLs). We thank K. Naruse and T. Yamashita of the University of Tokyo for the OLc08.02d EST clone and for fish care, respectively. We also thank Y. Takei of the University of Tokyo for experimental advice. This study is supported by grants-in-aid from the Ministry of Agriculture, Forestry and Fisheries of Japan and by a Grant-in-Aid for Scientific Research on Priority Area (Genome Science), Ministry of Education, Sports, and Science (MEXT), Japan. S.F. is supported by Research Fellowships of the Japan Society for the Promotion of Science for Young Scientists (no. 17-10821).
References
- Amemiya, Y., Y. Sogabe, M. Nozaki, A. Takahashi and H. Kawauchi, 1999. Somatolactin in the white sturgeon and African lungfish and its evolutionary significance. Gen. Comp. Endocrinol. 114: 181–190. [DOI] [PubMed] [Google Scholar]
- Calduch-Giner, J. A., C. Pendon, M. M. Valdivia and J. Perez-Sanchez, 1998. Recombinant somatolactin as a stable and bioactive protein in a cell culture bioassay: development and validation of a sensitive and reproducible radioimmunoassay. J. Endocrinol. 156: 441–447. [DOI] [PubMed] [Google Scholar]
- Calduch-Giner, J. A., H. Duval, F. Chesnel, G. Boeuf, J. Perez-Sanchez et al., 2001. Fish growth hormone receptor: molecular characterization of two membrane-anchored forms. Endocrinology 142: 3269–3273. [DOI] [PubMed] [Google Scholar]
- Calduch-Giner, J. A., M. Mingarro, S. Vega-Rubin de Celis, D. Boujard and J. Perez-Sanchez, 2003. Molecular cloning and characterization of gilthead sea bream (Sparus aurata) growth hormone receptor (GHR). Assessment of alternative splicing. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 136: 1–13. [DOI] [PubMed] [Google Scholar]
- Cavari, B., B. Funkenstein and H. Kawauchi, 2000. Cloning of a second variant of somatolactin-encoding cDNA from the gilthead seabream Sparus aurata. Fish. Physiol. Biochem. 22: 145–150. [Google Scholar]
- Cheng, K. W., Y. H. Chan, Y. D. Chen, K. L. Yu and K. M. Chan, 1997. Sequence of a cDNA clone encoding a novel somatolactin in goldfish, Carassius auratus. Biochem. Biophys. Res. Commun. 232: 282–287. [DOI] [PubMed] [Google Scholar]
- Dinerstein, H., F. Lago, L. Goujon, F. Ferrag, N. Esposito et al., 1995. The proline-rich region of the GH receptor is essential for JAK2 phosphorylation, activation of cell proliferation, and gene transcription. Mol. Endocrinol. 9: 1701–1707. [DOI] [PubMed] [Google Scholar]
- Folch, J., M. Lees and G. H. Sloane-Stanley, 1957. A simple method for the isolation and purification of total lipids from animal tissues. J. Biol. Chem. 226: 497–509. [PubMed] [Google Scholar]
- Fukada, H., Y. Ozaki, A. L. Pierce, S. Adachi, K. Yamauchi et al., 2004. Salmon growth hormone receptor: molecular cloning, ligand specificity, and response to fasting. Gen. Comp. Endocrinol. 139: 61–71. [DOI] [PubMed] [Google Scholar]
- Fukada, H., Y. Ozaki, A. L. Pierce, S. Adachi, K. Yamauchi et al., 2005. Identification of the salmon somatolactin receptor, a new member of the cytokine receptor family. Endocrinology 146: 2354–2361. [DOI] [PubMed] [Google Scholar]
- Fukamachi, S., M. Sugimoto, H. Mitani and A. Shima, 2004. Somatolactin selectively regulates proliferation and morphogenesis of neural-crest derived pigment cells in medaka. Proc. Natl. Acad. Sci. USA 101: 10661–10666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffin, V., and P. A. Kelly, 1997. The prolactin/growth hormone receptor family: structure/function relationships. J. Mammary Gland Biol. Neoplasia 2: 7–17. [DOI] [PubMed] [Google Scholar]
- Hansen, J. A., L. H. Hansen, X. Wang, J. J. Kopchick, F. Gouilleux et al., 1997. The role of GH receptor tyrosine phosphorylation in Stat5 activation. J. Mol. Endocrinol. 18: 213–221. [DOI] [PubMed] [Google Scholar]
- Kajimura, S., N. Kawaguchi, T. Kaneko, I. Kawazoe, T. Hirano et al., 2004. Identification of the growth hormone receptor in an advanced teleost, the tilapia (Oreochromis mossambicus) with special reference to its distinct expression pattern in the ovary. J. Endocrinol. 181: 65–76. [DOI] [PubMed] [Google Scholar]
- Kakizawa, S., T. Kaneko, S. Hasegawa and T Hirano, 1995. Effects of feeding, fasting, background adaptation, acute stress, and exhaustive exercise on the plasma somatolactin concentrations in rainbow trout. Gen. Comp. Endocrinol. 98: 137–146. [DOI] [PubMed] [Google Scholar]
- Kaneko, T., S. Kakizawa and T. Yada, 1993. Pituitary of “cobalt” variant of the rainbow trout separated from the hypothalamus lacks most pars intermedial and neurohypophysial tissue. Gen. Comp. Endocrinol. 92: 31–40. [DOI] [PubMed] [Google Scholar]
- Kelsh, R. N., C. Inoue, A. Momoi, H. Kondoh, M. Furutani-Seiki et al., 2004. The Tomita collection of medaka pigmentation mutants as a resource for understanding neural crest cell development. Mech. Dev. 121: 841–859. [DOI] [PubMed] [Google Scholar]
- Kumar, S., K. Tamura and M. Nei, 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5: 150–163. [DOI] [PubMed] [Google Scholar]
- Lee, L. T., G. Nong, Y. H. Chan, D. L. Tse and C. H. Cheng, 2001. Molecular cloning of a teleost growth hormone receptor and its functional interaction with human growth hormone. Gene 270: 121–129. [DOI] [PubMed] [Google Scholar]
- Lobie, P. E., G. Allevato, J. H. Nielsen, G. Norstedt and N. Billestrup, 1995. Requirement of tyrosine residues 333 and 338 of the growth hormone (GH) receptor for selected GH-stimulated function. J. Biol. Chem. 270: 21745–21750. [DOI] [PubMed] [Google Scholar]
- Lu, M., P. Swanson and J. L. Renfro, 1995. Effect of somatolactin and related hormones on phosphate transport by flounder renal tubule primary cultures. Am. J. Physiol. 268: R577–R582. [DOI] [PubMed] [Google Scholar]
- Miya, M., H. Takeshima, H. Endo, N. B. Ishiguro, J. G. Inoue et al., 2003. Major patterns of higher teleostean phylogenies: a new perspective based on 100 complete mitochondrial DNA sequences. Mol. Phylogenet. Evol. 26: 121–138. [DOI] [PubMed] [Google Scholar]
- Nakao, N., Y. Higashimoto, T. Ohkubo, H. Yoshizato, N. Nakai et al., 2004. Characterization of structure and expression of the growth hormone receptor gene of the Japanese flounder (Paralichtys olivaceus). J. Endocrinol. 182: 157–164. [DOI] [PubMed] [Google Scholar]
- Oguri, M., 1992. Renal hypertrophy in the “cobalt” variant of rainbow trout. Nippon Suisan Gakkaishi 58: 803. [Google Scholar]
- Rand-Weaver, M., T. G. Pottinger and J. P. Sumpter, 1993. Plasma somatolactin concentrations in salmonid fish are elevated by stress. J. Endocrinol. 138: 509–515. [DOI] [PubMed] [Google Scholar]
- Santos, C. R., P. M. Ingleton, J. E. Cavaco, P. A. Kelly, M. Edery et al., 2001. Cloning, characterization, and tissue distribution of prolactin receptor in the sea bream (Sparus aurata). Gen. Comp. Endocrinol. 121: 32–47. [DOI] [PubMed] [Google Scholar]
- Tse, D. L., M. C. Tse, C. B. Chan, L. Deng, W. M. Zhang et al., 2003. Seabream growth hormone receptor: molecular cloning and functional studies of the full-length cDNA, and tissue expression of two alternatively spliced forms. Biochim. Biophys. Acta 1625: 64–76. [DOI] [PubMed] [Google Scholar]
- Vega-Rubin de Celis, S., P. Gomez, J. A. Calduch-Giner, F. Medale and J. Perez-Sanchez, 2003. Expression and characterization of European sea bass (Dicentrarchus labrax) somatolactin: assessment of in vivo metabolic effects. Mar. Biotechnol. 5: 92–101. [DOI] [PubMed] [Google Scholar]
- Yada, T., and F. Ito, 1998. Sexual difference in acid tolerance in medaka Oryzias latipes. Fish. Sci. 64: 694–699. [Google Scholar]
- Yada, T., S. Moriyama, Y. Suzuki, T. Azuma, A. Takahashi et al., 2002. Relationships between obesity and metabolic hormones in the “cobalt” variant of rainbow trout. Gen. Comp. Endocrinol. 128: 36–43. [DOI] [PubMed] [Google Scholar]
- Yang, B. Y., and T. T. Chen, 2003. Identification of a new growth hormone family protein, somatolactin-like protein, in the rainbow trout (Oncorhynchus mykiss) pituitary gland. Endocrinology 144: 850–857. [DOI] [PubMed] [Google Scholar]
- Zhu, Y., and P. Thomas, 1995. Red drum somatolactin: development of a homologous radioimmunoassay and plasma levels after exposure to stressors or various backgrounds. Gen. Comp. Endocrinol. 99: 275–288. [DOI] [PubMed] [Google Scholar]
- Zhu, Y., J. W. Stiller, M. P. Shaner, A. Baldini, J. L. Scemama et al., 2004. Cloning of somatolactin alpha and beta cDNAs in zebrafish and phylogenetic analysis of two distinct somatolactin subtypes in fish. J. Endocrinol. 182: 509–518. [DOI] [PubMed] [Google Scholar]