Abstract
Epistasis contributes significantly to intrapopulation variation in floral morphology, development time, and male fitness components of Mimulus guttatus. This is demonstrated with a replicated line-cross experiment involving slightly over 7000 plants. The line-cross methodology is based on estimates for means. It thus has greater power than the variance partitioning approaches historically used to estimate epistasis within populations. The replication of the breeding design across many pairs of randomly extracted, inbred lines is necessary given the diversity of multilocus genotypes residing within an outbred deme. Male fitness is shown to exhibit synergistic epistasis, an accelerating decline in fitness with inbreeding. Synergism is a necessary, but not sufficient, condition for a mutational deterministic hypothesis for the evolutionary maintenance of sexual reproduction. Unlike male fitness measures, flower morphology and development time yield positive evidence of epistasis but not of synergism. The results for these traits suggest that epistatic effects are variable across genetic backgrounds or sets of interacting loci.
EPISTASIS is the interaction between different genetic loci in determining phenotype. The prevalence and nature of epistasis are central to a range of questions in evolutionary biology. Sewall Wright developed his shifting balance theory to provide a mechanism for the evolution of favorable gene combinations (Wright 1931; Wade 1992). The evaluation of this theory, and more generally of the interaction between epistasis and population structure, has been a major impetus to experimental work (Mallet and Singer 1987; Husband and Barrett 1992a,b; see reviews by Coyne et al. 1997; Wade and Goodnight 1998). Ronald Fisher, the principal antagonist to the shifting balance model, himself invoked “modifiers” in a number of evolutionary hypotheses (e.g., Fisher 1958, Chap. 3). Modifiers, which are epistatic by definition, were also a recurring theme of Fisher's colleagues in the ecological genetics school (Ford 1971). More recently, evolutionary geneticists have explored the role of gene interactions on the evolution of sex and recombination (Peters and Lively 2000), developmental integration (Rice 1998), speciation (Rieseberg et al. 1996; Johnson 2000), and the rate and pattern of adaptive diversification (Wade 2000).
There is a great deal of evidence that epistasis contributes to the genetic differences among species (e.g., Doebley et al. 1995; Fishman and Willis 2001) and among populations within a species (e.g., Hard et al. 1992; Burton et al. 1999; Fenster and Galloway 2000). This has been established by comparing the average phenotype of various hybrids (the F1, F2, or backcross progeny) to the corresponding values of the parental species or lines. Without epistasis, the mean phenotype of F2 individuals should be exactly intermediate between the F1 and midparent means (the midparent is the average of the parental species). The often observed “F2 breakdown,” where the average F2 phenotype is substantially lower than expected, is thus an example of epistasis (Grant 1975; Burton 1990). Fine mapping of the variants responsible for hybrid failure in interspecies crosses has subsequently revealed complicated patterns of gene interaction (Cabot et al. 1994; Palopoli and Wu 1994; but also see Zeng et al. 2000).
The finding of epistasis in interspecies crosses is not compelling evidence that selection acted on gene combinations to produce the phenotypic divergence between these species (Whitlock et al. 1995, pp. 612–613). Nor does it directly imply that epistatic variation was present within the ancestral species. In fact, the loci interacting to cause hybrid failure may never have been simultaneously polymorphic within the ancestral population. The classical Dobzhansky-Muller model of hybrid breakdown yields interspecies epistasis without selection ever acting directly on gene combinations (Dobzhansky 1936; Muller 1939; Orr 1995). We need to consider the genetic variability present within species to evaluate the evolutionary consequences of epistasis.
Barker (1979) reviewed attempts to estimate the contribution of epistasis to quantitative trait variation within contiguous natural populations, i.e., demes. Considering studies of different types (e.g., variance partitioning, response to selection and inbreeding, and linkage disequilibria), he concluded that the evidence was “equivocal and unsatisfying.” This could be interpreted to mean that epistasis is simply more important in determining differences among highly divergent genotypes (species or lines) than the more modest differences among individuals within demes. Alternatively, the differing results from inter- and intraspecific studies may simply reflect differences in statistical power (Fenster et al. 1997). The line-cross method used in agricultural and interspecies studies is based on statistical comparisons of mean phenotypic values. In contrast, the variance-partitioning approaches frequently used to estimate epistasis within populations (e.g., Miller et al. 1963; Barker 1974) are based on second moments, variances, and covariances. For a given sample size, means are typically estimated with far greater accuracy than variances or covariances.
The traditional method of model evaluation in line-cross studies is the joint scaling test (Mather 1949; Anderson and Kempthorne 1954; Hayman 1960; Mather and Jinks 1982). Following Lynch and Walsh (1998, Chap. 9), I refer to the various categories of individuals such as the parental lines, F1, F2, and backcrosses as “line-cross derivatives.” In a joint scaling test, the means for each line-cross derivative are used to calibrate and then test models of inheritance. Analysis generally begins by considering the simplest model, additive inheritance. With strict additivity, the means of all line-cross derivatives can be predicted given only two parameters, the overall mean (M) and the additive effect (A). A set of expected values for each line-cross derivative is obtained by estimating these parameters. The correspondence between observed and expected means, and hence the sufficiency of the model, can then be evaluated using the chi-square test. If the strictly additive model is rejected, one then proceeds to evaluate a model allowing dominance (three parameters: M, A, and D, the dominance deviation). The rejection of this model implies epistasis. With a sufficient number of distinct line-cross derivates, increasingly complicated models of gene interaction can be tested (e.g., Jinks and Perkins 1969).
A frequently cited shortcoming of line-cross studies is that the results, estimates for dominance and epistasis, are specific to the particular pair of inbred lines chosen as parentals. This is not a serious difficulty for many agricultural applications, because a limited number of cultivars/varieties/genotypes are widely used by farmers. However, there are an enormous number of distinct multilocus genotypes in an outbred deme. It is difficult, perhaps even impossible, to realistically concentrate this variability into a single pair of inbred lines.
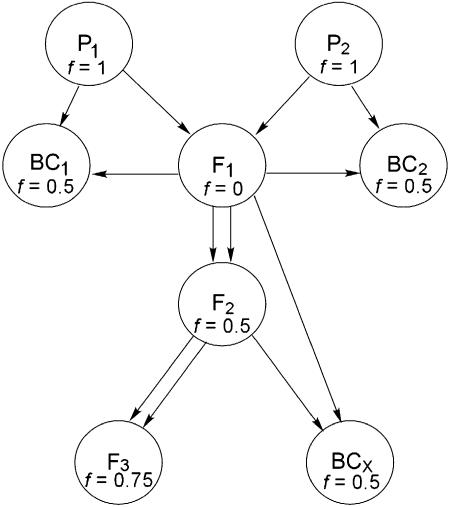
This article describes a study using the line-cross methodology to investigate variation resident within a single natural population. I replicate the joint scaling test across a large collection of inbred lines that are representative of the ancestral outbred population in terms of allele frequencies at quantitative trait loci. Lines were randomly paired without consideration of their phenotypic attributes and each pair of lines was used to synthesize a set of line-cross derivatives (Figure 1). The progeny produced from this array of crosses were grown under common conditions and measured for development time, flower morphology, and male fitness components. This experiment addresses two basic questions: Do these traits exhibit significant epistasis? If so, are epistatic effects consistent in their direction across sets of interacting loci?
Figure 1.
A diagram of the crossing scheme used to synthesize line-cross families. Each circle encloses a line-cross derivative and arrows denote the transmission of a gamete. The inbreeding coefficient (f) relative to the ancestral population is given for each derivative.
The characters chosen for study, flower morphology, development time, and pollen production, are primary determinants of plant fitness in the wild. The genetic analysis of measurements on these traits consists of two parts, both of which are based on comparisons of mean phenotypic values. The first considers the phenotypic response to inbreeding. In the context of the ancestral population, line-cross derivatives have inbreeding coefficients (f) that range from 0 to 1 (Figure 1). A simple test for epistasis within a diploid population is to determine whether the relationship between the average trait value and f is nonlinear. If loci contribute additively to trait variation, the mean phenotype should change linearly with f (Wright 1951; Kempthorne 1957). In contrast, consistently positive or consistently negative interactions among loci predict a nonlinear relationship between mean phenotype and f (Crow and Kimura 1970, pp. 77–85).
The failure to detect nonlinearity in the relationship between mean phenotype and f does not conclusively indicate the absence of epistasis. An approximately linear relationship may be obtained if epistatic effects are important, but variable in direction across loci (Elena and Lenski 1997; Phillips et al. 2000). However, such variable epistatic effects should be evident as deviations between observed and expected means within individual cross-families. For example, the mean value of F2 plants may be significantly greater than the expected values in some families, but less than those in others. These deviations may cancel when considering the population as a whole because all F2 plants have the same inbreeding coefficient. However, these fluctuating deviations are appropriately integrated in the Replicated joint scaling test to produce a population-wide test for epistasis.
MATERIALS AND METHODS
Study system:
Mimulus guttatus is rapidly becoming one of the most thoroughly studied plants in ecology and evolutionary biology (Grant 1924; Kiang 1972, 1973; Vickery 1978; Macnair and Cumbes 1989; Fenster and Ritland 1992, 1994a,b; Willis 1996, 1999a,b; Dudash and Carr 1998). Yellow monkeyflower is a self-compatible hermaphrodite that occurs throughout western North America. It reproduces by a mixture of outcrossing and self-fertilization, but the selfing rate differs greatly among natural populations (Ritland and Ganders 1987a,b; Dudash and Ritland 1991; Dole and Ritland 1993; Willis 1993b). Local populations also differ extensively in floral morphology (Waser et al. 1982), in the amount of genetic variation for floral traits (Carr and Fenster 1994; Robertson et al. 1994; Kelly and Arathi 2003), and in the timing of selfing over the life span of the flower (Dole 1990; Dudash and Ritland 1991; Leclerc-Potvin and Ritland 1994; Arathi et al. 2002).
This experiment investigates genetic variation within one particular natural population called “Iron Mountain,” which is located in Oregon's Western Cascades (see Willis 1996, 1999a,b). Iron Mountain contains several hundred thousand individuals continuously distributed over an area of ∼400 square meters. The population contains a tremendous amount of variation at microsatellite loci (Kelly and Willis 1998, 2002). Some loci have 20–30 alleles and expected heterozygosities >0.8, indicating that the population has not suffered a recent bottleneck in size. Despite limitations on both seed and pollen dispersal, microsatellite polymorphisms exhibit little or no spatial structure within this population (Sweigart et al. 1999). The implication is that gene flow is sufficient so that Iron Mountain can be considered a cohesive genetic population for evolutionary studies. However, the microsatellite data do not imply that spatial structure is absent at loci that might be under stronger selection.
John H. Willis initiated ∼1200 independent lines from Iron Mountain in August of 1995. Each line was founded from the seed set of a separate field-collected plant. Each line was subsequently maintained by single-seed descent (self-fertilization) for seven to nine generations. Over the course of these generations, most lines went extinct due to the random fixation of lethal and/or sterile alleles. However, over 300 survived to at least generation seven. The progeny of these surviving lines now have an inbreeding coefficient of >0.99. As expected, the surviving lines are almost completely homozygous at highly polymorphic microsatellite loci with different lines fixed for different alleles (Willis 1999a; L. Holeski, unpublished results).
Crossing design:
Two hundred of these lines were used as parental plants in a breeding design (Figure 1). Pairs of lines, P1 and P2, were randomly selected to initiate independent “line-cross families.” These lines were crossed to produce an F1 family. The F1 individuals were self-fertilized to produce F2 families and also crossed to each parental line to produce backcross families: Backcross (BC)1 is the family derived from the cross between P1 and F1 while BC2 is the family derived from P2 and F1. Seeds from the F2 family were grown to maturity, self-fertilized to produce an F3 family, and backcrossed to an F1 individual. I denote progeny from the last cross as BCX. Due to failure of crosses or seed germination, only 76 line-cross families had at least 4 line-cross derivatives, and for reasons described below, the replicated joint scaling tests are limited to these data.
The controlled matings and self-fertilizations described in Figure 1 were conducted over the span of three plant generations, distinct from the generations in which plants were measured (see Phenotypic measurements and plant growth). Most of the line-cross derivatives within a line-cross family were synthesized from multiple crosses, each from a pairing of distinct individuals of the parental types. Many of the crosses were done reciprocally, with both parental line-cross derivatives serving as the maternal plant. For example, the collection of F1 plants in line-cross family 84 consisted of five full-sib families, each derived from a distinct pairing of P1 and P2 individuals. Multiple crosses were done for a number of reasons. First, they effectively eliminated any association between the age of seed when sown and the inbreeding coefficient (Lynch and Walsh 1998, pp. 262–263). Seeds for P1 and P2 were generated in the first generation of the mating scheme, simultaneously with the first set of F1 plants. A second set of P1 and P2 seeds was created in the second generation of the mating scheme, simultaneously with the synthesis of BC1, BC2, and F2 plants; and a third set was created in the final generation, simultaneously with the synthesis of BCX and F3 plants. Seeds for several of the other derivates, BC1, BC2, F1, and F2, were also generated during different generations of the breeding scheme.
Synthesizing line-cross derivatives from multiple crosses also alleviates potential biases caused by maternal effects. The phenotype of an individual may depend not only on its own inbreeding coefficient, but also on that of its mother (Vogler et al. 1999). The maternal inbreeding effect can be estimated by comparing plants from reciprocal crosses in which the parents differ in their respective inbreeding coefficients. In the present crossing design (Figure 1), each backcross family was derived from a cross between a completely inbred plant (P1 or P2) and an outbred F1. The two sets of progeny in the reciprocal cross have the same individual inbreeding coefficients (f = 0.5), but different maternal inbreeding coefficients (either 0 or 1). A consistent difference in phenotype across line-cross families, say reduced pollen production by plants with inbred moms, would indicate an effect of maternal inbreeding. In addition to the backcrosses, the cross generating BCX was done reciprocally between parents with differing inbreeding coefficients.
Three of the eight line-cross derivatives (P1, P2, and F1) are genetically homogenous. All individuals within these categories are genetically identical, both within and among full-sib families (at least for the autosomal genome within a particular line-cross family). The F1 plants may be heterogeneous in the cytoplasmic genome, as it may be descended from different parental lines in different full-sib families. The other five line-cross derivatives (BC1, BC2, F2, F3, and BCX) will contain a mixture of genotypes even in the autosomal genome. The parents are genetically homogeneous for the first three of these derivates, BC1, BC2, and F2. Thus, for the autosomal genome, there is no greater genetic relatedness of individuals within full-sib families than among full-sib families (again we are referring to the different progeny sets within these line-cross derivatives of a particular line-cross family). However, for the last two derivates, F3 and BCX, at least one of the parents is an F2 individual. Since the F2 is genetically heterogeneous, there is increased genetic relatedness of individuals within full-sibs of the F3 and BCX line-cross derivatives. These considerations do not directly impact the calculations described below, because they are based entirely on the mean values of line-cross derivatives. However, they are relevant to the more rigorous treatment that includes the variance within line-cross derivatives to be described in a future article (J. K. Kelly, unpublished results).
Phenotypic measurements and plant growth:
Phenotypic data were collected from plants grown in three successive intervals. The first “block” was initiated on May 8, 2003, using only six of the line-cross derivatives within each line-cross family, the P1, P2, F1, BC1, BC2, and F2. The second block of plants, initiated on July 30, 2003, also consisted of plants from the first six line-cross derivatives. The third block, the seeds of which were sown on Jan 20, 2004, consisted of plants from all eight line-cross derivatives. Seeds were germinated under standard conditions in the greenhouse (see Kelly and Arathi 2003), and 2 weeks later, individual seedlings were transplanted into 98-well plug trays. These trays were maintained in a growth room with 18-hr days and 6-hr nights. Plants were fertilized once per week by bottom watering using a high P nutrient mixture. Trays were rotated daily. I noted the day on which each plant produced its first flower and then measured corolla width, corolla length, pistil length, and anther length. Stigma-anther separation is calculated as the difference between the latter two measurements.
Anthers were collected from the first flower into a microcentrifuge tube and stored. Pollen counts were later done using a Coulter (Miami) counter model Z1 dual. The sample from each tube was then diluted into 20 ml of electrolyte solution and run through the machine to produce particle counts within two size ranges. My estimate for total pollen per flower is the sum of these two counts × 20 (since only 1 ml of solution is processed in a run of the machine). The pollen size index (PSI) is the proportion of grains in the larger size category. There is a strong positive correlation between PSI and pollen viability as measured through standard staining techniques (Kelly et al. 2002). The proportion of viable grains (PV) is accurately estimated from the equation: PV = 0.l + PSI. For the rare samples in which PSI > 0.9, I set PV = 1.0. Viable pollen per flower was calculated as the product of PV and pollen number. In data analysis, I treat pollen traits as characteristics of the parental plant (the sporophyte).
ANALYSIS AND RESULTS
Table 1 summarizes the means, standard deviations, and sample sizes for each trait in each of the three blocks. Since each block contains different proportions of inbred and outbred individuals, the differences in mean trait values are only partly reflective of a “block effect.” A substantial number of measurements are missing from the 7053 plants in the study. Day of first flower was noted for all plants and the great majority were measured for corolla diameter. The remaining measurements were taken on most, but not on all plants.
TABLE 1.
The mean, standard deviation (in parentheses), and sample size (n) for each measured trait are given for each block and for all plants pooled
| Trait | Block 1 | Block 2 | Block 3 | Total |
|---|---|---|---|---|
| Corolla width | 15.02 (2.37) | 14.56 (2.56) | 16.42 (2.74) | 15.34 (2.70) |
| n = 1943 | n = 2545 | n = 2426 | n = 6914 | |
| Days to flower | 31.36 (2.91) | 28.42 (4.15) | 30.48 (2.79) | 29.95 (3.61) |
| n = 1956 | n = 2633 | n = 2464 | n = 7053 | |
| Corolla length | 18.98 (2.36) | 18.23 (2.56) | 19.89 (2.58) | 19.19 (2.58) |
| n = 1511 | n = 949 | n = 1739 | n = 4199 | |
| Pistil length | 13.79 (1.46) | 13.21 (1.37) | 14.16 (1.47) | 13.81 (1.49) |
| n = 1597 | n = 994 | n = 1749 | n = 4340 | |
| Anther length | 11.01 (1.17) | 10.90 (1.21) | 11.37 (1.16) | 11.13 (1.19) |
| n = 1520 | n = 987 | n = 1771 | n = 4278 | |
| Stigma-anther separation | 2.77 (1.23) | 2.30 (1.04) | 2.78 (1.03) | 2.66 (1.13) |
| n = 1491 | n = 976 | n = 1728 | n = 4195 | |
| Total pollen | 8889 (3932) | 6388 (3112) | 6759 (3412) | 7254 (3635) |
| n = 1578 | n = 1766 | n = 2126 | n = 5470 | |
| Pollen size index (PSI) | 0.55 (0.21) | 0.64 (0.18) | 0.60 (0.19) | 0.60 (0.19) |
| n = 1578 | n = 1766 | n = 2126 | n = 5470 | |
| Viable pollen | 5958 (3438) | 4951 (2931) | 5016 (3061) | 5267 (3165) |
| n = 1578 | n = 1766 | n = 2126 | n = 5470 |
Morphological trait values are given in millimeters.
Each trait was subjected to two analyses, first the combined population analysis and then the replicated joint scaling test. Both involve comparisons of different statistical models. The model parameters are estimated from means and standard errors using generalized least squares. The fitted model is then used to establish a set of “expected values” for each mean. Observed and expected values are compared, using chi-square statistics to assess significance. I wrote C programs to conduct the relevant calculations following the theory described in Lynch and Walsh (1998, pp. 198–204 and 216–219). The code for these programs and a sample input file (plant data) are provided in the supplemental materials (http://www.genetics.org/supplemental/).
Combined population analysis:
This procedure determines the effects of block, individual inbreeding, and maternal inbreeding on each character. Let μ denote the outbred grand mean for a particular trait and Bi represent the effect of block i. I assume that, if maternal inbreeding matters, the effect (v) is linearly related to the inbreeding coefficient of the maternal plant (Fm). Finally, δj represents the effect of individual inbreeding coefficient j on trait values. Individual inbreeding coefficients were 0 (F1 plants), 0.5 (BC1, BC2, F2, and BCX), 0.75 (F3), or 1 (P1and P2) in this experiment (Figure 1). The general model for the expected mean phenotype can then be written as
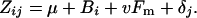 |
(1) |
There are seven distinct parameters in this model for this study (μ, B2, B3, v, δ0.5, δ0.75, and δ1.0). The parameter μ essentially estimates the mean of outbred plants with outbred mothers when grown in block 1.
Of the 36 possible combinations of block, individual inbreeding, and maternal inbreeding, 14 were actually represented in this study. By generalized least squares, I estimated the seven parameters in Equation 1 from this set of means and standard errors. A chi-square value was calculated from the observed and expected means, and this statistic was used in tests for phenotypic responses to both maternal and individual inbreeding. Both of the null hypotheses described below are defined as simplified versions of Equation 1.
To test whether the level of maternal inbreeding had an effect on a character, I set v = 0 and then reestimated the remaining parameters of Equation 1. The chi-square value produced when the expected values from this reduced model are compared to the observed means must be greater than or equal to the corresponding chi-square from the full model. A sufficiently large difference indicates a significant effect of maternal inbreeding. Since the two models differ by a single parameter (1 d.f.), the appropriate critical value from the chi-square distribution is 3.84. To test whether the response to individual inbreeding is linear, I estimated a model in which δ0.5 = 0.5b, δ0.75 = 0.75b, and δ1.0 = b, where b is the slope. This reduces the number of parameters by 2 and the appropriate critical value is 5.99. By similar means, one could address additional hypotheses, e.g., for an effect of block or for whether an individual inbreeding coefficient has any effect on trait values. I do not present these analyses because the effects are obvious, have been demonstrated previously, or are tangential to the purposes of this article.
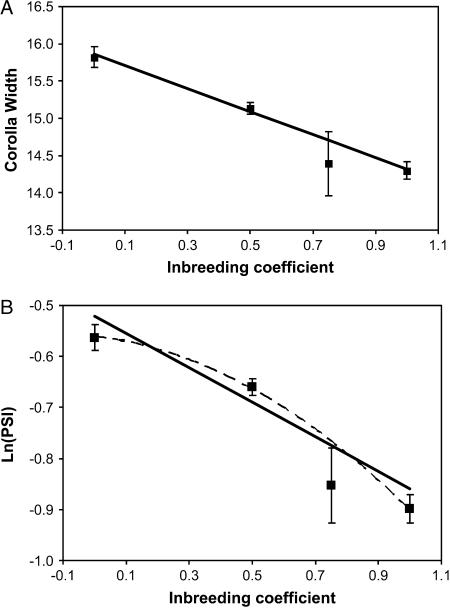
Table 2 summarizes model tests for each trait on its original scale of measurement and also for log-transformed male fitness measures. The pollen traits (pollen number, PSI, and viable pollen) were log-transformed because, in many situations, the “null model” for fitness assumes that loci and environmental effects combine multiplicatively (Crow and Kimura 1970). These dependences become additive after log-transformation. Only number of days to flower exhibits a significant effect of maternal inbreeding, although log-transformed viable pollen is only marginally nonsignificant. Response to individual inbreeding is approximately linear for each morphological trait and for total pollen number (Figure 2A). However, both PSI and viable pollen show a significantly nonlinear response to inbreeding. Both fitness measures exhibit “synergistic” or “reinforcing” epistasis (Crow and Kimura 1970, pp. 80–81), an accelerating decline in value as inbreeding coefficient increases (Figure 2B).
TABLE 2.
The chi-square values and test results from the combined population analyses (see text for details)
| Null hypothesis
|
||
|---|---|---|
No maternal effect: 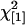
|
No epistasis: 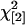
|
|
| Untransformed trait values | ||
| Corolla width | 1.39 | 4.06 |
| Days to flower | 3.96* | 2.25 |
| Corolla length | 0.00 | 1.79 |
| Pistil length | 0.76 | 0.82 |
| Anther length | 0.73 | 0.07 |
| Stigma-anther separation | 0.15 | 0.32 |
| Pollen no. | 0.16 | 4.49 |
| PSI | 1.86 | 45.19*** |
| Viable pollen | 0.34 | 8.03* |
| Log-transformed pollen values | ||
| Ln(pollen no.) | 2.03 | 3.78 |
| Ln(PSI) | 2.33 | 42.53*** |
| Ln(viable pollen) | 3.85 | 16.51*** |
The test for the maternal effect has 1 d.f. while the test for epistasis has 2 d.f. Significance levels: *0.01 < P < 0.05, ***P < 0.001.
Figure 2.
The mean value of plants for (A) corolla width and (B) log-transformed PSI is given as a function of individual inbreeding coefficient. The means for each category are given with error bars denoting ±2 SE. Each phenotypic value was corrected for effect of block prior to calculations. The dashed line in B represents a quadratic fit with y = −0.56 − 0.07x − 0.27x2 (see discussion).
Replicated joint scaling test:
For each character, I first applied a standard joint scaling test to each line-cross family. Prior to this analysis, the data were corrected for the effects of maternal inbreeding (for days to flower) and block effects (for all characters) as estimated in the Combined population analysis. Then, by generalized least squares, I estimated the three parameters of the additive-dominance model from the observed means and standard errors of each line-cross derivative. Using the formulation of Lynch and Walsh (1998, p. 209), the expected values are given in Table 3. The parameters M, A, and D are specific to each line-cross family, their values defined by the genotypes of the parental lines.
TABLE 3.
The expected values for each line-cross derivative as a function of genetic parameters
| Line-cross derivative | Expected mean phenotype |
|---|---|
| P1 | M + A − D |
| P2 | M − A − D |
| F1 | M + D |
| F2 | M |
| BC1 | M + ½ A |
| BC2 | M − ½ A |
| F3 | M − ½ D |
| BCX | M |
M is the overall mean (standardized to the F2 generation), A is the additive effect, and D is the dominance deviation.
If the number of estimated line-cross derivative means exceeds the number of estimated parameters, then a test of model sufficiency is possible. Thus, we require only four line-cross derivatives to test the additive-dominance model, but more derivatives (up to eight here) increase power to detect epistasis. The hypothesis test for a particular line-cross family is similar to that used previously. I first established expected values for each line-cross derivative using the estimated values for M, A, and D. Deviations between observed and expected means were measured using a chi-square statistic. This statistic was then compared to the chi-square distribution with the number of degrees of freedom equal to the number of line-cross derivatives minus the number of estimated parameters (three).
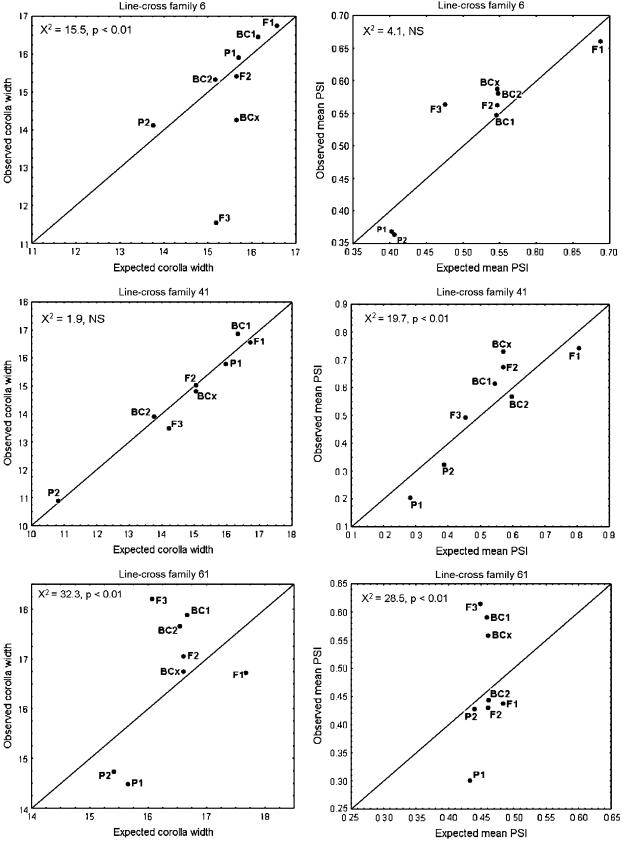
Figure 3 provides a sample of results for corolla width and PSI within three line-cross families. The degree of correspondence between the observed means of line-cross derivatives and those predicted from the nonepistatic model is highly variable, both among line-cross families for a particular character and among traits within a line-cross family. For most characters, the magnitude and direction of deviations for particular derivatives are idiosyncratic. For example, the observed mean corolla width of F3 plants is substantially less than expected in line-cross family 6, slightly less than expected in family 41, but substantially greater than expected in family 61 (Figure 3, left column). However, the nonlinear relationship between PSI and inbreeding coefficient (Figure 2) does yield some patterns for this trait. Consistent with a convex relationship, observed means are often less than expected for the line-cross derivatives at the ends of the range of inbreeding coefficients (P1, P2, and F1), but greater than expected for derivates with intermediate inbreeding coefficients (Figure 3, right column).
Figure 3.
The correspondence between observed and expected phenotypic values is depicted for corolla width (left column) and PSI (right column) within three different line-cross families. Each point denotes a particular line-cross derivative. The chi-square and P-values are based on that trait within that line-cross family.
Results from the full collection of line-cross families can be synthesized into a single test by noting that each family is independent. Each one is based on measurements of distinct plants derived from distinct lines, randomly extracted from the ancestral population. As a consequence, I can simply sum the chi-square values and degrees of freedom across families. This provides an overall test for each trait and eliminates any “multiple-tests problem” associated with attempting to draw conclusions from the collection of P-values from individual line-cross families. The cumulative chi-square values and degrees of freedom are given in Table 4. In the full data set, all traits exhibit highly significant deviations from the nonepistatic model.
TABLE 4.
Results from the replicated joint scaling test (see text for details)
| d.f. total | Cumulative χ2 | |
|---|---|---|
| Untransformed trait values | ||
| Corolla width | 306 | 628.5*** |
| Days to flower | 307 | 573.3*** |
| Corolla length | 296 | 411.3*** |
| Pistil length | 295 | 451.3*** |
| Anther length | 297 | 495.0*** |
| Stigma-anther separation | 295 | 477.9*** |
| Pollen no. | 304 | 496.0*** |
| PSI | 304 | 785.5*** |
| Viable pollen | 304 | 598.2*** |
| Log-transformed pollen values | ||
| Ln(pollen no.) | 304 | 475.9*** |
| Ln(PSI) | 304 | 731.4*** |
| Ln(viable pollen) | 304 | 600.5*** |
Significance levels: ***P < 0.001.
The complete collection of estimated values (across line-cross families) allows a range of comparisons. For example, the correlations of F1 means for different traits are given in Table 5. Measures of flower size (corolla width, corolla length, anther length, and pistil length) exhibited strong, positive correlations. The number of pollen grains and the estimated number of viable grains showed lower, but still significantly positive correlations with flower size. There was a significant positive relationship between number of pollen grains and PSI (see also Willis 1999b). Stigma-anther separation was negatively correlated with PSI. Development time was not significantly correlated with any other measure, at least not for F1 mean values.
TABLE 5.
The correlations of F1 means between different traits (each line-cross family contributes a single value for each trait)
| Days to flower | Corolla length | Pistil length | Anther length | SA separation | Pollen no. | PSI | Viable pollen | |
|---|---|---|---|---|---|---|---|---|
| Corolla width | 0.11 | 0.76 | 0.68 | 0.50 | 0.37 | 0.28 | 0.04 | 0.24 |
| Days to flower | 0.14 | 0.12 | 0.09 | 0.06 | −0.02 | −0.04 | 0.00 | |
| Corolla length | 0.82 | 0.77 | 0.23 | 0.23 | −0.04 | 0.16 | ||
| Pistil length | 0.77 | 0.51 | 0.24 | −0.09 | 0.13 | |||
| Anther length | −0.15 | 0.32 | 0.07 | 0.26 | ||||
| SA separation | −0.07 | −0.27 | −0.19 | |||||
| Pollen no. | 0.30 | 0.87 | ||||||
| PSI | 0.71 |
Italic entries indicate that the correlation is significantly different from zero.
SA, stigma-anther.
Given the variability among line-cross families, it is worthwhile to consider the factors most closely related to the magnitude of deviations from the nonepistatic model. One might expect the chi-square value to increase with the amount of phenotypic differentiation between parental lines (given the greater evidence for epistasis in inter- vs. intraspecific crosses, see Introduction). However, this expectation is not born out in these data: There was no relationship between chi square and the amount of difference between P1 and P2 for any character.
The majority of estimates for D, the dominance deviation, were negative for days to flower, but positive for other traits. These observations are consistent with “heterosis,” the tendency for crossing of lines to increase fitness or yield. Moll et al. (1965) noted that the magnitude of heterosis in corn appears to be greatest in crosses between lines that are divergent to an intermediate degree (Lynch and Walsh 1998, pp. 225–226). I plotted estimates of D vs. the absolute phenotypic difference between P1 and P2 for each character. As with chi-square values, the relationships between line differentiation and D-estimates were weak and generally nonsignificant.
Scale transformation:
The logarithm is only one of many nonlinear but monotonic functions that can be applied to measurements prior to analysis. The simplest form of epistasis is scalar. Loci affecting a trait contribute additively to a genetic value, g. The phenotype, z, is equal to F(g + e), where e is the environmental effect and F(*) is a simple nonlinear, but generally monotonic, function. Scalar epistasis can be “removed” by applying a transformation to the measurements: z′ = F−1(z) = g + e. The standard model accurately describes inheritance of the transformed variable, z′.
It is thus useful to determine if the significant deviations from the additive-dominance model (documented in Table 6) can be eliminated by applying scale transformations. To evaluate this possibility, I applied the power series transformations over the four orders of magnitude spanning the original scale of measurement (Box and Cox 1964; Sokal and Rohlf 2000, p. 419). Letting Z denote the original values, the following series of transformations were applied to each trait: 1/Z, 1/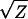 , Ln(Z),
, Ln(Z), 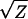 , Z2, and Z3. While scale transformation did reduce cumulative chi-square values of some traits, all replicated joint scaling tests remained highly significant.
, Z2, and Z3. While scale transformation did reduce cumulative chi-square values of some traits, all replicated joint scaling tests remained highly significant.
TABLE 6.
The cumulative χ2 values are given from replicated joint scaling tests on transformed values of each trait
| Trait | 1/Z | 1/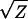
|
Ln(Z) | 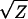 |
Z | Z2 | Z3 |
|---|---|---|---|---|---|---|---|
| Corolla width | 593 | 601 | 610 | 618 | 629 | 656 | 692 |
| Days to flower | 609 | 601 | 593 | 583 | 573 | 551 | 526 |
| Corolla length | 414 | 414 | 414 | 413 | 411 | 408 | 403 |
| Pistil length | 439 | 445 | 449 | 451 | 452 | 448 | 439 |
| Anther length | 526 | 520 | 513 | 505 | 495 | 474 | 449 |
| SA separation | 439 | 458 | 469 | 471 | 478 | 492 | 504 |
| Pollen no. | 372 | 436 | 476 | 494 | 496 | 471 | 425 |
| PSI | 605 | 674 | 731 | 769 | 786 | 776 | 738 |
| Viable pollen | 372 | 508 | 601 | 620 | 598 | 512 | 437 |
The function used for transformation is given at the top of each column. The degrees of freedom for each trait are the same as those in Table 4. For SA separation and PSI, a constant was added to all measurements (to ensure that all values were positive) prior to transformation. All χ2 values are statistically significant; P < 0.01 in all cases.
SA, stigma-anther.
The same set of scale transformations can also be applied to measurements prior to the combined population analysis. For most characters this is unwarranted given the approximately linear response to inbreeding on the original scale of measurement. The reciprocal (1/Z) and 1/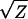 transformations do reduce the magnitude of nonlinearity in PSI and viable pollen, but deviations remain significant. Moreover, since scale of measurement is not arbitrary for fitness components (or at least not for their evolutionary interpretation), an explicit nonlinear regression is more informative (Figure 2B, see discussion).
transformations do reduce the magnitude of nonlinearity in PSI and viable pollen, but deviations remain significant. Moreover, since scale of measurement is not arbitrary for fitness components (or at least not for their evolutionary interpretation), an explicit nonlinear regression is more informative (Figure 2B, see discussion).
DISCUSSION
Epistasis seems inevitable when one considers the interconnected nature of biochemical pathways and gene networks. Yet evidence of epistasis in quantitative character variation is surprisingly weak, at least within contiguous natural populations (demes). This study supports the view that our failure to see epistasis is not due to its absence. Statistical power is a central issue in practically all studies of quantitative variation. Attempts to distinguish the contributions of additive effects, dominance deviations, and gene interactions to trait variation are confounded by the large statistical error associated with variance estimates. This analysis of M. guttatus, based on comparisons of phenotypic means, provides clear evidence of epistasis in development time, floral morphology, and male fitness components.
The increasing use of genomic techniques, e.g., QTL mapping, is frequently cited as the key to elucidating the contribution of epistasis to quantitative trait variation (e.g., Erickson et al. 2004). The measured genotype approach, in which phenotype is determined for collections of individuals sharing the same molecular genotypes, has yielded both positive and negative results regarding the importance of epistasis (positive results: Long et al. 1995; Clark and Wang 1997; Elena and Lenski 1997; Routman and Cheverud 1997; Lukens and Doebley 1999; Templeton 2000) (negative results: Tanksley 1993 and references therein). In some studies (e.g., Weber et al. 1999), results have been ambiguous despite extensive replication. The ability to isolate particular genomic regions and directly associate phenotype with a limited collection of genotypes is certainly a favorable feature of measured genotype studies. However, their power to detect epistasis is largely a consequence of the fact that these studies are simple comparisons of mean values, those associated with different multilocus genotypes.
While based on phenotypic values, the line-cross methodology provides a surprisingly simple and direct test for epistasis (Mather and Jinks 1982). The method capitalizes on the essential simplification of the additive/dominance model, the direct extrapolation from individual loci to the whole genotype. For a single locus with two alleles, there are three distinct genotypes and thus three mean values to be estimated. Given estimates for these genotypic values, one can predict the mean phenotype of all descendants from a cross between the alternative homozygotes. Under the additive/dominance model, the contributions of different loci sum and single-locus effects conveniently distill into aggregate parameters (Table 3, M, A, and D). These are sufficient to predict the means of all line-cross derivatives. If there is epistasis, single-locus effects do not combine in a simple fashion and the aggregate parameters will not accurately predict the means of all line-cross derivatives. This is the basis of the test.
When epistasis is detected in line-cross studies, the general practice is to parse deviations from the nonepistatic model into components, e.g., the additive-by-additive and additive-by-dominance components (Mather and Jinks 1982; Clark and Wang 1997; Routman and Cheverud 1997). I have not done this here. The main practical difficulty is that many of the line-cross families are incomplete, with one or more line-cross derivatives absent. Because the type and complexity of model that can be fit to the data depend on the number and type of derivatives that are present, different sorts of model would have to be fit to different line-cross families. This hinders interpretation because the same genetic effect would likely be absorbed into different terms (e.g., additive-by-additive instead of additive-by-dominance) in different families.
The larger conceptual difficulty is that, beyond documenting the existence of epistasis, interaction variance statistics have limited inferential value. They are useful in predicting how a population will respond to inbreeding or genetic drift (Kempthorne 1957; Goodnight 1988), but essentially uninformative about the nature of gene interactions (Mackay 2001). Phillips et al. (2000, p. 26) go as far at to state that the “epistatic variance is so far removed from the actual interactions as to be basically useless for determining the impact of gene interactions on evolutionary processes.” There does not seem to be any simple, general relationship between interaction variance statistics and response to selection. In contrast, the standard additive genetic variance is useful for predicting response to selection even when there is epistasis (Turelli and Barton 1994, p. 933; see below).
The nature of gene interactions:
The standard nonepistatic model of quantitative genetics is often considered a linear approximation of the complicated mapping from genotype to phenotype. It is thus natural to attempt to characterize gene interactions with an explicit nonlinear function that predicts phenotype from one or more genetic metrics. These simple kinds of gene interaction, denoted “directional epistasis” by Phillips et al. (2000), have been considered in a variety of evolutionary models. Epistatic interactions among deleterious mutations have been characterized using a function with fitness as the dependent variable and the total number of deleterious mutations (summed across loci) as the independent variable. Different treatments have considered a quadratic function (Kimura and Maruyama 1966), an exponential quadratic function (Charlesworth 1990; Charlesworth et al. 1991), or a step function (Kondrashov 1985, 1988). Rice (1998, 2000) models the phenotype as a nonlinear function of multiple “underlying factors.” These factors may then be treated as genetic and environmental effects in the standard quantitative genetic model. Turelli and Orr (1995) model hybrid breakdown by allowing loci to contribute additively to a “breakdown scale” and assume that fitness is a monotonically decreasing function of this breakdown value (see also Turelli and Orr 2000). Finally, Hansen and Wagner (2001) analyze a model in which the effects of allelic differences at a particular locus are a linear function of the genetic background.
The utility of these approaches, in which the interaction of many loci is distilled into a simple mathematical function, has become a subject of debate. These models will probably not prove to be mechanistically accurate in most cases. However, they may nonetheless capture essential features of the genotype-to-phenotype mapping, allowing theorists to consider otherwise intractable problems and providing experimentalists with meaningful parameters to estimate. Countering this view, Phillips et al. (2000) argue that the multidimensional nature of genotypic space cannot be justifiably reduced to simple one- or two-dimensional representations.
The main purpose of the combined population analysis (Table 2, Figure 2) is to identify simple directional forms of gene interaction, at least types that generate a nonlinear relationship between average trait values and f, the inbreeding coefficient of plants. If loci contribute additively to trait variation, the mean phenotype should change linearly with f (Wright 1951; Kempthorne 1957). If loci contribute multiplicatively, the relationship should be log-linear. Floral morphology and pollen number vary in an approximately linear way with f, while PSI and viable pollen exhibited significant deviations from linearity. The relationship between log-transformed PSI and f is more accurately described by a quadratic function (Figure 2B, dashed line) than by a line. This suggests that pollen viability corresponds more closely to the exponential quadratic model than to either the additive or the multiplicative model.
The response to inbreeding test for epistasis has been applied to a substantial number of species, both natural and agricultural (Lynch and Walsh 1998, Chap. 10). Both linear (e.g., Hallauer and Sears 1973) and nonlinear (e.g., Carr and Dudash 1997) responses have been documented, but much of the evidence is equivocal. Most experiments fail to include appropriate controls or suffer statistical flaws (Lynch 1988; Lynch and Walsh 1998, p. 272). Oftentimes, studies have been conducted over multiple generations with different levels of inbreeding assessed in different generations, which makes it impossible to distinguish inbreeding effects from environmental effects (e.g., differences in growth conditions in different generations). Statistical power is also limited when the maximum inbreeding coefficient is low (<0.25). A linear model will likely provide a good fit within a limited range of values for the inbreeding coefficient, regardless of the level of epistasis.
Perhaps the most serious difficulty in interpreting previous results concerns the potential effects of genetic purging. The linear prediction is based on the assumption that experimental inbreeding changes genotype frequencies without changing allele frequencies. Random fluctuations in allele frequencies can be accounted for in a statistical analysis, but deterministic changes are assumed to be absent. Unfortunately, some allele frequency changes are inevitable during line formation. Lines that become homozygous for mutations that are lethal or cause sterility in homozygous form will go extinct and eliminate these alleles from the experimental population. Selection within lines will have similar effects even in lineages that survive. As a consequence, the frequencies of deleterious mutations will differ between inbred and outbred individuals. Depending on circumstances, genetic purging of this kind can either produce artificial nonlinearities or obscure genuine epistasis.
This experiment was designed to avoid this list of difficulties. An advantage of using fully inbred lines as parents in a biometric breeding design is that they are already fully purged. Subsequent inbreeding or outcrossing should result in predictable changes in homozygosity without changes in allele frequency (apart from the random changes due to finite sample sizes). This experiment also grew plants spanning the full range of inbreeding coefficients simultaneously. The multigenerational crossing scheme eliminated any association between seed age and inbreeding coefficient. Reciprocal crosses between plants inbred to differing degrees disentangle individual and maternal inbreeding effects.
Lynch (1988) also noted that many statistical analyses of response to inbreeding have failed to account for the genetic relatedness of individuals within an inbreeding lineage. This can elevate the type I error rate, increasing the likelihood of “significant” nonlinearity even when there is no epistasis. The family structure of the present experiment is fully accounted in Replicated joint scaling test, but not in Combined population analysis. However, the nonlinearity apparent for log-transformed male fitness measures (Table 2, Figure 2B) is not a statistical artifact. Seventy-three line-cross families produced at least one plant that was fully outbred (f = 0), inbred to an intermediate level (f = 0.5), and fully inbred (f = 1). We can distill each of these families into a pair of values: Δ1 is the difference in mean trait value between plants with f = 0 and f = 0.5 and Δ2 is the difference between mean values for plants with f = 0.5 and f = 1. If the true relationship between trait values and inbreeding coefficient is linear, we expect that Δ1 will be >Δ2 in about half of the families. With synergistic epistasis, Δ2 should be >Δ1 in most line-cross families. For Ln[PSI] in this study, 52 of 73 cross families exhibited a greater decline at higher inbreeding coefficients (Δ2 > Δ1). For Ln[viable pollen], Δ2 > Δ1 in 48 of 73 families. Both trends are statistically significant (Wilcoxon sign rank test, P < 0.01).
Synergism in monkeyflowers:
The response of M. guttatus to inbreeding has been investigated extensively (Willis 1993a,b, 1996, 1999a,b; Carr and Dudash 1995, 1996, 1997; Carr et al. 1997; Dudash and Carr 1998; Kelly and Willis 2001; Carr and Eubanks 2002; Carr et al. 2003). Willis (1993a) and Carr and Dudash (1997) measured fitness-related characters of plants with a range of inbreeding coefficients and their results are thus comparable to those in Table 2. Carr and Dudash (1997) generated plants of six different inbreeding levels (f = 0, 0.5, 0.75, 0.875, 0.938, and 0.969) from two California populations, “S” and “T.” They estimated the relationship between inbreeding coefficient and mean trait values to be approximately linear for female fitness measures in both populations. However, male fitness in population T exhibited an accelerating decline with f, a result consistent with synergistic epistasis. Their measure of male fitness is most directly comparable to viable pollen of this study.
Willis (1993a) synthesized plants with four levels of inbreeding (f = 0, 0.25, 0.5, and 0.75) from two Oregon populations, “Cone Peak” and the Iron Mountain population that I consider here. Pollen viability exhibited synergistic epistasis for the Cone Peak population, but the relationship with f was not significantly nonlinear for the Iron Mountain plants. In contrast, the present study of Iron Mountain provides compelling evidence for synergism in log-transformed values of both pollen viability (as estimated by PSI) and total viable pollen (Figure 2B). The difference may simply reflect statistical power. Willis (1993a) did note a nonsignificant trend toward synergism in his study (see Figure 1f of that article) and the present study of Iron Mountain considers a larger range of inbreeding coefficients with ∼10 times as many plants.
Perhaps the simplest explicit genetic model that yields synergism is complementary gene action (Brookfield 1997). Consider two loci involved in pollen production. Each locus has recessive mutations that reduce pollen viability. If the two loci are complementary (functional duplicates in the extreme case), pollen viability is significantly reduced only in individuals that are homozygous for mutant alleles at both loci. Loss or reduction of function at only one locus is effectively compensated by full function of the complementary locus. This mode of action has been demonstrated for some chlorophyll deficiency mutations in M. guttatus (Willis 1992). More generally, the high levels of genetic redundancy apparent in the Arabidopsis genome suggest that complementary gene action may not be uncommon.
Variable epistatic effects:
Flower traits, development time, and pollen number did not exhibit nonlinear responses to inbreeding. However, all yielded highly significant evidence of epistasis in the replicated joint scaling tests (Figure 3, Table 4). Moreover, the results from each character remained significant even after a full series of scale transformations (Table 6), which indicates gene interactions are not scalar in nature. This difference in outcomes between the two analyses suggests that epistatic interactions may be variable across the loci influencing these traits (see, for example, de Visser et al. 1997). Certain pairs of loci may interact strongly and others may act more or less independently. However, it would be premature to conclude that these data are inconsistent with more complicated models of directional epistasis (e.g., Rice 1998, 2000; Hansen and Wagner 2001)only on the basis of Table 6.
Scale transformations could often eliminate significant deviations between observed and expected values within individual line-cross families. For example, cubing values for corolla width reduces chi square for line-cross family 6 from 15.5 to 9.7, which is not significant given 5 d.f. (see Figure 3, top left). Unfortunately, the same transformation exaggerates deviations in other families such that the cumulative chi square actually increases. This result is reminiscent of Powers' studies of tomato fruit characters, in which transformations useful in some crosses (yielding approximate additivity) were detrimental in others (Powers 1950, 1951).
Comparisons among line-cross families are also informative about the nature of gene interactions. Noting the greater evidence of epistasis in interspecies crosses, one might expect that line-cross families in which P1 and P2 are very different would yield higher chi-square values than line-cross families constituted from phenotypically similar lines. This prediction may also follow from the idea that the nonepistatic model is a “linear approximation” of the nonlinear mapping from genotype to phenotype. Linear approximations are generally taken around a specific point on the phenotypic axis and are most accurate close to that point. It is thus noteworthy that, in contrast to these expectations, I failed to find any relationship between the level of phenotypic differentiation of parental lines and the strength of evidence for epistasis.
Maternal effects:
In both plants and animals, an organism's phenotype is often substantially influenced by the characteristics of its mother. Seed provisioning, in terms of both nutrients and genetic instruction (mRNAs), is a likely avenue for maternal effects in plants. Since inbreeding clearly influences individual traits, it is reasonable to expect that it will also affect the quality of maternal effects. In fact, this has been clearly demonstrated in both animals (White 1972) and plants (Melchinger et al. 1986; Vogler et al. 1999).
Surprisingly, I found little evidence of maternal inbreeding effects in this study, despite both large sample sizes and strong comparisons (Table 2). The latter refers to line-cross derivatives synthesized by reciprocal crosses between parents with substantially different inbreeding coefficients (BC1, BC2, and BCX). Which parent served as mother seemed to have minimal effects on most traits. Only days to flower exhibited a significant effect. All else being equal, plants with fully inbred mothers produced their first flower an average of 0.25 days later than plants with outbred mothers.
Maternal effects may be significant but unrelated to level of inbreeding or at least only weakly related. For this reason, the plants derived from a single cross (a full-sib family) may exhibit a consistent deviation from the expected value of their line-cross derivative even without epistasis. These deviations average out if each line-cross derivative is synthesized from a substantial number of distinct crosses (e.g., Jinks and Perkins 1969). However, with a limited number of crosses per derivative, maternal effects must be accounted statistically to distinguish them from epistasis. The sequel to this article develops the theory necessary for a more rigorous treatment of the data, on the basis of not only the means of line-cross derivatives but also patterns of variability within and between derivatives (J. K. Kelly, unpublished results). A maximum-likelihood-based application of this theory to the present data set confirms that both maternal effects and epistasis contribute to deviations between observed and expected line-cross means. However, epistasis remains the primary cause.
Evolutionary implications:
The purpose of this study was to determine if gene interactions influence characters of M. guttatus, and if they do, to investigate the nature of these interactions. Regarding the first part, there does seem to be significant epistasis affecting development time, floral morphology, and male fitness components. Male fitness involves synergism, a tendency for alleles at different loci that reduce pollen viability to have greater effects when combined than in isolation (Figure 2B). Synergism among deleterious mutations is a central tenet of the mutational deterministic hypothesis for the evolutionary maintenance of sexual reproduction (Charlesworth 1990; Kondrashov 1993). Synergistic interactions can substantially reduce the mutational load in a sexual population, but not in an asexual population (Kimura and Maruyama 1966). This, combined with a sufficiently high rate of deleterious mutation, can allow sexual genotypes to outcompete asexual genotypes in the face of the twofold cost of sex.
Despite the results of Figure 2B, support for the mutational deterministic model from M. guttatus remains equivocal. First, the quantitative relationship between viable pollen per flower (as measured here) and evolutionary fitness in the wild has yet to be determined empirically. Synergism between mutations affecting the latter is necessary for the model. Second, estimates for the genomic deleterious mutation rate in M. guttatus fall largely within the range 0.1–1.0 (Willis 1999a; Kelly 2003). These estimates are somewhat lower than what is thought necessary to favor sex and, for a variety of reasons, are more likely to be overestimates than underestimates.
Flower morphology and development time yielded negative results in the combined population analysis, but positive evidence of epistasis in the replicated joint scaling test. Such a difference is expected if epistatic effects are variable across genetic backgrounds or sets of interacting loci. Similar results have been obtained in measured genotype studies (de Visser et al. 1997; Elena and Lenski 1997). These findings are intriguing in light of theoretical studies exploring the implications of variable interactions among loci (Phillips et al 2000). To consider an example noted previously, consistent synergistic epistasis among deleterious mutations can favor increased rates of recombination (Charlesworth 1990; Barton 1995). If interactions are variable, however, modifiers that reduce the rate of recombination may actually be favored (Otto and Feldman 1997).
An important question is how the existence of epistasis affects the application of quantitative genetics methods. Should we disregard analyses or conclusions based on the additive/dominance model if there is epistasis? Perhaps the most fundamental of such applications is the “breeder's equation” for predicting response to selection (Lush 1937). Under this model, the change in mean phenotype, 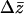 , equals the product of the narrow sense heritability (
, equals the product of the narrow sense heritability (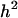 ) and the selection differential (S). When selection is imposed on multiple traits, the predicted response is proportional to the additive genetic variance/covariance matrix (Lande and Arnold 1983; Falconer and Mackay 1996). In either case, the genetic quantities can be estimated from the resemblance of relatives, independent of the observed response to selection.
) and the selection differential (S). When selection is imposed on multiple traits, the predicted response is proportional to the additive genetic variance/covariance matrix (Lande and Arnold 1983; Falconer and Mackay 1996). In either case, the genetic quantities can be estimated from the resemblance of relatives, independent of the observed response to selection.
The breeder's equation, and its multitrait generalization, is one of the most important predictive models in evolutionary biology (Grant and Grant 1995) and it is worth considering whether it fails when there is epistasis. This can be directly evaluated for corolla width of M. guttatus, one of the traits shown to exhibit epistasis here (Table 4). We have estimated the heritability of this character for the Iron Mountain population (Kelly and Arathi 2003) and, in other experiments, have imposed selection on corolla diameter (e.g., Kelly and Willis 2001). In the largest of these studies, bidirectional truncation selection was imposed for seven generations. The observed divergence between the means of high- and low-selected populations (5.2 phenotypic standard deviations) differs from the prediction of the breeder's equation by <6% (J. K. Kelly, unpublished results). Given the myriad of factors other than epistasis that can generate deviations, one has to conclude that the model performs quite well.
These results from M. guttatus are complementary to those from previous studies. Clayton et al. (1957) demonstrated the accuracy of the breeder's equation in predicting short-term evolution of bristle number in Drosophila melanogaster. While this trait was often cited as an example of additive inheritance, mapping studies have recently demonstrated that variation is substantially influenced by interactions among QTL (Long et al. 1995, 1996; Mackay 2001). Two complementary conclusions emerge from these selection experiments. First, short-term responses to selection that are consistent with the additive/dominance model (breeder's equation) do not imply the absence of epistasis in the selected traits. Second, the existence of epistasis does not necessarily invalidate conclusions derived from simpler, nonepistatic models.
It seems likely that the additive genetic variance is itself a function of genetic interactions among QTL. In fact, theorists have begun to explore the interaction of epistasis, mutation, and natural selection in determining the genetic architecture of quantitative characters (e.g., Hermisson et al. 2003). Experimentalists will continue to identify and describe particular gene interactions, at least in model species (Templeton 2000). At present, there is a substantial gap between theory and experiment. I suggest that this is due, at least in part, to the absence of meaningful statistics, measurable quantities with clear relevance to evolutionary questions. Identifying such statistics, if they do exist, should facilitate the synthesis of theory and experiment.
Acknowledgments
I thank T. Marriage, J. Gleason, L. Holeski, M. Orive, D. Houle, and two anonymous reviewers for comments on this manuscript. Kim Milne, L. Holeski, B. Harris, R. How, J. Schaller, D. Deole, L. Richman, H. Nelson, and S. Jablonski each helped to complete the enormous volume of crossing and plant measurements.
References
- Anderson, V. L., and O. Kempthorne, 1954. A model for the study of quantitative inheritance. Genetics 30: 883–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arathi, H. S., A. Rasch, C. Cox and J. K. Kelly, 2002. Autogamy and floral longevity in Mimulus guttatus. Int. J. Plant Sci. 163: 567–573. [Google Scholar]
- Barker, J. S. F., 1974. The state of information concerning deviations from additivity of gene effects. Proceedings of the 1st World Congress on Genetics Applied to Livestock Production, Madrid, Vol. 1, pp. 373–383.
- Barker, J. S. F., 1979. Inter-locus interactions: a review of experimental evidence. Theor. Popul. Biol. 16: 323–346. [DOI] [PubMed] [Google Scholar]
- Barton, N. H., 1995. A general model for the evolution of recombination. Genet. Res. 65: 123–144. [DOI] [PubMed] [Google Scholar]
- Box, G. E., and D. R. Cox, 1964. An analysis of transformations. J. R. Stat. Soc. B 26: 211–252. [Google Scholar]
- Brookfield, J. F. Y., 1997. Genetic redundancy. Adv. Genet. 36: 137–155. [DOI] [PubMed] [Google Scholar]
- Burton, R. S., 1990. Hybrid breakdown in development time in the copepod Tigriopus californicus. Evolution 44: 1814–1822. [DOI] [PubMed] [Google Scholar]
- Burton, R. S., P. D. Rawson and S. Edmands, 1999. Genetic architecture of physiological phenotypes: empirical evidence for coadapted gene complexes. Am. Zool. 39: 451–462. [Google Scholar]
- Cabot, E. L., A. W. Davis, N. A. Johnson and C. I. Wu, 1994. Genetics of reproductive isolation in the Drosophila simulans clade: complex epistasis underlying hybrid male sterility. Genetics 137: 175–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr, D. E., and M. R. Dudash, 1995. Inbreeding depression under a competitive regime in Mimulus guttatus: consequences for potential male and female function. Heredity 75: 437–445. [Google Scholar]
- Carr, D. E., and M. R. Dudash, 1996. Inbreeding depression in two species of Mimulus (Scrophulariaceae) with contrasting mating systems. Am. J. Bot. 83: 586–593. [Google Scholar]
- Carr, D. E., and M. R. Dudash, 1997. The effects of five generations of enforced selfing on potential male and female function in Mimulus guttatus. Evolution 51: 1797–1807. [DOI] [PubMed] [Google Scholar]
- Carr, D. E., and M. D. Eubanks, 2002. Inbreeding alters resistance to insect herbivory and host plant quality in Mimulus guttatus (Scrophulariaceae). Evolution 56: 22–30. [DOI] [PubMed] [Google Scholar]
- Carr, D. E., and C. B. Fenster, 1994. Levels of genetic variation and covariation for Mimulus (Scrophulariaceae) floral traits. Heredity 72: 606–618. [Google Scholar]
- Carr, D. E., C. B. Fenster and M. R. Dudash, 1997. The relationship between mating-system characters and inbreeding depression in Mimulus guttatus. Evolution 51: 363–372. [DOI] [PubMed] [Google Scholar]
- Carr, D. E., J. F. Murphy and M. D. Eubanks, 2003. The susceptibility and response of inbred and outbred Mimulus guttatus to infection by Cucumber mosaic virus. Evol. Ecol. 17: 85–103. [Google Scholar]
- Charlesworth, B., 1990. Mutation-selection balance and the evolutionary advantage of sex and recombination. Genet. Res. 55: 199–221. [DOI] [PubMed] [Google Scholar]
- Charlesworth, B., M. T. Morgan and D. Charlesworth, 1991. Multilocus models of inbreeding depression with synergistic selection and partial self-fertilization. Genet. Res. 57: 177–194. [Google Scholar]
- Clark, A. G., and L. Wang, 1997. Epistasis in measured genotypes: Drosophila P-element insertions. Genetics 147: 157–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton, G. A., J. A. Morris and A. Robertson, 1957. An experimental check on quantitative genetic theory. I. Short term responses to selection. J. Genet. 5: 131–151. [Google Scholar]
- Coyne, J. A., N. H. Barton and M. Turelli, 1997. Perspective: a critique of Sewall Wright's shifting balance theory of evolution. Evolution 51: 643–671. [DOI] [PubMed] [Google Scholar]
- Crow, J. F., and M. Kimura, 1970. An Introduction to Population Genetics Theory. Harper & Row, New York.
- de Visser, J. A. G. M., R. F. Hoekstra and H. van den Ende, 1997. Test of interaction between genetic markers that affect fitness in Aspergillus niger. Evolution 51: 1499–1505. [DOI] [PubMed] [Google Scholar]
- Dobzhansky, T. H., 1936. Studies on hybrid sterility. II. Localization of sterility factors in Drosophila pseudoobscura hybrids. Genetics 21: 113–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley, J., A. Stec and C. Gustus, 1995. teosinte branched1 and the origin of maize: evidence for epistasis and the evolution of dominance. Genetics 141: 333–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dole, J. A., 1990. Role of corolla abscission in delayed self-pollination of Mimulus guttatus (Scrophulariaceae). Am. J. Bot. 77: 1505–1507. [Google Scholar]
- Dole, J. A., and K. Ritland, 1993. Inbreeding depression in 2 Mimulus taxa measured by multigenerational changes in the inbreeding coefficient. Evolution 47: 361–373. [DOI] [PubMed] [Google Scholar]
- Dudash, M. R., and D. E. Carr, 1998. Genetics underlying inbreeding depression in Mimulus with contrasting mating systems. Nature 393: 682–684. [Google Scholar]
- Dudash, M. R., and K. Ritland, 1991. Multiple paternity and self-fertilization in relation to floral age in Mimulus guttatus (Scrophulariaceae). Am. J. Bot. 78: 1746–1753. [Google Scholar]
- Elena, S. F., and R. E. Lenski, 1997. Test of synergistic interactions among deleterious mutations in bacteria. Nature 390: 395–398. [DOI] [PubMed] [Google Scholar]
- Erickson, D. L., C. B. Fenster, H. K. Stenoien and D. Price, 2004. Quantitative trait locus analyses and the study of evolutionary process. Mol. Ecol. 13: 2505–2522. [DOI] [PubMed] [Google Scholar]
- Falconer, D. S., and T. F. C. Mackay, 1996. Introduction to Quantitative Genetics. Prentice Hall, London. [DOI] [PMC free article] [PubMed]
- Fenster, C. B., and L. F. Galloway, 2000. Population differentiation in an annual legume: genetic architecture. Evolution 54: 1157–1172. [DOI] [PubMed] [Google Scholar]
- Fenster, C. B., and K. Ritland, 1992. Chloroplast Dna and isozyme diversity in 2 Mimulus species (Scrophulariaceae) with contrasting mating systems. Am. J. Bot. 79: 1440–1447. [Google Scholar]
- Fenster, C. B., and K. Ritland, 1994. a Evidence for natural-selection on mating system in Mimulus (Scrophulariaceae). Int. J. Plant Sci. 155: 588–596. [Google Scholar]
- Fenster, C. B., and K. Ritland, 1994. b Quantitative genetics of mating system divergence in the yellow monkeyflower species complex. Heredity 73: 422–435. [Google Scholar]
- Fenster, C. B., L. F. Galloway and L. Chao, 1997. Epistasis and its consequences for the evolution of natural populations. Trends Ecol. Evol. 12: 282–286. [DOI] [PubMed] [Google Scholar]
- Fisher, R. A., 1958. The Genetical Theory of Natural Selection. Dover, New York.
- Fishman, L., and J. Willis, 2001. Evidence for Dobzhansky-Muller incompatibilites contributing to the sterility of hybrids between Mimulus guttatus and M. nasutus. Evolution 55: 1932–1942. [DOI] [PubMed] [Google Scholar]
- Ford, E. B., 1971. Ecological Genetics. Chapman & Hall, London.
- Goodnight, C. J., 1988. Epistasis and the effect of founder events on the additive genetic variance. Evolution 42: 441–454. [DOI] [PubMed] [Google Scholar]
- Grant, A. L., 1924. A monograph of the genus Mimulus. Ann. Mo. Bot. Gard. 11: 99–398. [Google Scholar]
- Grant, P. R., and B. R. Grant, 1995. Predicting microevolutionary responses to directional selection on heritable variation. Evolution 49: 241–251. [DOI] [PubMed] [Google Scholar]
- Grant, V., 1975. Genetics of Flowering Plants. Cambridge University Press, New York.
- Hallauer, A. R., and J. H. Sears, 1973. Changes in quantitative traits associated with inbreeding in a synthetic variety of maize. Crop Sci. 13: 327–330. [Google Scholar]
- Hansen, T. F., and G. Wagner, 2001. Modeling genetic architecture: a multilinear theory of gene interaction. Theor. Popul. Biol. 59: 61–86. [DOI] [PubMed] [Google Scholar]
- Hard, J. J., W. E. Bradshaw and C. M. Holzapfel, 1992. Epistasis and the genetic divergence of photoperiodism between populations of the pitcher-plant mosquito, Wyeomyia smithii. Genetics 131: 389–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayman, B. I., 1960. The separation of epistatic from additive and dominance variation in generation means. Genetica 31: 371–390. [DOI] [PubMed] [Google Scholar]
- Hermisson, J., T. F. Hansen and G. P. Wagner, 2003. Epistasis in polygenic traits and the evolution of genetic architecture under stabilizing selection. Am. Nat. 161: 708–734. [DOI] [PubMed] [Google Scholar]
- Husband, B. C., and S. C. H. Barrett, 1992. a Effective population size and genetic drift in tristylous Eichornia paniculata (Pontederiaceae). Evolution 46: 1875–1890. [DOI] [PubMed] [Google Scholar]
- Husband, B. C., and S. C. H. Barrett, 1992. b Genetic drift and the maintenance of the style length polymorphism in tristylous populations of Eichornia paniculata (Pontederiaceae). Heredity 69: 440–449. [Google Scholar]
- Jinks, J. L., and J. M. Perkins, 1969. The detection of linked epistatic genes for a metrical trait. Heredity 24: 465–475. [DOI] [PubMed] [Google Scholar]
- Johnson, N. A., 2000. Gene interactions and the origin of species, pp. 197–212 in Epistasis and the Evolutionary Process, edited by J. B. Wolf, E. D. Brodie and M. J. Wade. Oxford University Press, New York.
- Kelly, A. J., and J. H. Willis, 1998. Polymorphic microsatellite loci in Mimulus guttatus and related species. Mol. Ecol. 7: 769–774. [Google Scholar]
- Kelly, J. K., 2003. Deleterious mutations and the genetic variance of male fitness components in Mimulus guttatus. Genetics 164: 1071–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly, J. K., and H. S. Arathi, 2003. Inbreeding and the genetic variance of floral traits in Mimulus guttatus. Heredity 90: 77–83. [DOI] [PubMed] [Google Scholar]
- Kelly, J. K., and J. H. Willis, 2001. Deleterious mutations and genetic variation for flower size in Mimulus guttatus. Evolution 55: 937–942. [DOI] [PubMed] [Google Scholar]
- Kelly, J. K., and J. H. Willis, 2002. A manipulative experiment to estimate bi-parental inbreeding in monkeyflowers. Int. J. Plant Sci. 163: 575–579. [Google Scholar]
- Kelly, J. K., A. Rasch and S. Kalisz, 2002. A method to estimate pollen viability from pollen size variation. Am. J. Bot. 89: 1021–1023. [DOI] [PubMed] [Google Scholar]
- Kempthorne, O., 1957. An Introduction to Genetic Statistics. Wiley, New York.
- Kiang, Y. T., 1972. Pollination study in a natural population of Mimulus guttatus. Evolution 26: 308–310. [DOI] [PubMed] [Google Scholar]
- Kiang, Y. T., 1973. Floral structure, hybridization and evolutionary relationships of two species of Mimulus. Rhodora 75: 225–238. [Google Scholar]
- Kimura, M., and T. Maruyama, 1966. The mutation load with epistatic gene interactions in fitness. Genetics 54: 1337–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov, A. S., 1985. Deleterious mutations as an evolutionary factor. 2. Facultative apomixis and selfing. Genetics 111: 635–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov, A. S., 1988. Deleterious mutations and the evolution of sexual reproduction. Nature 336: 435–440. [DOI] [PubMed] [Google Scholar]
- Kondrashov, A. S., 1993. Classification of hypotheses on the advantage of amphimixis. J. Hered. 84: 372–387. [DOI] [PubMed] [Google Scholar]
- Lande, R., and S. Arnold, 1983. The measurement of selection on correlated characters. Evolution 37: 1210–1226. [DOI] [PubMed] [Google Scholar]
- Leclerc-Potvin, C., and K. Ritland, 1994. Modes of self-fertilization in Mimulus-guttatus (Scrophulariaceae)—a field experiment. Am. J. Bot. 81: 199–205. [Google Scholar]
- Long, A. D., S. L. Mullaney, L. A. Reid, J. D. Fry, C. H. Langley et al., 1995. High-resolution mapping of genetic factors affecting abdominal bristle number in Drosophila melanogaster. Genetics 139: 1273–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, A. D., S. L. Mullaney, T. F. C. Mackay and C. H. Langley, 1996. Genetic interactions between naturally occurring alleles at quantitative trait loci and mutant alleles at candidate loci affecting bristle number in Drosophila melanogaster. Genetics 144: 1497–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukens, L. N., and J. Doebley, 1999. Epistatic and environmental interactions for quantitative trait loci in maize evolution. Genet. Res. 74: 291–302. [Google Scholar]
- Lush, J. L., 1937. Animal Breeding Plans. Iowa State Press, Ames, IA.
- Lynch, M., 1988. Design and analysis of experiments on random drift and inbreeding depression. Genetics 120: 791–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, M., and B. Walsh, 1998. Genetics and Analysis of Quantitative Characters. Sinauer Associates, Sunderland, MA.
- Mackay, T. F. C., 2001. The genetic architecture of quantitative traits. Annu. Rev. Genet. 35: 303–339. [DOI] [PubMed] [Google Scholar]
- Macnair, M. R., and Q. J. Cumbes, 1989. The genetic architecture of interspecific variation in Mimulus. Genetics 122: 211–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallet, J., and M. C. Singer, 1987. Individual selection, kin selection and the shifting balance in the evolution of warning colours in butterflies. Biol. J. Linn. Soc. 32: 337–350. [Google Scholar]
- Mather, K., 1949. Biometrical Genetics. Methuen, London.
- Mather, K., and J. L. Jinks, 1982. Biometrical Genetics. Chapman & Hall, New York.
- Melchinger, A. E., H. H. Geiger and F. W. Schnell, 1986. Epistasis in maize (Zea mays L.) 2. Genetic effects in crosses among early flint and dent lines determined by three methods. Theor. Appl. Genet. 72: 231–239. [DOI] [PubMed] [Google Scholar]
- Miller, R. H., J. E. Legates and C. C. Cockerham, 1963. Estimation of nonadditive hereditary variance in traits of mice. Genetics 48: 177–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll, R. H., J. H. Longquist, J. V. Fortuno and E. C. Johnson, 1965. The relationship of heterosis and genetic divergence in maize. Genetics 52: 139–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, H. J., 1939. Reversibility in evolution considered from the standpoint of genetics. Biol. Rev. 14: 185–268. [Google Scholar]
- Orr, H. A., 1995. The population genetics of speciation: the evolution of hybrid incompatibilities. Genetics 139: 1805–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto, S. P., and M. W. Feldman, 1997. Deleterious mutations, variable epistatic interactions, and the evolution of recombination. Theor. Popul. Biol. 51: 134–147. [DOI] [PubMed] [Google Scholar]
- Palopoli, M. F., and C.-I Wu, 1994. Genetics of hybrid male-sterility between Drosophila sibling species—a complex web of epistasis is revealed in interspecific studies. Genetics 138: 329–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters, A. D., and C. M. Lively, 2000. Epistasis and the maintenance of sex, pp. 99–112 in Epistasis and the Evolutionary Process, edited by J. B. Wolf, E. D. Brodie and M. J. Wade. Oxford University Press, New York.
- Phillips, P. C., S. P. Otto and M. C. Whitlock, 2000. Beyond the average: the evolutionary importance of gene interactions and variability of epistatic effects, pp. 20–38 in Epistasis and the Evolutionary Process, edited by J. B. Wolf, E. D. Brodie and M. J. Wade. Oxford University Press, New York.
- Powers, L., 1950. Determining scales and the use of transformations in studies on weight per locule of tomato fruit. Biometrics 6: 145–163. [PubMed] [Google Scholar]
- Powers, L., 1951. Gene analysis by the partitioning method when interactions of genes are involved. Bot. Gaz. 113: 1–23. [Google Scholar]
- Rice, S. H., 1998. The evolution of canalization and the breaking of von Baer's laws: modeling the evolution of development with epistasis. Evolution 52: 647–656. [DOI] [PubMed] [Google Scholar]
- Rice, S. H., 2000. The evolution of developmental interactions, pp. 82–98 in Epistasis and the Evolutionary Process, edited by J. B. Wolf, E. D. Brodie and M. J. Wade. Oxford University Press, Oxford.
- Rieseberg, L. H., B. Sinervo, C. R. Linder, M. Ungerer and D. M. Arias, 1996. Role of gene interactions in hybrid speciation: evidence from ancient and experimental hybrids. Science 272: 741–745. [DOI] [PubMed] [Google Scholar]
- Ritland, K., and F. R. Ganders, 1987. a Covariation of selfing rates with parental gene fixation indexes within populations of Mimulus-guttatus. Evolution 41: 760–771. [DOI] [PubMed] [Google Scholar]
- Ritland, K., and F. R. Ganders, 1987. b Crossability of Mimulus-guttatus in relation to components of gene fixation. Evolution 41: 772–786. [DOI] [PubMed] [Google Scholar]
- Robertson, A. W., A. Diaz and M. R. MacNair, 1994. The quantitative genetics of floral characters in Mimulus guttatus. Heredity 72: 300–311. [Google Scholar]
- Routman, E. J., and J. M. Cheverud, 1997. Gene effects on a quantitative trait: two-locus epistatic effects measured at microsatellite markers and at estimated QTL. Evolution 51: 1654–1662. [DOI] [PubMed] [Google Scholar]
- Sokal, R. R., and F. J. Rohlf, 2000. Biometry. W. H. Freeman, New York.
- Sweigart, A., K. Karoly, A. Jones and J. H. Willis, 1999. The distribution of individual inbreeding coefficients and pairwise relatedness in a population of Mimulus guttatus. Heredity 83: 625–632. [DOI] [PubMed] [Google Scholar]
- Tanksley, S. D., 1993. Mapping polygenes. Annu. Rev. Genet. 27: 205–233. [DOI] [PubMed] [Google Scholar]
- Templeton, A. R., 2000. Epistasis and complex traits, pp. 41–57 in. Epistasis and the Evolutionary Process, edited by J. B. Wolf, E. D Brodie and M. J. Wade. Oxford University Press, Oxford.
- Turelli, M., and N. H. Barton, 1994. Genetic and statistical-analyses of strong selection on polygenic traits—what, me normal. Genetics 138: 913–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli, M., and H. A. Orr, 1995. The dominance theory of Haldane's rule. Genetics 140: 389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli, M., and H. A. Orr, 2000. Dominance, epistasis and the genetics of postzygotic isolation. Genetics 154: 1663–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickery, R. K., 1978. Case studies in the evolution of species complexes in Mimulus. Evol. Biol. 11: 405–507. [Google Scholar]
- Vogler, D. W., K. Filmore and A. G. Stephenson, 1999. Inbreeding depression in Campanula rapunculoides L. I. A comparison of inbreeding depression in plants derived from strong and weak self-incompatibility phenotypes. J. Evol. Biol. 12: 483–494. [Google Scholar]
- Wade, M. J., 1992. Sewall Wright: gene interaction and the shifting balance theory. Oxf. Surv. Evol. Biol. 8: 35–62. [Google Scholar]
- Wade, M. J., 2000. Epistasis as a genetic constraint within populations and an accelerant of adaptive divergence among them, pp. 213–231 in Epistasis and the Evolutionary Process, edited by J. B. Wolf, E. D. Brodie and M. J. Wade. Oxford University Press, New York.
- Wade, M. J., and C. J. Goodnight, 1998. The theories of Fisher and Wright in the context of metapopulations: when nature does many small experiments. Evolution 52: 1537–1553. [DOI] [PubMed] [Google Scholar]
- Waser, N. M., R. K. Vickery and M. V. Price, 1982. Patterns of seed dispersal and population differentiation in Mimulus guttatus. Evolution 36: 753–761. [DOI] [PubMed] [Google Scholar]
- Weber, K., R. Eisman, L. Morey, A. Patty, J. Sparks et al., 1999. An analysis of polygenes affecting wing shape on chromosome 3 in Drosophila melanogaster. Genetics 153: 773–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, J. M., 1972. Inbreeding effects upon growth and maternal ability in laboratory mice. Genetics 70: 307–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock, M. C., P. C. Phillips, F. B. G. Moore and S. J. Tonsor, 1995. Multiple Fitness Peaks and Epistasis. Annu. Rev. Ecol. Syst. 26: 601–629. [Google Scholar]
- Willis, J. H., 1992. Genetic analysis of inbreeding depression caused by cholorphyll-deficient lethals in Mimulus guttatus. Heredity 69: 562–572. [Google Scholar]
- Willis, J. H., 1993. a Effects of different levels of inbreeding on fitness components in Mimulus guttatus. Evolution 47: 864–876. [DOI] [PubMed] [Google Scholar]
- Willis, J. H., 1993. b Partial self fertilization and inbreeding depression in two populations of Mimulus guttatus. Heredity 71: 145–154. [Google Scholar]
- Willis, J. H., 1996. Measures of phenotypic selection are biased by partial inbreeding. Evolution 50: 1501–1511. [DOI] [PubMed] [Google Scholar]
- Willis, J. H., 1999. a Inbreeding load, average dominance, and the mutation rate for mildly deleterious alleles in Mimulus guttatus. Genetics 153: 1885–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis, J. H., 1999. b The role of genes of large effect on inbreeding depression in Mimulus guttatus. Evolution 53: 1678–1691. [DOI] [PubMed] [Google Scholar]
- Wright, S., 1931. Evolution in Mendelian populations. Genetics 16: 97–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, S., 1951. The genetical structure of populations. Ann. Eugen. 15: 323–354. [DOI] [PubMed] [Google Scholar]
- Zeng, Z.-B., J. Liu, L. F. Stam, C.-H. Kao, J. M. Mercer et al., 2000. Genetic architecture of a morphological shape difference between two Drosophila species. Genetics 154: 299–310. [DOI] [PMC free article] [PubMed] [Google Scholar]