Abstract
Reproductive isolation is often caused by the disruption of genic interactions that evolve in geographically separate populations. Identifying the genomic regions and genes involved in these interactions, known as “Dobzhansky-Muller incompatibilities,” can be challenging but is facilitated by the wealth of genetic markers now available in model systems. In recent years, the complete genome sequence and thousands of single nucleotide polymorphisms (SNPs) from laboratory mice, which are largely genetic hybrids between Mus musculus and M. domesticus, have become available. Here, we use these resources to locate genomic regions that may underlie reproductive isolation between these two species. Using genotypes from 332 SNPs that differ between wild-derived strains of M. musculus and M. domesticus, we identified several physically unlinked SNP pairs that show exceptional gametic disequilibrium across the lab strains. Conspecific alleles were associated in a disproportionate number of these cases, consistent with the action of natural selection against hybrid gene combinations. As predicted by the Dobzhansky-Muller model, this bias was differentially attributable to locus pairs for which one hybrid genotype was missing. We assembled a list of potential Dobzhansky-Muller incompatibilities from locus pairs that showed extreme associations (only three gametic types) among conspecific alleles. Two SNPs in this list map near known hybrid sterility loci on chromosome 17 and the X chromosome, allowing us to nominate partners for disrupted interactions involving these genomic regions for the first time. Together, these results indicate that patterns produced by speciation between M. musculus and M. domesticus are visible in the genomes of lab strains of mice, underscoring the potential of these genetic model organisms for addressing general questions in evolutionary biology.
IDENTIFYING the genes that contribute to reproductive isolation between diverging populations and thus may underlie speciation is a formidable challenge. This goal is becoming increasingly feasible with the rapid accumulation of molecular markers, particularly in genetic model organisms. Over the past several years, genes causing hybrid inviability (Wittbrodt et al. 1989; Barbash et al. 2003; Presgraves et al. 2003) and hybrid sterility (Ting et al. 1998) have been located and characterized, leading to new insights about the molecular details of speciation (Orr et al. 2005).
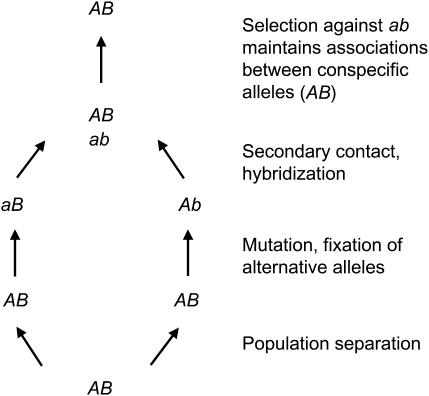
These advances were facilitated by the realization that reproductive isolation often involves the disruption of genic interactions. After populations separate geographically, mutations arise and are fixed by genetic drift or natural selection. Although these substitutions need not reduce fitness in the populations in which they originate, the combination of mutations at interacting loci that is formed when populations hybridize can lead to reproductive isolation (Figure 1). This allopatric speciation process (Bateson 1909; Dobzhansky 1936, 1937; Muller 1940, 1942), termed the “Dobzhansky-Muller model,” has received considerable attention and support from both theoretical (Orr 1995, 1996; Gavrilets 1997; Barton 2001; Orr and Turelli 2001; Turelli et al. 2001; Welch 2004) and empirical (Hollingshead 1930; Dobzhansky 1936; Wu and Beckenbach 1983; Christie and Macnair 1984; Orr 1987; Pantazidis and Zouros 1988; Perez and Wu 1995; True et al. 1996; Coyne and Orr 1998; Fishman and Willis 2001; Presgraves 2003; Tao and Hartl 2003) studies.
Figure 1.
A simple example of a Dobzhansky-Muller incompatibility that involves two loci. Genotypes are shown as haploid for simplicity.
The notion that speciation is often driven by Dobzhansky-Muller incompatibilities suggests several testable predictions about empirical patterns, ranging from the rate at which isolation accumulates (Orr 1995; Mendelson et al. 2004) to the molecular evolution of participating loci (Kondrashov et al. 2002; Welch 2004). One such prediction is that natural selection will remove unfavorable combinations of alleles generated by hybridization between species. When epistatic selection purges heterospecific allelic combinations in this manner, gametic disequilibrium can result, even in the face of recurrent recombination. This rationale can be used to identify genomic regions or genes harboring mutations that maintain reproductive barriers between diverging populations (Gardner et al. 2002).
The house mouse is an excellent subject for locating genomic regions underlying reproductive isolation using interlocus associations. First, the success and promise of efforts to connect genotypic variation with phenotypic variation among inbred strains of mice have fueled the identification of large numbers of molecular polymorphisms that distinguish these lines (Silver 1995; Lindblad-Toh et al. 2000; Pletcher et al. 2004), and the recent completion of the genome sequence (Mouse Genome Sequencing Consortium 2002) has rapidly increased the discovery rate of polymorphic markers. For example, Pletcher et al. (2004) reported the genotypes for >10,000 single nucleotide polymorphisms (SNPs) in 48 inbred strains, and efforts to locate hundreds of thousands of SNPs are under way.
Second, commonly used inbred strains of mice are genetic hybrids—descendants of crosses between a few wild species of house mice (Bonhomme et al. 1987; Guenet and Bonhomme 2003). Strains are primarily derived from two species, M. domesticus and M. musculus, with smaller contributions from M. castaneus (these taxa are also referred to as M. musculus domesticus, M. musculus musculus, and M. musculus castaneus in the literature). Surveys of microsatellite polymorphism indicate that the relative contribution of M. domesticus vs. other species to lab strain genomes is ∼3:1 (Sakai et al. 2005). Most inbred strains carry a Y chromosome from M. musculus (Bishop et al. 1985) and a mitochondrial genome from M. domesticus (Yonekawa et al. 1980; Ferris et al. 1983). Additionally, many SNPs among the inbred strains appear to reflect divergence between M. domesticus and M. musculus or M. castaneus (Wade et al. 2002). A high-density SNP study of chromosome 16 suggested that ∼20% of the differences between lab strains derive from divergence between species and that the remaining 80% of the SNPs come from variation segregating within M. domesticus (Zhang et al. 2005).
Third, the genetic basis of speciation between the ancestors of inbred mouse strains has been studied for decades, primarily by following the introgression of molecular markers across naturally occurring contact zones (Boursot et al. 1993; Sage et al. 1993). In a hybrid zone between M. domesticus and M. musculus that stretches across Europe, allele frequency clines at most surveyed loci are narrow relative to the ranges of these species (Hunt and Selander 1973; Vanlerberghe et al. 1986; Tucker et al. 1992; Dod et al. 1993; Munclinger et al. 2002; Payseur et al. 2004). This observation suggests that the hybrid zone is maintained by a balance between selection against hybrids and dispersal (Barton and Gale 1993). Secondary contact between M. domesticus and M. musculus has occurred recently (5000–10,000 years ago; Auffray et al. 1990) relative to estimates of divergence time (500,000 years ago; Boursot et al. 1996; Din et al. 1996), consistent with the accumulation of reproductive isolation in allopatry according to a Dobzhansky-Muller model.
Crosses between mouse strains have also demonstrated reproductive isolation. Male hybrids between wild M. musculus and some lab strains are sterile (and female hybrids are fertile, consistent with Haldane's 1922 rule; Forejt and Ivanyi 1975; Storchova et al. 2004), and similar patterns have been observed in crosses between wild species (Alibert et al. 1997; Britton-Davidian et al. 2005). Furthermore, natural hybrids between M. domesticus and M. musculus harbor more parasites than do pure-species animals, suggesting that these species may also be isolated by hybrid inviability (Sage et al. 1986; Moulia et al. 1993; Moulia et al. 1995). Finally, mate choice experiments suggest some behavioral isolation between these species (Smadja and Ganem 2002), which may be disrupted in hybrids.
Here, we use the hybrid status of inbred mouse strains and available genotypes from across the genome to search for Dobzhansky-Muller incompatibilities in the form of unlinked SNPs that show unusual levels of gametic disequilibrium. Specifically, we use genotypes from wild-derived strains to select markers that measure gene flow between species in the lab strains. We show that patterns of genetic diversity in the lab strains have been affected by reproductive isolation between M. domesticus and M. musculus, and we identify several candidate regions for this isolation.
MATERIALS AND METHODS
Selection of strains and loci:
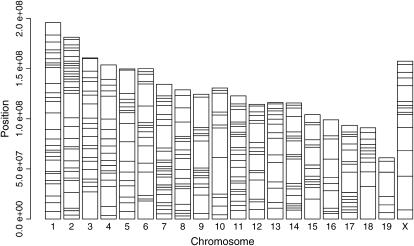
Pletcher et al. (2004) reported genotypes for 48 lab strains of mice at 10,990 evenly spaced SNPs, a subset of those discovered by sequence comparisons between five strains (DBA/2J, A/J, C57BL/6J, 129S1/SvImJ, and 129X1/SvJ) at Celera (Mural et al. 2002). Before conducting analyses, we filtered SNPs and strains using several conservative criteria. First, to increase the probability of correctly inferring the ancestral (species) source of alleles, we focused on SNPs that featured complete data for, and differed between, multiple wild-derived strains of M. domesticus and M. musculus and showed no variation among three strains from each species (M. domesticus—WSB/Ei, PERA/Ei, ZALENDE/Ei; M. musculus—PWD/Ph, CZECHII/Ei, MAI/Pas; number of SNPs, n = 869). After eliminating other wild-derived strains (due to excessive divergence in comparison with lab strains) and substrains (due to a lack of divergence), we further filtered the SNPs to those with complete genotype data for the remaining 22 strains (Table 1; n = 658) to avoid difficulties in comparing across sites with different amounts of missing data. Next, we polarized each SNP on the basis of comparison to the genotypes of the wild-derived strains of M. domesticus and M. musculus (which were subsequently removed from the data set). We removed all SNPs for which only one strain differed from all the other strains (n = 606). Finally, motivated by the observation that the average size of haplotype blocks in previous work using subsets of the lab strains was ∼1.5 Mb (Wade et al. 2002; Wiltshire et al. 2003; Frazer et al. 2004; Ideraabdullah et al. 2004), we used only SNPs whose nearest neighbors were at least 2 Mb away (n = 332). Three hundred thirty-two SNPs that differ between wild-derived strains of M. domesticus and M. musculus and feature complete genotype data from across the culled set of 22 strains (Table 1) composed the final data set used for analyses; the chromosomal positions of these loci are shown in Figure 2. Estimates of the total genetic length (1373.7 cM; Dietrich et al. 1996) and physical length (2577.3 Mb; Mouse Genome Sequencing Consortium 2002) of the mouse genome indicate that the average distances between SNPs used in this study were 4.1 cM and 7.8 Mb. The 22 strains ranged from 26.8 to 49.1% allelic identity to M. musculus at these SNPs (Table 1).
TABLE 1.
Strains and species composition for 332 SNPs used in analyses
| Strain | % of SNPs that match M. musculus genotype |
|---|---|
| A/J | 39.2 |
| AKR/J | 30.1 |
| BTBR T+ tf/J | 38.6 |
| BUB/BnJ | 29.8 |
| C3H/HeJ | 33.7 |
| C57BL/10J | 39.2 |
| DBA/1J | 32.8 |
| FVB/NJ | 26.8 |
| I/LnJ | 35.5 |
| KK/HIJ | 38.6 |
| LG/J | 31.9 |
| LP/J | 41.9 |
| MA/MyJ | 35.2 |
| NOD/LtJ | 30.7 |
| NON/LtJ | 31.6 |
| NZB/BINJ | 38.9 |
| PL/J | 32.2 |
| RIIIS/J | 30.7 |
| SEA/GnJ | 31.9 |
| SJL/J | 32.8 |
| ST/bJ | 36.4 |
| 129X1/SvJ | 49.1 |
Figure 2.
Map of SNP markers used in analyses. Each horizontal line represents one SNP. Total chromosome sizes were taken from Mouse Genome Sequencing Consortium (2002). Moving up the plot, chromosomes run from proximal to distal.
Analyses:
Because of perpetual inbreeding, each of the strains has an essentially homozygous genome (Beck et al. 2000). This lack of intrastrain polymorphism allowed us to use measures of gametic disequilibrium. Interlocus associations were estimated using R2 (Hill and Robertson 1968) and D′ (Lewontin 1964). R2 ranges from 0 to 1, and D′ ranges from –1 to 1. Although both gametic disequilibrium measures are affected by allele frequencies, D′ is more sensitive to them. Both metrics are expected to decay as a function of recombination frequency and time.
Our goal was to identify physically unlinked SNP pairs showing clear associations despite generations of recombination. To maximize the frequency of recombination events between assayed SNPs during the history of the strains, we estimated gametic disequilibrium between loci that mapped to different chromosomes. The statistical significance of gametic disequilibrium values for individual SNP pairs was estimated by randomly shuffling strain genotypes at one of the two SNPs, calculating disequilibrium values for this randomized data set, performing this randomization procedure 10,000 times, and estimating the P-value as the fraction of replicates with disequilibrium values exceeded by the observed association (a one-tailed test). We accounted for the performance of a large number of tests using the false discovery rate (FDR; Storey and Tibshirani 2003) with a q-value cutoff of 5% and a Bonferroni correction. We predicted that if selection against combinations of alleles derived from M. domesticus and M. musculus (due to reproductive isolation) has affected patterns of polymorphism in the lab strains, we would discover a higher fraction of SNP pairs showing associations between conspecific alleles than those showing associations between heterospecific alleles among tests with extreme P-values.
RESULTS
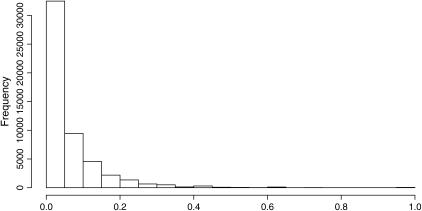
Using genotypes from 22 strains at 332 SNPs selected to be enriched for differences between M. domesticus and M. musculus, we estimated gametic disequilibrium values for 51,991 pairs of sites lying on different chromosomes (Figure 3). Average values of R2 (0.06) and D′ (−0.06) suggested some departure from the equilibrium expectation for unlinked SNPs, reflecting substantial power to detect shifts in the mean. Analyses of D′ revealed many locus pairs with values of –1 and 1, a result that reflects the sensitivity of this disequilibrium measure to allele frequency (and the absence of one gametic type due to sampling variance). Because of this finding and because our general conclusions are similar regardless of the disequilibrium measure we use, we focus on results for R2.
Figure 3.
Genomic distribution of R2 for all pairwise comparisons (n = 51,991). R2 is on the x-axis. Average R2 for the genome was 0.06.
We asked whether SNP pairs in which the associated alleles were derived from the same ancestral species exhibited more extreme P-values than SNP pairs in which the associated alleles were derived from different ancestral species. Locus pairs with P < 0.01 were enriched for conspecific associations (Fisher's exact test; P < 6 × 10−14; odds ratio = 2.32). This pattern was also observed when significance thresholds of P < 0.005 (Fisher's exact test; P < 6 × 10−7; odds ratio = 2.14), P < 0.001 (Fisher's exact test; P < 0.023; odds ratio = 2.36), and P < 0.0005 (Fisher's exact test; P < 0.018; odds ratio = 3.63) were applied. Both Bonferroni and FDR procedures indicated that only disequilibrium values that exceeded those for all 10,000 randomized data sets showed strong evidence of statistical significance. One test satisfied this requirement, preventing us from conducting the comparisons described above at this significance level.
A further prediction of the Dobzhansky-Muller model is that the effects of incompatibilities should be asymmetrical, particularly at early stages of species divergence (Muller 1942). Among the four gametic types, natural selection on hybrids should specifically target one (heterospecific) combination of alleles because the other three combinations represent ancestral stages of divergence (see Figure 1). We tested this prediction by repeating the above analyses separately for locus pairs showing three gametic types and locus pairs showing four gametic types (Table 2). Tests with extreme P-values (P < 0.001) were again biased toward conspecific associations for loci showing three gametic types (Fisher's exact test; P < 0.006), but no such bias was seen for loci showing four gametic types (P < 0.64). In this comparison, only four tests were significant among locus pairs with four gametic types, raising the possibility that this disparity was caused by differences in power. However, similar results were obtained using a higher P-value threshold (0.005; Table 2), where the number of significant tests in the four-gametic-type category was large enough to detect a deviation from the predicted odds ratios. Hence, the subset of locus pairs showing three gametic types was differentially responsible for the skew toward conspecific associations among tests with low P-values in the complete data set, as predicted by the Dobzhansky-Muller model.
TABLE 2.
Categorization of gametic disequilibrium tests by nature of association (conspecific vs. heterospecific) and P-value
| Conspecific | Heterospecific | |
|---|---|---|
| All locus pairs* | ||
| P < 0.005 | 127 | 61 |
| P ≥ 0.005 | 25,564 | 26,239 |
| Locus pairs with three gametic types** | ||
| P < 0.005 | 72 | 36 |
| P ≥ 0.005 | 4,375 | 8,668 |
| Locus pairs with four gametic types***a | ||
| P < 0.005 | 20 | 22 |
| P ≥ 0.005 | 21,183 | 17,571 |
Fisher's exact test, P < 6 × 10−7.
Fisher's exact test, P < 5 × 10−12.
Fisher's exact test, P < 0.44.
A significance threshold of P = 0.005 is used to retain power.
Because the X chromosome is expected to play a disproportionate role in reproductive isolation (Coyne and Orr 1989), we also compared the fraction of SNPs showing high disequilibrium values on the X to that on the autosomes. On the genomic scale, locus pairs that included one X-linked SNP and those that did not showed no difference in R2 values (average, including X = 0.05; average, not including X = 0.06; P > 0.05; Mann-Whitney U).
The 14 SNP pairs that showed conspecific associations, only three gametic types and P < 0.001 are listed in Table 3. These locus pairs featured an average R2-value of 0.70, a remarkable amount of association for physically unlinked markers. Although this subset of locus pairs did not satisfy strict significance criteria after corrections for multiple tests, this subset seemed likely to contain Dobzhansky-Muller incompatibilities, given the results presented above. Among these candidate incompatibilities, four SNPs, located on chromosomes 2, 3, 6, and 14, each showed extreme disequilibria with multiple partners on different chromosomes. Allele frequencies of SNPs included among the candidate incompatibilities ranged from 0.18 to 0.73, suggesting that sufficient power to find associations from across the frequency spectrum existed. Pairs of SNPs involved in candidate incompatibilities also showed more similar allele frequencies to one another (average difference is 0.07; average difference across the genome is 0.23; P < 3 × 10−5; Mann-Whitney U), as might be expected if selection acts against two-locus genotypes.
TABLE 3.
Pairs of SNPs showing three gametic types, conspecific associations, and P < 0.001 (sorted by position of site 1)
| R2 | Site 1 chromosome | Site 1 position (bp) | Named genes in 2-Mb window | Site 2 chromosome | Site 2 position (bp) | Named genes in 2-Mb window |
|---|---|---|---|---|---|---|
| 0.58 | 1 | 125936917 | Slc35f5, Actr3, Gpr39, Lypdc1 | 7 | 93165258 | Trim3, Olfr690, Olfr677, Olfr697, Olfr678, Olfr693, Arfip2, Olfr701, Olfr667, Olfr665, Olfr698, Olfr695, Olfr694, Olfr706, Dub6, Olfr659, Olfr669, Olfr700, Olfr703, Olfr692, Smpd1, Dub1, Cln2, Cckbr, Olfr702, Dchs1, Mrpl17, Hpxn, Iigs1, Olfr666, Olfr686, Olfr670, Dub1a, Olfr668, Olfr705, Olfr672, Olfr676, Olfr691, Olfr658, Cnga4, Olfr679, Olfr681, Olfr684, Olfr689, Olfr661, Olfr683, Fxc1, Ilk, Prkcdbp, Taf10, Olfr688, Olfr704, Olfr699, Olfr685, Olfr687, Olfr663, Olfr671, Olfr675, Olfr664 |
| 0.76 | 2 | 26442964 | Fcna, Lcn4, Lcn5, Adamts13, Notch1, Traf2, Vav2, Slc2a6, Inpp5e, Surf6, Surf2, Rpl7a, Dbh, Wdr5, Bmyc, Lcn11, Egfl7, Lcn8, Lcn9, Brd3, Snapc4, Qscn6l1, Lcn3, Sardh, Pmpca, Gpsm1, Surf4, Lhx3, Lcn13, Ubadc1, Surf1, Surf5, Edf1, Phpt1, Agpat2, Lcn12, Gm711, Btbd14a, Lcn10, Fbxw5, Gm110, Lcn6, Gm996, Gm111, Camsap1 | 3 | 160118223 | Rpe65, Depdc1a |
| 0.76 | 2 | 26442964 | a | 18 | 64784116 | |
| 0.66 | 2 | 153094785 | H13, Dnmt3b, Dnmt3b, Gm123, Bpil3, Spag4l, Tpx2, Tm9sf4, Psp, Bcl2l1, Csnk2a1, Bpil1, Bpil1, Tcf15, Hck, Cox4i2, Rya3, Trib3, Sox12, Kif3b, Idb1, Tomm20, Fkhl18, Dusp15, Rem1, Asxl1, Plagl2, Zcchc3, Commd7, Defb19, Defb36, Pdrg1, Npn3, Pofut1 | 15 | 93935842 | Pdzrn4, Pphln1, Prickle1, Adamts20, Yaf2 |
| 0.76 | 3 | 95116767 | Pip5k1b, Selenbp1, Ecm1, Oaz3, Golph3l, Anp32e, Tarsl1, Snx27, Ctsk, Setdb1, Lass2, Cgn, Tmod4, Tcfl1, Rorc, Selenbp2, Ctss, Bnipl, Tnrc4, Prpf3, Scnm1, Za20d1, Mcl1, Arnt, Tdrkh, Tsrc1, Lrrn6d, Anxa9, Rfx5, Vps45, Car14, Ensa, Cdc42se1, Gabpb2, Pogz, Tuft1, Hormad1, Prune, Mrps21, Mrpl9 | 6 | 134673836 | Ddx47, Rai3, Emp1, Lrp6, Etv6, Gpr19, Dusp16, Cdkn1b, Mansc1, Bcl2l14, Pbp2, Hebp1, Kap, Gsg1 |
| 0.76 | 3 | 160118223 | a | 15 | 31266214 | Catnd2, Ropn1l, Dap |
| 0.76 | 5 | 16224015 | Cd36, Gnai1, Speer4f | 18 | 86188576 | Hmga1, Nars, Siat8c, Nedd4l, Fech, Txnl1, Wdr7, Cndp2, Neto1, Cyb5 |
| 0.54 | 5 | 111872715 | Selpl, Mvk, Oasl2, Git2, Acads, Coro1c, Usp30, Cmklr1, Tcf1, Ube3b, Pop5, Dao1, Acacb, Foxn4, Sppl3, Rnf10, Sfrs, Kctd10, Sart3, Oasl1, Msi1h, Gltp, Ssh1, Ung, Cabp1, Mmab, Myo1h | 17 | 26327773 | Rpl10a, Bak1, Bnip1, Anks1, Phf1, Kifc5a, Tead3, Fkbp5, Fance, Syngap1, Def6, Srpk1, Lemd2, Mapk14, Kifc1, Grm4, Tulp1, Tcp11, Pacsin1, Slc26a8, Ppard, Taf11, Rps10, Nudt3, Spdef, Clps, Zfp523, Snrp1c, Hmga1 |
| 0.76 | 6 | 55769693 | Crhr2, Adcyap1r1, Neurod6, Aqp1, Ghrhr, Gars, Card4, Kbtbd2, Pde1c, Gsbs, Lsm5 | 14 | 77885410 | Diap3 |
| 0.76 | 6 | 134673836 | a | 7 | 117314860 | Fgfr2, Tacc2, Wdr11, Ate1, Etos1 |
| 0.58 | 7 | 16267788 | Fbl, Nalp9a, Sertad1, Nalp4a, Adck4, Nalp9c, Rab4b, Dyrk1b, Dll3, Sertad3, Blvrb, Akt2, Egln2, Ltbp4, Numbl, Shkbp1, Spnb4, Map3k10, Zfp59, Zfp60, Pld3, Snrpa, Prx, Mia1, Timm50, Cri2 | 12 | 95557943 | Fbln5, Gpr68, Tdp1, Rps6ka5, Calm1, Ttc7b, Mtac2d1 |
| 0.76 | 7 | 104427524 | Xylt1, Plekha7, Nucb2, Arl6ip1, Rps13, Rps15a | 14 | 77885410 | a |
| 0.82 | 8 | 96761667 | 10 | 128349364 | Mip, Prim1, Itga7, Erbb3, Rdhs, Olfr768, Pa2g4, Suox, Admr, Shmt2, Rdh9, Il23a, Tmem4, Gdf11, Rab5b, Rdh1, Olfr770, Olfr769, Rnf41, Rdh7, Olfr771, Olfr765, Sdro, Stat2, Hsd17b9, Si, Mbc2, Inhbc, Olfr772, Dctn2, Olfr774, Mmp19, Stat6, Gls2, Lrp1, Rdh5, Kif5a, Apof, Slc395, Olfr763, Inhbe, Nab2, Baz2a, Ddit3, Dgka, Cd63, Usp52, Rbms2, Tebp, Mars, Rdh6, Cs, Cdk2, Olfr9, Ormdl2, Nxph4, Tac2, Bloc1s1, Rps26, Arhgap9, Gli1, Apon, Rpl41, Naca, Olfr766 | |
| 0.66 | 9 | 77840500 | Ick, Lrrc1, Gclc, Elovl5, Fbxo9, Tinag, Gcm1, Gsta2, Gsta1, Gsta4, Mosg | X | 68140394 | Tbl1x, Pls3, Magea7 |
See first appearance in table for list of genes.
We used the PANTHER classification system (https://panther.appliedbiosystems.com/) to ask whether particular functional categories of genes were overrepresented in the candidate regions listed in Table 3 (using a 2-Mb window surrounding each SNP). The results of this analysis are displayed in Table 4. Gametogenesis, steroid metabolism, olfaction, and chemosensory perception were among the biological processes found to be overrepresented in the candidate regions.
TABLE 4.
Functional categories of genes that were overrepresented in incompatibility candidate regions from Table 3
| Biological process | Genome | Regions | Expected | P |
|---|---|---|---|---|
| Steroid metabolism | 165 | 15 | 4.69 | 0.0001 |
| Olfaction | 936 | 44 | 26.61 | 0.0009 |
| Chemosensory perception | 972 | 45 | 27.63 | 0.0017 |
| Electron transport | 296 | 17 | 8.41 | 0.0057 |
| Asymmetric protein localization | 13 | 3 | 0.37 | 0.0064 |
| Lipid, fatty acid and steroid metabolism | 706 | 31 | 20.07 | 0.013 |
| Induction of apoptosis | 176 | 11 | 5 | 0.013 |
| Cell proliferation and differentiation | 689 | 30 | 19.59 | 0.016 |
| Embryogenesis | 134 | 9 | 3.81 | 0.016 |
| Sensory perception | 1404 | 54 | 39.91 | 0.017 |
| Other steroid metabolism | 7 | 2 | 0.2 | 0.017 |
| Other coenzyme and prosthetic group metabolism | 7 | 2 | 0.2 | 0.017 |
| Nitric oxide biosynthesis | 9 | 2 | 0.26 | 0.028 |
| Osmosensing | 1 | 1 | 0.03 | 0.028 |
| Amino acid activation | 41 | 4 | 1.17 | 0.031 |
| Gametogenesis | 267 | 13 | 7.59 | 0.045 |
| Other carbon metabolism | 48 | 4 | 1.36 | 0.050 |
Genes were found within a 2-Mb window centered on each SNP, and overrepresentation of particular functions was detected using the PANTHER gene classification system. The observed number of genes in the genome, the observed number of genes in candidate regions, and the expected number of genes in candidate regions are denoted by Genome, Regions, and Expected, respectively, for each functional class.
DISCUSSION
Signatures of speciation in patterns of gametic disequilibrium:
We located several candidate Dobzhansky-Muller incompatibilities between M. domesticus and M. musculus by searching for strongly associated pairs of SNPs lying on different chromosomes in lab strains of house mice. Pairs of loci showing the most extreme disequilibrium values were biased toward associations between conspecific alleles, as predicted by the Dobzhansky-Muller model, revealing a signal of ancestral reproductive isolation in the genomes of these strains. The history of the lab strains complicates interpretations of gametic disequilibrium, and we discuss these issues below.
Patterns of disequilibrium and the history of the mouse inbred strains:
In a large, panmictic population at demographic equilibrium, allelic associations among unlinked loci are expected to decay rapidly: the level of gametic disequilibrium should be reduced by 50% after just one generation (Hedrick 2000). Therefore, cases of extreme association among SNPs on different chromosomes must be caused by departures from this idealized population. Several such departures are likely to characterize the collection of strains on which we focused, including admixture between differentiated genomes, genetic drift, and inbreeding, in addition to selection against Dobzhansky-Muller incompatibilities. Allelic frequency differentiation among the wild-derived ancestors that were crossed to start the lab strains (and related differences in subsequent rounds of backcrossing) likely generated associations among unlinked SNPs in generations immediately following these crosses (Barton 1983). Using small numbers of individuals to found and propagate these strains also could have increased disequilibrium levels (Hill and Robertson 1968; Ohta and Kimura 1969). Finally, mice were eventually inbred for dozens of generations to remove heterozygosity, a process that could retard the approach to gametic equilibrium across strains (Hedrick 2000).
These nonequilibrium conditions challenge attempts to precisely predict patterns of gametic disequilibrium across the genomes of inbred mouse strains. Indeed, haplotypes among subsets of these strains appear to be less numerous and extend further than originally anticipated (Wade et al. 2002; Wiltshire et al. 2003; Frazer et al. 2004; Ideraabdullah et al. 2004). Precise predictions about the shape of the distribution of gametic disequilibrium require historical information about the inbred strains. Although many details of the relationships between the strains are known (Atchley and Fitch 1991, 1993; Beck et al. 2000), the precise nature and number of crosses between the time the common ancestors of the strains were formed and the beginning of perpetual within-strain inbreeding are unclear (Atchley and Fitch 1993). This time period was an especially important contributor to the patterns of gametic disequilibrium among the extant strains observed here because most of the recombination among alleles inherited from different species probably occurred during this period. Other research goals that are affected by the history of the strains, such as the association between haplotypes and phenotypes (Grupe et al. 2001; Pletcher et al. 2004), also depend critically on these generations of recombination. However, the complex history of inbred strains should be manifested as genome-wide departures from equilibrium, and therefore we focus on those pairs of loci that show extreme gametic disequilibrium relative to the average across all loci.
Selection unrelated to speciation:
A second caveat to our analysis is that forms of selection other than Dobzhansky-Muller-type selection are also predicted to maintain gametic disequilibrium in the face of recombination. For example, if researchers selected for combinations of phenotypes derived from M. domesticus or M. musculus during the history of the lab strains, gametic disequilibrium between conspecific alleles at unlinked genomic regions could be maintained as a result. This form of selection may be unrelated to reproductive isolation between the ancestral species, suggesting some caution in the designation of incompatibilities. Although some of the locus pairs we identified may be tracking artificial selection in this manner, the overall patterns we observe are difficult to explain under this scenario. First, this explanation requires a bias among researchers in selecting for combinations of traits inherited from the same ancestral species. This bias seems unlikely because hybridization between ancestral species was presumably used to obtain suites of phenotypes that had not been observed before. Second, to the extent that the strains have endured independent selection during their histories, selection pressures would need to be similar across many different lines to explain the observed patterns. Although such convergence may have occurred, it is unlikely to be widespread. Finally, we observe an excess of conspecific associations among locus pairs that show extreme disequilibria, and this pattern is differentially caused by SNP pairs with three gametic types. This asymmetry is predicted by the Dobzhansky-Muller model but is not an obvious consequence of simple two-trait artificial selection schemes.
Evidence for disrupted functional interactions between genomic regions in inbred mouse strains:
Despite some uncertainty about strain history and the causes of genomic departures from equilibrium, the observed average level of gametic disequilibrium indicates that a sufficient number of generations of independent assortment occurred during the history of the inbred strains for strong associations between alleles on different chromosomes to decay. This result suggests that extreme disequilibrium values such as those in Table 3 are not simple consequences of processes expected to affect the entire genome (such as admixture, genetic drift, and inbreeding) but instead reflect forces that target specific genomic regions. Investigations of haplotype diversity have also uncovered clear signs of historical recombination since the origins of the lab strains (Zhang et al. 2005).
The observation that only one test was statistically significant after correcting for the performance of multiple tests suggests caution in designating locus pairs with low P-values as Dobzhansky-Muller incompatibilities. However, these corrections may be conservative: because each SNP participates in hundreds of tests, some tests were partially correlated, while the Bonferroni and FDR procedures assumed that all tests were completely independent. Stronger evidence that the pairs of SNPs we have identified as showing strong associations mark functional interactions among genomic regions comes from the fulfillment of several biological predictions.
Under the simple (two-locus) Dobzhansky-Muller model, natural selection is expected to specifically target one allelic combination in hybrids because the other three gametic types represent ancestral stages of divergence (Muller 1942). This prediction is supported by empirical and theoretical studies (Wu and Beckenbach 1983; Orr 1995; Coyne and Orr 2004). If selection against a particular heterospecific combination of alleles is strong, we expect this gametic type to be absent. In agreement with this prediction, we demonstrated that the bias toward extreme associations among conspecific alleles was driven by cases in which only three gametic types were present.
Because genes often interact with multiple partners, substitutions in one gene may generate incompatibilities with several different loci. This prediction was supported by the candidate incompatibilities listed in Table 3: four SNPs exhibited extreme gametic disequilibrium with multiple, unlinked genomic regions. This observation suggests the existence of multiple incompatibilities in a pathway and seems unlikely to arise in the absence of epistatic selection (as a result of neutral departures from equilibrium, for example). Locus pairs in our list of incompatibilities also showed significantly more similarity in allele frequencies than the genomic average. Natural selection against two-locus genotypes derived from different species is expected to produce this pattern.
Tentative evidence that Dobzhansky-Muller incompatibilities are represented in our list of extreme associations also comes from the known/inferred functions of genes mapping to the corresponding genomic regions. Twelve of 28 SNPs from our incompatibility list map within 1 Mb of genes involved in gametogenesis, a statistical excess relative to the remainder of the genome. Six genes within this subset have functions in spermatogenesis. This SNP set includes the markers on chromosome 17, the X chromosome, and their potential partners. Because M. domesticus and M. musculus may be primarily (postzygotically) isolated by hybrid male sterility, these genes represent reasonable candidates for reproductive isolation. Olfaction and chemosensory perception, processes that play crucial roles in mating behavior in mice (Bronson 1979), were overrepresented in the genomic regions we identified, suggesting that some markers may be tracking prezygotic isolation between the ancestral species. Such isolation has been documented in mate choice experiments between wild-derived M. domesticus and M. musculus (Smadja and Ganem 2002), where mating cues are present in the urine (Ganem et al. 2005). These kinds of genes may be targeted by sexual selection, facilitating rapid functional divergence and resulting in Dobzhansky-Muller incompatibilities. Genes involved in steroid metabolism, another process related to reproduction, were also overrepresented in the candidate regions. Two caveats accompany these interpretations. First, genes with similar functions often map near one another in the mouse genome (Mouse Genome Sequencing Consortium 2002); some functional categories may be overrepresented merely because few regions contain clusters of coregulated genes. Second, we have focused on a few functional categories from Table 3 because they seem likely to participate in Dobzhansky-Muller incompatibilities. However, other overrepresented groups showing no obvious relationship to reproductive isolation were also identified, with stronger statistical support than the gametogenesis category.
Correspondence between SNPs in our list of candidate incompatibilities and loci experimentally demonstrated to affect reproductive isolation would provide the most compelling evidence that patterns of gametic disequilibrium among the lab strains contain information about reproductive isolation between species of house mice. Two genomic regions have been repeatedly associated with hybrid male sterility in crosses between lab strains and wild-derived strains: one on the proximal part of chromosome 17 and one on the middle part of the X chromosome. Matings between wild-derived M. musculus and some lab strains yield sterile hybrid males, while crosses with other lab strains produce fertile male offspring (Forejt and Ivanyi 1975). The difference between two of the lab strains (C57BL/10 and C3H) in hybrid male sterility with wild-derived M. musculus, which presumably reflects an M. musculus-M. domesticus incompatibility still segregating within M. domesticus, maps to a 360-kb region on the proximal part of chromosome 17 (Hst1; Forejt and Ivanyi 1975; Forejt et al. 1991; Gregorova et al. 1996; Trachtulec et al. 1997, 2005). Four loci that cause hybrid male sterility in crosses between lab strains and the more phylogenetically distant M. spretus also map to this region (Forejt 1996). Furthermore, a quantitative trait locus (QTL) that explains variation in fertility and testes weight in crosses between lab strains and M. macedonicus (Forejt 1996; Elliott et al. 2004) is located in this region. This part of chromosome 17 is also the location of t-haplotypes, variants composed of four recombination-suppressing inversions that segregate in wild mouse populations and are maintained partly by a severe transmission bias in males (Silver 1985).
One SNP located in this proximal part of chromosome 17, mapping to 26.3 Mb, was included in our list of candidate incompatibilities, showing a strong association (R2 = 0.54; P = 0.0009; only three gametic types present) with an SNP located at 111.9 Mb on chromosome 5. This candidate incompatibility may not match Hst1: it is located about 10 Mb distal to Hst1, and the missing gametic type is an M. musculus chromosome 17 allele with an M. domesticus chromosome 5 allele (the nature of the crosses used to identify Hst1 suggests that the alternative heterospecific combination should be absent). However, hybrid males produced by some crosses between M. spretus and lab strains show defects in sperm flagellar assembly and curvature (Pilder et al. 1993), and these phenotypes map near the chromosome 17 SNP (to a locus known as Hst6) in our list of incompatibilities. Recently, a candidate gene corresponding to this locus was identified (Fossella et al. 2000). Dnahc8 encodes an axonemal dynein heavy chain, the kind of molecule that forms the basis for the “defective dynein” model of t-haplotype-mediated hybrid male sterility (Harrison et al. 1998). The chromosome 5 SNP that showed extreme disequilibrium with the chromosome 17 SNP in our study maps near another dynein gene (Dnclc1), which also functions in gametogenesis.
Another genomic region likely to have played an important role in speciation between M. musculus and M. domesticus is the X chromosome. Molecular markers on the X chromosome show reduced introgression (relative to the autosomal loci surveyed) across the European hybrid zone between M. domesticus and M. musculus (Tucker et al. 1992; Dod et al. 1993; Munclinger et al. 2002), and the X chromosome has been associated with hybrid male sterility in crosses involving lab strains (Guenet et al. 1990; Elliott et al. 2001; Storchova et al. 2004). In particular, an X-linked QTL for hybrid male sterility (Hstx1) was recently localized to an interval between 64.0 and 70.1 Mb by introgressing pieces of the wild M. musculus X chromosome on to the autosomal background of C57BL/6 (Storchova et al. 2004). One SNP mapping to this region (68.1 Mb) was included in our list of candidate incompatibilities, exhibiting strong disequilibrium among conspecific alleles with an SNP mapping to 77.8 Mb on chromosome 9 (R2 = 0.66; P = 0.0004) and showing only three gametic types. The missing allelic combination was an M. musculus X-linked locus with an M. domesticus chromosome 9 locus, as predicted if the Dobzhansky-Muller incompatibility corresponds to that identified by Storchova et al. (2004). Additionally, a neighboring X-linked SNP (located at 68.4 Mb), while not included in our gametic disequilibrium survey, was fixed for the M. domesticus allele across all strains, suggesting that the M. musculus allele at this locus may reduce fitness when combined with M. domesticus alleles at other loci. Two genes involved in spermatogenesis are located near the SNPs for this candidate incompatibility, Magea7 (X chromosome) and Ick (chromosome 9).
Although one X-linked region was included in our list of incompatibilities, we uncovered little sign of increased involvement of the X chromosome overall. This result can be explained by considering the nature of the incompatibilities we have identified. The prediction that the X chromosome will be enriched for incompatibilities derives from the observation of Haldane's rule in crosses between mouse strains (Forejt and Ivanyi 1975; Storchova et al. 2004). However, the incompatibilities underlying Haldane's rule are recessive (X chromosome) dominant (autosome). Because we have identified exclusively recessive-recessive incompatibilities (all SNPs in this study are assumed to be homozygous), the prediction under Haldane's rule does not apply.
Coverage of the X chromosome was also relatively sparse in our study (see Figure 2). Part of this bias reflects the smaller number of X-linked SNPs overall (Mouse Genome Sequencing Consortium 2002), a pattern presumably related to the lower neutral mutation rate on the X chromosome (McVean and Hurst 1997).
The relevance of wild mice to studies of lab strains:
The hybrid origins of the lab strains of mice, as well as the reliance of biomedical research on the genetics of these strains, emphasize the importance of understanding the contribution of evolutionary history in wild mouse species to present patterns of molecular diversity in the lab strains (Guenet and Bonhomme 2003). For example, individual genetic effects on phenotypic variation may erroneously appear to map to multiple chromosomes containing Dobzhansky-Muller incompatibilities between wild species due to gametic disequilibrium between these regions. Conversely, the lab strains provide exciting opportunities to unravel the genetics of speciation among their ancestors. Reproductive isolation between M. musculus and M. domesticus may be at an early stage (given that different populations show different levels of postzygotic isolation; Vyskocilova et al. 2005), providing a glimpse of speciation in progress. Additionally, more detailed characterizations of sequence diversity among the lab strains, including the discovery of additional SNPs, the completion of genome sequences for multiple lab strains, and improved genome annotation, will facilitate identification of genomic features that may correlate with incompatibilities (Payseur and Nachman 2005), such as accelerated interspecific divergence. The combination of these resources with tools for functional characterization of genomic regions underlying reproductive isolation and opportunities to measure introgression of these regions in natural hybrid zones suggests that the house mouse has much to tell us about the genetics of speciation.
Acknowledgments
We thank Tim Wiltshire and Mathew Pletcher for providing access to the data. We thank Gary Churchill for sharing an unpublished manuscript. Aida M. Andrés, Erik Dopman, Rick Harrison, Mohamed Noor, Daven Presgraves, Todd Schlenke, and Scott Williamson provided useful input during the project. We thank Matt Dean, Jeff Good, Michael Nachman, Mohamed Noor, David Rand, Leslie Turner, Tim Wiltshire, and one anonymous reviewer for helpful comments on the paper. This work was supported in part by NSF–DEB0344710 to H.E.H.
References
- Alibert, P., F. Fel-Clair, K. Manolakou, J. Britton-Davidian and J.-C. Auffray, 1997. Developmental stability, fitness, and trait size in laboratory hybrids between European subspecies of the house mouse. Evolution 51: 1284–1295. [DOI] [PubMed] [Google Scholar]
- Atchley, W. R., and W. M. Fitch, 1991. Gene trees and the origins of inbred strains of mice. Science 254: 554–558. [DOI] [PubMed] [Google Scholar]
- Atchley, W. R., and W. M. Fitch, 1993. Genetic affinities of inbred mouse strains of uncertain origin. Mol. Biol. Evol. 10: 1150–1169. [DOI] [PubMed] [Google Scholar]
- Auffray, J.-C., F. Vanlerberghe and J. Britton-Davidian, 1990. The house mouse progression in Eurasia: a palaeontological and archaeozoological approach. Biol. J. Linn. Soc. 41: 13–25. [Google Scholar]
- Barbash, D. A., D. F. Siino, A. M. Tarone and J. Roote, 2003. A rapidly evolving MYB-related protein causes species isolation in Drosophila. Proc. Natl. Acad. Sci. USA 100: 5302–5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton, N. H., 1983. Multilocus clines. Evolution 37: 454–471. [DOI] [PubMed] [Google Scholar]
- Barton, N. H., 2001. The role of hybridization in evolution. Mol. Ecol. 10: 551–568. [DOI] [PubMed] [Google Scholar]
- Barton, N. H., and K. S. Gale, 1993. Genetic analysis of hybrid zones, pp. 13–45 in Hybrid Zones and the Evolutionary Process, edited by R. G. Harrison. Oxford University Press, Oxford.
- Bateson, W., 1909. Heredity and variation in modern lights, pp. 85–101 in Darwin and Modern Science, edited by A. C. Seward. Cambridge University Press, Cambridge, UK.
- Beck, J. A., S. Lloyd, M. Hafezparast, M. Lennon-Pierce, J. T. Eppig et al., 2000. Genealogies of mouse inbred strains. Nat. Genet. 24: 23–25. [DOI] [PubMed] [Google Scholar]
- Bishop, C. E., P. Boursot, B. Baron, F. Bonhomme and D. Hatat, 1985. Most classical Mus musculus domesticus laboratory mouse strains carry a Mus musculus musculus Y chromosome. Nature 325: 70–72. [DOI] [PubMed] [Google Scholar]
- Bonhomme, F., J. L. Guenet, B. Dod, K. Moriwaki and G. Bulfield, 1987. The polyphyletic origin of laboratory inbred mice and their rate of evolution. Biol. J. Linn. Soc. 30: 51–58. [Google Scholar]
- Boursot, P., J.-C. Auffray, J. Britton-Davidian and F. Bonhomme, 1993. The evolution of house mice. Ann. Rev. Ecol. Syst. 24: 119–152. [Google Scholar]
- Boursot, P., W. Din, R. Anand, D. Darviche, B. Dod et al., 1996. Origin and radiation of the house mouse: mitochondrial DNA phylogeny. J. Evol. Biol. 9: 391–415. [Google Scholar]
- Britton-Davidian, J., F. Fel-Clair, J. Lopez, P. Alibert and P. Boursot, 2005. Postzygotic isolation between the two European subspecies of the house mouse: estimates from fertility patterns in wild and laboratory-bred hybrids. Biol. J. Linn. Soc. 84: 379–393. [Google Scholar]
- Bronson, F. H., 1979. The reproductive ecology of the house mouse. Quart. Rev. Biol. 54: 265–299. [DOI] [PubMed] [Google Scholar]
- Christie, P., and M. R. Macnair, 1984. Complementary lethal factors in two North American populations of the yellow monkeyflower. J. Hered. 75: 510–511. [Google Scholar]
- Coyne, J. A., and H. A. Orr, 1989. Two rules of speciation, pp. 180–207 in Speciation and Its Consequences, edited by D. Otte and J. Endler. Sinauer Associates, Sunderland, MA.
- Coyne, J. A., and H. A. Orr, 1998. The evolutionary genetics of speciation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 353: 287–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne, J. A., and H. A. Orr, 2004. Speciation. Sinauer Associates, Sunderland, MA.
- Dietrich, W. F., J. Miller, R. Steen, M. A. Merchant, D. Damron-Boles et al., 1996. A comprehensive genetic map of the mouse genome. Nature 380: 149–152. [DOI] [PubMed] [Google Scholar]
- Din, W., R. Anand, P. Boursot, D. Darviche, B. Dod et al., 1996. Origin and radiation of the house mouse: clues from nuclear genes. J. Evol. Biol. 9: 519–539. [Google Scholar]
- Dobzhansky, T., 1936. Studies on hybrid sterility. II. Localization of sterility factors in Drosophila pseudoobscura hybrids. Genetics 21: 113–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky, T., 1937. Genetics and the Origin of Species. Columbia University Press, New York.
- Dod, B., L. S. Jermiin, P. Boursot, V. H. Chapman, J. Tonnes-Nielsen et al., 1993. Counterselection on sex chromosomes in the Mus musculus European hybrid zone. J. Evol. Biol. 6: 529–546. [Google Scholar]
- Elliott, R. W., D. R. Miller, R. S. Pearsall, C. Hohman, Y. Zhang et al., 2001. Genetic analysis of testis weight and fertility in an interspecies hybrid congenic strain for Chr X. Mamm. Genome 12: 45–51. [DOI] [PubMed] [Google Scholar]
- Elliott, R. W., D. Poslinski, D. Tabaczynski, C. Hohman and J. Pazik, 2004. Loci affecting male fertility in hybrids between Mus macedonicus and C57BL/6. Mamm. Genome 15: 704–710. [DOI] [PubMed] [Google Scholar]
- Ferris, S. D, R. D. Sage, E. M. Prager, U. Ritte and A. C. Wilson, 1983. Mitochondrial DNA evolution in mice. Genetics 105: 681–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman, L., and J. H. Willis, 2001. Evidence for Dobzhansky-Muller incompatibilities contributing to the sterility of hybrids between Mimulus guttatus and M. nasutus. Evolution 55: 1932–1942. [DOI] [PubMed] [Google Scholar]
- Forejt, J., 1996. Hybrid sterility in the mouse. Trends Genet. 12: 412–417. [DOI] [PubMed] [Google Scholar]
- Forejt, J., and P. Ivanyi, 1975. Genetic studies on male sterility of hybrids between laboratory and wild mice (Mus musculus L.). Genet. Res. 24: 189–206. [DOI] [PubMed] [Google Scholar]
- Forejt, J., V. Vincek, J. Klein, H. Lehrach and M. Loudova-Mickova, 1991. Genetic mapping of the t-complex region on mouse chromosome 17 including the Hybrid sterility-1 gene. Mamm. Genome 1: 84–91. [DOI] [PubMed] [Google Scholar]
- Fossella, J., S. A. Samant, L. M. Silver, S. M. King, K. T. Vaughan et al., 2000. An axonemal dynein at the Hybrid Sterility 6 locus: implications for t haplotype-specific male sterility and the evolution of species barriers. Mamm. Genome 11: 8–15. [DOI] [PubMed] [Google Scholar]
- Frazer, K. A., C. M. Wade, D. A. Hinds, N. Patil, D. R. Cox et al., 2004. Segmental phylogenetic relationships of inbred mouse strains revealed by fine-scale analysis of sequence variation across 4.6 Mb of mouse genome. Genome Res. 14: 1493–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem, G., C. Ginane, M.-F. Ostrowski and A. Orth, 2005. Assessment of mate preference in the house mouse with reference to investigations on assortative mating. Biol. J. Linn. Soc. 84: 461–472. [Google Scholar]
- Gardner, K., A. Buerkle, J. Whitton and L. H. Rieseberg, 2002. Inferring epistasis in wild sunflower hybrid zones, pp. 264–279 in Epistasis and the Evolutionary Process, edited by J. B. Wolf, E. D. Brodie III and M. J. Wade. Oxford University Press, Oxford.
- Gavrilets, S., 1997. Hybrid zones with Dobzhansky-type epistatic selection. Evolution 51: 1027–1035. [DOI] [PubMed] [Google Scholar]
- Gregorova, S., M. Mnukova-Fajdelova, Z. Trachtulec, J. Capkova, M. Loudova et al., 1996. Sub-milli-Morgan map of the proximal part of mouse Chromosome 17 including the hybrid sterility 1 gene. Mamm. Genome 7: 107–113. [DOI] [PubMed] [Google Scholar]
- Grupe, A., S. Germer, J. Usuka, D. Aud, J. K. Belknap et al., 2001. In silico mapping of complex disease-related traits in mice. Science 292: 1915–1918. [DOI] [PubMed] [Google Scholar]
- Guenet, J. L., and F. Bonhomme, 2003. Wild mice: an ever-increasing contribution to a popular mammalian model. Trends Genet. 19: 24–31. [DOI] [PubMed] [Google Scholar]
- Guenet, J. L., C. Nagamine, D. Simon-Chazottes, X. Montagutelli and F. Bonhomme, 1990. Hst-3: an X-linked hybrid sterility gene. Genet. Res. 56: 163–165. [DOI] [PubMed] [Google Scholar]
- Haldane, J. B. S., 1922. Sex-ratio and unisexual sterility in hybrid animals. J. Genet. 12: 101–109. [Google Scholar]
- Harrison, A., P. Olds-Clarke and S. M. King, 1998. Identification of the t-complex-encoded cytoplasmic dynein light chain Tctex1 in inner arm I1 supports the involvement of flagellar dyneins in meiotic drive. J. Cell Biol. 140: 1137–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick, P. W., 2000. Genetics of Populations. Jones & Bartlett, London.
- Hill, W. G., and A. Robertson, 1968. Linkage disequilibrium in finite populations. Theor. Appl. Genet. 38: 226–231. [DOI] [PubMed] [Google Scholar]
- Hollingshead, L., 1930. A lethal factor in Crepis effective only in interspecific hybrids. Genetics 15: 114–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt, W. G., and R. K. Selander, 1973. Biochemical genetics of hybridisation in European house mice. Heredity 31: 11–33. [DOI] [PubMed] [Google Scholar]
- Ideraabdullah, F. Y., E. de la Casa-Esperon, T. A. Bell, D. A. Detwiler, T. Magnuson et al., 2004. Genetic and haplotype diversity among wild-derived mouse inbred strains. Genome Res. 14: 1880–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov, A. S., S. Sunyaev and F. A. Kondrashov, 2002. Dobzhansky-Muller incompatibilities in protein evolution. Proc. Natl. Acad. Sci. USA 99: 14878–14883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewontin, R. C., 1964. The interaction of selection and linkage. I. General considerations; heterotic models. Genetics 49: 49–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindblad-Toh, K., E. Winchester, M. J. Daly, D. G. Wang, J. N. Hirschhorn et al., 2000. Large-scale discovery and genotyping of single-nucleotide polymorphisms in the mouse. Nat. Genet. 24: 381–386. [DOI] [PubMed] [Google Scholar]
- McVean, G. T., and L. Hurst, 1997. Evidence for a selectively favourable reduction in the mutation rate of the X chromosome. Nature 386: 388–392. [DOI] [PubMed] [Google Scholar]
- Mendelson, T. C., B. D. Inouye and M. D. Rausher, 2004. Quantifying patterns in the evolution of reproductive isolation. Evolution 58: 1424–1433. [DOI] [PubMed] [Google Scholar]
- Moulia, C., N. LeBrun, J. Dallas, A. Orth and F. Renaud, 1993. Experimental evidence of genetic determinism in high susceptibility to intestinal pinworm infection in mice: a hybrid zone model. Parasitology 106: 387–393. [DOI] [PubMed] [Google Scholar]
- Moulia, C., N. LeBrun, C. Loubes, R. Marin and F. Renaud, 1995. Hybrid vigor in parasites of interspecific crosses between two mice species. Heredity 74: 48–52. [DOI] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium, 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520–562. [DOI] [PubMed] [Google Scholar]
- Muller, H. J., 1940. Bearing of the Drosophila work on systematics, pp. 185–268 in The New Systematics, edited by J. S. Huxley. Clarendon Press, Oxford.
- Muller, H. J., 1942. Isolating mechanisms, evolution, and temperature. Biol. Symp. 6: 71–125. [Google Scholar]
- Munclinger, P., E. Bozikova, M. Sugerkova, J. Pialek and M. Macholan, 2002. Genetic variation in house mice (Mus, Muridae, Rodentia) from the Czech and Slovak Republics. Folia Zool. 51: 81–92. [Google Scholar]
- Mural, R. J., M. D. Adams, E. W. Myers, H. O. Smith, G. L. Gabor Miklos et al., 2002. A comparison of whole-genome shotgun-derived mouse chromosome 16 and the human genome. Science 296: 1661–1671. [DOI] [PubMed] [Google Scholar]
- Ohta, T., and M. Kimura, 1969. Linkage disequilibrium due to random genetic drift. Genet. Res. 13: 47–55. [Google Scholar]
- Orr, H. A., 1987. Genetics of male and female sterility in hybrids of Drosophila pseudoobscura and Drosophila persimilis. Genetics 116: 555–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr, H. A., 1995. The population genetics of speciation: the evolution of hybrid incompatibilities. Genetics 139: 1805–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr, H. A., 1996. Dobzhansky, Bateson, and the genetics of speciation. Genetics 144: 1331–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr, H. A., and M. Turelli, 2001. The evolution of postzygotic isolation: accumulating Dobzhansky-Muller incompatibilities. Evolution 55: 1085–1094. [DOI] [PubMed] [Google Scholar]
- Orr, H. A., J. P. Masly and D. C. Presgraves, 2005. Speciation genes. Curr. Op. Genet. Dev. 14: 675–679. [DOI] [PubMed] [Google Scholar]
- Pantazidis, A. C., and E. Zouros, 1988. Location of an autosomal factor causing sterility in Drosophila mojavensis males carrying the Drosophila arizonensis Y chromosome. Heredity 60: 299–304. [DOI] [PubMed] [Google Scholar]
- Payseur, B. A., and M. W. Nachman, 2005. The genomics of speciation: investigating the molecular correlates of X chromosome introgression across the hybrid zone between Mus domesticus and Mus musculus. Biol. J. Linn. Soc. 84: 523–534. [Google Scholar]
- Payseur, B. A., J. G. Krenz and M. W. Nachman, 2004. Differential patterns of introgression across the X chromosome in a hybrid zone between two species of house mice. Evolution 58: 2064–2078. [DOI] [PubMed] [Google Scholar]
- Perez, D. E., and C.-I Wu, 1995. Further characterization of the Odysseus locus of hybrid sterility in Drosophila: one gene is not enough. Genetics 140: 201–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilder, S. H., P. Olds-Clarke, D. M. Phillips and L. M. Silver, 1993. Hybrid Sterility 6: a mouse t complex locus controlling sperm flagellar assembly and movement. Dev. Biol. 159: 631–642. [DOI] [PubMed] [Google Scholar]
- Pletcher, M. T., P. McClurg, S. Batalov, A. I. Su, S. W. Barnes et al., 2004. Use of a dense single nucleotide polymorphism map for in silico mapping in the mouse. PLOS Biol. 2: 2159–2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves, D. C., 2003. A fine-scale genetic analysis of hybrid incompatibilities in Drosophila. Genetics 163: 955–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves, D. C., L. Balagopalan, S. M. Abymayr and H. A. Orr, 2003. Adaptive evolution drives divergence of a hybrid inviability gene between two species of Drosophila. Nature 423: 715–719. [DOI] [PubMed] [Google Scholar]
- Sage, R. D., D. Heyneman, K.-C. Lim and A. C. Wilson, 1986. Wormy mice in a hybrid zone. Nature 324: 60–63. [DOI] [PubMed] [Google Scholar]
- Sage, R. D., W. R. Atchley and E. Capanna, 1993. House mice as models in systematic biology. Syst. Biol. 42: 523–561. [Google Scholar]
- Sakai, T., Y. Kikkawa, I. Miura, T. Inoue, K. Moriwaki et al., 2005. Origins of mouse inbred strains deduced from whole-genome scanning by polymorphic microsatellite loci. Mamm. Genome 16: 11–19. [DOI] [PubMed] [Google Scholar]
- Silver, L. M., 1985. Mouse t haplotypes. Annu. Rev. Genet. 19: 179–208. [DOI] [PubMed] [Google Scholar]
- Silver, L. M., 1995. Mouse Genetics. Oxford University Press, New York.
- Smadja, C., and G. Ganem, 2002. Subspecies recognition in the house mouse: a study of two populations from the border of a hybrid zone. Behav. Ecol. 13: 312–320. [Google Scholar]
- Storchova, R., S. Gregorova, D. Buckiova, V. Kyselova, P. Divina et al., 2004. Genetic analysis of X-linked hybrid sterility in the house mouse. Mamm. Genome 15: 515–524. [DOI] [PubMed] [Google Scholar]
- Storey, J. D., and R. Tibshirani, 2003. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 100: 9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao, Y., and D. L. Hartl, 2003. Genetic dissection of hybrid incompatibilities between Drosophila simulans and D. mauritiana. III. Heterogeneous accumulation of hybrid incompatibilities, degree of dominance, and implications for Haldane's rule. Evolution 57: 2580–2598. [DOI] [PubMed] [Google Scholar]
- Ting, C. T., S. C. Tsaur, M. L. Wu and C.-I Wu, 1998. A rapidly evolving homeobox at the site of a hybrid sterility gene. Science 282: 1501–1504. [DOI] [PubMed] [Google Scholar]
- Trachtulec, Z., M. Mnukova-Fajdelova, R. M. Hamvas, S. Gregorova, W. E. Mayer et al., 1997. Isolation of candidate hybrid sterility 1 genes by cDNA selection in a 1.1 megabase pair region on mouse chromosome 17. Mamm. Genome 8: 312–316. [DOI] [PubMed] [Google Scholar]
- Trachtulec, Z., O. Mihola, C. Vlcek, H. Himmelbauer, V. Paces et al., 2005. Positional cloning of the Hybrid sterility 1 gene: fine genetic mapping and evaluation of two candidate genes. Biol. J. Linn. Soc. 84: 637–641. [Google Scholar]
- True, J. R., B. S. Weir and C. C. Laurie, 1996. A genome-wide survey of hybrid incompatibility factors by the introgression of marked segments of Drosophila mauritiana chromosomes into Drosophila simulans. Genetics 142: 819–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker, P. K., R. D. Sage, J. Warner, A. C. Wilson and E. M. Eicher, 1992. Abrupt cline for sex chromosomes in a hybrid zone between two species of mice. Evolution 46: 1146–1163. [DOI] [PubMed] [Google Scholar]
- Turelli, M., N. H. Barton and J. A. Coyne, 2001. Theory and speciation. Trends Ecol. Evol. 16: 330–343. [DOI] [PubMed] [Google Scholar]
- Vanlerberghe, F., B. Dod, P. Boursot, M. Bellis and F. Bonhomme, 1986. Absence of Y chromosome introgression across the hybrid zone between Mus musculus domesticus and Mus musculus musculus. Genet. Res. 48: 191–197. [DOI] [PubMed] [Google Scholar]
- Vyskocilova, M., Z. Trachtulec, J. Forejt and J. Pialek, 2005. Does geography matter in hybrid sterility in house mice? Biol. J. Linn. Soc. 84: 663–674. [Google Scholar]
- Wade, C. M., E. J. Kulbokas III, A. W. Kirby, M. C. Zody, J. C. Mullikin et al., 2002. The mosaic structure of variation in the laboratory mouse genome. Nature 420: 574–578. [DOI] [PubMed] [Google Scholar]
- Welch, J. J., 2004. Accumulating Dobzhansky-Muller incompatibilities: reconciling theory and data. Evolution 58: 1145–1156. [DOI] [PubMed] [Google Scholar]
- Wiltshire, T., M. T. Pletcher, S. Batalov, S. W. Barnes, L. M. Tarantino et al., 2003. Genome-wide single nucleotide polymorphism analysis defines haplotype patterns in mouse. Proc. Natl. Acad. Sci. USA 100: 3380–3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittbrodt, J., D. Adam, B. Malitschek, W. Mqaueler, F. Raulf et al., 1989. Novel putative receptor tyrosine kinase encoded by the melanoma-inducing Tu locus in Xiphophorus. Nature 341: 415–421. [DOI] [PubMed] [Google Scholar]
- Wu, C.-I, and A. T. Beckenbach, 1983. Evidence for extensive genetic differentiation between the sex-ratio and the standard arrangement of Drosophila pseudoobscura and D. persimilis and identification of hybrid sterility factors. Genetics 105: 71–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekawa, H., K. Moriwaki, O. Gotoh, J. Watanabe, J.-I. Hayashi et al., 1980. Relationship between laboratory mice and subspecies Mus musculus domesticus based on restriction endonuclease cleavage patterns of mitochondrial DNA. Jpn. J. Genet. 55: 289–296. [Google Scholar]
- Zhang, J., K. W. Hunter, M. Gandolph, W. L. Rowe, R. P. Finney et al., 2005. A high-resolution multistrain haplotype analysis of laboratory mouse genome reveals three distinctive genetic variation patterns. Genome Res. 15: 241–249. [DOI] [PMC free article] [PubMed] [Google Scholar]