Abstract
We mapped quantitative trait loci (QTL) controlling differences in seed oil content and composition between cultivated and wild sunflower and used the results, along with those of a previous study of domestication-related QTL, to guide a genome-wide analysis of genetic variation for evidence of past selection. The effects of the seed oil QTL were almost exclusively in the expected direction with respect to the parental phenotypes. A major, oil-related QTL cluster mapped near a cluster of domestication-related QTL on linkage group six (LG06), the majority of which have previously been shown to have effects that are inconsistent with the parental phenotypes. To test the hypothesis that this region was the target of a past selective sweep, perhaps resulting in the fixation of the antagonistic domestication-related QTL, we analyzed simple sequence repeat (SSR) diversity from 102 markers dispersed throughout the sunflower genome. Our results indicate that LG06 was most likely the target of multiple selective sweeps during the postdomestication era. Strong directional selection in concert with genetic hitchhiking therefore offers a possible explanation for the occurrence of numerous domestication-related QTL with apparently maladaptive phenotypic effects.
THE derivation of crop plants from their wild ancestors has typically involved rapid phenotypic evolution in response to strong directional selection (Harlan 1992). Biologists dating back at least as far as Darwin (1859) have argued that these dramatic, human-mediated transformations provide a model for studying phenotypic evolution. This is due, in part, to the fact that studies of evolution under domestication often enjoy a historical backdrop that is unavailable to many investigators studying phenotypic divergence in the wild. Not only do we know the types of traits that were likely under selection during crop domestication and improvement, but also the timescale over which this evolution occurred is often well documented. These factors, when combined with the recent development of genetic tools for the analysis of many domesticated plants and animals, translate into unique opportunities for studying the genetic and phenotypic consequences of strong directional selection.
One approach that has been widely applied to questions about the genetics underlying phenotypic divergence has been quantitative trait locus (QTL) mapping. While QTL mapping has produced a tremendous amount of information on the genetic architecture of trait differences, this approach is largely agnostic regarding the evolutionary forces causing these differences. The one exception to this is the QTL sign test of Orr (1998), which uses data on the direction of QTL effects to detect the footprint of directional selection. Unfortunately, the utility of this test is closely associated with the number of loci detected. As such, the QTL sign test is of little use for traits with relatively few detectable QTL.
A complementary approach to QTL mapping is to use population genetic data to identify regions of the genome that harbor selectively important genes. This idea, which was first proposed by Cavalli-Sforza (1966), was later formalized by Lewontin and Krakauer (1973), who used variation in the inbreeding coefficient across loci in an attempt to detect selection. Although this approach was subsequently criticized on a variety of grounds (e.g., Nei and Maruyama 1975; Robertson 1975a,b), the underlying logic remains valid. While the effects of migration, inbreeding, and genetic drift are manifested throughout the genome, selection acts in a locus-specific manner. Thus, selective sweeps can reduce genetic variation at both the target locus and the linked neutral loci while leaving the remainder of the genome unaffected (Maynard-Smith and Haigh 1974; Slatkin 1995). Since Lewontin and Krakauer's (1973) initial publication on the subject, a number of authors have proposed improved methods for using population genetic data to detect selection (e.g., Tsakas and Krimbas 1976; Bowcock et al. 1991; Beaumont and Nichols 1996; Vitalis et al. 2001; Schlötterer 2002), and recent years have seen the increasingly frequent use of such methods to successfully identify selectively important loci in a wide variety of taxa, including plants (e.g., Vigouroux et al. 2002), microbes (e.g., Wootton et al. 2002), insects (e.g., Schöfl and Schlötterer 2004), mollusks (e.g., Wilding et al. 2001), fishes (e.g., Campbell and Bernatchez 2004), and mammals (e.g., Akey et al. 2002; Storz and Nachman 2003; Storz et al. 2004).
When used in combination, QTL mapping and population genetic analyses provide an especially powerful approach to the study of phenotypic divergence in response to selection. While QTL-based approaches can identify genomic regions that harbor genes underlying phenotypic differences, population genetic data can be used to investigate the role of selection in producing the observed differences. Conversely, QTL mapping can provide an initial framework for investigating the phenotypic effects of selectively important regions of the genome. Here we describe the joint application of these techniques in a study of the evolution of cultivated sunflower (Helianthus annuus L.).
Derived from the wild, common sunflower (also H. annuus) ∼4000 years ago (Crites 1993), cultivated sunflower stands as the only major food crop that is native to temperate North America (Harter et al. 2004). Despite being considered members of the same species, cultivated and wild sunflower exhibit a number of morphological differences that trace back to the original domestication event. For example, wild sunflower is characterized by a highly branched growth form with numerous, small flowering heads and relatively small achenes (i.e., single-seeded fruits) that are released upon maturation. Cultivated sunflower, on the other hand, is completely unbranched, producing a single large head as well as relatively large achenes that are held until harvest.
Following its domestication, cultivated sunflower served as an important source of food, pigment, and medicine for Native Americans (Heiser 1951). Like many crop plants, however, sunflower has experienced a complex evolutionary history involving multiple population bottlenecks accompanied by periods of presumably intense selection (Putt 1997; Tang and Knapp 2003). The most recent of these occurred in the mid-20th century, when plant breeders transformed sunflower into one of the world's most important sources of edible oil (Putt 1997). However, our recent analysis of domestication-related traits in a cross between wild and cultivated sunflower (Burke et al. 2002) failed to provide evidence of consistent directional selection for most traits. In some cases, there was insufficient power (i.e., too few QTL) to reject the null hypothesis of neutral divergence (Orr 1998). In other cases, there were sufficient numbers of QTL but antagonistic effects (i.e., cultivar alleles producing a more wild-like phenotype and vice versa) were common, suggesting that the observed trait differences often were not the result of consistent directional selection.
The one exceptional trait was achene size, which showed clear evidence of past directional selection. This result led us to conclude that “strong directional selection for increased achene size appears to have played a central role in sunflower domestication” (Burke et al. 2002, p. 1257). For 9 of 10 genomic regions influencing achene size, the cultivar allele produced larger achenes. The 10th region, which carried QTL for both achene width and achene weight, was located on LG06 and was embedded within a cluster of other, apparently maladaptive (i.e., antagonistic) QTL (Figure 1). While it is possible that this region could have arisen through the chance fixation of a maladaptive chromosomal block during domestication, we previously hypothesized that strong selection favoring one or more QTL underlying a presumably unmeasured trait, and with antagonistic effects on other (domestication-related) traits, might be responsible for the abundance of antagonistic QTL on LG06. Given the recent history of sunflower, oil-related characters are the most obvious candidate traits to investigate. In this article, we report the results of a QTL analysis of seed oil content and composition in sunflower and use our results to guide a genome-wide analysis of patterns of simple sequence repeat (SSR) variation for evidence of past selection.
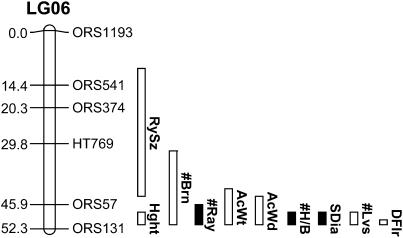
Figure 1.
Genetic map of sunflower LG06 depicting the occurrence of numerous domestication-related QTL with effects in the “wrong” direction. Map distances (in centimorgans) are listed to the left of each linkage group, whereas marker names are listed to the right. Vertical bars to the right of each linkage group reflect the 1-LOD support intervals for the location of each QTL. Solid bars indicate QTL with effects in the expected direction, whereas open bars indicate QTL with effects in the wrong direction. DFlr, days to first flower; #Lvs, number of main stem leaves; SDia, stem diameter; #H/B, number of heads/branch; AcWd, mean achene width; AcWt, mean achene weight; #Ray, number of ray flowers; #Brn, number of lateral branches; RySz, size of ray flowers; Hght, height at flowering.
MATERIALS AND METHODS
Development and genotypic characterization of the mapping population:
The mapping population, which was previously described by Burke et al. (2002), consisted of 374 F3 individuals derived from a single, self-pollinated F1 hybrid of an elite oilseed cultivar (cmsHA89) crossed with wild H. annuus (ANN1238). Each individual in the mapping population was initially genotyped for 106 SSRs and two morphological markers (hypocotyl/disc pigmentation and restoration of male fertility). For the purposes of this study, five dominant markers were removed from the analysis because data from subsequent mapping populations suggest that they were initially misplaced (Tang et al. 2002; Yu et al. 2003). In addition, a single EST-based SSR (HT769) was added to the map to fill a 20-cM gap in the middle of LG06.
Phenotypic characterization of the mapping population:
Achenes (i.e., single-seeded fruits) were collected from the self-pollinated primary head of each plant. For all plants with sufficient yield (125 total), seed bulks were phenotyped for percentage of oil content and fatty acid composition following standard protocols as follows. The oil concentrations of clean, physiologically mature achenes were measured by pulsed nuclear magnetic resonance (NMR) analysis on an Oxford 4000 NMR (Concord, MA). Flat-bottomed sample tubes were filled with achene samples weighing between 0.5 and 1.0 g each. The NMR was calibrated using standards developed from the achenes of low-oil (285.9 g/kg oil) and high-oil (444.5 g/kg oil) recombinant inbred lines (RIL 54 and 75, respectively) developed from the cmsHA89 × ANN1238 mapping population (Burke et al. 2002).
Fatty acid concentrations were measured by gas chromatography of fatty acid methyl esters. For each individual, samples were prepared by grinding 20 achenes (seeds) in 10 ml of HPLC-grade hexane using a Polytron (Brinkmann Instruments, Westbury, NY). The mixture was allowed to settle for 20–30 min before transferring 0.5 ml of the supernatant to a 16 × 100-mm glass tube. Capped samples were heated for 15 min at 50° in a heat block. The hexane was evaporated under a gentle stream of nitrogen gas before adding 0.1 ml of ethyl ether and 0.1 ml 0.1 m of KOH in methanol and heating the samples for 5 min at 50°. The transesterification reaction was neutralized by adding 0.1 ml of 0.15 m HCl to each tube, followed by 2.0 ml of hexane. Samples were then mixed by swirling and allowed to settle. Using a disposable glass Pasteur pipette, 0.5 ml of the upper phase (hexane) was transferred to a gas chromatography vial and capped. We injected 1.0-μl samples onto an Agilent Technologies (Palo Alto, CA) DB-23 micrometer column mounted in an HP6890 gas chromatograph (Hewlett-Packard, Wilmington, DE) using a split ratio of 1:80. The initial oven temperature was 50°. Oven temperatures were ramped up from an initial temperature of 50° to 185° in 30°/min increments and held at 185° for 4.5 min. Total run time was 10 min. Fatty acid concentrations were calculated using ChemStation software (Agilent Technologies). Palmitic, stearic, oleic, and linoleic acid peaks were identified using standards purchased from NU-CHEK PREP Prep (Elysian, MN).
QTL analysis:
All five oil-related traits were analyzed via composite-interval mapping (CIM; Zeng 1993, 1994) as implemented by the software package QTL Cartographer version 1.17 (Basten et al. 2003). Tests for the presence of a QTL were performed at 2-cM intervals using a 10-cM window and five background cofactors, which were selected via forward-backward stepwise regression. For each trait, genome-wide threshold values (α = 0.05) for declaring the presence of a QTL were estimated from 1000 permutations of the data (Churchill and Doerge 1994; Doerge and Churchill 1996). The 1-LOD support limits for the position of each QTL were calculated from the CIM results. Also note that the positions of the previously mapped domestication-related QTL were reestimated following the removal of the aberrantly placed dominant markers and the addition of HT769.
In addition to testing for the presence/absence of QTL, Zmapqtl also provides an estimate of the additive (a) and dominance (d) effects of the QTL. The degree of dominance of the cultivar allele at each locus was calculated as d/a, such that the expected value of a perfectly additive locus is 0. Completely dominant or recessive loci have expected values of 1.0 and −1.0, respectively. Values of >1.0 or <−1.0 are due to over/underdominance. Following Burke et al. (2002), the mode of gene action of each QTL was classified as follows: underdominant <−1.25 ≤ recessive <−0.75 ≤ partially recessive <−0.25 ≤ additive ≤ 0.25 < partially dominant ≤ 0.75 < dominant ≤ 1.25 < overdominant.
Analyses of genetic diversity:
On the basis of the results of the QTL analysis (see below for details), we wanted to test the hypothesis that LG06 has been the target of recent selection. To test for the signature of selection on LG06, we analyzed patterns of SSR variation across the sunflower genome using the software package POPGENE (version 1.31; Yeh et al. 1999). This analysis was based on genotypic data from a total of 102 single-locus, codominant SSR loci (supplemental Table 1S at http://www.genetics.org/supplemental/) that were run on a panel consisting of one individual from each of 15 wild sunflower populations, 13 Native American landraces and open-pollinated, primitive cultivars (hereafter referred to as the “exotic” lines), and 16 highly improved, elite oilseed inbred lines (supplemental Table 2S at http://www.genetics.org/supplemental/; see Tang and Knapp 2003 and Yu et al. 2002 for additional details regarding both the markers employed and the individuals surveyed). The number of markers per linkage group ranged from three to eight, with a total of eight loci (ORS57, ORS339, ORS349, ORS381, ORS608, ORS678, ORS1193, and HT769) residing on LG06 (Burke et al. 2002, 2004; Tang et al. 2002; Yu et al. 2003). Of these, three (ORS57, ORS1193, and HT769) were already on our map from the QTL analysis, three (ORS 339, ORS 381, and ORS678) were subsequently added to our map to investigate the actual target(s) of selection, and two (ORS349 and ORS608) could not be mapped in our population but are known to reside on LG06 on the basis of their locations on other sunflower genetic maps (Tang et al. 2002; Yu et al. 2003, Burke et al. 2004).
RESULTS
QTL analysis:
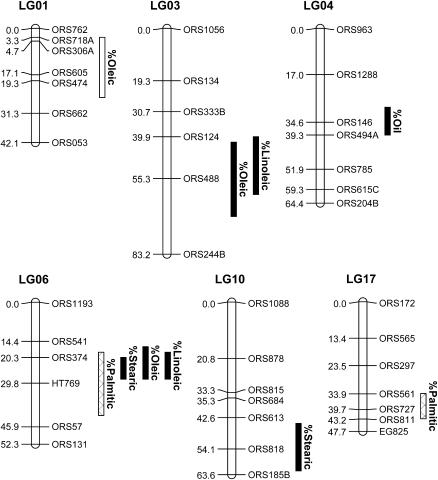
Results of the QTL analysis are summarized in Table 1 and presented graphically in Figure 2. CIM revealed the presence of 9 QTL affecting the five traits analyzed. The 1-LOD support limits, which provide an approximate confidence interval for the location of each QTL, ranged from 8.0 to 27.4 cM (mean = 15.5 cM), and the number of QTL per trait ranged from 1 to 3. These 9 loci were distributed across six linkage groups, one of which (LG06) carried a cluster of 4 oil-related QTL. Because two of these traits (% oleic and % linoleic) exhibit a strong negative correlation (r2 = 0.99), they were further analyzed via multiple-trait CIM (Jiang and Zeng 1995). This analysis, which was performed using the JZmapqtl module of QTL Cartographer, resulted in the identification of an additional % oleic QTL on LG03, bringing the total number of detected QTL to 10 (Table 1).
TABLE 1.
Putative QTL positions, effect of magnitudes/directions, and modes of action for seed oil content and composition using CIM in an F3 population of cultivated (cmsHA89) × wild (H. annuus var. annuus) sunflower
| Trait | Linkage group | Positiona | 1-LOD intervalb | PVEc | Direction of effectd | Degree of dominanced | Mode of actione | Method of detectionf |
|---|---|---|---|---|---|---|---|---|
| % oil content | LG04 | 36.6 | 29.1–39.3 | 14.5 | + | −0.51 | r | CIM |
| % palmitic acid | LG06 | 26.3 | 18.4–41.7 | 15.8 | − | −0.14 | A | CIM |
| LG17 | 39.6 | 33.8–43.1 | 10.7 | + | 1.23 | D | CIM | |
| % stearic acid | LG06 | 24.3 | 20.3–28.3 | 35.9 | + | −0.29 | r | CIM |
| LG10 | 50.7 | 44.7–62.2 | 11.5 | + | −0.36 | r | CIM | |
| % oleic acid | LG01 | 19.5 | 3.3–25.5 | 10.1 | − | 0.72 | d | CIM |
| LG03 | 63.3 | 41.9–69.3 | 12.7 | + | 1.31 | O | MCIM | |
| LG06 | 22.3 | 16.4–28.3 | 24.0 | + | 0.25 | d | CIM/MCIM | |
| % linoleic acid | LG03 | 47.9 | 39.9–61.2 | 15.1 | − | 0.34 | d | CIM/MCIM |
| LG06 | 24.3 | 18.4–28.3 | 28.9 | − | 0.24 | A | CIM/MCIM |
Absolute position from left telomere in centimorgans.
The region flanking each QTL peak in which LOD scores decline by 1.
Percentage of phenotypic variation explained by each QTL using CIM.
The direction of the additive effect and degree of dominance of the cmsHA89 allele.
Mode of action of the cmsHA89 allele: r, partially recessive; A, additive; d, partially dominant; D, dominant; and O, overdominant.
The statistical methodology used to detect each QTL. CIM, composite-interval mapping; MCIM, multi trait composite-interval mapping.
Figure 2.
Genetic map of six linkage groups showing the locations of QTL underlying various measures of seed oil content and composition in a cross between cultivated and wild sunflower. Map distances (in centimorgans) are listed to the left of each linkage group, whereas marker names are listed to the right. Vertical bars to the right of each linkage group reflect the 1-LOD support intervals for the location of each QTL. Solid bars indicate QTL with effects in the expected direction, whereas open bars indicate QTL with effects in the wrong direction. The cross-hatched bars reflect the fact that no expectation could be established for palmitic acid content.
Individual loci explained 10.1–35.9% of the phenotypic variation of any given trait (Table 1) and, for the most part, had effects in the direction expected on the basis of the trait differences outlined in Table 2. The exceptions to this were the QTL for palmitic acid content as well as 1 QTL for oleic acid content. The former trait does not differ significantly between cultivated and wild sunflower such that no expectation could be established, and the 2 QTL were split, with one causing an increase and the other a decrease in palmitic acid. With regard to oleic acid content, the cultivar allele at 1 of the 3 QTL (located on LG01) produced a decrease in oleic acid. In terms of gene action, 2 of the 10 QTL behaved in an additive fashion, whereas the cultivar allele was partially recessive at 3 loci and partially or completely dominant at 4 loci. One locus showed evidence of overdominance.
TABLE 2.
Comparison of seed oil content and composition between cultivated (cmsHA89) and common sunflower (H. annuus)
| Trait | Cultivated sunflower | Common sunflower |
|---|---|---|
| % oil contenta | 40 | 26 |
| % palmitic acid | 6 | 5 |
| % stearic acida | 6 | 3 |
| % oleic acida | 50 | 25 |
| % linoleic acida | 38 | 63 |
Data were obtained from the United States Department of Agriculture Germplasm Resources Information Network (GRIN) database at http://www.ars-grin.gov/.
A statistically significant difference (P < 0.01) between cultivated and common sunflower.
As noted above, the fact that LG06 harbors a majority of domestication-related QTL with effects in the opposite direction of the cultivar phenotype (Figure 1) led us to hypothesize that strong selection favoring one or more previously unidentified QTL with opposing effects on other (domestication-related) traits might be responsible for the abundance of antagonistic QTL on LG06. We further hypothesized that such selectively important QTL would likely be of large effect. The finding of relatively large, oil-related QTL [percentage of phenotypic variation explained (PVE) is 15.8, 35.9, 24.0, and 28.9; Table 1] on this linkage group is consistent with our hypothesis of selective fixation and prompted us to analyze this region of the genome for population genetic evidence of a past selective sweep. Note that no other genomic region had oil QTL with PVE > 15.1 (Table 1) or affected more than two seed oil traits.
Patterns of genetic diversity:
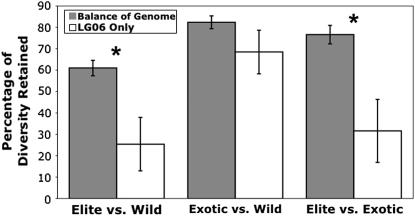
As previously documented (Tang and Knapp 2003), wild sunflower populations (He = 0.81 ± 0.13, mean ± SD), exotic lines (He = 0.64 ± 0.15), and elite inbred lines (He = 0.45 ± 0.22) contain progressively less genetic diversity (calculated for each locus as He = 1 − Σpi2, summing across i alleles at each locus), presumably due to a series of population bottlenecks associated with domestication and subsequent improvement. To test the hypothesis that LG06 was the target of selection during the evolution of cultivated sunflower, we compared the reduction in expected heterozygosity along LG06 that occurred during the wild-elite transition with the reduction experienced across the balance of the genome. We did this by calculating ln(RH) for each locus (where RH is the ratio of gene diversity in the elite vs. wild screening panels) and comparing the values for markers located on LG06 against those found elsewhere in the genome. This statistic has approximately the same expected value when compared between populations, and simulations indicate that it is largely independent of the mutation rate and robust to population expansions, bottlenecks, admixture, and the predominant mode of mutation (Schlötterer and Dieringer 2004). Consistent with the hypothesis that LG06 experienced strong directional selection during the evolution of cultivated sunflower, markers located on this linkage group exhibited a significantly greater loss of genetic diversity than did markers located elsewhere in the genome (t = −3.35, d.f. = 100, P = 0.001; Wilcoxon χ2 = 7.44, d.f. = 1, P = 0.006; Figure 3). To investigate when during the evolution of cultivated sunflower this pattern arose (i.e., during domestication or subsequent improvement), we performed a similar analysis of both the wild-exotic transition and the exotic-elite transition (Figure 3). These analyses revealed that, while there is no detectable difference between LG06 and the remainder of the genome when comparing the exotic lines against the wild populations (t = −1.47, d.f. = 100, P > 0.10; Wilcoxon χ2 = 2.12, d.f. = 1, P > 0.10), the significantly greater drop in genetic diversity on LG06 reappears when comparing the highly improved elite lines against the exotic lines (t = −3.70, d.f. = 100, P = 0.0004; Wilcoxon χ2 = 7.85, P = 0.005).
Figure 3.
Graphical representation of the percentage of genetic diversity (He) retained by SSR markers located on LG06 vs. those located elsewhere in the genome in comparisons between wild, domesticated (exotic), and improved (elite) sunflower lines. Error bars indicate ± 1 SE.
As a further testament to the extreme loss of genetic diversity on LG06, it is worth noting that only 9 of the 102 markers that we surveyed had ln(RH) values (on the basis of the elite-wild transition) that were >2 standard deviations below the mean, yet 4 of these 9 loci are located on LG06. On the basis of the results of a permutation test wherein we randomly shuffled markers across linkage groups, we can conclude that this number is significantly greater than expected by chance (P < 0.001). Indeed, not even 1 of the 1000 permutations that we ran resulted in 4 or more such markers on LG06. Again, this result is consistent with the hypothesis that LG06 was a major target of selection during the evolution of cultivated sunflower.
In terms of the partitioning of genetic variation within and among our “population” samples, markers on LG06 exhibited significantly higher FST values than did markers from the balance of the genome when the wild and elite gene pools were compared [FST = 0.226 ± 0.028 vs. 0.152 ± 0.008, respectively; t = 2.08, d.f. = 100, P = 0.04; FST values were Box-Cox transformed (Box and Cox 1964) to restore normality]. This difference disappeared entirely when comparing between the wild and exotic gene pools (FST = 0.095 ± 0.020 vs. 0.093 ± 0.006, respectively; t = 0.13, d.f. = 100, P > 0.80) only to reemerge (albeit weakly and nonsignificantly) when the exotic and elite gene pools were compared (FST = 0.110 ± 0.022 vs. 0.076 ± 0.007; t = 1.266, d.f. = 100, P = 0.21). Again, these results are consistent with the hypothesis that LG06 was a major target of selection during the evolution of cultivated sunflower, most likely during the postdomestication era.
DISCUSSION
Unlike our previous analysis of domestication-related traits in sunflower (Burke et al. 2002), this study of seed oil content and composition (which was performed in the same cultivated × wild mapping population) revealed the occurrence of a number of loci with moderate to large effects. Indeed, QTL underlying oil-related traits explained significantly more of the phenotypic variation segregating in the mapping population (17.9 ± 2.9%, mean ± SE) than did those corresponding to other domestication-related traits (10.5 ± 1.0%; t = 2.50, d.f. = 86, P = 0.02). This result suggests that, while the phenotypic transition from wild to domesticated sunflower may have been rather smooth, the transformation of sunflower into an oilseed crop was likely more punctuated, resulting from the accumulation of alleles of comparatively large effect. It should be noted, however, that even the larger oil-related QTL are still relatively small when compared to those derived from other studies of crop plant evolution (e.g., Doebley and Stec 1991; Koinange et al. 1996). Thus, even this latter phase of sunflower evolution may have involved relatively few major phenotypic leaps. In terms of gene action, the derived (i.e., cultivar) allele was, on average, slightly (but not significantly) more dominant for oil-related compared to domestication-related QTL (d/a = 0.27 ± 0.88, mean ± SE, vs. 0.14 ± 0.34, respectively; t = 0.14, d.f. = 86, P > 0.80).
With regard to the direction of QTL effects, just over one-third of the domestication-related QTL that we previously identified had antagonistic effects, with the cultivar allele producing a wild-like phenotype (and vice versa). In contrast, the effects of the oil-related QTL identified here were mostly in the expected direction. The only possible exceptions to this were the two QTL for palmitic acid content, for which no expectation could be established (Table 2), and a single oleic acid content QTL on LG01. While the occurrence of selection on seed oil content during the transformation of sunflower into an oilseed crop is well established (Putt 1997), the role of selection in producing the observed differences in seed oil composition is less clear. Thus, following the methods of Rieseberg et al. (2002), we pooled our results across the four oil-related traits for which an expected direction of effect could be established and applied Orr's (1998) QTL sign test. Although the results are only marginally significant (P = 0.086), owing at least in part to the relatively small number of QTL detected, the outcome of this test suggests that this suite of traits, unlike most domestication-related traits, has experienced a history of directional selection.
As noted in the Introduction, LG06 is unique with respect to the occurrence of antagonistic QTL (Figure 1). Seven of the 10 domestication-related QTL on this linkage group have effects that oppose the cultivar phenotype, including 2 that influence the only trait for which there is strong evidence of selection during domestication (i.e., achene size). This pattern, when combined with our finding that this same linkage group carries comparatively large oil-related QTL with effects in the expected direction, motivated our analysis of LG06 for population genetic evidence of a past selective sweep. Not surprisingly, there was an overall drop in diversity across the sunflower genome across successive stages of domestication and improvement (Figure 3). More interestingly, however, LG06 experienced a significantly greater drop in diversity than did the remainder of the genome; similarly, this linkage group exhibits significantly stronger partitioning of genetic variation than does the balance of the genome. Both of these results are consistent with the hypothesis that LG06 has been the subject of recent selection. Moreover, there is no evidence of selection on LG06 during the transition from wild to domesticated sunflower (see “Exotic vs. Wild” in Figure 3), whereas the pattern reemerges when looking at the exotic-to-elite transition. Thus, this signature of selection appears to have arisen during the postdomestication era.
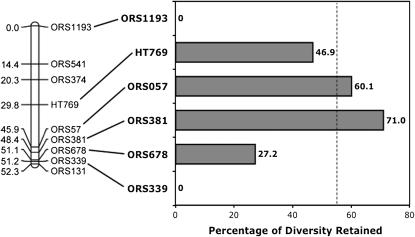
Although LG06 appears to have been under very recent and strong selection, the causes of this selection are less clear. Inspection of the pattern of diversity loss across LG06 reveals the presence of two distinct “diversity valleys,” with markers at (or near) each end of the linkage group showing a complete loss of diversity across the elite lines (ORS339 and ORS1193; Figure 4). In contrast, the four more centrally located markers retained considerably more diversity. As with ORS339 and ORS1193, both the two remaining markers (ORS349 and ORS608) from our diversity scan that are known to map to LG06, but which were unmappable in our population, show a complete loss of diversity across the elite lines. Data from other sunflower mapping populations (e.g., Tang et al. 2002; Yu et al. 2003; Burke et al. 2004) indicate that ORS349 maps in very close proximity to ORS339 near the bottom of the linkage group. In contrast, ORS608 appears to fall somewhere in the central portion of the linkage group. Although the precise location of this latter marker relative to our other mapped markers remains somewhat unclear, these results suggest the presence of a third diversity valley.
Figure 4.
Graphical representation of the percentage of genetic diversity (He) retained by SSR markers spanning LG06 vs. those located elsewhere in the genome in a comparison between wild and improved (elite) sunflower lines. The vertical broken line represents the average percentage of diversity retained across the genome as a whole (55.6%).
The simplest explanation for the presence of three distinct diversity valleys on LG06 is that this linkage group has experienced three independent selective sweeps, with selection on the oil-related QTL detected here responsible for the central sweep (marked by ORS608) only. This does not, however, provide an explanation for the other two sweeps and further implies that major QTL for other, as of yet unmapped, traits (e.g., seed dormancy) may be located near the ends of LG06. Alternatively, the widespread loss of diversity across LG06 might be the result of a single, extremely rapid selective sweep. Although it seems very unlikely that a single selective sweep could influence diversity across much of a linkage group, it must be kept in mind that the modern era of sunflower breeding has involved intensive inbreeding, and it may be that reduced recombination resulting from such inbreeding has played an important role in producing the pattern documented here. Indeed, the average inbreeding coefficient across loci on the nonswept linkage groups (i.e., all but LG06) is 0.98 in the elite lines, resulting in a nearly complete suppression of effective recombination within lines. If the pattern documented here was, in fact, the result of only a single sweep, then the heterogeneity of diversity loss across LG06 would have to be explained by other factors, such as (1) errors in marker ordering, perhaps due to structural rearrangements; (2) variable mutation rates across loci combined with genetic drift following the selective bottleneck; and/or (3) the presence of loci on LG06 that cause inbreeding depression and concomitant selection for the maintenance of genetic diversity at those loci. While errors in marker ordering remain a possibility, the latter two explanations seem rather unlikely because of the short time frame available for mutation accumulation and the fact that what variation remains along LG06 is predominantly among lines with very little heterozygosity being maintained within lines.
Taken together, the results of this study indicate that strong selection can have a profound effect on the level of standing genetic variation in a population and can influence much more than the frequencies of linked, neutral alleles. Indeed, the occurrence of numerous antagonistic domestication-related QTL in the immediate vicinity of a presumptive sweep indicates that linked, maladaptive alleles can be brought along for the ride. This may account, at least in part, for the widespread occurrence of QTL with opposing effects, although other evolutionary processes likely contribute as well (Rieseberg et al. 2002, 2003). Several studies are currently under way to further investigate the possible causes of the pattern reported here. First, a dense genetic map of LG06 is currently being generated in a larger and more diverse exotic × wild mapping population to verify marker orders. Second, additional markers along LG06 are being surveyed for allelic diversity to better understand the scale of diversity variation along this linkage group. Finally, we are continuing to map QTL for additional domestication and/or improvement-related traits in this and other mapping populations, and the positions of any such loci on LG06 will be analyzed with reference to the regions of low diversity.
Acknowledgments
This work was supported by grants from the United States Department of Agriculture (00-52100-9609 to L.H.R. and S.J.K., 03-35300-13104 to J.M.B. and 03-39210-13958 to L.H.R. and J.M.B.) and the National Science Foundation (DBI-0332411 to J.M.B.).
References
- Akey, J. M., G. Zhang, K. Zhang, L. Jin and M. D. Shriver, 2002. Interrogating a high-density SNP map for signatures of natural selection. Genome Res. 12: 1805–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basten, C. J., B. S. Weir and Z-B. Zeng, 2003. QTL Cartographer, Version 1.17. Department of Statistics, North Carolina State University, Raleigh, NC.
- Beaumont, M. A., and R. A. Nichols, 1996. Evaluating loci for use in the genetic analysis of population structure. Proc. R. Soc. Lond. B Biol. Sci. 263: 1619–1626. [Google Scholar]
- Bowcock, A. M., J. R. Kidd, J. L. Mountain, J. M. Hebert, L. Carotenuto et al., 1991. Drift, admixture, and selection in human evolution: a study with DNA polymorphisms. Proc. Natl. Acad. Sci. USA 88: 839–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Box, G. E. P., and D. R. Cox, 1964. An analysis of transformations. J. R. Stat. Soc. Ser. B 26: 211–243. [Google Scholar]
- Burke, J. M., S. Tang, S. J. Knapp and L. H. Rieseberg, 2002. Genetic analysis of sunflower domestication. Genetics 161: 1257–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke, J. M., Z. Lai, M. Salmaso, T. Nakazato, S. Tang et al., 2004. Comparative mapping and rapid karyotypic evolution in the genus Helianthus. Genetics 167: 449–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell, D., and L. Bernatchez, 2004. Generic scan using AFLP markers as a means to assess the role of directional selection in the divergence of sympatric whitefish ecotypes. Mol. Biol. Evol. 21: 945–956. [DOI] [PubMed] [Google Scholar]
- Cavalli-Sforza, L. L., 1966. Population structure and human evolution. Proc. R. Soc. Lond. B Biol. Sci. 164: 362–379. [DOI] [PubMed] [Google Scholar]
- Churchill, G. A., and R. W. Doerge, 1994. Empirical threshold values for quantitative trait mapping. Genetics 138: 963–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crites, G. D., 1993. Domesticated sunflower in fifth millennium B.P. temporal context: new evidence from Middle Tennessee. Am. Antiq. 58: 146–148. [Google Scholar]
- Darwin, C., 1859. On the Origin of Species. Harvard University Press, Cambridge, MA.
- Doebley, J., and A. Stec, 1991. Genetic analysis of the morphological differences between maize and teosinte. Genetics 129: 285–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerge, R. W., and G. A. Churchill, 1996. Permutation tests for multiple loci affecting a quantitative character. Genetics 142: 285–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlan, J. R., 1992. Crops and Man. American Society of Agronomy, Madison, WI.
- Harter, A. V., K. A. Gardner, D. Falush, D. L. Lentz, R. Bye et al., 2004. Origin of extant domesticated sunflowers in eastern North America. Nature 430: 201–205. [DOI] [PubMed] [Google Scholar]
- Heiser, C. B., 1951. The sunflower among North American Indians. Proc. Am. Philos. Soc. 95: 432–448. [Google Scholar]
- Jiang, C., and Z-B. Zeng, 1995. Multiple trait analysis of genetic mapping for quantitative trait loci. Genetics 140: 1111–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koinange, E. M. K., S. P. Singh and P. Gepts, 1996. Genetic control of the domestication syndrome in common bean. Crop Sci. 36: 1037–1045. [Google Scholar]
- Lewontin, R. C., and J. Krakauer, 1973. Distribution of gene frequency as a test of the theory of the selective neutrality of polymorphism. Genetics 74: 175–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard-Smith, J., and J. Haigh, 1974. The hitch-hiking effect of a favorable gene. Genet. Res. 23: 23–35. [PubMed] [Google Scholar]
- Nei, M., and T. Maruyama, 1975. Lewontin-Krakauer test for neutral genes. Genetics 80: 395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr, H. A., 1998. Testing natural selection vs. genetic drift in phenotypic evolution using quantitative trait locus data. Genetics 149: 2099–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putt, E. D., 1997. Early history of sunflower, pp. 1–19 in Sunflower Production and Technology, edited by A. A. Scheiter. American Society of Agronomy, Madison, WI.
- Rieseberg, L. H., A. Widmer, A. M. Arntz and J. M. Burke, 2002. Directional selection is the primary cause of phenotypic diversification. Proc. Natl. Acad. Sci. USA 99: 12242–12245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg, L. H., A. Widmer, A. M. Arntz and J. M. Burke, 2003. The genetic architecture necessary for transgressive segregation is common in both natural and domesticated populations. Philos. Trans. R. Soc. Lond. B Biol. Sci. 358: 1141–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson, A., 1975. a Gene frequency distributions as a test of selective neutrality. Genetics 81: 775–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson, A., 1975. b Remarks on the Lewontin-Krakauer test. Genetics 80: 396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlötterer, C., 2002. A microsatellite-based multilocus screen for the identification of local selective sweeps. Genetics 160: 753–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlötterer, C., and D. Dieringer, 2004. A novel test statistic for the identification of local selective sweeps based on microsatellite gene diversity, pp. 55–64 in Selective Sweep, edited by D. Nurminsky. Landes Bioscience, Georgetown, TX.
- Schöfl, G., and C. Schlötterer, 2004. Patterns of microsatellite variability among X chromosomes and autosomes indicate a high frequency of beneficial mutations in non-African D. simulans. Mol. Biol. Evol. 21: 1384–1390. [DOI] [PubMed] [Google Scholar]
- Slatkin, M., 1995. Hitchhiking and associative overdominance at a microsatellite locus. Mol. Biol. Evol. 12: 473–480. [DOI] [PubMed] [Google Scholar]
- Storz, J. F., and M. W. Nachman, 2003. Natural selection on protein polymorphism in the rodent genus Peromyscus: evidence from interlocus contrasts. Evolution 57: 2628–2635. [DOI] [PubMed] [Google Scholar]
- Storz, J. F., B. A. Payseur and M. W. Nachman, 2004. Genome scans of DNA variability in humans reveal evidence for selective sweeps outside of Africa. Mol. Biol. Evol. 21: 1800–1811. [DOI] [PubMed] [Google Scholar]
- Tang, S., and S. J. Knapp, 2003. Microsatellites uncover extraordinary diversity in native American land races and wild populations of cultivated sunflowers. Theor. Appl. Genet. 106: 990–1003. [DOI] [PubMed] [Google Scholar]
- Tang, S., J.-K. Yu, M. B. Slabaugh, D. K. Shintani and S. J. Knapp, 2002. Simple sequence repeat map of the sunflower genome. Theor. Appl. Genet. 105: 1124–1136. [DOI] [PubMed] [Google Scholar]
- Tsakas, S., and C. B. Krimbas, 1976. Testing heterogeneity of F values: a suggestion and a correction. Genetics 84: 399–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigouroux, Y., J. S. Jaqueth, Y. Matsuoka, O. S. Smith, W. D. Beavis et al., 2002. Rate and pattern of mutation at microsatellite loci in maize. Mol. Biol. Evol. 19: 1251–1260. [DOI] [PubMed] [Google Scholar]
- Vitalis, R., K. Dawson and P. Boursot, 2001. Interpretation of variation across marker loci as evidence of selection. Genetics 158: 1811–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilding, C. S., R. K. Butlin and J. Grahame, 2001. Differential gene exchange between parapatric morphs of Littorina saxatilis detected using AFLP markers. J. Evol. Biol. 14: 611–619. [Google Scholar]
- Wootton, J. C., X. Feng, M. T. Ferdig, R. A. Cooper, J. Mu et al., 2002. Genetic diversity and chloroquine selective sweeps in Plasmodium falciparum. Nature 418: 320–323. [DOI] [PubMed] [Google Scholar]
- Yeh, F. C., R.-C. Yang and T. Boyle, 1999. POPGENE, version 1.32: the user friendly software for population genetic analysis. Molecular Biology and Biotechnology Centre, University of Alberta, Edmonton, AB, Canada.
- Yu, J. K., J. Mangor, L. Thompson, K. J. Edwards, M. B. Slabaugh et al., 2002. Allelic diversity of simple sequence repeats among elite inbred lines of cultivated sunflower. Genome 45: 652–660. [DOI] [PubMed] [Google Scholar]
- Yu, J. K., S. Tang, M. B. Slabaugh, A. Heesacker, G. Cole et al., 2003. Towards a saturated molecular genetic linkage map for cultivated sunflower. Crop Sci. 43: 367–387. [Google Scholar]
- Zeng, Z-B., 1993. Theoretical basis of separation of multiple linked gene effects on mapping quantitative trait loci. Proc. Natl. Acad. Sci. USA 90: 10972–10976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, Z-B., 1994. Precision mapping of quantitative trait loci. Genetics 136: 1457–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]