Abstract
Cytogenetic maps of sorghum chromosomes 3–7, 9, and 10 were constructed on the basis of the fluorescence in situ hybridization (FISH) of ∼18–30 BAC probes mapped across each of these chromosomes. Distal regions of euchromatin and pericentromeric regions of heterochromatin were delimited for all 10 sorghum chromosomes and their DNA content quantified. Euchromatic DNA spans ∼50% of the sorghum genome, ranging from ∼60% of chromosome 1 (SBI-01) to ∼33% of chromosome 7 (SBI-07). This portion of the sorghum genome is predicted to encode ∼70% of the sorghum genes (∼1 gene model/12.3 kbp), assuming that rice and sorghum encode a similar number of genes. Heterochromatin spans ∼411 Mbp of the sorghum genome, a region characterized by a ∼34-fold lower rate of recombination and ∼3-fold lower gene density compared to euchromatic DNA. The sorghum and rice genomes exhibit a high degree of macrocolinearity; however, the sorghum genome is ∼2-fold larger than the rice genome. The distal euchromatic regions of sorghum chromosomes 3–7 and 10 are ∼1.8-fold larger overall and exhibit an ∼1.5-fold lower average rate of recombination than the colinear regions of the homeologous rice chromosomes. By contrast, the pericentromeric heterochromatic regions of these chromosomes are on average ∼3.6-fold larger in sorghum and recombination is suppressed ∼15-fold compared to the colinear regions of rice chromosomes.
GRASSES are ecologically well adapted, covering ∼20% of the earth's land surface (Schantz 1954). This group of plants is especially important to agriculture, contributing a large portion of the calories consumed in the human diet (Evans 1998). The grass family originated ∼55–70 million years ago (MYA) and today includes ∼10,000 species (Kellogg 2001). Advances in our understanding of grass phylogeny have helped place important functional differentiation that occurs within the grass family into evolutionary context (Grass Phylogeny Working Group 2000). Phylogenetic information is also useful for guiding the selection and utilization of reference species for comparative genome research (Thornton and DeSalle 2000; Ureta-Vidal et al. 2003). For example, grass species such as rice, wheat, barley, and oats carry out C3 photosynthesis as do other members of Pooideae, Ehrhartoideae, and Bambusoideae (Kellogg 2001). Rice was targeted for intense grass genomics research because of its relatively small genome (<490 Mbp), technologies for analysis of gene function (i.e., Shimamoto and Kyozuka 2002; An et al. 2003; Li et al. 2005), and its agricultural importance (Cantrell and Reeves 2002). A multinational effort has reported a nearly complete sequence of the rice genome, for which annotations and other sources of information indicate a nontransposable element-related protein-coding gene complement of ∼37,500–∼50,000 genes (Rice Full-Length cDNA Consortium 2003, http://www.tigr.org/tdb/e2k1/osa1/pseudomolecules/info.shtml; Bennetzen et al. 2004; International Rice Genome Sequencing Project 2005).
Sorghum, maize, sugarcane, and millet are grass species of the PACC clade that includes the Panicoideae (Grass Phylogeny Working Group 2000; Kellogg 2001) that diverged from rice ∼50 MYA (Doebley et al. 1990). These grass species carry out C4 photosynthesis, an important adaptation that increases the efficiency of CO2 fixation in plants. C4 species contribute disproportionately to global productivity and agriculture, in that they compose only ∼3% of angiosperm species (Edwards et al. 2004). The C4 grasses are particularly well adapted to regions of lower latitude that have higher average temperatures and are prone to drought (Edwards et al. 2004). Among the C4 grasses, sorghum has a relatively small genome containing 818 Mbp of DNA distributed among 10 chromosomes (Price et al. 2005). The importance of sorghum as a subsistence cereal crop in the semiarid tropics (Doggett 1988; National Research Council 1996), potential importance in biofuel production (Gnansounou et al. 2005), adaptation to drought (Doggett 1988; National Research Council 1996), diverse germplasm (i.e., Menz et al. 2004), and close relationship to maize (Kellogg 2001; Swigonova et al. 2004) make this species a valuable target for grass genome research.
Extensive resources have been developed for sorghum genomics (Sorghum Genomics Planning Workshop Participants 2005). Several linkage maps have been constructed on the basis of interspecific (i.e., Bowers et al. 2003) and intraspecific crosses (i.e., Menz et al. 2002). The sorghum genome map has been aligned to the genome maps of other cereals revealing extensive macrocolinearity, especially between sorghum, rice, and maize (Peng et al. 1999; Wilson et al. 1999; Klein et al. 2003; Paterson et al. 2004; Devos 2005). Approximately 200,000 sorghum ESTs have been collected revealing ∼22,000 unique transcript clusters (L. H. Pratt, personal communication; http://www.fungen.org/). Microarrays and qRT-PCR assays based on these sequences have been used to collect information on sorghum gene expression modulated by plant hormones involved in plant protection (Salzman et al. 2005) and osmotic stress (Buchanan et al. 2005). In addition, the collection of ∼500,000 methyl-filtered sorghum sequences tagged >90% of the sorghum genes (Bedell et al. 2005). The architecture of sorghum chromosomes has also been characterized in several studies. A molecular karyotype of the sorghum genome was developed on the basis of fluorescence in situ hybridization (FISH) of BACs derived from each sorghum chromosome (Kim et al. 2002). Karyotype-aided analysis of sorghum chromosome size and DNA content recently allowed the establishment of a unified sorghum chromosome numbering system (Kim et al. 2005a). In addition, the molecular cytology of three sorghum chromosomes has been analyzed in detail using genetically mapped BAC clones and FISH (Islam-Faridi et al. 2002; Kim et al. 2005b). These sorghum chromosomes were found to contain distal regions of euchromatin and pericentromeric regions of heterochromatin (Islam-Faridi et al. 2002; Kim et al. 2005b).
Genome sizes and chromosome numbers of grasses range widely (http://www.rbgkew.org.uk/cval/homepage.html). For example, rice has an ∼370- to 490-Mbp genome distributed among 12 chromosomes and wheat has an ∼16,900-Mbp genome distributed among three sets of 7 chromosomes (http://www.rbgkew.org.uk/cval/homepage.html). Moreover, even within the genus Sorghum (Poaceae), chromosome numbers (2n = 10, 20, 30, 40) and genome size vary considerably (∼8.1-fold) (Price et al. 2005). Variation in genome size among the grasses is due primarily to differences in polyploidy, segmental duplications, and accumulation of repetitive sequences (Saghai Maroof et al. 1996; Tikhonov et al. 1999; Gaut 2002; Levy and Feldman 2002; Vandepoele et al. 2003). The S. bicolor genome is approximately twofold larger in size and has two fewer chromosomes relative to Oryza sativa. The difference in sorghum's chromosome number relative to rice is due to two chromosome fusion events that occurred prior to the divergence of sorghum from maize and Pennisetum (Wilson et al. 1999; Kellogg 2001). The sorghum and rice genomes are thought to encode a similar number of genes (http://fungen.botany.uga.edu/Projects/Sorghum/SorghumUnigeneSet.htm, http://www.tigr.org/tdb/e2k1/osa1/pseudomolecules/info.shtml). Moreover, rice and sorghum chromosomes are largely colinear indicating that sorghum's genome expansion relative to that of rice is not due to a large-scale genome duplication event (Peng et al. 1999; Wilson et al. 1999; Klein et al. 2003). These results suggest that the difference in S. bicolor and O. sativa genome size could be due primarily to differential accumulation of repetitive sequences in sorghum.
In this study, the molecular cytogenetic architecture of 7 sorghum chromosomes was analyzed, thereby completing characterization of all 10 sorghum chromosomes. Colinear regions of sorghum and rice chromosomes were compared and the nature of the genome expansion of sorghum is detailed and discussed.
MATERIALS AND METHODS
Mitotic metaphase chromosome spreads were prepared from root tips of BTx623 seedlings according to the protocol of Kim et al. (2002), and pachytene chromosomes were prepared from immature anthers according to the protocol of Kim et al. (2005b). BACs used for FISH were derived from libraries prepared by Woo et al. (1994), Tao and Zhang (1998), and Klein et al. (2003). The BACs were located on the sorghum linkage map as described by Klein et al. (2003), and BAC DNA used for FISH was isolated as previously described (Kim et al. 2002). A centromere-associated sequence (pCEN38) (Zwick et al. 2000) was used to locate sorghum centromeres as previously described (Islam-Faridi et al. 2002). In situ hybridization techniques were a modification of Islam-Faridi et al. (2002). The chromosomal DNA on the glass slide was denatured at 70° in 70% formamide, 2× SSC for 1.5 min, followed by dehydration in 70, 85, 95, and 100% ethanol for 2 min each at –20°. The hybridization mixture (25 μl/slide) contained 50 ng of labeled probe DNA, a 10-fold excess of blocking DNA (200- to 300-bp-digested genomic DNA), 50% formamide, and 10% dextran sulfate in 2× SSC. The mixture was denatured at 90° for 10 min, chilled on ice, added to the slide, and covered with a glass coverslip. Following overnight incubation at 37°, slides were rinsed three times at 37° in 2× SSC for 5 min each. Slides were blocked 5 min at room temperature with 5% (w/v) BSA in 4× SSC plus 0.2% Tween 20. Biotin-labeled probes were detected with 1% Cy3-conjugated streptavidin and digoxygenin-labeled probes with 1% fluorescein isothiocyanate (FITC)-conjugated anti-digoxygenin antibody. Slides were washed three times in 4× SSC plus 0.2% Tween 20 for 5 min at 37°. DAPI (4′,6-diamidino-2-phenylindole) in Vectashield antifade solution (Vector Laboratories, Burlingame, CA) was applied for counterstaining of chromosomes. Images were viewed through an Olympus AX-70 epifluorescence microscope equipped with standard filter cubes. Images from a Peltier-cooled 1.3-megapixel Sensys camera (Roper Scientific, Trenton, NJ) were captured with the MacProbe v. 4.2.3 digital image system (Applied Imaging, Santa Clara, CA). To assess the location of FISH signals, blue (DAPI signal from chromosomal DNA), green (FITC from BAC probes), and red (Cy3 from BAC probes) signals were measured from digital images using Optimas v. 6.0 (Media Cybernetics, Carlsbad, CA). Image capture and data analysis were done as previously described (Kim et al. 2005a).
Sample sequencing of BAC DNA was performed as described previously (Klein et al. 2003). DNA from nine phase III-sequenced sorghum BACs was isolated and fingerprinted using a modified version of five-color-based high information content fingerprinting (HICF; Klein et al. 2003; Luo et al. 2003). The nine BACs correspond to GenBank accession nos. AF114171, AF466199, AF466200, AF466201, AF369906, AY661656, AY661657, AY661658, and AY661659. To calculate the number of 75- to 500-bp DNA bands generated per kilobase pair of BAC DNA, all common vector bands were removed from each dye lane and the number of remaining unique bands was divided into the total kilobase pairs in each BAC insert. This analysis showed that a 75- to 500-bp DNA fingerprint band is generated on average every 1.405 ± 0.124 kbp of BAC-insert DNA analyzed by HICF. Using this information, two BAC contigs from the distal euchromatic part of the long arm of sorghum chromosome 3 spanning BAC sbb20220 (211e12) to BAC sbb16652 (174d8) and BAC sbb22184 (232a8) to BAC sbb8411 (88e11) were calculated to contain ∼2.19 and ∼2.36 Mbp of DNA, respectively.
RESULTS
Molecular architecture of sorghum chromosomes:
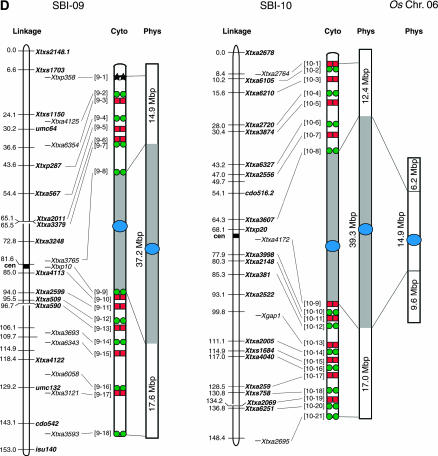
In previous work the architecture of sorghum chromosomes 1, 2, and 8 was characterized using mapped BACs and FISH analysis (Islam-Faridi et al. 2002; Kim et al. 2005b). In this study the molecular cytogenetic architecture of the remaining seven sorghum chromosomes, 3–7, 9, and 10, was characterized by FISH analysis using 18–30 BACs per chromosome. The BACs used in this study mapped at regular intervals across the linkage maps of each chromosome (supplemental Table 1 at http://www.genetics.org/supplemental/) (Klein et al. 2003). The relative order of BACs along each chromosome arm was determined in some cases by dual-color FISH using pairs of adjacent BACs that map incrementally closer to the centromere as previously described (Islam-Faridi et al. 2002). In addition, multiprobe FISH cocktails were used to visualize the spacing and order of groups of BACs that mapped and hybridized sequentially along the chromosomes. Several examples of multiprobe BAC FISH on a pachytene bivalent corresponding to sorghum chromosome 3 are shown in Figure 1, A and F. Similar results were obtained when BAC-FISH cocktails corresponding to each of the remaining six chromosomes were analyzed on pachytene bivalents (data not shown). For each bivalent examined, the cytogenetic location of each fluorescent signal was determined by locating the peak luminance value along the segmented line that collectively spanned the FISH-adorned bivalent. The peaks were then used to assign linear positions along the pachytene bivalent and create a cytogenetic map. The cytogenetic maps corresponding to the seven sorghum chromosomes in this study are shown in Figure 2 (chromosome diagrams denoted as Cyto). For comparative purposes, the cytogenetic maps of chromosomes 1, 2, and 8 characterized previously (Islam-Faridi et al. 2002; Kim et al. 2005b) are also shown in Figure 2. Overall, the order of BAC-FISH signals along the seven chromosomes analyzed was concordant with the location of these BACs on the sorghum linkage maps (Figure 2, supplemental Table 1 at http://www.genetics.org/supplemental/). In several cases, FISH analysis helped resolve the order of DNA markers and their associated BAC clones that were located in the same “bin” on the linkage map. In addition, the analysis showed that the linkage maps of the seven chromosomes span nearly the entire length of each chromosome arm (Figure 2).
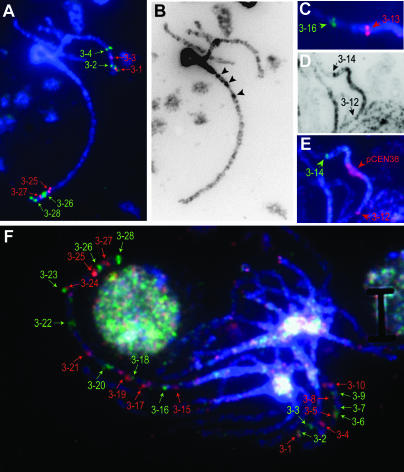
Figure 1.
Cytogenetic analysis of sorghum chromosome 3 using genetically mapped BACs and FISH. (A) FISH signals on sorghum pachytene bivalents using an eight-probe cocktail with BACs mapped to the termini of sorghum chromosome 3. (B) Pachytene bivalents showing a central darkly staining pericentromeric region of heterochromatin plus interspersed regions of heterochromatin (arrows) and euchromatin. (C) FISH analysis showing hybridization of BAC probe 3-13 within the heterochromatic region. (D and E) Localization of BAC probes 3-12 and 3-14 at the euchromatin:heterochromatin boundaries and DNA associated with the centromere using the pCEN38 probe. (F) FISH signals on pachytene bivalents using a multiprobe cocktail composed of BACs mapping across sorghum chromosome 3. BAC numbers refer to those listed in supplemental Table 1 (http://www.genetics.org/supplemental/).
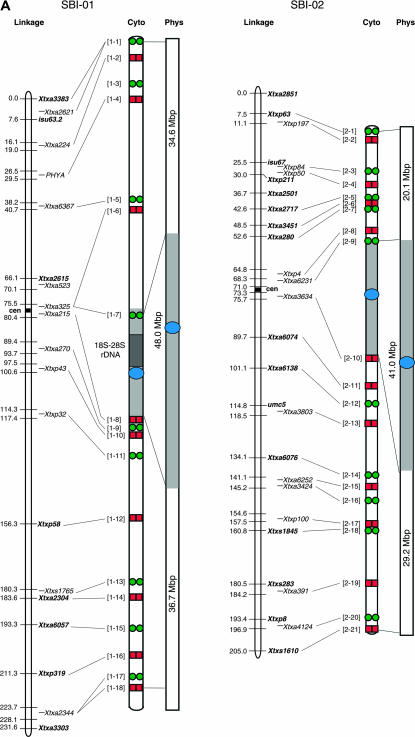
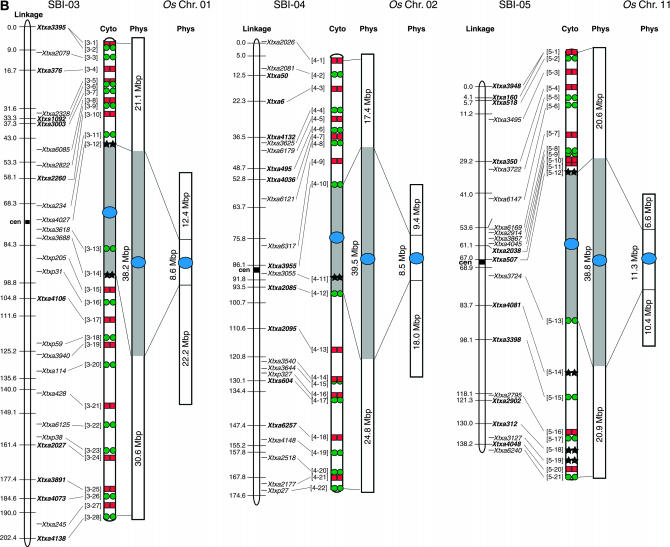
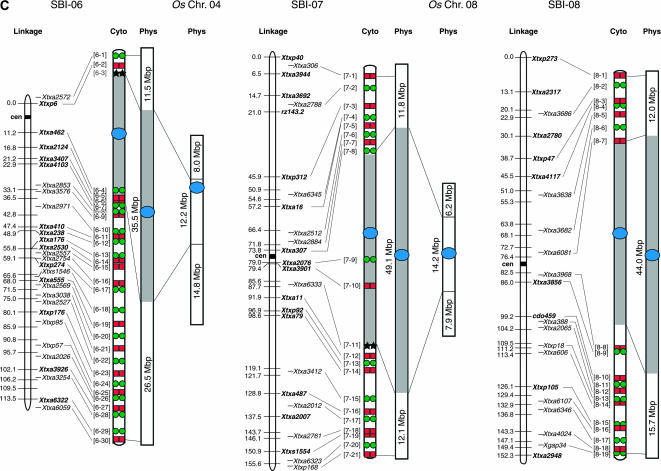
Figure 2.
Integrated linkage, cytogenetic, and physical maps of the sorghum genome and alignment to the physical maps of rice chromosomes 1, 2, 11, 4, 8, and 6. Sorghum linkage maps at the left are drawn using data from Menz et al. (2002) and cytogenetic maps are drawn on the basis of BAC signals observed on pachytene bivalents. Centromeres of chromosomes mapped using the pCEN38 probe are shown as blue circles. Red squares and green circles represent the Cy3 and FITC signals, respectively, used in the detection of multiprobe BAC-FISH. Black stars represent additional BACs used in single-probe FISH detection. Shaded boxes represent heterochromatin and open boxes represent euchromatin in the sorghum cytogenetic and physical maps. Numbers to the left of the cytogenetic maps correspond to BAC numbers shown in supplemental Table 1 (http://www.genetics.org/supplemental/) used for FISH analysis. Physical maps of sorghum are drawn on the basis of calculations using the distribution of DNA content in heterochromatic and euchromatic regions as determined from BAC-FISH measurements of mitotic metaphase cells (supplemental Table 2 at http://www.genetics.org/supplemental/). Physical maps of rice were drawn using data based on the TIGR Osa1 version 3 pseudomolecule (http://www.tigr.org/tdb/e2k1/osa1/pseudomolecules/info.shtml). The sorghum BACs at the boundaries of sorghum euchromatin:heterochromatin were aligned to each rice physical map on the basis of gene sequence alignments obtained from BAC sample sequencing (supplemental Table 3 at http://www.genetics.org/supplemental/).
In sorghum pachytene chromosomes, there is a significant difference in the intensity of DAPI staining of heterochromatin and euchromatin, making the boundaries of these regions relatively easy to detect. For example, on chromosome 3, lightly staining euchromatic DNA was present in the distal portion of each arm (Figure 1, A and B). In addition, a punctate staining pattern was observed on this chromosome near the transition region between euchromatin and heterochromatin (Figure 1B, dark-staining regions marked by arrows). This indicates the presence of small, interspersed regions of heterochromatin within the euchromatic DNA near this boundary. Overall, only 4 of the 162 BACs used for FISH analysis were located in regions of heterochromatin (e.g., Figure 1C, BAC probe 3-13 on chromosome 3). This was due in part to the preferential selection of BACs with higher gene content (Klein et al. 2003) and because most BACs from heterochromatic regions hybridized nonspecifically, due to their high content of repetitive DNA (data not shown; Kim et al. 2005b). The location of the centromeres in each sorghum chromosome was determined using the probe pCEN38 (Zwick et al. 2000). We have previously shown that the pCEN38 probe hybridizes to a region in the heterochromatin of sorghum chromosome 1 adjacent to the nucleolus organizing region (NOR) and that the heterochromatic region of the long arm of sorghum chromosome 1 is ∼1.9-fold larger than the heterochromatic region of the short arm (Islam-Faridi et al. 2002; Figure 2). By contrast, hybridization of the pCEN38 probe to pachytene bivalents corresponding to the other nine chromosomes was observed near the midpoints of the respective heterochromatic regions (Figure 1E, Figure 2).
The relative size and DNA content of each sorghum chromosome were quantified in a previous study on the basis of measurements of mitotic chromosomes at metaphase, when DNA density is most uniform (Kim et al. 2005a). One objective of this study was to estimate the amount of DNA located in the euchromatic and heterochromatic regions of each sorghum chromosome using a similar approach. For this determination, BACs were identified that hybridize at the boundaries between euchromatic and heterochromatic DNA in each pachytene bivalent. For example, BAC probes 3-12 and 3-14 hybridized to the boundaries between heterochromatin and euchromatin on the short and long arms of chromosome 3, respectively (Figure 1, D and E). The BAC probes marking the euchromatin:heterochromatin boundaries on chromosome 3 were subsequently hybridized to mitotic metaphase chromosomes and the relative size of the euchromatic and heterochromatic regions was determined by analysis of multiple chromosomes (N = 20) (supplemental Table 2 at http://www.genetics.org/supplemental/). The amount of DNA in the euchromatic and heterochromatic regions delimited by the BAC probes was then calculated on the basis of the relative size of each region and the previous determination of the DNA content in sorghum chromosome 3 (Kim et al. 2005a). This analysis indicated that the euchromatic portion of the short arm of sorghum chromosome 3 contains ∼21.1 Mbp of DNA, the euchromatic portion of the long arm contains ∼30.6 Mbp of DNA, and the heterochromatic region contains ∼38.2 Mbp of DNA (Table 1, Figure 2).
TABLE 1.
Sorghum chromosome size and distribution of euchromatic and heterochromatic DNA
| Chromosome | Chromosome size (Mbp) | Euchromatin short arm (Mbp) | Euchromatin long arm (Mbp) | Total euchromatin (Mbp) | Heterochromatin (Mbp) | Total euchromatin (%) |
|---|---|---|---|---|---|---|
| SBI-01 | 119.3 | 34.6 ± 0.945a | 36.7 ± 0.730 | 71.3 ± 1.264 | 48.1 ± 1.264 | 59.8 |
| SBI-02 | 90.3 | 20.1 ± 1.008 | 29.2 ± 0.924 | 49.3 ± 1.189 | 41.1 ± 1.189 | 54.7 |
| SBI-03 | 89.9 | 21.1 ± 0.399 | 30.6 ± 0.701 | 51.7 ± 0.826 | 38.2 ± 0.826 | 57.5 |
| SBI-04 | 81.7 | 17.4 ± 0.900 | 24.8 ± 0.710 | 42.2 ± 1.421 | 39.5 ± 1.421 | 51.7 |
| SBI-05 | 80.3 | 20.6 ± 0.779 | 20.9 ± 0.567 | 41.5 ± 0.667 | 38.8 ± 0.667 | 51.7 |
| SBI-06 | 73.5 | 11.5 ± 0.811 | 26.5 ± 1.044 | 38.0 ± 1.077 | 35.5 ± 1.077 | 51.8 |
| SBI-07 | 73.1 | 11.8 ± 0.503 | 12.1 ± 0.765 | 23.9 ± 0.884 | 49.2 ± 0.884 | 33.0 |
| SBI-08 | 71.7 | 12.0 ± 0.543 | 15.7 ± 0.862 | 27.7 ± 0.999 | 44.0 ± 0.999 | 38.6 |
| SBI-09 | 69.6 | 14.9 ± 0.712 | 17.6 ± 0.724 | 32.5 ± 0.828 | 37.2 ± 0.828 | 46.6 |
| SBI-10 | 68.6 | 12.4 ± 0.718 | 17.0 ± 0.809 | 29.4 ± 1.274 | 39.3 ± 1.274 | 42.7 |
| Total: | 818 | 177 | 231 | 407 | 411 | 49.9 |
The sample size for measurements was 20.
The estimates of DNA content of the euchromatic and heterochromatic regions from measurement of metaphase chromosomes are based on the assumption that sorghum chromosomes are uniformly compacted at this stage. To test this assumption and our method for estimating DNA content, we used a second approach that relies on a combination of BAC contig fingerprinting (HICF) and BAC-FISH analysis to determine the amount of euchromatic DNA present in sorghum chromosome 3. To calibrate the analysis, HICF was carried out on nine phase III-sequenced BAC clones containing DNA derived from euchromatic regions of the sorghum genome. The average number of ∼75- to 500-bp DNA bands detected by HICF per megabase pair of sorghum DNA was determined from this analysis. This information was then used to calculate the amount of DNA in two BAC contigs that spanned portions of the euchromatic region on the long arm of sorghum chromosome 3 (see materials and methods). Next, BACs from the ends of each of these contigs were used in FISH analysis and the relative length of the chromosomal interval defined by the BAC-FISH signals was determined by measurement of 14 SBI-03 pachytene bivalents. The combined HICF and FISH analysis showed that on average each relative unit length of euchromatic DNA contained ∼0.648 Mbp (see supplemental Table 1 at http://www.genetics.org/supplemental/). Using this conversion factor, the euchromatic portion of the short arm of sorghum chromosome 3 was estimated to contain ∼19.4 Mbp of DNA (chromosome end to BAC probe 3-12) and the euchromatic portion of the long arm was estimated to contain ∼33.2 Mbp DNA (chromosome end to BAC probe 3-14). Thus, if sorghum chromosome 3 contains ∼89.9 Mbp of DNA overall (Kim et al. 2005a), then the BAC-FISH/HICF method indicates that ∼52.6 Mbp of DNA is located in the distal euchromatic regions and ∼37.3 Mbp of DNA is present in the pericentromeric heterochromatic region. These estimates based on HICF and BAC FISH differ by <3% from the estimates based on BAC-FISH measurements on metaphase chromosomes (∼51.7 Mbp for the euchromatic region and ∼38.2 Mbp for the heterochromatic region). Thus, despite the inherent limitations associated with the two methods used for quantification of chromosomal DNA in this study, both approaches gave similar estimates of the DNA content of the euchromatic and pericentromeric regions of sorghum chromosome 3.
The analysis of DNA content of euchromatic and heterochromatic regions for sorghum chromosome 3 described above was extended to the other nine sorghum chromosomes. On the basis of previous estimates of the DNA content of each sorghum chromosome (Kim et al. 2005a), the amount of DNA in the euchromatic and pericentromeric heterochromatic regions of each chromosome was determined (supplemental Table 2 at http://www.genetics.org/supplemental/, Table 1, Figure 2). Overall, the estimates of DNA in the euchromatic portion of the 10 sorghum chromosomes ranged from ∼71.3 Mbp for chromosome 1 to ∼23.9 Mbp for chromosome 7 (Table 1). The amount of DNA in heterochromatic regions of the sorghum chromosomes was estimated to vary from ∼49.2 Mbp in chromosome 7 to ∼35.5 Mbp in chromosome 6 (Table 1). Overall, euchromatin and heterochromatin each spanned ∼50% of the sorghum genome (Table 1, ∼407 and ∼411 Mbp, respectively).
Comparative size, architecture, and gene density of sorghum and rice chromosomes:
A genome size of 818 Mbp (Price et al. 2005) indicates that the sorghum genome is approximately twofold larger than the rice genome (370–490 Mbp) (http://www.rbgkew.org.uk/cval/homepage.html; Uozu et al. 1997). This analysis can be extended to individual chromosomes using our data and rice genome sequence data (http://www.tigr.org/tdb/e2k1/osa1/pseudomolecules/info.shtml). We compared the sizes of established homeologous sorghum and rice chromosomes (SBI-03:Os-01, SBI-04:Os-02, SBI-05:Os-11, SBI-06:Os-04, SBI-07:Os-08, SBI-08:Os-12, SBI-09:Os-05, SBI-10:Os-06) (i.e., Moore et al. 1995; Wilson et al. 1999). In addition, sorghum chromosome 1 contains DNA corresponding to rice chromosomes 3 and 10, while sorghum chromosome 2 contains DNA corresponding to rice chromosomes 7 and 9 (Wilson et al. 1999; Kellogg 2001). Sizes of the homeologous sorghum and rice chromosomes (or chromosomal fusions) were calculated on the basis of the estimated DNA content of each sorghum chromosome (Kim et al. 2005a; Table 1) vs. the chromosome-specific rice sequence data (http://www.tigr.org/tdb/e2k1/osa1/pseudomolecules/info.shtml). This showed that sorghum chromosomes contain ∼1.7- to ∼2.8-fold more DNA than the homeologous rice chromosome sequences.
The increased size of sorghum chromosomes compared to homeologous rice chromosomes could be due to a proportional or disproportional increase in euchromatic chromosome arms and pericentromeric heterochromatic regions. To begin to address this question we compared the relative size of the euchromatic and heterochromatic regions of six sorghum chromosomes to the colinear regions of the corresponding homeologous rice chromosomes. In related studies, sorghum BACs distributed across all 10 sorghum chromosomes were sequence scanned and the resulting sorghum genes used to align the sorghum and rice chromosomes (Klein et al. 2003; P. E. Klein, personal communication). In this study, unique colinear gene sequences were obtained from BACs mapped to the euchromatin:heterochromatin boundaries of sorghum chromosomes 3–7 and 10 (supplemental Table 3 at http://www.genetics.org/supplemental/), allowing us to align the euchromatic and heterochromatic regions from these six sorghum chromosomes to the corresponding homeologous rice chromosomes. The amounts of DNA in the rice segments colinear to the euchromatic region of each sorghum chromosomal arm and to the pericentromeric heterochromatin were determined using data based on the TIGR Osa1 version 3 pseudomolecule (http://www.tigr.org/tdb/e2k1/osa1/pseudomolecules/info.shtml) and compared to the amount of DNA present in the colinear regions of sorghum (Table 2). Among the six chromosome pairs analyzed, the euchromatic regions of sorghum averaged ∼1.8-fold larger in size compared to the colinear regions of rice (ranging from ∼1.4- to ∼3.1-fold). In contrast to the euchromatic regions, sorghum pericentromeric heterochromatic regions were on average ∼3.6-fold larger in size relative to the colinear regions of rice (ranging from ∼2.6- to ∼4.6-fold) (Table 2).
TABLE 2.
Ratio of DNA in colinear regions of homeologous sorghum and rice chromosomes
| Chromosome pairs | Chromosome Sb:Osa | Euchromatin Sb-SA/Os-SA | Euchromatin Sb-LA/Os-LA | Euchromatin Sb/Os | Heterochromatin Sb/Os |
|---|---|---|---|---|---|
| SBI-03:Os-01 | 2.1 | 1.7 | 1.4 | 1.5 | 4.4 |
| SBI-04:Os-02 | 2.3 | 1.9 | 1.4 | 1.5 | 4.6 |
| SBI-05:Os-11 | 2.8 | 3.1 | 2.0 | 2.4 | 3.4 |
| SBI-06:Os-04 | 2.1 | 1.5 | 1.8 | 1.7 | 2.9 |
| SBI-07:Os-08 | 2.6 | 1.9 | 1.5 | 1.7 | 3.5 |
| SBI-10:Os-06 | 2.2 | 2.0 | 1.8 | 1.9 | 2.6 |
| Average | 2.4 | 2.0 | 1.7 | 1.8 | 3.6 |
SA, chromosome short arm; LA, chromosome long arm.
Data for Mbp/sorghum chromosome were taken from Kim et al. (2005a). Data for Mbp/rice chromosome were taken from the TIGR Osa1 version 3 pseudomolecule (http://www.tigr.org/tdb/e2k1/osa1/pseudomolecules/info.shtml).
Currently, ∼37,500–47,000 nontransposable element (non-TE) gene models have been identified in the rice genome sequence depending on the methods used to annotate gene models (International Rice Genome Sequencing Project 2005; http://www.tigr.org/tdb/e2k1/osa1/pseudomolecules/info.shtml). Using the upper estimate as a reference, the number of non-TE gene models/chromosome ranges from ∼2600 for rice chromosome 9 to ∼6100 for rice chromosome 1 (http://www.tigr.org/tdb/e2k1/osa1/pseudomolecules/info.shtml). Using these estimates, the number of non-TE gene models present in regions of the rice genome that are colinear with the euchromatic and heterochromatic regions of the six sorghum chromosomes analyzed was determined. Approximately 25,400 non-TE gene models have been identified in the six rice chromosomes that are colinear to sorghum chromosomes 3–7 and 10 with ∼18,500 of these in regions colinear to the sorghum euchromatic arms and the remaining ∼6900 located in regions colinear to sorghum heterochromatin. Assuming there are a similar number of genes in colinear regions of sorghum and rice, then the average gene density of the sorghum euchromatic regions is predicted to be ∼81 gene models/Mbp (18,500/227 Mbp) or ∼1 gene model/12.3 kbp and the average gene density in the sorghum heterochromatic regions is predicted to be ∼29 gene models/Mbp (6,895 gene models/240.5Mbp) or ∼1 gene model/34.5 kbp. If the lower estimate of the rice gene complement (∼37,500 genes) is used in this calculation, then the overall gene numbers and gene density predicted for sorghum are lower by ∼20%.
Recombination in sorghum and rice chromosomes:
Alignment of the sorghum linkage and cytogenetic maps and identification of the euchromatic and heterochromatic regions in each of the sorghum chromosomes allowed average rates of recombination in these regions to be determined (Table 3). In general, heterochromatic regions of sorghum chromosomes showed much lower rates of recombination (∼8.7 Mbp/cM) compared to euchromatic regions (∼0.25Mbp/cM) (Table 3). In addition, recombination in heterochromatic regions of different sorghum chromosomes varied ∼15-fold from ∼2 Mbp/cM (SBI-06) to ∼31 Mbp/cM (SBI-01). The euchromatic region on the short arm of sorghum chromosome 6 showed a relatively low rate of recombination compared to other regions of euchromatin (∼2.3 Mbp/cM vs. an overall average of ∼0.25 Mbp/cM) (Table 3). The rate of recombination across euchromatic DNA of the other 19 chromosome arms varied ∼2.8-fold from ∼0.44 Mbp/cM (SBI-01) to ∼0.16 Mbp/cM (SBI-07).
TABLE 3.
Recombination in sorghum chromosomes
| Chromosome | Euchromatin short arm (Mbp/cM) | Euchromatin long arm (Mbp/cM) | Total euchromatin (Mbp/cM) | Heterochromatin (Mbp/cM) |
|---|---|---|---|---|
| SBI-01 | 0.444 | 0.241 | 0.310 | 31.0 |
| SBI-02 | 0.289 | 0.224 | 0.246 | 8.4 |
| SBI-03 | 0.278 | 0.249 | 0.260 | 11.2 |
| SBI-04 | 0.203 | 0.306 | 0.253 | 5.2 |
| SBI-05 | 0.308 | 0.308 | 0.308 | 11.6 |
| SBI-06 | 2.29 | 0.286 | 0.364 | 2.0 |
| SBI-07 | 0.160 | 0.176 | 0.167 | 3.8 |
| SBI-08 | 0.161 | 0.230 | 0.194 | 4.5 |
| SBI-09 | 0.193 | 0.252 | 0.221 | 6.1 |
| SBI-10 | 0.192 | 0.240 | 0.218 | 2.9 |
| Average | 0.254 | 8.7 |
The relative rate of recombination in colinear regions of the six pairs of homeologous sorghum and rice chromosomes was compared (Table 4). The rate of recombination in the short arm of sorghum chromosome 4 was slightly higher than that in the colinear region of rice chromosome 2, whereas the rate of recombination in most of the euchromatic arms of the sorghum chromosomes analyzed was slightly lower than that in the colinear regions of rice (∼1.1- to ∼1.7-fold lower recombination in sorghum). In contrast, the euchromatic portion of the short arm of sorghum chromosome 6 showed an ∼5.6-fold lower rate of recombination compared to the colinear region of rice. Moreover, rates of recombination in the heterochromatic regions of the sorghum chromosomes analyzed were ∼15.4-fold lower than those in the colinear regions of rice (ranging from ∼6.4- to ∼36.3-fold) (Table 4).
TABLE 4.
Ratio of recombination in colinear regions of homeologous sorghum and rice chromosomes
| Chromosome pair | Euchromatin short arm (Mbp/cM) | Euchromatin long arm (Mbp/cM) | Total euchromatin (Mbp/cM) | Heterochromatin (Mbp/cM) |
|---|---|---|---|---|
| SBI-03:Os-01 | 1.5 | 1.1 | 1.2 | 25.9 |
| SBI-04:Os-02 | 0.9 | 1.7 | 1.3 | 9.2 |
| SBI-05:Os-11 | 1.6 | 1.4 | 1.5 | 36.3 |
| SBI-06:Os-04 | 5.6 | 1.4 | 1.8 | 6.4 |
| SBI-07:Os-08 | 1.2 | 1.1 | 1.1 | 7.3 |
| SBI-10:Os-06 | 1.2 | 1.3 | 1.2 | 7.2 |
| Average | 1.4 | 15.4 |
DISCUSSION
FISH analysis utilizing mapped BACs helped elucidate the molecular architecture of sorghum chromosomes 3–7, 9, and 10. These results, combined with prior analysis of sorghum chromosomes 1, 2, and 8 (Islam-Faridi et al. 2002; Kim et al. 2005b), completes an overall cytogenetic analysis of the organization of S. bicolor chromosomes. In total, these studies utilized 220 BACs for FISH analysis ranging from 18 to 30 BACs per chromosome. The BACs were selected to span the sorghum linkage map and for their ability to generate strong, localized FISH signals. As a consequence of this selection, most of the BACs were low in repetitive sequence content and located in euchromatin (Kim et al. 2005b), where gene density and recombination rate are higher. A smaller number of the BACs were located within heterochromatic regions indicating that while more difficult, these regions can also be analyzed by BAC-FISH analysis. The analysis demonstrated that the sorghum linkage map derived from an intraspecific cross of BTx623 and IS3620C (Menz et al. 2002) provides excellent coverage of the sorghum genome. Overall, the order of BACs along the sorghum chromosomes was concordant with the location of these BACs on the sorghum linkage map, thereby confirming the high level of accuracy that was achieved during integration of the linkage and physical maps. This study enables information on the architecture of the sorghum chromosomes to be cross-referenced to linkage, physical, and comparative maps of the sorghum genome (Klein et al. 2000; Menz et al. 2002; Klein et al. 2003) and to an assessment of sorghum diversity (Menz et al. 2004). This integrated set of information provides a solid foundation for genome research on this important C4 grass species and should help provide a framework for the S. bicolor sequencing project (Sorghum Genomics Planning Workshop Participants 2005; http://www.jgi.doe.gov/News/news_5_12_05.html). The integration of genetic and cytogenetic information was an important step in the rice and Medicago truncatula genome sequencing projects because it provided information on the distribution of heterochromatin, euchromatin, centromeres, and genes across these genomes (Cheng et al. 2001b; Kulikova et al. 2001). For example, analysis of pachytene stage chromosomes and FISH mapping of two to five BACs per M. truncatula linkage group indicated that the majority of the genes in this genome are located in distal chromosomal regions of euchromatic DNA (Kulikova et al. 2001).
Sorghum chromosomes contain pericentromeric regions of heterochromatin and regions of euchromatin located in the distal portions of each chromosome arm. A similar arrangement of heterochromatin and euchromatin is found in most chromosomes of tomato (Peterson et al. 1999), M. truncatula (Kulikova et al. 2001), and rice (Cheng et al. 2001a). In contrast, heterochromatin is distributed across a much greater portion of the maize and wheat chromosomes (i.e., Gill et al. 1991; Chen et al. 2000). Overall, euchromatin spans ∼50% of the sorghum genome ranging from ∼60% of sorghum chromosome 1 (∼71 Mbp) to ∼33% of sorghum chromosome 7 (∼24 Mbp). The amount of heterochromatin per chromosome is more uniform varying ∼1.4-fold from ∼49 Mbp on sorghum chromosome 7 to ∼35.5 Mbp on sorghum chromosome 6. DNA associated with sorghum centromeres was located near the midpoint of the heterochromatin except for chromosome 1 where more heterochromatic DNA was present on the long arm than on the short arm (Islam-Faridi et al. 2002). This distinctive feature could reflect an ancestral chromosome rearrangement, the possibility of which is suggested by the relative locations of the NOR in S. propinquum vs. S. bicolor (Bowers et al. 2003).
The sorghum genome has been estimated to be ∼2-fold larger than the genome of rice (Price et al. 2005). However, rice and sorghum chromosomes are largely colinear indicating that sorghum genome expansion relative to rice is not due to a large-scale genome duplication event (Peng et al. 1999; Wilson et al. 1999; Klein et al. 2003) even though there is strong evidence for segmental and possibly a whole-genome duplication in the common progenitor of rice and sorghum (Paterson et al. 2003; Yu et al. 2005). Several studies have identified and aligned eight homeologous pairs of sorghum and rice chromosomes and shown that sorghum chromosomes 1 and 2 represent fusions of DNA corresponding to rice chromosomes 3 and 10 and 7 and 9, respectively (Kellogg 2001). Estimates of DNA content per chromosome based on a genome size of 818 Mbp show that sorghum chromosomes are ∼1.7- to ∼2.8-fold larger than the corresponding homeologous rice chromosomes or chromosomal regions. However, the increase in DNA content is not uniform across the homeologous sorghum and rice chromosomes even though they show a high degree of macrocolinearity (Peng et al. 1999; Wilson et al. 1999; Klein et al. 2003). An analysis of six pairs of homeologous sorghum and rice chromosomes showed that the euchromatic portion of these sorghum chromosomes is ∼1.8-fold larger than the colinear regions of the homeologous rice chromosomes, whereas the pericentromeric heterochromatic regions are ∼3.6-fold larger.
The increased size of euchromatic and heterochromatic regions of S. bicolor relative to O. sativa raises questions about how and when this difference was established and its functional significance. Numerous studies have shown that variation in genome size not related to polyploidy is due primarily to differences in repetitive DNA (Flavell et al. 1974; Uozu et al. 1997; Bennetzen et al. 2005). In plants, retroelement DNA accounts for a large portion of the repeat fraction. For example, in rice, retroelement sequences account for at least ∼13% of the genome (Sasaki et al. 2002) compared to ∼33% for sorghum (Bedell et al. 2005) and at least ∼60% for the maize genome (Messing et al. 2004; SanMiguel and Bennetzen 1999). While single LTR-retrotransposons are sometimes inserted in close proximity to genes located in euchromatic regions, these sequences are preferentially located in pericentromeric regions of heterochromatin (Kumar and Bennetzen 1999). This is consistent with sequence scanning of sorghum BACs mapping to heterochromatic regions, which showed lower gene density and a corresponding increase in retroelement-related sequences compared to BACs from euchromatic regions (Klein et al. 2003). The increase in retroelement content in sorghum compared to rice is also consistent with the ∼3.6-fold increase in the size of the sorghum heterochromatic regions compared to the colinear regions of rice.
The increase in size of sorghum heterochromatic regions compared to rice could be due to expansion and/or dispersion of heterochromatin along the chromosomes. Two different methods indicated that the pericentromeric heterochromatic region of sorghum chromosome 3 spans ∼38 Mbp. This ∼38-Mbp heterochromatic region of sorghum chromosome 3 aligns to a ∼8.6-Mbp colinear region of rice chromosome 1 that spans the centromere (Klein et al. 2003). Earlier cytogenetic analysis of rice chromosomes revealed that heterochromatin covers ∼18% of rice chromosome 1 pachytene-stage bivalents or a minimum of ∼8.2 Mbp of DNA (estimated on the basis of data in Cheng et al. 2001a). Therefore, the ∼4-fold increase in the size of the pericentromeric heterochromatic region of sorghum chromosome 3 relative to the colinear region of rice chromosome 1 appears to be due primarily to an increase in the size of this region rather than to heterochromatin dispersion along the chromosome. However, the boundaries between heterochromatin and euchromatin are less distinct in rice than in sorghum. Therefore, further analysis of the location of heterochromatin on rice chromosomes, like that conducted by Li et al. (2005), will be needed to determine more precisely the relative distribution of heterochromatin in sorghum and rice. This comparison may also contribute to an understanding of important differences in gene expression in these regions of the genomes (Li et al. 2005).
Retroelement sequences are often nested due to multiple rounds of localized LTR-retrotransposon insertion (SanMiguel et al. 1996). The preferential insertion of retroelements into related sequences already present in heterochromatic regions could have contributed to localized expansion of heterochromatin and to a decrease in gene density in these regions observed in sorghum relative to rice. It is also possible that the current differences in the size and distribution of heterochromatin in sorghum compared to rice and maize are the result of processes that decrease the DNA content of genomes (Bennetzen et al. 2005). Recent studies have demonstrated that unequal homologous recombination and illegitimate recombination play major roles in eliminating DNA from genomes (Shirasu et al. 2000; Devos et al. 2002). For example, it was estimated that ∼190 Mbp of DNA has been removed from the rice genome over the past 5 million years (Bennetzen et al. 2005). Differential action of retrotransposons could also have caused differential genome expansion in this same time frame because dating of LTR divergence in rice, sorghum, and maize indicates that the average age of these elements is ∼1.5–2.5 MY in all three species (Ma et al. 2004; Bennetzen et al. 2005). Overall, the amount of heterochromatic DNA in different sorghum chromosomes ranged from ∼35.5 to ∼49 Mbp, suggesting that the processes adding and removing DNA from these regions are acting somewhat uniformly on all chromosomes.
The results obtained in this study were also used to estimate the number of genes in the euchromatic and heterochromatic regions of the sorghum genome. In one approach, gene density in sorghum was predicted on the basis of alignment of sorghum chromosomal regions to rice because the organization of genes across homeologous pairs of sorghum and rice chromosomes shows a high degree of macrocolinearity. In this study, the euchromatic and heterochromatic regions of six sorghum chromosomes were aligned to colinear portions of the corresponding homeologous rice chromosomes. If the gene content of rice and sorghum is similar in colinear regions, then the euchromatic portions of the six sorghum chromosomes analyzed are predicted to have an average gene density of 1 gene model/12.3 kbp on the basis of an estimated gene complement of ∼47,000. This measurement of gene density is in reasonable agreement with published results on the annotation of a small number of BAC sequences from sorghum euchromatic regions (Morishige et al. 2002; Lai et al. 2004; Swigonova et al. 2004; Klein et al. 2005). Our measurements indicate that euchromatic DNA spans ∼50% of the sorghum genome (∼407 Mbp). If a gene density of 1 gene model/12.3 kbp is present throughout the euchromatic regions of sorghum, then we predict that ∼70% of the sorghum genes are located in the euchromatic portion of the sorghum genome. Assuming sorghum and rice encode a similar set of genes, ∼30% of the sorghum genes reside in heterochromatic regions at ∼2.7-fold lower gene density (∼1 gene model/34.5 kbp). We note that the estimates above are based on a prediction of ∼47,000 rice gene models (http://www.tigr.org/tdb/e2k1/osa1/pseudomolecules/info.shtml) and that a lower estimate of ∼37,500 rice gene models has been predicted by another group using somewhat different criteria (International Rice Genome Sequencing Project 2005). Current procedures for predicting gene models likely result in an overestimate of actual gene content in both sorghum and rice (Bennetzen et al. 2004). Therefore, a precise understanding of the sorghum gene complement will require a complete sorghum genome sequence and validation of predicted gene models. However, given these caveats, the current analysis predicts that ∼70% of the sorghum genes are located in the distal euchromatic regions that span ∼50% of the sorghum genome with a gene density on average only ∼1.6-fold lower than that of rice. Therefore, sequencing these regions by a combination of whole-genome shotgun and BAC-by-BAC sequencing methods appears straightforward. Small blocks of heterochromatin are located within the euchromatic regions, and knowledge of their location based on BAC-FISH analysis will be useful during sequence assembly. In contrast, the ∼50% of the sorghum genome represented by heterochromatin will be more challenging to sequence and assemble. These regions are predicted to have at least ∼2.7-fold lower gene density than the euchromatic arms. This is consistent with the reduced gene density observed in sequence scan data from BACs that map to the heterochromatic regions (Klein et al. 2003). While the distribution of genes within the heterochromatin is unknown, islands of genic sequences do appear to exist in the pericentromeric region. Most BACs from this region used in FISH analysis resulted in a “chromosome painting” pattern consistent with the presence of interspersed repetitive elements (data not shown). However, several BACs produced a strong, localized signal consistent with the existence of small sectors of relatively high gene density (Figure 1C, BAC probe 3-13). Nevertheless, sequences from most of the BACs mapped to the pericentromeric region are rich in repetitive sequences, especially retrotransposon-derived sequences, relative to BACs mapped to euchromatic DNA (Klein et al. 2003). Because recombination is suppressed in the heterochromatic regions and sequence repeat content is high, construction of robust BAC-based physical maps and sequence assembly in this part of the sorghum genome will be much more challenging.
The rate of recombination across the euchromatic portion of the sorghum genome is relatively high (∼0.25 Mbp/cM), similar to the average rate of recombination across rice chromosomes outside of centromeric regions (Wu et al. 2003). There was an ∼2.8-fold variation in average rate of recombination in the euchromatic regions of different chromosomes, excluding the short arm of sorghum chromosome 6 where average recombination was suppressed significantly (∼5.6 Mbp/cM). This chromosome is acrocentric, and pericentromeric heterochromatin spans most of the short arm. A similar organization of heterochromatin is present in the homeologous rice chromosome 4 that is also associated with reduced recombination (Cheng et al. 2001a). The average rate of recombination across the heterochromatic portion of the sorghum genome was ∼34-fold lower than recombination within euchromatic regions. This is not surprising because the general suppression of recombination in heterochromatic regions is a long-standing observation (Mather 1939). Suppression of recombination in heterochromatin is associated with accumulation of repeated sequences and the formation of modified chromatin structure in these regions (Avramova 2002). Recombination in the heterochromatic region of sorghum was ∼15-fold lower than that in colinear regions of rice. This is consistent with an increase in repetitive DNA and associated heterochromatin in sorghum compared to the colinear regions of rice. However, it was surprising that the rate of recombination in heterochromatic regions of different sorghum chromosomes also varied ∼15-fold. This may be explained in part by differences in gene density within the heterochromatic regions of different chromosomes. However, other factors including differences in the arrangement of DNA segments and specific genes, and differential expansion of repetitive DNA in the two parental lines used in linkage map construction, could also contribute to the observed variation (Brunner et al. 2005). At present we know little about the order and distribution of genes and repetitive DNA within sorghum heterochromatin and the relationship between this and local rates of recombination. Studies similar to those conducted in maize (Fu et al. 2001; Fu et al. 2002; Yao et al. 2002) will be required to more fully explain the variation in recombination observed here. While the exact causes of low and variable recombination in pericentromeric heterochromatin remain to be clarified, the impact of low recombination on the evolution and linkage disequilibrium of the sorghum genes encoded by this portion of the genome is of great interest and practical significance to sorghum geneticists and breeders.
Acknowledgments
We thank W. L. Rooney for providing plant material for chromosome preparations. This work was supported in part by National Science Foundation Plant Genome Research Grants DBI-0077713 (P.E.K. and J.E.M.) and DBI-0321578 (P.E.K., R.R.K., and J.E.M.), by the Perry Adkisson Chair in Agricultural Biology (J.E.M.), and by the U.S. Department of Agriculture's Agricultural Research Service (R.R.K.).
References
- An, S. Y., S. Park, D. H. Jeong, D. Y. Lee, H. G. Kang et al., 2003. Generation and analysis of end sequence database for T-DNA tagging lines in rice. Plant Physiol. 133: 2040–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avramova, Z. V., 2002. Heterochromatin in animals and plants. Similarities and differences. Plant Physiol. 129: 40–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedell, J. A., M. A. Budiman, A. Nunberg, R. W. Citek, D. Robbins et al., 2005. Sorghum genome sequencing by methylation filtration. PLoS. Biol. 3: 103–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen, J. L., C. Coleman, R. Y. Liu, J. X. Ma and W. Ramakrishna, 2004. Consistent over-estimation of gene number in complex plant genomes. Curr. Opin. Plant Biol. 7: 732–736. [DOI] [PubMed] [Google Scholar]
- Bennetzen, J. L., J. X. Ma and K. Devos, 2005. Mechanisms of recent genome size variation in flowering plants. Ann. Bot. 95: 127–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers, J. E., C. Abbey, S. Anderson, C. Chang, X. Draye et al., 2003. A high-density genetic recombination map of sequence-tagged sites for Sorghum, as a framework for comparative structural and evolutionary genomics of tropical grains and grasses. Genetics 165: 367–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner, S., K. Fengler, M. Morgante, S. Tingey and A. Rafalski, 2005. Evolution of DNA sequence nonhomologies among maize inbreds. Plant Cell 17: 343–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan, C. D., S. Lim, R. A. Salzman, I. Kagiampakis, D. T. Morishige et al., 2005. Sorghum bicolor's transcriptome response to dehydration, high salinity and ABA. Plant Mol. Biol. 58: 699–720. [DOI] [PubMed] [Google Scholar]
- Cantrell, R. P., and T. G. Reeves, 2002. The rice genome—the cereal of the world's poor takes center stage. Science 296: 53. [DOI] [PubMed] [Google Scholar]
- Chen, C. C., C. M. Chen, F. C. Hsu, C. J. Wang, J. T. Yang et al., 2000. The pachytene chromosomes of maize as revealed by fluorescence in situ hybridization with repetitive DNA sequences. Theor. Appl. Genet. 101: 30–36. [Google Scholar]
- Cheng, Z., C. R. Buell, R. A. Wing, M. Gu and J. Jiang, 2001. a Toward a cytological characterization of the rice genome. Genome Res. 11: 2133–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, Z., G. G. Presting, C. R. Buell, R. A. Wing and J. Jiang, 2001. b High-resolution pachytene chromosome mapping of bacterial artificial chromosomes anchored by genetic markers reveals the centromere location and the distribution of genetic recombination along chromosome 10 of rice. Genetics 157: 1749–1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos, K. M., 2005. Updating the “Crop circle.” Curr. Opin. Plant Biol. 8: 155–162. [DOI] [PubMed] [Google Scholar]
- Devos, K. M., J. K. M. Brown and J. L. Bennetzen, 2002. Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis. Genome Res. 12: 1075–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley, J., M. Durbin, E. M. Golenberg, M. T. Clegg and D. P. Ma, 1990. Evolutionary analysis of the large subunit of carboxylase (rbcL) nucleotide sequence among the grasses (Gramineae). Evolution 44: 1097–1108. [DOI] [PubMed] [Google Scholar]
- Doggett, H., 1988. Sorghum. John Wiley, New York.
- Edwards, G. E., V. R. Franceschi and E. V. Voznesenskaya, 2004. Single-cell C4 photosynthesis versus the dual-cell (Kranz) paradigm. Ann. Rev. Plant Biol. 55: 173–196. [DOI] [PubMed] [Google Scholar]
- Evans, L. T., 1998. Feeding the Ten Billion: Plants and Population Growth. Cambridge University Press, Cambridge, UK.
- Flavell, R. B., M. D. Bennett, J. B. Smith and D. B. Smith, 1974. Genome size and proportion of repeated nucleotide DNA sequence in plants. Genetics 12: 257–269. [DOI] [PubMed] [Google Scholar]
- Fu, H. H., W. K. Park, X. H. Yan, Z. W. Zheng, B. Z. Shen et al., 2001. The highly recombinogenic bz locus lies in an unusually gene-rich region of the maize genome. Proc. Natl. Acad. Sci. USA 98: 8903–8908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, H. H., Z. W. Zheng and H. K. Dooner, 2002. Recombination rates between adjacent genic and retrotransposon regions in maize vary by 2 orders of magnitude. Proc. Natl. Acad. Sci. USA 99: 1082–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut, B. S., 2002. Evolutionary dynamics of grass genomes. New Phytol. 154: 15–28. [Google Scholar]
- Gill, B. S., B. Friebe and T. R. Endo, 1991. Standard karyotype and nomenclature system for description of chromosome bands and structural aberrations in wheat (Triticum aestivum). Genome 34: 830–839. [Google Scholar]
- Gnansounou, E., A. Dauriat and C. E. Wyman, 2005. Refining sweet sorghum to ethanol and sugar: economic trade-offs in the context of North China. Bioresour. Technol. 96: 985–1002. [DOI] [PubMed] [Google Scholar]
- Grass Phylogeny Working Group, 2000. A phylogeny of the grass family (Poaceae), as inferred from eight character sets, pp. 3–7 in Grasses: Systematics and Evolution, edited by S. W. L. Jacobs and J. E. Everett. Commonwealth Scientific and Industrial Research Organization, Collingwood, Victoria, Australia.
- International Rice Genome Sequencing Project, 2005. The map-based sequence of the rice genome. Nature 436: 793–800. [DOI] [PubMed] [Google Scholar]
- Islam-Faridi, M. N., K. L. Childs, P. E. Klein, G. Hodnett, M. A. Menz et al., 2002. A molecular cytogenetic map of sorghum chromosome 1: fluorescence in situ hybridization analysis with mapped bacterial artificial chromosomes. Genetics 161: 345–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg, E. A., 2001. Evolutionary history of the grasses. Plant Physiol. 125: 1198–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J. S., K. L. Childs, N. I. Faridi, M. A. Menz, R. R. Klein et al., 2002. Integrated karyotyping of sorghum by in situ hybridization of landed BACs. Genome 45: 402–412. [DOI] [PubMed] [Google Scholar]
- Kim, J. S., P. E. Klein, R. R. Klein, H. J. Price, J. E. Mullet et al., 2005. a Chromosome identification and nomenclature of Sorghum bicolor. Genetics 169: 1169–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J. S., P. E. Klein, R. R. Klein, H. J. Price, J. E. Mullet et al., 2005. b Molecular cytogenetic maps of sorghum linkage groups 2 and 8. Genetics 169: 955–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein, P. E., R. R. Klein, S. W. Cartinhour, P. E. Ulanch, J. Dong et al., 2000. A high-throughput AFLP-based method for constructing integrated genetic and physical maps: progress toward a sorghum genome map. Genome Res. 10: 789–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein, P. E., R. R. Klein, J. Vrebalov and J. E. Mullet, 2003. Sequence-based alignment of sorghum chromosome 3 and rice chromosome 1 reveals extensive conservation of gene order and one major chromosomal rearrangement. Plant J. 34: 605–621. [DOI] [PubMed] [Google Scholar]
- Klein, R. R., P. E. Klein, J. E. Mullet, P. Minx, W. L. Rooney et al., 2005. Fertility restorer locus Rf1 of sorghum (Sorghum bicolor L.) encodes a pentatricopeptide repeat protein not present in the colinear region of rice chromosome 12. Theor. Appl. Genet. 111: 994–1012. [DOI] [PubMed] [Google Scholar]
- Kulikova, O., G. Gualtieri, R. Geurts, D. J. Kim, D. Cook et al., 2001. Integration of the FISH pachytene and genetic maps of Medicago truncatula. Plant J. 27: 49–58. [DOI] [PubMed] [Google Scholar]
- Kumar, A., and J. L. Bennetzen, 1999. Plant retrotransposons. Annu. Rev. Genet. 33: 479–532. [DOI] [PubMed] [Google Scholar]
- Lai, J., J. Ma, Z. Swigonova, W. Ramakrishna, E. Linton et al., 2004. Gene loss and movement in the maize genome. Genome Res. 14: 1924–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy, A. A., and M. Feldman, 2002. The impact of polyploidy on grass genome evolution. Plant Physiol. 130: 1587–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L., X. Wang, M. Xia, V. Stole, N. Su et al., 2005. Tiling microarray analysis of rice chromosome 10 to identify the transcriptome and relate its expression to chromosomal architecture. Genome Biol. 6: R52.51–R52.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, M. C., C. Thomas, F. M. You, J. Hsiao, S. Ouyang et al., 2003. High-throughput fingerprinting of bacterial artificial chromosomes using the SNaPshot labeling kit and sizing of restriction fragments by capillary electrophoresis. Genomics 82: 378–389. [DOI] [PubMed] [Google Scholar]
- Ma, J., K. M. Devos and J. L. Bennetzen, 2004. Analyses of LTR-retrotransposon structures reveal recent and rapid genomic DNA loss in rice. Genome Res. 14: 860–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mather, K., 1939. Crossing over and heterochromatin in the X chromosome of Drosophila melanogaster. Genetics 24: 413–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menz, M. A., R. R. Klein, J. E. Mullet, J. A. Obert, N. C. Unruh et al., 2002. A high-density genetic map of Sorghum bicolor (L.) Moench based on 2926 AFLP(R), RFLP and SSR markers. Plant Mol. Biol. 48: 483–499. [DOI] [PubMed] [Google Scholar]
- Menz, M. A., R. R. Klein, N. C. Unruh, W. L. Rooney, P. E. Klein et al., 2004. Genetic diversity of public inbreds of sorghum determined by mapped AFLP and SSR markers. Crop Sci. 44: 1236–1244. [Google Scholar]
- Messing, J., A. K. Bharti, W. M. Karlowski, H. Gundlach, H. R. Kim et al., 2004. Sequence composition and genome organization of maize. Proc. Natl. Acad. Sci. USA 101: 14349–14354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, G., K. M. Devos, Z. Wang and M. D. Gale, 1995. Grasses, line up and form a circle. Curr. Biol. 5: 737–739. [DOI] [PubMed] [Google Scholar]
- Morishige, D. T., K. L. Childs, L. D. Moore and J. E. Mullet, 2002. Targeted analysis of orthologous phytochrome A regions of the sorghum, maize, and rice genomes using gene-island sequencing. Plant Physiol. 130: 1614–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Research Council, 1996. Lost Crops of Africa, Volume 1: Grains. National Academy Press, Washington, DC.
- Paterson, A. H., J. E. Bowers, D. G. Peterson, J. C. Estill and B. A. Chapman, 2003. Structure and evolution of cereal genomes. Curr. Opin. Genet. Dev. 13: 644–650. [DOI] [PubMed] [Google Scholar]
- Paterson, A. H., J. E. Bowers and B. A. Chapman, 2004. Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc. Natl. Acad. Sci. USA 101: 9903–9908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, Y., K. F. Schertz, S. Cartinhour and G. E. Hart, 1999. Comparative genome mapping of Sorghum bicolor (L.) Moench using an RFLP map constructed in a population of recombinant inbred lines. Plant Breed. 118: 225–235. [Google Scholar]
- Peterson, D. G., N. L. V. Lapitan and S. M. Stack, 1999. Localization of single- and low-copy sequences on tomato synaptonemal complex spreads using fluorescence in situ hybridization (FISH). Genetics 152: 427–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price, H. J., S. L. Dillon, G. Hodnett, W. L. Rooney, L. Ross et al., 2005. Genome evolution in the genus Sorghum (Poaceae). Ann. Bot. 95: 219–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice Full-length cDNA Consortium, 2003. Collection, mapping, and annotation of over 28,000 cDNA clones from japonica rice. Science 301: 376–379. [DOI] [PubMed] [Google Scholar]
- Saghai Maroof, M. A., G. P. Yang, R. M. Biyashev, P. J. Maughan and Q. Zhang, 1996. Analysis of the barley and rice genomes by comparative RFLP linkage mapping. Theor. Appl. Genet. 92: 541–551. [DOI] [PubMed] [Google Scholar]
- Salzman, R. A., J. A. Brady, S. A. Finlayson, C. D. Buchanan, E. J. Summer et al., 2005. Transcriptional profiling of sorghum induced by methyl jasmonate, salicylic acid, and aminocyclopropane carboxylic acid reveals cooperative regulation and novel gene responses. Plant Physiol. 138: 352–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SanMiguel, P., and J. L. Bennetzen, 1999. Evidence that a recent increase in maize genome size was caused by the massive amplification of intergene retrotransposons. Ann. Bot. 82: 37–44. [Google Scholar]
- SanMiguel, P., A. Tikhonov, Y.-K. Jin, N. Motchoulskaia, D. Zakharov et al., 1996. Nested retrotransposons in the intergenic regions of the maize genome. Science 274: 765–768. [DOI] [PubMed] [Google Scholar]
- Sasaki, T., T. Matsumoto, K. Yamamoto, K. Sakata, T. Baba et al., 2002. The genome sequence and structure of rice chromosome 1. Nature 420: 312–316. [DOI] [PubMed] [Google Scholar]
- Schantz, H. L., 1954. The place of grasslands in the earth's cover of vegetation. Ecology 35: 143–145. [Google Scholar]
- Shimamoto, K., and J. Kyozuka, 2002. Rice as a model for comparative genomics of plants. Ann. Rev. Plant Biol. 53: 399–419. [DOI] [PubMed] [Google Scholar]
- Shirasu, K., A. H. Schulman, T. Lahaye and P. Schulze-Lefert, 2000. A contiguous 66-kb barley DNA sequence provides evidence for reversible genome expansion. Genome Res. 10: 908–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorghum Genomics Planning Workshop Participants, 2005. Toward sequencing the sorghum genome: a US National Science Foundation-sponsored workshop report. Plant Physiol. 138: 1898–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swigonova, Z., J. Lai, J. Ma, W. Ramakrishna, V. Llaca et al., 2004. Close split of sorghum and maize genome progenitors. Genome Res. 14: 1916–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao, Q., and H. B. Zhang, 1998. Cloning and stable maintenance of DNA fragments over 300 kb in Escherichia coli with conventional plasmid-based vectors. Nucleic Acids Res. 26: 4901–4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton, J. W., and R. DeSalle, 2000. Gene family evolution and homology: genomics meets phylogenetics. Annu. Rev. Genomics Hum. Genet. 1: 41–73. [DOI] [PubMed] [Google Scholar]
- Tikhonov, A. P., P. J. SanMiguel, Y. Nakajima, N. M. Gorenstein, J. L. Bennetzen et al., 1999. Colinearity and its exceptions in orthologous adh regions of maize and sorghum. Proc. Natl. Acad. Sci. USA 96: 7409–7414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uozu, S., H. Ikehashi, N. Ohmido, H. Ohtsubo, E. Ohtsubo et al., 1997. Repetitive sequences: cause for variation in genome size and chromosome morphology in the genus Oryza. Plant Mol. Biol. 35: 791–799. [DOI] [PubMed] [Google Scholar]
- Ureta-Vidal, A., L. Ettwiller and E. Birney, 2003. Comparative genomics: genome-wide analysis in metazoan eukaryotes. Nat. Rev. Genet. 4: 251–262. [DOI] [PubMed] [Google Scholar]
- Vandepoele, K., C. Simillion and Y. Van de Peer, 2003. Evidence that rice and other cereals are ancient aneuploids. Plant Cell 15: 2192–2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, W. A., S. E. Harrington, W. L. Woodman, M. Lee, M. E. Sorrells et al., 1999. Inferences on the genome structure of progenitor maize through comparative analysis of rice, maize and the domesticated panicoids. Genetics 153: 453–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo, S. S., J. M. Jiang, B. S. Gill, A. H. Paterson and R. A. Wing, 1994. Construction and characterization of bacterial artificial chromosome library of Sorghum bicolor. Nucleic Acids Res. 22: 4922–4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J., H. Mizuno, M. Hayashi-Tsugane, Y. Ito, Y. Chiden et al., 2003. Physical maps and recombination frequency of six rice chromosomes. Plant J. 36: 720–730. [DOI] [PubMed] [Google Scholar]
- Yao, H., Q. Zhou, J. Li, H. Smith, M. Yandeau et al., 2002. Molecular characterization of meiotic recombination across the 140-kb multigenic a1-sh2 interval of maize. Proc. Natl. Acad. Sci. USA 99: 6157–6162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J., J. Wang, W. Lin, S. G. Li, H. Li et al., 2005. The genomes of Oryza sativa: a history of duplications. PLoS. Biol. 3: 266–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwick, M. S., M. N. Islam-Faridi, H. B. Zhang, G. L. Hodnett, M. I. Gomez et al., 2000. Distribution and sequence analysis of the centromere-associated repetitive element CEN38 of Sorghum bicolor (Poaceae). Am. J. Bot. 87: 1757–1764. [PubMed] [Google Scholar]