Abstract
The LmR1 locus, which controls seedling resistance to the blackleg fungus Leptosphaeria maculans in the Brassica napus cultivar Shiralee, was positioned on linkage group N7. Fine genetic mapping in a population of 2500 backcross lines identified three molecular markers that cosegregated with LmR1. Additional linkage mapping in a second population colocalized a seedling resistance gene, ClmR1, from the cultivar Cresor to the same genetic interval on N7 as LmR1. Both genes were located in a region that showed extensive inter- and intragenomic duplications as well as intrachromosomal tandem duplications. The tandem duplications seem to have occurred in the Brassica lineage before the divergence of B. rapa and B. oleracea but after the separation of Brassica and Arabidopsis from a common ancestor. Microsynteny was found between the region on N7 carrying the resistance gene and the end of Arabidopsis chromosome 1, interrupted by a single inversion close to the resistance locus. The collinear region in Arabidopsis was assayed for the presence of possible candidate genes for blackleg resistance. These data provided novel insights into the genomic structure and evolution of plant resistance loci and an evaluation of the candidate gene approach using comparative mapping with a model organism.
BLACKLEG caused by the fungus Leptosphaeria maculans (Desm) Ces. & de Not is a devastating disease of canola (Brassica napus L. and B. rapa L.) and has caused extensive yield losses in Europe, Australia, and Canada (Gugel and Petrie 1992). Since chemical control of the disease is difficult, is uneconomical, and poses risks to the producer and the environment (Ahlers 1991), the introduction of genetic resistance to blackleg has become a major objective of canola breeding programs. Brassica crop species comprise primary diploids containing the A, B, or C genome, respectively, and amphidiploids, like B. napus (AACC), carrying a combination of two diploid genomes (U 1935). A complete hypersensitive resistance to L. maculans has been found in Brassica species containing the B genome, which is effective throughout the life of the plant (Roy 1984; Rimmer and Van Den Berg 1992). Such resistance loci have been identified and mapped in B. nigra, B. juncea, and B. carinata and a number of attempts have been made to transfer them into B. napus breeding lines through interspecific hybridization (Roy 1984; Sjödin and Glimelius 1988; Rimmer and Van Den Berg 1992; Struss et al. 1992, Chèvre et al. 1996; Pang and Halloran 1996a). The results have been promising; however, cotransfer of unwanted traits and low frequencies of recombination between the different species have complicated the development of blackleg-resistant oilseed cultivars by conventional breeding (Chèvre et al. 1997).
Most modern commercial canola varieties carry sources of seedling and adult plant resistance to L. maculans, which originate from the A genome of B. napus and B. rapa. While adult plant resistance in the field is ultimately the trait that is essential for large-scale oilseed production, the wide pathogen variability and the constraints of field testing make it difficult to assess this resistance genetically. Monogenic and polygenic inheritance of adult resistance under controlled and field conditions has been reported in the literature (Cargeeg and Thurling 1979; Sippell et al. 1991; Stringam et al. 1992; Dion et al.1995; Ferreira et al. 1995; Pang and Halloran 1996b,c; Pilet et al. 1998; Li and Cowling 2003). In contrast, cotyledon or seedling-stage resistance is generally believed to be controlled by single dominant genes, which have been shown to confer resistance to specific races of the pathogen. Although numerous studies suggest that seedling and adult blackleg resistances are under different genetic controls, many authors have observed a significant correlation between the two (Newman and Bailey 1987; Mcnabb et al. 1993; Bansal et al. 1994; Li and Cowling 2003). This could be due to tight linkage of a cotyledon resistance gene to one of several major adult resistance quantitative trait loci (QTL) in a particular variety or allelic variation at a single locus could control both adult and cotyledon resistance. Closely linked loci for adult and seedling resistance were identified in the canola cultivar Major and in the B. napus breeding accessions RB87-62 and DH88-752 (Ferreira et al. 1995; Zhu and Rimmer 2003). Similarly, the race-specific cotyledon resistance gene Rlm1 has been shown to explain 70% of the variation for adult plant resistance in the French cultivar Maxol (Delourme et al. 2004). In fact, the strong correlation between seedling and field resistance in the Australian canola cultivar Maluka was exploited to expedite the development of the cultivar Quantum for Western Canadian field conditions (Stringam et al. 1995). Therefore, markers for seedling resistance can be a great asset for accelerating oilseed breeding programs.
Loci controlling cotyledon resistance to L. maculans have been mapped in various B. napus cultivars using molecular markers. Single dominant genes were identified in the cultivars Major, RB87-62, Shiralee, and Maluka (Ferreira et al. 1995; Mayerhofer et al. 1997; Rimmer et al. 1999) and mapped to the same linkage group LG6, which has been shown subsequently to be equivalent to the A genome linkage group N7 (Ferreira et al. 1995; Parkin et al. 1995). Interestingly, a single major locus controlling field disease resistance in the cultivar Cresor has also been mapped to the same region of N7 (Dion et al. 1995). More recently, race-specific resistance genes identified in the cultivars Maxol, Quinta, Glacier, Samourai, and Darmor have been mapped to two genomic regions on linkage groups LG10 and LG16 of a consensus linkage map (Delourme et al. 2004). A cluster of five cotyledon resistance genes was suggested on LG10. Due to the absence of common molecular markers this linkage group cannot be associated directly with the previously cited mapping studies but on the basis of comparisons of resistance gene and avirulence gene specificities Delourme et al. (2004) suggest that LG10 corresponds to LG6 of Ferreira et al. (1995) and hence to N7. A concentration of blackleg resistance loci appears to be on N7; however, little is known about the relationship of these genes and much less is known about their role in the defense response. Cloning of such resistance genes would not only allow analysis of the function and structural organization of these loci but also expedite the transfer of resistance into elite breeding lines without the associated linkage drag that is unavoidable in conventional breeding. Moreover, different cloned R genes could be used to “pyramid” the resistance in one cultivar to enhance the durability and range of the resistance.
Map-based cloning has been successfully employed to isolate disease resistance genes from a number of plant species (Bent 1996). This strategy involves fine-scale mapping of the gene in a large segregating population and identification of tightly linked flanking markers that allow quick chromosome walking toward the gene or, preferably, chromosome landing on a large insert clone carrying the resistance gene (Tanksley et al. 1995). For this approach to be successful, an ample supply of polymorphic markers is required to delimit the gene within a sufficiently small genetic interval of <1 cM.
Brassica species represent the closest crop plant relatives to the model plant Arabidopsis thaliana and the presence of chromosomal collinearity between these two species has been established in several studies (reviewed in Schmidt et al. 2001; Schmidt 2002). Large-scale ancestral genome duplications in both Arabidopsis and the amphidiploid B. napus, as well as the occurrence of rearrangements and insertions/deletions, complicate this collinear relationship (Schmidt 2002). Nevertheless, the extensive resources developed for the model plant, including the genome sequence, can be exploited for fine mapping in B. napus (Snowdon and Friedt 2004). Muangprom and Osborn (2004) found almost perfect collinearity of 13 RFLP markers between Arabidopsis and B. rapa around a dwarf gene, enabling them to identify a candidate gene in Arabidopsis and subsequently to clone and characterize the corresponding gene in B. rapa (Muangprom et al. 2005). Since Arabidopsis accessions are usually blackleg resistant (Bohman et al. 2004) and sequence homology between the two species in coding regions can be >85% (Cavell et al. 1998), candidate resistance genes could be present in collinear regions of Arabidopsis and be identified by similarity to known resistance gene motifs. Indeed, it was shown by somatic fusion experiments that Arabidopsis harbors blackleg resistance genes that are functional in B. napus (Bohman et al. 2002).
In this article we present a fine-scale map of the LmR1 locus of the Australian canola cultivar Shiralee, which is positioned in the same genetic interval as a cotyledon resistance gene originating from the French cultivar Cresor. The region was shown to be genetically complex yet displayed strong collinearity with the end of Arabidopsis chromosome 1. The intricacies of mapping in a highly duplicated genome and the feasibility of a candidate gene approach are discussed.
MATERIALS AND METHODS
Plant material and mapping populations:
Two different mapping populations were used to map the blackleg resistance genes of the canola cultivars Shiralee and Cresor, respectively. All parental lines were doubled-haploid (DH) lines of B. napus. A common susceptible parent was used for both mapping crosses: a newly resynthesized B. napus line PSA12 [M. Beschorner and D. Lydiate, Agriculture and Agri-Food Canada (AAFC) Saskatoon Research Centre], produced by embryo rescue from an interspecific cross between B. oleracea ssp. alboglabra line A12DH (G. King, Horticultural Research International) and B. rapa line Parkland Sunshine (K. Falk, AAFC Saskatoon Research Centre). DH12075 was derived from a cross between the blackleg-resistant canola cultivar Cresor and the susceptible cultivar Westar and was shown to be blackleg resistant in Canadian field trials (G. Seguin Schwartz and G. Rakow, AAFC Saskatoon Research Centre). Shiralee is an Australian canola cultivar resistant to the prevalent blackleg isolate found in Canada. Single resistant F1 plants from the cross DH12075 × PSA12 as well as from Shiralee × PSA12 were backcrossed to PSA12 to generate two backcross (BC)1 populations. Initially 90 and 100 BC1 lines from the DH12075 and Shiralee populations, respectively, were genotyped and assessed for blackleg resistance to identify markers flanking the resistance genes. Another 250 and 2448 lines from each mapping population, respectively, were screened for the presence of recombination events across the interval carrying the resistance loci. This maximized the resolution of marker order within the interval carrying the resistance gene.
Greenhouse screening for blackleg resistance:
The cotyledon resistance response to blackleg was determined for the BC1 populations as described in Mayerhofer et al. (1997). The resistance phenotype of each backcross line was determined using 16 self progeny from each BC1.
DNA isolations, Southern hybridizations, PCR reactions, and sequencing:
Genomic DNA preparations from fresh or lyophilized leaf tissue, Southern analyses, and RAPD reactions were carried out as described in Sharpe et al. (1995) and Mayerhofer et al. (1997). PCR reactions for sequence-characterized amplified region (SCAR) markers were performed in a Gene Amp 9700 thermocycler (Perkin-Elmer, Norwalk, CT) in 25-μl reaction volumes containing 10 mm Tris-HCl (pH 8.0); 50 mm KCl; 0.01% gelatin; 2.5 mm MgCl; 0.4 mm each of dATP, dCTP, dGTP, and dTTP; 0.2 μm each of primer; 0.5 units/reaction Taq DNA polymerase (Perkin-Elmer); and 25 ng of template DNA. The cycle parameters were 94°/2 min; 12 cycles of 94°/30 sec, 72° (−0.1°/cycle)/30 sec, 72°/1 min; and 23 cycles of 94°/30 sec, 60°/30 sec, 72°/1 min, followed by 72°/5 min. Sequencing reactions were carried out using a BigDye Terminator v.3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA).
Molecular markers:
DNA probes with the prefix “est” designate Arabidopsis EST clones purchased from the Arabidopsis Biological Resource Centre (http://www.arabidopsis.org). Markers rapdD3 and rapd184 are RFLP probes cloned from gel-purified RAPD bands generated by the primers OPD-03 (5′-GTCGCCGTCA-3′; Operon Technologies, Alameda, CA) and 184 (5′-CAAACGGCAC-3′; University of British Columbia), respectively. RFLP markers with prefixes “mi,” “pO,” “pW,” and “ec” are genomic or cDNA clones from Arabidopsis and Brassica species, respectively, and have been described earlier (Ferreira et al. 1995; Sharpe et al. 1995; Liu et al. 1996). Markers starting with the designation “BL,” “LL,” “ML,” “RL,” and “es” are B. napus EST clones, described on the Brassica/Arabidopsis Genomics Initiative web site (http://Brassica.agr.gc.ca/). The SCAR marker AO39 was kindly provided by Daryl Somers (AAFC Saskatoon Research Centre). “BNID,” “BNIH,” BNIB,” and “TH” designate end probes (from the T7 and R ends) generated from DH12075-derived BAC clones (S. Edes and I. Parkin, unpublished data). All markers were employed as RFLP probes in Southern analyses except when followed by the designation “(SCAR),” in which case their sequence was used to generate allele-specific PCR primers.
Linkage analysis:
All polymorphic loci detected with RFLP- or PCR-based markers were analyzed independently in the two mapping populations. Linkage analysis was carried out using Mapmaker v3 with a LOD score of 4.0 (Lander et al. 1987). Linkage groups generated in the two populations were subsequently aligned through the identification of common alleles at several loci. The loci were assigned to the linkage groups of B. napus through the identification of common alleles with parents of the mapping populations described in Parkin et al. (1995) and Sharpe et al. (1995). Brassica-derived sequences were analyzed for homology to the Arabidopsis genome using the BLASTN program of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/).
B. napus BAC library screening:
The BAC clones were derived from DH12075 genomic DNA partially digested with either HindIII (TH, BNID, and BNIH clones) or BamHI (BNIB) and ligated into pINDIGOBAC-5 (except those named TH, which were ligated into pBELOBAC11). The BAC clones were screened either by hybridization of probes to doubled-spotted macro-arrayed clones as described in Ryder et al. (2001) or by PCR amplification from four-dimensional pools of BAC DNA.
RESULTS
Strategy for fine mapping blackleg resistance loci:
Initially, the genotype and resistance phenotype were determined for a small number of individuals from each of the two backcross populations. As expected, in both populations the resistance phenotype mapped as a single dominant locus to linkage group N7 (Mayerhofer et al. 1997; Rimmer et al. 1999). Further mapping in each population was targeted to the resistance gene regions by assessing only the phenotype of lines with recombination events between the markers flanking the resistance loci, with the most extensive mapping carried out in the Shiralee population. Global comparative mapping between B. napus and Arabidopsis had shown that the identified region of N7 displayed some homology to the bottom of Arabidopsis chromosome 1 (Parkin et al. 2005). Arabidopsis EST clones from this region and Brassica EST clones homologous to this region of Arabidopsis chromosome 1 were assayed as potential markers in an attempt to saturate the interval on N7 containing the resistance loci (see below). Concurrently B. napus BACs were identified from a library of DH12075 using markers mapped to the region and probes generated from BAC end sequences were used to further target the region. Similarly, clones from a genomic library of Shiralee in λ-DASHII were identified and used to generate probes from both end sequences. Altogether 91 Arabidopsis ESTs, 23 B. napus ESTs, 54 B. napus BAC end sequences, and 19 B. napus λ-DASHII clone end sequences for a total of 187 markers were assayed for their ability to identify polymorphic loci on N7 in the target interval.
Inter- and intragenomic duplication of the R gene region:
Of the 187 markers tested, 50 identified 109 polymorphic loci in the Shiralee population that were mapped to B. napus linkage groups. The majority of the polymorphic loci (57) were mapped to linkage group N7 (Table 1, Figure 1). The subsequent selection of BC1 lines with recombination events between the flanking markers rapdD3 and AO39 allowed the locus order between these markers to be more accurately resolved compared to that of homologous loci mapped to duplicated regions of the B. napus genome (Figure 1). Twenty-nine of the RFLP markers that identified loci on N7 (58%) also revealed duplicated loci. The duplicate loci were mapped to a second location on linkage group N7 (tandem repeat), to three other linkage groups, N2, N12, and N16, and several remained unlinked (Table 1, Figure 1). All the RFLP markers hybridized to additional monomorphic bands that presumably represented further duplicated loci. The relatively insensitive process of Southern hybridization means that several more loci could remain undetected due to nucleotide divergence or small insertion/deletions of the duplicated sequences. N2 and N7 are derived from the B. rapa A-genome progenitor and N12 and N16 are derived from the B. oleracea C-genome progenitor of B. napus (Parkin et al. 1995). N2 and N7 shared eight homologous marker loci and N12 and N16 shared four loci detected by the same marker, which indicated the presence of intragenomic duplications within the A and C genomes, respectively. Intergenomic duplications between the A and C genomes were also evident. N2/N12 and N7/N16 have been shown previously to be homeologous (Parkin et al. 1995, 2003; Sharpe et al. 1995) and shared 8 and 11 homologous marker loci, respectively. However, the most extensive homeology was detected between N7 and N12. A total of 17 markers detected duplicated loci between the two linkage groups and both regions had undergone an intrachromosomal tandem duplication event. An intrachromosomal duplication event has previously been identified for the same region of linkage group O6 (equivalent to N16) of B. oleracea (Ryder et al. 2001). On the basis of these data, including the tandem duplication, it appears that the R gene region is present in at least three copies in the A genome and possibly in four copies in the C genome of Brassica species. However, the inability to map all detected loci due to lack of polymorphism precludes the absolute level of genome duplication from being determined.
TABLE 1.
Location and number of marker loci on Brassica linkage groups
| No. of loci per linkage group
|
|||||
|---|---|---|---|---|---|
| Markers | N7 | N2 | N12 | N16 | Unlinked |
| BNID37E13R | 1 | ||||
| BL2665 | 1 | ||||
| BNIH148M04T7(SCAR) | 1 | ||||
| RL2534 | 2 | 1 | |||
| rapdD3 | 2 | ||||
| TH8L20R | 2 | 1 | |||
| pO10 | 2 | ||||
| est179D16 | 1 | 1 | |||
| estM79E02 | 1 | ||||
| est148G3 | 1 | 1 | 1 | ||
| est119E6 | 1 | ||||
| est126M9 | 2 | 1 | |||
| est32A6 | 2 | 1 | 1 | ||
| est149M9 | 2 | 1 | 1 | 1 | 1 |
| BNID116A5T7 | 1 | ||||
| BNID116A5R | 1 | 1 | 1 | 1 | |
| rapd184 | 1 | ||||
| BNID7804T7 | 1 | 1 | |||
| es1450 | 1 | 1 | 3 | ||
| ec2h2 | 1 | 1 | |||
| est187O3 | 1 | ||||
| AO39(SCAR) | 1 | ||||
| est118L9 | 1 | 1 | 1 | ||
| est151K13 | 1 | ||||
| estM64L03 | 1 | ||||
| LL2348 | 1 | 1 | 2 | ||
| BNID97N1T7 | 1 | 1 | |||
| BNID92M21T7(SCAR) | 1 | ||||
| estM70H09 | 1 | ||||
| BNID12B17T7 | 1 | 1 | |||
| TH28E21T7 | 1 | 3 | |||
| ML853R(SCAR) | 1 | ||||
| estYAY175 | 1 | 1 | 2 | 1 | |
| est147H3 | 1 | 1 | |||
| I-D2 | 1 | ||||
| est200P8 | 1 | 1 | 2 | 1 | 1 |
| est32F9 | 1 | 1 | 2 | ||
| pW134 | 1 | ||||
| pW150 | 1 | 1 | |||
| est144I14 | 1 | 1 | |||
| est149P9 | 1 | 1 | |||
| RL1856 | 1 | 1 | |||
| mi157 | 1 | ||||
| est160P22 | 1 | ||||
| est205F17 | 1 | ||||
| estM67J04 | 1 | 1 | |||
| estM66B02 | 1 | 1 | |||
| est155E11 | 1 | 1 | |||
| est103P6 | 1 | ||||
| BNID22D17T7 | 1 | 1 | |||
Figure 1.
Linkage map of Shiralee × PSA12 population. Marker loci linked to the blackleg resistance gene LmR1 as well as duplicated loci are shown. Vertical bars show linkage groups N2, N7, N12, and N16 of Brassica napus. Map distances are given in recombination units. Markers with tandem duplications on linkage groups N7 and N12 are underlined and are adjacent to hatched boxes.
Intrachromosomal tandem duplication of R gene region:
Seven of the markers linked to the resistance gene on linkage group N7 had duplicated loci that formed a repeat unit contiguous with the interval defined by the flanking markers rapdD3 and AO39 (SCAR). The loci in this second unit cosegregated as a result of the selection of mapping lines (Figure 1). Consequently the orientation of the two repeats with respect to each other could not be established, although the duplication previously identified on the homeologous region of linkage group O6 (Ryder et al. 2001) suggests that the tandem duplication may be part of a larger inverted repeat. Five duplicated loci indicated the presence of a similar tandem duplication event on linkage group N12 (Figure 1).
Correspondence of Cresor and Shiralee R gene region:
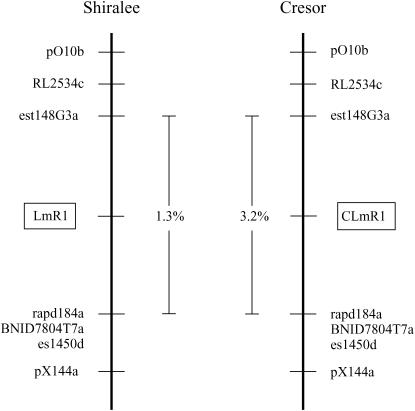
Cotyledon resistance to blackleg was mapped independently in the Shiralee- and Cresor-derived mapping populations that were generated using a common susceptible parent. Alignment of the two maps using common marker loci demonstrated that the Shiralee (LmR1) and Cresor (CLmR1) resistance genes mapped to the same genomic location on N7 and were flanked by common loci (Figure 2a). However, the Shiralee resistance gene cosegregated with three markers, BNID116A5T7, BNID116A5R, and est149M9, which could not be mapped to this region in the Cresor population. The closest flanking markers with common alleles between the two mapping populations were est148G3 and a cluster of markers, rapd184, BNID7804T7, and es1450, which spanned a distance of 1.3 recombination units in Shiralee and 3.2 recombination units in Cresor (Figure 2b). These data suggest that the resistance genes from Shiralee and Cresor could be allelic or members of an R gene cluster.
Figure 2.
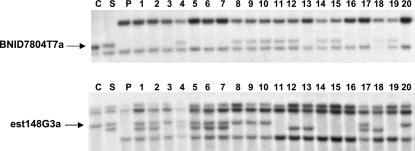
(a) Comparison of flanking markers of the blackleg resistance genes from Shiralee (LmR1) and Cresor (CLmR1) on linkage group N7. Only marker loci with identical Southern bands in both populations are shown. Genetic distances are given in recombination units. (b) Southern analysis of flanking markers BNID7804T7 and est148G3 in segregating populations derived from Shiralee and Cresor (DH12075). The arrows show the bands for loci BNID7804T7a and est148G3a, which are tightly linked to the resistance genes LmR1 and CLmR1. Lanes C, S, and P show parental DNA from DH12075, Shiralee, and PSA12, respecively. Lanes 1–10 show DNA of 10 BC1 lines derived from Shiralee × PSA12. Lanes 11–20 show DNA from 10 BC1 lines derived from DH12075 × PSA12. The two Southern analyses were generated using different BC1 lines. All DNAs were digested with EcoRV.
Comparative mapping with A. thaliana—search for candidate genes:
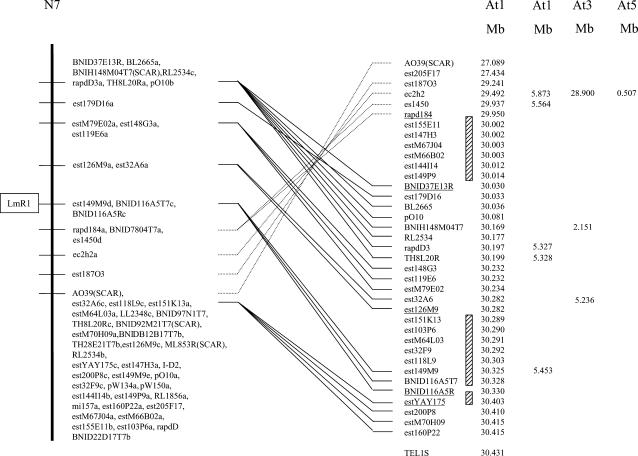
Sequences from all the markers linked to the resistance locus were subjected to BLASTN searches against the Arabidopsis genomic sequence. Using a score value of ≥82 as a cutoff for putative orthology, all the sequences had homologs at the bottom of Arabidopsis chromosome 1 (At1) (Figure 3, Table 2). The 16 markers between BNID116A5R and BNID37E13R were almost perfectly collinear between B. napus and Arabidopsis, except for marker est179D16. The order of markers below the R gene from rapd184 to AO39 suggests inversion of a collinear block between the two genomes. The extent of collinearity between this region of At1 and the second duplicated region on N7 could not be accurately determined since the markers in this region were cosegregating.
Figure 3.
Comparison of the LmR1 locus on linkage group N7 of Brassica napus with the physical maps of Arabidopsis chromosomes At1, At3, and At5. Averaged positions of markers on Arabidopsis chromosomes are given in megabases as described on the Arabidopsis Information Resource web site (http://www.Arabidopsis.org/). Only orthologous loci on the repeat unit carrying the resistance gene are connected by lines for better clarity. Dotted lines depict the inversion of marker orders between the two genomes. Hatched boxes flanked by underlined markers show the areas on At1 that were analyzed for possible candidate resistance genes.
TABLE 2.
Location and score values of markers on Arabidopsis chromosomes
| Markers | Arabidopsis chromosome | Arabidopsis BAC clone | Arabidopsis BAC position (Mb) | BLASTN score |
|---|---|---|---|---|
| BNID37E13R | 1 | F20B17 | 29.942–30.032 | 143 |
| BL2665 | 1 | F19K16 | 29.99–30.109 | 218 |
| BNIH148M04T7(SCAR) | 3 | F3E22 | 2.102–2.166 | 125 |
| 1 | F18B13 | 30.068–30.185 | 119 | |
| RL2534 | 1 | F5I6 | 30.177–30.310 | 129 |
| rapdD3 | 1 | F18B13 | 30.068–30.185 | 133 |
| 1 | T16N11 | 5.296–5.387 | 103 | |
| TH8L20R | 1 | F5I6 | 30.177–30.310 | 287 |
| 1 | T16N11 | 5.296–5.387 | 145 | |
| pO10 | 1 | F18B13 | 30.068–30.185 | 216 |
| est179D16 | 1 | F19K16 | 29.99–30.109 | 482 |
| estM79E02 | 1 | F5I6 | 30.177–30.310 | 212 |
| est148G3 | 1 | F5I6 | 30.177–30.310 | 256 |
| est119E6 | 1 | F5I6 | 30.177–30.310 | 218 |
| est126M9 | 1 | F5I6 | 30.177–30.310 | 618 |
| est32A6 | 1 | F5I6 | 30.177–30.310 | 777 |
| 1 | MJK13 | 5.185–5.268 | 712 | |
| est149M9 | 1 | F23A5 | 30.320–30.430 | 228 |
| 1 | F7H2 | 5.387–5.464 | 123 | |
| BNID116A5T7 | 1 | F23A5 | 30.320–30.430 | 222 |
| BNID116A5R | 1 | F23A5 | 30.320–30.430 | 434 |
| rapd184 | 1 | F20B17 | 29.942–30.032 | 115 |
| BNID7804T7 | 1 | F25O15 | 15.315–15.425 | 86 |
| 1 | F8L2 | 14.294–14.386 | 86 | |
| 3 | T28G19 | 13.689–13.810 | 84 | |
| es1450 | 1 | F3O9 | 5.544–5.659 | 254 |
| 1 | T8K14 | 29.864–29.942 | 149 | |
| ec2h2 | 1 | F3F9 | 29.435–29.531 | 196 |
| 1 | F20D23 | 5.808–5.923 | 76 | |
| 5 | T1E22 | 0.451–0.547 | 208 | |
| 3 | F3L24 | 2.803–2.910 | 94 | |
| est187O3 | 1 | T32E8 | 29.197–29.279 | 761 |
| AO39(SCAR) | 1 | F17M19 | 27.031–27.111 | 86 |
| est118L9 | 1 | F5I6 | 30.177–30.310 | 517 |
| est151K13 | 1 | F5I6 | 30.177–30.310 | 609 |
| estM64L03 | 1 | F5I6 | 30.177–30.310 | 480 |
| LL2348 | 1 | F5I6 | 30.177–30.310 | 480 |
| BNID97N1T7 | 1 | F5I6 | 30.177–30.310 | 168 |
| BNID92M21T7(SCAR) | 1 | F23A5 | 30.320–30.430 | 82 |
| estM70H09 | 1 | F23A5 | 30.320–30.430 | 246 |
| BNID12B17T7 | 1 | F18B13 | 30.068–30.185 | 218 |
| TH28E21T7 | 1 | T21F11 | 30.233–30.323 | 121 |
| 3 | T28G19 | 13.689–13.810 | 80 | |
| 1 | T18N24 | 14.196–14.294 | 80 | |
| 1 | T25F15 | 13.592–13.673 | 80 | |
| 1 | F25O15 | 15.315–15.425 | 80 | |
| 3 | T18B3 | 13.822–13.900 | 80 | |
| 1 | F8L2 | 14.294–14.386 | 80 | |
| 1 | T17D15 | 21.320–21.445 | 80 | |
| 1 | T2P3 | 17.102–17.181 | 80 | |
| ML853R(SCAR) | 1 | F5I6 | 30.177–30.310 | 113 |
| estYAY175 | 1 | F23A5 | 30.320–30.430 | 706 |
| 1 | T24D18 | 5.464–5.545 | 280 | |
| est147H3 | 1 | F20B17 | 29.942–30.032 | 676 |
| est200P8 | 1 | F23A5 | 30.320–30.430 | 533 |
| est32F9 | 1 | F16N3 | 17.396–17.568 | 394 |
| 1 | F5I6 | 30.177–30.310 | 505 | |
| 5 | MUA22 | 4.524–4.589 | 82 | |
| 1 | F28K20 | 11.088–11.165 | 80 | |
| pW134 | 1 | F1N21 | 25.140–25.255 | 157 |
| pW150 | 1 | F5A8 | 25.050–25.152 | 147 |
| est144I14 | 1 | F20B17 | 29.942–30.032 | 519 |
| est149P9 | 1 | F20B17 | 29.942–30.032 | 202 |
| RL1856 | 1 | F23A5 | 30.320–30.430 | 68 |
| mi157 | 1 | F23A5 | 30.320–30.430 | 1203 |
| est160P22 | 1 | F23A5 | 30.320–30.430 | 462 |
| est205F17 | 1 | F3N23 | 27.401–27.518 | 638 |
| estM67J04 | 1 | F20B17 | 29.942–30.032 | 468 |
| estM66B02 | 1 | F20B17 | 29.942–30.032 | 515 |
| est155E11 | 1 | F20B17 | 29.942–30.032 | 535 |
| est103P6 | 1 | F5I6 | 30.177–30.310 | 490 |
| BNID22D17T7 | 1 | F18B13 | 30.068–30.185 | 113 |
| 1 | T18N24 | 14.196–14.294 | 78 | |
| 3 | T28G19 | 13.689–13.810 | 78 | |
| 1 | T17D15 | 21.320–21.445 | 78 | |
| 3 | T25F15 | 13.592–13.673 | 76 | |
| 1 | F25O15 | 15.315–15.425 | 76 | |
| 1 | T18B3 | 13.822–13.900 | 76 | |
| 1 | F8L2 | 14.294–14.386 | 76 | |
| 1 | T2P3 | 17.102–17.181 | 76 |
The LmR1 gene from Shiralee was found to cosegregate with three markers: est149M9, BNID116A5R, and BNID116A5T7. The sequences of two, est149M9 and BNID116A5R, were homologous to two adjacent annotated Arabidopsis genes, At1g80670 (WD-40 repeat family protein) and At1g80680 (nucleoporin family protein), neither of which has been implicated previously in a plant's response to pathogens. Due to the inversion directly flanking the map position of LmR1, extending the search for candidate genes to an area delimited by the flanking markers, est126M9 and rapd184, identifies three regions on At1 (Figure 3). The first is a collinear region between markers est126M9 and BNID116A5R covering an interval of ∼43 kb on At1 and containing 15 annotated genes. Two of the 15 predicted genes contain protein motifs that have been associated with disease resistance genes, namely At1g80630 (leucine-rich repeat motif) and At1g80640 (protein kinase motif). The second interval on At1 that could contain candidate genes lies between markers BNID116A5R and YAY175 and the third lies between rapd184 and BNID37E13R on Arabidopsis chromosome At1. These latter two regions contain 26 and 24 annotated Arabidopsis genes, respectively. Although no characterized R genes have been mapped to these regions, two protein kinases (At1g80870 and At1g79640), two wall-associated kinases (At1g79670 and At1g79680), and one LRR transmembrane protein kinase (At1g79620) were found in these regions.
DISCUSSION
A. thaliana is a close relative of Brassica species and it has been recognized that the sequence information and genomic resources of the extensively studied model organism could be used for marker development, map-based gene cloning, and candidate gene identification in Brassica crops (reviewed in Pflieger et al. 2001; Schmidt et al. 2001, 2002; Snowdon and Friedt 2004). The anticipated duplications due to the amphidiploid nature of the B. napus genome and the ancestral duplications observed in its diploid progenitors (Parkin et al. 2003), as well as the occurrence of rearrangements within collinear regions, can pose impediments to such applications. In this study, the fine mapping of the blackleg R gene was further complicated by ancient duplication of the collinear genomic region in Arabidopsis and tandem intrachromosomal duplication of the region containing the R gene in B. napus.
We detected intergenomic duplications of A-genome N7 loci on N12 and N16 of the C genome. Previous analyses showed that N7 and N16 are primary homeologs, but in the region containing the blackleg resistance locus we found that the highest number of duplicated loci was shared between N7 and N12. A more extensive interval from these two linkage groups has also been shown to be homologous by identifying the collinear region on Arabidopsis chromosome 1 (Parkin et al. 2005).
A further layer of complexity was added through intragenomic duplication of marker loci within the A and C genomes. Such duplications within the diploid Brassica genomes have been described in previous mapping studies and suggest recent evolution of the diploid genomes from a common polyploid ancestor (Lagercrantz 1998; Lan et al. 2000; O'Neill and Bancroft 2000; Parkin et al. 2002). The majority of markers, linked to the blackleg resistance locus, were localized on two linkage groups of the A or C genome (N2/N7 and N12/N16, respectively), suggesting chromosomal duplication of the region. A large number of monomorphic loci remained unmapped, which may reflect the choice of mapping parents and/or the fact that only a small genetic interval was analyzed; however, this made it difficult to estimate the copy number for this region. Additionally, the R gene was found to lie within an intrachromosomal tandem duplication on N7. The presence of these internal duplications at homologous regions on the A and C genome (N7 and N12) indicates that they took place before the divergence of B. rapa and B. oleracea into separate species. Tandem and segmental duplications of resistance genes have frequently been observed in other plants. Genomewide analyses of the organization of NBS-LRR genes in Arabidopsis suggest that tandem duplications distribute and separate this class of resistance genes in the genome and contribute to the diversification of gene families (Arabidopsis Genome Initiative 2000; reviewed in Leister 2004). Linkage group N7 (alternative nomenclature LG6) has already been shown to be a valuable source of blackleg resistance loci, harboring at least five different loci (Ferreira et al. 1995; Mayerhofer et al. 1997; Rimmer et al. 1999). It seems likely that the tandem duplication could harbor a homolog of LmR1, which although nonfunctional in Shiralee, might reveal the identity of one of the additional blackleg loci already assigned to N7. The detected tandem segments are restricted to a small genetic interval and were uncovered only during the fine-scale mapping of the LmR1 locus. It is expected that the genomic structure around the resistance locus will prove to be even more complex; this will be determined through sequencing of B. napus BAC clones spanning the R gene.
Collinearity between the region carrying the resistance gene and the TEL1S end of chromosome 1 of Arabidopsis was established and this information was exploited to saturate the R gene region with markers. However, due to limited polymorphism in the Brassica populations only 26% of the analyzed markers could be genetically mapped and only a fraction could be separated by recombination events, resulting in a cluster of markers on the N7 linkage group. Ancient segmental duplications of large portions of the genome exist in Arabidopsis and the bottom of Arabidopsis chromosome 1 shares homology with a region on the upper arm of chromosome 1 (Arabidopsis Genome Initiative 2000). This duplicated region on chromosome 1 has been shown to be collinear with six different regions of the B. napus genome, but not with N2/N12 or N7/N16 (Parkin et al. 2005), and no comparable tandem repeat of the region has been identified in Arabidopsis. These data suggest that the tandem duplication took place in the Brassica lineage after the separation of the two genera. Although the presence of such duplications has been suggested previously (Parkin et al. 2002), this is the first substantial evidence of a segmental duplication that postdates the evolution of the Brassica and Arabidopsis lineages. It is possible that the presence of such tandem duplications will become more evident as further physical mapping of the Brassica genomes takes place. Such events could contribute not only to the expansion of the genome size in Brassicas but also to the extensive phenotypic variation found within the diploid Brassicas.
Arabidopsis-derived resistance against L. maculans has been transferred to B. napus through somatic hybridization (Bohman et al. 2002). Two regions on chromosome 3 of Arabidopsis were found to be associated with adult-leaf resistance in B. napus. An Arabidopsis EMS mutant, severely compromised in its resistance to the blackleg fungus, was identified in the same laboratory (Bohman et al. 2004). Genetic analysis suggested the presence of a recessive gene linked to an area of Arabidopsis chromosome 2, which did not contain any previously characterized disease resistance genes. The resistance gene from Shiralee was mapped to a small region collinear with Arabidopsis chromosome 1, which has not been implicated previously in blackleg resistance. Although no characterized R genes were identified in the region, several genes were found that encode proteins containing either protein kinase or leucine-rich repeat domains; such structural motifs have been suggested as characteristic features that can be used to identify plant resistance genes. T-DNA insertional mutants for most of the 65 genes in the collinear region are available and were obtained from Arabidopsis Biological Resource Center. None of the mutant lines analyzed to date, including knockouts for two of the protein kinase domain proteins, have shown susceptibility to blackleg infection compared to wild type (data not shown).
Since both B. napus and Arabidopsis have highly duplicated genomes it is crucial that orthologous rather than paralogous sequences are compared when looking for regions with the same gene content. Although some Brassica loci from the R gene region identified sequences on other regions of the Arabidopsis genome, all the Brassica sequences linked to the resistance locus were homologous to an area between 27 and 30 Mb on At1 with a minimum BLASTN score of 82, an empirically determined cutoff for orthology as determined by Lukens et al. (2003). This supports the assumption that candidate genes should be found in this region of the Arabidopsis genome; however, it is probable that fine-scale rearrangements of this complex Brassica locus will have occurred with respect to Arabidopsis. Brown et al. (2003) exploited collinearity with Arabidopsis to clone the Rfo restorer gene from radish. Interestingly the Rfo-equivalent gene was not present at the collinear location in Arabidopsis but was found ∼40 kb from this site. Similar events could be imagined at plant resistance loci that have been shown to contain multiple, tightly linked genes that are under constant adaptive pressure to create novel resistance specificities and are prone to gene duplication and intergenic recombination. Thus, the blackleg R gene may have originated from another region of the Arabidopsis genome and been transposed into the region on N7 that is homologous to Arabidopsis chromosome 1. Such changes in the microsynteny will be determined only through sequencing of B. napus BAC clones spanning the region. Another possibility for the absence of a classical candidate R gene for blackleg resistance in the region could be more fundamental, that Arabidopsis in fact displays a nonhost response to L. maculans. Bohman et al. (2004) found that susceptibility among Arabidopsis ecotypes is extremely rare and the disease symptoms were delayed compared to those of B. napus, suggesting a resistance mechanism independent from known host defense pathways.
Single resistance genes corresponding to the prevalent avirulent race within the pathogen population can be effective in the field but the resistance usually breaks down fairly quickly due to the rapid evolution of the matching L. maculans race or directed selection of a virulent race. “Pyramiding” of resistance genes could delay such a breakdown since more mutational events will be needed within the pathogen population to overcome the new resistance and there will be no selective advantage for any one race. We originally intended to use the markers generated during the fine mapping of the Shiralee and Cresor R genes for this purpose. However, our results indicated a very close proximity for the two genes, which makes such an approach impractical. This find is perhaps not surprising since both the adult-plant resistance of Cresor and the cotyledon resistance of Shiralee have been mapped to linkage group LG6/N7 in separate studies (Mayerhofer et al. 1997; Rimmer et al. 1999), and although of very different geographical origin, their breeding pedigrees suggest that there may be a common link. Since Cresus*3 was used in the generation of the French variety Cresor (Canbra × Cresus*3) and Cresus-o-Precose, a seed stock derived from Cresus, was used in the generation of the Australian variety Shiralee (Haya// Zephyr/ Bronowski/3/RC33/// BJ168/Cresus-o-Precose) (Sernyk 1994), our data suggest that the resistance genes from Shiralee and Cresor could be alleles of the same resistance gene or members of the same gene cluster. Classical and molecular genetic studies on a wide array of plant-pathogen systems have demonstrated that R loci can be single genes with multiple alleles (like the 13 alleles for the L locus in flax) or, more commonly, clusters of genes that can affect resistance to different pathogens (reviewed in Michelmore and Meyers 1998). A similar close relationship has been suggested in earlier reports between cotyledon resistance genes from Shiralee and Maluka (Mayerhofer et al. 1997), between those from Maluka and RB87-62 (Rimmer et al. 1999), and between the resistance genes Rlm3, Rlm4, Rlm7, and Rlm9 present in the B. napus lines Maxol, Quinta, 23.1.1., and Darmor, respectively (Delourme et al. 2004).
A sequence inversion between the collinear regions of the B. napus and Arabidopsis genomes close to the resistance gene, together with the recent intrachromosomal duplications on N7, caused complications in the early stages of the mapping project, frustrating the efforts to identify suitable markers. Once these fine-scale rearrangements were elucidated, Arabidopsis proved to be an invaluable source of markers, and the saturation of the R locus was greatly accelerated. This emphasizes the need for fine-scale mapping before collinear relationships can be exploited for chromosome walking or candidate gene approaches.
We have initiated physical mapping of the blackleg resistance locus from Cresor and have identified BAC clones carrying the flanking markers and are in the process of sequencing these clones. Sequence data from the Cresor region and from the equivalent region from Shiralee will help to elucidate the structure, evolution, and specificities of R gene complexes in Brassica species. The duplication of the region containing LmR1 on linkage group N7 and the clustering of independent blackleg resistance genes on N7 suggest that the cloning of LmR1 and the subsequent capture of LmR1 homologs may identify additional blackleg resistance genes.
Acknowledgments
We thank M. Depauw for providing the Shiralee genomic library and D. Sillito and S. Edes for excellent technical assistance. This research was funded by the Alberta Agricultural Research Institute grant 2001J527 to A.G.
References
- Ahlers, D., 1991. Krankheiten und Schädlinge in Raps im Anbaujahr 90/91 aus norddeutscher Sicht. RAPS, Fachzeitschrift für Öl-und Eiweisspflanzen 9: 191–192. [Google Scholar]
- Arabidopsis Genome Initiative, 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 797–815. [DOI] [PubMed] [Google Scholar]
- Bansal, V. K., P. D. Kharbanda, G. R. Stringam, M. R. Thiagarajah and J. P. Tewari, 1994. A comparison of greenhouse and field screening methods for blackleg resistance in doubled haploid lines of Brassica napus. Plant Dis. 78: 276–281. [Google Scholar]
- Bent, A. F., 1996. Plant disease resistance genes: function meets structure. Plant Cell 8: 1757–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohman, S., M. Wang and C. Dixelius, 2002. Arabidopsis thaliana-derived resistance against Leptosphaeria maculans in a Brassica napus genomic background. Theor. Appl. Genet. 105: 498–504. [DOI] [PubMed] [Google Scholar]
- Bohman, S., J. Staal, B. P. H. J. Thomma, M. Wang and C. Dixelius, 2004. Characterisation of an Arabidopsis-Leptosphaeria maculans pathosystem: resistance partially requires camalexin biosynthesis and is independent of salicylic acid, ethylene and jasmonic acid signalling. Plant J. 37: 9–20. [DOI] [PubMed] [Google Scholar]
- Brown, G. G., N. Formanova, H. Jin, R. Wargachuk, C. Dendy et al., 2003. The radish Rfo restorer gene of Ogura cytoplasmic male sterility encodes a protein with multiple pentatricopeptide repeats. Plant J. 35: 262–272. [DOI] [PubMed] [Google Scholar]
- Cargeeg, L. A., and N. Thurling, 1979. Seedling and adult plant resistance to blackleg [Leptosphaeria maculans (Desm.) Ces et de Not.] in spring rape (Brassica napus L). Aust. J. Agric. Res. 30: 37–46. [Google Scholar]
- Cavell, A. C., D. J. Lydiate, I. A. P. Parkin, C. Dean and M. Trick, 1998. Collinearity between a 30-centimorgan segment of Arabidopsis thaliana chromosome 4 and duplicated regions within the Brassica napus genome. Genome 41: 62–69. [PubMed] [Google Scholar]
- Chèvre, A. M., F. Eber, P. This, P. Barret, X. Tanguy et al., 1996. Characterization of Brassica nigra chromosomes and of blackleg resistance in B. napus-B. nigra addition lines. Plant Breed. 115: 113–118. [Google Scholar]
- Chèvre, A. M., P. Barret, F. Eber, P. Dupuy, H. Brun et al., 1997. Selection of stable Brassica napus-B. juncea recombinant lines resistant to blackleg (Leptosphaeria maculans). 1. Identification of molecular markers, chromosomal and genomic origin of the introgression. Theor. Appl. Genet. 95: 1104–1111. [Google Scholar]
- Delourme, R., M. L. Pilet-Nayel, M. Archipiano, R. Horvais, X. Tanguy et al., 2004. A cluster of major specific resistance genes to Leptosphaeria maculans in Brassica napus. Phytopathology 94: 578–583. [DOI] [PubMed] [Google Scholar]
- Dion, Y., R. K. Gugel, G. F. W. Rakow, G. Seguin-Swartz and B. S. Landry, 1995. RFLP mapping of resistance to the blackleg disease [causal agent, Leptosphaeria maculans (Desm.) Ces. et de Not.] in canola (Brassica napus). Theor. Appl. Genet. 91: 1190–1194. [DOI] [PubMed] [Google Scholar]
- Ferreira, M. E., S. R. Rimmer, P. H. Williams and T. C. Osborn, 1995. Mapping loci controlling Brassica napus resistance to Leptosphaeria maculans under different screening conditions. Phytopathology 85: 213–217. [Google Scholar]
- Gugel, R. K., and G. A. Petrie, 1992. History, occurrence, impact and control of blackleg of rapeseed. Can. J. Plant Pathol. 14: 36–45. [Google Scholar]
- Lagercrantz, U., 1998. Comparative mapping between Arabidopsis thaliana and Brassica nigra indicates that Brassica genomes have evolved through extensive genome replication accompanied by chromosome fusions and frequent rearrangements. Genetics 150: 1217–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan, T. H., T. A. Delmonte, K. P. Reischmann, J. Hyman, S. P. Kowalski et al., 2000. An EST-enriched comparative map of Brassica oleracea and Arabidopsis thaliana. Genome Res. 10: 776–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander, E. S., J. Abrahamson, A. Barlow, M. Daley, S. Lincoln et al., 1987. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1: 174–181. [DOI] [PubMed] [Google Scholar]
- Leister, D., 2004. Tandem and segmental gene duplication and recombination in the evolution of plant disease resistance genes. Trends Genet. 20: 116–122. [DOI] [PubMed] [Google Scholar]
- Li, C. X., and W. A. Cowling, 2003. Identification of a single dominant allele for resistance to blackleg in Brassica napus ‘Surpass 400.’ Plant Breed. 122: 485–488. [Google Scholar]
- Liu, Y. G., N. Mitsukawa, C. Lister, C. Dean and R. F. Whittier, 1996. Isolation and mapping of a new set of 129 RFLP markers in Arabidopsis thaliana using recombinant inbred lines. Plant J. 10: 733–736. [DOI] [PubMed] [Google Scholar]
- Lukens, L., F. Zou, D. Lydiate, I. Parkin and T. Osborn, 2003. Comparison of a Brassica oleracea genetic map with the genome of Arabidopsis thaliana. Genetics 164: 359–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayerhofer, R., V. K. Bansal, M. R. Thiagarajah, G. R. Stringam and A. G. Good, 1997. Molecular mapping of resistance to Leptosphaeria maculans in Australian cultivars of Brassica napus. Genome 40: 294–301. [DOI] [PubMed] [Google Scholar]
- Mcnabb, W. M., C. G. J. Van Den Berg and S. R. Rimmer, 1993. Comparison of inoculation methods for selection of plant resistance to Leptosphaeria maculans in Brassica napus. Can. J. Plant Sci. 73: 1199–1207. [Google Scholar]
- Michelmore, R. W., and B. C. Meyers, 1998. Clusters of resistance genes in plants evolve by divergent selection and a birth-and-death process. Genome Res. 8: 1113–1130. [DOI] [PubMed] [Google Scholar]
- Muangprom, A., and T. C. Osborn, 2004. Characterization of a dwarf gene in Brassica rapa, including the identification of a candidate gene. Theor. Appl. Genet. 108: 1378–1384. [DOI] [PubMed] [Google Scholar]
- Muangprom, A., S. G. Thomas, T. P. Sun and T. C. Osborn, 2005. A novel dwarfing mutation in a green revolution gene from Brassica rapa. Plant Physiol. 137: 931–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman, P. L., and D. J. Bailey, 1987. Screening for resistance to canker (Leptosphaeria maculans) in winter oilseed rape (Brassica napus ssp. oleifera). Plant Pathol. 36: 346–354. [Google Scholar]
- O'Neill, C. M., and I. Bancroft, 2000. Comparative physical mapping of segments of the genome of Brassica oleracea var. alboglabra that are homoeologous to sequenced regions of chromosomes 4 and 5 of Arabidopsis thaliana. Plant J. 23: 233–243. [DOI] [PubMed] [Google Scholar]
- Pang, E. C. K., and G. M. Halloran, 1996. a The genetics of adult-plant blackleg (Leptosphaeria maculans) resistance from Brassica juncea in B. napus. Theor. Appl. Genet. 92: 382–387. [DOI] [PubMed] [Google Scholar]
- Pang, E. C. K., and G. M. Halloran, 1996. b The genetics of blackleg [Leptosphaeria maculans (Desm.) Ces. et de Not.] resistance in rapeseed (Brassica napus L). 1. Adult plant resistance in F2 and first-backcross populations. Theor. Appl. Genet. 93: 932–940. [DOI] [PubMed] [Google Scholar]
- Pang, E. C. K., and G. M. Halloran, 1996. c The genetics of blackleg [Leptosphaeria maculans (Desm.) Ces. et de Not.] resistance in rapeseed (Brassica napus L). II. Seedling and adult-plant resistance as quantitative traits. Theor. Appl. Genet. 93: 941–949. [DOI] [PubMed] [Google Scholar]
- Parkin, I. A. P., A. G. Sharpe, D. J. Keith and D. J. Lydiate, 1995. Identification of the A and C genomes of amphidiploid Brassica napus (oilseed rape). Genome 38: 1122–1131. [DOI] [PubMed] [Google Scholar]
- Parkin, I. A. P., D. J. Lydiate and M. Trick, 2002. Assessing the level of collinearity between Arabidopsis thaliana and Brassica napus for A. thaliana chromosome 5. Genome 45: 356–366. [DOI] [PubMed] [Google Scholar]
- Parkin, I. A. P., A. G. Sharpe and D. J. Lydiate, 2003. Patterns of genome duplication within the Brassica napus genome. Genome 46: 291–303. [DOI] [PubMed] [Google Scholar]
- Parkin, I. A. P., S. M. Gulden, A. G. Sharpe, L. Lukens, M. Trick et al., 2005. Segmental structure of the Brassica napus genome based on comparative analysis with Arabidopsis thaliana. Genetics 171: 765–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pflieger, S., V. Lefebvre and M. Causse, 2001. The candidate gene approach in plant genetics: a review. Mol. Breed. 7: 275–291. [Google Scholar]
- Pilet, M. L., R. Delourme, N. Foisset and M. Renard, 1998. Identification of loci contributing to quantitative field resistance to blackleg disease, causal agent Leptosphaeria maculans (Desm.) Ces. et de Not., in Winter rapeseed (Brassica napus L.). Theor. Appl. Genet. 96: 23–30. [Google Scholar]
- Rimmer, S. R., and C. G. J. Van Den Berg, 1992. Resistance of oilseed Brassica spp. to blackleg caused by Leptosphaeria maculans. Can. J. Plant Pathol. 14: 56–66. [Google Scholar]
- Rimmer, S. R., M. H. Borhan, B. Zhu and D. Somers, 1999. Mapping resistance genes in Brassica napus to Leptosphaeria maculans. Proceedings of the 10th International Rapeseed Congress, Canberra, Australia, p. 262.
- Roy, N. N., 1984. Interspecific transfer of Brassica juncea-type high blackleg resistance to Brassica napus. Euphytica 33: 295–303. [Google Scholar]
- Ryder, C. D., L. B. Smith, G. R. Teakle and G. J. King, 2001. Contrasting genome organisation: two regions of the Brassica oleracea genome compared with collinear regions of the Arabidopsis thaliana genome. Genome 44: 808–817. [PubMed] [Google Scholar]
- Schmidt, R., 2002. Plant genome evolution: lessons from comparative genomics at the DNA level. Plant Mol. Biol. 48: 21–37. [PubMed] [Google Scholar]
- Schmidt, R., A. Acarkan and K. Boivin, 2001. Comparative structural genomics in the Brassicaceae family. Plant Physiol. Biochem. 39: 253–262. [Google Scholar]
- Sernyk, L., 1994. Catalogue of oilseed rape cultivars. Mycogen Plant Sciences, Madison, WI.
- Sharpe, A. G., I. A. P. Parkin, D. J. Keith and D. J. Lydiate, 1995. Frequent nonreciprocal translocations in the amphidiploid genome of oilseed rape (Brassica napus). Genome 38: 1112–1121. [DOI] [PubMed] [Google Scholar]
- Sippell, D. W., W. Mcnabb, R. Hall and J. Patel, 1991. Inheritance of the resistance to blackleg (Leptosphaeria maculans) of canola. Proceedings of the 8th International Rapeseed Congress, Saskatoon, Saskatchewan, Canada, pp. 232–237.
- Sjödin, C., and K. Glimelius, 1988. Screening for resistance to Phoma lingam (Tode ex Fr.) Desm. within Brassicaceae. J. Plant Phytopathol. 123: 322–332. [Google Scholar]
- Snowdon, R. J., and W. Friedt, 2004. Molecular markers in Brassica oilseed breeding: current status and future possibilities. Plant Breed. 123: 1–8. [Google Scholar]
- Stringam, G. R., V. K. Bansal, M. R. Thiagarajah and J. P. Tewari, 1992. Genetic analysis of blackleg (Leptosphaeria maculans) resistance in Brassica napus L. using the doubled haploid method. Proceedings of the 13th International EUCARPIA Congress, Angers, France, pp. 213–214.
- Stringam, G. R., V. K. Bansal, M. R. Thiagarajah, D. F. Degengardt and J. P. Tewari, 1995. Development of an agronomically superior blackleg resistant canola cultivar in Brassica napus L. using doubled haploidy. Can. J. Plant Sci. 75: 437–439. [Google Scholar]
- Struss, D., C. F. Quiros and G. Röbbelen, 1992. Mapping of molecular markers on Brassica B-genome chromosomes. Plant Breed. 108: 320–323. [Google Scholar]
- Tanksley, S. D., M. W. Ganal and G. B. Martin, 1995. Chromosome landing: a paradigm for map-based gene cloning in plants with large genomes. Trends Genet. 11: 63–68. [DOI] [PubMed] [Google Scholar]
- U, N., 1935. Genome analysis in Brassica with special reference to the experimental formation of Brassica napus and peculiar mode of fertilization. Jpn. J. Bot. 7: 389–452. [Google Scholar]
- Zhu, B., and S. R. Rimmer, 2003. Inheritance of resistance to Leptosphaeria maculans in two accessions of Brassica napus. Can. J. Plant Pathol. 25: 98–103. [Google Scholar]