Abstract
An algorithm for detecting local clusters of homologous genes was applied to the genome of Caenorhabditis elegans. Clusters of two or more homologous genes are abundant, totaling 1391 clusters containing 4607 genes, over one-fifth of all genes in C. elegans. Cluster genes are distributed unevenly in the genome, with the large majority located on autosomal chromosome arms, regions characterized by higher genetic recombination and more repeat sequences than autosomal centers and the X chromosome. Cluster genes are transcribed at much lower levels than average and very few have gross phenotypes as assayed by RNAi-mediated reduction of function. The molecular identity of cluster genes is unusual, with a preponderance of nematode-specific gene families that encode putative secreted and transmembrane proteins, and enrichment for genes implicated in xenobiotic detoxification and innate immunity. Gene clustering in Drosophila melanogaster is also substantial and the molecular identity of clustered genes follows a similar pattern. I hypothesize that autosomal chromosome arms in C. elegans undergo frequent local gene duplication and that these duplications support gene diversification and rapid evolution in response to environmental challenges. Although specific gene clusters have been documented in C. elegans, their abundance, genomic distribution, and unusual molecular identities were previously unrecognized.
IT is widely appreciated that genes of related function tend to reside in clusters in eubacteria and archaea, often arranged in coregulated operons. Functional clusters in eukaryotes are less common, although there are various indications that such clusters may be more common than was first apparent from genetic studies. For example, in Caenorhabditis elegans ∼15% of genes are members of cotranscribed operons (Blumenthal et al. 2002). The two to eight genes in each of the ∼1050 operons are subject to similar transcriptional regulation (Lercher et al. 2003), and genes within an operon are often involved in related biological processes (Blumenthal et al. 2002). Such operon clusters are not generally composed of homologous genes, but instead seem to group distinct gene sequences for transcriptional coregulation (Blumenthal et al. 2002). In addition to such functional clustering, specific examples of clusters of homologous genes have also been reported in C. elegans (e.g., Gotoh 1998; Robertson 1998, 2000; Sluder et al. 1999) and in a wide variety of other metazoans (e.g., Fritsch et al. 1980; Akam 1989; Hofker et al. 1989; Del Punta et al. 2000; Glusman et al. 2001). A cursory global analysis of homologous gene clusters was reported in the C. elegans genome sequence report (C. elegans Sequencing Consortium 1998). To investigate homologous gene clusters systematically, I developed an algorithm for scanning the genome for locally abundant gene families. This method identified 1391 cases of local clusters of two or more homologous genes, 216 of which had five or more members. The larger families tend to share a variety of interesting properties, including striking clustering on autosomal arms, an abundance of nematode-specific gene families, and probable involvement in environmental and pathogen interactions.
MATERIALS AND METHODS
Sliding-window clusters:
Frozen WormBase data set WS120 (http://ws120.wormbase.org/) was used for all analysis except as noted. Downloaded WormBase GFF files were parsed to produce a set of usable Java objects for computer analysis, including all exon positions, coding regions, and other features of interest for every gene, including all alternative splice forms. A set of 22,234 gene products were translated on the basis of this information and the matching chromosome sequences. For genes with multiple splice forms, all but the longest splice form were discarded from this set, which resulted in a set of 19,874 proteins. Keeping a single splice form eliminated blastp matches among splice forms and simplified subsequent analysis. The longest splice form was kept with the idea that this form was most likely to encode a complete functional gene product, but this choice might affect clustering accuracy in some cases. An all-by-all blastp search was conducted for all proteins from each chromosome, using the “–m 8” tabular output option (NCBI Blast 2004 at ftp://ftp.ncbi.nlm.nih.gov/blast/executables/release/2.2.9/). For cluster analysis, a window of 20 genes was moved along the chromosome and cases were collected in which at least one pair of genes in the window were blastp matches with an E-value ≤ 0.001 and with a blast alignment that extended over at least 80% of the query protein. For each such pair, the blast bit score was divided by the query length to generate a bit-score-per-residue value. All such pair values for the window were summed and divided by the number of genes in the window. A histogram bar was plotted at the position of the window centroid (the mean position of the coding start for each gene in the window). This plot is shown in Figure 1, after scaling and annotation of some of the major clusters. Note that this approach produces a histogram bar that reflects the total local gene clustering, regardless of whether this results from one or more gene families. It also weights high-scoring blast matches more heavily than low-scoring ones. The window was moved one gene at a time, so a specific local cluster will contribute values to several histogram bars in its region. Search window sizes ranging from 10 to 50 and various blast match criteria were tested with broadly similar results to those reported here.
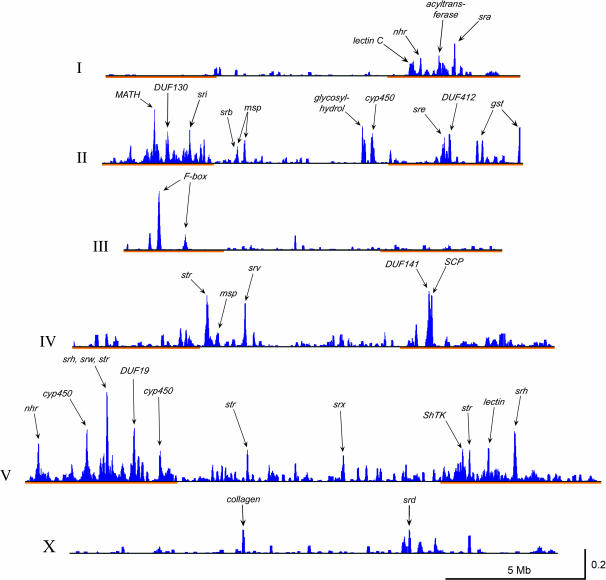
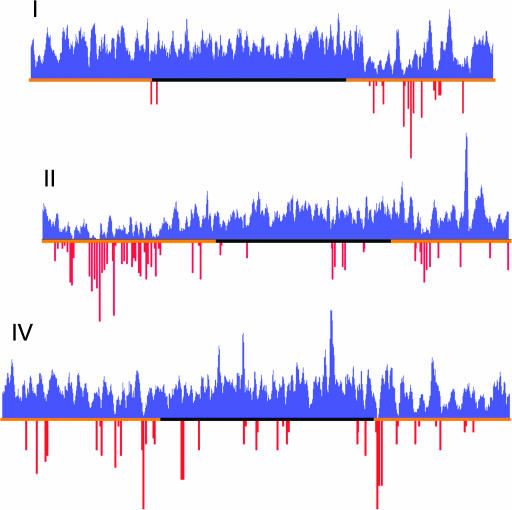
Figure 1.
Positions of homologous gene clusters. The results of a 20-gene window cluster scan were plotted on scaled chromosome schematics. Blue peaks indicate gene clusters, scored by the measure of blastp bit-score per amino acid residue and summed locally as described in materials and methods. Chromosomal lengths and peak heights were scaled to match their physical size and their comparable cluster-scan scores. Chromosomal arms are marked in orange, as defined by increased meiotic recombination (Barnes et al. 1995). Some of the more prominent cluster peaks are labeled to indicate the major homologous gene group that contributes to the peak (see supplemental data 3 and 4 at http://www.genetics.org/supplemental/ for a complete listing).
Cluster accuracy:
Because the clustering algorithm depends on comparing predicted gene products, there will inevitably be some inaccuracy in the clusters assigned. The most likely inaccuracy is underinclusion since gene prediction errors may cause failure to pass the clustering criteria. Because the clustering algorithm requires 80% blast alignment only of query (not of target), it should be relatively insensitive to modest inaccuracies. Such underinclusion is likely to be more severe for genes with less experimental transcript validation, which largely derives from EST data. Cluster genes and autosomal arm genes are transcribed at lower levels than other genes; I anticipate that more accurate gene predictions will modestly increase the number and size of gene clusters and that their genomic location will remain arm biased (and possibly will be more arm biased).
Cluster number and size statistics:
To test whether the distribution of clusters was nonrandom, I used a position-randomizing approach. For each chromosome to test, gene order was randomized and an identical clustering algorithm was run with the new gene order. This was repeated multiple times to acquire a statistical sampling. The number of clusters in randomized tests fit a normal distribution and a one-sample t-test was performed to determine whether the real cluster number deviated from this distribution. Significance of the size of clusters was determined by a nonparametric test because the distribution of real cluster sizes deviated sharply from normal. A list of real cluster sizes was compared to a concatenated list of cluster sizes from multiple randomized tests and the lists were compared by the Mann-Whitney U-test. For both cluster number and cluster size, the P-value was two-tailed and was determined using InStat 3.05 (GraphPad Software).
Merged clusters:
The sliding-window approach arbitrarily limited clusters to the local 20 gene window, which is useful for plotting genomic distributions. For subsequent analysis, these local clusters were merged by joining clusters whenever they shared at least one gene. The result is a set of merged clusters, each of which represents a regional sequence family. More than one cluster of a particular sequence family will be assigned on the same chromosome only when their nearest genomic neighbors are separated by at least 20 unrelated genes. In principle, this might result in undesirable merging of extended groups of genes that are scattered sparsely across long regions. In practice, no such cases were found and clusters defined in this manner were remarkably tight, in the sense that most genes in each cluster region belonged to the cluster family, interspersed with a modest number of unrelated genes. Figure 6 shows an example, albeit an unusually dense one.
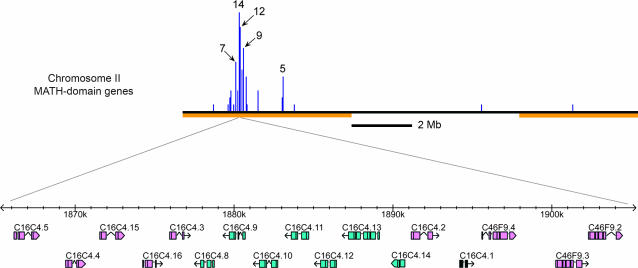
Figure 6.
Schematic of a large MATH-domain gene cluster. (Top) The entire chromosome II with a histogram of positions of all MATH-domain genes in 50-kb bins. Chromosome arms are marked as in Figure 1. (Bottom) An expanded view of a 40-kb segment of chromosome II, derived from the WormBase display (Release WS120; http://ws120.wormbase.org/). Sixteen of the 50 MATH-domain genes in this cluster are located in this segment, with one unrelated gene (C16C4.1, solid black). The first MATH gene in the complete cluster is F36H5.3 and the last is B0047.5. Of the 94 predicted genes in this 285.4-kb interval, 50 are MATH genes that passed the cluster-detecting criteria.
Unclustered gene families:
The same set of longest-splice gene predictions used to define gene clusters was classified into merged gene families using exactly the same blast match and merging criteria as for clustering, except that genome position was ignored. The resulting gene families were ranked by size and were manually inspected for molecular identity using the University of California at Santz Cruz Family Browser (UCSC Gene Sorter, May 2003 C. elegans data set; http://genome.ucsc.edu/) and WormBase Release WS120 (http://ws120.wormbase.org/).
Data records:
An HTML table, which lists all gene clusters and documents the identity and size of each cluster with five or more genes (supplementary data 4 at http://www.genetics.org/supplemental/), was created for each chromosome. These tables include entries for each gene in the cluster and links to stable UCSC Family Browser pages for one member of each cluster. The linked UCSC page is the family browser output keyed to the protein encoded by the link gene and sorted by protein similarity. To provide a stable available data set, all of the UCSC family tables were saved and the links are to these saved files. In addition, two tables that merged clusters from all chromosomes were made. One was sorted by genome position and has selected annotations (supplemental data 3A at http://www.genetics.org/supplemental/); the second was sorted by cluster size and includes more extensive annotations, including annotations for every cluster of five or more genes (supplemental data 3B at http://www.genetics.org/supplemental/). Members of a gene family were defined by gene products on the UCSC Family Browser (UCSC Gene Sorter, May 2003 C. elegans data set; http://genome.ucsc.edu/) that had blastp E-values <10−6 and at least 20.0% blastp identity with reference members of the family. In a few cases, additional information that was more recent than the May 2003 data release was incorporated, notably for the seven-pass receptor (SR) families and the insulin-like gene family. All of these files are available in supplemental data 2 and 3 at http://www.genetics.org/supplemental/.
Conservation in other phyla:
All-by-all blastp searches were conducted with the most current predicted protein sets from Drosophila melanogaster, Saccharomyces cerevisiae, and Homo sapiens, using the WS120 version of C. elegans WormPep as query. Quality scores as a function of genome position were computed by averaging the blastp score for a sliding window of 20 genes. The score was computed by dividing the blastp bit score by the length of the query protein. The general feature of higher conservation on autosomal arms was first reported in C. elegans Sequencing Consortium (1998). My results showed a smaller difference between autosomal arms and centers, probably because the protein query set used previously was from an earlier genome curation, which may have tended to exclude genes on chromosomal arms.
Cluster annotations:
Most annotations derive from reports on the Pfam and InterPro websites, which are based on conserved domains noted on the UCSC family browser [Pfam release 16 (http://www.sanger.ac.uk/Software/Pfam/); InterPro release 8.1 (http://www.ebi.ac.uk/interpro/); UCSC Gene Sorter, May 2003 C. elegans data set (http://genome.ucsc.edu/)]. In a few cases, blastp or Ψ-blast searches were conducted (NCBI_Blast 2004 at http://www.ncbi.nlm.nih.gov/BLAST/) and manual inspection of hits was used to further confirm or reinterpret these annotations. The UCSC data set was from May 2003 (the most recent available at the time) and updated annotations of the SR and insulin gene superfamilies were abstracted from WormBase WS120 (March 2004) because I was aware that improved annotations had occurred in the interim.
Signal sequence and transmembrane domain analysis:
All 19,874 proteins analyzed for clustering were submitted to the SignalP 3.0 and TMHMM 2.0 servers (Nielsen et al. 1997; Remm and Sonnhammer 2000; Bendtsen et al. 2004). A protein was assigned as secreted according to the SignalP HMM method. A protein was assigned as transmembrane (TM) if the TMHMM short report predicted one or more TM domains and the protein did not have a predicted signal sequence (these are often mistermed “N-terminal transmembrane domains”). Lists of the one-line summary SignalP and TMHMM outputs for the entire protein set are available in supplemental materials (supplemental data 7 and 8 at http://www.genetics.org/supplemental/).
Gene counts and nematode specificity:
Gene counts in C. elegans and C. briggsae were assessed mostly on the basis of blastp searches on WormPep and BriggPep with WormBase data set WS123 (Release WS123; http://wormbase.org/). The gene numbers are presented as rough estimates based on a relatively arbitrary E-value cutoff of 10−4 and a consensus hit count from several different queries from the family. No attempt was made to determine how many members are unpredicted or how many of the predicted members are likely to be pseudogenes. Representation outside of nematodes was assessed from a combination of Pfam and InterPro annotations and a Ψ-blast search in June 2004 on the NCBI nr data set with a persistent threshold E-value of 10−6 (http://www.ncbi.nlm.nih.gov/BLAST/). This threshold appeared to serve well in preventing convergence on short domain matches that do not represent near-full-length homologs. For small proteins, these criteria were relaxed somewhat because of their lower information content. Prior to choosing Ψ-blast search query proteins, family members were recursively aligned and culled in an attempt to discard gene-prediction artifacts. Ψ-blast searches were initiated with proteins that appeared typical for the family as a whole, without any large insertions, deletions, or terminal extensions (these are common gene-finding artifacts in C. elegans).
Protein alignment, phylogenetics, and hydropathy plots:
Protein alignments for specifically investigated families were computed with ClustalX using BLOSUM matrices and otherwise default settings (Thompson et al. 1994, 1997). Phylogenetic trees were generated by Bonsai 1.1.4 (J. H. Thomas, March 2004 at http://calliope.gs.washington.edu/software/index.html) using the neighbor-joining distance method (Saitou and Nei 1987) and by PHYLIP proml using the maximum-likelihood method (Felsenstein 1993). Composite hydropathy plots were generated from ClustalX multiple alignments using Bonsai 1.1.4 and a window of nine amino acids. This method determines average hydropathy in aligned columns and is otherwise the same as Kyte-Doolittle hydropathy on single proteins (Kyte and Doolittle 1982).
Codon analysis for positive selection:
Codon analysis was performed only with members that appeared typical for the family as a whole, with no large insertions, deletions, or terminal extensions. Sets of 5–10 closely related proteins were selected and aligned using ClustalX with BLOSUM matrices and otherwise default settings. This protein alignment was used to construct a maximum-likelihood phylogenetic tree with proml and to make a corresponding codon alignment. These were provided to the codeml program in the PAML package (Yang 1997). Models 7 and 8, using at least three starting dN/dS values to avoid local optima during maximum-likelihood analysis were run (Yang 1997). Statistical significance was assessed using a chi-square test of twice the difference in likelihood scores for the two models, with 2 d.f. (Yang 1997).
Clusters in D. melanogaster:
Analysis of clusters in D. melanogaster was the same as for C. elegans except that a gene window of 30, blastp cutoffs of 0.0001, and an alignment length of ≥70% were used. Statistical tests were similar to those for C. elegans.
RESULTS
Detection of gene clusters:
A general method was developed for detecting physically linked clusters of genes that encode related protein sequences. The method uses a sliding gene window and all-by-all blastp results to locate regions of local protein-coding gene duplications. A variety of specific window sizes, blast match, and cluster scoring criteria was explored and the general pattern of clustering was robust to these changes. The results reported here are for a window of 20 genes with a blast E-value cutoff of 10−3 and at least 80% query alignment (see materials and methods). For the 19,874 genes analyzed, 4607 were located in 1391 local clusters of 2 or more genes. Of these, 1819 genes were in 216 clusters of 5 or more genes. For simplicity, I will refer to these as cluster-2 genes and cluster-5 genes. The set of proteins used for the analysis and sets of proteins found in clusters are found in supplemental data 1 and 2 at http://www.genetics.org/supplemental/). Summaries of the highest-scoring gene clusters are in Figure 1, Table 1, and supplemental data 3 at http://www.genetics.org/supplemental/). This overview has two striking features. First, the frequency and size of gene clusters varies greatly among the six C. elegans chromosomes. The most abundant and largest gene clusters occur on chromosomes II, IV, and V, with a notable paucity of clusters on the X chromosome. Second, gene clusters have a strong tendency to reside on autosomal arms. All but two autosomal arms are enriched in clustered genes, and some arms consist predominantly of clusters. Chromosomal arms in C. elegans are defined by a higher frequency of meiotic recombination (Barnes et al. 1995). Arms also have higher densities of simple and complex repeat DNA and more divergent gene products when compared to other phyla (Barnes et al. 1995; C. elegans Sequencing Consortium 1998). The SR superfamily of G-protein-coupled receptors is a major contributor to gene clusters, especially on chromosome V where most SR genes reside (Robertson 2000, 2001). However, similar genomic cluster patterns were seen even when all SR genes were removed prior to analysis (shown for chromosome V in Figure 2; data not shown). As shown in Table 2, the degree of gene clustering is statistically significant, since randomized gene orders subjected to the same clustering procedure reproducibly gave fewer and smaller clusters. The randomizing method was conservative since it randomized gene order only within a chromosome. Since a substantial feature of clustering is chromosome specificity, the degree of clustering from randomizing the entire genome would be substantially lower.
TABLE 1.
Chromosomal cluster summary
| Chromosome | No. of genes | No. of all clusters | No. of large clusters | % genes in all clusters (N) | Mean cluster size (genes) |
|---|---|---|---|---|---|
| I | 2,850 | 139 | 14 | 13.8 (393) | 2.83 |
| II | 3,452 | 233 | 43 | 26.1 (902) | 3.83 |
| III | 2,633 | 107 | 6 | 11.8 (312) | 2.92 |
| IV | 3,235 | 222 | 25 | 22.4 (726) | 2.66 |
| V | 4,959 | 537 | 115 | 40.0 (1,983) | 3.66 |
| X | 2,745 | 99 | 13 | 11.8 (323) | 2.54 |
| Summed | 19,874 | 1,037 | 216 | 23.3 (4,639) | 3.34 |
Large clusters are defined as clusters with five or more genes from a particular family. See materials and methods for the definition of a cluster. The percentage of genes in all clusters is based on all clusters (of two or more genes).
Figure 2.
Chromosome V homologous gene clusters with SR genes removed. See Figure 1 for specifics. All known members of the SR chemoreceptor superfamily (including the new srt family described here) were removed from the chromosome V gene set prior to clustering.
TABLE 2.
Clustering significance in C. elegans and D. melanogaster
| Real gene order
|
Random gene order (100 repeats)
|
|||||
|---|---|---|---|---|---|---|
| Chromosome | No. of clusters | Mean size | Maximum size | Mean no. of clusters (SD) | Mean size (SD) | Maxium size (SD) |
| ce I | 139 | 2.83a | 10 | 28.5 (4.37) | 2.08 (0.06) | 3.08 (0.61) |
| ce II | 233 | 3.83a | 50 | 67.6 (6.32) | 2.14 (0.05) | 4.02 (0.88) |
| ce III | 107 | 2.92a | 32 | 27.8 (4.11) | 2.25 (0.11) | 4.43 (1.12) |
| ce IV | 222 | 2.66a | 13 | 52.1 (6.85) | 2.11 (0.05) | 3.71 (0.77) |
| ce V | 537 | 3.66a | 23 | 246.3 (10.07) | 2.69 (0.06) | 12.57 (2.60) |
| ce X | 99 | 2.54a | 7 | 23.6 (4.17) | 2.08 (0.06) | 2.99 (0.75) |
| dm 2L | 89 | 2.40a | 5 | 27.7 (4.42) | 2.14 (0.08) | 3.59 (0.85) |
| dm 2R | 104 | 2.68 | 11 | 41.1 (4.71) | 2.26 (0.09) | 4.87 (1.15) |
| dm 3L | 103 | 2.77a | 17 | 37.7 (4.43) | 2.27 (0.11) | 4.82 (1.22) |
| dm 3R | 145 | 2.63a | 9 | 54.5 (5.83) | 2.19 (0.06) | 4.35 (1.10) |
| dm X | 71 | 2.35 | 10 | 27.9 (4.09) | 2.16 (0.09) | 3.85 (1.27) |
Clusters are of all sizes (two or more genes). The randomizing of gene order and clustering was carried out 100 times independently and both the mean and the SD (standard deviation) were averaged over all 100 runs. In all cases, the real number of clusters deviated from the randomized distribution by a two-tailed t-test (P < 0.0001). In addition, the mean and maximum cluster sizes (in genes) in the randomized trials were always smaller than those for real clusters.
In these cases the real cluster size distribution was significantly larger than that for the randomized sets by a Mann-Whitney U-test (P < 0.001). A t-test was inappropriate because cluster sizes do not have a normal distribution.
The distribution of the number of genes per cluster for the entire genome is summarized in Figure 3. The number of clusters drops rapidly with cluster size, but the distribution has a long tail with a small number of very large clusters. Most clusters are compactly arranged in the genome, with relatively few unrelated genes interspersed among the homologous genes that define the cluster. The cluster with the largest genome span is also the one with the most genes; it spans 286 kb on chromosome II and contains 50 homologous MATH-domain-containing genes interspersed irregularly with 44 other genes.
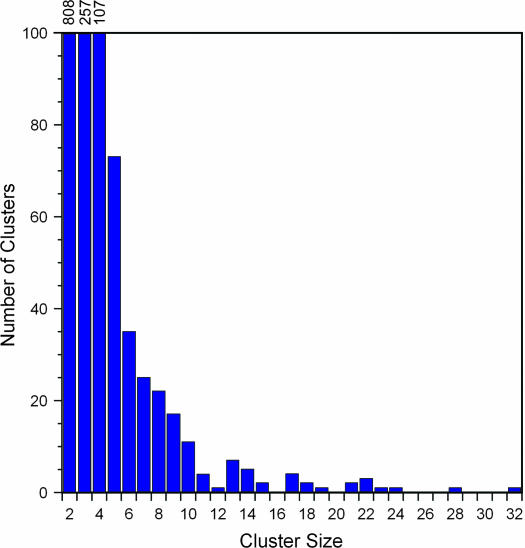
Figure 3.
Cluster size histogram for entire genome. Local clusters for the entire genome were merged if they shared any genes, and the size of all merged clusters was plotted as a histogram. One cluster of size 50 was removed to show the smaller sizes more clearly.
Some of the largest gene families that are represented in clusters have very biased genomic distributions; prominent examples are mentioned here and histograms of gene positions for the 12 largest gene clusters are available in supplemental data 5 at http://www.genetics.org/supplemental/. In the MATH family, 78 of 80 genes are on chromosome II and 50 of these are in one cluster near the middle of the left arm. In the DUF19 family, 33 of 37 genes are on chromosome V and most of these are in one large cluster. In the DUF227 family, 21 of 23 genes are on chromosome V and half of these are in two clusters. In the DUF750 family, 15 of 21 genes are on chromosome V and nearly all of these are in one large cluster. As previously noted, nearly all of the SR families are enriched on chromosome V, with smaller numbers of genes scattered on other autosomes. Regardless of chromosome, these SR families tend to be in large clusters on chromosomal arms. In the cytochrome P450 family, 43 of 76 genes are on chromosome V, mostly in two large clusters on the left arm. Finally, 154 of 253 nuclear hormone receptor genes are on chromosome V, again mostly in clusters on the arms.
Relationship of phylogenetic conservation to gene clusters:
The clustering method has the potential to find families with a wide range of conservation properties. Although the number of clusters makes a full description of their nature difficult, investigation of specific families made it clear that most or all meet a reasonable standard for constituting a gene family. A sampling of alignments among members of four families is shown in Figure 4. Apart from choosing families of sufficient size to provide abundant alignment material, these four families were arbitrarily chosen and appear typical. In all four families, a significant fraction of predicted proteins aligned dubiously with other family members, with large insertions, deletions, or extensions on one or both ends. Some of these are likely to be nonfunctional genes, but preliminary investigation suggests that many are due to errors in ab initio gene finding, since improved gene models were readily identified by manual curation (data not shown). The alignments in Figure 4 were made with proteins that appear typical for their family and that appear to have satisfactory gene models (no large deletions or insertions). In addition to good alignments within cluster families, extensive blast searches and annotation with the UCSC Gene Sorter (May 2003 C. elegans data set; http://genome.ucsc.edu/) showed that the sequences of cluster families are well separated from each other and from unclustered genes. A good test case was the SR families; for all previously identified families and one new family (see below), the clustering algorithm correctly grouped specific families in local clusters, even though members of different SR gene families are often close to each other and sometimes interspersed in the genome.
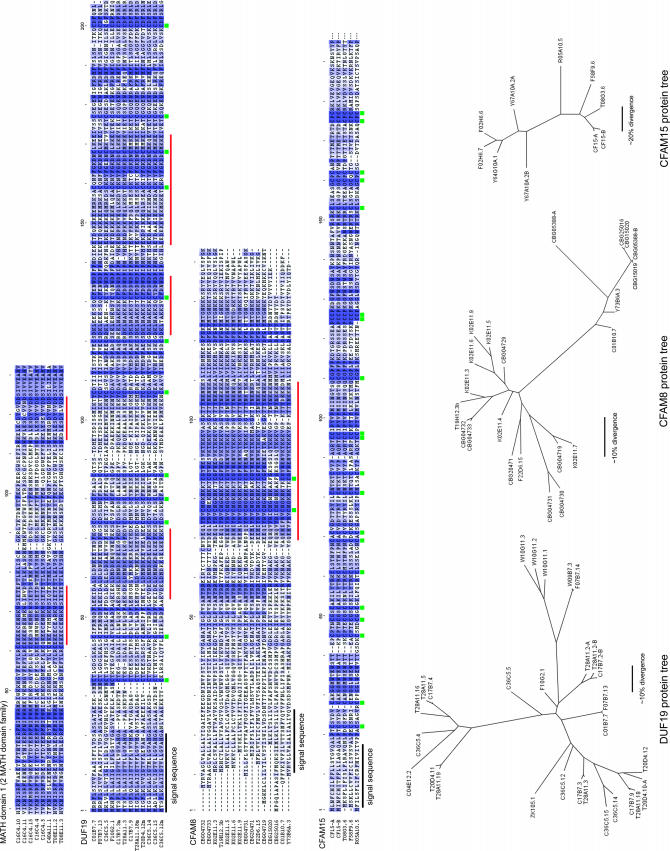
Figure 4.
Examples of cluster family protein alignments and trees. Members of the MATH domain, DUF19, CFAM8, and CFAM15 families are shown aligned, with color shading proportional to amino acid conservation. Only about half of each MATH-domain protein is shown, corresponding to the first MATH repeat in proteins with two such repeats. Signal sequences are indicated in black, conserved Cys residues are marked with green squares, and regions with high charge density are marked with red bars. Unrooted maximum-likelihood trees for 29 DUF19 proteins, 22 CFAM8 proteins, and 10 CFAM15 proteins are shown at the bottom, with approximate amino acid divergence scale bars. The CFAM8 proteins include 12 from C. briggsae (names starting with CBG); the other families also have C. briggsae relatives but they are not shown. Several proteins were based on corrected gene predictions, and two in CFAM15 are from completely new gene predictions (CF15-A and CF15-B). All modified gene predictions have been communicated to WormBase.
Many of the gene families identified by the clustering algorithm have been independently identified by various investigators and are the subject of published or current investigations into their patterns of duplication and divergence, including several of the SR families (Robertson 1998, 2000, 2001; Chen et al. 2005; Thomas et al. 2005), the nuclear receptor family (Maglich et al. 2001), the cytochrome P450 family (Gotoh 1998), the short-chain dehydrogenase family (Kallberg et al. 2002), the glutathione S-transferase family (Campbell et al. 2001), the C-type lectin family (Drickamer and Dodd 1999), and the NLP family of antifungal peptides (Couillault et al. 2004). In each case, the gene duplication patterns as inferred from protein relatedness suggest sporadic duplications, probably balanced by gene loss to produce a genetic complement in dynamic equilibrium. I conducted a limited analysis of three of the unstudied gene families identified by the clustering algorithm, the DUF19, CFAM8, and CFAM15 families. In each case, difficulties in gene prediction necessitated manual annotation of probable gene structures to analyze relationships among full-length family members. Unrooted protein trees for 29 DUF19 proteins, 22 CFAM8 proteins, and 10 CFAM15 proteins are shown in the lower part of Figure 4. For the CFAM8 family, 12 members from C. briggsae were annotated and included in the tree. In each case, the inferred patterns of duplication and divergence indicate that these families arose from sporadic duplication in patterns reminiscent of those documented for previously analyzed cluster families (see references above). All three families appear to include both ancient and recent duplications, producing protein divergences ranging from nearly identical to barely alignable. Further investigation of these families and the other new families is clearly required to obtain an adequate picture of their evolution.
As a preliminary assessment of the relationship of cluster size to divergence, I made a systematic analysis using blastp to compare proteins within each cluster size class. Clusters showed a weak correlation between cluster size and mean protein divergence within the cluster, with a slight trend toward greater divergence in large clusters. Specifically, genes had a mean length-normalized blastp score within their cluster as follows: cluster-2 (1.06), cluster-3 (0.96), cluster-4 (0.99), cluster-5 (0.95), and cluster-6 and greater (0.73). As expected, there was substantial variation among clusters, but on average small local clusters appear to result from duplications that are nearly as old as those in larger clusters. Complete lists of mean blastp values for each cluster are available in supplemental data 6 at http://www.genetics.org/supplemental/. Due to difficulties in gene prediction and the use of blast scores as a surrogate for proper distance measures, this analysis should be regarded as strictly provisional.
Finally, I analyzed the relationship of cluster proteins to proteins in other phyla. As previously noted (C. elegans Sequencing Consortium 1998), genes on autosomal arms in C. elegans tend to be less conserved in other phyla than genes in autosomal centers. This tendency is apparent in Figure 5, which graphs the best blastp match to D. melanogaster as a function of genome position for predicted proteins on three C. elegans chromosomes. Very similar patterns were observed for matches to S. cerevisiae and H. sapiens (data not shown). A substantial part of this trend appears to result from lower phylogenetic conservation of proteins in large gene clusters, which are concentrated on autosomal arms (Figure 5). The graphical correlation is striking and it stands up to quantitative scrutiny: cluster-5 proteins had a mean length-normalized best blastp score to D. melanogaster of 0.07, whereas cluster-2 proteins had a mean score of 0.146, and all other proteins had a mean score of 0.27 (see materials and methods). This trend is not due to cluster genes being part of gene families in C. elegans: when analyzed without regard to genome position, the half of C. elegans proteins with best self-blast hits are slightly more conserved in D. melanogaster than the lower half of proteins (not shown). This presumably results from the fact that unclustered gene families in C. elegans include many that are particularly well conserved phylogenetically (see discussion).
Figure 5.
Comparison of large gene clusters to phylogenetic conservation. The blue histogram reflects the mean best query-length normalized blastp hit to D. melanogaster for a sliding window of 20 genes. The red histogram reflects the number of cluster-5 genes for fixed bins of 80 kb. Chromosome arms are marked as in Figure 1.
Operon clusters and homology clusters:
To test whether genes within operons have sequence similarity to each other, I analyzed all 1048 assigned operons in the WS120 data release (Release WS120; http://ws120.wormbase.org/). There were 2821 genes in these operons (average operon size 2.69 genes). Using the same match criteria as for homologous gene clusters, there were 2 or more homologous genes within 9% of operons (95 of 1048), involving a total of 248 genes. Although 91% of operons contain only nonhomologous genes, the number of exceptions is statistically significant: when pseudo-operons were constructed from random genes with a gene number distribution matching real operons, only 0.5% of them contained homology matches. I also tested whether genes in homology clusters tend to reside in operons. Of the 1819 cluster-5 genes, only 98 (5.3%) were in a known operon, significantly less than the 14.2% of all genes that are in known operons. Conversely, of the 248 homology matches within operons, only 74 (29.8%) were cluster-5 genes, which is slightly below expectation since 39.5% of cluster-2 genes are in the cluster-5 set. I conclude that there is a modest negative correlation between membership in an operon and membership in large homology clusters.
Molecular identity of gene cluster proteins:
All gene clusters were documented as described in materials and methods and these data are available as supplemental data 3 and 4 at http://www.genetics.org/supplemetal/. Briefly, gene clusters with five or more members were annotated using the UCSC family browser, WormBase, Pfam, various blast resources, signal sequence and transmembrane domain predictors, and other resources. The records include overall family size, potential functional identity, links to specific genes in each cluster, links to additional data, and other notes. Brief summaries of the 24 largest gene clusters are shown in Table 3, and additional summaries of all clusters-5 with functionally obscure gene products are found in Table 4. The molecular identities of gene products encoded by clustered gene families are unusual in a variety of ways. I summarize these features first and then discuss each in more detail. First, most of the families are nematode specific, suggesting that they evolve more rapidly than the typical gene. Second, the families are enriched for predicted secreted and transmembrane proteins. Finally, cluster genes are enriched for genes implicated in environmental interactions, specifically those involved in chemosensation, xenobiotic detoxification, and antimicrobial response.
TABLE 3.
Top 24 merged gene clusters
| Cluster size (genes) | Family identifier | Pfam | Chromosome | Position | Nematode specific? |
|---|---|---|---|---|---|
| 50 | MATH/BTB domain | PF00917 | II | 1914430 | Yesa |
| 32 | F-box domain | PF00646 | III | 1291707 | Yesa |
| 28 | DUF141 | PF02408 | IV | 12939032 | Yes |
| 24 | str GPCR | PF01461 | IV | 4924426 | Yes |
| 23 | str GPCR | PF01461 | V | 15641267 | Yes |
| 22 | F-box domain | PF00646 | II | 1735306 | Yesa |
| 22 | srw GPCR | PF06976 | V | 2966739 | Yes |
| 22 | str GPCR | PF01461 | V | 17799674 | Yes |
| 21 | F-box domain | PF00646 | II | 945464 | Yesa |
| 21 | sre GPCR | PF03125 | II | 12386634 | Yes |
| 19 | srh GPCR | PF01604 | V | 16375464 | Yes |
| 18 | DUF130 | PF02343 | II | 2375376 | Yes |
| 18 | DUF19 | PF01579 | V | 3300295 | Yes |
| 17 | sri GPCR | PF01604 | II | 3139916 | Yes |
| 17 | Nuclear hormone receptor | PF00104, PF00105 | V | 528119 | Nob |
| 17 | str GPCR | PF01461 | V | 16107779 | Yes |
| 17 | Lectin C-type domains | PF00059 | V | 16821662 | No |
| 15 | F-box domain | PF00646 | III | 2255092 | Yesa |
| 15 | Nuclear hormone receptor | PF00104, PF00105 | V | 2249629 | Nob |
| 14 | srx GPCR | None | II | 3704818 | Yes |
| 14 | Glycosyl hydrolase, chitinase | PF00704 | II | 9454102 | No |
| 14 | srw GPCR | PF06976 | V | 16602427 | Yes |
| 14 | srh GPCR | PF01604 | V | 15421730 | Yes |
| 14 | srw GPCR | PF06976 | V | 15256608 | Yes |
Genes were clustered and merged as described in the text and in materials and methods. A variety of resources, especially the UCSC Family Browser, were used to derive a descriptive family identifier and the protein family (PF) as described on the Pfam site. The position is the location of the centroid of the gene cluster (see materials and methods) in nucleotide coordinates of the WS120 data set at WormBase (Release WS120; http://ws120.wormbase.org/). Later releases may differ slightly due to minor corrections to the genome sequence assembly.
These proteins contain domains that are found in characterized proteins from other phyla, but the overall structure of the protein family is specific to nematodes.
The nuclear hormone receptor family includes a large expanded branch that is specific to nematodes, although it still bears strong similarity to other family members.
TABLE 4.
Summary of 33 clustered novel gene families
| Family identifier | No. in large clusters | No. of predicted C. elegans | No. of predicted C. briggsae | Non-nematode | Approximate protein length | Notes |
|---|---|---|---|---|---|---|
| MATH | 62 | ∼100 | ∼70 | 0a | Varies | No signal sequence or TM domain, many contain an N-terminal MATH and a C-terminal BTB/POZ domain, others contain 2 or more MATH domainsb |
| F-box | 172 | ∼210 | ∼40 | 0a | 350 | No signal sequence or TM domain, nearly all have one short F-box motif and no other match to anything outside the family |
| DUF13 | 15 | ∼35 | ∼25 | 0 | 280 | Probable N-terminal TM domain, not a cleavable signal sequence |
| DUF18 | 40 | ∼60 | ∼60 | 0 | 240 | Probable Cys-rich secreted proteins |
| DUF19 | 34 | ∼40 | ∼30 | 0 | 180 | Probable Cys and Lys-rich secreted proteins |
| DUF23 | 13 | ∼30 | ∼30 | 0 | 500 | Probable large secreted proteins |
| DUF32 | 37 | ∼65 | ∼45 | 0 | 320 | New SR family (srt), G-protein-coupled receptor |
| DUF130 | 27 | ∼25 | ∼15 | 0 | 170 | Probable Cys-rich acidic secreted proteins |
| DUF141 | 28 | ∼40 | ∼30 | 0 | 460 | Probable large secreted proteins |
| DUF227 | 14 | ∼50 | ∼50 | Bacteria, fungi, insects | 420 | No TMs or signal sequence, large family in insects, related to choline kinases |
| DUF236 | 6 | ∼20 | ∼20 | 0 | 230 | Probable N-terminal TM domain, not a cleavable signal sequence |
| DUF272 | 10 | ∼15 | ∼10 | 0 | 400 | No TMs or signal sequence |
| DUF274 | 9 | ∼20 | ∼15 | 0 | 380 | Probable large secreted proteins |
| DUF278 | 13 | ∼15 | ∼5 | 0 | 320 | No TMs or signal sequence |
| DUF684 | 10 | ∼15 | ∼10 | 0 | 320–500 | No TMs or signal sequence |
| DUF713 | 5 | ∼15 | ∼15 | 0 | variable | No signal sequence, TM status unclear |
| DUF750 | 13 | ∼20 | 1c | Insects, chordates | 350 | One probable TM domain |
| DUF895 | 9 | ∼20 | ∼20 | Fungi, plants, insects, chordates | 440 | Multiple TM domains (10–14), major facillitator superfamily (membrane transport) |
| CFAM2 | 7 | ∼7 | ∼6 | 0 | 100 | Probable small secreted proteins, very basic conserved C-terminal domain |
| CFAM3 | 6 | ∼6d | ∼10d | 0 | 80 | No TMs or signal sequence, extremely His rich, some highly transcribed |
| CFAM4 | 6 | ∼6 | ∼6 | 0e | 320 | No TMs or signal sequence, very weak similarity to the DOM-3 family |
| CFAM5 | 6 | ∼10 | ∼10 | Bacteria, fungi, protozoa, insects, chordates | 400 | No TMs or signal sequence, Glu, Lys, and Arg rich, distantly related to laminins, metalloproteases, plectrins, and other proteins |
| CFAM6 | 6 | ∼10 | ∼10 | 0 | Variable | No TMs or signal sequence |
| CFAM7 | 6 | ∼8 | ∼4 | 0 | 150 | No TMs or signal sequence |
| CFAM8 | 6 | ∼10 | ∼12 | 0 | 120 | Probable small secreted basic proteins |
| CFAM9 | 5 | ∼9d | ∼8d | 0 | 100, some larger | Probable small secreted proteins, Pro and Gly rich, probably in ground-like family |
| CFAM10 | 5 | ∼14 | 0 | 0 | 250 | Multiple TM domains (probably SIX) |
| CFAM12 | 5 | ∼12 | ∼12 | 0 | 80 | Probable small secreted proteins, rich in Pro |
| CFAM13 | 5 | ∼10 | ∼10 | 0 | 130 | Probable small secreted proteins, Cys, Pro, and Asn rich |
| CFAM14 | 5 | ∼12 | ∼10 | 0 | 130 | Probable small secreted proteins |
| CFAM15 | 5 | ∼15 | ∼15 | Insects, chordates | 240 | Probable secreted proteins, related to brain-specific angiogenesis inhibitor, thrombospondin, hemicentin families |
| CFAM17 | 6 | ∼6 | ∼3 | 0 | 900 | No TMs or signal sequence, large proteins |
| CFAM18 | 10 | ∼15 | ∼15 | 0 | 100 | Probable small acidic secreted proteins, Cys rich, may be distantly related to defensin α- and β-families |
If most members of a family contained a PFAM domain of unknown function (DUF), the family is identified by that number. If not, an arbitrary CFAM number was assigned (cluster family). The approximate number of genes predicted in C. elegans and C. briggsae was based on the WormBase data set WS123. Representation outside of nematodes was assessed from a combination of InterPro annotations and a Ψ-blast search (see materials and methods). Notes were derived from a variety of prediction and annotation resources (see materials and methods).
These families contain well-studied domains but the arrangement of the domains in nematodes defines a new protein family.
The MATH-domain proteins fall into two or more families as described in the text.
In addition to a single hit in the predicted protein set, there is only one hit by tblastn; this family appears to be greatly expanded in C. elegans relative to C. briggsae.
Includes only near-full-length matches; the lower complexity region matches various other proteins.
There are very weak matches to the DOM-3 family, which is present broadly in metazoans.
The nematode specificity of cluster genes is dramatic. When all cluster-5 genes are classified by protein family, 51 of the 63 families are nematode specific (Tables 3 and 4 and supplemental data 3 at http://www.genetics.org/supplemetal/). Even when all known SR families are removed, this enrichment is clear (36 of 48 families). This property is not because cluster genes are parts of gene families: when gene families not represented in clusters were analyzed, none of the eight largest families were nematode specific (data not shown; also see discussion).
Table 5 documents the enrichment for secretion signals and transmembrane domains in cluster-2 and cluster-5 genes when compared to noncluster genes. The difference is not as dramatic as for nematode specificity, but this results in part from the presence of a few large cluster families with putative cytoplasmic or nuclear localization (the F-box domain, MATH domain, and nuclear hormone receptor proteins). When analyzed at the level of families, the trend is clearer: 34 of the 50 largest cluster families are predicted to be secreted or transmembrane.
TABLE 5.
Signal sequence and TM domain frequencies
| Gene set | Total genes | % signal sequence (N) | % TM domain(s) (N) | P-value |
|---|---|---|---|---|
| Cluster-2 | 4,607 | 25.2 (1,164) | 34.5 (1,590) | <0.0001 |
| Cluster-2 without SR | 3,637 | 30.1 (1,095) | 18.9 (689) | <0.0001 |
| Cluster-5 | 1,819 | 20.6 (375) | 43.6 (794) | <0.0001 |
| Cluster-5 without SR | 1,203 | 28.3 (341) | 17.6 (212) | <0.0001 |
| Noncluster | 15,627 | 17.6 (2,756) | 20.3 (3,179) | — |
P-values were computed from a 2 × 2 contingency table by a chi-square test, comparing the number of proteins with either a signal sequence or a TM domain for each cluster set to the noncluster set (all noncluster-2 genes). If a protein was predicted to have both a signal sequence and TM domains, it was counted only in the signal sequence class; many signal sequences are erroneously assigned as N-terminal TM domains by TMHMM.
Enrichment for genes implicated in environmental interactions cannot be documented as fully because there is no systematic way to classify genes in this way; analysis of gene ontology terms is inadequate because of inaccuracy and incompleteness. I took the alternative approach of identifying families known or inferred to be involved in specific processes and tested whether these specific families are present in clusters. Assignment of families to specific processes was done by a combination of blast searches, literature searches, and manual PFAM and WormBase browsing. These data are presented in Table 6. Putative environmental interaction genes in C. elegans can be divided into three groups. First, the SR superfamily of seven-pass receptors is implicated in chemosensation on the basis of function and tissue-specific expression in known chemosensory neurons (Troemel et al. 1995; Sengupta et al. 1996; Chen et al. 2005; C. Bargmann, personal communication). There is dramatic enrichment of SR superfamily members in gene clusters. A similar pattern is seen when each of the 15 families is analyzed individually (data not shown). Second, all 4 major gene families implicated in xenobiotic detoxification are highly enriched in clusters. Direct evidence that these genes function in detoxification in C. elegans is limited to the cytochrome P450 (Menzel et al. 2001) and glutathione S-transferase families (Tawe et al. 1998; Leiers et al. 2003); the other two are assigned on the basis of strong sequence similarity to families known to have such function in other organisms. Third, there is direct evidence for involvement in pathogen response for members of 9 specific gene families in C. elegans (Kato et al. 2002; Mallo et al. 2002; Couillault et al. 2004). Six of the 8 families with enough members to analyze are significantly enriched in cluster genes, 5 of them dramatically so. The ninth family (abf antimicrobial peptides) has only two members and they are in a cluster-2 (immediately adjacent genes). In addition to direct evidence for pathogen response, 3 of the pathogen-related cluster families have sequence similarity to proteins implicated in innate immune response in other organisms: two types of lectin proteins and lysozymes (Mallo et al. 2002).
TABLE 6.
Putative chemosensory, xenobiotic detoxification, and pathogen response families
| Gene family | No. of cluster-2 | No. of cluster-5 | Total | P-value |
|---|---|---|---|---|
| Putative chemosensory | ||||
| SR superfamily (15 families) | 970 | 616 | 1271 | <0.0001 |
| Xenobiotic detoxification | ||||
| Cytochrome P450 | 45 | 30 | 77 | <0.0001 |
| UDP-glycosyl transferase | 43 | 23 | 72 | <0.0001 |
| Short-chain dehydrogenase | 24 | 6 | 51 | <0.0001 |
| Glutathione S-transferase | 25 | 22 | 45 | <0.0001 |
| Pathogen response | ||||
| Lectin C-type | 45 | 21 | 62 | <0.0001 |
| DUF141 | 34 | 31 | 40 | <0.0001 |
| Ground-like | 7 | 5 | 29 | NS |
| Galectin | 11 | 0 | 24 | 0.016 |
| cnc antimicrobial peptide | 11 | 10 | 11 | <0.0001 |
| srp serpin protease inhibitor | 8 | 0 | 10 | 0.0001 |
| Lysozyme | 6 | 0 | 10 | 0.017 |
| Saposin-like | 7 | 5 | 10 | 0.0017 |
| Gastric lipase-related | 0 | 0 | 8 | NS |
| abf antibacterial peptide | 6 | 0 | 7 | 0.0005 |
P-values were computed from a 2 × 2 contingency table by a chi-square test, comparing the number of proteins from the family and not in the cluster-2 set relative to the entire genome (4607 cluster-2 proteins of 19,874 total proteins). NS, not significant.
Gene arrangement in homology clusters:
An example of the physical arrangement of homology cluster genes is shown in Figure 6. In Figure 6 (bottom), 16 cluster genes reside in a 40-kb region, with only 1 unrelated gene in their midst. Other clusters are not always this contiguous, but there is a strong tendency for them to reside in highly enriched blocks, and often most of the genes are homologous. The 16 cluster genes (Figure 6, bottom) reside in three blocks of genes and within each block all the genes are on the same strand. However, their evolution was not solely by repeated tandem duplication, since phylogenetic analysis suggests that several duplications and internal rearrangements gave rise to this cluster (data not shown). All of the genes shown have diverged sufficiently that it is difficult to discern which specific mechanism might underlie these rearrangements. Similarly, complex arrangements were found in a number of other clusters, with genes typically found on both strands.
MATH-domain and F-box domain families:
The two largest novel cluster families are the MATH-domain family and the F-box-FTH domain family, with ∼100 and 200 members, respectively. Both are predicted to encode cytoplasmic proteins and neither has yet been implicated in environmental interactions. However, both families appear to be subject to positive selection, a property often associated with changing selective pressure from the environment (Kamei et al. 2000; Choi and Lahn 2003; Thomas et al. 2005). Few of the MATH-domain and F-box-FTH domain genes have identified cDNAs and preliminary inspection of protein alignments and genomic sequences suggests that a substantial fraction of them are nonfunctional genes, perhaps as many as one-third. Although some are likely to be pseudogenes, there is no doubt that many of the genes are functional since there are large families of similar proteins in C. briggsae and dN/dS analysis shows that most of the protein sequence in both families is under strong purifying selection (data not shown). I carried out a preliminary evolutionary analysis of these two protein families based on the subset of predictions that align well with other members in the same family. Lists of proteins analyzed, schematics of protein structure, alignment, and dN/dS results are available in supplemental data 10 at http://www.genetics.org/supplemental/).
The MATH domain is ∼100 amino acids in length and is named for founding domain-containing members meprins and TRAF-C (Uren and Vaux 1996). The domain probably functions in protein-protein interactions (Sunnerhagen et al. 2002). C. elegans MATH-domain cluster-5 genes are predominantly of two sorts. In one type, nearly the entire protein is occupied by two or more repeats of the MATH domain. In the second type, there is a single N-terminal MATH domain followed by a BTB/POZ domain (Zollman et al. 1994). Like the MATH domain, the BTB domain is implicated in protein-protein interactions (Bardwell and Treisman 1994). Recent evidence indicates that some MATH-BTB proteins function as adapters to target other proteins to the ubiquitin-mediated proteolysis pathway (Furukawa et al. 2003; Pintard et al. 2003; Xu et al. 2003; Figueroa et al. 2005). An alignment of the first MATH domain from a sampling of the two-domain proteins is shown at the top of Figure 4. A minority of MATH-domain gene predictions consist largely of a single MATH domain; this fact, coupled with the criteria for clustering (see materials and methods), presumably explains why most MATH-domain genes were identified as members of the same merged clusters. Because of the paucity of confirmed gene structures, it is unclear whether there is real variability in the number of tandem MATH domains or whether the variability is an artifact of mispredicted genes or pseudogenes. Although neither the MATH nor the BTB domains are nematode specific, C. elegans has a hugely expanded number of MATH domains compared to other sequenced genomes (Pfam release 16 at http://www.sanger.ac.uk/Software/Pfam/). Analysis of codon alignments among closely related MATH-domain genes indicates that there is significant positive selection acting on specific sites. High dN/dS sites are concentrated largely in the MATH domain (supplemental data 10 at http://www.genetics.org/supplemetal/) and alignment with a solved MATH-domain protein structure suggests that the sites under positive selection are concentrated on one face of the domain in a region that interacts with one of its binding partners, CD40 (McWhirter et al. 1999; data not shown).
The F-box domain is ∼40 amino acids long and in some cases is known to act as an adapter to target other proteins to the ubiquitin-mediated proteolysis pathway (e.g., Bai et al. 1996; Schulman et al. 2000). In the C. elegans F-box-FTH family, the F-box domain occupies the N terminus followed by ∼250 amino acids called the FTH domain (Clifford et al. 2000; Nayak et al. 2005), which has no sequence relatives outside of nematodes. The entire protein aligns well among most members of the family in C. elegans, with the exceptions most likely being nonfunctional genes and gene prediction errors (supplemental data 10 at http://www.genetics.org/supplemetal/; data not shown). As with MATH-domain genes, analysis of codon alignments among closely related F-box genes shows clear indications of positive selection at specific sites in these proteins (supplemental data 10 at http://www.genetics.org/supplemental/). These sites are not in the F-box region and may cluster in specific regions in the rest of the protein. I speculate that F-box proteins in C. elegans function to target foreign proteins for proteolysis via binding sites in the regions under positive selection.
Chemosensory receptor families:
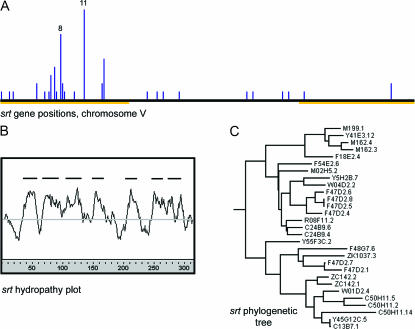
Members of multiple putative chemosensory receptor (SR) gene families are prominent contributors to gene clustering: 219 of the 1391 clusters contain genes in annotated SR families. These clusters range in size from 2 to 24 genes and contain a total of 1065 genes, including members of all previously described SR families. Extensive clustering of odorant, gustatory, and vomeronasal receptors is also found in vertebrates (e.g., Del Punta et al. 2000; Matsunami et al. 2000; Glusman et al. 2001) and, to a lesser extent, in Drosophila (Robertson et al. 2003), suggesting that local gene duplication and diversification is a phylogenetically conserved feature of chemoreceptor gene families. The specificity of the clustering algorithm in C. elegans is supported by the fact that each of many analyzed SR clusters contains genes from one specific SR family, despite the fact that most SR families have similar genome distributions and are often interspersed locally. One of the cluster families, with ∼75 predicted members, defines a new family in the SR superfamily. The new family is distantly related to the previously recognized srg, sru, srv, srh, and str SR families in C. elegans. Ψ-Blast searches started from two proteins from the new family (persistent E-value cutoff 10−4) also suggest a very distant relationship to melatonin receptors and opsins. A composite hydropathy plot and protein tree of 29 putative full-length predictions from the new family are shown in Figure 7. As with other SR families, the new family is concentrated on chromosome V (57 of 74 genes). Of the 57 genes on chromosome V, 45 are located on the left arm (Figure 7), including four clusters of 5 or more genes, all of which were identified by the clustering algorithm. The new family has been assigned the C. elegans gene designation srt (J. Hodgkin, personal communication). A full annotation of the srt family was completed and submitted to WormBase; a list of all known SR proteins, including the new srt family, is in supplemental data 9 at http://www.genetics.org/supplemental/.
Figure 7.
A new SR chemoreceptor family. (A) A histogram of the positions of all srt genes on chromosome V in 100-kb bins. Chromosomal arms are marked as in Figure 1. (B) A composite hydropathy plot of 29 full-length SRT proteins. (C) A maximum-likelihood tree of the same 29 proteins, rooted with respect to other SR families.
Gene clusters in D. melanogaster are similar:
To investigate whether the extent of homologous gene clustering in metazoans is nematode specific, I performed a similar analysis with the nearly completed sequence of D. melanogaster (Adams et al. 2000). Gene clusters in this genome appear to be slightly less compact and gene-product heterogeneity was more problematic for blast comparisons, so slightly different cluster parameters were used (see materials and methods). Although results were less dramatic than with C. elegans, it is clear that there are homology gene clusters in Drosophila as well (Table 2). A more limited analysis of the molecular identities of large clusters indicated fewer families with no known function, although several were found (DUF227, DUF243, DUF1091, PF02448, DUF725, PF02756, DUF733, and PF03207 in order of descending cluster size). As in C. elegans, there was enrichment for genes implicated in xenobiotic detoxification and pathogen response. For xenobiotic detoxification, all four families that clustered in C. elegans are also clustered in Drosophila: cytochrome P450, glutathione S-transferase, UDP-glycosyl transferase, and short-chain dehydrogenase. Most pathogen response genes are sufficiently divergent to make it difficult to compare Drosophila directly with C. elegans, but Drosophila has clusters for many insect gene families implicated in pathogen response, including the persephone protease family (Ligoxygakis et al. 2002), γ-thionin defensins (PF00304, FlyBase 2004 at http://flybase.bio.indiana.edu; UCSC Gene Sorter, May 2003 C. elegans data set; http://genome.ucsc.edu/), lysozymes (Roxstrom-Lindquist et al. 2004), and serpin protease inhibitors (Levashina et al. 1999).
DISCUSSION
Drivers of cluster evolution:
Chromosome arms in C. elegans have features that might contribute to a higher frequency of gene duplication, including higher densities of simple and complex DNA repeats and approximately threefold higher rates of meiotic recombination. If unequal crossing over or some other homology-based mechanism were prominent drivers of gene duplication, these properties would produce more primary duplication material on chromosome arms. In agreement with this idea, DNA repeat sequences and gene clusters are less abundant on the X chromosome. However, DNA repeats are abundant on the left arm of chromosome I and on the right arm of chromosome III, regions nearly devoid of gene clusters as detected by the algorithm reported here. In addition, there is no immediately obvious correlation within chromosomal arms between the local frequency of DNA repeats and gene clusters (data not shown). Nevertheless, genome rearrangements probably do occur more frequently on arms, since regions of synteny between C. elegans and C. briggsae are reported to be shorter on autosomal arms than in centers and the X chromosome (Stein et al. 2003). However, this synteny analysis is more likely to detect large inversions, transpositions, and translocations than local duplications. Preliminary analysis of very recent gene duplications in the C. elegans genome suggests that most duplications are modest in size (a few genes or less in length) and cause local tandem or inverted repeats (data not shown; also see Katju and Lynch 2003). I speculate that this class of duplications is the primary source material for gene clusters. Further analysis is necessary to understand the mechanism by which these duplications arise and how they relate to the evolutionarily persistent gene clusters reported here.
Gene clusters contain unusual genes:
By various measures, the gene families identified by the clustering algorithm are unusual. Perhaps the most marked of these is the preponderance of nematode-specific gene products. In contrast to clustered gene families, unclustered homologous families in C. elegans are dramatically different. Using the same family-finding algorithm without regard to genome position, the largest gene families (after removal of cluster-5 families) encoded protein kinases, ligand-gated ion channels, ras/rab family G proteins, two types of transposases, transmembrane tyrosine kinases, protein phosphatases, and phosphoesterases. All of these families are well known, none are nematode specific, and all are the subject of thousands of research articles. To the extent that I investigated, their genome distribution lacks the autosomal arm bias that characterizes nearly every cluster-5 family. Other distinctive features of cluster-5 genes, when compared to the rest of C. elegans genes, are listed in Table 7. These include a dramatically reduced frequency of assigned phenotypes in RNA interference tests of gene function, shorter introns, reduced expression levels by two measures, and increased divergence from their closest predicted C. briggsae relative. All of these features, except shorter introns, are readily rationalized on the basis of functional redundancy and higher rates of evolution for cluster genes. Shorter introns may be an indirect result of the fact that genes with very low expression levels have a smaller average intron size (data not shown).
TABLE 7.
Global properties of clustered genes
| Clusters ≥2
|
Clusters ≥5
|
Clusters ≥10
|
||||
|---|---|---|---|---|---|---|
| Property | Clustered (N) | All other (N) | Clustered (N) | All other (N) | Clustered (N) | All other (N) |
| Lethal or sterile phenotype | 1.0% (3,615) | 8.2% (12,040) | 0.3% (1,422) | 7.2% (14,233) | 0.2% (585) | 6.8% (15,070) |
| Visible phenotype | 1.4% (3,615) | 9.0% (12,040) | 0.3% (1,422) | 8.0% (14,233) | 0.2% (585) | 7.6% (15,070) |
| Intron length | 190 (19,535) | 312 (82,366) | 181 (7,274) | 297 (94,627) | 174 (2,942) | 292 (98,959) |
| mRNA expression | 10.8 (3,835) | 13.9 (12,984) | 5.1 (1,525) | 14.0 (15,294) | 1.9 (632) | 13.6 (16,187) |
| EST number | 2.18 (4,607) | 6.25 (15,267) | 0.88 (1,819) | 5.75 (18,055) | 0.32 (745) | 5.50 (19,129) |
| Best C. briggsae blast match | 0.98 (4,607) | 1.21 (15,267) | 0.87 (1,819) | 1.19 (18,055) | 0.77 (745) | 1.17 (19,129) |
For “Lethal or sterile phenotype” and “visible phenotype,” the value is the percentage of genes in each class that had the indicated phenotypes; for all other properties, the value is the mean for all genes in the class. Parentheses give the number of genes (or the number of introns for “intron length”) in the class for which data were available for a specific property. Merged gene clusters were grouped according to the number of genes that they contained inclusively (i.e., “Clusters ≥2” contains all the genes from clusters ≥5 and 10). For each set, the properties of all the cluster genes were compared against all other genes (i.e., “All Other” for clusters ≥5 includes genes in smaller clusters). “Best C. briggsae blast match” was derived from an all-by-all blastp search with the cb25 hybrid protein data set; the number is the blastp score divided by query protein length. All other results were derived from data sets found in the WS120 release at WormBase (http:ws120.wormbase.org/). Phenotype data are from only Fraser et al. (2000) to ensure uniformity of results. mRNA expression is derived from averaged Affymetrix hybridization data (Hill et al. 2000) after subtraction of a background value of 1.95, which was obtained from WormBase.
Apart from these features, do homology cluster genes have any common biological thread? Since the families are nearly all nematode specific and very few have members of known biological function, this is a difficult question to answer. Nevertheless, the enrichment for secretion signal sequences, transmembrane domains, putative chemoreceptors, xenobiotic detoxification genes, and pathogen response genes strongly suggests that environmental interactions are a prominent feature of cluster genes. In addition to these documented features, a surprising number of the novel families appear to encode secreted proteins with peculiar amino acid frequencies, properties shared by many antimicrobial peptides (Boman 2003). These families include DUF19 (34 cluster-5 genes), DUF130 (27 cluster-5 genes), CFAM2 (7 cluster-5 genes), CFAM8 (6 cluster-5 genes), CFAM12 (5 cluster-5 genes), CFAM13 (5 cluster-5 genes), and CFAM18 (10 cluster-5 genes). Finally, one cluster family encodes glycosyl hydrolases, among which are chitinases, potential antifungal enzymes (Leah et al. 1991). I speculate that many of these families are as yet uncharacterized elements of innate immunity in nematodes.
Nuclear hormone receptors:
The genome distribution of nuclear hormone receptor (nhr) genes is particularly telling because it includes both phylogenetically conserved genes and a large expanded family of nematode-specific relatives (Sluder et al. 1999). C. elegans possesses ∼30 nhr genes that belong to families with broad phylogenetic representation, including members of five of the six recognized chordate nhr families (Sluder and Maina 2001). The phylogenetically conserved subset of the nhr genes is distributed widely in the genome, with no obvious chromosome or arm bias. There is also a large expansion of nhr genes in C. elegans, including >200 that define expanded nematode-specific families. Presumably, these genes duplicated and diversified during nematode evolution. The phylogenetic tree for these genes indicates that these duplications occurred over a long period with no obvious indication of temporal clustering. The expanded nhr genes are distributed very nonuniformly in the genome, with 149 of 197 tested genes residing on chromosome V and with a strong bias toward clusters on autosomal arms. I speculate that this segment of the nhr family is specialized for transcriptional response to environmental challenges and that the genes duplicate and diversify on chromosomal arms in concert with this selection. One member of the nhr family, nhr-8, is experimentally implicated in xenobiotic response (Lindblom et al. 2001); however, this gene is not in a homology cluster and appears to be a member of the phylogenetically conserved class of C. elegans nhr genes. I speculate that some of these genes also participate in environmental responses.
Operons and homology clusters:
Genes that are found in large homology clusters tend not to be found in operons and vice versa. Why is this true? I speculate that operons, in their role as transcriptional regulatory units, tend to group genes that are unrelated in sequence but that function together in shared processes (as suggested by Blumenthal et al. 2002). In contrast, homology clusters exist as a consequence of evolutionary patterns of duplication and divergence rather than shared transcriptional regulation. Evolutionary theory indicates that duplicate genes that persist over time must acquire at least partially distinct functions to permit natural selection to act in retaining both gene copies (Ohno et al. 1968). It is likely that some of the functional distinctness acquired by such duplicate genes occurs at the transcriptional level, for example, when each of the duplicates is expressed in a subset of the tissues that expressed their ancestor. This mechanism of divergence is unlikely to be consistent with the duplicates residing in the same operon. In addition, the duplications that give rise to gene clusters might disrupt operon structure, favoring persistence of genes with their own promoters. The genomic distribution of operons is also different from homologous gene clusters; operons are relatively evenly distributed by chromosome (although reduced on the X chromosome) and they are less common on autosomal arms where most homology clusters reside (Blumenthal et al. 2002; data not shown).
Acknowledgments
I thank Hugh Robertson for inspiring me to analyze gene clusters and Bob Waterston, Emily Rocke, Zhirong Bao, Willie Swanson, Phil Green, Evan Eichler, Colin Manoil, and members of the Thomas lab for helpful discussions of this work. The work was supported by National Institutes of Health grant RO1GM48700.
References
- Adams, M. D., S. E. Celniker, R. A. Holt, C. A. Evans, J. D. Gocayne et al., 2000. The genome sequence of Drosophila melanogaster. Science 287: 2185–2195. [DOI] [PubMed] [Google Scholar]
- Akam, M., 1989. Hox and HOM: homologous gene clusters in insects and vertebrates. Cell 57: 347–349. [DOI] [PubMed] [Google Scholar]
- Bai, C., P. Sen, K. Hofmann, L. Ma, M. Goebl et al., 1996. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 86: 263–274. [DOI] [PubMed] [Google Scholar]
- Bardwell, V. J., and R. Treisman, 1994. The POZ domain: a conserved protein-protein interaction motif. Genes Dev. 8: 1664–1677. [DOI] [PubMed] [Google Scholar]
- Barnes, T. M., Y. Kohara, A. Coulson and S. Hekimi, 1995. Meiotic recombination, noncoding DNA and genomic organization in Caenorhabditis elegans. Genetics 141: 159–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendtsen, J., H. Nielsen, G. von Heijne and S. Brunak, 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340: 783–795. [DOI] [PubMed] [Google Scholar]
- Blumenthal, T., D. Evans, C. D. Link, A. Guffanti, D. Lawson et al., 2002. A global analysis of Caenorhabditis elegans operons. Nature 417: 851–854. [DOI] [PubMed] [Google Scholar]
- Boman, H. G., 2003. Antibacterial peptides: basic facts and emerging concepts. J. Intern. Med. 254: 197–215. [DOI] [PubMed] [Google Scholar]
- Campbell, A. M., P. H. Teesdale-Spittle, J. Barrett, E. Liebau, J. R. Jefferies et al., 2001. A common class of nematode glutathione S-transferase (GST) revealed by the theoretical proteome of the model organism Caenorhabditis elegans. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 128: 701–708. [DOI] [PubMed] [Google Scholar]
- C. elegans Sequencing Consortium, 1998. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282: 2012–2018. [DOI] [PubMed] [Google Scholar]
- Chen, N., S. Pai, Z. Zhao, A. Mah, R. Newbury et al., 2005. Identification of a nematode chemosensory gene family. Proc. Natl. Acad. Sci. USA 102: 146–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, S. S., and B. T. Lahn, 2003. Adaptive evolution of MRG, a neuron-specific gene family implicated in nociception. Genome Res. 13: 2252–2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifford, R., M. H. Lee, S. Nayak, M. Ohmachi, F. Giorgini et al., 2000. FOG-2, a novel F-box containing protein, associates with the GLD-1 RNA binding protein and directs male sex determination in the C. elegans hermaphrodite germline. Development 127: 5265–5276. [DOI] [PubMed] [Google Scholar]
- Couillault, C., N. Pujol, J. Reboul, L. Sabatier, J. F. Guichou et al., 2004. TLR-independent control of innate immunity in Caenorhabditis elegans by the TIR domain adaptor protein TIR-1, an ortholog of human SARM. Nat. Immunol. 5: 488–494. [DOI] [PubMed] [Google Scholar]
- Del Punta, K., A. Rothman, I. Rodriguez and P. Mombaerts, 2000. Sequence diversity and genomic organization of vomeronasal receptor genes in the mouse. Genome Res. 10: 1958–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drickamer, K., and R. B. Dodd, 1999. C-type lectin-like domains in Caenorhabditis elegans: predictions from the complete genome sequence. Glycobiology 9: 1357–1369. [DOI] [PubMed] [Google Scholar]
- Felsenstein, J., 1993. PHYLIP (Phylogeny Inference Package), Version 3.6a2. Department of Genome Sciences, University of Washington, Seattle.
- Figueroa, P., G. Gusmaroli, G. Serino, J. Habashi, L. Ma et al., 2005. Arabidopsis has two redundant Cullin3 proteins that are essential for embryo development and that interact with RBX1 and BTB proteins to form multisubunit E3 ubiquitin ligase complexes in vivo. Plant Cell 17: 1180–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser, A. G., R. S. Kamath, P. Zipperlen, M. Martinez-Campos, M. Sohrmann et al., 2000. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature 408: 325–330. [DOI] [PubMed] [Google Scholar]
- Fritsch, E. F., R. M. Lawn and T. Maniatis, 1980. Molecular cloning and characterization of the human beta-like globin gene cluster. Cell 19: 959–972. [DOI] [PubMed] [Google Scholar]
- Furukawa, M., Y. J. He, C. Borchers and Y. Xiong, 2003. Targeting of protein ubiquitination by BTB-Cullin 3-Roc1 ubiquitin ligases. Nat. Cell Biol. 5: 1001–1007. [DOI] [PubMed] [Google Scholar]
- Glusman, G., I. Yanai, I. Rubin and D. Lancet, 2001. The complete human olfactory subgenome. Genome Res 11: 685–702. [DOI] [PubMed] [Google Scholar]
- Gotoh, O., 1998. Divergent structures of Caenorhabditis elegans cytochrome P450 genes suggest the frequent loss and gain of introns during the evolution of nematodes. Mol. Biol. Evol. 15: 1447–1459. [DOI] [PubMed] [Google Scholar]
- Hill, A. A., C. P. Hunter, B. T. Tsung, G. Tucker-Kellogg and E. L. Brown, 2000. Genomic analysis of gene expression in C. elegans. Science 290: 809–812. [DOI] [PubMed] [Google Scholar]
- Hofker, M. H., M. A. Walter and D. W. Cox, 1989. Complete physical map of the human immunoglobulin heavy chain constant region gene complex. Proc. Natl. Acad. Sci. USA 86: 5567–5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallberg, Y., U. Oppermann, H. Jornvall and B. Persson, 2002. Short-chain dehydrogenase/reductase (SDR) relationships: a large family with eight clusters common to human, animal, and plant genomes. Protein Sci. 11: 636–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamei, N., W. J. Swanson and C. G. Glabe, 2000. A rapidly diverging EGF protein regulates species-specific signal transduction in early sea urchin development. Dev. Biol. 225: 267–276. [DOI] [PubMed] [Google Scholar]
- Katju, V., and M. Lynch, 2003. The structure and early evolution of recently arisen gene duplicates in the Caenorhabditis elegans genome. Genetics 165: 1793–1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato, Y., T. Aizawa, H. Hoshino, K. Kawano, K. Nitta et al., 2002. abf-1 and abf-2, ASABF-type antimicrobial peptide genes in Caenorhabditis elegans. Biochem. J. 361: 221–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte, J., and R. F. Doolittle, 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157: 105–132. [DOI] [PubMed] [Google Scholar]
- Leah, R., H. Tommerup, I. Svendsen and J. Mundy, 1991. Biochemical and molecular characterization of three barley seed proteins with antifungal properties. J. Biol. Chem. 266: 1564–1573. [PubMed] [Google Scholar]
- Leiers, B., A. Kampkotter, C. G. Grevelding, C. D. Link, T. E. Johnson et al., 2003. A stress-responsive glutathione S-transferase confers resistance to oxidative stress in Caenorhabditis elegans. Free Radic. Biol. Med. 34: 1405–1415. [DOI] [PubMed] [Google Scholar]
- Lercher, M. J., T. Blumenthal and L. D. Hurst, 2003. Coexpression of neighboring genes in Caenorhabditis elegans is mostly due to operons and duplicate genes. Genome Res. 13: 238–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levashina, E. A., E. Langley, C. Green, D. Gubb, M. Ashburner et al., 1999. Constitutive activation of toll-mediated antifungal defense in serpin-deficient Drosophila. Science 285: 1917–1919. [DOI] [PubMed] [Google Scholar]
- Ligoxygakis, P., N. Pelte, J. A. Hoffmann and J. M. Reichhart, 2002. Activation of Drosophila Toll during fungal infection by a blood serine protease. Science 297: 114–116. [DOI] [PubMed] [Google Scholar]
- Lindblom, T. H., G. J. Pierce and A. E. Sluder, 2001. A C. elegans orphan nuclear receptor contributes to xenobiotic resistance. Curr. Biol. 11: 864–868. [DOI] [PubMed] [Google Scholar]
- Maglich, J. M., A. Sluder, X. Guan, Y. Shi, D. D. McKee et al., 2001. Comparison of complete nuclear receptor sets from the human, Caenorhabditis elegans and Drosophila genomes. Genome Biol. 2: RESEARCH0029. [DOI] [PMC free article] [PubMed]
- Mallo, G. V., C. L. Kurz, C. Couillault, N. Pujol, S. Granjeaud et al., 2002. Inducible antibacterial defense system in C. elegans. Curr. Biol. 12: 1209–1214. [DOI] [PubMed] [Google Scholar]
- Matsunami, H., J. P. Montmayeur and L. B. Buck, 2000. A family of candidate taste receptors in human and mouse. Nature 404: 601–604. [DOI] [PubMed] [Google Scholar]
- McWhirter, S. M., S. S. Pullen, J. M. Holton, J. J. Crute, M. R. Kehry et al., 1999. Crystallographic analysis of CD40 recognition and signaling by human TRAF2. Proc. Natl. Acad. Sci. USA 96: 8408–8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzel, R., T. Bogaert and R. Achazi, 2001. A systematic gene expression screen of Caenorhabditis elegans cytochrome P450 genes reveals CYP35 as strongly xenobiotic inducible. Arch. Biochem. Biophys. 395: 158–168. [DOI] [PubMed] [Google Scholar]
- Nayak, S., J. Goree and T. Schedl, 2005. fog-2 and the evolution of self-fertile hermaphroditism in Caenorhabditis. PLoS Biol. 3: e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, H., J. Engelbrecht, S. Brunak and G. von Heijne, 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10: 1–6. [DOI] [PubMed] [Google Scholar]
- Ohno, S., U. Wolf and N. B. Atkin, 1968. Evolution from fish to mammals by gene duplication. Hereditas 59: 169–187. [DOI] [PubMed] [Google Scholar]
- Pintard, L., J. H. Willis, A. Willems, J. L. Johnson, M. Srayko et al., 2003. The BTB protein MEL-26 is a substrate-specific adaptor of the CUL-3 ubiquitin-ligase. Nature 425: 311–316. [DOI] [PubMed] [Google Scholar]
- Remm, M., and E. Sonnhammer, 2000. Classification of transmembrane protein families in the Caenorhabditis elegans genome and identification of human orthologs. Genome Res. 10: 1679–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson, H. M., 1998. Two large families of chemoreceptor genes in the nematodes Caenorhabditis elegans and Caenorhabditis briggsae reveal extensive gene duplication, diversification, movement, and intron loss. Genome Res. 8: 449–463. [DOI] [PubMed] [Google Scholar]
- Robertson, H. M., 2000. The large srh family of chemoreceptor genes in Caenorhabditis nematodes reveals processes of genome evolution involving large duplications and deletions and intron gains and losses. Genome Res. 10: 192–203. [DOI] [PubMed] [Google Scholar]
- Robertson, H. M., 2001. Updating the str and srj (stl) families of chemoreceptors in Caenorhabditis nematodes reveals frequent gene movement within and between chromosomes. Chem. Senses 26: 151–159. [DOI] [PubMed] [Google Scholar]
- Robertson, H. M., C. G. Warr and J. R. Carlson, 2003. Molecular evolution of the insect chemoreceptor gene superfamily in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 100(Suppl. 2): 14537–14542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roxstrom-Lindquist, K., O. Terenius and I. Faye, 2004. Parasite-specific immune response in adult Drosophila melanogaster: a genomic study. EMBO Rep. 5: 207–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou, N., and M. Nei, 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406–425. [DOI] [PubMed] [Google Scholar]
- Schulman, B. A., A. C. Carrano, P. D. Jeffrey, Z. Bowen, E. R. Kinnucan et al., 2000. Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex. Nature 408: 381–386. [DOI] [PubMed] [Google Scholar]
- Sengupta, P., J. H. Chou and C. I. Bargmann, 1996. odr-10 encodes a seven transmembrane domain olfactory receptor required for responses to the odorant diacetyl. Cell 84: 899–909. [DOI] [PubMed] [Google Scholar]
- Sluder, A. E., and C. V. Maina, 2001. Nuclear receptors in nematodes: themes and variations. Trends Genet. 17: 206–213. [DOI] [PubMed] [Google Scholar]
- Sluder, A. E., S. W. Mathews, D. Hough, V. P. Yin and C. V. Maina, 1999. The nuclear receptor superfamily has undergone extensive proliferation and diversification in nematodes. Genome Res. 9: 103–120. [PubMed] [Google Scholar]
- Stein, L. D., Z. Bao, D. Blasiar, T. Blumenthal, M. R. Brent et al., 2003. The genome sequence of Caenorhabditis briggsae: a platform for comparative genomics. PLoS Biol. 1: E45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunnerhagen, M., S. Pursglove and M. Fladvad, 2002. The new MATH: homology suggests shared binding surfaces in meprin tetramers and TRAF trimers. FEBS Lett. 530: 1–3. [DOI] [PubMed] [Google Scholar]
- Tawe, W. N., M. L. Eschbach, R. D. Walter and K. Henkle-Duhrsen, 1998. Identification of stress-responsive genes in Caenorhabditis elegans using RT-PCR differential display. Nucleic Acids Res. 26: 1621–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, J. H., J. L. Kelley, H. M. Robertson, K. Ly and W. Swanson, 2005. Adaptive evolution in the SRZ chemoreceptor families of Caenorhabditis elegans and Caenorhabditis briggsae. Proc. Natl. Acad. Sci. USA 102: 4476–4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J. D., D. G. Higgins and T. J. Gibson, 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin and D. G. Higgins, 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troemel, E. R., J. H. Chou, N. D. Dwyer, H. A. Colbert and C. I. Bargmann, 1995. Divergent seven transmembrane receptors are candidate chemosensory receptors in C. elegans. Cell 83: 207–218. [DOI] [PubMed] [Google Scholar]
- Uren, A. G., and D. L. Vaux, 1996. TRAF proteins and meprins share a conserved domain. Trends Biochem. Sci. 21: 244–245. [PubMed] [Google Scholar]
- Xu, L., Y. Wei, J. Reboul, P. Vaglio, T. H. Shin et al., 2003. BTB proteins are substrate-specific adaptors in an SCF-like modular ubiquitin ligase containing CUL-3. Nature 425: 316–321. [DOI] [PubMed] [Google Scholar]
- Yang, Z., 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13: 555–556. [DOI] [PubMed] [Google Scholar]
- Zollman, S., D. Godt, G. G. Prive, J. L. Couderc and F. A. Laski, 1994. The BTB domain, found primarily in zinc finger proteins, defines an evolutionarily conserved family that includes several developmentally regulated genes in Drosophila. Proc. Natl. Acad. Sci. USA 91: 10717–10721. [DOI] [PMC free article] [PubMed] [Google Scholar]