Abstract
Mutations in Surf1, a human gene involved in the assembly of cytochrome c oxidase (COX), cause Leigh syndrome, the most common infantile mitochondrial encephalopathy, characterized by a specific COX deficiency. We report the generation and characterization of functional knockdown (KD) lines for Surf1 in Drosophila. KD was produced by post-transcriptional silencing employing a transgene encoding a dsRNA fragment of the Drosophila homolog of human Surf1, activated by the UAS transcriptional activator. Two alternative drivers, Actin5C–GAL4 or elav–GAL4, were used to induce silencing ubiquitously or in the CNS, respectively. Actin5C–GAL4 KD produced 100% egg-to-adult lethality. Most individuals died as larvae, which were sluggish and small. The few larvae reaching the pupal stage died as early imagos. Electron microscopy of larval muscles showed severely altered mitochondria. elav–GAL4-driven KD individuals developed to adulthood, although cephalic sections revealed low COX-specific activity. Behavioral and electrophysiological abnormalities were detected, including reduced photoresponsiveness in KD larvae using either driver, reduced locomotor speed in Actin5C–GAL4 KD larvae, and impaired optomotor response as well as abnormal electroretinograms in elav–GAL4 KD flies. These results indicate important functions for SURF1 specifically related to COX activity and suggest a crucial role of mitochondrial energy pathways in organogenesis and CNS development and function.
IN humans, mutations in mitochondrial or nuclear genes encoding mitochondrial components are associated with a wide range of multisystem disorders affecting tissues highly dependent on mitochondrial energy, such as brain, muscle, heart, and the sensorineural epithelia (Fadic and Johns 1996). An example of such a disorder is Leigh syndrome (LS, OMIM 256000) which is a progressive mitochondrial encephalopathy characterized by the presence of symmetric necrotizing lesions in subcortical areas of the central nervous system (CNS), including brain stem, cerebellum, diencephalon, and corpus striatum (Leigh 1951; Farina et al. 2002). The disease onset is in infancy, generally after a symptom-free interval of several months. By and large, neurological signs are correlated to the functional anatomy of the brain structures involved in the lesions and include generalized hypotonia, dystonia, ataxia, oculomotor abnormalities, disturbances of swallowing, breathing, and other brain-stem-related neurological functions and less frequently seizures, sensory neural deafness, and optic atrophy (for a review see Di Mauro and Schon 2003). The syndrome has a relentlessly progressive course, with death usually occurring within the first decade of life, although protracted cases are known.
The single most prevalent cause of LS are mutations in the Surf1 gene, which encodes a 31-kDa protein located in the inner membrane of mitochondria (Tiranti et al. 1998; Zhu et al. 1998). Mammalian Surf1 belongs to the surfeit locus, which comprises a cluster of six nonhomologous housekeeping genes (Surf1-6) (Colombo et al. 1996) showing a strong conservation from birds to mammals, the function of which is relatively little known. Surf1 is ubiquitously expressed in human tissues, in particular in aerobic tissues such as heart, skeletal muscle, and kidney (Yao and Shoubridge 1999). The protein (SURF1), which is highly conserved from recent prokaryotes to humans (Poyau et al. 1999), is characterized by an N-terminal mitochondrial targeting sequence and by two transmembrane domains and it is actively imported into mitochondria where it localizes to the inner membrane (Tiranti et al. 1999a; Yao and Shoubridge 1999). More than 50 mutations of human Surf1 have been reported, most of which are nonsense mutations predicting the expression of a prematurely truncated and/or aberrant protein (Péquignot et al. 2001).
The biochemical hallmark of LSSurf1 mutant patients is a marked decrease in the activity of mitochondrial respiratory chain complex IV (COX) (Tiranti et al. 1999b). COX is the terminal enzyme in both the eukaryotic and prokaryotic respiratory chain, catalyzing the reduction of molecular oxygen to water with concomitant proton pumping from the matrix to the intermembrane space. Studies in Surf1−/− cells from humans (Tiranti et al. 1999a), mice (Agostino et al. 2003), and yeast (Nijtmans et al. 2001) indicate that SURF1 is involved in COX assembly. However, in human Surf1−/− cells low amounts of apparently fully assembled COX can still be detected by 2D-blue native electrophoresis, suggesting redundancy of the SURF1 function via unknown mechanism(s) (Coenen et al. 1999; Tiranti et al. 1999a; Hanson et al. 2001).
Since the molecular mechanism leading to COX deficiency and the pathogenesis of neurodegeneration that characterize human LS are still poorly understood, we sought a nonhuman model of the disease, ideally tractable at the genetic, biochemical, molecular, and physiological levels. We thus set up a Drosophila melanogaster model by inducing the post-transcriptional silencing (PTS) of the human Surf1 homolog by transgenic double-stranded RNA interference. The D. melanogaster Surf1 homolog (CG9943-PA; Surf1) maps to chromosome 3L 65D4, and while the vertebrate homolog is part of the surfeit gene cluster, Drosophila Surf genes are dispersed throughout the genome. The Surf1 gene spans 1200 bp, and the 900-bp coding sequence consists of 4 exons. The inferred 300-aa protein features a 41-aa N-terminal mitochondrial targeting sequence and two transmembrane domains (included between aa 62–80 and 296–314, respectively). Following Surf1 PTS, our findings show that this gene plays a fundamental role in the completion of the pupal-to-adult molt during development as well as in the accomplishment of oculomotor and sensory neural functions in the adult fly.
MATERIALS AND METHODS
Fly strains:
Flies were raised on a standard yeast–glucose–agar medium (Roberts and Standen 1998) and were maintained at 23°, 70% relative humidity, in 12-hr light/12-hr dark cycles. The w1118 strain was microinjected with the transformation plasmid. The dominantly-marked, multiply-inverted balancer chromosomes w; CyO/Sco; MKRS/TM6B stock was used for determining the chromosomal locations of the transgenes and for subsequent manipulations of the transgenic lines. The following GAL4 lines were used to drive the expression of the UAS–Surf1–IR construct: y, w; Act–GAL4/TM6B, Tb (Bloomington Stock Center; genotype: y[1] w[*]; P{w[+mC]=Act5C–GAL4}17bFO1/TM6B, Tb[1]) and elav–GAL4/CyO (a gift from the Garçia Bellido laboratory). To study elav–GAL4 Surf1 KD larvae, a fluorescent balancer was introduced in the elav–GAL4/CyO strain by crossing it with the w;L2Pin1/CyO, Kr–GAL4, UAS–GFP strain (Bloomington Stock Center; genotype: w[*]; L[2] Pin[1]/CyO, P{w[+mC]=GAL4-Kr.C}DC3, P{w[+mC]=UAS–GFP.S65T} DC7) and then selecting green fluorescent individuals with the curly wing phenotype. Observations were made with a Leica MZFLIII stereoscope equipped with an ebq100 UV source. Wild-type Canton-S and UAS–Surf1–IR strains were used as controls.
Plasmid construction:
The procedure adopted is as in Piccin et al. (2001). In brief, an 858-bp fragment from the coding sequence of the Drosophila Surf1 gene (CG9943, Surf1) was amplified using the pair of primers 5′-ATACAGACTGTATTCCGAAAC-3′ and 5′-GTCAAGAGGATACCCTTCTGA-3′ and cloned in the T vector pDK101 to yield pDK–Surf1. A subfragment included between the endogenous EcoRI and PstI sites was then excised and cloned in the pBC KS+ vector (Stratagene, La Jolla, CA). In parallel, a 330-bp fragment of the green fluorescent protein (GFP)-coding sequence was amplified using primers 5′-ACGGCCTGCAGCTTCAGC-3′ and 5′-GAGCTGCAGGCTGCCGTCCT-3′. pDK–Surf1 was then linearized and recircularised with the 330-bp PstI–PstI GFP fragment and the Surf1 fragment included between the SalI and PstI sites excised from pBC. Finally, the EcoRI–EcoRI fragment containing the two Surf1–IR separated by the GFP spacer was cloned into the Drosophila transformation vector pUAST to give UAS–Surf1–IR.
P-element mediated transformation:
Transformation of Drosophila embryos was carried out according to Spradling and Rubin (1982). Three transgenic lines were obtained, each carrying single UAS–Surf1–IR autosomal insertions. Chromosomes harboring the UAS–Surf1–IR insert were balanced and later made homozygous. In situ hybridization was used to map the position and detect the number of inserts in each line as described in Piccin et al. (2001).
Egg-to-adult viability:
For each of the transgenic lines (23.4, 79.1, and 79.10) and for strain Canton-S, ∼300 fertilized eggs were collected on standard yeast–glucose–agar medium in a petri dish (60 × 15 mm) and the same procedure was adopted for the corresponding knockdown (KD) individuals, where Act–GAL4 was used to induce Surf1 gene silencing. The fertilized eggs were incubated at 23° and for each experimental condition the number of individuals reaching third instar larva, pupa, or adult, and the relative percentages, were calculated.
Whole-mount preparation of cuticular structures:
Cuticular details were studied in whole-mount larvae preparations as reported in Van Der Meer (1977). Specimens were analyzed with a Leica DMR microscope using 40× DIC optics and images were taken by a Leica DFC 480 digital camera and Image Cast imaging software.
Whole-mount embryo in situ analysis of Surf1 expression
In situ expression analysis was conducted essentially as in Lehmann and Tautz (1994). The RNA probe was designed to include the whole Surf1 sequence excluding the mitochondrial target sequence. The corresponding cDNA was cloned using the PCR II topo-cloning kit (Invitrogen), transcribed, and DIG-labeled with an RNA labeling kit (Roche).
Northern blot analysis:
Total RNA was extracted from 50 mg of larvae using the TRIZOL reagent (Life Technologies) according to the manufacturer's instructions. Total RNA (25 μg and 50 μg for each sample) was subjected to electrophoresis through a 1% agarose-formaldehyde gel, blotted onto nylon membrane (Magna Nylon Transfer Membrane; Stepbio), and hybridized with DNA probes labeled with [α-32P]dCTP by random priming (New England Biolabs, Beverly, MA). Probes included (1) the full-length Surf1 cDNA, (2) the full-length cDNA of the rp49 gene (as a standard), and (3) a 330-bp fragment of the green fluorescent protein coding sequence. Hybridization conditions were: 0.25 m Na2HPO4 pH 7.2 and 7% SDS at 65° overnight. The blots were then washed twice for 20 min at 65° in 3× SSC and 0.5% SDS and exposed at −80° to X-ray film with an intensifying screen.
RNA extraction and real-time PCR:
RNA was treated with RQ1 RNAse-free DNAse (Promega, Madison, WI) and a cleaning step with 1:1 phenol-chloroform. For each sample, 1 μg of RNA was used for the first-strand cDNA synthesis, employing oligonucleotides dT−20 and M-MLV Reverse Transcriptase Rnase H Minus, Point Mutant (Promega) according to the manufacturer's instructions. The primers used for the real-time PCR were designed with the Primer 3 Software available at (http://www.basic.nwu.edu/biotools/Primer3.html). Real-time PCR was performed on a Rotor Gene 3000 (Corbett Research) by using the following primers: 5′-TATGATACGTTTGGGGAACC-3′ and 5′-ACCATCCCAAAGGAGCTATC-3′ for Surf1 mRNA (positions 87–106 and 264–283, respectively) and 5′-ATCGGTTACGGATCGAACAA-3′ and 5′-GACAATCTCCTTGCGCTTCT-3′ for the housekeeping rp49 mRNA (positions 169–188 and 314–333 in the sequence, respectively). The primers used for Surf1, were designed so as not to amplify the ds interfering RNA. SYBR Green (Applied Biosystems, Foster City, CA) fluorescent marker was used to generate semiquantitative data. PCR premixes containing all reagents except for target cDNAs were prepared and aliquoted by a Robotic Liquid Handling System (CAS-1200, Corbett Robotics) into PCR tubes (Corbett Research). The PCR reaction was carried out following the program: 95° for 30 sec, 60° for 30 sec, 72° for 45 sec, for 40 cycles, followed by a final 10 min at 72°. The amplification efficiencies (E = 10−1/slope) for each sample were calculated on the basis of the results of three amplification reactions, each with a different quantity of the template (6, 18, or 54 ng of total reverse transcribed RNA), according to Pfaffl (2001). Each reaction was performed in duplicate. The amplification efficiency, obtained for each sample, was used to estimate the relative level of expression between the normal sample we chose as the “calibrator” (w1118 larvae or parental line brains) and the silenced samples (called “X conditions”): R = EtΔCt/EhΔCh where Et is the amplification efficiency of Surf1 mRNA; Eh is the amplification efficiency of rp49 mRNA; ΔCt = Ctc − Ctx is the difference between cycle threshold (Ct) of the calibrator and the Ct of X condition for Surf1 mRNA; ΔCh = Ctc − Ctx is the difference between Ct of the calibrator and the Ct of X condition for rp49 mRNA.
Western blot analysis
Antibodies:
Polyclonal antibodies against D. melanogaster SURF1 (324 anti-DmSURF1 and 325 anti-DmSURF1) were produced in rabbits using two synthetic peptides (VNWTTSIPNQAAKDKEKC and RGRFLHDKEMRLGPR) corresponding to residues 42–60 and 115–131 of the Drosophila putative SURF1 protein (Neosystem, Strasbourg, France).
Preparation of mitochondria:
Approximately 70 mg of sample was placed in 600 μl of buffer pH 7.4 containing 20 mm HEPES, 100 mm KCl, 1 mm EDTA, 0.3% BSA, and 2 mm mercaptoethanol and homogenized with a tight-fitting glass potter homogenizer. The solution was spun at 800 × g for 10 min and the precipitate discarded. The supernatant was spun at 10000 × g for 15 min and the crude mitochondrial pellet was suspended in 150 μl of MSEM buffer (220 mm mannitol, 70 mm sucrose, 1 mm EDTA and 5 mm MOPS pH 7.2).
Protein extraction and SDS–PAGE:
The mitochondrial suspension was sonicated twice for 5 sec, and frozen and thawed in liquid nitrogen three times; a protease inhibitor cocktail and detergents were added (2% Triton X-100, 0.25% SDS, 1 mm DTT, 0.5 mm PMSF, 20 mg/ml aprotinin, 5 mg/ml leupeptin, 5 mg/ml pepstatin A) and the solution was left 1 hr and 30 min on ice to allow membrane solubilization. Protein quantification was conducted using the Bradford assay (Bio-Rad, Hercules, CA), and equal amounts of total proteins (60 μg) for each sample were resolved on a 12% SDS-polyacrylamide gel. Following separation and Western blotting onto nitrocellulose membrane (Trans-Blot Transfer Medium; Bio-Rad), the membrane was blocked for 1 hr in 1 × TBST (0.01 m Tris-Cl pH 7.5, 0.14 m NaCl, 0.1% Tween-20) with 10% dry milk (Carnation nonfat dry milk) and incubated overnight at 4° with rabbit anti-DmSURF1 antibody (1:500, Neosystem). An anti-rabbit IgG-HRP (1:3000; Bio-Rad) was used as secondary antibody. Positive immunoreactivity was visualized using a chemiluminescent system. The immunological specificity of anti-DmSURF1 antibody 325 was confirmed by two types of negative controls: (1) omission of the primary antibody and (2) performing a peptide/antigen competition test for the antibody. For the peptide competition experiment the primary antibody (diluted 1:500) was incubated with the peptide (300 μm) at 4° for 30 min before incubation with the membrane. For quantization of the immunodetected signals, SURF1 signal was compared with that of an unknown protein signal (∼70 kDa).
Behavioral and physiological analyses
Survival of adult flies:
Single adults were placed into polystyrene test tubes (7.5 × 1.2 cm) containing ∼1 ml of standard culture medium and plugged with cotton wool. For each genotype we analyzed the survival of 50 males and 50 virgin females that were collected (using brief CO2 anesthesia) on the day of eclosion and kept at 23° under a 12-hr light/12-hr dark cycle. Dead flies were counted each day and all surviving flies were transferred into clean medium-containing tubes every 14 days without anesthesia.
Larval locomotor speed:
The locomotor speed of third instar larvae was analyzed in 100 × 15-mm petri dishes, with 1% agar covering the base to a depth of 0.5 cm. Individual larvae were placed into a narrow linear groove produced with the aid of a glass capillary (diameter 1 mm) on the surface of the agar layer and allowed to acclimatize for 60 sec. Testing was performed using a fiberoptic lamp (100 W) with a photon throughput of 60 μm/m2/sec (light intensity was measured by a Basic Quantum Meter, model QMSW-SS; Apogee Instruments). The time necessary to cover a 1-cm distance was recorded and larval locomotor speed was expressed as centimeters per second. Statistical analyses were done using Student's t-test.
Photobehavioral assay:
Measurements of larval photobehavior were conducted using the checker assay (Hassan et al. 2000). Petri dishes (140 × 15 mm) containing 30 ml of 1% agar were used. Data were collected excluding the agar surface area included between a circular boundary at 1 cm from the plate edge and the plate edge itself. Testing was performed using a fiberoptic lamp (100 W) with a photon throughput of 60 μm/m2/sec. Four-day-old (after egg deposition) larvae were placed on a pretest plate in dark conditions (illuminated only by a darkroom red light) for a period of 30 min. Each larva was then positioned in the center of the checkered test plate. The test plate was lit from below by positioning on a suitable light box. Larval movement was checked by eye. Larval behavior was recorded either until individuals reached the 1-cm boundary or after a total test time of 180 sec. For each genotype at least 20–30 larvae were tested. Residence time on the dark and light quadrants was obtained. A response index (RI) [(time on dark squares – time on light squares)/total time of test], with lights on (RI ON) was calculated on a per larva basis and an average of these individual indices was taken.
Walking optomotor test:
The optomotor response was tested as in Sandrelli et al. (2001), with modifications as in Zordan et al. (2005). For each genotype 20 individuals (10 for each sex) were tested. The significance of the differences between the behavior shown by the KD individuals and random behavior (assumed as 50% of correct turns) was determined by a χ2 test.
Physiological investigations
Muscle-force measurement:
Muscle-force measurement experiments were performed at 20–21° as described in Beramendi et al. (2005). Briefly, body-wall preparations of third instar larvae were cut to obtain a narrow strip comprising longitudinal muscles 6 and 7 on both sides of the midline still attached to the cuticle. The tail and head ends were tied to the hook of a micromanipulator and the hook of a force transducer (AME-801; SensorOne, Sausalito, CA). After A/D conversion, the signal from the force transducer was fed into a personal computer. For data storage and analysis, Spike 2 software (CED, Cambridge, UK) was used. Tension was calculated by dividing force by cross-sectional area. The latter was approximated by multiplying the width of the muscle preparation (i.e., the distance between the outer edges of each of the two muscle 6 fibers present in each segment) by a thickness value obtained from measurements of transverse sections of fixed tissue. The contractile performance was analyzed by recording for each preparation:
Single twitch: the preparation was stimulated using two platinum electrodes connected to a Grass S88 stimulator. Single stimuli (10 msec duration) were delivered by increasing the voltage until threshold voltage for force development was reached. The comparison between control and preparations obtained from Surf1 KD larvae was based on force measured at the same voltage in each preparation.
Tetanus: the preparations were stimulated with trains of electrical stimuli (train duration 1.5 sec, frequency 15 Hz, pulse duration 10 msec). Force was measured during the tetanic force plateau.
Caffeine contracture: contraction was induced by quickly immersing the preparation in physiological saline containing 25 mm caffeine. Caffeine causes maximal calcium release from the sarcoplasmic reticulum through the opening of ryanodine receptors (Vazquez-Martinez et al. 2003). Whereas electrical stimulation (twitch and tetanus) induced only limited activations, caffeine induced a maximal contractile response; the amplitude of the caffeine contracture might be limited by the amount of calcium present in the intracellular stores or by the amount of myofibrillar proteins.
On average, 10 larvae for each genotype were tested.
Evoked neurotransmitter release:
Experiments were performed at room temperature (20–22°) on third instar larvae body wall as described in Beramendi et al. (2005). Single segmental nerves were stimulated (square wave stimuli, 0.15 msec duration × 1.5 threshold voltage) using a suction electrode connected to a Grass S88 stimulator. Excitatory junctional potentials (EJPs, evoked by segmental nerve stimulation) were recorded intracellularly in muscle fibers 6 and 7 of abdominal segments A2–A4 using glass microelectrodes (0.5 μm tip diameter, 10–15 MΩ resistance). Single-pulse stimulations were given at 0.3 Hz frequency (0.1 msec duration). Recordings were amplified with a current-voltage clamp amplifier (NPI, Tamm, Germany), conditioned with a signal conditioner (Cyber Amp; Axon Instruments, Foster City, CA) and fed to a computer for subsequent analyses via an A/D interface (Digidata 1200; Axon Instruments). Digitized data were analyzed using appropriate software (MiniAnalysis, Synaptosoft, PClamp 6.04; Axon Instruments). An average of five larvae for each genotype was used for these experiments.
Electroretinogram:
All experiments were done as described in Zordan et al. (2005). Recorded signals were amplified with an intracellular amplifier (705; WPI Instruments), fed to a signal conditioner (CyberAmp; Axon Instruments), lowpass filtered (3 kHz), and then fed to a personal computer through an A/D converter (Digidata 1200; Axon Instruments). For each fly, the amplitude of the first two electroretinogram (ERG) responses was measured using appropriate software (PClamp 6.04; Axon Instruments). Data were analyzed for statistical significance using one-way ANOVA tests.
Morphological and ultrastructural investigations
Histochemistry:
COX was visualized cytochemically in tissue sections of adult heads. Adult heads were included in OCT (CellPath), frozen in liquid nitrogen, and sectioned at a thickness of 12 μm with a cryostat (Leica CM1850). Sections were made to adhere onto gelatinized glass slides and stained for COX activity using a kit supplied by Bio-Optica according to the manufacturer's instructions. Images were then acquired by means of a digital camera mounted on a microscope and images were analyzed by means of ImageJ software (Research Services Branch, National Institute of Mental Health, National Institute of Neurological Disorders and Stroke; available at http://rsb.info.nih.gov/ij/index.html). Briefly, regions of interest (ROI) were identified in the digitized images (i.e., the single neuropils), and pixel counts as well as gray level distributions were performed within these ROIs. The mean gray level/pixel for each ROI was then used as an estimate of the level of enzyme activity being determined (i.e., high mean gray level/pixel equals high enzyme activity). Tissue sections of adult heads were also tested for succinic dehydrogenase activity as described in Nachlas et al. (1957) and images aquired and analyzed as described above.
Histology of adult fly cephalic sections:
Cephalic serial horizontal sections (7 μm) were cut from flies following formalin fixation, inclusion in paraffin, and staining with hematoxylin-eosin. A total of 100 flies were analyzed (50 7-day-old flies and 50 28-day-old flies, equally subdivided between males and females, for each of the following genotypes: w1118; elav–GAL4/CyO; elav–GAL4/+; UAS-Surf1 23.4 IR and elav-GAL4-Surf1 KD).
Phalloidine-rhodamine staining:
Third instar larvae were dissected and treated as described in Beramendi et al. (2005). Samples were then observed with a fluorescence confocal microscope (Radiance 2000; Bio-Rad), and measurements were made on digitized images using ImageJ software. In control and KD larvae four parameters relating to muscle fiber 6 were measured: length, width of the two extremities, and length of the central region of the fiber.
Electron microscopy:
For transmission electron microscopy, Actin5C–GAL4–Surf1 KD and third instar wild-type larvae were pierced and immediately transferred to ice-cold fixation solution containing 3% paraformaldehyde, 2% glutaraldehyde, 100 mm sucrose, and 2 mm EGTA in 0.1 m sodium phosphate buffer at pH 7.2. Samples were fixed for 6 hr and subsequently washed overnight at 4° in 0.1 m phosphate buffer, pH 7.2. The next day larvae were treated for 2 hr with cold (4°) postfixative solution (1% OsO4 in 0.1 m sodium cacodylate buffer pH 7.2), rinsed three to four times (5 min each) in 0.1 m sodium phosphate buffer pH 7.2, and then dehydrated through ethanol series and finally by three 15-min washes in propylene oxide. Samples were embedded in Epon resin mixture (14 ml EPOXY, 7 ml DDSA, 9 ml MNA) to which 2% DMP30 solidification accelerator was added. The following gradual embedding procedure was utilized: three 45-min infiltrations in resin:propylene oxide, 1:2, 1:1, and 2:1, respectively; embedding in Epon within plastic capsules; and polymerization at 60° for 3 days. Ultrathin (400 Å) cross sections of larval body-wall muscles were cut with a diamond knife and stained for 20 min in 2% aqueous uranyl acetate solution, followed by 30 sec staining in 5% aqueous lead citrate solution, and finally rinsed in distilled water. Sections were examined and photographed with a Philips 200 electron microscope.
RESULTS
Generation of Surf1 KD lines:
We generated three independent Drosophila transgenic lines, 23.4, 79.10, and 79.1. For each line obtained the insert map position is given in parentheses: 23.4 (2R, 54BC), 79.1 (3R, 88D) and 79.10 (2R, 50C). Each line carried a single UAS–Surf1 inverted repeat (IR) autosomal insertion, which allowed the post-transcriptional gene silencing of Surf1 via dsRNA interference (dsRNAi), following activation by GAL4.
Ubiquitous KD of Surf1:
The Actin5C (Act–GAL4) driver was used for early, ubiquitous expression of dsRNAi-mediated KD.
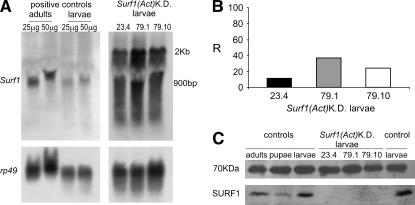
Northern blot analysis on the Act–GAL4 KD larvae showed the presence of an approximately 2-kb band corresponding to the RNAisurf1 transcript, as well as a smear starting from 0.9 kb, the expected size of the Surf1 transcript (clearly visible in the controls on the left in Figure 1A), corresponding to degradation products originating from the interference of the Surf1 gene transcript (Figure 1A). Controls were larvae and adults from strain w1118, which was used to generate the UAS–Surf1 IR transgenic lines. The same 2-kb band was visualized by rehybridization of the blot with a probe complementary to the GFP spacer used in the RNAisurf1 construct (data not shown).
Figure 1.
Expression analysis in Actin5C–GAL4 Surf1 KD larvae. (A) Northern blot analysis using as a probe the complete cDNA of the Surf1 gene. The 2-kb band is relative to the transcription of the hairpin construct while the intense smear, below the band at 0.9 kb (which corresponds to the Surf1 transcript), is indicative of degradation products resulting specifically from dsRNAi. This pattern of degradation is not seen when the Northern blots are hybridized with a probe against the heterologous GFP spacer which separates the two arms of the IR (data not shown). Signs of very slight aspecific mRNA degradation (probably artifactual) are also visible below the 2-kb band, as well as below the rp49 band in the samples collected from larvae, but not in those from adults. Positive controls consisted in mRNA extracted from larvae of the w1118 strain, which was used to generate the UAS–Surf1 inverted repeat transgenic lines, for which two different quantities of mRNA (25 and 50 μg) were loaded on the gels. The experiments with the KD individuals were done using 50 μg of mRNA. In each case a housekeeping gene, i.e., rp49 (which encodes for Drosophila ribosomal protein 49), served as an external reference. (B) Real-time PCR estimate of the relative percentage of Surf1 mRNA in KD individuals from each of the three lines analyzed compared to those of the respective parental lines bearing only the UAS–Surf1-inverted repeat (controls). In each case controls are assigned an arbitrary value of 100%. Histograms represent the mean of two independent experiments. Values obtained for each independent experiment were: 32 and 43 for Act–GAL4 KD79.1, 12.5 and 36 for Act–GAL4 KD79.10, 7.5 and 12.2 for Act–GAL4 KD23.4. (C) Western blot analysis on KD and control (see B) individuals. The 70-kDa band corresponds to an unknown protein, which was used as a reference signal for the quantization of the SURF1 signal. Controls consisted of mitochondrial protein extracts from larvae, pupae or adults of the w1118 strain (see above).
Real-time PCR-based quantitative analysis confirmed a drastic reduction of Surf1 mRNA (Figure 1B), which corresponded to the virtual absence of SURF1 protein, as demonstrated by Western blot analysis (Figure 1C).
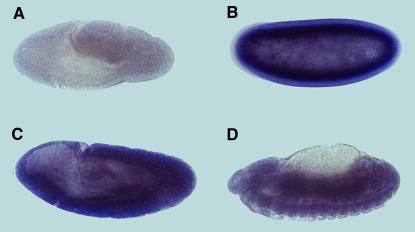
In situ analysis on whole-mount embryo preparations showed that Surf1 mRNA is expressed ubiquitously from early to late embryonic stages (Figure 2).
Figure 2.
In situ expression analysis of Surf1 mRNA in whole-mount preparations of wild-type embryos. (A) Stage 9, negative control. (B) Stage 4–5, blastoderm. (C) Stage 9–10. (D) Stage 14–15. In all developmental stages analyzed expression is fairly strong and ubiquitous, with a tendency to become weaker toward the end of embryonic development (see D).
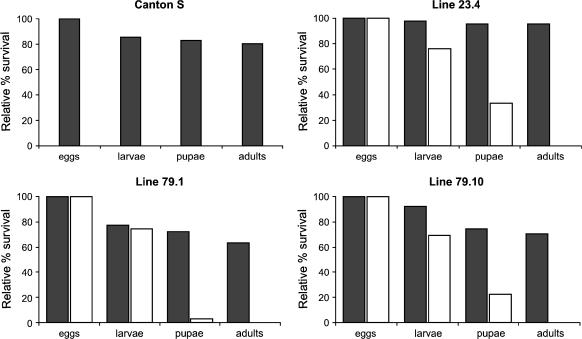
Larval lethality in Act-GAL4 Surf1 KD individuals:
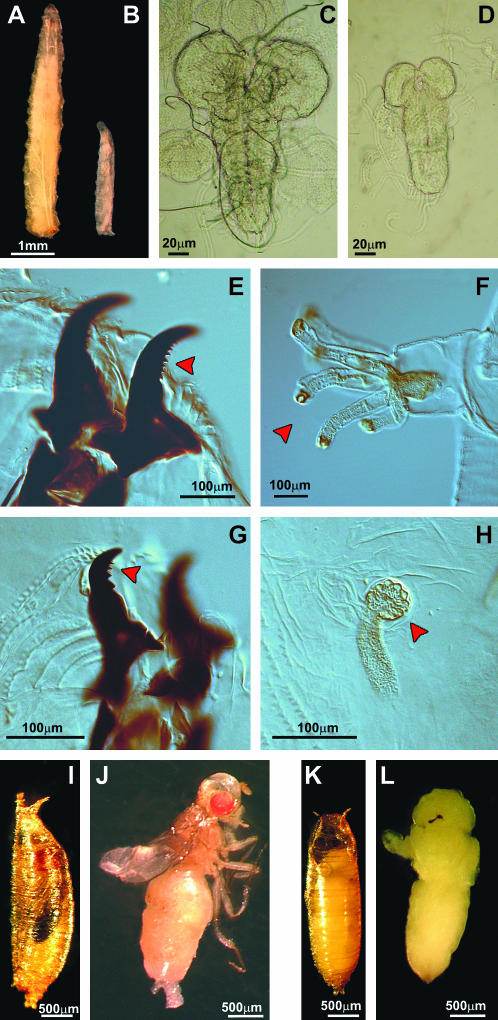
All KD individuals derived from the three transgenic KD lines (Act–GAL4 KD23.4, Act–GAL4 KD79.10, and Act–GAL4 KD79.1) showed 100% egg-to-adult lethality (Figure 3). Most individuals died as larvae, which were very sluggish and showed impaired development. In all three strains, the 7-day-old KD larvae appeared smaller than age-matched controls (Figure 4, A and B), with undersized optic lobes (Figure 4, C and D). The Act–GAL4 KD23.4 and Act–GAL4 KD79.10 individuals had the typical cuticular features of third-stage larvae (Figure 4, E and F) while in Act–GAL4 KD79.1 larvae the morphological features were typical of the second instar stage (Figure 4, G and H). Only a few larvae, in particular Act–GAL4 KD23.4 and Act–GAL4 KD79.10, reached the pupal stage but they did not progress any further in development. Dissection of the KD pupae showed that these individuals died at early imago stages (Figure 4, K and L) as compared to controls (Figure 4, I and J).
Figure 3.
Relative percentage of egg-to-adult viability, calculated at each of four developmental stages, i.e., eggs, larvae, pupae and adults, in controls (black) and Actin5C–GAL4-driven Surf1 knockdown flies (white). For each line, the control consisted of individuals from the line bearing the non-activated UAS–Surf1 inverted repeat. CS, Canton-S, a reference wild-type strain.
Figure 4.
Most of the Actin5C–GAL4 Surf1 KD individuals die as larvae, which show an impaired development: 7 day-old KD larvae (B) appear drastically smaller than controls (A) and have undersized optic lobes (C is a control brain, D is a brain from Actin5C–GAL4-driven Surf1 KD). KD larvae from lines 23.4 and 79.10 have all the distinctive characters of the third instar (i.e., the cuticular structures: mouth hooks (E) and anterior spiracles (F); see red arrows). On the other hand, in Actin5C–GAL4 Surf1 KD79.1 larvae, cuticular structures are typical of the second instar (G and H); see red arrows. Some KD larvae reach the pupal stage and do not progress any further in development. Dissection of the dead pupae shows that individuals die at early imago stages, when adult cuticular structures have only just everted. I and J are control pupae, respectively, with and without puparium; K and L are Actin5C–GAL4 Surf1 KD pupae.
Locomotor and photobehavioral defects in Act–GAL4 Surf1 KD larvae:
To better characterize the sluggishness observed in Surf1 Act–GAL4 KD larvae, we applied the checker test paradigm (see materials and methods), which measures both the speed of spontaneous locomotion (i.e., in the absence of external stimuli) and the locomotor response to light. The KD larvae showed a highly significant reduction in locomotor speed (Figure 5A) and a profoundly altered (i.e., less responsive) photobehavioral response (Figure 5B), which was particularly evident in Act–GAL4 KD79.10 individuals. The photobehavioral test consists of the evaluation of the light-avoidance reaction, which is normally shown by wild-type larvae. In the checker test this is evaluated by the ratio, expressed in the form of a normalized relative index (RI), of the time spent by a larva on the black (opaque to light) squares with respect to the white (transparent to light) ones of a checkerboard illuminated from beneath.
Figure 5.
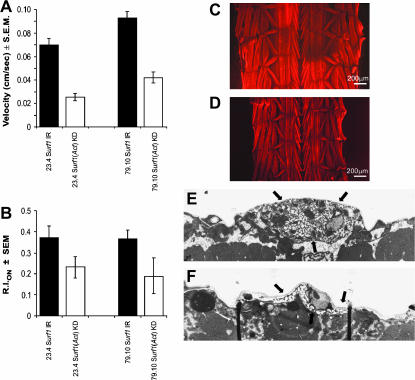
(A) The speed of locomotion (cm/sec) measured in Actin5C–-GAL4 Surf1 KD larvae is significantly different from that of the respective controls for both lines analyzed (23.4 vs. control, P < 0.0001, n = 23 and 21, respectively; 79.10 vs. control, P = 0.0366, n = 20 and 10, respectively). (B) Response in the photobehavioral assay of Surf1 KD and control larvae tested using the checker test (see materials and methods). Briefly, the photobehavioral test consists of the evaluation of the larval light-avoidance reaction. In the checker test paradigm this is evaluated by the ratio, expressed in the form of a normalized relative index (RI), of the time spent by a larva on the black (opaque to light) squares with respect to the white ones (transparent to light) of a checkerboard illuminated from beneath. KD individuals from both lines showed a significant decrease in their ability to avoid the light stimulus [Act–GAL4 KD23.4 vs. control (UAS–IR23.4); P = 0.032] and [Act–GAL4 KD79.10 vs. control (UAS–IR79.10); P = 0.02]. (C and D) An example of phalloidin-rhodamine staining of whole-mount larval body-wall preparations shows no major anomalies in muscle structure and arrangement in Actin5C–GAL4 Surf1 KD79.1 individuals (D), although a clear reduction in size is evident with respect to controls (C). (E and F) Cross sectional ultrastructure of glutamatergic synapses from third segment muscles 6 or 7 of stage three larvae. (E) Electron micrograph of a Type 1 bouton in a control (UAS–IR23.4) larva, showing the well-developed subsynaptic reticulum (SSR); see arrows. (F) Representative micrograph of a Type 1 bouton in an Actin5C–GAL4 Surf1 KD23.4 larva, showing a reduction in the complexity of the SSR, which is more typical of a mid first to second stage wild type larva. Magnification (E and F), ×2842.
Morphological and physiological examination of muscle:
To understand whether the altered locomotion of Act–GAL4 KD larvae was due to structural defects of the muscular apparatus, we examined body-wall preparations by fluorescence microscopy. Rhodamine/phalloidin-stained preparations showed that muscle fibers appeared normal in structure and arrangement, but they were much smaller in size, compared to age- and development-matched controls (Figure 5, C and D).
Furthermore, the measurement of the specific force output (i.e., force/muscle fiber unit section area), developed by longitudinal muscles following a single electrical stimulus, gave similar values in Act–GAL4 KD vs. control UAS–IR23.4 (i.e., 0.397 ± 0.083 vs. 0.548 ± 0.085 mN/mm2, determined in 8 and 20 individuals, respectively). Specific force output was also determined, again without significant differences between KD individuals and the respective controls. Following tetanic stimulation the observed values were Act–GAL4 KD = 1.97 ± 0.37 vs. UAS–IR23.4 = 2.25 ± 0.37 mN/mm2, and following caffeine-induced contraction the observed values were Act–GAL4 KD = 8.94 ± 1.68 vs. UAS–IR23.4 = 7.78 ± 0.73 mN/mm2. Altogether, these results allow exclusion of the existence of muscular impairments involving signal transduction, calcium release, or structural abnormalities of the contractile apparatus.
To further clarify the nature of the larval locomotor defects, we examined the neuromuscular junction on muscle fibers 6 and 7 by confocal and electron microscopy. We found no abnormality in the number, size, or type of synaptic boutons, except that the structure of the subsynaptic reticulum (SSR) showed a lower complexity in KD larvae (Figure 5F) than in controls (Figure 5E). Nonetheless, no abnormality in the evoked junction potential (EJPs) was found in the KD larvae compared to age-matched controls (control EJP amplitude, UAS–IR23.4 = 23.01 ± 4.10; Act-GAL4 KD23.4 EJP amplitude = 25.72 ± 6.57, amplitudes expressed in mV).
Electron microscopic analysis was also conducted on third-stage larval body-wall sections to check the distribution and morphology of mitochondria in muscle tissue of Act–GAL4 KD23.4 individuals. Figure 6 shows that in control individuals' mitochondria are rather small, homogeneously stained, and are present in clusters in the intermyofibrillar spaces (Figure 6, A and C). On the other hand, the mitochondria in KD individuals are much larger and more electron dense, with irregularly stained crests and matrix. In addition, they present a rounded shape, do not appear to be restricted to the intermyofibrillar spaces, and do not show a tendency to cluster (Figure 6, B and D).
Figure 6.
Electron micrograph of larval body-wall muscle fibers showing mitochondria (arrows) in control larvae bearing only the UAS–Surf1 inverted repeat and in Actin5C–GAL4 KD individuals. Cross section of control (A) and Actin5C–GAL4 KD (B) larva. Longitudinal section of control (C) and Actin5C–GAL4 KD (D) larva. Magnification for all figures: ×10,000. Mitochondria in controls tend to be rather small, elongated, and homogeneously stained, forming clusters within the intermyofibrillar spaces, whereas mitochondria in KD individuals are much larger and round with an apparently disorganized internal structure (i.e., matrix and cristae). Furthermore, the latter mitochondria are not typically found in clusters within the muscle fiber.
CNS-restricted KD of Surf1 using an elav–GAL4 driver:
To bypass the larval lethality caused by Act–GAL4-driven KD of Surf1, we restricted the expression of the dsRNAi to the CNS by using a pan-neuronal elav–GAL4 driver. The elav–GAL4 driver induces dsRNAi in the developing CNS and in the eye and antenna disc from early larval stages, and continues to be active in the corresponding areas of adult individuals (Yao and White, 1994).
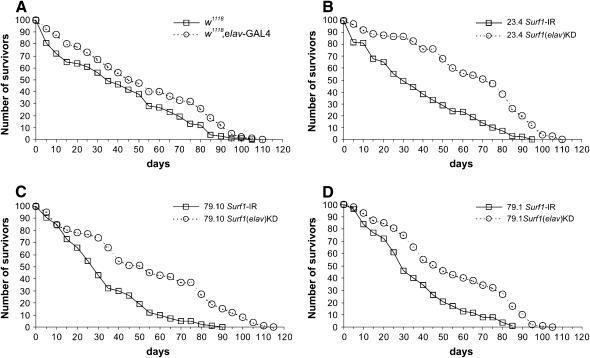
elav–GAL4 KDSurf1 dsRNAi had no effects on egg-to-adult viability. Nonetheless, such KD individuals (from lines 23.4 and 79.10), show a striking and significant increase in lifespan with respect to controls (Figure 7).
Figure 7.
Graphs showing the number of adult flies surviving after up to 115 days. (A) w1118 (the background used for transgenesis) and elav–GAL4 x w1118. (B) 23.4 Surf1 IR and elav–GAL4 KD23.4. (C) 79.10 Surf1 IR and elav–GAL4 KD79.10. (D) 79.1 Surf1 IR and elav–GAL4 KD79.1. For each graph the statistical comparison of the pairs of curves (i.e., KD vs. parental IR line) was done using the Wilcoxon test. (A) P = NS; (B) P = 0.008; (C) P = 0.02; (D) P = NS.
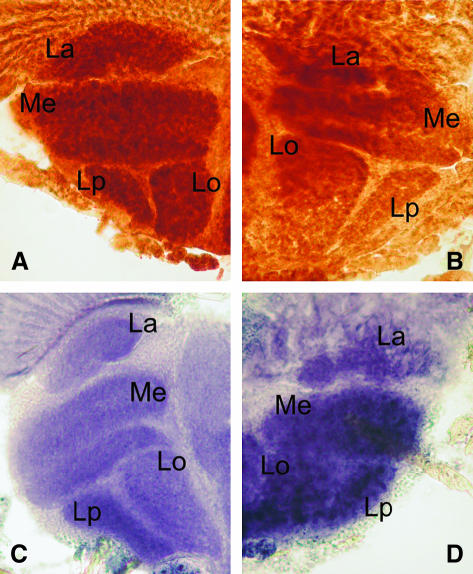
Histological sections obtained from heads of elav–GAL4 KD23.4 flies showed a significant (Student's t-test, P < 0.05) reduction in COX activity (KD23.4 = 177.98 ± 22.03 vs. controls = 189.23 ± 15.43) and a highly significant (Student's t-test, P < 0.001) increase in succinate dehydrogenase (SDH) activity (KD23.4 = 171.514 ± 25.34 vs. controls = 140.60 ± 31.27). Controls were age-matched individuals from the UAS–IR23.4 parental line (Figure 8, A–D). SDH is encoded by a nuclear gene and the enzyme is localized to the internal mitochondrial membrane. Thus the above values allow determination of the contribution of COX activity relative to SDH, which is an independent indicator of mitochondrial mass (Maier et al., 2001).
Figure 8.
(A) COX activity in adult control flies bearing only the UAS–Surf1 inverted repeat. (B) COX activity in elav–GAL4 Surf1 KD adult flies from line 23.4. (C) Succinate dehydrogenase (SDH) activity in adult control flies bearing only the UAS–Surf1 inverted repeat. (D) SDH activity in elav–GAL4 Surf1 KD adult flies from line 23.4. Nuclear-encoded SDH is localized to the internal mitochondrial membrane. Thus, comparison of COX levels to those of SDH provide an indication of the contribution of COX activity relative to SDH, an independent indicator of mitochondrial mass. In each image analyzed, the ROI were limited to the optic neuropils (i.e., lamina, medulla, lobula, and lobula plate). ROIs were identified in the digitized images (i.e., the single neuropils), and pixel counts and gray level distributions were performed within these ROIs. The mean gray level/pixel for each ROI was then used as an estimate of the level of enzyme activity being determined (i.e., high mean gray level/pixel = high enzyme activity). Sections obtained from heads of elav–GAL4 KD23.4 flies showed a significant (Student's t-test, P < 0.05) reduction in COX activity [KD23.4 = 177.98 ± 22.03 vs. controls (UAS–IR23.4) = 189.23 ± 15.43] and a highly significant (Student's t-test, P < 0.001) increase in SDH activity [KD23.4 = 171.514 ± 25.34 vs. controls (UAS–IR23.4) = 140.60 ± 31.27]. In particular SDH activity in KD individuals is greater than that observed in controls, suggesting an increase in mitochondrial mass in the former. La, lamina; Me, medulla; Lo, lobula; Lp, lobula plate.
No evident signs of KD and/or age-dependent neurodegeneration were evident following the analysis of hematoxilin-eosin-stained cephalic sections of 7- and 28-day-old elav–GAL4 KD adult flies (Figure 9).
Figure 9.
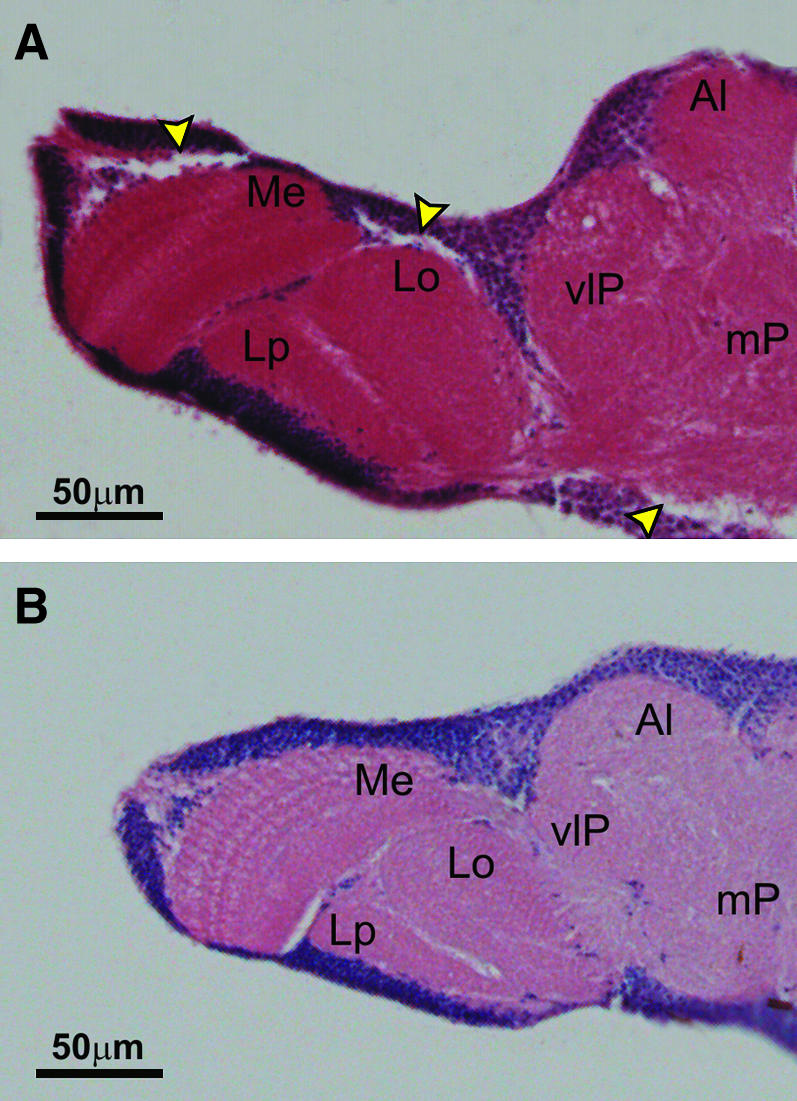
Hematoxilin-eosin stains of paraffin-embedded frontal cephalic sections of adult flies. (A) Control flies bearing only the UAS-Surf1 inverted repeat. (B) elav–GAL4 Surf1 KD adult flies from line 23.4. Me, medulla; Lo, lobula; Lp, lobula plate; Al, antennal lobe; vlP, ventrolateral protocerebrum; mP, medial protocerebrum. Arrows indicate sectioning artifacts.
The locomotion of elav–GAL4 KD larvae was significantly reduced relative to controls only in the case of line elav–GAL4 KD79.10 (0.078 ± 0.02 vs. 0.09 ± 0.02 cm/sec; P = 0.0366), while the photobehavioral responses were significantly altered for elav–GAL4 KD23.4 individuals [RI ON = 0.19 ± 0.26 vs. control (UAS–IR23.4) RI ON = 0.37 ± 0.29; P = 0.01] but they were not for elav–GAL4 KD79.10 flies [RI ON = 0.30 ± 0.28 vs. control (UAS–IR79.10) RI ON = 0.37 ± 0.22; P = 0.3].
The EJP amplitudes, recorded in larvae following single segmental nerve stimulation, showed no significant difference between KD and the respective controls, which in each case consisted of the parental line bearing the IR construct. Thus, control (UAS–IR23.4) amplitude = 23 ± 4.098 vs. Act–GAL4 KD23.4 = 22 ± 7.510; control (UAS–IR79.1) amplitude = 25.447 ± 7.617 vs. Act–GAL4 KD79.1 = 23.963 ± 4.746; all amplitudes are expressed in mV.
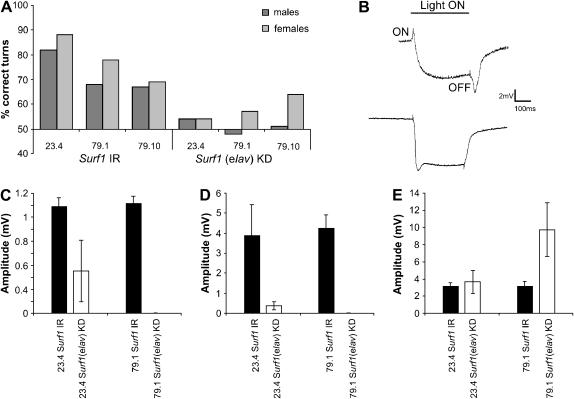
In the optomotor test, which measures locomotor and photobehavioral proficiency, adult control flies tended to turn in the direction of the rotating stripes. By contrast, elav–GAL4 KD23.4 and elav–GAL4 KD79.1 flies, as well as the elav–GAL4 KD79.10 males, but not the elav–GAL4 KD79.10 females, turned at random, meaning that they exhibited a number of correct turns which was not significantly different from 50%, indicating an impairment in the capacity to react to the rotating environment (Figure 10A). None of the KD flies produced a null response (i.e., was unable to move at all), indicating the absence of gross motor impairments.
Figure 10.
(A) Optomotor behavior in elav–GAL4 Surf1 KD and control (UAS–IR) adults. KD flies (males and females) from lines 23.4 and 79.1 and only KD males from line 79.10 turned at random, giving mean values of correct turns not significantly different from 50%. On the other hand, controls (UAS–IR23.4, UAS–IR79.1 and UAS–IR79.10) turned in the direction of the moving environment 70–80% of the time. For each genotype 20 individuals (10 males and 10 females) were tested. Each fly was given 10 trials, and each time the direction of rotation of the stripes was changed. (B, top) An example of a wild-type ERG response showing the major features, the ON and OFF transients with the sustained response between the two transients. (B, bottom) Example of an ERG response from an elav–GAL4 KD individual from line 79.1; in this case the ON and OFF transients are completely missing. (C, D, and E) results of ERGs recorded from elav–GAL4 Surf1 KD individuals from lines 23.4 and 79.1 and controls (as in A). Graphs represent mean amplitudes (mV ± SD) of ON (C) and OFF (D) transients (due to synaptic activation of second order neurons of the visual pathway) and of the sustained response (E) (due to photoreceptor light-induced depolarization). Mean amplitude of ON and OFF transients for both lines of KD vs. control (as above) individuals were significantly different (P < 0.05). However, only the sustained response of KD individuals from line 79.1 was significantly different to the relative control (as above). The graphs showing the ERG data are relative to 16–29 individuals for line 23.4 and 11–14 individuals in the case of line 79.1.
To verify whether the incorrect response to the optomotor test was due to visual impairment, the response to light of the neuronal retinal pathway was measured by ERG. A normal ERG recording under a photic stimulus is shown in Figure 10B. We observed a significant reduction of both ON (Figure 10C) and OFF (Figure 10D) transients in elav–GAL4 KD23.4 individuals, and their complete disappearance in KD79.1 elav–GAL4 individuals (Figure 10, C and D), while the sustained response was increased (Figure 10E). ON/OFF ERG transients are due to synaptic activation of second-order neurons of the visual pathway, while the sustained response is due to photoreceptor light-induced depolarization.
DISCUSSION
In this article we explored the functional consequences resulting from post-transcriptional silencing of the Drosophila homolog of human Surf1, a COX assembler gene whose mutations are responsible for human LS.
Post-transcriptional silencing was obtained by recombinant expression of a hairpin-loop dsRNA composed of two IRs, separated by a spacer. This tool has previously been proven to induce a very efficient and highly specific attenuation in the expression of a target gene (Piccin et al. 2001). The GAL4–UAS binary system (Brand and Perrimon 1993) was exploited to drive the knockdown of the target gene transcript under the control of two alternative promoters, namely those of Actin5C, an early-expressed housekeeping gene, or of elav, an early expressed neuron-specific gene.
Severe, global impairment of larval development and reduced spontaneous and light-induced locomotion were the main phenotypic features in ubiquitous Actin5C–GAL4-driven Surf1 KD individuals. The CNS in such individuals was markedly underdeveloped, particularly the optic lobes. In spite of the smaller size of muscle fibers and immaturity of the subsynaptic reticulum of the neuromuscular junction, we showed that the impaired locomotor behavior in KD larvae was not due to structural and/or functional abnormalities of the segmental muscle fibers or reduction in contractile efficiency. Taken together, these data suggest that the altered motor patterns observed in the constitutive Surf1 KD larvae are not due to specific functional defects in the motor effector structures (i.e., muscle fibers or the neuromuscular junction), but are likely caused by defective energy supply determined by Surf1 silencing. A generalized defect in respiration and ATP synthesis may also be responsible for the developmental delay and late larval lethality observed in KD individuals. End-stage larval development, and larval-to-adult metamorphosis, which occurs during the pupal phase, is an energy-requiring process (Singh and Singh 1999). Thus, defects leading to the depletion of key elements involved in respiration are expected to impinge strongly on the successful completion of this critical developmental phase. Nonetheless, in KD individuals, egg to early larval development by and large appears to proceed, even if such individuals show developmental delay and neuromotor impairments. This suggests that the early larval stages of development can progress probably thanks to a maternal contribution to the developing embryo of the Surf1 gene product as well as of mitochondria. Our conclusions are supported by the phenotype observed in several Drosophila mutants for genes related to mitochondrial functions. For instance, mutant phenotypes either for the tamas gene, which encodes for the mitochondrial DNA polymerase catalytic subunit (Pol-γA) (Iyengar et al. 1999, 2001), or for the gene encoding the Pol-γB accessory subunits, cause both lethality during early pupation, concomitant with loss of mtDNA and mitochondrial mass, and reduced cell proliferation in the CNS. A similar phenotype is presented by Drosophila mutants for the lopo (low power) gene, which encodes the mitochondrial single-stranded DNA-binding protein, a key component in mtDNA replication and maintenance (Maier et al. 2001). The lopo mutants die late in the third instar before completion of metamorphosis, because of a failure in cell proliferation. Molecular, histochemical, and physiological experiments show a drastic decrease in mtDNA content that is coupled with the loss of respiration in these mutants. Pleiotropic phenotypes are linked to different mutations in the cyclope gene, which encodes a COX subunit VIc homolog (Szuplewski and Terracol 2001), including embryonic lethality, several body growth abnormalities, and aberrant development of the eye disc and ommatidia. Larval lethal phenotypes were also obtained in mutants for the alpha subunit of mitochondrial ATP-synthase (complex V) (Talamillo et al. 1998) and for subunit 9 of the mitochondrial ubiquinol-cytochrome c reductase (complex III) (Frolov et al. 2000). Taken together, these studies on Drosophila mutants show early or late larval lethality often associated with neuromotor and/or photobehavioral defects, suggesting that these functions are energy demanding and are exquisitely sensitive to reductions in the energy supply provided by mitochondrial oxidative phosphorylation.
CNS-wide silencing of Surf1 was obtained by driving transcription of the target gene-specific dsRNA with elav–GAL4, and allowed us to bypass the larval lethality observed with the Actin5C–GAL4 driver. In this case, individuals survived to the adult phase with no overt signs of impairment. Intriguingly, such individuals actually showed a strikingly enhanced survival with respect to controls. The larval stages of the elav–GAL4 Surf1 KD individuals were morphologically comparable to the corresponding parental line controls (i.e., UAS–IR lines), including development of the CNS, although a slight impairment in locomotor activity and photobehavior was noted also in these lines. Once again, electrophysiological analyses produced no evidence of functional synaptic impairments. However, the photobehavior of adult elav-GAL4–KD flies, measured by the optomotor test paradigm, was clearly impaired, suggesting an abnormality in the visual pathway, which was confirmed by alterations in the ERG response. These alterations consisted in a reduction or disappearance of ON and OFF transients and by an increase of the sustained response. The ERG consists in an extracellular recording from the Drosophila eye that measures light-induced depolarization of photoreceptors (the sustained response) and synaptic activation of second-order neurons in the visual pathway (Hotta and Benzer 1969; Pak et al. 1969; Heisenberg 1971). The latter synaptic events occur at the onset and termination of a light pulse and are represented by the ON and OFF transients, which are generated at the level of the R1 to L1/L2 histaminergic synapses (Wu and Wong 1977). Several mutations known to block synaptic transmission, including synaptotagmin (DiAntonio and Schwarz 1994), rop (Harrison et al. 1994), and csp (Zinsmaier et al. 1994), decrease or abolish the ON/OFF transients (Heisenberg 1971).
Taken together, the optomotor and ERG results in our elav–GAL4 KD flies are likely to be due to complex alterations in the eye and CNS, involving several visual circuits. This conclusion is also supported by the demonstration that the neural foci responsible for the ERG transients are different from those mediating the optomotor response (Buchner et al. 1978; Sandrelli et al. 2001). As in the case of the Actin5C–GAL4 KD larvae, the morphological and functional analysis of the elav–GAL4 KD larvae and adult flies indicate that the behavioral abnormalities are not due to functional failure of single or specific components of the neuromotor and/or photoreceptive structures, but are the consequence of subtle widespread nervous system defects. Furthermore, following a detailed histological analysis, we were unable to show any overt signs of neurodegenerative damage in the brains of such individuals. A direct effect of Surf1 silencing on mitochondrial respiration was demonstrated by the significant reduction in the histochemical reaction to COX observed in the optic lobes of the elav–GAL4 KD adult flies. In addition, the parallel strong increase in the SDH levels measured in the same tissues suggests that there is an increase in mitochondrial mass, which may be tentatively interpreted as a compensatory reaction to the decrease in COX. Interestingly, electron microscopic observation of larval body-wall muscle fibers, shows that in Act–GAL4 KD individuals, mitochondria are much larger than normal, with evident morphological alterations (in particular regarding the organization of the internal membranes) while also showing an altered distribution within the muscle tissue (i.e., in controls mitochondria tend to form clusters within the intermyofibrillar spaces, while in KD individuals they do not). Although we do not have any direct evidence, it is tempting to speculate that perhaps the increased size of the mitochondria in KD individuals could be due to mitochondrial fusion, a process which is thought to occur as a response to mitochondrial damage and which could protect cells from entering the apoptotic pathway (Meeusen and Nunnari 2005). Altogether, the above results strongly support the involvement of respiratory deficiency in determining the developmental and functional impairments observed in KD individuals.
The structural and functional abnormalities at the nervous system level, as well as the global developmental arrest, observed following Surf1 KD in Drosophila are largely concordant with the human LSSurf1−/− phenotype, which predominantly affects the brain and, to a lesser extent, the skeletal muscle apparatus, and is also frequently associated with poor growth and failure to thrive. In the light of these considerations it will be of great interest to conduct further studies on the molecular and biochemical effects of Surf1 KD in the adult fly, as well as at the level of specific organs and structures. These investigations will contribute to our understanding on the specific involvement of the SURF1 protein and, more broadly, of mitochondrial energy metabolism, to body growth, neural development, and neurodegeneration.
Acknowledgments
Thanks go to Giuliano Carlesso (Department of Human Anatomy and Physiolology, University of Padova) for providing hematoxilin/eosin-stained sections of adult fly CNS. This study was supported by the collaborative program between CNR and MIUR “Legge 449/97” (Grant N° CU04.00067 to R.C.), Fondazione Pierfranco e Luisa Mariani to M.Z., Italian Ministry of Health grant RF-2002/158 to M.Z., Fondazione Cariplo to M.Z., EUMITOCOMBAT network grant from the EU-FP6 to M.Z., and by grant Telethon N. GP0048Y01 to R.C.
References
- Agostino, A., F. Invernizzi, C. Tiveron, G. Fagiolari, A. Prelle et al., 2003. Constitutive knockout of Surf1 is associated with high embryonic lethality, mitochondrial disease and cytochrome c oxidase deficiency in mice. Hum. Mol. Genet. 12: 1–15. [DOI] [PubMed] [Google Scholar]
- Beramendi, A., S. Peron, A. Megighian, C. Reggiani and R. Cantera, 2005. The inhibitor kappaB-ortholog Cactus is necessary for normal neuromuscular function in Drosophila melanogaster. Neuroscience 134: 397–406. [DOI] [PubMed] [Google Scholar]
- Brand, A. H, and N. Perrimon, 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415. [DOI] [PubMed] [Google Scholar]
- Buchner, E., K. G. Gotz and C. Straub, 1978. Elementary detectors for vertical movement in the visual system of Drosophila. Biol. Cybern. 31: 235–242. [DOI] [PubMed] [Google Scholar]
- Coenen, M., L. P. van den Heuvel, L. G. Nijtmans, E. Morava, I. Marquardt et al., 1999. SURFEIT-1 gene analysis and two-dimensional blue native gel electrophoresis in cytochrome c oxidase deficiency. Biochem. Biophys. Res. Commun. 265: 339–344. [DOI] [PubMed] [Google Scholar]
- Colombo, P., J. Yon, K. Garson and M. Fried, 1996. Conservation of the organization of five tightly clustered genes over 600 million years of divergent evolution. Proc. Natl. Acad. Sci. USA. 89: 6358–6362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiAntonio, A., and T. L. Schwarz, 1994. The effect on synaptic physiology of synaptotagmin mutations in Drosophila. Neuron 12: 909–920. [DOI] [PubMed] [Google Scholar]
- DiMauro, S., and E. A. Schon, 2003. Mitochondrial respiratory-chain diseases. New Engl. J. Med. 348: 2656–2668. [DOI] [PubMed] [Google Scholar]
- Fadic, R., and D. R. Johns, 1996. Clinical spectrum of mitochondrial diseases. Semin. Neurol. 16: 11–20. [DOI] [PubMed] [Google Scholar]
- Farina, L., L. Chiapparini, G. Uziel, M. Bugiani and M. Zeviani, 2002. MR findings in Leigh syndrome with COX deficiency and Surf-1 mutations. AJNR Am J Neuroradiol. 23: 1095–1100. [PMC free article] [PubMed] [Google Scholar]
- Frolov, M. V., E. V. Benevolenskaya and J. A. Birchler, 2000. The oxen gene of Drosophila encodes a homolog of subunit 9 of yeast ubiquinol-cytochrome c oxidoreductase complex: evidence for modulation of gene expression in response to mitochondrial activity. Genetics 156: 1727–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson, B. J., R. Carrozzo, F. Piemonte, A. Tessa, B. H. Robinson et al., 2001. Cytochrome c oxidase-deficient patients have distinct subunit assembly profiles. J. Biol. Chem. 276: 16296–16301. [DOI] [PubMed] [Google Scholar]
- Harrison, S. D., K. Broadie, J. van de Goor and G. M. Rubin, 1994. Mutations in the Drosophila rop gene suggest a function in general secretion and synaptic transmission. Neuron 13: 555–566. [DOI] [PubMed] [Google Scholar]
- Hassan, J., M. Busto, B. Iyengar and A. R. Campos, 2000. Behavioural characterization and genetic analysis of the Drosophila melanogaster larval response to light as revealed by a novel individual assay. Behav. Genet. 30: 59–69. [DOI] [PubMed] [Google Scholar]
- Heisenberg, M., 1971. Separation of receptor and lamina potentials in the electroretinogram of normal and mutant Drosophila. J. Exp. Biol. 55: 85–100. [DOI] [PubMed] [Google Scholar]
- Hotta, Y., and S. Benzer, 1969. Abnormal electroretinograms in visual mutants of Drosophila. Nature 222: 354–356. [DOI] [PubMed] [Google Scholar]
- Iyengar, B., J. Roote and A. R. Campos, 1999. The tamas gene, identified as a mutation that disrupts larval behavior in Drosophila melanogaster, codes for the mitochondrial DNA polymerase catalytic subunit (DNApol-gamma125). Genetics 153: 1809–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyengar, B., J. Roote and A. R. Campos, 2001. Erratum. Genetics 159: 1867. [Google Scholar]
- Leigh, D., 1951. Subacute necrotizing encephalomyelopathy in an infant. J. Neurol. Neurosurg. Psychiatr. 14: 216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann, R., and D. Tautz, 1994. in situ hybridization to RNA. Methods Cell Biol. 44: 575–598. [DOI] [PubMed] [Google Scholar]
- Maier, D., C. L. Farr, B. Poeck, A. Alahari, M. Vogel et al., 2001. Mitochondrial single-stranded DNA-binding protein is required for mitochondrial DNA replication and development in Drosophila melanogaster. Mol. Biol. Cell 12: 821–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeusen, S. L., and J. Nunnari, 2005. How mitochondria fuse. Curr. Opin. Cell Biol. 17: 389–394. [DOI] [PubMed] [Google Scholar]
- Nachlas, M. M., A. C. Young and A. M. Seligman, 1957. Problems of enzymatic localization by chemical reactions applied to tissue sections. J. Histochem. Cytochem. 5: 565–583. [DOI] [PubMed] [Google Scholar]
- Nijtmans, L. G. J., M. Artal Sanz, M. Bucko, M. H. Farhoud, M. Feenstra et al., 2001. Shy1p occurs in a high molecular weight complex and is required for efficient assembly of cytochrome c oxidase in yeast. FEBS Lett. 498: 46–51. [DOI] [PubMed] [Google Scholar]
- Pak, W. L., J. Grossfield and N. V. White, 1969. Nonphototactic mutants in a study of vision of Drosophila. Nature 222: 351–354. [DOI] [PubMed] [Google Scholar]
- Péquignot, M. O., R. Dey, M. Zeviani, V. Tiranti, C. Godinot et al., 2001. Mutations in the SURF1 gene associated with Leigh syndrome and cytochrome c oxidase deficiency. Hum. Mutat. 17: 374–381. [DOI] [PubMed] [Google Scholar]
- Pfaffl, M. W., 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccin, A., A. Salameh, C. Benna, F. Sandrelli, G. Mazzotta et al., 2001. Efficient and heritable functional knock-out of an adult phenotype in Drosophila using a GAL4-driven hairpin RNA incorporating a heterologous spacer. Nucleic Acids Res. 29: E55–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poyau, A., K. Buchet and C. Godinot, 1999. Sequence conservation from human to prokaryotes of Surf1, a protein involved in cytochrome c oxidase assembly, deficient in Leigh syndrome. FEBS Lett. 462: 416–420. [DOI] [PubMed] [Google Scholar]
- Roberts, D. B., and G. N. Standen, 1998. The elements of Drosophila biology and genetics, pp. 1–54 in Drosophila: A Practical Approach, edited by D. B. Roberts. IRL Press, Oxford.
- Sandrelli, F., S. Campesan, M. G. Rossetto, C. Benna, E. Zieger et al., 2001. Molecular dissection of the 5′ region of no-on-transientA of Drosophila melanogaster reveals cis-regulation by adjacent dGpi sequences. Genetics 157: 765–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, K. and R. N. Singh, 1999. Metamorphosis of the central nervous system of Drosophila melanogaster Meigen (Diptera: Drosophilidae) during pupation. J. Biosci. 24: 345–360. [Google Scholar]
- Spradling, A. C., and G. M. Rubin, 1982. Transposition of P elements into Drosophila germ line chromosomes. Science 218: 341–352. [DOI] [PubMed] [Google Scholar]
- Szuplewski, S., and R. Terracol, 2001. The cyclope gene of Drosophila encodes a cytochrome c oxidase subunit VIc homolog. Genetics 158: 1629–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talamillo, A., A. A. Chisholm, R. Garesse and H. T. Jacobs, 1998. Expression of the nuclear gene encoding mitochondrial ATP synthase subunit alpha in early development of Drosophila and sea urchin. Mol. Biol. Rep. 25: 87–94. [DOI] [PubMed] [Google Scholar]
- Tiranti, V., C. Galimberti, L. Nijtmans, S. Bovolenta, M. P. Perini et al., 1999. a Characterization of SURF-1 expression and Surf-1p function in normal and disease conditions. Hum. Mol. Genet. 8: 2533–2540. [DOI] [PubMed] [Google Scholar]
- Tiranti, V., M. Jaksh, S. Hofman, C. Galimberti, K. Hoertnagel et al., 1999. b Loss-of-function mutations of SURF1 are specifically associated with Leigh syndrome with cytochrome c oxidase deficiency. Ann. Neurol. 46: 161–166. [DOI] [PubMed] [Google Scholar]
- Tiranti, V., K. Hoertnagel, R. Carrozzo, C. Galimberti, M. Munaro et al., 1998. Mutations of SURF-1 in Leigh disease associated with cytochrome c oxidase deficiency. Am. J. Hum. Genet. 63: 1609–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Meer, J. M., 1977. Optically clean and permanent whole mount preparation for phase-contrast microscopy of cuticular structures of insect larvae. Dros. Inf. Serv. 52: 160. [Google Scholar]
- Vazquez-Martinez, O., R. Canedo-Merino, M. Diaz-Munoz and J. R. Riesgo-Escovar, 2003. Biochemical characterization, distribution and phylogenetic analysis of Drosophila melanogaster ryanodine and IP3 receptors, and thapsigargin sensitive Ca2+ ATPase. J. Cell. Sci. 116: 2483–2494. [DOI] [PubMed] [Google Scholar]
- Wu, C. F., and F. Wong, 1977. Frequency characteristics in the visual system of Drosophila: genetic dissection of electroretinogram components. J. Gen. Physiol. 69: 705–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, K-M, and K. White, 1994. Neural specificity of elav expression: defining a Drosophila promoter for directing expression to the nervous system. J. Neurochem. 63: 41–51. [DOI] [PubMed] [Google Scholar]
- Yao, J., and E. A. Shoubridge, 1999. Expression and functional analysis of SURF1 in Leigh syndrome patients with cytochrome c oxidase deficiency. Hum. Mol. Genet. 8: 2541–2549. [DOI] [PubMed] [Google Scholar]
- Zhu, Z., J. Yao, T. Johns, K. Fu, I. De Bie et al., 1998. SURF1, encoding a factor involved in the biogenesis of cytochrome c oxidase, is mutated in Leigh syndrome. Nat. Genet. 20: 337–343. [DOI] [PubMed] [Google Scholar]
- Zinsmaier, K. E., K. K. Eberle, E. Buchner, N. Walter and S. Benzer, 1994. Paralysis and early death in cysteine string protein mutants of Drosophila. Science 263: 977–980. [DOI] [PubMed] [Google Scholar]
- Zordan, M. A., M. Massironi, M. G. Ducato, G. Te Kronnie, R. Costa et al., 2005. Drosophila CAKI/CMG protein, a homolog of human CASK, is essential for regulation of neurotransmitter vesicle release. J. Neurophysiol. 94: 1074–1083. [DOI] [PubMed] [Google Scholar]