Abstract
Protein ADP ribosylation catalyzed by cellular poly(ADP-ribose) polymerases (PARPs) and tankyrases modulates chromatin structure, telomere elongation, DNA repair, and the transcription of genes involved in stress resistance, hormone responses, and immunity. Using Drosophila genetic tools, we characterize the expression and function of poly(ADP-ribose) glycohydrolase (PARG), the primary enzyme responsible for degrading protein-bound ADP-ribose moieties. Strongly increasing or decreasing PARG levels mimics the effects of Parp mutation, supporting PARG's postulated roles in vivo both in removing ADP-ribose adducts and in facilitating multiple activity cycles by individual PARP molecules. PARP is largely absent from euchromatin in PARG mutants, but accumulates in large nuclear bodies that may be involved in protein recycling. Reducing the level of either PARG or the silencing protein SIR2 weakens copia transcriptional repression. In the absence of PARG, SIR2 is mislocalized and hypermodified. We propose that PARP and PARG promote chromatin silencing at least in part by regulating the localization and function of SIR2 and possibly other nuclear proteins.
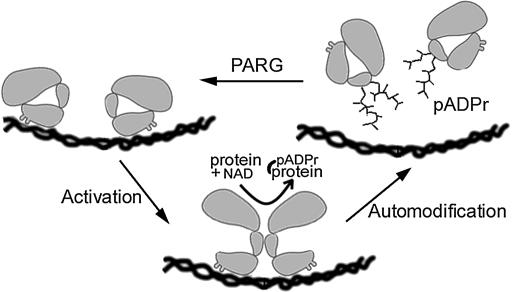
ADP-ribose modification of nuclear proteins mediates DNA repair, gene transcription, telomere elongation, and chromatin structure (reviewed in de Murcia and Shall 2000; Ziegler and Oei 2001; Tulin et al. 2003). Protein ADP-ribosylation levels are ultimately determined by the location and activity of poly(ADP-ribose) polymerase (PARP) and tankyrase enzymes that utilize NAD to add such residues, as well as poly(ADP-ribose) glycohyrolase (PARG) enzymes that remove them. Although a great deal has been learned about the biochemical properties of these enzymes in vitro, exactly how they function in vivo remains poorly known. Most of the time, the vast majority of PARP molecules are enzymatically inactive, unmodified, and thought to act only during brief bursts of activity (Figure 1). Damaged or altered DNA conformation, along with other uncharacterized signals, can cause nearby PARP molecules within small chromosome regions to dimerize and become transiently active before they are dissociated and shut off by automodification with long poly(ADP-ribose) chains (pADPr). Histones and other chromosomal proteins in the affected chromatin domain adopt a looser configuration, either by binding avidly to PARP-linked poly(ADP-ribose) polymers or by direct modification, thereby facilitating repair or gene activation (reviewed in Tulin et al. 2003). When PARG eventually removes all the ADP-ribosyl groups from a PARP monomer, the cycle can repeat until the inducing conditions are no longer present (see Figure 1). Other mechanisms of PARP action have been proposed as well, including some that do not require PARP enzymatic function (Tulin et al. 2002; Ju et al. 2004; Kim et al. 2004).
Figure 1.
Model of action of PARP and PARG on chromatin structure.
Genetic analysis of this system is greatly facilitated in Drosophila, which contains a single Parp gene located in 3R heterochromatin that encodes an enzyme with the same domain structure as that of the major mammalian PARP1 protein (Hanai et al. 1998; Tulin et al. 2002). Drosophila also contains a single tankyrase gene (Adams et al. 2000) and a single gene (Parg) predicted to encode a PARG (Hanai et al. 2004). Parp mutations are lethal and drastically alter many aspects of developmental physiology (Tulin et al. 2002; Tulin and Spradling 2003). These include the ability to activate and maintain nucleoli, to form polytene chromosome puffs, and to activate genes located therein that respond to stress, infection, or steroid hormones.
Heterochromatin forms in early embryonic cells and additional chromatin domains are silenced as individual cell types differentiate (reviewed in Gerasimova and Corces 2001; Fischle et al. 2003; Orlando 2003). The ability to compact heterochromatin and to silence specific gene regions also requires Parp. For example, the 30–50 normally quiescent genomic copies of the copia retrotransposon are overexpressed >50-fold in Parp mutants (Tulin et al. 2002). Normally, copia transcription is suppressed by a chromatin-based mechanism related to gene silencing in other regions (see Stapleton et al. 2001). Thus, in addition to its role as an activator, PARP contributes to the repression of at least some chromatin domains.
The evolutionarily conserved silent information repressor protein 2 (SIR2) protein contributes to heterochromatin formation through the action of its NAD-dependent histone deacetylase activity (Landry et al. 2000). NAD is cleaved in conjunction with removal of acetyl groups from the target, forming nicotinamide and O-acetyl-ADP-ribose (reviewed in Denu 2003). In addition, many SIR2 protein family members catalyze protein ADP ribosylation (Frye 1999). Drosophila contains five genes related to yeast SIR2, but the Sir2 gene residing at 34A7 shows the highest level of conservation (see Rosenberg and Parkhurst 2002) and exhibits NAD-dependent histone deacetylase activity (Barlow et al. 2001; Newman et al. 2002). While nonessential, Sir2 participates in chromatin silencing (Newman et al. 2002; Astrom et al. 2003; Furuyama et al. 2004).
To better understand how poly(ADP)-ribose metabolism regulates chromatin activity, we have characterized the Drosophila Parg gene (see also Hanai et al. 2004). Our findings reinforce previous evidence that PARP-catalyzed ADP ribosylation plays widespread roles in the nucleus, which are not limited to DNA repair. They support the view that PARP acts in vivo by undergoing bursts of activation limited by automodification and reversed by PARG action (Figure 1). In addition, we find that PARG controls the localization of other nuclear proteins. In Parg mutants, SIR2 protein is mislocalized and hypermodified; endogenous copia retrotransposon expression is elevated, suggesting that chromatin silencing is compromised. These experiments further document important roles played by ADP-ribose modification in controlling chromatin structure and activity and suggest that some of these effects are mediated through SIR2.
MATERIALS AND METHODS
Drosophila strains and genetics:
Genetic markers are described in FlyBase (1999) and stocks were obtained from the Bloomington Stock Center except as indicated. The ParpCH1 mutant stock was described in Zhang and Spradling (1994) and Tulin et al. (2002). Parg27.1 was constructed by Hanai et al. (2004). Sir205237 was constructed by Karpen and Spradling (1992) and is described by Rosenberg and Parkhurst (2002). We also used a Sir217 mutant stock in which background lethal mutations were removed (Astrom et al. 2003). pP{w+, UAST-PARP-I-DsRed}, called UAS-Parp-I-dsRed, was described in Tulin et al. (2002). The following GAL4 driver strains were used: da-GAL4 (gift of A. Veraksa), 69B-GAL4 (Manseau et al. 1997), hs-GAL4 (gift of G. Cavalli lab), and arm-GAL4 (Bloomington stock no. 1560). To induce expression from the hs-GAL4 driver, Drosophila were heat-shocked for 1 hr at 37° twice daily for 4 days prior to the late third instar stage. Balancer chromosomes carrying Kr-GFP, i.e., TM3, Sb, P{w+, Kr-GFP}, and CyO, P{w+, Kr-GFP} (Casso et al. 2000), were used to identify heterozygous and homozygous Parg27.1, Sir205237, and Sir217 mutant animals and those expressing PargRNAi and appropriate drivers.
Construction of transgenic Drosophila:
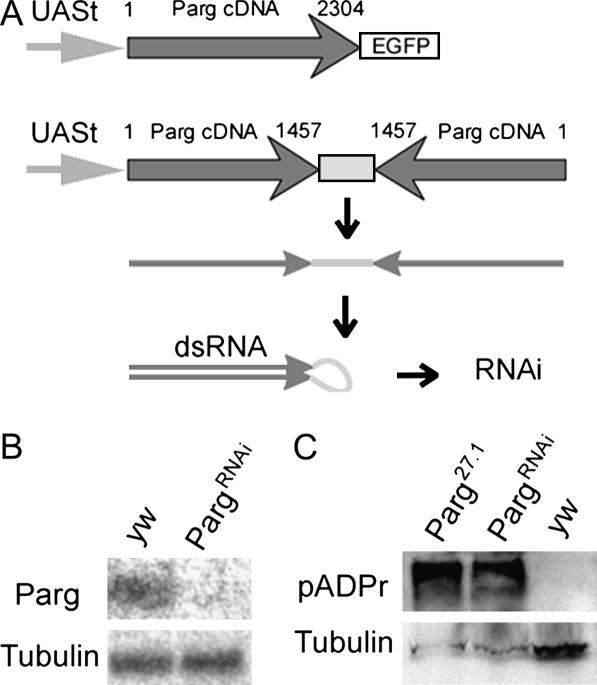
To construct UAS-Parg-EGFP, a full-length Parg cDNA, LD42380 (purchased from Research Genetics, Huntsville, AL), was fused to enhanced green fluorescent protein (EGFP) (CLONTECH Laboratories, Palo Alto, CA) in pP{w+, UAST-PARG-EGFP} (see Figure 2A). To construct UAS-Sir2-DsRed, a full-length Sir2 cDNA, LD38188 (from Research Genetics) was fused in frame within pP{w+, UAST-Sir2-DsRed}. To construct the anti-Parg “snap back,” we cloned a 1457-bp 5′-end fragment of Parg cDNA (from LD42380) in direct and inverted orientation within the pUASt vector. As a spacer between inverted repeats we used an 825-bp fragment (Figure 2A). Transformation was as described (Spradling and Rubin 1982), with modifications (Prokhorova et al. 1994).
Figure 2.
Structure and effects of constructs that alter Parg expression. (A) Structures of UAS-Parg-EGFP and PargRNAi transgenes. Nucleotide positions in PARG cDNA are indicated. (B) Effect of PargRNAi on PARG protein expression. Third instar larvae expressing the PargRNAi transgene lack a band ∼90 kDa that is detected in Western blots from wild-type animals using anti-PARG antibodies. (C) Reducing PARG increases the steady-state levels of pADPr. The labeling of diverse proteins, including a prominent band shown here the size of automodified-Parp, is greatly increased in Parg27.1 or PargRNAi third instar mutant larvae compared to wild type. A Western blot was probed with anti-pADPr antibody 10H.
Antibodies and immunohistochemistry:
Tissues were fixed and stained with primary and secondary antibodies as described previously (Grieder et al. 2000) and examined by confocal microscopy using a Leica TCS-NT microscope. Primary antibodies were anti-FIBRILLARIN (1:200) (from J. Gall), rabbit anti-EGFP antibodies (Torrey Pines Biolab), monoclonal anti-dsRed (CLONTECH Laboratories), rabbit anti-hyperacetylated histone H4 (Upstate Biologicals, Lake Placid, NY), and mouse mAb 10H (1:20-50) from Manfred Frey (Steinbeis-Transferzentrum fur Angewandte Biologische Chemie). Antibody 10H specifically recognizes branch points in ADP-ribose polymers (Kawamitsu et al. 1984). Monoclonal antibodies against PARG (A0683) were purchased from the Center for Biomedical Inventions of the Utah Southwestern Medical Center. Mouse Alexa-488 or Alexa-568 (Molecular Probes, Eugene, OR) were used as secondary antibodies (1:400).
Protein extract preparation:
Two grams of fresh pupae/larvae of the appropriate genotypes were homogenized in ice-cold lysis buffer (50 mm Tris-HCl, pH 7.5; 125 mm NaCl; 5% glycerol; 0.2% NP40; 1.5 mm MgCl2; 25 mm NaF; 1 mm Na3VO4; 1 mm EDTA; 1 mm DTT; Roche complete inhibitor, 1 tablet/25 ml of buffer) using a prechilled Potter homogenizer. Samples were filtered through two layers of Miracloth (Calbiochem, La Jolla, CA), homogenized using a Dounce homogenizer, and then treated with DNAse at 4° for 20 min. The supernatant fraction was collected after two subsequent centrifugations at 4°: 1000 × g for 10 min and 28,000 rpm for 25 min in an SW60Ti rotor.
Western blots:
Western blots were carried out as follows. After SDS-PAGE, proteins were transferred to nitrocellulose using a semidry method. Membranes were blocked in PBT buffer (pH 7.2, 137 mm NaCl, 2.7 mm KCl, 4.3 mm Na2HPO4, 1.8 mm KH2PO4, 0.2% Tween-20) with 10% nonfat dry milk and then incubated with primary antibodies (A0683 at 1:500 dilution; 10H at 1:50; anti-hyperacetylated histone H4 at 1:100; anti-α-tubulin at 1:100). Proteins were detected by chemiluminescence using HRP-conjugated goat anti-mouse or goat anti-rabbit IgG (Jackson Laboratories) diluted 1:2000 in PBT buffer with 10% nonfat dry milk.
RESULTS
Generation of reagents for analyzing Parg:
To analyze Parg function, we utilized Parg27.1, an imprecise excision derivative of strain EP0351 that partially deletes the Parg coding region (Hanai et al. 2004). We also built constructs that express epitope-tagged PARG or that reduce PARG levels using in vivo-produced RNA interference (RNAi) (Figure 2A). Strains with insertions at favorable sites where these constructs are expressed and function effectively were identified. For example, when combined with the weak, uniform 69B GAL4 driver, the UAS-Parg-EGFP transgene rescues the lethality of Parg27.1 mutants, verifying that its defects are caused by reduced PARG expression. These rescue constructs have no phenotype in a wild-type background. Strains expressing PargRNAi lose detectable expression of endogenous PARG, as indicated using a specific antibody (Figure 2B), and of UAS-Parg-EGFP driven by GAL4-69B in the same animals (data not shown). Driving Parg-EGFP with strong drivers such as da-GAL4 greatly elevates levels of PARG and EGFP immunoreactivity as assayed by both immunofluorescence and Western blots (data not shown).
Disrupting Parg function mimics Parp mutation:
With these tools in hand, we first examined the effects of reducing Parg function. Parg27.1 mutants at 25° hatch but subsequently arrest at a variety of developmental time points, including the pupal arrest described by Hanai et al. (2004). More detailed examination revealed that up to 30% of the mutant animals arrest at the L2/L3 larval molt, similar to ParpCH1 homozygotes. Loss of Parg may drastically reduce Parp activity by allowing each PARP protein to function only once before it is trapped in its inactive, automodified state. To look for an accumulation of automodified PARP, we utilized the H10 antibody (Kawamitsu et al. 1984) that recognizes a specific epitope associated with pADPr polymer branch points. As predicted, Western blots of protein from Parg27.1 flies probed with H10 antisera revealed greatly elevated levels of a band that corresponds in size to automodified PARP (Figure 2C). Expressing PargRNAi using a da-GAL4 driver also causes lethality, although the level of residual PARG activity is probably higher, as some animals survive up to the adult stage. However, PargRNAi expression is sufficient to elevate pADPr levels extensively, as with the Parg27.1 mutation (Figure 2C).
Excess PARG blocks PARP function:
To further investigate the Parg gene, we characterized its expression during development and within individual cells using PARG-EGFP and the 69B-GAL4 driver. These experiments revealed that PARG-EGFP is located in the nucleoplasm of all cell types examined, including the cells of the L3 larval salivary gland (Figure 3, A and C). In contrast, PARP-DsRed is predominantly found along chromosomes and in nucleoli (Figure 3, B and C; Tulin et al. 2002). PARG may bind only to automodified PARP and other proteins containing ADP-ribose adducts whose pools on chromatin may be small due to their release following modification. However, some of the nucleoplasmic PARG may correspond to molecules produced at higher-than-normal levels from the expression constructs.
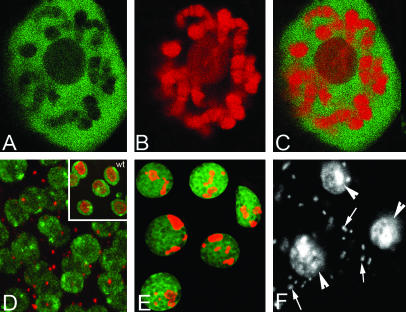
Figure 3.
PARG and PARP proteins are found in different nuclear compartments but effect similar processes. (A) PARG-EGFP is localized throughout the nucleoplasm, but very little is associated with chromosomes or nucleoli. (B) PARP-DsRed is associated with chromosomes and nucleoli in the same salivary gland cells. (C) Overlay of PARG-EGFP and PARP-DsRed demonstrates the complementary localization of those proteins. (D) In PARPCH1 animals, most cells lack nucleoli and FIBRILLARIN is confined to a cytoplasmic body. Inset: wild-type control. DNA, green; FIBRILLARIN, red. (E) da-GAL4∷UAS-Parg-EGFP animals that overexpress PARG show fragmented nucleoli. PARG-EGFP, green; FIBRILLARIN, red. (F) Overexpression of PARG causes intracellular bacterial invasion (arrows). DAPI stain was used; arrowheads indicate nuclei.
Producing PARG in great excess might disrupt normal ADP-ribose metabolism (and again mimic Parp mutation) by altering PARG localization and the balance of PARG and PARP activity. We found that overexpressing PARG using the UAS-PARG-EGFP transgene driven by strong GAL4 drivers such as da-GAL4 is toxic. The level of overexpression and the exact lethal phase depended on the particular driver used, on its insertion site, and on the gene dosage of both construct and driver. The lethality of excess PARG could be rescued by simultaneous overexpression of PARP-DsRed along with PARG-EGFP. Excess PARG, like Parp mutation, disrupts nucleoli (Figure 3E). However, the nature of the disruption is not identical, because instead of losing all nucleolar structure and driving nucleolar proteins such as FIBRILLARIN into the cytoplasm (Figure 3D), the nuclei of animals with excess PARG contain nucleolar proteins such as FIBRILLARIN in discrete nucleolar fragments.
The PARG-overexpressing animals share two other distinctive phenotypes with Parp mutants. First, they are hypersensitive to heat shocks. Up to 40% of PARG-EGFP overexpressing third instar larvae did not recover after a 40-min heat shock (compared to 1% lethality of wild-type controls and 90% lethality in the case of ParpCH1 mutants). In addition, tissues of PARG-EGFP overexpressing animals become extensively contaminated with intracellular bacteria (Figure 3F). Together these observations strongly support our previous conclusions concerning the role of PARP-mediated protein ADP-ribosyl modification during the Drosophila life cycle (Tulin et al. 2002; Tulin and Spradling 2003) and demonstrate that an appropriate level of Parg activity is required for Parp-dependent processes in vivo.
Normal levels of PARG are required for PARP localization on chromosomes:
When automodified, PARP is thought to leave its normal position on chromosomes due to the negative charges present on its ADP-ribose chains. Photobleaching experiments using PARP-EGFP in wild-type cells confirmed that it cycles rapidly within a bleached segment (data not shown). We next investigated whether reducing PARG levels would alter the steady-state distribution of PARP among chromosomes and other nuclear compartments. PARP relocalization, as well as inactivation, might contribute to the nucleolar disruption that is observed when Parg function is reduced (Figure 3).
When we examined the localization of PARP in salivary gland nuclei from PargRNAi animals, we observed that its association with chromosomes and active nucleoli is weakened (compare Figure 4, D–F, with Figure 4, A–C). Under these conditions, PARP accumulates in dense, nucleoplasmic bodies. Such bodies may be involved in recycling ADP-ribose-modified proteins. An even greater relocalization of PARP is apparent in larvae bearing the stronger Parg27.1 allele. In these animals, PARP can no longer be detected in association with the chromosome arms. Instead, PARP molecules are found primarily in the nucleoplasmic bodies, as well as in the heterochromatin of the polytene chromocenter (Figure 4I, outlined region).
Figure 4.
Parg is required to maintain PARP protein in euchromatin and for nucleolar integrity. (A–C) A wild-type salivary gland nucleus. (A) FIBRILLARIN (green) labels the nucleolus; (B) PARP-DsRed (red) is found on chromosomes and within the nucleolus; (C) overlay of A and B. DNA is blue. (D–F) Salivary gland nucleus from PargRNAi showing destruction of nucleolar integrity, removal of PARP-DsRed from chromatin, and accumulation of PARP and nucleolar components in nucleoplasmic bodies. (G–I) The same immunostaining of the Parg27.1 mutant demonstrates complete removal of PARP-DsRed from the euchromatic part of the genome, but retention in heterochromatin. FIBRILLARIN and PARP could still be detected in the nucleolus but the nucleolar chromatin is condensed.
These experiments imply that the normal location of PARP protein depends on its modification state. Blocking ADP-ribose removal would inactivate PARP molecules by by trapping them in their inactive, automodified state. There are several possible reasons why the modified molecules accumulate in large nuclear bodies that also contain FIBRILLARIN and probably many other molecules. Normally, automodified PARP is thought to remain tethered in the vicinity of its chromosomal origin and, by the action of PARG, to release a cargo of local chromatin proteins bound to its ADP-ribose polymers (Tulin et al. 2003). When present at high levels, automodified PARP and stripped chromosomal proteins such as FIBRILLARIN may simply aggregate. However, we favor the idea that the nucleoplasmic bodies represent a pool of modified proteins arrested in a normal recycling process. A lower normal level of PARP recycling in heterochromatin and condensed nucleolar regions may explain why these zones were relatively less affected.
Parg is required to suppress copia transcription:
In addition to its requirement for gene activation, Parp is needed to silence the transcriptional activity of repetitive elements such as the copia retrotransposon (Tulin et al. 2002). There are several possible mechanisms for this paradoxical action. PARP-mediated chromatin loosening might comprise an early transient step in heterochromatin formation, for example, by making histones and other proteins available to heterochromatin-promoting histone deacetylases and methylases (Tulin et al. 2003). Alternatively, PARP might modify or directly interact with silencing proteins. Finally, Parp might suppress copia transcription by a mechanism independent of PARP enzymatic activity. It is known that a catalytically inactive isoform of PARP is produced by differential splicing (Tulin et al. 2002).
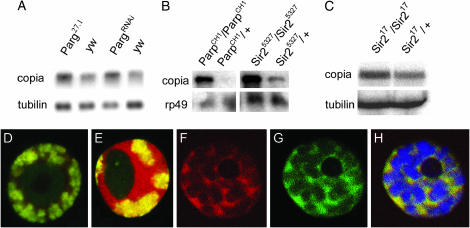
If Parp requires its ADP-ribosylation activity to silence copia, then reducing PARG should shift PARP to the inactive state and upregulate copia levels. This is what we observed. Parg27.1 and PargRNAi larvae contain significantly elevated levels of copia transcripts (Figure 5A). These experiments suggest that ADP-ribose modification of a target protein(s) by PARP plays a critical role in suppressing copia. Parg mutation would act indirectly by affecting active PARP levels, but might also act directly by removing ADP-ribose moieties from silencing molecules. If so, however, the targets of such action remain unknown.
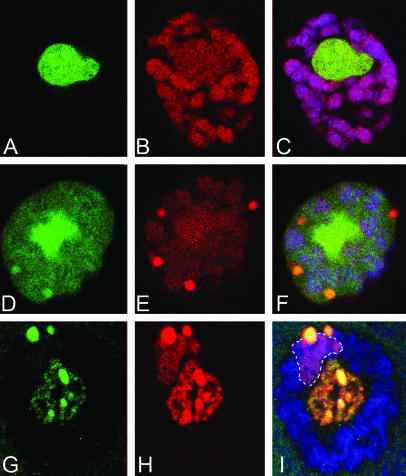
Figure 5.
PARG and SIR2 colocalize and are required for copia silencing. (A) RNA from larvae of the indicated genotypes is shown after probing with copia and control sequences. The 5.5-kb copia primary transcript is elevated in RNA from Parg27.1 and PargRNAi animals. (B) A similar experiment to A, revealing that copia RNA is greatly elevated in Sir25327 animals. ParpCH1 is shown for comparison. (C) Sir217 mutant animals also show elevated levels of copia RNA. (D) An L3 salivary gland cell expressing Sir2-dsRed from a 69B GAL4 driver and costained for DNA (green). Sir2-dsRed is localized in the nucleoplasm. (E) An L3 salivary gland cell expressing Sir2-dsRed from a da-GAL4 driver, costained for DNA (green). Sir2-DsRed levels are elevated, chromatin is overcondensed, and nucleolar size is reduced. (F–H) Colocalization of PARG-EGFP and Sir2-DsRed in the nucleoplasm: PARG-EGFP, green; Sir2-DsRed, red; DNA, blue.
Sir2 is required to suppress copia transcription and to promote histone deacetylation:
To identify potential PARP and PARG target proteins involved in gene silencing, we examined whether strains with mutations in candidate silencing genes contain elevated copia RNA levels. We found that mutations in the Drosophila Sir2 gene elevate copia transcript levels, as in Parp or Parg mutations (Figure 5, B and C). This effect is seen in two different Sir2 alleles (Figure 5, B and C); hence it is the result of reductions in Sir2 function and not due to background mutations (Astrom et al. 2003). To further investigate the role of SIR2 in copia silencing, we constructed flies that express an epitope-tagged SIR2 (Tulin et al. 2002). Expressing SIR2-DsRed from the strong da-Gal4 driver causes chromatin compaction, reduces nucleolar size (Figure 5, D and E), and induces variegation (data not shown). When expressed at moderate levels using a weakly expressed 69B GAL4 driver, SIR2-dsRed is localized in the nucleoplasm (Figure 5F) in a pattern indistinguishable from that of PARG (compare Figure 5, G and H). Consequently, SIR2 represents a candidate target of PARG in facilitating copia silencing.
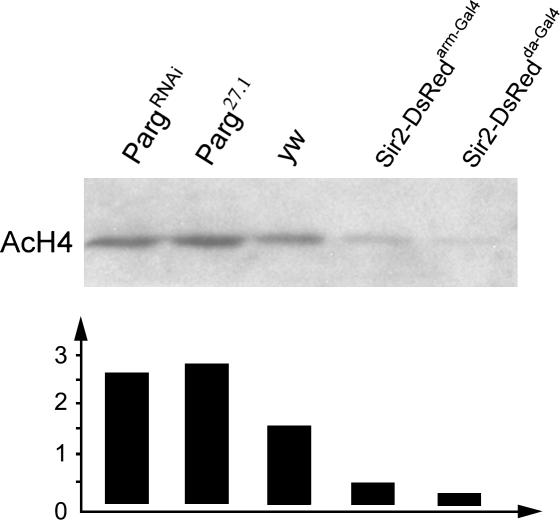
SIR2 proteins, including Drosophila SIR2, exhibit an NAD-dependent histone deacetylase activity. We used specific antibodies that recognize an acetylated form of H4 to see if the general levels of this modification are affected under conditions where copia expression is increased. Relative to control animals, Western blots probed with the modification-specific antibodies showed a strong increase in the levels of H4 modification in Parg27.1 and PargRNAi mutants (Figure 6), which overexpress copia. In contrast, the levels of modification were reduced in flies expressing elevated levels of SIR2-DsRed (Figure 6).
Figure 6.
Parg mutations and SIR2 overexpression affect levels of histone H4 acetylation. Western blot of proteins from Parg27.1, PargRNAi, wild type (wt), and SIR2-DsRed-overexpressing animals were probed with antibodies specific for the hyperacetylated form of histone H4. The amounts of material loaded onto the gel were normalized to the amount of DNA in the samples. Quantitation of the relative band intensities (arbitrary units) shows that the level of histone H4 hyperacetylation is elevated in the Parg mutant genotypes and diminished in animals overexpressing Sir2.
PARG levels affect SIR2 localization and modification:
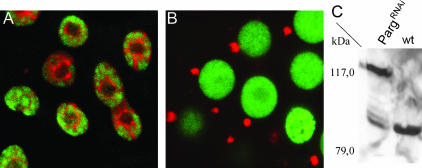
To investigate whether SIR2 requires PARG for its normal function, we examined the distribution of SIR2-dsRed in the PargRNAi strain. The localization of SIR2 protein was dramatically altered in many but not all cells (due to some variegation of the GAL4/UAS constructs). SIR2 protein left the nucleus and accumulated only within a single, discrete round structure in the perinuclear cytoplasm (Figure 7, A and B). Thus, reducing PARG levels and poly(ADP-ribose) cycling is correlated with and may directly alter SIR2 localization.
Figure 7.
Parg disruption affects SIR2 localization and modification. L3 salivary gland cell from wild type (A) and from UAS-PargRNAi; hsGAL4; UAS-Sir2-dsRed (B). Strong reduction of Parg activity causes SIR2 to leave the nucleus and accumulate in a small peri-nuclear structure. DNA, green. (C) Western blot of proteins from hs-GAL4; UAS-Sir2-dsRed; UAS-PargRNAi (PargRNAi) and hs-GAL4; UAS-Sir2-dsRed flies (wt) flies that was probed with anti-dsRed antibodies. A 79-kDa band corresponding to unmodified SIR2-dsRed is seen in wild type. In the PargRNAi flies, higher-molecular-weight bands are apparent, including a prominent band at 117 kDa.
The simplest way to explain this result would be that SIR2 is a direct target of an ADP-ribose modification that controls SIR2 nuclear localization. To look for evidence of ADP ribosylation of Drosophila SIR2, we carried out Western blots of SIR2-dsRed extracts in wild-type flies where it is localized normally and in PargRNAi flies where much of SIR2 is cytoplasmic. In wild type we observed a single prominent band at 79 kDa, the expected molecular weight of SIR2-dsRed (Figure 7C). In contrast, in extracts from PargRNAi flies the intensity of the 79-kDa band was reduced. A faint “ladder” of additional bands migrating more slowly was observed (Figure 7C), including a prominent band located at a nominal molecular weight of 117 kDa. These results suggest that, in the absence of PARG activity, modified forms of SIR2 protein accumulate in the cell. Although these experiments were not sufficient to definitively determine the nature of the PARG-dependent SIR2 modification, our results are consistent ADP-ribose moieties. These experiments suggest that SIR2 localization and/or activity are regulated by a cycle of ADP-ribose modification and that the effects of Parp and Parg mutations on copia silencing may be due in whole or in part to disruption of this pathway.
DISCUSSION
PARG and PARP act in common nuclear pathways:
A previous study (Hanai et al. 2004) found that Parg mutations were lethal or semilethal and reported an effect of mutations on pADPr levels in neural tissue and on organismal life span. However, the relationship between PARG and the roles played by PARP (Tulin et al. 2002; Tulin and Spradling 2003) remained unclear. Here we have shown that reducing or increasing PARG causes phenotypic effects very similar to disrupting Parp on nucleolar function, chromatin structure, immunity, and heat-shock sensitivity. This shows that PARG acts in many common pathways with PARP, presumably by virtue of its enzymatic action on ADP-ribose groups.
The phenotypic similarity that we observed among Parg disruption, PARG overproduction, and Parp mutation argues that protein ADP ribosylation, rather than a direct structural function, underlies many of the reported actions of PARP. Molecular studies of Parg mutants directly verified the predicted increase in ADP-ribose modification levels. In the absence of PARG, newly synthesized PARP molecules would still be expected to function until they became automodified. Thus, the strong phenotypic effects of Parg mutation imply that recycling of automodified PARP molecules is quantitatively important, at least locally. For example, the developmental delays that we observed prior to each molt are probably caused by the extra time needed to synthesize de novo enough new PARP to support molting gene expression. PARG overproduction would also be expected to interfere with PARP action. Poly(ADP-ribose) chains on automodified PARP might remain too short to function, and protein recycling that depends on the kinetics of poly(ADP-ribose) modification might be disrupted. In support of these interpretations, we found that the phenotypic effects of PARG overproduction are completely suppressed by simultaneously producing extra PARP.
Consequently, our studies strongly support the view that localized episodes of poly(ADP)-ribose modification under the control of PARP, and recycling of ADP-ribose-modified proteins under the control of PARG, play a major role in controlling chromatin structure, gene activity and nuclear function in vivo (Figure 1). However, it still remains unclear how PARP is incorporated into chromatin in an inactive state, how it becomes locally activated (except in the case of DNA damage), and to what extent other chromatin proteins in addition to PARP itself are important substrates for PARP and PARG enzymatic activities in vivo. Moreover, our work does not rule out the possibility that PARP also acts via other mechanisms, including some that do not require enzymatic function. Drosophila oocytes and early embryos contain an essential isoform, PARP-e, that lacks a catalytic domain (Tulin et al. 2002). In vitro, human PARP-1 represses chromatin by binding to nucleosomes, displacing histone H1, and compacting its local architecture independently of PARP enzymatic activity (Kim et al. 2004). In addition, PARP may form a stable component of repressive chromatin complexes on target genes (Ju et al. 2004).
Parg facilitates multiple nuclear events that require ADP-ribose modification:
The phenotype of Parg disruption is not identical to Parp mutation, suggesting that PARG carries out some functions independently of PARP. In particular, PARP protein and FIBRILLARIN accumulate in large nucleoplasmic and peri-centromeric bodies in Parg mutant cells (Figure 4, D–I), in contrast to the FIBRILLARIN-rich cytoplasmic body seen in Parp mutants (Figure 3D). We suggest that these structures correspond to intermediates in a process that normally recycles ADP-ribose-modified proteins within the cell. In this respect, they are reminiscent of Cajal bodies, which have been postulated to serve as staging, storage, or assembly sites of factors involved in transcript production and processing (reviewed by Gall 2003). The normal rate of this recycling may be greater in regions of high gene activity. When ADP-ribose groups cannot be removed, the recycling process backs up, causing the observed breakdown of the nucleolus and loss of PARP from eukaryotic chromosome regions. The nucleolus might be particularly sensitive if ongoing ADP ribosylation is needed to maintain rDNA genes, which do not exhibit a normal nucleosomal organization, in an active state (see Dammann et al. 1993). The phenotypic differences between Parp and Parg mutants may result from different arrest points within a common recycling pathway or because PARG also reverses the action of other poly(ADP-ribose) polymerases in addition to PARP.
ADP-ribose metabolism and SIR2-dependent silencing:
PARP activation or PARG reduction might block SIR2 action simply by depleting cellular NAD pools (Zhang 2003). However, PARG overexpression should not reduce NAD pools, and yet nucleolar structure was disrupted in animals with elevated PARG. Instead of acting via NAD, our observation that, in the absence of PARG, a higher-molecular-weight form of SIR2 accumulates in the cell cytoplasm suggests that SIR2 is modified by an ADP-ribose addition as part of its function. Many SIR2 protein family members themselves exhibit protein ADP-ribosylation activity (Frye 1999; Furuyama et al. 2004), and mouse SIRT6, a predominantly nuclear protein, can direct its own mono-(ADP) ribosylation (Liszt et al. 2005). PARG may be needed to remove ADP-ribose groups from SIR2 that are added by these or other mechanisms that are independent of PARP.
Taken together, our experiments suggest a model in which PARP, PARG, and SIR2 cooperate to silence specific chromosomal domains. We propose that activation and ADP ribosylation of PARP molecules (and possibly other local chromatin proteins) loosen chromatin early in the silencing process and that this facilitates SIR2 access to acetylated histone tails. In some cases this process would transiently strip the target chromatin proteins off the affected region and transfer them in an organized fashion to the branched ADP-ribose polymers on auto-inactivated PARP molecules within the immediately adjacent nucleoplasm. Here the ADPr/chromatin complex would encounter PARG and SIR2, possibly in conjunction with other proteins involved in chromatin remodeling (Furuyama et al. 2004). SIR2 molecules would undergo autoADP-ribosylation and deacetylate histones such as H4 within the complex, while PARG begins to cleave their ADP-ribose moieties. As the ADPr tails shorten, the chromatin proteins would be driven to reassemble onto their former chromosome region. PARG action on SIR2 might also help coordinate these events.
When Parp is mutated or when PARG activity becomes too high or too low, chromatin activation and silencing would be drastically disrupted. Without PARP or in the presence of excess PARG, chromatin proteins would fail to loosen and become accessible to modification. The state of chromatin would become “frozen” at whatever state it had reached when the deficiency became acute (i.e., when maternal PARP is depleted in the case of a zygotic Parp mutant, or when expressed PARG reaches a critical level). When PARG levels are too low, in contrast, excess levels of ADP-ribose-modified SIR2 would build up, driving it into the cytoplasm. Following a single activation, PARP molecules would be trapped in the inactive automodified state. Large amounts of pADPr would accumulate, shunting chromatin proteins into remodeling complexes that cannot break down. We can now look forward to obtaining a more detailed understanding of these events using the genetic tools available for the study of chromatin organization in Drosophila.
Acknowledgments
We thank J. Gall, P. Harte, A. Veraksa, and G. Cavalli for providing materials.
References
- Adams, M. D., S. E. Celniker, R. A. Holt, C. A. Evans, J. D. Gocayne et al., 2000. The genome sequence of Drosophila melanogaster. Science 287: 2185–2195. [DOI] [PubMed] [Google Scholar]
- Astrom, S. U., T. W. Cline and J. Rine, 2003. The Drosophila melanogaster Sir2(+) gene is nonessential and has only minor effects on position-effect variegation. Genetics 163: 931–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow, A. L., C. M. van Drunen, C. A. Johnson, S. Tweedie, A. Bird et al., 2001. dSIR2 and dHDAC6: two novel, inhibitor-resistant deacetylases in Drosophila melanogaster. Exp. Cell Res. 265: 90–103. [DOI] [PubMed] [Google Scholar]
- Casso, D., F. Ramirez-Weber and T. B. Kornberg, 2000. GFP-tagged balancer chromosomes for Drosophila melanogaster. Mech. Dev. 91: 451–454. [DOI] [PubMed] [Google Scholar]
- Dammann, R., R. Lucchini, T. Koller and J. M. Sogo, 1993. Chromatin structures and transcription of rDNA in yeast Saccharomyces cerevisiae. Nucleic Acids Res. 10: 2331–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Murcia, G., and S. Shall, 2000. From DNA Damage and Stress Signaling to Cell Death: Poly(ADP-Ribosylation) Reactions. Oxford University Press, New York.
- Denu, J. M., 2003. Linking chromatin function with metabolic networks: Sir2 family of NAD(+)-dependent deacetylases. Trends Biochem. Sci. 28: 41–48. [DOI] [PubMed] [Google Scholar]
- Fischle, W., Y. Wang and C. D. Allis, 2003. Histone and chromatin cross-talk. Curr. Opin. Cell Biol. 15: 172–183. [DOI] [PubMed] [Google Scholar]
- FlyBase, 1999. The FlyBase database of the Drosophila genome projects and community literature. Nucleic Acids Res. 27: 85–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye, R. A., 1999. Characterization of five human cDNAs with homology to the yeast SIR2 gene: Sir2-like proteins (sirtuins) metabolize NAD and may have protein ADP-ribosyltransferase activity. Biochem. Biophys. Res. Commun. 260: 273–279. [DOI] [PubMed] [Google Scholar]
- Furuyama, T., R. Banerjee, T. R. Breen and P. J. Harte, 2004. SIR2 is required for Polycomb silencing and is associated with an E(Z) histone methyltransferase complex. Curr. Biol. 14: 1812–1821. [DOI] [PubMed] [Google Scholar]
- Gall, J. G., 2003. The centennial of the Cajal body. Nat. Rev. Mol. Cell Biol. 4: 975–980. [DOI] [PubMed] [Google Scholar]
- Gerasimova, T., and V. Corces, 2001. Chromatin insulators and boundaries: effects on transcription and nuclear organization. Annu. Rev. Genet. 35: 193–208. [DOI] [PubMed] [Google Scholar]
- Grieder, N. C., M. de Cuevas and A. C. Spradling, 2000. The fusome organizes the microtubule network during oocyte differentiation in Drosophila. Development 127: 4253–4264. [DOI] [PubMed] [Google Scholar]
- Hanai, M., M. Uchida, S. Kobayashi, M. Miwa and K. Uchida, 1998. Genomic organization of Drosophila poly(ADP-ribose) polymerase and distribution of its mRNA during development. J. Biol. Chem. 273: 11881–11886. [DOI] [PubMed] [Google Scholar]
- Hanai, S., M. Kanai, M. Ohashi, M. Okamoto, M. Yamada et al., 2004. Loss of poly(ADP-ribose) glycohydrolase causes progressive neurodegeneration in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 101: 82–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju, B.-J., S. Solum, E. J. Song, K.-J. Lee, K. W. Rose et al., 2004. Activating the PARP-1 sensor component of the Groucho/TLE1 corepressor complex mediates a CaMKinase II-dependent neurogenic gene activation pathway. Cell 119: 815–829. [DOI] [PubMed] [Google Scholar]
- Karpen, G., and A. C. Spradling, 1992. Analysis of subtelomeric heterochromatin in the Drosophila minichromosome Dp1187 by single P element insertional mutagenesis. Genetics 132: 737–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamitsu, H., H.-O. Hoshino, H. Okada, M. Miwa, H. Momoi et al., 1984. Monoclonal antibodies to poly(adenosine diphosphate ribose) recognize different structures. Biochemistry 23: 3771–3777. [DOI] [PubMed] [Google Scholar]
- Kim, M. Y., S. Mauro, N. Gévry, J. T. Lis and W. L. Kraus, 2004. NAD+-dependent modulation of chromatin structure and transcription by nucleosome binding properties of PARP-1. Cell 119: 803–814. [DOI] [PubMed] [Google Scholar]
- Landry, J., A. Sutton, S. T. Tafrov, R. C. Heller, J. Stebbins et al., 2000. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc. Natl. Acad. Sci. USA 97: 5807–5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liszt, G., E. Ford, M. Kurtev and L. Guarente, 2005. Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. J. Biol. Chem. 280: 21313–21320. [DOI] [PubMed] [Google Scholar]
- Manseau, L., A. Baradaran, D. Brower, A. Budhu, F. Elefant et al., 1997. GAL4 enhancer traps expressed in the embryo, larval brain, imaginal discs, and ovary of Drosophila. Dev. Dyn. 209: 310–322. [DOI] [PubMed] [Google Scholar]
- Newman, B. L., J. R. Lundblad, Y. Chen and S. M. Smolik, 2002. A Drosophila homologue of Sir2 modifies position-effect variegation but does not affect life span. Genetics 162: 1675–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando, V., 2003. Polycomb, epigenomes, and control of cell identity. Cell 112: 599–606. [DOI] [PubMed] [Google Scholar]
- Prokhorova, A. V., M. A. Voloshina, N. G. Shostak, V. E. Barskii and M. D. Golubovskii, 1994. Preparation and primary genetic analysis of Drosophila melanogaster transformants line w′lz(b)/XXywf, containing mini-white genes, integrated in the genome during P-element-dependent transformation. Genetika 30: 874–878. [PubMed] [Google Scholar]
- Rosenberg, M. I., and S. M. Parkhurst, 2002. Drosophila Sir2 is required for heterochromatic silencing and by euchromatic Hairy/E(Spl) bHLH repressors in segmentation and sex determination. Cell 109: 447–458. [DOI] [PubMed] [Google Scholar]
- Spradling, A. C., and G. M. Rubin, 1982. Transposition of cloned P elements into Drosophila germ line chromosomes. Science 218: 341–347. [DOI] [PubMed] [Google Scholar]
- Stapleton, W., S. Das and B. D. McKee, 2001. A role of the Drosophila homeless gene in repression of Stellate in male meiosis. Chromosoma 110: 228–240. [DOI] [PubMed] [Google Scholar]
- Tulin, A., and A. Spradling, 2003. Chromatin loosening by poly(ADP-ribose) polymerase (PARP) at Drosophila puff loci. Science 299: 560–562. [DOI] [PubMed] [Google Scholar]
- Tulin, A., D. Stewart and A. C. Spradling, 2002. The Drosophila heterochromatic gene encoding poly(ADP-ribose) polymerase (PARP) is required to modulate chromatin structure during development. Genes Dev. 16: 2108–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulin, A., Y. Chinenov and A. C. Spradling, 2003. Regulation of chromatin structure and gene activity by poly(ADP-ribose) polymerases. Curr. Top. Dev. Biol. 56: 55–83. [DOI] [PubMed] [Google Scholar]
- Zhang, J., 2003. Are poly(ADP-ribosylation) by PARP-1 and deacetylation by Sir2 linked? BioEssays 25: 808–814. [DOI] [PubMed] [Google Scholar]
- Zhang, P., and A. C. Spradling, 1994. Insertional mutagenesis of Drosophila heterochromatin with single P elements. Proc. Natl. Acad. Sci. USA 91: 3539–3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler, M., and S. L. Oei, 2001. A cellular survival switch: poly(ADP-ribosyl)ation stimulates DNA repair and silences transcription. BioEssays 23: 543–548. [DOI] [PubMed] [Google Scholar]