Abstract
High mutation rate in mammalian mitochondrial DNA generates a highly divergent pool of alleles even within species that have dispersed and expanded in size recently. Phylogenetic analysis of 277 human mitochondrial genomes revealed a significant (P < 0.01) excess of rRNA and nonsynonymous base substitutions among hotspots of recurrent mutation. Most hotspots involved transitions from guanine to adenine that, with thymine-to-cytosine transitions, illustrate the asymmetric bias in codon usage at synonymous sites on the heavy-strand DNA. The mitochondrion-encoded tRNAThr varied significantly more than any other tRNA gene. Threonine and valine codons were involved in 259 of the 414 amino acid replacements observed. The ratio of nonsynonymous changes from and to threonine and valine differed significantly (P = 0.003) between populations with neutral (22/58) and populations with significantly negative Tajima's D values (70/76), independent of their geographic location. In contrast to a recent suggestion that the excess of nonsilent mutations is characteristic of Arctic populations, implying their role in cold adaptation, we demonstrate that the surplus of nonsynonymous mutations is a general feature of the young branches of the phylogenetic tree, affecting also those that are found only in Africa. We introduce a new calibration method of the mutation rate of synonymous transitions to estimate the coalescent times of mtDNA haplogroups.
MITOCHONDRIAL DNA (mtDNA) encodes for 13 proteins, two ribosomal genes, and 22 tRNAs that are essential in the energy production of the human cell. Variation in the sequence of mtDNA has provided significant insights into the maternal history of anatomically modern humans (Giles et al. 1980; Denaro et al. 1981), complementing the paternal legacy of the Y chromosome (Underhill et al. 2000). Studies based on restriction fragment length polymorphism (RFLP) of the coding and direct sequencing of the noncoding control region have formed the basis of a hierarchical classification of distinct geographic and ethnic affinities (Torroni et al. 1993, 1996; Chen et al. 1995; Watson et al. 1997; Macaulay et al. 1999; Forster et al. 2001). Studies addressing sequence variation in the mtDNA coding region have suggested that natural selection has significantly shaped the course of human mtDNA evolution (Cann et al. 1984; Nachman et al. 1996; Ingman and Gyllensten 2001; Mishmar et al. 2003; Moilanen et al. 2003; Moilanen and Majamaa 2003; Elson et al. 2004; Ruiz-Pesini et al. 2004). These studies have disagreed, however, upon whether the distribution of specific human mtDNA clades or haplogroups is due to an adaptation to different climates or if their distribution is a function of random genetic drift assisted by purifying selection that eliminates nonsynonymous changes. In an attempt to clarify this disagreement and to study the mode of natural selection in mtDNA variation in human populations, we provide here a phylogenetic analysis of a global sample of mtDNAs and investigate the position, chemical nature, and geographic distribution of recurrent and frequent mutations in the coding region.
MATERIALS AND METHODS
DNA samples:
The ascertainment set comprised 277 individuals from the five continents, including genomic DNA samples from 129 Africans (10 Biaka Pygmy, 15 Mbuti Pygmy, two Lisongo, six San, two Mandenka, four Ethiopian Jews, nine Sudanese, one Eritrean, one Ghanan, three Herero, one Ovambo, one Pedi, one Sotho, two Tswana, two Zulu, 10 Fulbe, 10 Mossi, 10 Rimaibe, one Berta, one Tuareg, 37 Dominicans), 43 Asians (one Arab, one Kazak, one Druze, four Bedouin, one Sepharadim, one Yemenite Jew, two Pathan, five Sindhi, two Burushaski, one Baluchi, one Brahui, two Makran, two Hazara, one Tamil, two Cambodians, one Hmong, one Atayal, one Ami, four Han Chinese, five Japanese, four Koreans), 76 Europeans (five Northern Europeans, 12 Italians, one Greek, two Finns, two Ashkenazi, one Georgian, 17 Hungarians, three Icelanders, three Czechs, one Sardinian, five Basque, one Iberian, 23 Dutch), 13 Oceanians (four New Guineans, three Melanesians, six Australian Aborigines), and 16 Native Americans (one Auca, one Guarani, five Brazilian Indians, three Colombian Indians, two Mayan, one Piman, one Muskogee, one Navaho, one Quechua) (supplemental Table 1 at http://www.genetics.org/supplemental/). A subset of 103 sequences from these populations has been reported elsewhere (Shen et al. 2005). DNA was extracted using the QIAamp DNA blood kit (QIAGEN, Valencia, CA). Immortalized cell lines have been established for all individuals with the exception of the 17 Hungarians, the 23 Dutch, the 37 Dominicans, the 10 Mossi, the 10 Rimaibe, and the 10 Fulbe.
PCR and DNA sequencing:
The 41 primer pairs used for bidirectional sequencing of mtDNA nucleotides 435–16,023, the PCR conditions, and the determined complete coding region sequence information for 277 individual samples are available at http://insertion.stanford.edu/primers_mitogenome.html. Amplicons were purified with QIAGEN QIAquick spin columns and sequenced with the Applied Biosystems (Foster City, CA) Dye Terminator Cycle sequencing kit and a model 3700 DNA sequencer.
Phylogenetic and statistical analyses:
An unrooted tree from a median-joining network (Bandelt et al. 1999) was drawn and labeled following existing mtDNA haplogroup nomenclature (Torroni et al. 1996, 2001; Macaulay et al. 1999; Kivisild et al. 2002, 2004; Salas et al. 2002; Yao et al. 2002; Kong et al. 2003, 2004; Shen et al. 2005). The tree was rooted using nuclear inserts of mtDNA retrieved from human genomic sequence and the consensus sequence of the three chimpanzee mitochondrial genomes. The accession numbers, mtDNA positional range, and identity (ID) measures of the genomic contigs containing the inserts that were used for rooting are as follows: NT_006713.14 (bp 341–2697; ID 94%); NT_009237.17 (bp 521–2976; ID 94%); NT_006316.15 (bp 2899–3050; ID 94%); NT_077913.3 (bp 3914–9756; ID 98%); and NT_034772.5 (bp 10,269–15,487; ID 94%). The assembled sequence of the inserts is available at http://insertion.stanford.edu/mtDNA.html. The GenBank accession numbers of the two Pan troglodytes and one Pan paniscus sequences that were used are D38113, X93335, and D38116, respectively. Haplogroup divergence estimates ρ and their error ranges were calculated as averages of the distances from the tips to the most recent common ancestor of the haplogroup (Forster et al. 1996; Saillard et al. 2000). Two separate measures of nonsynonymous (N) to synonymous (S) substitution ratios were used: first, the MN/MS ratio estimates the number of mutational changes inferred from the phylogenetic tree (Figure 1), and second, the dN/(dS + constant) refers as in Mishmar et al. (2003) to the ratio of the average pairwise distances of N and S changes in the given sample. Statistical significance was determined from binomial or χ2 probabilities. Disease-implicated substitutions were excluded from these analyses. For interspecies comparisons, mammalian mtDNA sequences were retrieved from the Mitochondriome website (http://bighost.area.ba.cnr.it/mitochondriome/Mt_chordata.htm).
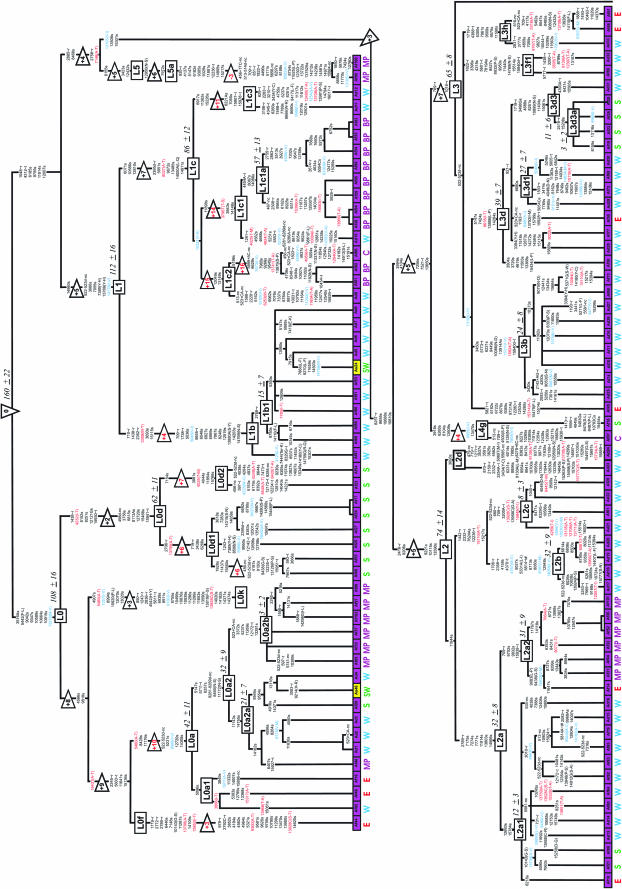
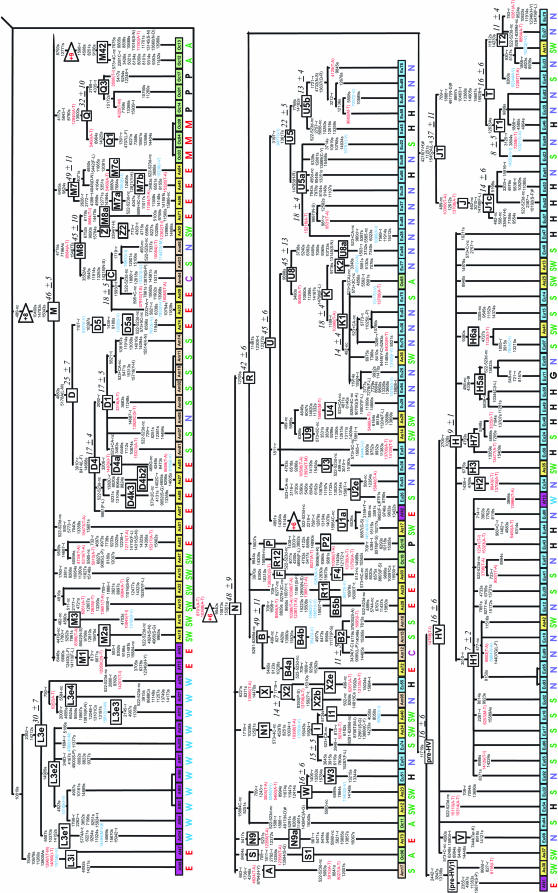
Figure 1.
Maximum parsimony tree of 277 human mtDNA coding region sequences. Locations of branch-defining mutations are listed relative to the revised reference sequence (Andrews et al. 1999). Transversions are specified in capital letters, del indicates deletion, and + indicates insertion. Amino acid replacements are specified in parentheses with threonine- and valine-affecting changes highlighted in red and blue, respectively. ∼t, change in tRNA; ∼r, change in rRNA gene. Haplogroup labels follow existing classifications (Chen et al. 1995; Torroni et al. 1993, 1996, 2001; Watson et al. 1997; Macaulay et al. 1999; Bandelt et al. 2001; Forster et al. 2001; Kivisild et al. 2002, 2004; Kong et al. 2003; Achilli et al. 2004; Palanichamy et al. 2004; Shen et al. 2004; Tanaka et al. 2004; Friedlaender et al. 2005). Substitutions at np 709, 3693, and 4715 in haplogroups B4, L2, and Z, respectively, are reconstructed less parsimoniously, yet in accordance with the phylogenetic relations suggested by additional data (Finnilä et al. 2001; Torroni et al. 2001; Kivisild et al. 2002; Kong et al. 2003). Coalescent estimates of haplogroups, shown in thousands of years in italics next to clade labels, are based on the average number of synonymous transitions to the root of the clade. The tree was rooted using as outgroup the nuclear inserts of mtDNA and the majority consensus of complete sequences of two P. troglodytes and one P. paniscus (for details see materials and methods). The numbers in triangles indicate the additional mutational steps required in comparison to the optimally rooted tree using only the chimpanzee outgroup with red type indicating the root that was rejected (P < 0.01) under the assumption of equal evolutionary rate over branches. The positions at which the derived character states in humans and the chimpanzee consensus match are shown above the triangles. Disease-implicated substitutions confirmed by two independent studies (http://www.mitomap.org) are indicated by “#.” Ad, Af, Am, As, Eu, and Oc correspond to African descent, African, Amerindian, Asian, European, and Oceanian, respectively. The continent of origin is distinguished for each individual sample by box color and subcontinental affiliation is indicated with letters below the boxes where N, E, W, S, C, and SW refer to north, east, west, south, central, and southwest parts of the continents, respectively. BP, Biaka Pygmies; MP, Mbuti Pygmies; A, Australian; P, Papuan; M, Melanesian; H, Hungarian LHON patients; G, Georgian. The two African mtDNAs (L0a2 and L1b1) found in West Asia (Pakistan) are likely due to recent admixture as indicated by their low genetic distance from related African samples presented in the tree and by the fact that they were detected in a Makrani and a Sindhi. According to Quintana-Murci et al. (2004), the Makrani harbor an extremely high frequency (39%) of the African haplogroups L3d, L3b, L2a, and L1a, most likely as a result of the forced migration of slave women from Africa that began in the seventh century and increased considerably during the Omani Empire. European gene flow, on the other hand, may account for the presence of European K and X sequences in one Aboriginal Australian and in one Native American (Muskogee), respectively.
Tests for positive selection:
Seven primate taxa, namely Homo sapiens, P. troglodytes, Gorilla gorilla, Papio hamadryas, Hylobates lar, Pongo pygmaeus, and Macaca sylvanus were chosen from GenBank (gi|17981852, gi|5835121, gi|5835149, gi|5835638, gi|5835820, gi|5835163, gi|14010693) and aligned using clustalW (Thompson et al. 1994) to test for the historic occurrence of positive, directional selection on the 13 coding regions of the primate mitochondrion using the program codeml of the PAML package (Yang 2002). In these tests, maximum-likelihood ratios of nonsynonymous-to-synonymous mutations (ω) exceeding 1 are consistent with the hypothesis of positive selection, while values close to 1 indicate selective neutrality, and values converging on 0 suggest strong purifying selection. We conducted both lineage and site-specific tests. For the lineage-specific tests, we used a model in which all lineages have the same ω (hereafter referred to as M0) and compared that with a model in which ω is estimated for each lineage (hereafter referred to as M1). To test for the action of selection among amino acid sites within a specific lineage, we compared a model that allows for heterogeneity in ω among sites, but not among lineages, with a model that allows for variation in ω along a predefined lineage (as in Yang and Nielsen 2002). We assumed the following unrooted phylogeny (troglodytes, ((((macaca, papio), hylobates), pongo), gorilla), troglodytes), human). However, results of our analyses were robust to minor fluctuations in the tree.
RESULTS
The deepest splits of the phylogeny constructed from 277 mtDNA complete coding region sequences (Figure 1) were sustained by African mtDNAs, which belonged to previously defined haplogroups L0–L5 (Torroni et al. 2001; Salas et al. 2002; Mishmar et al. 2003; Kivisild et al. 2004; Shen et al. 2005). A number of new subclades were identified among these (see Figure 1. Haplogroup sharing between distinct geographic regions was generally low. All European sequences could be assigned to clades N1, X, W, HV, TJ, and U (Torroni et al. 1996; Macaulay et al. 1999; Finnilä et al. 2001; Herrnstadt et al. 2002). Asian, Amerindian, Oceanian, and Australian Aborigine sequences belonged to region-specific haplogroups nested within macro-clades M and N (Kivisild et al. 2002; Yao et al. 2002; Kong et al. 2003, 2004; Friedlaender et al. 2005). All Australian Aborigine M sequences (two from this study and one from Ingman et al. (2000) share six mutations that define the new haplogroup M42. The majority of Australian N and R sequences (Ingman and Gyllensten 2003) belong to clades S and P defined by transitions at nucleotide positions (np) 8404 and 15607, respectively (Forster et al. 2001; Friedlaender et al. 2005).
The most parsimonious root of the mtDNA tree using nuclear inserts of mtDNA and the chimpanzee consensus sequence as outgroups appeared between haplogroup L0 and the rest of the phylogeny (Figure 1). Extensive interspecies homoplasy and mutational saturation was highlighted by the fact that for more than one-third (417/1292) of the variable sites, regardless of their phylogenetic position on the tree, the derived allele among humans corresponded to the chimpanzee allele. In agreement with noncoding region information (Aquadro and Greenberg 1983), a high ratio (21.5 on average, 34.8 in synonymous positions) of transitions to transversions was observed in the coding region (577–16023).
Interspecies calibration of the molecular clock over the complete mtDNA sequence (Ingman et al. 2000; Mishmar et al. 2003) is problematic because of saturation of transitions at silent positions and the effect of selection on the fixation rate of amino acid replacement mutations (Ho et al. 2005). Assuming 6 million years for the human–chimp species split (Goodman et al. 1998) and 6.5 million years for the most recent common ancestor of their mtDNA lineages (Mishmar et al. 2003), we estimated the average transversion rate at synonymous and rRNA positions as 2.1 × 10−9 and 4.1 × 10−10/year/position, respectively. Using the observed relative rates of different substitution types in humans (Table 1), the average transition rate at 4212 synonymous positions is 3.5 × 10−8 (SD 0.1 × 10−8)/year/position. Over all genes in mtDNA this would be equivalent to accumulation of one synonymous transition/6764 (SD 140) years on average. The coalescent date of the human mitochondrial DNA tree using this rate is 160,000 (SD 22,000) years. This coalescent date is broadly consistent with the dates of the Homo sapiens fossils recognized so far from Ethiopia (Clark et al. 2003; White et al. 2003; McDougall et al. 2005). The most recent common ancestor of all the Eurasian, American, Australian, Papua New Guinean, and African lineages in clade L3 dates to 65,000 ± 8000 years while the average coalescent time of the three basic non-African founding haplogroups M, N, and R is 45,000 years. These estimates, bracketing the time period for the recent out-of-Africa migration (Stringer and Andrews 1988), are younger than those based on calibrations involving all coding region sites (Ingman et al. 2000; Mishmar et al. 2003) but are still in agreement with the earliest archaeological signs of anatomically modern humans outside Africa (Mellars 2004). The differences between the date estimates of previous studies are most likely due to the overrepresentation of possibly slightly deleterious nonsynonymous mutations in the younger branches of the tree (Elson et al. 2004) that introduces a bias to the coalescent approach if all the sites of the coding region are used.
TABLE 1.
Distribution of mtDNA mutations by recurrence
| Nonsynonymous | rRNA | tRNA | Synonymous | |
|---|---|---|---|---|
| Length in base pairs | 8812 | 2513 | 1486 | 4212a |
| No. of observed substitutions (per site) | 413 (0.047) | 173 (0.069) | 110 (0.074) | 1037 (0.246) |
| Transition/transversion ratio | 12.4 | 23.7 | 12.8 | 34.8b |
| Invariable sites | 8506 | 2404 | 1409 | 3427 |
| Sites with single hit | 241 | 80 | 57 | 617 |
| Sites with one recurrent hit | 47 | 21 | 12 | 111 |
| Sites with two recurrent hits | 9 | 2 | 4 | 40 |
| Sites with three recurrent hits | 3 | 0 | 3 | 12 |
| Sites with four or more recurrent hits | 6 | 6 | 1 | 5 |
| No. of variable sites (proportion) | 306 (0.035) | 109 (0.043) | 77 (0.052) | 785 (0.186) |
Includes 2039 sites that are allowed to carry synonymous transversions.
Effectively, Ts/Tv = 16.8, when taking into account the number of sites that are allowed to vary.
Of the 1788 mutations depicted in the tree, 1758 occurred at 1292 variable sites in the coding region between np 577 and 16023. Consistent with previous reports (Mishmar et al. 2003; Moilanen and Majamaa 2003; Elson et al. 2004), there was a significant excess of synonymous mutations in all genes coded by mtDNA, especially among those positions that defined the deeper branches of the tree (Tables 2 and 3). In contrast to Ruiz-Pesini et al. (2004), we did not observe any significant regional (climatic) differences in the rate of nonsynonymous changes for mtDNA haplogroups. This discrepancy likely results from the fact that Ruiz-Pesini et al. (2004) compared region-specific haplogroups of different diversity levels: e.g., the “old” paragroup L in Africans vs. “young” Arctic haplogroups (Table 4). Populations of Asian, European, and West African origin showed significantly negative Tajima's D values (Table 3), consistent with selection, population growth, and/or population subdivision (Ray et al. 2003). That population substructure accounts at least for part of the deviation from neutrality is obvious from the observation that it decreases upon partitioning of West Africans into a sample from Burkina Faso and one from the Dominican Republican.
TABLE 2.
Distribution of mutations as a function of derived-allele frequency in 277 mtDNAs
| Frequency (%) | Nonsynonymous | Synonymous | Nonsynonymous/ synonymous | tRNA | rRNA | rRNA/ synonymous |
|---|---|---|---|---|---|---|
| <1 | 221 | 514 | 0.43 | 49 | 63 | 0.12 |
| 1–5 | 67 | 197 | 0.34 | 24 | 28 | 0.14 |
| 5–10 | 14 | 39 | 0.38 | 1 | 8 | 0.21 |
| >10 | 5 | 32 | 0.16* | 3 | 10 | 0.31* |
*P < 0.05.
TABLE 3.
Ratios of numbers and rates of nonsynonymous over synonymous sites, nucleotide diversity, Tajima's D, Fu and Li's D and F, and number of changes from and to valine and threonine for the 13 protein-coding mitochondrial genes according to continental affiliation
| Continent
|
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Africa | South Africa | West Africa | Burkina Fasoa | Dominican Republicb | Pygmy | East Africa | Non-African | Southwest Asia | Oceania | East Asia | America | Southern Europe | Northern Europe | Global | ||
| Gene | No. of samples: | 129 | 16 | 70 | 33 | 37 | 25 | 15 | 148 | 23 | 12 | 18 | 15 | 20 | 38 | 277 |
| ND1 | ||||||||||||||||
| MN/MS | 13/46 | 3/14 | 11/26 | 5/15 | 8/20 | 1/9 | 1/11 | 15/39 | 3/8 | 2/6 | 2/9 | 4/4 | 2/5 | 3/8 | 19/74 | |
| dN/(dS+ const.) | 0.058 | 0.052 | 0.073 | 0.073 | 0.071 | 0.037 | 0.014 | 0.116 | 0.102 | 0.080 | 0.028 | 0.193 | 0.089 | 0.119 | 0.074 | |
| ND2 | ||||||||||||||||
| MN/MS | 17/42 | 2/10 | 13/25 | 6/12 | 11/20 | 3/10 | 3/13 | 19/53 | 8/10 | 4/5 | 5/16 | 3/6 | 1/8 | 4/16 | 30/84 | |
| dN/(dS+ const.) | 0.129 | 0.079 | 0.118 | 0.086 | 0.140 | 0.108 | 0.064 | 0.138c | 0.196 | 0.239 | 0.107 | 0.079 | 0.056 | 0.121 | 0.136 | |
| COI | ||||||||||||||||
| MN/MS | 11/58 | 3/14 | 9/37 | 4/15 | 8/33 | 2/19 | 3/15 | 10/60 | 2/17 | 0/4 | 0/11 | 1/7 | 1/9 | 5/15 | 18/100 | |
| dN/(dS+ const.) | 0.022 | 0.050 | 0.060 | 0.060 | 0.060 | 0.042 | 0.055 | 0.028c | 0.030 | 0 | 0 | 0.018 | 0.024 | 0.06 | 0.043 | |
| COII | ||||||||||||||||
| MN/MS | 7/31 | 0/10 | 4/16 | 1/8 | 3/15 | 3/9 | 0/5 | 6/27 | 0/5 | 1/4 | 2/5 | 2/2 | 1/3 | 0/9 | 10/49 | |
| dN/(dS+ const.) | 0.045 | 0 | 0.035 | 0.008 | 0.053 | 0.093 | 0 | 0.025c | 0 | 0.044 | 0.070 | 0.080 | 0.030 | 0 | 0.073 | |
| ATP8 | ||||||||||||||||
| MN/MS | 8/9 | 5/6 | 4/8 | 0/5 | 4/7 | 1/3 | 1/5 | 6/8 | 2/1 | 0/1 | 1/1 | 1/0 | 0/0 | 1/2 | 12/14 | |
| dN/(dS+ const.) | 0.050 | 0.102 | 0.049 | 0 | 0.014 | 0.039 | 0.020 | 0.061c | 0.055 | 0 | 0.101 | 0.168 | 0 | 0.008 | 0.054 | |
| ATP6 | ||||||||||||||||
| MN/MS | 25/23 | 5/4 | 15/16 | 9/5 | 10/13 | 4/9 | 7/3 | 23/23 | 8/6 | 3/7 | 7/5 | 4/0 | 2/2 | 6/6 | 42/39 | |
| dN/(dS+ const.) | 0.174 | 0.191 | 0.187 | 0.212 | 0.168 | 0.079 | 0.300 | 0.236c | 0.245 | 0.134 | 0.352 | 0.367 | 0.066 | 0.148 | 0.259 | |
| COIII | ||||||||||||||||
| MN/MS | 8/38 | 2/10 | 3/20 | 0/11 | 3/18 | 3/14 | 2/14 | 11/37 | 2/9 | 0/8 | 2/10 | 0/3 | 2/6 | 3/11 | 16/59 | |
| dN/(dS+ const.) | 0.024 | 0.055 | 0.014 | 0 | 0.025 | 0.027 | 0.019 | 0.062c | 0.033 | 0 | 0.037 | 0 | 0.057 | 0.124 | 0.037 | |
| ND3 | ||||||||||||||||
| MN/MS | 5/9 | 1/1 | 3/6 | 2/4 | 3/4 | 2/2 | 2/2 | 5/10 | 1/3 | 1/3 | 1/5 | 2/1 | 1/1 | 2/1 | 7/18 | |
| dN/(dS+ const.) | 0.118 | 0.037 | 0.075 | 0.058 | 0.088 | 0.197 | 0.103 | 0.109c | 0.087 | 0.055 | 0.045 | 0.103 | 0.045 | 0.108 | 0.157 | |
| ND4L | ||||||||||||||||
| MN/MS | 1/10 | 0/3 | 1/9 | 0/6 | 1/7 | 0/4 | 0/4 | 1/10 | 0/1 | 0/1 | 0/3 | 0/0 | 0/2 | 1/3 | 2/16 | |
| dN/(dS+ const.) | 0.004 | 0 | 0.009 | 0 | 0.015 | 0 | 0 | 0.005 | 0 | 0 | 0 | 0 | 0 | 0.018 | 0.005 | |
| ND4 | ||||||||||||||||
| MN/MS | 9/67 | 2/14 | 6/46 | 0/20 | 6/38 | 3/19 | 0/20 | 14/56 | 4/15 | 2/8 | 3/13 | 3/5 | 1/12 | 2/16 | 21/103 | |
| dN/(dS+ const.) | 0.023 | 0.030 | 0.015 | 0 | 0.026 | 0.034 | 0 | 0.029c | 0.049 | 0.041 | 0.037 | 0.101 | 0.027 | 0.016 | 0.024 | |
| ND5 | ||||||||||||||||
| MN/MS | 35/75 | 5/17 | 25/38 | 12/21 | 21/26 | 9/23 | 11/25 | 38/60 | 8/18 | 7/6 | 8/10 | 4/10 | 3/12 | 8/17 | 56/112 | |
| dN/(dS+ const.) | 0.181 | 0.154 | 0.193 | 0.161 | 0.220 | 0.106 | 0.220 | 0.098c | 0.094 | 0.171 | 0.135 | 0.084 | 0.046 | 0.086 | 0.145 | |
| ND6 | ||||||||||||||||
| MN/MS | 9/26 | 2/7 | 7/16 | 5/8 | 3/16 | 1/7 | 0/8 | 7/23 | 1/7 | 1/2 | 2/7 | 1/4 | 1/4 | 1/4 | 14/42 | |
| dN/(dS+ const.) | 0.059 | 0.060 | 0.061 | 0.079 | 0.047 | 0.054 | 0 | 0.033c | 0.014 | 0.048 | 0.067 | 0.064 | 0.031 | 0.015 | 0.047 | |
| Cytb | ||||||||||||||||
| MN/MS | 31/41 | 5/12 | 18/31 | 12/14 | 12/26 | 3/8 | 8/8 | 29/41 | 5/11 | 1/10 | 8/11 | 2/9 | 5/4 | 8/10 | 47/65 | |
| dN/(dS+ const.) | 0.145 | 0.080 | 0.163 | 0.213 | 0.126 | 0.111 | 0.193 | 0.199c | 0.138 | 0.035 | 0.127 | 0.054 | 0.240 | 0.351 | 0.166 | |
| No. of recurrent synonymous sites | 81 | 2 | 28 | 5 | 14 | 4 | 2 | 71 | 8 | 3 | 4 | 2 | 1 | 1 | 241 | |
| No. of recurrent nonsynonymous sites | 35 | 2 | 14 | 5 | 6 | 1 | 3 | 30 | 3 | 0 | 2 | 0 | 0 | 3 | 106 | |
| π (×10−3) | 3.79 | 3.59 | 3.22 | 2.69 | 3.68 | 4.26 | 3.16 | 1.85 | 1.78 | 1.93 | 1.99 | 1.66 | 1.01 | 1.41 | 3.06 | |
| ±SD | ±0.12 | ±0.23 | ±0.17 | ±0.18 | ±0.23 | ±0.16 | ±0.37 | ±0.08 | ±0.14 | ±0.23 | ±0.15 | ±0.19 | ±0.18 | ±0.09 | ±0.10 | |
| θ (×10−3) | 10.55 | 4.12 | 7.54 | 4.34 | 7.01 | 3.99 | 4.61 | 10.02 | 3.75 | 2.54 | 3.79 | 2.11 | 2.24 | 3.44 | 15.36 | |
| ±SD | ±2.45 | ±1.49 | ±1.96 | ±1.31 | ±2.08 | ±1.30 | ±1.69 | ±2.28 | ±1.25 | ±1.00 | ±1.34 | ±0.79 | ±0.78 | ±1.03 | ±3.14 | |
| Tajima's D | −2.139* | −0.552 | −2.014* | −1.455 | −1.806* | 0.265 | −1.385 | −2.681**** | −2.117* | −1.121 | −2.012* | −0.933 | −2.249** | −2.198** | −2.528**** | |
| Fu and Li's D | −4.203*** | −0.668 | −4.186*** | −2.524* | −2.744* | 0.963 | −1.015 | −8.572*** | −3.300*** | −1.407 | −2.847*** | −1.272 | −3.138*** | −4.440*** | −7.738*** | |
| Fu and Li's F | −3.883*** | −0.735 | −3.956*** | −2.560* | −2.871* | 0.870 | −1.291 | −6.894*** | −3.437*** | −1.519 | −3.024*** | −1.357 | −3.349*** | −4.339*** | −5.872*** | |
| To threonine and valine | 78 | 10 | 36 | 13 | 20 | 7 | 12 | 74 | 10 | 6 | 14 | 9 | 3 | 14 | 158 | |
| From threonine and valine | 47 | 2 | 34 | 10 | 22 | 2 | 3 | 52 | 9 | 2 | 13 | 3 | 5 | 11 | 99 | |
*P < 0.05; **P < 0.01; ***P < 0.02; ****P < 0.001.
Rimaibe (N = 10), Foulbe (10), Mossi (10), all from Burkina Faso, plus 2 Mandenka and 1 Ghanan (subset of West Africa).
Individuals of African descent from the Dominican Republic (subset of West Africa).
Significant (P < 0.0001) difference between African and non-African distribution of dN/(dS + constant) values.
TABLE 4.
The rate of nonsynonymous/synonymous changes in relation to continental geography and haplo-group diversity
| Clade | Geography | Diversity (ρ)a | MN | MS | MN/ MS | SD |
|---|---|---|---|---|---|---|
| L1c | West and Central Africa | 12.7 | 25 | 65 | 0.38 | |
| L0d | South Africa | 9.1 | 15 | 42 | 0.36 | |
| M7 | Southeast Asia | 7.3 | 8 | 21 | 0.38 | |
| M8(CZ) | Northeast Asia | 6.7 | 12 | 31 | 0.39 | |
| U | Europe | 6.6 | 40 | 111 | 0.36 | |
| “Old” haplogroups | >6 | 0.37 | 0.01 | |||
| K | Europe | 2.6 | 10 | 23 | 0.43 | |
| C | Northeast Asia | 2.6 | 8 | 13 | 0.62 | |
| D1 | Native American | 2.5 | 5 | 16 | 0.31 | |
| L1b1 | West Africa | 2.2 | 10 | 11 | 0.91 | |
| L2a1 | West Africa | 1.8 | 14 | 16 | 0.88 | |
| H1 | Europe | 1.2 | 11 | 19 | 0.58 | |
| “Young” haplogroups | <3 | 0.62 | 0.24 | |||
Average number of synonymous substitutions to the ancestral sequence of the haplogroup. One or two of the most frequent haplogroups for (a) Africa, (b) Europe, (c) East Asia, and Native Americans are displayed for the upper and lower range of sequence diversity. Using the average mutation rate of synonymous transitions, the threshold of six synonymous transitions for the “older” haplogroups indicates coalescent times of the haplogroups >40 thousand years, while the threshold of fewer than synonymous transitions for the “younger” haplogroups indicates <20 thousand years of divergence.
Significant (P < 0.05) mutational bias toward specific (NNG to NNA and NNU to NNC) codon usage was observed in 27 of 32 pairs of codons that differed by a transition in the third codon position (Table 5). This relative preference of G-to-A and T-to-C mutations (per existing nucleotide pool in the light strand) extends over the nonsilent positions and is characteristic of the noncoding D-loop region (Aquadro and Greenberg 1983; Malyarchuk and Rogozin 2004). However, the general strand bias, known to be reversed in some Metazoan genera, can be related to asymmetric mutational constraints involving deaminations of A and C nucleotides during the replication and/or transcription processes (Hassanin et al. 2005). Importantly, the ND6 gene, encoded by the heavy strand showed the opposite mutational bias, suggesting that the differences of codon usage in human mtDNA might be primarily a function of strand asymmetry rather than of differences in the tRNA pools, as generally expected (Tanaka and Ozawa 1994). Notably, 16/18 nucleotide positions (Table 6) that had undergone five or more recurrent changes involved the transition of guanine to adenine in the light strand.
TABLE 5.
Significant differences (P < 0.05) in mutational direction in synonymous sites
| Amino acid | Codon change (count in CRS) | No. of observed changesa to/from | P | P′ |
|---|---|---|---|---|
| Ala | GCG (8) to GCA (80) | 9/5 | 3 × 10−7 | 2 × 10−4 |
| GCU (43) to GCC (123) | 19/8 | 2 × 10−6 | 0.002 | |
| Asn | AAU (32) to AAC (132) | 13/13 | 7 × 10−4 | 0.042 |
| Asp | GAU (15) to GAC (51) | 12/5 | 2 × 10−4 | 0.004 |
| Gln | CAG (8) to CAA (82) | 7/9 | 0.001 | 0.018 |
| Glu | GAG (24 )to GAA (64) | 18/10 | 4 × 10−4 | 0.046 |
| Gly | GGG (34) to GGA (67) | 44/37 | 0.005 | |
| GGU (24) to GGC (87) | 11/12 | 0.014 | ||
| Ile | AUU (124) to AUC (196) | 38/26 | 0.002 | |
| Leu | CUG (45) to CUA (276) | 39/59 | 1 × 10−7 | 0.006 |
| UUG (19) to CUG (45) | 4/3 | 0.099 | ||
| UUA (73) to CUA (276) | 28/18 | 4 × 10−8 | 2 × 10−4 | |
| CUU (65) to CUC (167) | 25/18 | 1 × 10−4 | 0.032 | |
| UUG (19) to UUA (73) | 14/11 | 5 × 10−4 | 0.048 | |
| Lys | AAG (10) to AAA (85) | 6/10 | 0.004 | |
| Met | AUG (40) to AUA (167) | 31/29 | 5 × 10−6 | 0.008 |
| Phe | UUU (77) to UUC (139) | 24/17 | 0.006 | |
| Pro | CCG (7) to CCA (52) | 14/10 | 1 × 10−5 | 3 × 10−4 |
| CCU (41) to CCC (119) | 22/14 | 3 × 10−4 | 0.008 | |
| Ser | AGU (14) to AGC (39) | 7/3 | 0.02 | |
| UCG (7) to UCA (83) | 5/10 | 0.02 | ||
| UCU (32) to UCC (99) | 14/7 | 9 × 10−5 | 0.008 | |
| Thr | ACU (52) to ACC (155) | 27/16 | 6 × 10−7 | 0.002 |
| ACG (10) to ACA (134) | 24/11 | 9 × 10−16 | 9 × 10−13 | |
| Try | UGG (11) to UGA (93) | 18/36 | 5 × 10−4 | 0.031 |
| Tyr | UAU (46) to UAC (89) | 22/17 | 0.01 | |
| Val | GUU (31) to GUC (48) | 15/6 | 0.008 | |
| Total | NNG (223) to NNA (1256) | 229/237 | 4.6 × 10−52 | 5.3E-19 |
| NNU (596) to NNC (1444) | 249/162 | 2.8 × 10−34 | 1.8E-27 | |
| ND6 | NNG (62) to NNA (35) | 14/19 | 0.03 inverse | |
| NNU (72) to NNC (6) | 10/6 | 1.1 × 10−3 |
P is a chi-square probability assuming equal rates of codon exchange and estimates the difference from the expected number of changes, given the codon frequencies in the reference mtDNA sequence (Andrews et al. 1999). P′ is binomial probability taking into account additional transitional biases observed over the whole mitochondrial genome favoring transitions G to A over A to G and T to C over C to T by factors of 2.33 and 1.93, per respective nucleotides.
Number of changes corresponds to mutations (including multiple hits per site) inferred in phylogenetic analysis (Figure 1).
TABLE 6.
Number and location of recurrent mutations in the mtDNA coding region (577–16023)
| No. of recurrences | Nucleotide positions |
|---|---|
| 14 | 709 ∼r |
| 10 | 13708 (A to T) |
| 8 | 1888 ∼r, 8251 |
| 7 | 11914, 10398 (A ↔ T) |
| 5 | 1438 ∼r, 5460 (A to T), 13105 (I to V) |
| 4 | 1598 ∼r, 1719 ∼r, 3010 ∼r, 13928C (S to T), 13966 (T to A), 15930 ∼t, 5147, 13368, 15217 |
| 3 | 3394 (Y to H), 5821 ∼t, 12172 ∼t, 14110 (F to L), 15110 (A to T), 15924 ∼t, 5231, 6182, 6221, 7055, 8790, 9545, 9554, 9950, 12007, 12501, 13359, 15514 |
| 2 | 930 ∼r, 1503 ∼r, 2768 ∼r, 3434 (Y to C), 4025 (T to M), 4048 (D to N), 5046 (V to I), 5773 ∼t, 8027 (A to T), 10084 (I to T), 12236 ∼t, 12950 (N to S), 13759 (A to T), 14687 ∼t, 15758 (I to V), 15927 ∼t, 3666, 3915, 4562, 4580, 4688, 4703, 5417, 5471, 5585-nc, 6260, 6446, 6680, 6752, 7076, 7388, 8020, 8152, 8155, 8392, 8964, 8994, 9254, 9266, 9509, 9755, 9824, 9932, 10685, 10790, 11260, 11944, 12354, 12477, 12810, 14007, 14034, 14148, 14182, 14905, 15115, 15301, 15784, 15884-nc |
Amino acid changes are indicated in parentheses. ∼r, change in rRNA; ∼t, change in tRNA sequences; nc, change in noncoding position. The following positions showed a single recurrence, having mutated twice: 593 ∼t, 597 ∼t, 719 ∼r, 813 ∼r, 827 ∼r, 1018 ∼r, 1193 ∼r, 1243 ∼r, 1694 ∼r, 1811 ∼r, 1822 ∼r, 2245 ∼r, 2332 ∼r, 2352 ∼r, 2416 ∼r, 2706 ∼r, 2757 ∼r, 2772 ∼r, 2789 ∼r, 2885 ∼r, 3203 ∼r, 3206 ∼r, 3505 (T to A), 4500 (S to P), 4596 (V to I), 4824 (T to A), 4917 (N to D), 5442 (F to L), 5910 (A to T), 6253 (M to T), 6261 (A to T), 6480 (V to I), 7389 (Y to H), 7444 (Ter to K), 7569 ∼t, 7673 (I to V), 7805 (V to I), 7853 (V to I), 8329G ∼t, 8387 (V to M), 8393 (P to S), 8566 (I to V), 8584 (A to T), 9095 (L to P), 9139 (A to T), 9438 (G to S), 9477 (V to I), 9861 (F to L), 9966 (V to I), 10031 ∼t, 10143 (G to S), 10321 (V to A), 10463 ∼t, 11016 (S to N), 11025 (L to P), 12142 ∼t, 12248 ∼t, 12346 (H to Y), 12358 (T to A), 12397 (T to A), 12940 (A to T), 13135 (A to T), 13145 (S to N), 13651 (T to A), 13879 (S to P), 13889 (C to Y), 14129 (T to I), 14180 (Y to C), 14315 (S to N), 14798 (F to L), 15287 (F to L), 15314 (A to T), 15317 (A to T), 15323 (A to T), 15326 (T to A), 15479 (F to L), 15907 ∼t, 15928 ∼t, 15939 ∼t, 15951 ∼t, 3483, 3591, 3693, 3777, 3834, 3852, 4038, 4117, 4200, 4248, 4655, 4715, 4823, 4883, 4907, 4916, 4937, 5054C, 5162, 5237, 5393, 5580-nc, 5581-nc, 5656-nc,6026, 6179, 6392, 6431, 6455, 6827, 7184, 7337, 7424, 7861, 8050, 8104, 8227, 8269, 8277-nc,8383, 8485, 8697, 8856, 9150, 9180, 9299, 9305, 9365, 9377, 9449, 9716, 9758, 9899, 10238, 10389, 10586, 10589, 10688, 11002, 11257, 11299, 11332, 11350, 11353, 11383, 11404, 11437, 11452, 11812, 11854, 12372, 12432, 12540, 12609, 12630, 12720, 12771, 13020, 13104, 13116, 13215, 13263, 13470, 13590, 13680, 13827, 13980, 14094, 14212, 14233, 14323, 14364, 14470, 14560, 14581, 14620, 14668, 15043, 15061, 15106, 15148, 15172, 15289, 15313, 15346, 15394, 15454, 15466, 15550, 15607, 15670, 15697, 15883, 15886-nc.
Of the 1292 variable sites, 288 (22.2%) had mutated recurrently. Unexpectedly, the hotspots that had mutated five or more times were predominantly within mitochondrial rRNA (P < 5 × 10−15) and showed a significantly higher ratio of nonsynonymous-to-silent mutations (90:32 hits, respectively) than polymorphic sites with lower recurrence (608:1004) (Table 6). Finally, these hotspots of mutational activity included positions where the human-derived allele predominates in the mammalian consensus sequences (e.g., np 709, 3010, 10398, and 13928), implying the effect of site-specific positive selection. Among the six nonsynonymous substitutions that have recurred four or more times, five involved threonine (P < 6.1 × 10−7). Overall, threonine and valine codons were involved in 259 of the 414 amino acid replacements observed on the tree.
Lineage-specific tests failed to detect significant positive selection along any unique lineage in the seven-taxon phylogeny of primates. A model fixing a single ratio of ω to all lineages (M0) could not be rejected in favor of a model of different ω's on specified lineages (M1). The ω estimated across all lineages in the phylogeny was 0.35. A test of the previous model against a model enforcing neutral selection, where ω is expected to be equal to 1, showed that these data do not deviate significantly from neutrality (M0 was rejected in favor of model where ω = 1; P ≈ 0, d.f. = 1). Further tests for lineage-specific variation in ω, including a model that assigned a different ω to the human lineage from the remaining primates, did not fit the data as well as M1 did. However, site-specific model testing revealed significant positive selection across regions of the primate mitochondrion. A model enforcing a single ω ratio on all codon sites was rejected in favor of a model allowing for three ratios across sites with three site classes (P ≈ 0, d.f. = 5). The three-ratio model identified 16 codon sites to be under significant (posterior probabilities > 0.95; dN/dS = 2.02) positive selection (Table 7). Among these, four codon sites appeared to be among the nonsynonymous sites with recurrent mutation (particularly no. 114 in the ND3 gene, np 10398 with seven recurrences) in human–human comparisons (Table 6).
TABLE 7.
Codon sites found to be under positive selection in the mitochondrion-encoded protein genes in primates
| Site | Gene | Codon no. | Nucleotideposition | Nh | Posterior probabilities |
|---|---|---|---|---|---|
| 1 | ND2 | 218 | 5121-3 | 0 | 0.9732 |
| 2 | ND2 | 265 | 5262-4 | 2 | 0.9603 |
| 3 | ATP6 | 10 | 8554-6 | 0 | 0.9521 |
| 4 | ATP6 | 188 | 9088-0 | 0 | 0.9809 |
| 5 | ND3 | 9 | 10083-5 | 3 | 0.9628 |
| 6 | ND3 | 44 | 10188-0 | 0 | 0.9823 |
| 7 | ND3 | 107 | 10377-9 | 0 | 0.9666 |
| 8 | ND3 | 114 | 10398-0 | 8 | 0.9542 |
| 9 | ND4L | 9 | 10494-6 | 0 | 0.9641 |
| 10 | ND4 | 55 | 10922-4 | 0 | 0.9897 |
| 11 | ND4 | 424 | 12029-1 | 0 | 0.9738 |
| 12 | ND5 | 109 | 12661-3 | 0 | 0.9514 |
| 13 | ND5 | 202 | 12940-2 | 2 | 0.983 |
| 14 | ND5 | 459 | 13711-3 | 1 | 0.9525 |
| 15 | ND5 | 492 | 13810-2 | 0 | 0.9617 |
| 16 | ND6 | 11 | 14641-3 | 0 | 0.954 |
The test of positive selection (dN/dS ≫ 1) was applied to 13 protein-coding genes of mtDNA in the phylogenetic tree involving seven primate species: H. sapiens, P. troglodytes, G. gorilla, P. hamadryas, H. lar, P. pygmaeus, and M. sylvanus) using PAML (Yang 2002). A model of neutral selection on codon sites was rejected in favor of a model allowing for three ratios across sites with K = 3 site classes (P ≈ 0, d.f. = 5). The three-ratio model identified the listed 16 sites to be under significant (posterior probabilities > 0.95) positive selection (dN/dS = 2.02). Nucleotide positions and gene names are given as in the human reference sequence (GenBank accession no. NC_001807.3). Nh, Number of nonsynonymous mutations per respective codon observed in the tree of 277 human mtDNA sequences (Figure 1).
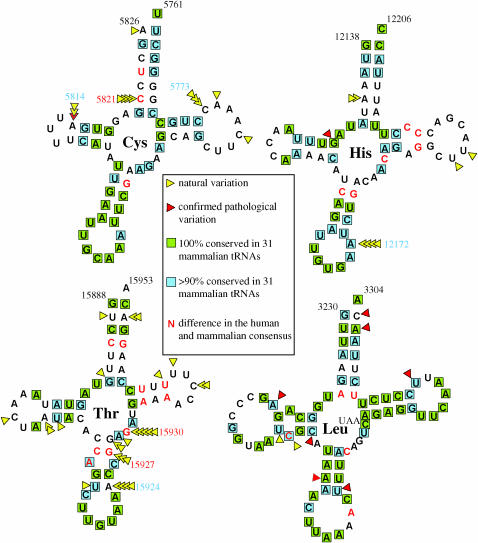
A majority of the mitochondrial disease-related mutations have been detected in the tRNA genes, and they mainly affect the secondary structure of the molecule (McFarland et al. 2004 and references therein). Our global survey of natural variation in the tRNA genes showed a sevenfold excess of tRNAThr mutations (N = 28) over other tRNA genes (P < 10−19). This finding would suggest, at first glance, that this gene might have become nonfunctional in the mitochondrion and that its encoded tRNA needs to be imported from the nucleus. Evidence suggesting nuclear tRNA import into mitochondria in marsupials has been obtained previously (Dörner et al. 2001). However, plotting the observed mutations in the tRNAThr gene against the mammalian consensus sequences (Helm et al. 2000) showed that none of the mutations that we have observed in 277 humans fell within the 100% conserved regions of the tRNA (Figure 2). Most pathological mutations affecting tRNAs cluster in highly conserved regions (McFarland et al. 2004), as illustrated in the case of tRNALeu in Figure 2. Four private mutations changed the nucleotide that is >90% conserved in mammalian tRNAThr, while most of the frequent and recurrent mutations in the data set affected the minor fraction of the sites that are not highly conserved in mammalian species. This argues against the proposition that human mitochondrial tRNAThr has lost function. A large fraction (12/28) of the mutations affecting tRNAThr occurred at three positions, two of which have a different allele in consensus humans as compared to the 31 mammalian species analyzed by Helm et al. (2000). Similarly, a mutational hotspot at position 5821 in tRNACys showed a majority allele in humans different from that found in consensus mammalians (Figure 2). Surprisingly, in the latter tRNA we observed two parallel mutations at position 5814 that have been previously reported pathogenic. Yet, because this position is not highly conserved in other mammalian species, its pathological role has to be questioned. No other tRNA site that has been confirmed as associated with mitochondrial disease was found to be variable in our data set. The only mutational hotspot (12,172) affecting the human allele that matches the allele conserved in >90% of mammalian tRNA-s was found in tRNAHis.
Figure 2.
Natural and pathological variation in human mitochondrial species of tRNAs. Mutations observed in this study are in yellow; those with a status of confirmed pathological variation (http://www.mitomap.org) are in red triangles. Nucleotide positions that are 100% and 90% conserved in mammalian species (Helm et al. 2000) are highlighted in green and blue rectangles, respectively. Nucleotide positions at which the human and mammalian consensus differ from each other are in red type.
A comparison of the amino acid substitutions in the mtDNA-encoded proteins in humans, primates, carnivores, and artiodactyls revealed that substitutions between threonine and alanine are significantly overrepresented in humans while changes between methionine and leucine are most common in other mammalian species (Table 8). The direction of threonine and valine substitution with other amino acids was significantly different among populations with neutral and significantly negative Tajima's D values, respectively (Table 3), and among haplogroups: in H1 sequences sampled broadly from Europe and the Near East, 7 of 11 nonsynonymous mutations resulted in the replacement of threonine and valine with alanine and isoleucine, while only three mutations resulted in a change toward threonine or valine (Figure 1). In contrast to this pattern, in haplogroup V sequences from Finland (Finnilä et al. 2001), where populations continued to rely largely on hunting and fishing for subsistence even after the first contacts with farmers, six of seven replacement polymorphisms resulted in a change to threonine and valine, and none resulted in the replacement of the latter two amino acids (P < 0.01). Similarly, L3 sequences of West African origin showed a significantly lower (P < 0.001) ratio of gains to losses of threonine and valine residues (13/14) than haplogroup L0 sequences from East and South Africa (22/2; Figure 1). East Asian sequences showed an increase of valine codons (9 mutations to and 2 from valine codons), but also a significant (P < 0.01) decrease of threonine, with 11 mutations from and 5 to threonine codons, while Native Americans had 1 mutation from and 7 to threonine, respectively. Over all haplogroups and genes, the direction of amino acid change was significantly (P < 0.02) biased toward replacement of isoleucine and methionine to valine, even when considering the transitional preferences observed in the mitochondrial D-loop (Tamura and Nei 1993). The strand-specific mutational biases are unlikely to explain this pattern because of the observed excess of mutations involving valine codons (8/16 in our data set and 13/16 in the list of ND6 polymorphic sites in MITOMAP) in the ND6 gene that is encoded by the opposite strand.
TABLE 8.
Predominant amino acid replacement types in mitochondrial genes by intra- and interspecies comparisons
| Noc | Ala ↔ Thr | Ile ↔ Val | Ile ↔ Thr | Phe ↔ Leu | Asn ↔ Ser | Met ↔ Thr | Ser ↔ Thr | Ile ↔ Leu | Met ↔ Leu | Other | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human–humana | 414 | 0.273 | 0.145 | 0.08 | 0.07 | 0.058 | 0.029 | 0.01 | 0.005 | 0.005 | 0.326 |
| Human–chimpanzeeb | 167 | 0.18 | 0.132 | 0.078 | 0.042 | 0.066 | 0.072 | 0.042* | 0.012 | 0.012 | 0.365 |
| Chimpanzee–orangutan | 452 | 0.119* | 0.071* | 0.082 | 0.066 | 0.031 | 0.053 | 0.04* | 0.029* | 0.042* | 0.467 |
| Cat–dog | 96 | 0.055* | 0.107 | 0.047 | 0.025* | 0.02* | 0.03 | 0.07* | 0.045* | 0.087* | 0.515 |
| Pig–cow | 479 | 0.054* | 0.09 | 0.035* | 0.033 | 0.027 | 0.05 | 0.044* | 0.063* | 0.117* | 0.486 |
Difference from the human-human pattern; χ2 probability P < 0.01. Only the most frequent (>5% in at least one comparison) amino acid replacement types are reported.
Proportions of specified amino acid changes as reported in the tree of 277 human sequences (Figure 1).
Proportions of changes between rCRC (NC_001807.3) and P. troglodytes (D38113): orangutan (D38115); pig (AJ002189); dog (AY729880); cat (NC_001700); and cow (V00654) sequences.
Total number of observed amino acid replacements.
DISCUSSION
In phylogenetic analysis of human mitochondrial DNA-coding region sequences, two different spheres of character evolution can be distinguished (Ho et al. 2005; Penny 2005). First, within our species, at the population level, a relatively low level of parallel mutations—as compared to the mtDNA control-region-based phylogenies—enables the reconstruction of the unrooted tree from individual sequences without significant ambiguity. This tree is determined by a substantial fraction of amino acid replacement mutations whose proportion to synonymous substitutions increases from the average of 0.37 in “older” clades to 0.62 in “younger” ones. In the second sphere, a high level of homoplasy with chimpanzees, affecting at least one-third of the variable sites in humans, complicates detailed phylogenetic analyses at the interspecies level. Approximately 930 synonymous mutations that can be observed between human and chimpanzee mtDNA represent only the visible component of variation between the species while the effective ratio of nonsynonymous-to-silent mutations is expectedly significantly less than the observed value of 0.2 due to the hidden load of synonymous mutations. These differences between the two spheres imply that, even among the substitutions that define the deepest branches of the human mtDNA tree, a significant excess of nonsynonymous mutations have not yet been eliminated by purifying selection—assuming, of course, that they are generally deleterious, after all.
More than half of the amino acid replacements observed in the human mtDNA tree involved threonine and valine codons. Adaptive correlation with the elevated mutability in the mitochondrion-encoded tRNAThr, in principle, could be considered as one explanation for the excess of mutations involving threonine codons. However, none of the highly conserved sites in the tRNAThr gene was found to be different in humans from that of the consensus mammalians and, instead, the excessive variability in this gene could be ascribed largely to the presence of three hotspot positions. Furthermore, no such general molecular phenomenon or the characteristic G-to-A and T-to-C mutational bias on the light strand of mtDNA would explain the pattern of differences of amino acid replacement directions that were observed among human populations.
One factor that could explain, theoretically at least, the different amino acid replacement patterns observed among populations and between humans and other mammals is diet. Threonine and valine, essential amino acids that must be taken in the diet, are abundant in meats, fish, peanuts, lentils, and cottage cheese, but deficient in most grains. Alternatively, or in combination with dietary restriction, other constraints of selection on slightly deleterious positions during the phases of population expansion and contraction may be involved. Because of the specific compositional bias in mtDNA induced by characteristic mutational preferences different from those observed in the nuclear genome, additional inter- and intraspecies comparisons of mtDNA-encoded amino acid replacement patterns should be examined to gain deeper insights into the nonsynonymous character evolution in metazoan mitochondria, particularly in taxa with shifted strand symmetry (Hassanin et al. 2005).
Tests of neutrality based on the comparisons of the ratio of nonsynonymous and synonymous mutations across all sites can detect only major effects of purifying (KN/KS approaches 0) or directional selection (KN/KS is significantly >1), which affect simultaneously a large number of codon positions. Consistent with previous studies (Cann et al. 1984; Nachman et al. 1996; Ingman and Gyllensten 2001; Mishmar et al. 2003; Moilanen et al. 2003; Moilanen and Majamaa 2003; Elson et al. 2004; Ruiz-Pesini et al. 2004) human mtDNA-encoded proteins did not provide evidence of directional selection. However, several hotspots of mutational activity included nonsilent substitutions susceptible to site-specific positive selection. Comparing the mtDNA protein-encoding genes from several primates (Macaca, Papio, Hylobates, Pongo, Gorilla, and Pan) with human ones, we discovered significant positive selection in several regions, generally nonmatching, however, with the codons displaying a high KN/KS ratio in human–human comparisons. This difference might be explained by the dynamic polarity of the amino acid replacements at the intra- and interspecies levels whereby the constraint of selection is determined in each lineage by the ancestral state of each codon position.
In conclusion, we have provided new evidence for nonrandom processes affecting the evolution of the human mtDNA-encoded proteins. The potential role of selection in affecting fixation probabilities at different nonsilent positions undermines the appropriateness of using the average mitochondrial clock over all sites in dating events in human population history. Despite the evidence of departures from neutrality and high levels of homoplasy at the interspecies level, the phylogenetic approach for analyzing mtDNA sequence data at the intraspecies level remains viable because the reconstruction of the basic branches is robust and the excess of nonsynonymous substitutions affects mainly the terminal branches of the tree.
Acknowledgments
We thank Richard Villems for useful comments. This work was supported by National Institutes of Health grants GM28428, GM63883, and GM55273, European Commission research grant QLG2-CT-2002-90455, Progetto Consiglio Nazionale delle Ricerche-Ministero dell Istruzione, dell Universita e della Ricerca Genomica Funzionale-Legge 449/97, and Telethon-Italy E.0890. We thank Rita Horvath for providing DNA samples.
References
- Achilli, A., C. Rengo, C. Magri, V. Battaglia, A. Olivieri et al., 2004. The molecular dissection of mtDNA haplogroup H confirms that the Franco-Cantabrian glacial refuge was a major source for the European gene pool. Am. J. Hum. Genet. 75: 910–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews, R. M., I. Kubacka, P. F. Chinnery, R. N. Lightowlers, D. M. Turnbull et al., 1999. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat. Genet. 23: 147. [DOI] [PubMed] [Google Scholar]
- Aquadro, C. F., and B. D. Greenberg, 1983. Human mitochondrial DNA variation and evolution: analysis of nucleotide sequences from seven individuals. Genetics 103: 287–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandelt, H.-J., P. Forster and A. Röhl, 1999. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16: 37–48. [DOI] [PubMed] [Google Scholar]
- Bandelt, H. J., J. Alves-Silva, P. E. Guimaraes, M. S. Santos, A. Brehm et al., 2001. Phylogeography of the human mitochondrial haplogroup L3e: a snapshot of African prehistory and Atlantic slave trade. Ann. Hum. Genet. 65: 549–563. [DOI] [PubMed] [Google Scholar]
- Cann, R. L., W. M. Brown and A. C. Wilson, 1984. Polymorphic sites and the mechanism of evolution in human mitochondrial DNA. Genetics 106: 479–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y. S., A. Torroni, L. Excoffier, A. S. Santachiara-Benerecetti and D. C. Wallace, 1995. Analysis of mtDNA variation in African populations reveals the most ancient of all human continent-specific haplogroups. Am. J. Hum. Genet. 57: 133–149. [PMC free article] [PubMed] [Google Scholar]
- Clark, J. D., Y. Beyene, G. WoldeGabriel, W. K. Hart, P. R. Renne et al., 2003. Stratigraphic, chronological and behavioural contexts of Pleistocene Homo sapiens from Middle Awash, Ethiopia. Nature 423: 747–752. [DOI] [PubMed] [Google Scholar]
- Denaro, M., H. Blanc, M. J. Johnson, K. H. Chen, E. Wilmsen et al., 1981. Ethnic variation in Hpa 1 endonuclease cleavage patterns of human mitochondrial DNA. Proc. Natl. Acad. Sci. USA 78: 5768–5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dörner, M., M. Altmann, S. Pääbo and M. Morl, 2001. Evidence for import of a lysyl-tRNA into marsupial mitochondria. Mol. Biol. Cell 12: 2688–2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elson, J. L., D. M. Turnbull and N. Howell, 2004. Comparative genomics and the evolution of human mitochondrial DNA: assessing the effects of selection. Am. J. Hum. Genet. 74: 229–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnilä, S., M. S. Lehtonen and K. Majamaa, 2001. Phylogenetic network for European mtDNA. Am. J. Hum. Genet. 68: 1475–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster, P., R. Harding, A. Torroni and H.-J. Bandelt, 1996. Origin and evolution of Native American mtDNA variation: a reappraisal. Am. J. Hum. Genet. 59: 935–945. [PMC free article] [PubMed] [Google Scholar]
- Forster, P., A. Torroni, C. Renfrew and A. Röhl, 2001. Phylogenetic star contraction applied to Asian and Papuan mtDNA evolution. Mol. Biol. Evol. 18: 1864–1881. [DOI] [PubMed] [Google Scholar]
- Friedlaender, J., T. Schurr, F. Gentz, G. Koki, F. Friedlaender et al., 2005. Expanding southwest Pacific mitochondrial haplogroups P and Q. Mol. Biol. Evol. 22: 1506–1517. [DOI] [PubMed] [Google Scholar]
- Giles, R. E., H. Blanc, H. M. Cann and D. C. Wallace, 1980. Maternal inheritance of human mitochondrial DNA. Proc. Natl. Acad. Sci. USA 77: 6715–6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman, M., C. A. Porter, J. Czelusniak, S. L. Page, H. Schneider et al., 1998. Toward a phylogenetic classification of primates based on DNA evidence complemented by fossil evidence. Mol. Phylogenet. Evol. 9: 585–598. [DOI] [PubMed] [Google Scholar]
- Hassanin, A., N. Leger and J. Deutsch, 2005. Evidence for multiple reversals of asymmetric mutational constraints during the evolution of the mitochondrial genome of metazoa, and consequences for phylogenetic inferences. Syst. Biol. 54: 277–298. [DOI] [PubMed] [Google Scholar]
- Helm, M., H. Brule, D. Friede, R. Giege, D. Putz et al., 2000. Search for characteristic structural features of mammalian mitochondrial tRNAs. RNA 6: 1356–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrnstadt, C., J. L. Elson, E. Fahy, G. Preston, D. M. Turnbull et al., 2002. Reduced-median-network analysis of complete mitochondrial DNA coding-region sequences for the major African, Asian, and European haplogroups. Am. J. Hum. Genet. 70: 1152–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, S. Y., M. J. Phillips, A. Cooper and A. J. Drummond, 2005. Time dependency of molecular rate estimates and systematic overestimation of recent divergence times. Mol. Biol. Evol. 22: 1561–1568. [DOI] [PubMed] [Google Scholar]
- Ingman, M., and U. Gyllensten, 2001. Analysis of the complete human mtDNA genome: methodology and inferences for human evolution. J. Hered. 92: 454–461. [DOI] [PubMed] [Google Scholar]
- Ingman, M., and U. Gyllensten, 2003. Mitochondrial genome variation and evolutionary history of Australian and New Guinean aborigines. Genome Res. 13: 1600–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingman, M., H. Kaessmann, S. Pääbo and U. Gyllensten, 2000. Mitochondrial genome variation and the origin of modern humans. Nature 408: 708–713. [DOI] [PubMed] [Google Scholar]
- Kivisild, T., H.-V. Tolk, J. Parik, Y. Wang, S. S. Papiha et al., 2002. The emerging limbs and twigs of the East Asian mtDNA tree. Mol. Biol. Evol. 19: 1737–1751 (erratum: Mol. Biol. Evol. 20: 162). [DOI] [PubMed] [Google Scholar]
- Kivisild, T., M. Reidla, E. Metspalu, A. Rosa, A. Brehm et al., 2004. Ethiopian mitochondrial DNA heritage: tracking gene flow across and around the gate of tears. Am. J. Hum. Genet. 75: 752–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, Q.-P., Y.-G. Yao, C. Sun, H.-J. Bandelt, C.-L. Zhu et al., 2003. Phylogeny of East Asian mitochondrial DNA lineages inferred from complete sequences. Am. J. Hum. Genet. 73: 671–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, Q. P., Y. G. Yao, C. Sun, C. L. Zhu, L. Zhong et al., 2004. Phylogeographic analysis of mitochondrial DNA haplogroup F2 in China reveals T12338C in the initiation codon of the ND5 gene not to be pathogenic. J. Hum. Genet. 49: 414–423. [DOI] [PubMed] [Google Scholar]
- Macaulay, V. A., M. B. Richards, E. Hickey, E. Vega, F. Cruciani et al., 1999. The emerging tree of West Eurasian mtDNAs: a synthesis of control-region sequences and RFLPs. Am. J. Hum. Genet. 64: 232–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malyarchuk, B. A., and I. B. Rogozin, 2004. Mutagenesis by transient misalignment in the human mitochondrial DNA control region. Ann. Hum. Genet. 68: 324–339. [DOI] [PubMed] [Google Scholar]
- McDougall, I., F. H. Brown and J. G. Fleagle, 2005. Stratigraphic placement and age of modern humans from Kibish, Ethiopia. Nature 433: 733–736. [DOI] [PubMed] [Google Scholar]
- McFarland, R., J. L. Elson, R. W. Taylor, N. Howell and D. M. Turnbull, 2004. Assigning pathogenicity to mitochondrial tRNA mutations: when “definitely maybe” is not good enough. Trends Genet. 20: 591–596. [DOI] [PubMed] [Google Scholar]
- Mellars, P., 2004. Neanderthals and the modern human colonization of Europe. Nature 432: 461–465. [DOI] [PubMed] [Google Scholar]
- Mishmar, D., E. Ruiz-Pesini, P. Golik, V. Macaulay, A. G. Clark et al., 2003. Natural selection shaped regional mtDNA variation in humans. Proc. Natl. Acad. Sci. USA 100: 171–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moilanen, J. S., and K. Majamaa, 2003. Phylogenetic network and physicochemical properties of nonsynonymous mutations in the protein-coding genes of human mitochondrial DNA. Mol. Biol. Evol. 20: 1195–1210. [DOI] [PubMed] [Google Scholar]
- Moilanen, J. S., S. Finnila and K. Majamaa, 2003. Lineage-specific selection in human mtDNA: lack of polymorphisms in a segment of MTND5 gene in haplogroup J. Mol. Biol. Evol. 20: 2132–2142. [DOI] [PubMed] [Google Scholar]
- Nachman, M. W., W. M. Brown, M. Stoneking and C. F. Aquadro, 1996. Nonneutral mitochondrial DNA variation in humans and chimpanzees. Genetics 142: 953–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palanichamy, M., C. Sun, S. Agrawal, H.-J. Bandelt, Q.-P. Kong et al., 2004. Phylogeny of mtDNA macrohaplogroup N in India based on complete sequencing: implications for the peopling of South Asia. Am. J. Hum. Genet. 75: 966–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penny, D., 2005. Evolutionary biology: relativity for molecular clocks. Nature 436: 183–184. [DOI] [PubMed] [Google Scholar]
- Quintana-Murci, L., R. Chaix, S. Wells, D. Behar, H. Sayar et al., 2004. Where West meets East: the complex mtDNA landscape of the Southwest and Central Asian corridor. Am. J. Hum. Genet. 74: 827–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray, N., M. Currat and L. Excoffier, 2003. Intra-deme molecular diversity in spatially expanding populations. Mol. Biol. Evol. 20: 76–86. [DOI] [PubMed] [Google Scholar]
- Ruiz-Pesini, E., D. Mishmar, M. Brandon, V. Procaccio and D. C. Wallace, 2004. Effects of purifying and adaptive selection on regional variation in human mtDNA. Science 303: 223–226. [DOI] [PubMed] [Google Scholar]
- Saillard, J., P. Forster, N. Lynnerup, H.-J. Bandelt and S. Norby, 2000. mtDNA variation among Greenland Eskimos: the edge of the Beringian expansion. Am. J. Hum. Genet. 67: 718–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas, A., M. Richards, T. De la Fe, M. V. Lareu, B. Sobrino et al., 2002. The making of the African mtDNA landscape. Am. J. Hum. Genet. 71: 1082–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, P., T. Lavi, T. Kivisild, V. Chou, D. Sengun et al., 2004. Reconstruction of patri- and matri-lineages of Samaritans and other Israeli populations from Y-chromosome and mitochondrial DNA sequence variation. Hum. Mutat. 24: 248–260. [DOI] [PubMed] [Google Scholar]
- Shen, P., A. E. Hirsh, T. Kivisild, B. Do, S. Song et al., 2005. Population genetic implications from 103 pairs of globally representative Y-chromosome and mitochondrial DNA sequences. Am. J. Hum. Genet. (in press).
- Stringer, C. B., and P. Andrews, 1988. Genetic and fossil evidence for the origin of modern humans. Science 239: 1263–1268. [DOI] [PubMed] [Google Scholar]
- Tamura, K., and M. Nei, 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10: 512–526. [DOI] [PubMed] [Google Scholar]
- Tanaka, M., and T. Ozawa, 1994. Strand asymmetry in human mitochondrial DNA mutations. Genomics 22: 327–335. [DOI] [PubMed] [Google Scholar]
- Tanaka, M., V. M. Cabrera, A. M. Gonzalez, J. M. Larruga, T. Takeyasu et al., 2004. Mitochondrial genome variation in eastern Asia and the peopling of Japan. Genome Res. 14: 1832–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J. D., D. G. Higgins and T. J. Gibson, 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torroni, A., T. G. Schurr, M. F. Cabell, M. D. Brown, J. V. Neel et al., 1993. Asian affinities and continental radiation of the four founding Native American mtDNAs. Am. J. Hum. Genet. 53: 563–590. [PMC free article] [PubMed] [Google Scholar]
- Torroni, A., K. Huoponen, P. Francalacci, M. Petrozzi, L. Morelli et al., 1996. Classification of European mtDNAs from an analysis of three European populations. Genetics 144: 1835–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torroni, A., C. Rengo, V. Guida, F. Cruciani, D. Sellitto et al., 2001. Do the four clades of the mtDNA haplogroup L2 evolve at different rates? Am. J. Hum. Genet. 69: 1348–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill, P. A., P. Shen, A. A. Lin, L. Jin, G. Passarino et al., 2000. Y chromosome sequence variation and the history of human populations. Nat. Genet. 26: 358–361. [DOI] [PubMed] [Google Scholar]
- Watson, E., P. Forster, M. Richards and H. J. Bandelt, 1997. Mitochondrial footprints of human expansions in Africa. Am. J. Hum. Genet. 61: 691–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, T. D., B. Asfaw, D. DeGusta, H. Gilbert, G. D. Richards et al., 2003. Pleistocene Homo sapiens from Middle Awash, Ethiopia. Nature 423: 742–747. [DOI] [PubMed] [Google Scholar]
- Yang, Z., 2002. Likelihood and Bayes estimation of ancestral population sizes in hominoids using data from multiple loci. Genetics 162: 1811–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z., and R. Nielsen, 2002. Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol. Biol. Evol. 19: 908–917. [DOI] [PubMed] [Google Scholar]
- Yao, Y.-G., Q.-P. Kong, H.-J. Bandelt, T. Kivisild and Y.-P. Zhang, 2002. Phylogeographic differentiation of mitochondrial DNA in Han Chinese. Am. J. Hum. Genet. 70: 635–651. [DOI] [PMC free article] [PubMed] [Google Scholar]