Abstract
The extreme high-body-weight-selected mouse line DU6i is a polygenic model for growth research, harboring many small-effect QTL. We dissected the genome of this line into 19 autosomes and the Y chromosome by the construction of a new panel of chromosome substitution strains (CSS). The DU6i chromosomes were transferred to a DBA/2 mice genetic background by marker-assisted recurrent backcrossing. Mitochondria and the X chromosome were of DBA/2 origin in the backcross. During the construction of these novel strains, >4000 animals were generated, phenotyped, and genotyped. Using these data, we studied the genetic control of variation in body weight and weight gain at 21, 42, and 63 days. The unique data set facilitated the analysis of chromosomal interaction with sex and parent-of-origin effects. All analyzed chromosomes affected body weight and weight gain either directly or in interaction with sex or parent of origin. The effects were age specific, with some chromosomes showing opposite effects at different stages of development.
BODY weight at different stages of development and changes in weight between these growth stages are complex traits. As such, they reflect the effects of a complex net of gene actions under the influence of the environment. Crossbred populations have been widely used for the mapping of quantitative trait loci (QTL) influencing body weight and composition in polygenic mouse models (Brockmann and Bevova 2002). To identify genetic components underlying high body weight and growth, we have explored the genetically unique, extremely large and fat mouse line DU6i. This line is an inbred derivate of DU6, which has been selected for high body weight at the age of 42 days when animals have finished the period of fastest growth. Animals of this line differ from the unselected control by >100% in body weight and 300% in fat accumulation. To our knowledge, DU6 is the largest selected mouse line worldwide and therefore represents a unique resource for mapping growth genes (Bunger et al. 2001).
Recent genetic studies in the intercross population DU6i × DBA/2 have led to the identification of QTL with effects on body weight, obesity, and muscle weight on 11 chromosomes (Brockmann et al. 2000). Additional loci were mapped for factors that are involved in the regulation of body weight, namely leptin, insulin, IGF-I, and IGF-binding proteins. The contribution of every single QTL to the phenotypic variance was generally small, accounting for 2–6% of the observed phenotypic variance, except a QTL on chromosome 7 accounting for 12.3% of the phenotypic variance. The direct effects of all QTL accounted for one-third of the phenotypic variance of body weight and fat accumulation in the F2 population of DU6i × DBA/2 mice; another third was explained by epistatic effects (Brockmann et al. 2001).
Linkage analysis using F2 intercross progeny can resolve the position of a QTL to a region of ∼20 cM (Falconer and Mackay 1995). To obtain a more precise location and a higher resolution of QTL effects that would allow for the identification of putatively multiple genes influencing the trait in an identified QTL region requires different breeding strategies. These include the construction of recombinant inbred strains (Broman 2005), recombinant congenic strains, congenic strains, advanced intercross lines, and chromosome substitution strains (CSS) (Darvasi 1998; Nadeau et al. 2000). CSS have a single chromosome from a donor strain substituting for the corresponding chromosome of a recipient strain. The generation of CSS is particularly useful if many small-effect genes account for the phenotypic variance of a population. In such cases fine mapping by congenic strains is difficult because a high number of animals have to be produced to detect possibly small effects of the genes, which is a prerequisite for the subsequent selection decision to generate the congenic strains. CSS have the advantage that small effects on a chromosome are extremely unlikely to be lost because the selection decision for the generation of the subsequent generation is based solely on the genotype information. Furthermore, these strains are a validated source for fine mapping as the background is homogeneous.
So far, genomewide CSSs have been generated to study polygenic traits of the inbred mouse strain A/J on the genetic background of C57BL/6 J (Nadeau et al. 2000). Recently, these homozygous CSS have been phenotyped to show chromosome-wise effects on many traits (Singer et al. 2004). We constructed CSS for the high-body-weight selected mouse line DU6i, which is a model for polygenic body weight. From an initial cross of one DU6i male with three DBA/2 females 19 autosomes and the Y chromosome of DU6i were transferred to the background of the inbred line DBA/2. Here we report the results of the analysis of the recurrent backcross data from >4000 animals, which have been generated, phenotyped, and genotyped during the construction of CSS. The unique data gathered during the construction of CSS enabled us to analyze heterozygous carriers of the substituted chromosome. This approach allows estimation of complete or partial dominance or even heterosis effects (that is, the effect that heterozygous animals have more extreme phenotypes than either homozygous genotype), which have been detected in the QTL-mapping study in the DU6i × DBA/2 F2 population (Brockmann et al. 2000). Furthermore, heterozygous DU6i carriers in the pedigree allowed the analysis of paternal and maternal parent-of-origin effects of the transferred chromosomes.
MATERIALS AND METHODS
Mouse lines and husbandry:
The study was carried out with the mouse line DU6i, which is an inbred derivate from the line DU6 after 78 generations of selection. Line DU6 has been selected for high body weight from the base strain Fzt:DU, which is a systematic cross of four outbred (NMRI origin; Han: NMRI, CFW, CF1) and four inbred (CBA/Bln, AB/Bln, C57Bln/Bln, XVII/Bln) lines (Schüler 1985; Bunger et al. 2001). The age of selection was 42 days for all generations. The population size during selection was 80 pairs/generation; the litter size was standardized to nine. Offspring were weaned at the age of 21 days.
Mice of the strain DBA/2 OlaHsd (Harlan Winkelmann GmbH, Borchen, Germany) were used as a contrast strain for the construction of CSS. Recently this line was used for the mapping of QTL in the cross DU6i × DBA/2 (Brockmann et al. 2000, 2001).
All experimental animals were kept in polyvinyl chloride cages with 255 cm2 floor space. Mice were housed in a semibarrier system under conventional conditions in a windowless mouse laboratory. Air was exchanged 12 times/hr and coarsely filtered (no bacterial filter). The room temperature was 22.5° ± 0.3° and relative humidity was between 50% and 60%. The light cycle was 12 hr light:12 hr dark. All incoming materials or persons were sterilized or disinfected. Animals were fed ad libitum with a breeding diet containing 12.5 MJ/kg metabolic energy with an average content of 22.5% crude protein, 5.0% crude fat, 4.5% crude fiber, 6.5% crude ash, 13.5% water, 48% N-free extract, vitamins, trace elements, amino acids, and minerals (Altromin diet 1314, Lage, Germany).
Animals were mated at the minimum age of 6 weeks. Most matings were carried out between 8 and 9 weeks. Following successful mating, females were housed separately and checked daily for litters. Litters were not standardized at birth. All offspring were weaned at ∼3 weeks of age or later if they were too small. At weaning, individuals were given ear marker identification and separated by sex to prevent sib mating. A tail tip of ∼0.5 cm was cut from every mouse at the age of 21 days and frozen at −20° for subsequent DNA preparation and genotyping.
Pedigree design:
The construction of CSS was based on recurrent backcrossing of animals carrying the DU6i genome to DBA/2 as a recipient strain. Initially, one DU6i male was mated to three DBA/2 females. Subsequently, the F1 males were crossed back to female DBA/2 mice. This cross allowed the transfer of the DU6i autosomes and the DU6i Y chromosome, but not the mitochondrial genome or X chromosome of the DU6i line. Therefore, all generated individuals carried the mitochondria and X chromosome of the DBA/2 recipient strain. Beginning with the first backcross generation (BC1), animals harboring a nonrecombinant chromosome of interest from line DU6i, as assessed by microsatellite markers, were selected for mating to produce the subsequent generation. Repeated backcrosses to strain DBA/2 reduced the portion of the donor genome and increased the portion of the recipient genome. In this analysis, we used animals from generations F1 to BC8.
Phenotyping:
Parameters including personal identification number, identification numbers of mother and father, date of birth, litter size, sex, and body weight of the whole litter were recorded at birth. Number of alive offspring and their individual body weights were recorded at 21, 42, and 63 days. The age of 21 days corresponds to weaning and marks the independence of the young from their mother and the beginning of dependence on their own inherited ability to grow. The age of 42 days corresponds to the end of the juvenile phase of development when animals have finished the phase of the fastest growth and are postpubertal. Body weights at 63 days represent the adult stage of mice. Symbols for body weight at 21, 42, and 63 days are BW21, BW42, and BW63, respectively. The body weight gain between 21 and 42 days is termed BWG21.42 and that between 42 and 63 days, BWG42.63.
Genotyping:
The transfer of the chromosome of interest as well as the genetic background on the other chromosomes of the recipient line was controlled by 130 microsatellite markers. The markers were distributed over all chromosomes with an average distance of 11.6 cM between them. Two gaps were located between 12 and 46 cM on chromosome 4 and between 9 and 36 cM on chromosome 13. Beginning with the fifth backcross generation, the transfer of target chromosomes was controlled with an additional 32 markers. The full list of markers and their positions is given in supplemental Table 1 at http://www.genetics.org/supplemental/. Mouse MapPair primers were purchased from Research Genetics (Huntsville, AL).
DNA was extracted from mouse tail clips using NaOH extraction (the protocol for isolating DNA from embryo yolk sacs is at http://www.healthsystem.virginia.edu/internet/transgenic-mouse/DNAforPCR.cfm). DNA was amplified with Taq DNA polymerase (Promega, Madison, WI) with a modified standard PCR protocol (Brockmann et al. 1998). The amplifications were carried out in 96-well microtiter plates on a DNA thermal cycler (Biometra, Germany). PCR fragments were labeled by either IR 700 or IR 800 using 5′-labeled M13 primers. After PCR reaction, stop-loading buffer was added and plates with the dyes IR 700 and 800 were pooled using the automatic microdispenser Quadra 96 (Tomtec, Hamden, CT). PCR products were visualized using automatic fluorescent detection with the IR2 System (LI-COR, Lincoln, NE). Probes were separated on 25-cm-long 6.5% polyacrylamide gels. All genotyping results were scored twice and runs were repeated when there were discrepancies between reads.
Statistical analyses:
For the detection of genotyping errors we used the Loki program (Heath 1997), which is sensitive to single-locus inheritance incompatibilities. We also evaluated marker loci order and linkage groups by the use of Crimap 2.4 (Lander and Green 1987).
For simple descriptive statistics and detection of the influences of the fixed factors (generation, sex, litter size, year, and season of birth) we used the Statistical Package for Social Science (SPSS) software, v. 11.0. The residuals from regression analysis (difference between observed and predicted values) were tested for normality using the nonparametric Kolmogorov-Smirnov test.
The QTL coefficients (the probability of the DU6i line origin of chromosomal segments) were calculated with Loki-QTLc, v. 1.03 (Aulchenko et al. 2002). This software uses all pedigree information available for computation of QTL coefficients, which becomes essential if genotypes of some animals are missing. We ran a Markov chain Monte Carlo (MCMC) with a relaxation period of 10,000 steps and effective 10,000 steps. To ensure convergence, we ran two independent chains for every chromosome. Only one of them was used for the analysis, while the second was used to confirm the reliability of the first analysis. The QTL coefficients were calculated at the ends of a chromosome, at marker positions, at one position between adjacent markers, and between the end of the chromosome and the first or last marker. The chromosomal heterozygosity (the probability that one of the chromosomes came from DU6i) was estimated as the average of the QTL coefficients along the chromosome.
The chromosomal effects on a trait of interest were estimated using multiple linear regression (MLR). The trait value was regressed onto the chromosomal heterozygosity. All chromosomes were included in the analysis simultaneously to avoid spurious effects in a situation where several chromosomes were segregating together. We also considered fixed effects of generation (effects of F1 and BC1–BC8 were estimated), sex, litter size, and year and season of birth. For this analysis, we used the statistical package R v. 1.7.1 (the R Project for Statistical Computing at http://www.r-project.org). Statistical significance of our results was ensured by estimating empirical (bootstrap) P-values for the chromosomal effects. The following procedure was applied: Ten thousands of replicas were generated from the original data set by sampling with replacement using the “sample” procedure of R. These were reanalyzed and the proportion of times (pi) that the ith regression coefficient was >0 was recorded. The bootstrap P-value for an ith coefficient was estimated as min[2pi,2(1 − pi)].
Also within the MLR framework, the sex by chromosome interaction was tested by assessing the significance of the differences of chromosomal effects in both sexes. A similar procedure was applied for testing the parent-of-origin effects: The difference between the effects of chromosomes that have been inherited from the father or the mother was estimated with MLR.
In our testing procedure, we first tested if the total effect (of chromosome substitution, sex by chromosome interaction, or parent of origin) was significant. This led us to the test on 20 degrees of freedom (d.f.) for the test of total significance of effects of autosomes and the Y chromosome and to a 19-d.f. test in the case of sex-by-chromosome or parent-of-origin effects. If the total effect was significant, we considered the individual effects, which were tested with 1 d.f. to be significant if the individual P-value was <0.05.
The summary (net) direct effects and the effects of chromosomes by sex and chromosomes by parent of origin were characterized by the sum of individual effects. The standard error of the summary estimate was obtained using the complete variance-covariance matrix for the effects to be summarized.
A standard measure applied to characterize the effect of a genetic factor is the percentage of variance explained by variation of this factor in the experimental population. This measure cannot be applied to characterize a factor in a recurrent backcross, because it represents a series of experimental populations. For the purpose of comparison with other studies, we report the percentage of variation explained in the population of our first backcross.
As outcome variable, we analyzed body weight at 21 days (BW21), 42 days (BW42), and 63 days (BW63). We assessed body weight gain as the difference in body weight between maturity (42 days) and juvenile (21 days, end of weaning) phases (BWG21.42). Since we were interested in discovering genes acting in this period only, the weight at 21 days was included in the model as a covariate. Thus, by studying this trait we studied the growth from juvenile to maturity, which cannot be explained by baseline weight at 21 days. We also studied the difference between the weight at 63 and at 42 days, with weight at 42 days as covariate. This trait characterizes the growth from maturity to adulthood (BWG42.63).
RESULTS
Effect of fixed factors:
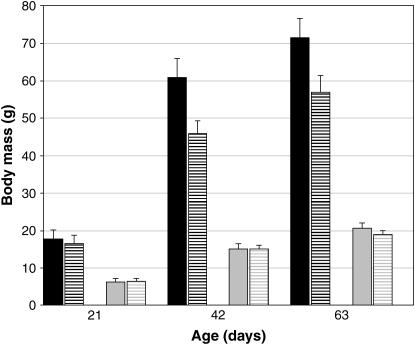
The two mouse lines DU6i and DBA/2 differed in body weight of males by a factor of 2.8, 4.1, and 3.5 at the ages of 21, 42, and 63 days, respectively (Figure 1). The differences between the two lines result from the direct genetic effects of loci and their interaction.
Figure 1.
Body weight of DU6i and DBA/2 mouse lines at different ages. Solid bars: body weight of DU6i males (mean ± SE). Shaded bars: mean body weight of DBA/2 males (mean ± SE). Solid striped bars: mean body weight of DU6i females (mean ± SE). Shaded striped bars: mean body weight of DBA/2 females (mean ± SE).
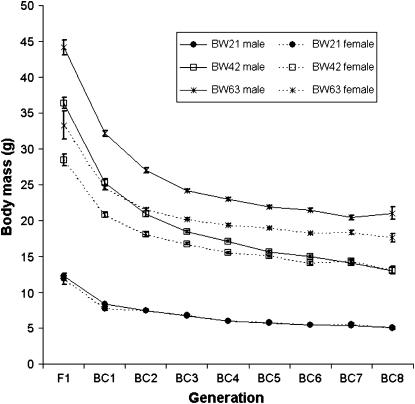
We have characterized 4166, 3235, and 2358 animals for body weight at 21, 42, and 63 days, respectively. As expected, with each generation of backcrossing, the mean of the body weight declines (Figure 2; supplemental Table 2 at http://www.genetics.org/supplemental/). A decrease in mean body weight is expected under the additive polygenic model of gene effects, as the portion of the DU6i genome is reduced by ∼50% in each generation. For all weights (at 21, 42, and 63 days), the decrease with every consecutive backcross generation roughly follows an exponential curve with the largest decrease observed between F1 and BC1. The decline becomes steadier at later backcross generations. The same holds for body weight gain from 21 to 42 days. For weight gain from 42 to 63 days, however, the decline does not follow the simple exponential model and is rather flat.
Figure 2.
Body weight at different backcross generations (mean ± SE). BW21, body weight at the age of 21 days; BW42, body weight at the age of 42 days; BW63, body weight at the age of 63 days. F1, first intercross generation. BC1–BC8, backcross generations 1–8.
For body weight at the age of 21 days, we did not detect any difference between sexes in any generation. However, the body weight gain between 21 and 42 days, and consequently, weight at 42 days, was significantly higher in males than in females in all generations except BC7 and BC8 where a comparatively small number of animals were studied (Figure 2; supplemental Table 2 at http://www.genetics.org/supplemental/). The sex differences in weight gain between day 21 and day 42 were maximal at generation F1 and decreased steadily with later backcrosses. For body weight at the age of 63 days we detected significant differences between females and males in all generations except F1. Also, from day 42 to day 63 the males gained significantly more weight than females. The difference between males and females in body weight gain from 42 to 63 days was almost constant across all generations.
We confirmed these results with a MLR using sex and generation as fixed effects: There was no significant effect of sex on BW21 (P > 0.1), while the effect of sex on weight at 42 and 63 days and weight gain at both periods was highly significant (P < 0.0001).
Starting with an MLR including generation and sex as predictor variables, we added other fixed effects. For BW42, the trait that DU6i was selected for, we also found significant effects of season (with highest weight at summer and lowest in winter), year of birth (the weight was lower for later years), and litter size. The same fixed effects were included in MLR for other traits. Fixed effects explained 26, 48, 43, 50, and 59% of the total variance for BW21, BWG21.42, BW42, BWG42.63, and BW63, respectively. For any of the five traits, the residuals were not distributed normally (all P < 0.01), and therefore we verified the significance with bootstrapping when analyzing chromosomal effects, sex by chromosome interaction, and parent-of-origin effects.
Chromosomal effects:
To test the direct genetic effects of a single chromosome in the set of recurrent backcrosses, we complemented our MLR model, which included significant fixed effects, by the effects of autosomal and Y chromosome heterozygosity. Chromosomal heterozygosity presented an important source of body weight variation: for all five traits, highly significant P-values of <0.0001 were obtained in overall significance tests using 20 d.f. In the BC1 generation, chromosomes explained 9.1, 8.1, 11, 12, and 11.1% of variation of BW21, BWG21.42, BW42, BWG42.63, and BW63, respectively.
The effects of the substitution of different chromosomes on body weight and weight gain are presented in Table 1. All autosomes except 9, 10, and 12 have shown significant effects on body weight (P < 0.05) at different stages of development. The chromosomes acted in an age-specific manner. Chromosomes 1, 3, 7, 8, 13, 15, 16, and 17 influenced body weight at all ages. Genes on chromosomes 19 had effect on body weight only during early growth until the age of 21 days, while chromosome 6 had effect on body weight until the age of 42 days. Chromosome 14 affected body weight at the early age of 21 days and at the adult age of 63 days. The influence of chromosome 18 was significant only at the age of 42 days. Effects on body weight after weaning at 21 days were found on chromosomes 2, 4, 5, 11, and Y. Effects on body weight gain were significant on chromosomes 2, 4, 5, 11, 17, and Y during the entire growth period. Chromosomes 1, 6, 8, 15, and 16 influenced body weight gain during the pubertal phase of development, while chromosome 14 had effect only on the postpubertal phase. DU6i chromosomes 1–8, 11, 13–16, 18, and Y demonstrated positive effects on body weight. Positive estimates of genetic effects for most DU6i chromosomes indicated that alleles from the selected line DU6i increase body weight. However, DU6i alleles showing negative effects as compared to the DBA/2 allele were detected on chromosomes 17 and 19. We also observed a switch of the effect of chromosome 17 between pre- and postweaning periods: The DU6i chromosome led to lower weight at 21 days, but, at later ages, the DU6i chromosome acted to increase weight. The total effect of all DU6i chromosomes was significantly positive (Table 1). Our main findings did not change when we applied bootstrap for the significance testing (supplemental Table 3 at http://www.genetics.org/supplemental/).
TABLE 1.
Estimated direct effects of DU6i chromosomes on body weight and body weight gain
| Body weight
|
Body weight gain
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 21 days
|
42 days
|
63 days
|
21–42 days
|
42–63 days
|
|||||||||||
| Chromosome | Effect (g) | SE | P-value | Effect (g) | SE | P-value | Effect (g) | SE | P-value | Effect (g) | SE | P-value | Effect (g) | SE | P-value |
| 1 | 0.42 | 0.14 | 0.003 | 1.29 | 0.32 | <0.001 | 0.87 | 0.29 | 0.003 | 0.57 | 0.22 | 0.011 | 0.06 | 0.19 | 0.753 |
| 2 | 0.36 | 0.23 | 0.118 | 1.87 | 0.50 | <0.001 | 2.68 | 0.44 | <0.001 | 1.53 | 0.35 | 0.000 | 1.54 | 0.28 | <0.001 |
| 3 | 0.80 | 0.16 | <0.001 | 1.48 | 0.34 | <0.001 | 1.12 | 0.32 | <0.001 | 0.44 | 0.24 | 0.067 | 0.12 | 0.20 | 0.560 |
| 4 | 0.14 | 0.17 | 0.397 | 0.53 | 0.36 | 0.135 | 0.72 | 0.31 | 0.020 | 0.66 | 0.25 | 0.008 | 0.43 | 0.20 | 0.029 |
| 5 | −0.12 | 0.18 | 0.504 | 1.18 | 0.41 | 0.004 | 1.77 | 0.36 | <0.001 | 0.72 | 0.28 | 0.011 | 1.10 | 0.23 | <0.001 |
| 6 | 0.27 | 0.11 | 0.012 | 0.73 | 0.23 | 0.002 | 0.15 | 0.22 | 0.489 | 0.33 | 0.16 | 0.040 | −0.19 | 0.14 | 0.159 |
| 7 | 0.45 | 0.18 | 0.012 | 1.07 | 0.39 | 0.006 | 0.88 | 0.34 | 0.009 | −0.09 | 0.27 | 0.751 | 0.01 | 0.21 | 0.959 |
| 8 | 0.33 | 0.15 | 0.031 | 0.96 | 0.33 | 0.004 | 0.91 | 0.32 | 0.005 | 0.49 | 0.23 | 0.035 | 0.36 | 0.20 | 0.074 |
| 9 | −0.22 | 0.15 | 0.137 | 0.22 | 0.33 | 0.511 | 0.18 | 0.29 | 0.545 | 0.24 | 0.23 | 0.293 | 0.13 | 0.19 | 0.471 |
| 10 | −0.19 | 0.18 | 0.289 | −0.48 | 0.39 | 0.221 | −0.28 | 0.34 | 0.408 | −0.33 | 0.27 | 0.223 | 0.06 | 0.21 | 0.768 |
| 11 | −0.21 | 0.23 | 0.379 | 1.45 | 0.51 | 0.005 | 1.79 | 0.44 | <0.001 | 1.82 | 0.36 | <0.001 | 0.68 | 0.28 | 0.015 |
| 12 | 0.12 | 0.14 | 0.386 | 0.36 | 0.31 | 0.245 | 0.20 | 0.28 | 0.469 | 0.12 | 0.22 | 0.592 | 0.04 | 0.18 | 0.841 |
| 13 | 0.36 | 0.15 | 0.016 | 1.06 | 0.33 | 0.001 | 0.89 | 0.30 | 0.003 | 0.33 | 0.23 | 0.145 | 0.02 | 0.19 | 0.933 |
| 14 | 0.33 | 0.13 | 0.010 | 0.28 | 0.28 | 0.326 | 0.66 | 0.26 | 0.009 | −0.35 | 0.20 | 0.077 | 0.33 | 0.16 | 0.039 |
| 15 | 0.48 | 0.14 | <0.001 | 1.37 | 0.31 | <0.001 | 0.90 | 0.31 | 0.004 | 0.67 | 0.21 | 0.002 | 0.14 | 0.19 | 0.483 |
| 16 | 0.26 | 0.11 | 0.023 | 1.02 | 0.26 | <0.001 | 0.51 | 0.24 | 0.037 | 0.54 | 0.18 | 0.003 | −0.22 | 0.15 | 0.159 |
| 17 | −0.35 | 0.13 | 0.009 | 0.64 | 0.30 | 0.033 | 1.00 | 0.28 | <0.001 | 1.05 | 0.21 | <0.001 | 0.71 | 0.18 | <0.001 |
| 18 | 0.22 | 0.12 | 0.069 | 0.62 | 0.26 | 0.019 | 0.24 | 0.25 | 0.335 | 0.19 | 0.18 | 0.298 | 0.00 | 0.16 | 0.991 |
| 19 | −0.37 | 0.16 | 0.020 | −0.51 | 0.36 | 0.159 | −0.53 | 0.33 | 0.116 | −0.32 | 0.25 | 0.209 | −0.04 | 0.21 | 0.833 |
| All | 3.08 | 0.46 | <0.001 | 15.15 | 1.20 | <0.001 | 14.69 | 1.03 | <0.001 | 8.63 | 0.84 | <0.001 | 5.28 | 0.67 | <0.001 |
| Y | −0.08 | 0.09 | 0.371 | 0.53 | 0.19 | 0.006 | 0.72 | 0.18 | <0.001 | 0.80 | 0.13 | <0.001 | 0.39 | 0.12 | 0.001 |
The effect is the estimated substitution effect of the DU6i chromosome vs. the DBA/2 chromosome. The P-value is given for the comparison between animals that are carriers of DU6i chromosomes vs. DBA/2 animals. P-values from bootstrapping can be found in supplemental Table 3 at http://www.genetics.org/supplemental/. Significant P-values <0.05 are in italics. SE, standard error.
Sex by chromosome interaction:
We tested whether the sex modified the effect of chromosomes (Table 2; supplemental Table 4 at http://www.genetics.org/supplemental/). At the age of 21 days, the total test of sex by chromosome interaction at 19 d.f. was significant (P = 0.029). For body weight at later ages and body weight gain, the total effect of sex by chromosome interaction became highly significant (all P < 0.0001). In generation BC1, the variance explained by chromosomes and sex by chromosome interactions was 17.2, 16.8, 16.1, 17.9, and 15.3% of the variation of BW21, BWG21.42, BW42, BWG42.63, and BW63, respectively.
TABLE 2.
Estimated sex-specific effects of DU6i chromosomes on body weight and body weight gain
| Body weight
|
Body weight gain
|
||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 21 days
|
42 days
|
63 days
|
21–42 days
|
42–63 days
|
|||||||||||||||||||||
| Males
|
Females
|
Males
|
Females
|
Males
|
Females
|
Males
|
Females
|
Males
|
Females
|
||||||||||||||||
| Chromosome | Effect (g) | SE | Effect (g) | SE | P-value | Effect (g) | SE | Effect (g) | SE | P-value | Effect (g) | SE | Effect (g) | SE | P-value | Effect (g) | SE | Effect (g) | SE | P-value | Effect (g) | SE | Effect (g) | SE | P-value |
| 2 | 0.09 | 0.33 | 0.49 | 0.32 | 0.387 | 2.47 | 0.71 | 0.93 | 0.68 | 0.115 | 3.35 | 0.63 | 1.70 | 0.60 | 0.053 | 2.58 | 0.49 | 0.35 | 0.47 | 0.001 | 1.55 | 0.40 | 1.45 | 0.38 | 0.863 |
| 4 | 0.25 | 0.23 | 0.07 | 0.24 | 0.573 | 1.06 | 0.49 | −0.07 | 0.50 | 0.103 | 1.61 | 0.42 | −0.34 | 0.44 | 0.001 | 0.98 | 0.34 | 0.24 | 0.34 | 0.124 | 0.91 | 0.27 | −0.08 | 0.28 | 0.009 |
| 5 | −0.57 | 0.26 | 0.22 | 0.25 | 0.026 | 0.21 | 0.60 | 1.64 | 0.55 | 0.074 | 1.18 | 0.52 | 1.93 | 0.48 | 0.293 | 0.37 | 0.41 | 0.75 | 0.38 | 0.496 | 1.20 | 0.33 | 0.88 | 0.31 | 0.478 |
| 8 | 0.60 | 0.20 | 0.10 | 0.23 | 0.101 | 1.62 | 0.44 | 0.46 | 0.51 | 0.082 | 1.63 | 0.41 | 0.20 | 0.49 | 0.023 | 0.76 | 0.30 | 0.41 | 0.35 | 0.449 | 0.86 | 0.26 | −0.17 | 0.31 | 0.011 |
| 9 | −0.14 | 0.21 | −0.28 | 0.21 | 0.613 | 0.70 | 0.45 | −0.45 | 0.47 | 0.071 | 0.62 | 0.39 | −0.49 | 0.41 | 0.048 | 0.65 | 0.31 | −0.38 | 0.32 | 0.019 | 0.31 | 0.25 | −0.18 | 0.26 | 0.171 |
| 11 | 0.40 | 0.32 | −0.77 | 0.33 | 0.009 | 1.59 | 0.70 | 0.72 | 0.71 | 0.372 | 2.37 | 0.60 | 0.76 | 0.61 | 0.053 | 0.87 | 0.48 | 2.21 | 0.49 | 0.045 | 1.10 | 0.38 | 0.26 | 0.39 | 0.113 |
| 14 | 0.31 | 0.19 | 0.34 | 0.17 | 0.920 | 0.31 | 0.41 | 0.14 | 0.37 | 0.760 | 1.34 | 0.37 | 0.03 | 0.33 | 0.008 | −0.33 | 0.28 | −0.45 | 0.26 | 0.748 | 0.80 | 0.24 | −0.07 | 0.21 | 0.005 |
| 17 | −0.26 | 0.19 | −0.43 | 0.18 | 0.530 | 1.11 | 0.44 | −0.01 | 0.40 | 0.056 | 0.88 | 0.41 | 0.71 | 0.36 | 0.754 | 1.38 | 0.30 | 0.57 | 0.28 | 0.046 | 0.41 | 0.26 | 0.74 | 0.23 | 0.330 |
| 18 | −0.02 | 0.16 | 0.48 | 0.17 | 0.031 | 0.38 | 0.37 | 0.83 | 0.36 | 0.386 | 0.10 | 0.36 | 0.39 | 0.34 | 0.542 | 0.32 | 0.25 | 0.03 | 0.25 | 0.396 | 0.09 | 0.23 | −0.07 | 0.21 | 0.589 |
| All | 3.58 | 0.49 | 2.51 | 0.51 | 0.130 | 16.99 | 1.22 | 10.32 | 1.34 | <0.001 | 16.80 | 1.04 | 8.95 | 1.17 | <0.001 | 10.12 | 0.85 | 4.51 | 0.93 | <0.001 | 6.51 | 0.69 | 2.75 | 0.76 | <0.000 |
| Y | −0.26 | 0.10 | −0.18 | 0.22 | −0.09 | 0.21 | 0.36 | 0.15 | 0.03 | 0.13 | |||||||||||||||
Only chromosomes showing significant interaction with sex are shown. The effect is the estimated effect of the DU6i chromosome vs. the DBA/2 chromosome in males and females. The P-value is given for the comparison between male and female carriers of DU6i chromosomes. P-values from bootstrapping can be found in supplemental Table 4 at http://www.genetics.org/supplemental/. Significant P-values <0.05 are in italics. SE, standard error.
For weight gain from day 21 to 42, we detected that chromosomes 2, 9, 11, and 17 acted differently in males and females. The effects of chromosomes 2 and 9 were not significant in females, while males had significant positive effect. The effect of chromosome 17 was almost three times larger in males as compared to females. The situation was opposite for chromosome 11. In total, the summary effect of sex by chromosome interactions led to weight increase of >6 g in male carriers of DU6i chromosomes as compared to female carriers. This sex-specific action of the genes might underlie the sex differences in weight gain between 21 and 42 days. We did not detect significant effects of sex by chromosome interaction for these chromosomes on weight at 42 days; the summary chromosomal effect, however, was highly significant and consisted of 6.4 g (Table 2). The analysis of body weight gain from 42 days to 63 days revealed more sex by genotype interactions. Significant sex effects were found on chromosomes 4, 8, and 14. Again, males gained significantly more than females (the summary effect of chromosomes 4, 8, and 14 was 2.9 g). The sex-specific effects of these chromosomes were also significant for body weight at 63 days. Interestingly, when sex-specific effects were included in the analysis, the effect of the Y chromosome remained significant only for early stages (body weight at 21 days, P = 0.008, and body weight gain from 21 to 42 days, P = 0.015; supplemental Table 4 at http://www.genetics.org/supplemental/).
Parent-of-origin effects:
For all traits, except body weight gain from 42 to 63 days, we detected that the total effect on body weight was dependent on whether the DU6i chromosomes were transmitted from mother or father (total significance test at 19 d.f., all P < 0.0001). In the BC1 generation, the variance explained by chromosomes and parent-of-origin effects was 22.3, 18.3, 17.7, 16.8, and 15.3% of the variation of BW21, BWG21.42, BW42, BWG42.63, and BW63, respectively. As expected for parent-of-origin effects, the proportion of the variance explained by these effects was highest at weaning and decreased with age, unlike direct chromosomal and sex by chromosome interaction effects, where the proportion of variance explained remained more or less constant with the age.
Specifically, at the age of 21 days significant parent-of-origin effects were found for chromosomes 3, 5, 8–12, 14–16, and 19 (Table 3; supplemental Table 5 at http://www.genetics.org/supplemental/). All these chromosomes, except chromosomes 9 and 14, had significant effects only when inherited maternally. Chromosome 14 had significant effect only when inherited paternally. The effects of maternally inherited DU6i chromosomes 5, 10, 11, and 19 were negative. The effect of DU6i chromosome 9 was significantly negative when transmitted from the father and significantly positive when transmitted from the mother.
TABLE 3.
Estimated parent-of-origin effects of DU6i chromosomes on body weight and body weight gain
| Body weight
|
Body weight gain
|
||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 21 days
|
42 days
|
63 days
|
21–42 days
|
42–63 days
|
|||||||||||||||||||||
| Paternal
|
Maternal
|
Paternal
|
Maternal
|
Paternal
|
Maternal
|
Paternal
|
Maternal
|
Paternal
|
Maternal
|
||||||||||||||||
| Chromosome | Effect (g) | SE | Effect (g) | SE | P-value | Effect (g) | SE | Effect (g) | SE | P-value | Effect (g) | SE | Effect (g) | SE | P-value | Effect (g) | SE | Effect (g) | SE | P-value | Effect (g) | SE | Effect (g) | SE | P-value |
| 1 | 0.33 | 0.18 | 0.55 | 0.23 | 0.437 | 1.37 | 0.40 | 1.26 | 0.55 | 0.875 | 0.94 | 0.35 | 1.05 | 0.54 | 0.871 | 1.01 | 0.28 | −0.11 | 0.38 | 0.017 | 0.03 | 0.22 | 0.34 | 0.34 | 0.449 |
| 3 | 0.22 | 0.20 | 1.45 | 0.27 | <0.001 | 0.90 | 0.44 | 2.25 | 0.58 | 0.068 | 0.74 | 0.40 | 1.82 | 0.56 | 0.124 | 0.52 | 0.31 | 0.41 | 0.41 | 0.824 | 0.14 | 0.25 | 0.28 | 0.36 | 0.755 |
| 4 | −0.07 | 0.21 | 0.56 | 0.29 | 0.088 | 0.38 | 0.45 | 0.90 | 0.62 | 0.506 | 0.26 | 0.39 | 1.69 | 0.54 | 0.036 | 0.73 | 0.31 | 0.45 | 0.43 | 0.604 | 0.07 | 0.25 | 1.13 | 0.34 | 0.014 |
| 5 | 0.36 | 0.25 | −0.72 | 0.26 | 0.003 | 1.71 | 0.57 | 0.40 | 0.61 | 0.121 | 1.89 | 0.51 | 1.60 | 0.54 | 0.701 | 0.58 | 0.40 | 0.81 | 0.42 | 0.688 | 1.11 | 0.32 | 0.98 | 0.34 | 0.788 |
| 8 | 0.09 | 0.17 | 1.00 | 0.34 | 0.016 | 0.41 | 0.38 | 3.27 | 0.73 | <0.001 | 0.59 | 0.37 | 2.03 | 0.65 | 0.051 | 0.34 | 0.26 | 1.59 | 0.51 | 0.029 | 0.23 | 0.23 | 0.90 | 0.41 | 0.153 |
| 9 | −0.78 | 0.19 | 0.67 | 0.24 | <0.001 | −0.54 | 0.44 | 1.35 | 0.52 | 0.005 | −0.54 | 0.40 | 1.33 | 0.45 | 0.002 | 0.62 | 0.30 | −0.24 | 0.36 | 0.065 | −0.12 | 0.25 | 0.43 | 0.28 | 0.138 |
| 10 | 0.24 | 0.22 | −0.92 | 0.28 | 0.001 | 0.44 | 0.50 | −2.08 | 0.63 | 0.002 | 0.74 | 0.43 | −1.80 | 0.55 | <0.001 | 0.03 | 0.34 | −1.04 | 0.44 | 0.052 | 0.32 | 0.28 | −0.34 | 0.35 | 0.131 |
| 11 | 0.27 | 0.29 | −1.14 | 0.41 | 0.005 | 2.75 | 0.63 | −1.00 | 0.88 | 0.001 | 2.73 | 0.53 | −0.35 | 0.80 | 0.001 | 2.59 | 0.44 | 0.29 | 0.61 | 0.002 | 1.14 | 0.34 | −0.33 | 0.51 | 0.016 |
| 12 | −0.24 | 0.20 | 0.60 | 0.21 | 0.003 | 0.13 | 0.43 | 0.74 | 0.46 | 0.338 | 0.18 | 0.39 | 0.22 | 0.41 | 0.952 | 0.22 | 0.30 | −0.13 | 0.32 | 0.437 | 0.31 | 0.25 | −0.24 | 0.26 | 0.130 |
| 14 | 0.71 | 0.20 | −0.14 | 0.18 | 0.001 | 0.86 | 0.42 | −0.23 | 0.40 | 0.060 | 0.95 | 0.40 | 0.50 | 0.35 | 0.392 | −0.27 | 0.29 | −0.19 | 0.28 | 0.838 | 0.39 | 0.25 | 0.37 | 0.22 | 0.966 |
| 15 | −0.22 | 0.18 | 1.28 | 0.23 | <0.001 | 0.54 | 0.43 | 2.47 | 0.50 | 0.005 | 0.41 | 0.40 | 1.72 | 0.54 | 0.061 | 0.77 | 0.30 | 0.85 | 0.35 | 0.870 | 0.24 | 0.26 | −0.02 | 0.34 | 0.549 |
| 16 | −0.20 | 0.15 | 0.58 | 0.18 | 0.001 | 0.42 | 0.34 | 1.55 | 0.40 | 0.029 | 0.28 | 0.33 | 0.51 | 0.36 | 0.637 | 0.42 | 0.24 | 0.83 | 0.28 | 0.261 | −0.32 | 0.21 | −0.21 | 0.23 | 0.717 |
| 19 | −0.23 | 0.18 | −1.27 | 0.41 | 0.024 | −0.30 | 0.41 | −1.54 | 0.91 | 0.219 | −0.17 | 0.37 | −2.50 | 0.87 | 0.015 | −0.35 | 0.28 | 0.17 | 0.63 | 0.455 | −0.03 | 0.24 | −0.36 | 0.55 | 0.589 |
| All | 1.80 | 0.57 | 3.69 | 0.52 | 0.015 | 15.44 | 1.32 | 14.02 | 1.48 | 0.477 | 14.67 | 1.14 | 13.40 | 1.28 | 0.457 | 10.95 | 0.92 | 5.18 | 1.04 | <0.001 | 5.27 | 0.74 | 5.27 | 0.82 | 0.996 |
| Y | 0.09 | 0.09 | 0.64 | 0.20 | 0.83 | 0.19 | 0.57 | 0.14 | 0.43 | 0.12 | |||||||||||||||
Only chromosomes showing significant interaction with the parent of origin are shown. The effect is the estimated effect of the DU6i chromosome transmitted by either the male or female parent. The P-value is given for the comparison between offspring that received the DU6i chromosome from either the mother or the father. P-values from bootstrapping can be found in supplemental Table 5 at http://www.genetics.org/supplemental/. Significant P-values <0.05 are in italics. SE, standard error.
The parent-of-origin effects, which were identified on chromosomes 9–11 at the age of 21 days, were also seen at 42 and 63 days. The effects of chromosomes 8, 15, and 16 were found until the age of 42 days. The pattern of action was the same at all ages, as expected.
Parent-of-origin effects of chromosomes 1, 8, and 11 influenced body weight gain from 21 to 42 days. The effect of chromosome 11 was also found for body weight gain between 42 and 63 days. The summary effect of the parent of origin was significant for body weight at 21 days. Animals with DU6i chromosomes transmitted paternally had ∼2 g less weight as compared with the maternal transmission (P = 0.015). However, paternally transmitted DU6i chromosomes determined an increase of body weight gain from 21 to 42 days by almost 6 g as compared to maternal transmission. The summary effect was not significant at other stages.
Thus, according to our data it seems that parent-of-origin effects acted preferentially at early stages of development (preweaning and to a much less extent from 21 to 42 days). The effects on weight observed at 42 and 63 days look like the consequence of the effect on weight at 21 days.
DISCUSSION
We have analyzed a pedigree generated from an extensive recurrent backcross between the extremely high-growth-selected mouse line DU6i and the inbred line DBA/2. The unique population consisted of heterozygous carriers of DU6i chromosomes and animals homozygous for DBA/2 chromosomes. This population allowed the study of direct chromosomal effects, which are the net effects of all genes contributing to body weight regulation on a substituted chromosome and which could be additive effects, dominance, or even overdominance (heterosis). Furthermore, the population structure facilitated the analysis of parent-of-origin effects, as we were able to distinguish between DU6i chromosomes, which were transmitted paternally and maternally. The large number of >4000 animals increased the power to detect direct chromosomal effects as well as effects of chromosomal interaction with sex and parent of origin.
In our study, we first tested the global significance and then presented the individual chromosomal effects that demonstrated nominal P-values of ≤0.05. As three models were evaluated on five traits, we performed multiple testing. Therefore, while many effects that we report are real, some of them may be false positives. However, the correction for multiple testing is not straightforward in our case as both traits and model parameters are far from being independent. As we want to report all potentially biologically important effects, we have chosen this relative relaxed procedure. We must emphasize, however, that many of our results would pass even the most stringent and conservative Bonferroni correction.
In our present study we found that almost all DU6i autosomes (except chromosomes 9, 10, and 12) contributed directly to body weight regulation at the age between 21 and 63 days. Direct effects on body weight were also found for the Y chromosome, which has not been assessed by our previous QTL study in an intercross population. Although we detected that chromosomes 1, 3, 7, 8, 13, 15, 16, and 17 acted throughout all stages, their effects changed with age. Other chromosomes acted age dependently. We found 9 chromosomes with increasing and 2 chromosomes with decreasing effects on body weight at 21 days. Eleven chromosomes promoted weight gain during the early postweaning period (21–42 days). Only 6 chromosomes did not affect body weight at 42 days, the age when animals of the DU6i line were selected. At the period from puberty until adulthood (42–63 days), animals carrying chromosomes 2, 4, 5, 11, 14, and 17 of DU6i origin gained weight significantly better. Only chromosomes 6, 9, 10, 12, 18, and 19 did not influence body weight at 63 days. Interesting results were obtained for chromosome 17, which exhibited antagonistic effects at different stages.
Previous linkage studies in the F2 pedigree DU6i × DBA/2 have identified genomic regions responsible for the drastic phenotypic differences in body weight at 42 days between these lines on chromosomes 1, 4, 5, 7, 9, and 11–13 (Brockmann et al. 2000). The genetic effects were additive and positive for the DU6i allele. In this study, we confirmed the highly significant effects on chromosomes 1, 5, 7, 11, and 13, but the effect of chromosomes 9 and 12 could not be detected. The QTL effect on chromosome 4 was seen only at 63 days and for weight gain in this study. The body-weight-enhancing effects of chromosomes 2, 3, 6, 8, and 14–19 were not detected in the linkage analysis.
Differences between the present comprehensive analysis of the dissected genome and our previous QTL-mapping study may result from the estimation of direct effects of whole chromosomes vs. chromosomal parts and the loss of favored allelic interactions, which might occur in the selection line DU6i. Furthermore, it is important to realize that this study had no power to detect recessive DU6i chromosomal effects, as animals homozygous for DU6i chromosomes were not presented in the analyzed backcross population.
Linkage analyses estimate most likely positions and effects of QTL and neglect nonsignificant effects of other parts of the chromosome. In this study, the overall net effect of all genes located on a given chromosome was estimated for each chromosome. The total effect of all DU6i autosomes and the Y chromosome was 15.68 g at the age of 42 days. This effect was 6.38 g lower than expected by total additive effects (half of the difference between parental strains DBA and DU6i). As our previous linkage analysis did not reveal a significant effect of the X chromosome on body weight, we assume that epistatic effects contribute to growth at 42 days. In our QTL study, epistatic effects account for about one-third of the phenotypic variance of body weight at 42 days within the analyzed F2 population (Brockmann et al. 2000).
Our results on age-dependent growth regulation are consistent with the findings that growth in rodents appears to occur via two general physiological mechanisms that act at different life stages (Falconer 1953). Quantitative genetic studies have indicated that individual genes may have opposite pleiotropic effects on early and late growth. It has been shown that genes causing relatively fast early growth may also cause slower later growth (Cheverud et al. 1983; Leamy and Cheverud 1984; Riska et al. 1984). The early growth effects tend to taper off at the age of 6 weeks. Indications for changing patterns of QTL expression with age were found in the mouse crosses LG/J × SM/J (Cheverud et al. 1996; Vaughn et al. 1999) and DBA/2 × NMRI (Brockmann et al. 2004). It was shown that two distinct sets of genes affect growth and body weight, with one set affecting early growth and another set affecting later growth.
Our large backcross population enabled us to search for sex and parent-of-origin specific effects. Although the net sex-specific effects of DU6i chromosomes on body weight was not significant between males and females at 21 days, at later ages, sex-specific chromosomal effects significantly affected both body weight and weight gain, with DU6i chromosomes acting with a significantly more weight-promoting effect in males. Obviously, the difference between sexes becomes very important between days 21 and 42, when animals have finished the period of fastest growth and are postpubertal. This is in agreement with the fact that we did not detect an effect of sex on weight at 21 days, but at later ages. For weight gain between 21 and 42 days, chromosomes 2, 9, and 17 were acting almost exclusively in males. For weight gain between 42 and 63 days, chromosomes 4, 8, and 14 showed significant interaction with sex and had a significant effect on weight when expressed in males. From our findings we conclude that genes on chromosomes 2, 9, and 17 and on 4, 8, and 14 may form the genetic basis of weight differences between sexes, in particular for the higher weight of males in comparison with females. The role of the large positive effect of chromosome 11 in females on weight gain at the age between 21 and 42 days remains open. This analysis of single chromosomal effects is much more powerful for the identification of sex-specific additive and dominance effects than our previous QTL study, where we did not detect significant sex-specific QTL effects (Brockmann et al. 2000). So far, sex-specific effects were preferentially found in QTL studies for obesity traits, in particular in the analysis of fat accumulation (Taylor et al. 1999, 2001; Brockmann et al. 2004). Therefore, we expect to identify even more sex-specific effects when chromosomal effects on fat deposition in the CSS lines will be analyzed.
We observed large parent-of-origin effects on early growth that also had an effect on body weight at later ages. The parent-of-origin effect may be explained by genetic imprinting or by metabolic imprinting during fetal or postnatal development, which may have long-term effects on growth regulation. It is well known that imprinting is important in genes regulating body weight in mice (Cattanach 1986; Cattanach and Beechey 1990). Currently, >40 imprinted genes have been identified in humans and mice (Morison et al. 2001). Significant variance components (up to 22% of the total phenotypic variance) due to paternally expressed genes were found, for example, in young bulls (Engellandt and Tier 2002). In our study, chromosome 9 was found to have a negative effect on body weight when inherited paternally and a positive effect when inherited maternally. The opposite effect of this chromosome was shown previously (Cattanach and Beechey 1990). Furthermore, we also detected that the summary effect of DU6i chromosomes on weight at 21 days was lower when inherited paternally as compared to maternally. A switch of the effects occurred at later stages. Concluding from our data, the parent of origin of a chromosome has a large effect on body weight, especially at early stages of development.
The results obtained in this study provide a partial chromosomal dissection of the complex traits of body weight and growth. The exact identification of the contribution of single chromosomes and regions on body weight and growth rate in mice is an important step toward a better understanding of the net of genetic factors that are involved in the regulation of body weight. Our article provides one of the few examples of using multi-generation recurrent backcross data for estimation of chromsomal effects. Moreover, the MCMC technique applied to estimate the proportion of founder line genomes at a chromosome is relatively new (Aulchenko 2002). MCMC methods have been used before for a long time to estimate genomic sharing in the framework of human genetics (Sobel and Lange 1996; Heath 1997) and have been proven to lead to consistent results. The methods for the estimation of chromosome-specific effects that we have applied in our analyses have not been tested via simulation. Therefore, the magnitudes of the estimated effects may be subject to some bias. The confirmation of the obtained results requires the analysis of the final CSS panel.
Our results also shed light on the mechanisms of selection, which may be important in future selection experiments and livestock improvement. The established mouse lines are a valuable source for further analyses of additional phenotypic measures related to body composition traits and for the generation of congenic lines. From a systematic screen for genetic interaction between genomic regions by statistical tests there are hints on epistatic effects influencing body weight and growth (Brockmann et al. 2000, 2004). Therefore, the CSS lines are particularly valuable for the targeted examination of interaction between chromosomes.
Acknowledgments
We thank Daniel Pomp and Kari Elo for extensive support in identifying an informative marker set and Pavel Borodin, Tatiana Axenovich, and Conelia van Duijn for helpful comments on this manuscript. This research was supported by the Deutsche Forschungsgemeinschaft, the German National Genome Research Network, the H. Wilhelm Schaumann Stiftung, and the Netherlands Organization for Scientific Research.
References
- Aulchenko, Y. S., F. Teuscher, H. H. Swalve and V. Guiard, 2002. Comparison of methods used for recovering the line origin of alleles in a cross between outbred lines. Genet. Res. 79: 75–83. [DOI] [PubMed] [Google Scholar]
- Brockmann, G. A., and M. R. Bevova, 2002. Using mouse models to dissect the genetics of obesity. Trends Genet. 18: 367–376. [DOI] [PubMed] [Google Scholar]
- Brockmann, G. A., C. S. Haley, U. Renne, S. A. Knott and M. Schwerin, 1998. Quantitative trait loci affecting body weight and fatness from a mouse line selected for extreme high growth. Genetics 150: 369–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockmann, G. A., J. Kratzsch, C. S. Haley, U. Renne, M. Schwerin et al., 2000. Single QTL effects, epistasis, and pleiotropy account for two thirds of the phenotypic F2 variance of growth and obesity in DU6i × DBA/2 mice. Genome Res. 10: 1941–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockmann, G. A., C. S. Haley, E. Wolf, S. Karle, J. Kratzsch et al., 2001. Genome-wide search for loci controlling serum IGF binding protein levels of mice. FASEB J. 15: 978–987. [DOI] [PubMed] [Google Scholar]
- Brockmann, G. A., E. Karatayli, C. S. Haley, U. Renne, O. J. Rottmann et al., 2004. QTLs for pre- and postweaning body weight and body composition in selected mice. Mamm. Genome 15: 593–609. [DOI] [PubMed] [Google Scholar]
- Broman, K.W., 2005. The genomes of recombinant inbred lines. Genetics 169: 1138–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunger, L., A. Laidlaw, G. Bulfield, E. J. Eisen, J. F. Medrano et al., 2001. Inbred lines of mice derived from long-term growth selected lines: unique resources for mapping growth genes. Mamm. Genome 12: 678–686. [DOI] [PubMed] [Google Scholar]
- Cattanach, B. M., 1986. Parental origin effects in mice. J. Embryol. Exp. Morphol. 97: 137–150. [PubMed] [Google Scholar]
- Cattanach, B. M., and C. V. Beechey, 1990. Autosomal and X-chromosome imprinting. Dev. Suppl.: 63–72. [PubMed]
- Cheverud, J. M., J. J. Rutledge and W. R. Atchley, 1983. Quantitative genetics of development: genetic correlation among age specific trait values and the evolution of ontogeny. Evolution 37: 895–905. [DOI] [PubMed] [Google Scholar]
- Cheverud, J. M., E. J. Routman, F. A. Duarte, B. van Swinderen, K. Cothran et al., 1996. Quantitative trait loci for murine growth. Genetics 142: 1305–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darvasi, A., 1998. Experimental strategies for the genetic dissection of complex traits in animal models. Nat. Genet. 18: 19–24. [DOI] [PubMed] [Google Scholar]
- Engellandt, T., and B. Tier, 2002. Genetic variances due to imprinted genes in cattle. J. Anim. Breed. Genet. 119: 154–165. [Google Scholar]
- Falconer, D. S., 1953. Selection for large and small size in mice. J. Genet. 51: 470–501. [Google Scholar]
- Falconer, D. S., and F. C. Mackay, 1995. Introduction to Quantitative Genetics. Longman, New York.
- Heath, S. C., 1997. Markov chain Monte Carlo segregation and linkage analysis for oligogenic models. Am. J. Hum. Genet. 61: 748–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander, E. S., and P. Green, 1987. Construction of multilocus genetic linkage maps in humans. Proc. Natl. Acad. Sci. USA 84: 2363–2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leamy, L., and J. M. Cheverud, 1984. Quantitative genetics and the evolution of ontogeny. II. Genetic and enviromental correlations among age-specific characters in randombred house mice. Growth 48: 339–353. [PubMed] [Google Scholar]
- Morison, I. M., C. J. Paton and S. D. Cleverley, 2001. The imprinted gene and parent-of-origin effect database. Nucleic Acids Res. 29: 275–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau, J. H., J. B. Singer, A. Matin and E. S. Lander, 2000. Analysing complex genetic traits with chromosome substitution strains. Nat. Genet. 24: 221–225. [DOI] [PubMed] [Google Scholar]
- Riska, B., W. R. Atchley and J. J. Rutledge, 1984. A genetic analysis of targeted growth in mice. Genetics 107: 79–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüler, L., 1985. Mouse strain Fzt:Du and its use as a model in animal breeding research. Arch. Tierz. 28: 357–363. [Google Scholar]
- Singer, J. B., A. E. Hill, L. C. Burrage, K. R. Olszens, J. Song et al., 2004. Genetic dissection of complex traits with chromosome substitution strains of mice. Science 18: 18. [DOI] [PubMed] [Google Scholar]
- Sobel, E., and K. Lange, 1996. Descent graphs in pedigree analysis: applications to haplotyping, location scores, and marker sharing statistics. Am. J. Hum. Genet. 58: 1323–1337. [PMC free article] [PubMed] [Google Scholar]
- Taylor, B. A., L. M. Tarantino and S. J. Phillips, 1999. Gender-influenced obesity QTLs identified in a cross involving the KK type II diabetes-prone mouse strain. Mamm. Genome 10: 963–968. [DOI] [PubMed] [Google Scholar]
- Taylor, B. A., C. Wnek, D. Schroeder and S. J. Phillips, 2001. Multiple obesity QTLs identified in an intercross between the NZO (New Zealand obese) and the SM (small) mouse strains. Mamm. Genome 12: 95–103. [DOI] [PubMed] [Google Scholar]
- Vaughn, T. T., L. S. Pletscher, A. Peripato, K. King-Ellison, E. Adams et al., 1999. Mapping quantitative trait loci for murine growth: a closer look at genetic architecture. Genet. Res. 74: 313–322. [DOI] [PubMed] [Google Scholar]