Abstract
The DDK syndrome is an early embryonic lethal phenotype observed in crosses between females of the DDK inbred mouse strain and many non-DDK males. Lethality results from an incompatibility between a maternal DDK factor and a non-DDK paternal gene, both of which have been mapped to the Ovum mutant (Om) locus on mouse chromosome 11. Here we define a 465-kb candidate interval for the paternal gene by recombinant progeny testing. To further refine the candidate interval we determined whether males from 17 classical and wild-derived inbred strains are interfertile with DDK females. We conclude that the incompatible paternal allele arose in the Mus musculus domesticus lineage and that incompatible strains should share a common haplotype spanning the paternal gene. We tested for association between paternal allele compatibility/incompatibility and 167 genetic variants located in the candidate interval. Two diallelic SNPs, located in the Schlafen gene cluster, are completely predictive of the polar-lethal phenotype. These SNPs also predict the compatible or incompatible status of males of five additional strains.
THE “DDK syndrome” (Babinet et al. 1990) is a parent of origin embryonic lethal phenotype that was first described in 1960 (Tomita 1960). Crosses between DDK females and many non-DDK males are infertile, but reciprocal crosses of non-DDK females mated to DDK males and intrastrain DDK crosses are fully viable (Wakasugi et al. 1967; Wakasugi 1973, 1974). Death has been shown to occur during early embryonic development, from the morula-blastocyst stage to around the time of implantation (Wakasugi 1974). Nuclear and cytoplasmic transfer studies have demonstrated that the embryos die as a result of an incompatible interaction between a maternal cytoplasmic factor that is present in the oocytes of DDK females and a non-DDK paternal gene (Renard and Babinet 1986; Babinet et al. 1990; Renard et al. 1994; Gao et al. 2005).
Both the maternal factor and the paternal gene are tightly linked and have been mapped to the Ovum mutant (Om) locus on mouse chromosome 11 (Wakasugi 1974; Baldacci et al. 1992, 1996; Sapienza et al. 1992; Cohen-Tannoudji et al. 1996; Pardo-Manuel de Villena et al. 1997). Progeny testing in BALB/c backcrosses identified a 1.5 Mb candidate interval for the paternal gene located between Scya2 and D11Pas18 (Baldacci et al. 1996; Cohen-Tannoudji et al. 2000). The candidate interval contains 33 known transcripts, 5 novel transcripts, 1 pseudogene, 9 ESTs, and 22 novel Genescan predictions (Le Bras et al. 2002; http://www.ensembl.org). Positional cloning of the paternal gene has been hindered by the large number of genes present in the candidate interval, the lack of obvious candidates, the early onset of the lethal phenotype, and the complex nature of the phenotype, requiring specific allelic contributions in a parent of origin-dependent manner.
Until recently, studies on the DDK syndrome have focused on “classical” laboratory inbred strains such as BALB/c, BS, CBA, C57BL/6, C3H, DDK, KK, and NC. All of these strains, with the exception of DDK, carry an incompatible allele at the paternal gene (Wakasugi et al. 1967; Wakasugi 1973, 1974; Wakasugi and Morita 1977; Mann 1986; Buehr et al. 1987; Baldacci et al. 1992, 1996; Sapienza et al. 1992; Leclerc et al. 1994; Pardo-Manuel de Villena et al. 1996, 1997, 1999), suggesting that the DDK strain carries rare mutations in both maternal and paternal components of the syndrome. Recently this view has been challenged by a report by Zhao et al. (2002) demonstrating that the alleles present at the paternal gene in two wild-derived inbred strains, MOM (Mus musculus molossinus) and CASP (Mus musculus castaneus) are fully compatible with the DDK maternal factor. Based on these data, they proposed that the DDK syndrome was an incompatibility between a mutated maternal DDK allele and a normal paternal Mus musculus domesticus allele at the Om locus. The view that compatible and incompatible paternal alleles represent a common polymorphism in M. musculus suggests that association mapping may be an efficient approach to identify the paternal gene.
To test this hypothesis we combined the characterization of the compatibility between the maternal DDK factor and the alleles present at the paternal gene in sires from 17 classical and wild-derived strains with our extensive sequencing data for a reduced candidate interval defined by progeny testing in the C57BL/6–DDK background. The DDK syndrome phenotype of each male was determined using the reproductive performance in matings to (C57BL/6 × DDK)F1 females. Our results demonstrate the value of combining multiple approaches in the identification of the genes involved in “complex” phenotypes. In particular, our study demonstrates that common functional genetic variation can be mapped with high resolution using panels of inbred strains with diverse phylogenetic histories.
MATERIALS AND METHODS
Mouse strains:
All mice and DNA from all strains were originally obtained from The Jackson Laboratory (Bar Harbor, ME) except for JF1/Ms and DDK/Pas that are maintained by Terry Magnuson and Fernando Pardo-Manuel de Villena, respectively, at the University of North Carolina at Chapel Hill. Based on their origin and known genetic make up, these strains were classified into three groups (http://www.jax.org; Ideraabdullah et al. 2004). The first group includes 11 wild-derived strains assigned to the following taxa: Mus spicilegus, PANCEVO/EiJ; M. m. castaneus, CAST/EiJ; M. m. molossinus, MOLC/RkJ; M. m. domesticus, PERA/EiJ, PERC/EiJ, ZALENDE/EiJ, TIRANO/EiJ, LEWES/EiJ, RBA/DnJ, and WSB/EiJ; M. m. domesticus × M. m. musculus hybrid, SKIVE/EiJ. The second group includes a single strain, JF1/Ms, with a complex history [although JF1/Ms is often described as M. m. molossinus, it is in fact a fancy mouse with considerable contribution of M. m. domesticus (http://www.jax.org)]. The last group includes seven classical strains: BALB/cJ, C57BL/6J, DBA/2J, A/J, 129X1/SvJ, C3H/HeJ, and DDK/Pas. All mice described in this report were treated according to the recommendations of the Institutional Animal Care and Use Committee (IACUC) of the University of North Carolina at Chapel Hill.
Selection of males:
Progeny testing:
Over 5000 meioses from C57BL/6 × DDK backcrosses and intercrosses were screened for the presence of recombination between D11Mit33 and D11Mit35. Animals carrying recombinant chromosomes were backcrossed to the parental strains or intercrossed to other C57BL/6–DDK mixed background mice to generate 40 experimental males. Recombinants were genotyped at the following 15 informative markers described previously: D11Mit33, D11Mit35, D11Mit37, D11Mit66, D11Mit97, D11Mit120, D11Mit283, D11Mi354, D11Pas18, D11Spn1, D11Spn2, D11Spn4, Cct6b-Ap2b1-29, Scya1, and Scya2 (Dietrich et al. 1996; Cohen-Tannoudji et al. 2000; Pardo-Manuel de Villena et al. 2000b; Ideraabdullah et al. 2004; Wu et al. 2005). In addition, recombinants were genotyped at 13 new microsatellite markers generated in our laboratories: D11Spn10, D11Spn31, D11Spn36, D11Spn39, D11Spn72, D11Spn78, D11Spn104, D11Spn128, D11Spn129, D11Spn173, D11Spn178, Scya6, and Scya7. Accession numbers at EMBL Nucleotide Sequence Database are AM075608–AM075620. The primers for these assays are provided in supplemental Table 1 at http://www.genetics.org/supplemental/. Genotypes were determined by PCR amplification and electrophoresis as described previously (Pardo-Manuel de Villena et al. 1996, 1997, 1999, 2000a,b). Only markers flanking the recombination events in the candidate region were used in the mapping analyses to maximize the information with the fewest number of markers (supplemental Table 2 at http://www.genetics.org/supplemental/).
Inbred males: Wild-derived strains were chosen to include a broad spectrum of M. musculus subspecies and a related species, M. spicilegus. We included an excess of M. m. domesticus strains because this is the only subspecies for which wild-derived strains have not been characterized previously for compatibility with the maternal DDK factor (Zhao et al. 2002). The classical strains included in this study are commonly used in mouse genetics studies.
Reproductive performance:
In all crosses described in this study, males were mated to 9- to 45-week-old identical (C57BL/6 × DDK)F1 females. The three crosses used as controls in this study, (C57BL/6 × DDK)F1 × C57BL/6, (C57BL/6 × DDK)F1 × DDK, and (C57BL/6 × DDK)F1 × (C57BL/6 × DDK)F1 (in all crosses described in this study the dam is always listed first and the sire second unless otherwise indicated) have been described previously (Sapienza et al. 1992; Pardo-Manuel de Villena et al. 1996, 1997, 1999, 2000a,b). Some of the crosses involving inbred males have been previously described in our efforts to characterize a meiotic drive phenotype (Kim et al. 2005; Wu et al. 2005). Briefly, each male was mated to identical (C57BL/6 × DDK)F1 females and monitored daily for newborn pups. The litter size was determined at birth to avoid biases due to postnatal lethality and cannibalization. The distribution of litter sizes produced by each type of male was used to determine the phenotype of the male.
Lower reproductive performance has been observed consistently in interspecific and intersubspecific crosses due to embryonic lethality that is not associated with the DDK syndrome (Alibert et al. 1997; Britton-Davidian et al. 2005). Therefore, it is necessary to correct for this effect based on the estimated phylogenetic divergence between the dam and the sire. Because the females used in this study are hybrids of two classical inbred strains whose genomes are derived mostly from M. m. domesticus, no correction is necessary in crosses involving males from classical strains, M. m. domesticus wild-derived strains, or JF1/Ms (see above). A second group includes a single strain, SKIVE/EiJ, a hybrid of M. m. musculus and M. m. domesticus, and the correction factor used was 1.09 (Table 1). Finally, a correction factor of 1.29 was used in crosses involving males from other subspecies (MOLC/RkJ and CAST/EiJ) and M. spicilegus (PANCEVO/EiJ). These correction factors were determined experimentally in crosses that cannot have DDK syndrome-related lethality and are the ratio between the average litter size in intrasubspecific crosses and the average litter size in interspecific and intersubspecific crosses (Table 1). The corrected values are used throughout our analyses.
TABLE 1.
Crosses used to determine the correction factors for reproductive performance in interspecific and intersubspecific crosses
| Type of cross | Dam | Sire | Average litter sizea | SD | No. of litters | References |
|---|---|---|---|---|---|---|
| Intersubspecific | (C57BL/6 × CASP)F1 | CASP | 7.3 | 1.20 | 9 | Zhao et al. (2002) |
| (C57BL/6 × MOM)F1 | C57BL/6 | 6.3 | 1.66 | 11 | Zhao et al. (2002) | |
| (C57BL/6 × MOM)F1 | MOM | 7.1 | 1.73 | 12 | Zhao et al. (2002) | |
| (CAST × C57BL/6)F1 | C57BL/6 | 8.27 | 1.57 | 66 | This study | |
| C57BL/6 | (C57BL/6 × CASP)F1 | 7.9 | 1.44 | 13 | Zhao et al. (2002) | |
| C57BL/6 | (C57BL/6 × MOM)F1 | 7.3 | 2.71 | 15 | Zhao et al. (2002) | |
| C3H | (C3H × MOM)F1 | 8 | 1.32 | 7 | Zhao et al. (2002) | |
| Subtotal | 7.45 (1.29) | |||||
| Hybrid | (C57BL/6 × SKIVE)F1 | C57BL/6 | 8.74 | 1.79 | 39 | This study |
| Subtotal | 8.74 (1.09) | |||||
| Intrasubspecific | (PERA × C57BL/6)F1 | C57BL/6 | 9.9 | 0.94 | 11 | This study |
| (C57BL/6 × PERA)F1 | C57BL/6 | 10.04 | 1.95 | 104 | This study | |
| (C57BL/6 × PERC)F1 | C57BL/6 | 8.9 | 2.33 | 50 | This study | |
| Subtotal | 9.61 (1) |
The correction factors for interspecific and hybrid crosses are the ratio between the average litter size in intrasubspecific crosses and the average litter size in intersubspecific and hybrid crosses, respectively, and are shown in parentheses.
Sequence variants:
Most of the sequence variants used in the in silico mapping and phylogenetic studies have been described previously [Cct6b-Ap2b1-14-20, 22-36 (Ideraabdullah et al. 2004)]. In addition, we have sequenced a fragment of 825 bp in the intergenic region between Slfn10 and Slfn2 (positions 82651885 to 82652710; forward 1, 5-TCCAGATGTATATAATTGAGGTG-3′; forward 2, 5′-GACCCAGAATAAGGCCTAAC-3′; reverse 1, 5′-CAAGCTGAAGATTTGAAATGAC-3′; reverse 2, 5′-GTGCAAGTTAGAAACTACAAGC-3′), and two fragments in the Slfn1 gene: a 677 bp fragment in exon 1 (positions 82729880 to 82730557; forward 1, 5′-GCTTCAACAGGTGCTCATGC-3′; forward 2, 5′-GGGGATTTGAGTGACGCTG-3′; reverse 1, 5′-CAGAGTTCCTAGAGAAGCACC-3′; reverse 2, 5′-CATTCTTGGTAGGCACTGG-3′) and a 1193 bp in exon 2 (positions 82734269 to 82735462; forward 1, 5′-CAAGCACATTTTGCTGTTAGC-3′; forward 2, 5′-TGGAAAATGAACATCACCG-3′; forward 3, 5′-CCCAACACTAATGTCTCTGTC-3′; reverse 1, 5′-CAACATCCCCAGCTAAACG-3′; reverse 2, 5′-AAGACATGAGGAGCTTGATCC-3′) (in each case multiple sets of primers were used to ensure complete coverage). Finally, we have also sequenced a 341 bp fragment, from 83005403 to 83004744, distal to the Ap2b1 gene (forward, 5′-AAAGGGCGACTGACCCTCATCAAA-3′; and reverse, 5′-ATGGGTGGAGCACCAAACTACGTA-3′) [all positions provided in this paper refer to the Ensembl gene build for the NCBI m33 mouse assembly (http://www.ensembl.org/Mus_musculus/)]. The genotypes in C3H/HeJ, WSB/EiJ, KK/HlJ, CBA/J, and PWK/PhJ at SNPs 82843176 and 82843476 were determined by PCR sequencing using the Cct6b-Ap2b1-26-F and R primers described previously (Ideraabdullah et al. 2004). The subspecific origin of the haplotypes of classical and hybrid strains in the D11Spn173–D11Spn129 region was determined on the basis of the presence of diagnostic alleles at 26 variants (supplemental Table 3 at http://www.genetics.org/supplemental/) distributed across the following fragments: Cct6b-Ap2b1-25-27 and Cct6b-Ap2b1-36 (Ideraabdullah et al. 2004). Sequences have been submitted to EMBL and access numbers will be provided.
Dot plot matrix:
The sequence flanked by D11Spn173 and D11Spn129 (positions 82560436 to 83025035 on chromosome 11) was retrieved from the Ensembl gene build for the NCBI m33 mouse assembly (http://www.ensembl.org/Mus_musculus/). The homologous region in the rat genome (positions 71,159,554 to 71,507,449 on chromosome 10, version 3.4 “November 2004 Update” of the rat genome assembly) was identified by BLAST search using the first and last 1 kb of the mouse interval. Dot plot analyses (Maizel and Lenk 1981; Pustell and Kafatos 1982; Quigley et al. 1984) were used to visualize the presence of duplications and inversions within M. musculus and to align homologous regions of the mouse and rat. These analyses were performed using a dedicated website at the University of Colorado (http://arbl.cvmbs.colostate.edu/molkit/dnadot/index.html). We used a 99 bp sliding window and 10 bp mismatch limit in the production of the matrix plots. The position and identity of genes in the mouse and rat regions was retrieved from Ensembl (http://www.ensembl.org), UCSC (http://genome.ucsc.edu) and NCBI (http://www.ncbi.nlm.nih.gov). In addition, mouse genes were used to identify additional putative homologous genes in the rat using BLAST search.
Phylogenetic analyses:
Sequences from the Slfn1 gene and fragments Cct6b-Ap2b1-22-27 (Cct6b-Ap2b1-25 was excluded in these analyses because the DDK strain has a deletion encompassing this fragment) were aligned as previously described (Ideraabdullah et al. 2004). In addition to the 17 inbred strains phenotyped, we also included PWK/PhJ and SPRET/EiJ to ensure complete representation of different lineages. All analyses were performed using the PHYLIP phylogeny inference software package, version 3.6 (Felsenstein 1989). For each set of sequences we generated 100 bootstrapped datasets using the SEQBOOT program. We then determined the phylogeny using a distance matrix method (NEIGHBOR), a maximum likelihood method (DNAML) and a maximum parsimony method (DNAPARS). The CONSENSE program was used to construct majority rule consensus trees.
Statistical Analysis and Mapping:
Litter size was analyzed using mixed and nested-mixed models within the computer program SAS version 9.1 (SAS Institute, Cary, NC). Three sets of statistical analyses were performed. In all analyses, to account for the correlation between litter sizes from the same male, “male” was treated as a random effect in our statistical models. In the first set of analyses, we tested for associations between genotype and litter size in 40 males with different C57BL/6–DDK backgrounds using a mixed model approach. In our second set of statistical models, we calculated the average litter size and their associated 99% confidence intervals for each of the 17 strains phenotyped. After accounting for the father, it was determined that the correlation in litter size due to the dam was not statistically significant. Finally, in our third set of statistical models we analyzed 167 markers in 17 inbred strains to determine whether genotype was associated with litter size using nested-mixed models. Genotype was treated as a fixed effect while strain was treated as a random effect to account for the correlation between litter sizes within strains, and father's identity was treated as a nested random effect within strain.
After accounting for genotype, the residuals were distributed in a symmetric, unimodal fashion, consistent with distributional assumptions necessary for the validity of the mixed model. The distribution of litter size was ordinal, with values ranging from 1 to 16. Given the discrete ordinal nature of the data, assumptions regarding the normality of the residuals in our statistical models were of some concern. However, the large sample size and the resulting asymptotics should make our P values fairly robust.
To validate our findings we performed permutation tests, based on 10,000 to 100,000 random replicate data sets, to assess the empirical statistical significance of our findings. For this application, permutation tests are very conservative because in order to maintain the correlation structure of the data and hence “exchangeability,” blocks consisting of all the litters for a given male (for the first set of analyses) or a given strain (for the third set of analyses) were permuted together rather than permuting individual litters. This constraint led to a relatively small number of possible permutations of the data and hence a conservative lower bound on the P values. In some instances it was feasible to calculate the empirical probability of our findings under the null hypothesis by determining the exact number of possible permutations of the genotype data that would result in as extreme or more extreme F statistics than the ones observed in our mixed models. In addition to protecting our conclusions from inaccurate P values due to erroneous distributional assumptions, the permutation test procedure offers some protection from overinterpreting results from a relatively small sample of males or strains, as the statistical tests are conditional on the observed phenotype and genotype data.
RESULTS
High-resolution recombination mapping of the paternal gene:
To map the paternal component of the syndrome requires that the females used in the experimental crosses produce oocytes with the maternal DDK factor. If this requirement is fulfilled, mapping the paternal gene is straightforward because the fate of the resulting embryos depends exclusively on whether they inherit a compatible or incompatible allele at the paternal gene (Sapienza et al. 1992). Therefore, there is an inverse proportional relationship between the number of incompatible alleles (0, 1, or 2) in a male and its reproductive performance. In addition, the presence of transmission ratio distortion against incompatible alleles can be used to determine whether males of mixed genetic background are homozygous or heterozygous for compatible and incompatible alleles at the paternal gene. In this study we use litter size as an indirect measure of the level of embryonic lethality (Pardo-Manuel de Villena et al. 1997, 1999). In crosses involving (C57BL/6 × DDK)F1 hybrid females the litter size is normally distributed around the mean and depends on the genotype of the sire at the paternal gene ((Pardo-Manuel de Villena et al. 1997, 1999). In contrast, the litter size is not normally distributed in crosses involving DDK females and most matings do not produce live pups (Pardo-Manuel de Villena et al. 1999). In these crosses, characterization of the DDK syndrome phenotype requires using both litter size and delivery ratio data and is subject to greater uncertainty (Pardo-Manuel de Villena et al. 1999).
We have characterized the reproductive performance of crosses between identical (C57BL/6 × DDK)F1 females and 40 males with mixed C57BL/6–DDK background (supplemental Table 2 at http://www.genetics.org/supplemental/). Crosses yielded an average of 29 litters (range 10–47). The males can be divided into two groups based on their recombination status in the 1.5 Mb interval defined previously (Baldacci et al. 1992, 1996). Twenty-two males carry nonrecombinant chromosomes in this interval while the remaining 18 males carry different combinations of five types of recombinant chromosomes defining six smaller intervals (Figure 1 and supplemental Table 2 at http://www.genetics.org/supplemental/).
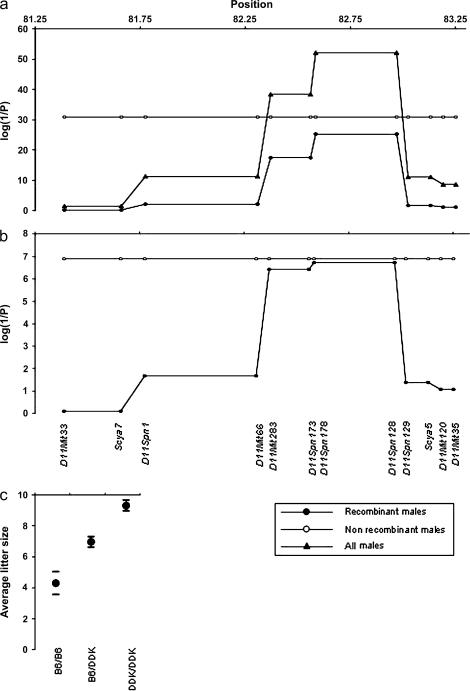
Figure 1.
Mapping by recombinant progeny testing in C57BL/6–DDK background. The reproductive performances of 40 males were used to map the paternal gene in the candidate interval defined previously (Baldacci et al.1992, 1996). The original interval was divided into six smaller intervals based on the presence of five recombinant chromosomes. The markers shown in the bottom of section b convey the complete genotypic information (supplemental Table 2 at http://www.genetics.org/supplemental/). In sections a and b the horizontal axes show distance in Mbp from the centromere of chromosome 11. The vertical axes show the statistical significance of the association findings. Triangles show the results using the entire set of 40 males. Open circles show the results for the 22 males carrying nonrecombinant chromosomes in the D11Mit33–D11Mit35 interval. Filled circles show the results in the 18 males carrying recombinant chromosomes in the D11Mit3–D11Mit35 interval (the inset in the bottom right shows this information visually). (a) The top plot shows the strength of the associations between genotype and litter size using the Fdistribution to assess the statistical significance of the mixed models. (b) The bottom plot shows the strength of the association between genotype and litter size using permutation tests to assess the statistical significance of the mixed models. Empirical results for the 22 nonrecombinant males and for the regions containing markers D11Spn173 and D11Spn178 among the 18 recombinant males were determined exactly. The statistical significance when analyzing all 40 males for the regions containing marker D11Mit33 (P = 0.045) was estimated using a permutation test while the statistical significance for D11Spn173 (P = 1.3 × 10−15) and D11Spn178 (P = 3.4 × 10−16) were calculated exactly (data not shown). It was not feasible to calculate the empirical statistical significance for the other regions (P < 1.0 × 10−5) using all 40 males. The statistical significance of the findings in these other regions were beyond the resolution of 100,000 random replications of the data and there were too many permutations of the data that would yield a more extreme value than the observed F-statistics to make it possible to determine the empirical significance exactly. (c) Circles represent the average litter size in males with the three possible genotypes in the D11Spn178–D11Spn128. The horizontal bars show the boundaries of the 99% confidence intervals.
First we confirmed that the 1.5 Mb interval analyzed contains the paternal gene using the reproductive performance of the 22 males with nonrecombinant chromosomes (Figure 1). Males homozygous for DDK alleles have the best reproductive performance (average litter size, 9.48 ± 0.17), males homozygous for C57BL/6 alleles have the worst reproductive performance (average litter size, 4.51 ± 0.49), and heterozygous males have an intermediate behavior (average litter size, 7.06 ± 0.16). These averages are as expected from mean and distribution of the litter sizes of the control crosses (Pardo-Manuel de Villena et al. 1997, 1999).
We then determined which of the six smaller intervals contains the paternal gene by testing for association between the genotypes present in these intervals and the reproductive performance of both the complete set of 40 males and the 18 recombinant males. Only two intervals, D11Spn178–D11Spn128 and D11Mit283–D11Spn173, show significant association in both the complete set of males and the recombinant males only (Figure 1). The most significant values were obtained for the D11Spn178–D11Spn128 interval. Again, males homozygous for DDK alleles in this interval have the best reproductive performance (average litter size, 9.07 ± 0.24), males homozygous for C57BL/6 alleles have the worst reproductive performance (average litter size, 4.19 ± 0.36), and heterozygous males have an intermediate behavior (average litter size, 6.81 ± 0.22). These results indicate that the paternal gene lies between the two recombinations defining this interval (Figure 1).
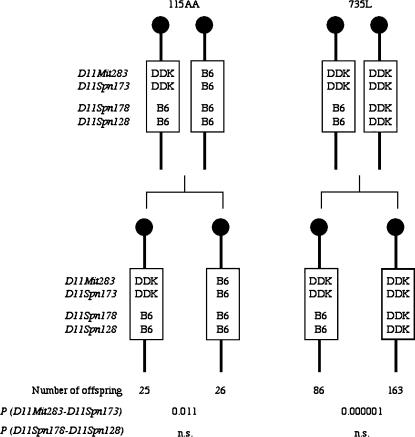
This conclusion was confirmed by analyzing the inheritance of paternal alleles in the surviving offspring of the two males, 115AA and 735L (Figure 2 and supplemental Table 2 at http://www.genetics.org/supplemental/), that have different genotypes in the D11Spn178–D11Spn128 and D11Mit283–D11Spn173 intervals. Each of these males is heterozygous for one interval and homozygous for the other. Because the DDK syndrome embryonic lethality requires inheritance of a paternal C57BL/6 allele, there should be an excess of offspring inheriting paternal DDK alleles in the progeny of the male that is heterozygous for the interval containing the paternal gene, and equal inheritance of paternal alleles in the male that is homozygous for the interval containing the paternal gene. As shown in Figure 2 there is a highly significant transmission ratio distortion against the inheritance of the recombinant chromosome among the offspring the 735L male while there is equal transmission of both chromosomes in the progeny of the 115AA male. These results are in agreement with the paternal gene lying in the D11Spn178–D11Spn128 interval while the proximal interval may be rejected (Figure 2).
Figure 2.
Transmission ratio of paternal haplotypes in the Om region to the progeny of sires with critical recombinant chromosomes. The haplotypes in the vicinity of Om of the two males, 115AA and 735L, that carry recombinant chromosomes between D11Spn173 and D11Spn178, and the number of offspring inheriting each paternal haplotype are shown. The figure also provides the level of significance for each interval using the chi-square test statistics under the null hypotheses of equal transmission of alleles in the progeny of homozygous males for that interval and 66% transmission of the DDK allele in the progeny of heterozygous males for that interval (Pardo-Manuel de Villena et al. 2000b). n.s., not significant.
In conclusion, we have defined a 465-kb interval that contains the paternal gene responsible for the DDK syndrome. This interval is flanked by the excluded markers D11Spn173 and D11Spn129 (Figure 1).
Characterization of the paternal allele compatibility of different inbred strains with the maternal DDK factor:
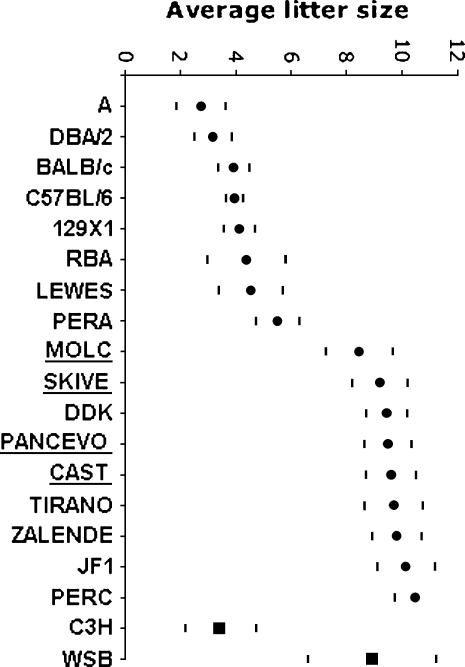
We have characterized the reproductive performance of (C57BL/6 × DDK)F1 females mated to males from 15 inbred strains (11 wild-derived strains: PANCEVO, CAST, JF1, MOLC, SKIVE, PERA, PERC, ZALENDE, TIRANO, LEWES, and RBA, and four classical strains: A, 129X1, DBA/2, and BALB/c). Crosses yielded an average of 47 litters (range 16–103). We have corrected the litter size in interspecific and intersubspecific crosses to account for any reductions in litter size (as compared to intrasubspecific crosses) that are unrelated to the DDK syndrome (see materials and methods). Figure 3 shows the mean litter size and 99% confidence intervals for these crosses. Figure 3 also includes the reproductive performance of C57BL/6 and DDK males obtained in previous studies (Pardo-Manuel de Villena et al. 1996, 1997, 1999). Inspection of Figure 3 reveals that there is considerable variation in the embryonic lethal phenotype depending on the inbred male used in the cross.
Figure 3.
Reproductive performance of males from different inbred strains. The vertical axis represents the average litter size observed in crosses between (C57BL/6 × DDK)F1 females and males from the inbred strain listed in the horizontal axis. Circles denote the strains used for in silico mapping and squares are the strains used to confirm the presence of complete linkage disequilibrium between the phenotype and selected SNPs (see text). The horizontal bars show the boundaries of the 99% confidence intervals adjusted for the correlation between litters from the same sire. Correction factors were applied to determine the reproductive performance of the underlined strains (see materials and methods).
Association mapping:
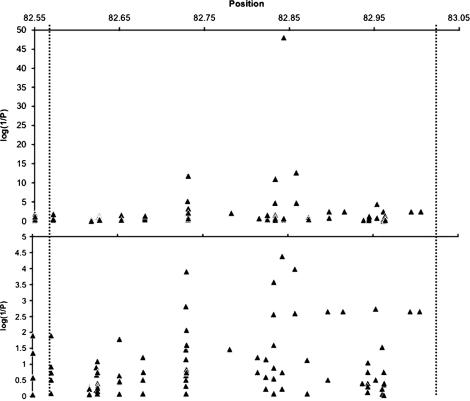
To further refine the location of the paternal gene we tested whether the reproductive performance (using the average litter size as a quantitative trait, see materials and methods) of the 17 inbred strains analyzed in the previous section is associated with the alleles present at any of 167 variants distributed across 26 regions spanning the entire D11Spn173–D11Spn129 candidate interval defined by recombinant progeny testing. The results of this analysis are shown in Figure 4. Several variants are significantly associated with reproductive performance. These variants are distributed across four fragments spanning 128 kb (Figure 5a). Two diallelic SNPs at positions 82,843,176 and 82,843,476 show the strongest association with the phenotype. Compatible strains carry G and T alleles, respectively, at these two SNPs while incompatible strains carry the A and G alleles (SNPs 10 and 11 in Table 2). The level of significance is exceptionally high and these variants are optimally associated with the reproductive performance phenotype (i.e., given the available phenotype data, it is not possible to construct any combination of genotypes for a SNP across the 17 strains that could better explain the phenotype than what we have observed for these two variants).
Figure 4.
In silico mapping. The association between the reproductive performance of males from the 17 inbred strains analyzed in Figure 3 and 167 diallelic variants distributed across the candidate interval defined by progeny testing is shown. Variants are shown as triangles and the position along the horizontal axis refers to the distance in megabasepairs from the centromere of chromosome 11. The vertical axes show the statistical significance of the association results while the dashed vertical lines denote the proximal and distal boundaries of the candidate interval. In some cases multiple variants with the same degree of association appear as a single triangle. The top plot shows the strength of the associations between genotype and litter size using the Fdistribution to assess the statistical significance of the nested-mixed models. The bottom plot shows the strength of the association between genotype and litter size using permutation tests to assess the statistical significance of our findings. The empirical significance estimates for the SNPs on the figure with the four most significant findings were determined exactly.
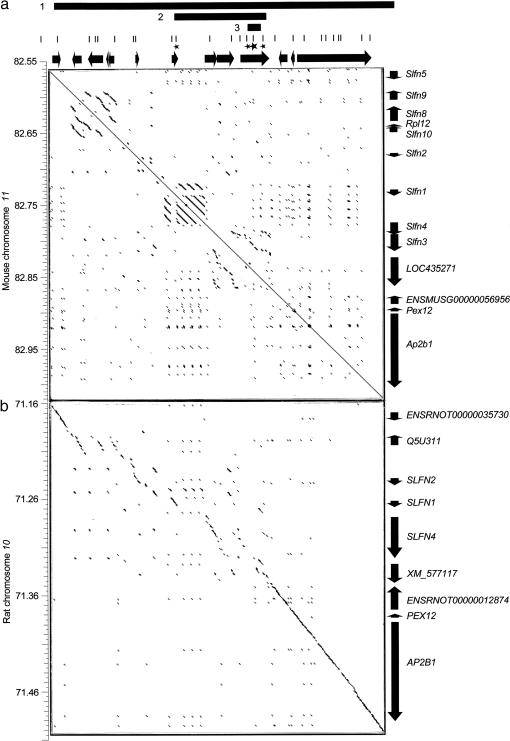
Figure 5.
Rapid evolution of the Schlafen gene cluster in mouse. The three bars shown at the top of the figure represent the candidate intervals defined by recombinant progeny testing (1), in silico mapping (2), and haplotype block in complete linkage disequilibrium with the DDK syndrome phenotype (3). The 26 fragments sequenced are shown as vertical bars directly underneath. Stars denote the four fragments containing the 12 variants most strongly associated with the phenotype. The larger star indicates the fragment containing the two variants that are in complete linkage disequilibrium with the DDK syndrome phenotype. Genes are shown as black arrows. Vertical axes provide distance in Mbp from the centromere in the appropriate chromosome. (a) Dot plot matrix comparing the candidate interval in the mouse against itself. (b) Dot plot matrix comparing the mouse candidate interval (horizontal axis) and the homologous region in rat (vertical axis). Each dot in the matrixes denotes an orthogonal 99 bp region with fewer than 10 mismatches. Consecutive dots form diagonal lines in regions of extended identity/similarity. For any given region on the horizontal or vertical axes the presence of multiple parallel diagonal lines denotes duplicated regions. Short scattered lines are for most part indicative of repetitive elements scattered across the region.
TABLE 2.
SNPs associated with the reproductive performance phenotype
| Strain
|
D11Spn173– D11Spn129
|
Paternal allele
|
SNP
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | Referencea | |||
| BS | domesticus? | I | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | Wakasugi and Morita (1977) |
| NC | domesticus? | I | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | Wakasugi et al. (1967) |
| A | domesticus | I | C | T | T | A | T | T | G | G | G | A | G | A | This study |
| DBA/2 | domesticus | I | C | T | T | A | T | T | G | G | G | A | G | A | This study |
| BALB/c | domesticus | I | C | T | T | A | T | T | G | G | G | A | G | A | This study |
| C57BL6 | domesticus | I | C | T | T | A | T | T | G | G | G | A | G | A | This study |
| 129X1 | domesticus | I | C | T | T | A | T | T | G | G | G | A | G | A | This study |
| PERA | domesticus | I | C | T | T | A | T | T | G | G | G | A | G | A | This study |
| RBA | domesticus | I | C | T | T | A | T | T | G | G | G | A | G | A | This study |
| CBAb | domesticus | I | ND | ND | ND | ND | T | T | ND | G | G | A | G | ND | Mann (1986) |
| C3Hb | domesticus | I | ND | ND | ND | ND | T | T | G | G | G | A | G | A | Buehret al. (1987); Leclercet al. (1994); this study |
| KKb | domesticus | I | ND | ND | ND | ND | A | C | A | A | A | A | G | G | Wakasugi et al. (1967) |
| LEWES | domesticus | I | C | T | T | A | A | C | A | A | A | A | G | G | This study |
| PWKb | musculus | C | C | T | T | A | A | C | A | A | A | G | T | G | Jiri Forejt (personal communication) |
| SKIVE | musculus | C | C | T | T | A | A | C | A | A | A | G | T | G | This study |
| WSBb | domesticus | C | ND | ND | ND | ND | ND | ND | ND | ND | ND | G | T | G | This study |
| DDK | domesticus | C | A | A | C | G | del | del | del | del | del | G | T | G | This study |
| MOLC | castaneus | C | A | A | C | G | A | C | A | A | A | G | T | G | This study |
| PANCEVO | M. spicilegus | C | A | A | C | G | A | C | A | A | A | G | T | G | This study |
| CAST | castaneus | C | A | A | C | G | A | C | A | A | A | G | T | G | This study |
| TIRANO | domesticus | C | A | A | C | G | A | C | A | A | A | G | T | G | This study |
| ZALENDE | domesticus | C | A | A | C | G | A | C | A | A | A | G | T | G | This study |
| JF1 | castaneus | C | A | A | C | G | A | C | A | A | A | G | T | G | This study |
| PERC | domesticus | C | A | A | C | G | A | C | A | A | A | G | T | G | This study |
| MOM | molossinus | C | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | Zhao et al. (2002) |
| CASP | castaneus | C | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | Zhao et al. (2002) |
The table provides the strain name, phylogenetic origin of the haplotype in the candidate interval for the paternal gene defined by progeny testing and whether the allele at the paternal gene is incompatible (I) or compatible (C) with the maternal DDK factor. In addition, the table provides the allele present in each strain at each of the 12 SNPs with the highest level of association with the DDK syndrome phenotype. The alleles at the two SNPs that are most strongly associated with phenotype are underlined. The positions of the SNPs are as follows: 1, 82734817; 2, 82734820; 3, 82735318; 4, 82735343; 5, 82833126; 6, 82833146; 7, 82833386; 8, 82833571; 9, 82833572; 10, 82843176; 11, 82843476; 12, 82857766. The phylogenetic origin of the haplotypes in wild-derived strains is shown as reported previously (http://www.jax.org, Ideraabdullah et al. 2004). The phylogenetic origin of the haplotypes in hybrid strains and classical strains was determined on the basis of the diagnostic alleles shown in supplemental Table 3 at http://www.genetics.org.supplemental/. del, deletion.
References support the assigment of the paternal allele.
Additional strains used to test whether SNP-10 and SNP-11 predict the reproductive performance.
To test whether the alleles at these two SNPs predict the DDK syndrome phenotype in other mouse strains, we genotyped four additional strains with known phenotype (C3H, KK, CBA, and PWK) and determined the phenotype of a strain of known genotype (WSB). In each case the alleles present at these two SNPs predicted accurately the compatible/incompatible phenotype of the strain, confirming strong association with the paternal gene (Table 2 and Figure 3).
DISCUSSION
We have used two methods for mapping the paternal gene responsible for the polar, embryonic-lethal phenotype known as the DDK syndrome. In the first approach, we identified chromosomes that were recombinant in the Om region and assayed the fertility of males carrying these chromosomes in crosses with females carrying the DDK maternal factor. In the second approach, we assayed the fertility of males from 17 inbred strains of diverse phylogenetic origin in crosses with females carrying the DDK maternal factor. These experiments have resulted in a dramatic reduction in the candidate region containing the paternal DDK syndrome gene, perhaps to a single gene, as well as more general conclusions on the evolutionary history of the Om region and the DDK syndrome.
Mapping the DDK syndrome paternal gene candidate region:
Over the course of our investigations (Pardo-Manuel de Villena et al. 1996, 1997, 1999, 2000a, 2000b; de La Casa-Esperón et al. 2002), we have screened over 5,000 meioses for chromosomes 11 that were recombinant in the Om region. We have now determined the paternal phenotype of 18 recombinant chromosomes (supplemental Table 2 at http://www.genetics.org/supplemental/ and Figures 1 and 2) and refined the candidate interval for the paternal gene of the DDK syndrome. Despite the substantial effort required to isolate and test these recombinant chromosomes, this traditional strategy allowed us to narrow the previous 1.5 Mb candidate interval (Baldacci et al. 1992, 1996; Cohen-Tannoudji et al. 2000) to a 385–465 kb interval that contains no fewer than 13 genes (Figure 5a).
Because we appeared to have reached the effective limit of mapping resolution by traditional methods, we attempted to refine further the candidate interval by in silico association mapping of DNA sequence variants against a quantitative measure of paternal reproductive compatibility (Figure 3). The power of this approach depends on the number of strains analyzed, the ratio between compatible and incompatible paternal alleles among the strains, and the level of linkage disequilibrium between nearby sequence variants. The high levels of genetic diversity present in wild-derived strains (Ideraabdullah et al. 2004) and the fact that wild-derived M. m. castaneus and M. m. molossinus strains carry compatible alleles at the paternal gene (Zhao et al. 2002) suggested that mapping of the paternal gene might benefit from the large number of historical recombination events captured by comparing a diverse collection of inbred strains. Therefore, we determined the paternal phenotype of 17 strains selected to maximize sampling across a diverse set of lineages. The males of these strains fall into one of two non-overlapping groups with respect to the reproductive performance (Figure 3). The group with the best reproductive performance includes nine strains (PANCEVO, CAST, JF1, MOLC, DDK, SKIVE, PERC, ZALENDE, and TIRANO). We conclude that these strains carry alleles at the paternal gene that are compatible with the maternal DDK factor. The remaining eight strains (A, 129X1, DBA/2, BALB/c, C57BL/6, PERA, LEWES, and RBA) have reduced litter sizes and are considered to be incompatible (Figure 3).
We had sequenced previously 7155 bp distributed across 22 fragments within the D11Spn173–D11Spn129 candidate interval in each of the 17 inbred strains (Ideraabdullah et al. 2004). Four additional fragments were sequenced to ensure a more uniform coverage of the region (Figures 4 and 5a). We focused our analysis on the 167 variants identified in the region that are polymorphic among strains with a M. m. domesticus haplotype based on our conclusion that the incompatible paternal allele arose in the M. m. domesticus lineage (see below) and, therefore, variants arising in other lineages are of little consequence for association mapping.
The 12 variants having the highest association with the reproductive performance phenotype are located in four clustered fragments (Table 2, Figures 4 and 5). This analysis suggests that the paternal gene lies within a 128-kb interval spanning four genes, from Slfn1 to LOC435271 (Figure 5a). Phylogenetic analyses of the sequenced fragments in the 128 kb region indicates that all incompatible strains share a common ancestor (Figure 6). This result is consistent with the M. m. domesticus origin of the incompatible allele and indicates that the paternal gene lies within the 128-kb interval containing the distal portion of the Schlafen gene cluster.
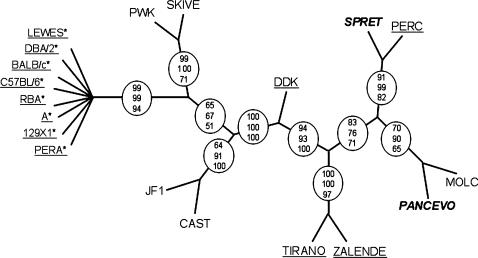
Figure 6.
Phylogenetic relationships of inbred strains in the 128 kb interval defined by in silico mapping. The tree depicted in the figure is a consensus cladogram from three consensus trees obtained by three different phylogenetic methods (materials and methods). Circles denote branches that are consistent among the three trees and the numbers in the circles represent the number of times out of 100 that the branch is observed in each method: top, DNAML; middle, NEIGHBOR; and bottom, DNAPARS. Underlined strains have M. m. domesticus haplotypes in that region. Strains in boldface and italics are derived from other species (SPRET, M. spretus and PANCEVO, M. spicilegus). Asterisks denote strains with incompatible alleles at the paternal gene. All incompatible strains are shown diverging from a single node because the internal branching order within this lineage is not consistent among trees obtained by different methods and the branching order is poorly supported within each method. The length of the branches is arbitrary.
The mouse Schlafen gene family is composed of 10 genes (Slfn1-5, 8-10, LOC435271, and ENSMUSG00000056956) (Figure 5a) and is thought to arise from a common ancestor through multiple unequal recombination events (Schwarz et al. 1998; Geserick et al. 2004). Previous studies classified the Schlafen genes into three groups on the basis of different levels of sequence identity (Geserick et al. 2004). Schlafen proteins share a divergent AAA domain thought to bind ATP (http://www.ensembl.org). Although the exact molecular functions of the Schlafen proteins are unknown, members of this family have been implicated in regulation of lymphocyte differentiation and growth control (Schwarz et al. 1998; Geserick et al. 2004).
The sequence variants having the highest significance are located in two genes, Slfn1 and LOC435271 (Figure 5a). Eight SNPs are found in introns of the LOC435271 gene. The four remaining SNPs are located in the coding region of the Slfn1 gene and result in 3 missense mutations. Given the location of these variants (Figure 5a) and the complete linkage disequilibrium between the phenotype and two SNPs in the LOC435271 gene (SNPs 10 and 11 in Table 2), we propose that the incompatible allele lies within the LOC435271 gene. This conclusion is supported by the successful a priori prediction of the paternal phenotype of the WSB strain on the basis of SNPs 10 and 11 (Table 2) as well as the successful prediction of the two SNPs present in the C3H, KK, CBA, and PWK strains on the basis of paternal phenotype (Table 2 and Figure 3). In addition, SNPs 10 and 11 define the only haplotype that discriminates between the KK and LEWES incompatible strains and the PWK and SKIVE compatible strains (Table 2). This haplotype block is in complete linkage disequilibrium to the DDK syndrome phenotype and may span a maximum of 23.2 kb (from variants 9 to 12 in Table 2).
Evolutionary history of the Om region and the DDK syndrome:
Strain phylogeny based on the Om region is incongruent with strain taxonomy:
Because the Om region includes two genetic factors (the maternal and the paternal components of the DDK syndrome) with a strong effect on reproductive performance and represents a genetic mechanism that could be linked to reproductive isolation and speciation, the evolutionary history of this region is of interest. A phylogenetic analysis of the sequence variants found within the 128-kb candidate interval defined by in silico mapping is shown in Figure 6. Although strains from well-defined monophyletic groups [such as M. musculus sp. and M. m. domesticus subspecies (Guenet and Bonhomme 2003)] are expected to cluster in a node that does not contain strains from other clades/taxa, these expectations are not met for the candidate region. For example, several M. m. castaneus and M. m. musculus strains separate two groups of M. m. domesticus strains (DDK, TIRANO, ZALENDE, and PERC in one group and LEWES, DBA/2, BALB/c, C57BL/6, RBA, A, 129X1, and PERA in the other). In the same vein, the location of the PERC and MOLC strains (two M. musculus strains, see materials and methods) in the tree is incongruent with a monophyletic origin of the M. musculus species. Figure 6 demonstrates that these observations are not due to a poorly supported tree and/or to a single strain. Homoplasy can be rejected because the incongruent phylogeny is supported by multiple variants in linkage disequilibrium. Hybridization between taxa is extremely unlikely because of the lack of concordance between phylogenetic clustering and geographical origin of the wild-derived strains. For example, MOLC, a M. m. molossinus wild-derived strain from Japan clusters with PANCEVO, a M. spicilegus strain from Serbia. The most likely explanation for the observed topology is the presence of multiple ancestral variants that have segregated across clades (Ideraabdullah et al. 2004). Although the presence of scattered ancestral variants is not surprising, per se, it is rare to find large numbers of ancestral variants that consistently contradict the expected phylogeny spanning extensive regions because normally recombination would erase the linkage disequilibrium between them. Rearrangements may suppress recombination and, therefore, in this situation sequence homology is dependent on the presence or absence of the rearrangement rather that on the overall phylogenetic relationship (Babcock and Anderson 1996). Rearrangements are possible candidates to explain the incongruent phylogenetic tree given the presence of multiple duplications and the fact that none of the 48 recombination uncovered between D11Mit33 and D11Mit35 occur in the 385 kb candidate interval defined by progeny testing (Figure 5a). Consistent with the idea of polymorphic rearrangements, we have also observed several-kilobase-long insertions/deletions that are polymorphic in the set of inbred strains analyzed here (data not shown).
Rapid evolution of the Om region:
Because the candidate region for the paternal gene appears to have evolved by gene duplication, we have attempted to obtain a more complete and accurate view of the organization of the Om region by aligning the sequence for the candidate interval defined by progeny testing against itself (Figure 5a). This analysis reveals three regions that have undergone tandem duplications. Based on the level of sequence identity these duplications arose at different times. The LOC435271 gene lies within the oldest detected duplication while the Slfn1 gene is flanked by the newest. Further evidence of a relatively fast evolutionary rate is provided by the comparison between the candidate interval in the mouse and the homologous rat sequence (Figure 5b). Importantly, only the central region of the Schlafen gene cluster has undergone rapid evolution (shown by the lack of a well-defined diagonal line between the Slfn1 and LOC435271 genes in Figure 5b), while the Slfn5, ENSMUSG00000056956, Pex12, and Ap2b1 genes have evolved at a significantly slower pace (Figure 5b). The former region spans the duplications and contains all the variants that are significantly associated with the reproductive performance phenotype. However, the deterioration of sequence identity in interspecies comparisons is not due to the duplications alone but is also due to enrichment for LTR and LINE repeats in the central region of the gene cluster (http://genome.ucsc.edu).
The compatible paternal DDK syndrome allele is ancestral:
Overall, a total of 26 strains have been characterized for paternal compatibility with the maternal DDK factor (Table 2). Compatible and incompatible alleles are represented equally among these strains (Table 3). However, a striking difference in the frequency of the incompatible allele emerges when one considers the phylogenetic origin of the D11Spn173–D11Spn129 candidate region (Test for independence between the allele at the paternal gene and phylogenetic origin of the strain, χ2 = 11.56, 1 d.f., P < 0.0007; Table 3). In strains with a M. m. domesticus haplotype only 28% of the strains examined are compatible. In contrast, 100% of strains with haplotypes from other subspecies and species are compatible (Table 3). These results strongly support the hypothesis that the compatible allele is ancestral and the incompatible allele arose in the M. m. domesticus lineage after the divergence of the subspecies (Zhao et al. 2002). Based on the allele frequencies observed in wild-derived M. m. domesticus inbred strains, we conclude that the incompatible allele appeared in the domesticus lineage shortly after the divergence of the M. musculus subspecies approximately 750,000 years ago (Guenet and Bonhomme 2003). However, the incompatible allele was not fixed in the ancestors of all extant mice originating in this lineage. Furthermore, compatible and incompatible alleles are present in M. m. domesticus strains derived from natural populations of three small and distant geographic areas. Briefly, among the three wild-derived inbred strains from Northern Italy and Switzerland, one strain, RBA, carries an incompatible allele while the other two strains, TIRANO and ZALENDE, have compatible alleles. Similarly, two strains from the Eastern US, WSB and LEWES, have compatible and incompatible alleles, respectively. Finally, two strains from Peru, PERA and PERC, also have discordant alleles for the DDK syndrome phenotype. Therefore, both compatible and incompatible alleles are found over a broad geographical range and have colonized the New World on more than one occasion. In contrast, only the incompatible allele is found among all classical inbred strains analyzed to date with the exception of the DDK strain. This might reflect a founder effect due to the modest number of progenitors that contributed to classical inbred strains (Guenet and Bonhomme 2003). Alternatively, selection operating in the derivation of classical inbred strains but not in the derivation of wild-derived inbred strains may be responsible for the fixation of the incompatible allele in the former. Although the incompatible allele is presumed to be neutral in the absence of the maternal DDK factor, it remains possible that this allele may have an effect on the fitness of its carriers based on the frequency and distribution of alleles in M. m. domesticus and classical inbred strains.
TABLE 3.
Compatible and incompatible paternal alleles in strains with haplotypes of different phylogenetic origin in the D11Spn173–D11Spn129 interval
|
D11Spn173–D11Spn129 haplotype
|
Allele at the paternal gene
|
||
|---|---|---|---|
| Compatible | Incompatible | Total | |
| M. m. domesticus | 5 | 13 | 18 |
| Other species and subspecies | 8 | 0 | 8 |
| Total | 13 | 13 | 26 |
The strains used in this analysis are listed in Table 2.
Our analysis also provides information for the location of the gene encoding the maternal DDK factor. We have mated C57BL/6 males to females carrying the chromosome that recombine between D11Spn173 and D11Spn178 (named “735L” in reference to the mouse in which this chromosome was initially identified). The reproductive performance observed in these crosses (supplemental Table 4 at http://www.genetics.org/supplemental/) indicates that the “735L” chromosome carries a C57BL/6 allele at the gene encoding the maternal factor. Specifically, the average litter size in mating between homozygous “735L” / “735L” females and C57BL/6 males (7.8 ± 2.6) is inconsistent with the presence of incompatible alleles (i.e., DDK) at the maternal factor. The “735L” chromosome carries DDK alleles in the proximal region and C57BL/6 alleles in the distal region and has a C57BL/6 allele at the paternal gene (Figures 1 and 2 and supplemental Table 2 at http://www.genetics.org/supplemental/). Therefore, both maternal and paternal components of the DDK syndrome lie in overlapping intervals. We wish to note that the use of homozygous “735L” / “735L” females forestalls the confounding effects of modifiers of the maternal contribution to the DDK syndrome that have been reported previously, because the mode of action of these modifiers is thought to require heterozygosity at Om in the dam (Pardo-Manuel de Villena et al. 1999; Le Bras et al. 2000; Zhao et al. 2000). These data represent the first step toward the definition of a candidate interval for the maternal gene.
Finally, although we have been unable to separate the paternal and maternal components of the DDK syndrome through recombination, the results from Zhao et al. (2002) and the results presented here demonstrate that they are different mutations. This conclusion is based on the type of combinations of compatible and incompatible alleles at the maternal factor and paternal gene found in inbred strains. Two types of combinations of alleles have been described previously, compatible maternal alleles and incompatible paternal alleles (found in strains such as C57BL/6, BALB/c, C3H, and PERA) and incompatible maternal alleles and compatible paternal alleles (found only in DDK). In this study we report a third combination of alleles in the PERC and CAST strains, compatible maternal alleles and compatible paternal alleles (Figure 3 and Table 1). The existence of three allelic combinations demonstrates conclusively that the maternal and the paternal components are nonallelic.
Acknowledgments
We are grateful to Tammi Briscoe, Brad Adey and Dale Bautista for technical assistance and to Terry Magnuson for the JF1/Ms inbred mice. This work was partly supported by grants from the National Institutes of Health (R01 HD34508 CS) and the National Science Foundation (MCB-0133526 FP-MV).
References
- Alibert, P., F. Fel-Clair, K. Manolakou, J. Britton-Davidian and J.-C. Auffray, 1997. Developmental stability, fitness, and trait size in laboratory hybrids between European subspecies of the house mouse. Evolution 5: 1284–1295. [DOI] [PubMed] [Google Scholar]
- Babinet, C., V. Richoux, J. L. Guenet and J. P. Renard, 1990. The DDK inbred strain as a model for the study of interactions between parental genomes and egg cytoplasm in mouse preimplantation development. Development (Suppl.), 81–87. [PubMed]
- Babcock, C. S., and W. W. Anderson, 1996. Molecular evolution of the sex-ratio inversion in Drosophila pseudoobscura: analysis of the Esterase 5 region. Mol. Biol. Evol. 13: 297–308. [DOI] [PubMed] [Google Scholar]
- Baldacci, P. A., V. Richoux, J. P. Renard, J. L. Guenet and C. Babinet, 1992. The locus Om, responsible for the DDK syndrome, maps close to Sigje on mouse chromosome 11. Mamm. Genome 2: 100–105. [DOI] [PubMed] [Google Scholar]
- Baldacci, P. A., M. Cohen-Tannoudji, C. Kress, S. Pournin and C. Babinet, 1996. A high-resolution map around the locus Om on mouse Chromosome 11. Mamm. Genome 7: 114–116. [DOI] [PubMed] [Google Scholar]
- Britton-Davidian, J., F. Fel-Clair, J. Lopez, P. Alibert and P. Boursot, 2005. Postzygotic isolation between two European subspecies of the house mouse: estimates from fertility patterns in wild and laboratory-bred hybrids. Biol. J. Linn. Soc. 84: 379–393. [Google Scholar]
- Buehr, M., S. Lee, A. McLaren and A. Warner, 1987. Reduced gap junctional communication is associated with the lethal condition characteristic of DDK mouse eggs fertilized by foreign sperm. Development 101: 449–459. [DOI] [PubMed] [Google Scholar]
- Cohen-Tannoudji, M., P. Baldacci, C. Kress, V. Richoux-Duranthon, J. P. Renard et al., 1996. Genetic and molecular studies on Om, a locus controlling mouse preimplantation development. Acta Genet. Med. Gemellol. 45: 3–14. [DOI] [PubMed] [Google Scholar]
- Cohen-Tannoudji, M., S. Vandormael-Pournin, S. Le Bras, F. Coumailleau, C. Babinet et al., 2000. A 2-Mb YAC/BAC-based physical map of the ovum mutant (Om) locus region on mouse chromosome 11. Genomics 68: 273–282. [DOI] [PubMed] [Google Scholar]
- de La Casa-Esperón, E., J. C. Loredo-Osti, F. Pardo-Manuel de Villena, T. L. Briscoe, J. M. Malette et al., 2002. X chromosome effect on maternal recombination and meiotic drive in the mouse. Genetics 161: 1651–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich, W. F., J. Miller, R. Steen, M. A. Merchant, D. Damron-Boles et al., 1996. A comprehensive genetic map of the mouse genome. Nature 380: 149–152. [DOI] [PubMed] [Google Scholar]
- Felsenstein, J., 1989. PHYLIP – Phylogeny inference package (version 3.2). Cladistics 5: 164–166. [Google Scholar]
- Gao, S., G. Wu, Z. Han, E. de la Casa-Esperón, C. Sapienza et al., 2005. Recapitulation of the ovum mutant (Om) phenotype and loss of Om locus polarity in cloned mouse embryos. Biol. Reprod. 72: 487–491. [DOI] [PubMed] [Google Scholar]
- Geserick, P., F. Kaiser, U. Klemm, S. H. Kaufmann and J. Zerrahn, 2004. Modulation of T cell development and activation by novel members of the Schlafen (slfn) gene family harbouring an RNA helicase-like motif. Int. Immunol. 16: 1535–1548. [DOI] [PubMed] [Google Scholar]
- Guenet, J. L., and F. Bonhomme, 2003. Wild mice: an ever-increasing contribution to a popular mammalian model. Trends Genet. 19: 24–31. [DOI] [PubMed] [Google Scholar]
- Ideraabdullah, F. Y., E. de la Casa-Esperón, T. A. Bell, D. A. Detwiler, T. Magnuson et al., 2004. Genetic and haplotype diversity among wild-derived mouse inbred strains. Genome Res. 14: 1880–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, K., S. Thomas, I. B. Howard, T. A. Bell, H. E. Doherty et al., 2005. Meiotic drive at the Om locus in wild-derived inbred mouse strains. Biol. J. Linn. Soc. 84: 487–492. [Google Scholar]
- Le Bras, S., M. Cohen-Tannoudji, C. Kress, S. Vandormael-Pournin, C. Babinet et al., 2000. BALB/c alleles at modifier loci increase the severity of the maternal effect of the “DDK syndrome.” Genetics 154: 803–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bras, S., M. Cohen-Tannoudji, V. Guyot, S. Vandormael-Pournin, F. Coumailleau et al., 2002. Transcript map of the Ovum mutant (Om) locus: isolation by exon trapping of new candidate genes for the DDK syndrome. Gene 296: 75–86. [DOI] [PubMed] [Google Scholar]
- Leclerc, C., D. Becker, M. Buehr and A. Warner, 1994. Low intracellular pH is involved in the early embryonic death of DDK mouse eggs fertilized by alien sperm. Dev. Dyn. 200: 257–267. [DOI] [PubMed] [Google Scholar]
- Mann, J. R., 1986. DDK egg-foreign sperm incompatibility in mice is not between the pronuclei. J. Reprod. Fertil. 76: 779–781. [DOI] [PubMed] [Google Scholar]
- Maizel, J. V., and R. P. Lenk, 1981. Enhanced graphic matrix analysis of nucleic acid and protein sequences. Proc. Natl. Acad. Sci. USA 78: 7665–7669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo-Manuel de Villena, F., C. Slamka, M. Fonseca, A. K. Naumova, J. Paquette et al., 1996. Transmission-ratio distortion through F1 females at chromosome 11 loci linked to Om in the mouse DDK syndrome. Genetics 142: 1299–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo-Manuel de Villena, F., A. K. Naumova, A. E. Verner, W. H. Jin and C. Sapienza, 1997. Confirmation of maternal transmission ratio distortion at Om and direct evidence that the maternal and paternal “DDK syndrome” genes are linked. Mamm. Genome 8: 642–646. [DOI] [PubMed] [Google Scholar]
- Pardo-Manuel de Villena, F., E. de la Casa-Esperon, A. Verner, K. Morgan and C. Sapienza, 1999. The maternal DDK syndrome phenotype is determined by modifier genes that are not linked to Om. Mamm. Genome 10: 492–497. [DOI] [PubMed] [Google Scholar]
- Pardo-Manuel de Villena, F., E. de la Casa-Esperon, T. L. Briscoe and C. Sapienza, 2000. a A genetic test to determine the origin of maternal transmission ratio distortion. Meiotic drive at the mouse Om locus. Genetics 154: 333–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo-Manuel de Villena, F., E. de La Casa-Esperon, J. W. Williams, J. M. Malette, M. Rosa et al., 2000. b Heritability of the maternal meiotic drive system linked to Om and high-resolution mapping of the Responder locus in mouse. Genetics 155: 283–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pustell, J., and F. C. Kafatos, 1982. A high speed, high capacity homology matrix: Zooming through SV40 and polyoma. Nucleic Acids Res. 10: 4765–4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley, G. J., L. Gehrke, D. A. Roth and P. E. Auron, 1984. Computer-aided nucleic acid secondary structure modeling incorporating enzymatic digestion data. Nucleic Acids Res. 12: 347–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renard, J. P., and C. Babinet, 1986. Identification of a paternal developmental effect on the cytoplasm of one-cell-stage mouse embryos. Proc. Natl. Acad. Sci. USA. 83: 6883–6886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renard, J. P., P. Baldacci, V. Richoux-Duranthon, S. Pournin and C. Babinet, 1994. A maternal factor affecting mouse blastocyst formation. Development 120: 797–802. [DOI] [PubMed] [Google Scholar]
- Sapienza, C., J. Paquette, P. Pannunzio, S. Albrechtson and K. Morgan, 1992. The polar-lethal Ovum mutant gene maps to the distal portion of mouse chromosome 11. Genetics 132: 241–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz, D. A., C. D. Katayama and S. M. Hedrick, 1998. Schlafen, a new family of growth regulatory genes that affect thymocyte development. Immunity 9: 657–668. [DOI] [PubMed] [Google Scholar]
- Tomita, T., 1960. One-side cross sterility between inbred strains of mice. Jpn. J. Genet. 35: 291. [Google Scholar]
- Wakasugi, N., T. Tomita and K. Kondo, 1967. Differences of fertility in reciprocal crosses between inbred strains of mice. DDK, KK and NC. Reprod. Fertil. 13: 41–50. [DOI] [PubMed] [Google Scholar]
- Wakasugi, N., 1973. Studies on fertility of DDK mice: reciprocal crosses between DDK and C57BL/6J strains and experimental transplantation of the ovary. J. Reprod. Fertil. 33: 283–291. [DOI] [PubMed] [Google Scholar]
- Wakasugi, N., 1974. A genetically determined incompatibility system between spermatozoa and eggs leading to embryonic death in mice. J. Reprod. Fertil. 41: 85–96. [DOI] [PubMed] [Google Scholar]
- Wakasugi, N., and M. Morita, 1977. Studies on the development of F1 embryos from inter-strain cross involving DDK mice. J. Embryol. Exp. Morphol. 38: 211–216. [PubMed] [Google Scholar]
- Wu, G., L. Hao, Z. Han, S. Gao, K. E. Latham et al., 2005. Maternal transmission ratio distortion at the mouse Om locus results from meiotic drive at the second meiotic division. Genetics 170: 327–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, W. D., H. J. Chung and N. Wakasugi, 2000. Modification of survival rate of mouse embryos developing in heterozygous females for ovum mutant gene. Biol. Reprod. 62: 857–863. [DOI] [PubMed] [Google Scholar]
- Zhao, W. D., A. Ishikawa, T. Yamagata, H. Bolor and N. Wakasugi, 2002. Female mice of DDK strain are fully fertile in the intersubspecific crosses with Mus musculus molossinus and M. m. castaneus. Mamm. Genome 13: 345–351. [DOI] [PubMed] [Google Scholar]