Abstract
Ts65Dn mice inherit a marker chromosome, T(1716)65Dn, producing segmental trisomy for orthologs of about half of the genes on human chromosome 21. These mice display a number of phenotypes that are directly comparable to those in humans with trisomy 21 and are the most widely used animal model of Down syndrome (DS). However, the husbandry of Ts65Dn mice is complicated. Males are sterile, and only 20–40% of the offspring of Ts65Dn mothers are trisomic at weaning. The lower-than-expected frequency of trisomic offspring has been attributed to losses at meiosis, during gestation and at postnatal stages, but no systematic studies support any of these suppositions. We show that the T(1716)65Dn marker chromosome is inherited at expected frequency and is fully compatible with development to midgestation. Disproportional loss of trisomic offspring occurs in late gestation and continues through birth to weaning. Different maternal H2 haplotypes are significantly associated with the frequency of trisomy at weaning in patterns different from those reported previously. The proportion of trisomic mice per litter decreases with age of the Ts65Dn mother. These results provide the first statistical and numerical evidence supporting the prenatal and perinatal pattern of loss in the Ts65Dn mouse model of DS.
DOWN syndrome (DS) is caused by trisomy 21 and is one of the most common genetic causes of cognitive impairment and congenital abnormalities. The incidence of DS is 1 in 800–1000 live births; the total prevalence of DS (number of affected cases in live births, stillbirths, and terminations) appears to be steady or rising (Leonard et al. 2000; Verloes et al. 2001; Forrester and Merz 2002; Bell et al. 2003; Roizen and Patterson 2003; Khoshnood et al. 2004). Improved detection methods including serum screening (maternal serum α-fetoprotein, estriol, and human chorionic gonadotropin) and fetal ultrasound may increase prenatal ascertainment. It has been conclusively shown that increasing maternal age strongly correlates with the incidence of DS (Verloes et al. 2001; Forrester and Merz 2002; Bell et al. 2003; Khoshnood et al. 2004). Trisomy 21 is the most frequently viable autosomal aneuploidy at birth. Nonetheless, up to 75% of all DS fetuses identified during the first trimester and around 50% of DS fetuses identified during the second trimester are lost before term (Morris et al. 1999; Spencer 2001). Although placental abnormalities in DS have been suspected, specific reasons for this extensive fetal loss have not been conclusively identified (Frendo et al. 2000; Debieve et al. 2001; Wright et al. 2004).
Neonatal mortality (death within 28 days of birth) for trisomy 21 is high, 1–2%. This is due primarily to congenital heart disease, which affects half of all children with DS (Halliday et al. 1995; Morris et al. 1999; Nembhard et al. 2001; Frid et al. 2004). Despite advances in neonatal care, DS survival is only 90–95% at 1 year and between 85 and 90% at 10 years (Leonard et al. 2000; Forrester and Merz 2002; Glasson et al. 2002; Bell et al. 2003; Frid et al. 2004; Marino et al. 2004). Premature births, low birth weight, and small size for gestational age are more common in DS individuals than in the general population and contribute to postnatal deaths and other sequelae of DS (Leonard et al. 2000; Frid et al. 2004). Although DS infants are usually underweight, they frequently become obese by 3–4 years of age (Bell et al. 2003; Roizen and Patterson 2003).
Mouse models have been used to understand the etiology of DS phenotypes and to correlate the genetic and phenotypic changes caused by trisomy. The mouse offers access to all tissues at all developmental stages and can be manipulated genetically. The most widely used DS model is Ts(1716)65Dn (hereafter Ts65Dn), which has segmental trisomy due to a small translocation chromosome containing the telomeric end of mouse chromosome 16 (Mmu16) attached to the centromeric end of Mmu17 (Davisson et al. 1993; Reeves et al. 1995). The Ts65Dn mouse is trisomic for orthologs of about half of the known genes on human chromosome 21 (Hattori et al. 2000; Mural et al. 2002; Gardiner et al. 2003). The triplicated region of Mmu16 corresponds to about 28 Mb of human chromosome 21 and is highly conserved between human and mouse in both gene order and content. This correspondence in gene content suggests that conserved genetic pathways will be disrupted in humans and mice with trisomy for this segment and supports investigation of the genetic basis for observed homologous DS phenotypes in the Ts65Dn mouse (Kola and Hertzog 1998; Reeves et al. 2001; Mural et al. 2002; Gardiner et al. 2003). Ts65Dn mice show DS-related phenotypes including developmental abnormalities, cognitive and neurological impairments, craniofacial abnormalities, and cerebellar maldevelopment (Davisson et al. 1993; Reeves et al. 1995; Holtzman et al. 1996; Baxter et al. 2000; Richtsmeier et al. 2000; Cooper et al. 2001; Belichenko et al. 2004; Hampton et al. 2004; Rueda et al. 2005). Analysis of Ts65Dn mice revealed a deficit in cerebellar granule cells which successfully predicted a corresponding phenotype in DS (Baxter et al. 2000).
Although Ts65Dn mice have been widely used to study DS, they are difficult to breed and propagate. They must be maintained on a genetically mixed background, and because Ts65Dn males are infertile, only female Ts65Dn mice can be used to generate offspring (Davisson et al. 1993). Although many anecdotal reports concerning Ts65Dn mice exist, few are supported by solid numerical and statistical evidence. We present data on the frequency of trisomy across gestation and early development from >1600 offspring of Ts65Dn females. This analysis of Ts65Dn mice from midgestation extends the model and suggests new applications to DS research as well as approaches to improve production of Ts65Dn mice.
MATERIALS AND METHODS
Mice:
Founder female B6EiC3Sn a/A–Ts(1716)65Dn (Ts65Dn) and male B6C3F1/J mice were purchased from The Jackson Laboratory (Bar Harbor, ME) and bred in our colonies at Johns Hopkins University School of Medicine. Offspring of Ts65Dn mothers were genotyped by FISH (Moore et al. 1999) and trisomic offspring were used to generate subsequent litters. Periodically, new trisomic females and B6C3F1/J males purchased from The Jackson Laboratory were introduced into our colonies. All data on weanlings are from crosses with B6EiC3Sn a/A males. Ts65Dn are maintained as an advanced intercross line and have approximately 50% C57BL6/J and C3H alleles. This study reports results from 1631 offspring of Ts65Dn mothers, continually generated over an approximate 3-year period. These offspring include 177 pups and 225 pups killed at the day of birth (postnatal day 0, P0) and at P6, respectively, and 108 embryos dissected at 9.5 days postcoitus (embryonic day 9.5, E9.5). E9.5 litters were taken from Ts65Dn × Wnt1–lacZ [B6CBA-Tg(Wnt1–lacZ)206Amc/J] crosses (The Jackson Laboratory). Ts65Dn mothers were removed from the colony when they ceased to produce litters, generally 8–12 months. Those mothers that abandoned two litters were preferentially but not exclusively used for prenatal studies. Mice were maintained on Labdiet Prolab RMH 2500 [24.9% protein, 4.8% fat (ether extract), 6.3% fat (acid hydrolysis), and 4.7 crude fiber] (Purina Mills, St. Louis) and acidified water ad libitum from weaning.
Genotyping:
Determination of ploidy was done by interphase FISH using BAC clones 401C2 and 433G17 as described (Moore et al. 1999). The genotype at H2 for Ts65Dn mothers and offspring was determined by PCR using both the D17Mit28 and D17Mit32 microsatellite markers (http://www.broad.mit.edu/cgi-bin/mouse/sts_info). D17Mit28 is found in the H2K (bm1) promoter region and D17Mit32 is found in the mouse C4 gene located inside the H2S region. PCR conditions were as follows: (1) 95° for 3 min, (2) 95° for 30 sec, (3) 55° for 30 sec, (4) 72° for 30 sec, and (5) 72° for 5 min (steps 2–4 were repeated 35 times). Products were separated on a 5% MetaPhor agarose gel (Cambrex, Rockland, ME) run for 2 hr at 150 V.
Statistical analyses:
Inheritance of the T(1716)65Dn chromosome was compared to Mendelian frequency using a χ2 goodness-of-fit test with a 1:1 or 1:2:1 null hypothesis. Multiple comparisons between E9.5, P0, P6, and weaned mice and comparisons among H2 types were determined by ANOVA and post-hoc LSD tests (α = 0.05). Weight differences between trisomic and euploid mice were found by using a t-test; a 1-tailed test was used for weights of P0, P6, and 1- to 3-month-old mice and a 2-tailed test was used for ages previously tested (4 and 5 months). A χ2 test was used to examine independence of ploidy and sex and of ploidy and H2 haplotype.
RESULTS
The frequency of trisomy is lower than Mendelian expectation at weaning:
Several laboratories have reported a lower-than-expected frequency of inheritance of the T(1716)65Dn marker chromosome from Ts65Dn mothers (Davisson et al. 1993; Davisson and Costa 1999; Paz-Miguel et al. 2001; Liu et al. 2003; Villar et al. 2005) (http://www.jax.org/cyto/ts65dncolony.html). We found a trisomy frequency of 36% among 1121 weaned offspring from 251 litters of Ts65Dn × B6C3F1 matings. This represented a highly significant deviation from Mendelian expectations (P = 8.73 × 10−19, Table 1). The non-Mendelian ratio of Ts65Dn mice has been attributed to losses immediately after birth, during gestation, or during gametogenesis (Davisson et al. 1993; Davisson and Costa 1999; Liu et al. 2003) (http://www.jax.org/cyto/ts65dncolony.html).
TABLE 1.
Offspring from Ts65Dn mothers
| Age | Euploid | Ts65Dn | Total mice | Trisomic (%) | χ2 | P |
|---|---|---|---|---|---|---|
| E9.5 | 52 | 53 | 108 | 50 | 0.01 | 0.92 |
| P0 | 96 | 75 | 177 | 44 | 2.58 | 0.11 |
| P6 | 125 | 85 | 225 | 40 | 7.62 | 0.006 |
| Weaning | 661 | 376 | 1121 | 36 | ≪0.001 |
Segregation, transmission and early developmental effects of Ts65Dn:
To determine when trisomic mice are lost, we first examined embryos of Ts65Dn mothers at midgestation (E9.5). Trisomy was assessed using FISH analysis on embryo-derived yolk sac cells. Fifty percent of the 108 embryos of Ts65Dn mothers were trisomic at E9.5 (Table 1). Two of the 21 litters examined at E9.5 consisted entirely of resorbing embryos and 13 additional resorbing embryos (11%) were found among the remaining 19 litters. Of the seven resorbing embryos that could be genotyped, four were found to be trisomic and three were euploid.
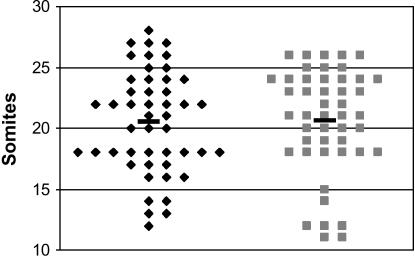
We compared the development of trisomic and euploid embryos at E9.5 by determining the number of somites (Figure 1). A substantial range was observed in both Ts65Dn and euploid embryos. However, the average number of somites was the same for both trisomic and euploid embryos (20.6 and 20.5 somites, respectively). Thus, we found no selection against trisomic embryos at midgestation, indicating that the T(1716)65Dn marker chromosome segregates freely and is not selected against during oogenesis and that this segmental trisomy does not cause early embryonic loss or gross developmental delay.
Figure 1.
No difference in size of midgestation trisomic and euploid embryos. Embryos taken from Ts65Dn mothers at embryonic day 9.5 (E9.5) were staged according to the number of somites present. Although there was a large range in the number of somites of both trisomic and euploid Ts65Dn offspring, there was no difference in the average number of somites in trisomic (20.6 somites, n = 47) and euploid (20.5 somites, n = 48) mice. Diamonds, euploid; squares, Ts65Dn.
Trisomic and euploid mice are lost after birth:
Next we determined the frequency of trisomic offspring at P0 and P6 (Table 1). We genotyped 177 pups that were found alive at P0 and found 44% to be trisomic, a suggestive but not significant reduction from the expected 50% (P = 0.11). Of 19 pups found dead at P0, 47% were trisomic (P = 0.82). At P6, however, only 40% of the 225 mice recovered were trisomic, a significant deviation from Mendelian ratios (P = 0.006). As discussed, the frequency of trisomy at weaning (4 weeks) was 36% of 1121 animals. The frequencies of trisomy at E9.5, P0, P6, and weaning show a pattern of loss of trisomic mice during late gestation, after birth, and in the first weeks of life. This may be due to problems stemming from trisomy, the poor mothering skills or uterine environment of Ts65Dn females, or a combination of these factors.
Litter size and postnatal weight in Ts65Dn litters:
Previous investigations report Ts65Dn litters of 3–7 mice, smaller than the 8–10 pups expected from B6C3F1 females (Davisson et al. 1993; Davisson and Costa 1999; Villar et al. 2005). Further, Ts65Dn females are reported anecdotally to be poor mothers, and while trisomic pups were lost disproportionately, euploid pups were lost as well. At E9.5, the average number of viable embryos per litter was 5.7 (19 litters). After birth, including only those litters that were cared for by the mother, the average number of live pups was 5.4 at P0 (142 litters), 5.3 at P6 (32 litters), and 4.1 at weaning (251 litters). During the last 15 months of this study, 28 of 166 litters seen at P0 (17%) had no viable pups remaining by P6.
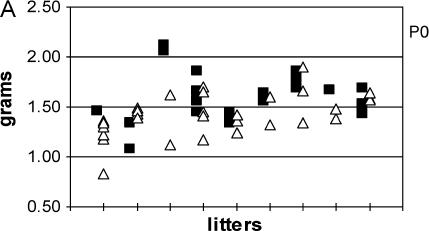
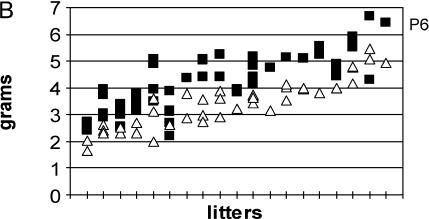
To determine whether disproportionate loss of trisomic pups correlated with their size, and to extend previous reports that noted that Ts65Dn mice are smaller and weigh less than euploid littermates at early ages but may become obese by the time they reach four to five months of age (Davisson et al. 1993; Holtzman et al. 1996), we determined the mass of pups from P0 through 5 months. At P0, Ts65Dn mice in our study weighed significantly less than euploid littermates (P < 0.0001, Table 2). The lower mass of Ts65Dn mice persisted at P6 (P < 0.0001, Table 2). While the average weights of trisomic mice were less at P0, Ts65Dn and euploid weights overlapped in 6 of 9 litters (67%). By P6, however, only 16% (3/19) of P6 litters included one or more trisomic mice that were larger than the smallest euploid mice (Figure 2, A and B). This suggests that, in addition to being smaller at birth, the trisomic pups grow more slowly than their euploid littermates. In this study, trisomic mice never caught up to their euploid littermates. On average, male Ts65Dn mice weighed significantly less than their euploid littermates from 1 to 5 months, the duration of the study (Table 1). Thus factors other than trisomy may be involved in the obesity reported in other colonies.
TABLE 2.
Weights of Ts65Dn offspring at P0, P6, and 1–5 months
| Euploid
|
Ts65Dn
|
||||
|---|---|---|---|---|---|
| Age (mo) | Average weight (SD) | N | Average weight (SD) | N | P |
| P0 | 1.56 (0.22) | 55 | 1.40 (0.17) | 57 | <0.0001 |
| P6 | 4.20 (0.97) | 83 | 3.30 (0.83) | 62 | <0.0001 |
| 1 | 18.48 (1.71) | 5 | 13.42 (4.27) | 6 | 0.0176 |
| 2 | 25.46 (3.27) | 14 | 22.75 (3.3) | 11 | 0.0257 |
| 3 | 29.72 (3.37) | 28 | 28.01 (3.52) | 23 | 0.0421 |
| 4 | 37.94 (9.8) | 13 | 28.76 (4.02) | 15 | 0.0027 |
| 5 | 35.44 (4.32) | 18 | 27.87 (4.69) | 7 | 0.0008 |
Figure 2.
Weight of Ts65Dn and euploid mice at (A) P0 and (B) P6, within litters. Squares, euploid; triangles, trisomic. In 6/9 Ts65Dn litters at P0, there was an overlap in weight between trisomic and eupolid mice. In addition, there was a significant overlap of weight in trisomic and euploid mice between litters (A). In P6 litters, trisomic mice generally weigh less than euploid mice and only 3/19 litters have overlap between weights of trisomic and euploid mice (B).
Maternal age and trisomy:
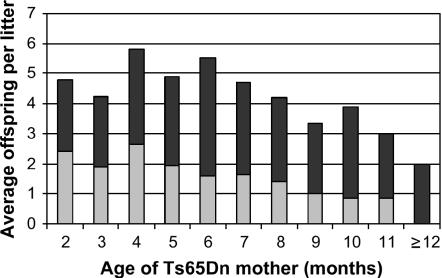
In contrast to trisomy 21, which arises from nondisjunction, the inheritance of the T(1716)65Dn marker chromosome is a malsegregation event, and the extra chromosome appears to distribute randomly to gametes on the basis of the occurrence of trisomy in 50% of embryos at E9.5. Thus we did not necessarily expect a change in the frequency of Ts65Dn mice with maternal age as happens for nondisjunction. Litter size generally decreased with maternal age in Ts65Dn mice (Figure 3 and Table 3). Surprisingly, the percentage of Ts65Dn offspring per litter at weaning also decreased as a function of the age of the Ts65Dn mothers (Figure 3). The timing of and reasons for this disproportionate decline are unclear, but the observation has practical implications for maintaining a Ts65Dn production colony.
Figure 3.
Average percentage of trisomic and euploid offspring by mother's age in months. The average total number and percentage of trisomic mice per litter generally decreases as the age of the mother increases. For 2 months n = 5 litters, 3 months n = 22, 4 months n = 19, 5 months n = 20, 6 months n = 19, 7 months n = 21, 8 months n = 15, 9 months n = 12, 10 months n = 8, 11 months n = 8, ≥ 12 months n = 6. Solid bars, euploid; shaded bars, Ts65Dn.
TABLE 3.
Litter size and composition of Ts65Dn offspring by age of mother
| Age (mo) | Euploida | Ts65Dna | Totala | Litters |
|---|---|---|---|---|
| 2 | 2.4 (1.7) | 2.4 (1.5) | 4.8 (1.1) | 5 |
| 3 | 2.3 (1.6) | 1.9 (1.5) | 4.2 (1.8) | 22 |
| 4 | 3.2 (1.4) | 2.6 (2.0) | 5.8 (2) | 19 |
| 5 | 2.9 (1.9) | 1.9 (2.0) | 4.7 (2.9) | 20 |
| 6 | 3.9 (1.5) | 1.6 (1.2) | 5.5 (1.8) | 19 |
| 7 | 3.1 (1.6) | 1.7 (1.5) | 4.5 (2.3) | 21 |
| 8 | 2.8 (1.7) | 1.4 (1.1) | 4.2 (1.8) | 15 |
| 9 | 2.3 (1.4) | 1 (1) | 3.3 (1.6) | 12 |
| 10 | 3 (2.3) | 1 (1.4) | 3.9 (2.6) | 8 |
| 11 | 2.1 (1.6) | 1 (1.1) | 3 (1.6) | 8 |
| 12 | 2 (2) | 0 (0) | 2 (2) | 6 |
Values are expressed as the average number in litter (standard deviation in parentheses).
Parental H2 linked to non-Mendelian ratios of Ts65Dn offspring:
Ts65Dn mice cannot be inbred and are maintained as an advanced intercross by breeding Ts65Dn females to B6C3F1 males. The C3H and B6 strains carry the H2k and H2b haplotypes, respectively, on Mmu17 (H2 is not included on the portion of Mmu17 on the T(1716)65Dn chromosome). Therefore Ts65Dn mothers may be H2b/b, H2b/k, or H2k/k. Paz-Miguel et al. (2001) reported that inheritance of the T(1716)65Dn chromosome was linked to H2 type in two crosses of Ts65Dn mice and that selection against the H2b haplotype was seen in trisomic pups. We determined H2 type among 401 weaned offspring from 97 Ts65Dn litters chosen at random from the colony. The progeny derived from three different crosses (with respect to H2) in which B6C3F1 (H2b/k) males were crossed to Ts65Dn females that were H2b/b, H2b/k, or H2k/k. Average maternal age was not significantly different among the H2 classes.
Overall, the frequency at weaning of trisomic pups was significantly less than euploid, as expected (41%, P < 0.0001). We observed fewer than expected H2k/k trisomic mice in litters from H2b/k mothers (Table 4). Further, H2k/k mice were obtained less frequently than H2b/k mice among the small number of progeny from H2k/k Ts65Dn mothers (27%, P = 0.005). The consistently smaller-than-expected frequency of H2k/k trisomic offspring in the two crosses where they could occur suggests that these animals are less robust than H2b/k or H2b/b trisomic mice.
TABLE 4.
Offspring by Ts65Dn mother's H2 genotype
| Mothera | Offspring | Euploid | Ts65Dn | Total |
|---|---|---|---|---|
| Ts65Dn H2b/b (n = 25) | H2b/b | 31 | 21 | 52 |
| H2b/k | 26 | 27 | 53 | |
| Total | 57 | 48 | 105 | |
| Ts65Dn H2b/k (n = 60) | H2b/b | 53 | 28 | 81 |
| H2b/k | 70 | 53 | 123 | |
| H2k/k | 43 | 18 | 61 | |
| Total | 166 | 99 | 265 | |
| Ts65Dn H2k/k (n = 12) | H2b/k | 13 | 14 | 27 |
| H2k/k | 4 | 6 | 10 | |
| Total | 17 | 20 | 37 |
n, number of litters born to Ts65Dn mothers with a particular genotype.
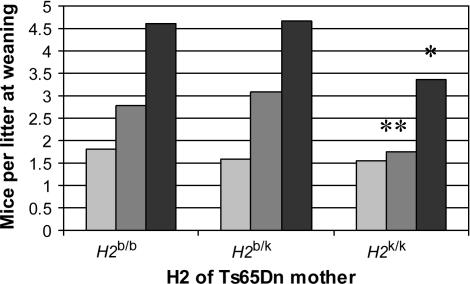
H2k/k mothers had the highest percentage of trisomic offspring (>50%). Both H2b/k and H2k/k trisomic pups occurred in near-Mendelian ratios in litters from H2k/k mothers (Table 4 and Figure 4). Unfortunately for production colony husbandry, H2k/k mothers also had the smallest litters, so they did not produce more total trisomic offspring than did mothers of the other H2 haplotypes (Figure 4).
Figure 4.
Composition of litters at weaning by H2 genotype of Ts65Dn mothers. Average number of trisomic, euploid and total mice per litter at weaning for Ts65Dn H2b/b (n = 38), H2b/k (n = 83), and H2k/k (n = 16) mothers. Using ANOVA and multiple comparisons, H2k/k have smaller average litter size (* F = 2.71, P = 0.07, α = 0.05) at weaning and less than average number of euploid pups (** F = 4.53, P = 0.01, α = 0.05) at weaning than H2b/k or H2b/b mothers. Light-shaded bars, Ts65Dn; dark-shaded bars, euploid; solid bars, total.
Gender difference in offspring of Ts65Dn mice:
DS infants are more frequently male than female (Leonard et al. 2000; Glasson et al. 2002). We evaluated the ratio of male and female offspring from Ts65Dn mothers. In contrast to the situation in DS, the frequency of Ts65Dn males (34% of male progeny) among 1037 weanlings was slightly reduced relative to trisomic females (39% of female progeny) (χ2 = 5.47, P = 0.06).
DISCUSSION
The Ts65Dn mouse is the most widely used model of DS, but research with these animals is limited because they are difficult to propagate and maintain. Males are sterile, and females are generally poor mothers. Lower than Mendelian inheritance of the T(1716)65Dn chromosome is reported in multiple studies, with ratios of trisomic offspring ranging widely from 20 to 40% (Davisson et al. 1993; Holtzman et al. 1996; Davisson and Costa 1999; Paz-Miguel et al. 2001; Villar et al. 2005). Despite their central role in DS research, little numerical or statistical evidence has been published to support claims of non-Mendelian transmission of the T(1716)65Dn chromosome, the timing of these losses, or several growth parameters relevant to survival and propagation of these mice.
We analyzed >1600 offspring from Ts65Dn dams produced in a large breeding colony over 3 years to establish basic parameters relevant to the use of this model. We established that the number of trisomic embryos meets Mendelian expectations at midgestation, indicating that the marker chromosome segregates freely. The finding of 50% trisomic embryos and the fact that the average somite number was the same in trisomic and euploid mice indicate that this segmental trisomy is not deleterious to early development. About twice as many genes are trisomic in DS as in Ts65Dn, and that additional genetic burden is likely a major part of the reported 50% prenatal lethality of conceptuses with trisomy 21. More genetically representative mouse models will be required to study this aspect of DS.
Assessment of trisomy frequencies at P0, P6, and weaning suggests that a small loss of trisomic pups may occur in the latter half of gestation, while most of the loss occurs postnatally. Both trisomic and euploid pups were lost between birth and weaning, but trisomic mice were lost disproportionately. We provide some evidence for the reputation of Ts65Dn females as “poor mothers” by showing that 17% of litters were neglected at birth with subsequent loss of all pups.
Ts65Dn mice have been reported to be smaller than euploid at birth but become obese by 3–4 months of age. We found that the average mass of trisomic mice was significantly reduced at all ages measured between P0 and 5 months. From P6 onward, there was no overlap between individual trisomic and euploid weights within most (16/19) litters; i.e., the largest trisomic mouse was smaller than the smallest euploid mouse. This was not true between litters, which showed considerable variation in average weight. Obesity of aging Ts65Dn in some colonies would appear to stem from reasons other than trisomy, including environmental influences.
We also examined a previously reported selection for and against specific H2 haplotypes in the advanced intercross used to maintain Ts65Dn. We found that the H2k/k genotype was underrepresented among trisomic progeny in the two crosses where it could occur. We saw no evidence for selection against H2b in trisomic progeny as reported in a previous study (Paz-Miguel et al. 2001). The opposing results may be due in part to the smaller size of the previous study (about half the number of mice that were considered here) and to the fact that only two of the three possible matings with respect to H2 haplotype were considered in that analysis. Although markers within H2 were utilized in both studies, the specificity of antibodies (previous study) and PCR (this study) in determining haplotype may differ and could also contribute to discrepancies between the two. It will be of interest to type flanking markers, as well, to determine whether the H2 locus or an adjoining gene(s) is responsible for the effects observed here. Given the complications associated with breeding Ts65Dn mice, a tightly linked gene may be difficult to distinguish.
The differences in selection against a particular haplotype in trisomic mice may also be due to the interaction of the H2 allele(s) with environmental or genetic factors. Environmental differences, including pathogens, could affect the survival of trisomic mice after birth. Our results demonstrate that loss of trisomic mice continues after birth, and the loss of a particular H2 haplotype could be due to the inability of a trisomic mouse with a particular haplotype to respond to an environmental (immunological) hazard specific to the location of the study. Alternatively, because Ts65Dn mice are maintained as an advanced intercross with approximately 50% C57BL6/J and C3H alleles, interactions between H2 and genes with allelic variation established in a particular environment may lead to selection against a particular haplotype. Such differences could be due to a founder effect of the Ts65Dn mice obtained in the two different studies. Variation in measured phenotypes within the Ts65Dn strain suggests that allelic effects are important in genotype–phenotype investigations and may have influenced differences seen in this and previous studies (Richtsmeier et al. 2000; Olson et al. 2004).
This is the largest population of Ts65Dn mice to be analyzed to date. Although the noninbred background of Ts65Dn mice and colony-specific environmental differences may influence the production of Ts65Dn mice, these observations suggest several ways to improve colony management for the production of this important model of Down syndrome. Our results also provide direct insight into the biology of trisomy in the mouse, suggesting both new areas for modeling DS and important differences between this model and DS, especially for studies of early prenatal development.
Acknowledgments
We thank Veronica Aquino, Donna Klinedinst, Nidhi Saran, and Valerie DeLeon for sharing samples and results from P0 and P6 Ts65Dn offspring. We also thank Grace Wan for helping obtain H2 haplotypes of Ts65Dn mothers and offspring. This work was supported by National Research Service Award fellowship HD-43614 (R.J.R.) and Public Health Service awards HD-38384 and HD-24605 (R.H.R.).
References
- Baxter, L. L., T. H. Moran, J. T. Richtsmeier, J. Troncoso and R. H. Reeves, 2000. Discovery and genetic localization of Down syndrome cerebellar phenotypes using the Ts65Dn mouse. Hum. Mol. Genet. 9: 195–202. [DOI] [PubMed] [Google Scholar]
- Belichenko, P. V., E. Masliah, A. M. Kleschevnikov, A. J. Villar, C. J. Epstein et al., 2004. Synaptic structural abnormalities in the Ts65Dn mouse model of Down Syndrome. J. Comp. Neurol. 480: 281–298. [DOI] [PubMed] [Google Scholar]
- Bell, R., J. Rankin and L. J. Donaldson, 2003. Down's syndrome: occurrence and outcome in the north of England, 1985–99. Paediatr. Perinat. Epidemiol. 17: 33–39. [DOI] [PubMed] [Google Scholar]
- Cooper, J. D., A. Salehi, J. D. Delcroix, C. L. Howe, P. V. Belichenko et al., 2001. Failed retrograde transport of NGF in a mouse model of Down's syndrome: reversal of cholinergic neurodegenerative phenotypes following NGF infusion. Proc. Natl. Acad. Sci. USA 98: 10439–10444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davisson, M. T., and A. C. S. Costa, 1999. Mouse models of Down Syndrome, pp. 297–327 in Advances in Neurochemisty, edited by B. Popko. Kluwer Academic/Plenum Publishers, New York.
- Davisson, M. T., C. Schmidt, R. H. Reeves, N. G. Irving, E. C. Akeson et al., 1993. Segmental trisomy as a mouse model for Down syndrome. Prog. Clin. Biol. Res. 384: 117–133. [PubMed] [Google Scholar]
- Debieve, F., A. Moiset, K. Thomas, S. Pampfer and C. Hubinont, 2001. Vascular endothelial growth factor and placenta growth factor concentrations in Down's syndrome and control pregnancies. Mol. Hum. Reprod. 7: 765–770. [DOI] [PubMed] [Google Scholar]
- Forrester, M. B., and R. D. Merz, 2002. Epidemiology of Down syndrome (Trisomy 21), Hawaii, 1986–97. Teratology 65: 207–212. [DOI] [PubMed] [Google Scholar]
- Frendo, J. L., M. Vidaud, J. Guibourdenche, D. Luton, F. Muller et al., 2000. Defect of villous cytotrophoblast differentiation into syncytiotrophoblast in Down's syndrome. J. Clin. Endocrinol. Metab. 85: 3700–3707. [DOI] [PubMed] [Google Scholar]
- Frid, C., G. Bjorkhem, A. Jonzon, J. Sunnegardh, G. Anneren et al., 2004. Long-term survival in children with atrioventricular septal defect and common atrioventricular valvar orifice in Sweden. Cardiol. Young 14: 24–31. [DOI] [PubMed] [Google Scholar]
- Gardiner, K., A. Fortna, L. Bechtel and M. T. Davisson, 2003. Mouse models of Down syndrome: how useful can they be? Comparison of the gene content of human chromosome 21 with orthologous mouse genomic regions. Gene 318: 137–147. [DOI] [PubMed] [Google Scholar]
- Glasson, E. J., S. G. Sullivan, R. Hussain, B. A. Petterson, P. D. Montgomery et al., 2002. The changing survival profile of people with Down's syndrome: implications for genetic counselling. Clin. Genet. 62: 390–393. [DOI] [PubMed] [Google Scholar]
- Halliday, J. L., L. F. Watson, J. Lumley, D. M. Danks and L. J. Sheffield, 1995. New estimates of Down syndrome risks at chorionic villus sampling, amniocentesis, and livebirth in women of advanced maternal age from a uniquely defined population. Prenat. Diagn. 15: 455–465. [DOI] [PubMed] [Google Scholar]
- Hampton, T. G., M. R. Stasko, A. Kale, I. Amende and A. C. Costa, 2004. Gait dynamics in trisomic mice: quantitative neurological traits of Down syndrome. Physiol. Behav. 82: 381–389. [DOI] [PubMed] [Google Scholar]
- Hattori, M., A. Fujiyama, T. D. Taylor, H. Watanabe, T. Yada et al., 2000. The DNA sequence of human chromosome 21. Nature 405: 311–319. [DOI] [PubMed] [Google Scholar]
- Holtzman, D. M., D. Santucci, J. Kilbridge, J. Chua-Couzens, D. J. Fontana et al., 1996. Developmental abnormalities and age-related neurodegeneration in a mouse model of Down syndrome. Proc. Natl. Acad. Sci. USA 93: 13333–13338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoshnood, B., C. De Vigan, V. Vodovar, J. Goujard and F. Goffinet, 2004. A population-based evaluation of the impact of antenatal screening for Down's syndrome in France, 1981–2000. BJOG 111: 485–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kola, I., and P. J. Hertzog, 1998. Down syndrome and mouse models. Curr. Opin. Genet. Dev. 8: 316–321. [DOI] [PubMed] [Google Scholar]
- Leonard, S., C. Bower, B. Petterson and H. Leonard, 2000. Survival of infants born with Down's syndrome: 1980–96. Paediatr. Perinat. Epidemiol. 14: 163–171. [DOI] [PubMed] [Google Scholar]
- Liu, D. P., C. Schmidt, T. Billings and M. T. Davisson, 2003. Quantitative PCR genotyping assay for the Ts65Dn mouse model of Down syndrome. Biotechniques 35: 1170–1174, 1176, 1178. [DOI] [PubMed] [Google Scholar]
- Marino, B., L. Diociaiuti, G. Calcagni and P. Mastroiacovo, 2004. Outcome in Down syndrome fetuses with cardiac anomalies. Am. J. Med. Genet. A 128: 101–102 (author reply, Am. J. Med. Genet A 128: 103). [DOI] [PubMed] [Google Scholar]
- Moore, C. S., J. S. Lee, B. Birren, G. Stetten, L. L. Baxter et al., 1999. Integration of cytogenetic with recombinational and physical maps of mouse chromosome 16. Genomics 59: 1–5. [DOI] [PubMed] [Google Scholar]
- Morris, J. K., N. J. Wald and H. C. Watt, 1999. Fetal loss in Down syndrome pregnancies. Prenat. Diagn. 19: 142–145. [PubMed] [Google Scholar]
- Mural, R. J., M. D. Adams, E. W. Myers, H. O. Smith, G. L. Miklos et al., 2002. A comparison of whole-genome shotgun-derived mouse chromosome 16 and the human genome. Science 296: 1661–1671. [DOI] [PubMed] [Google Scholar]
- Nembhard, W. N., D. K. Waller, L. E. Sever and M. A. Canfield, 2001. Patterns of first-year survival among infants with selected congenital anomalies in Texas, 1995–1997. Teratology 64: 267–275. [DOI] [PubMed] [Google Scholar]
- Olson, L. E., R. J. Roper, L. L Baxter, E. J. Carlson, C. J. Epstein et al., 2004. Down syndrome mouse models Ts65Dn, Ts1Cje, and Ms1Cje/Ts65Dn exhibit variable severity of cerebellar phenotypes. Dev. Dyn. 230: 581–589. [DOI] [PubMed] [Google Scholar]
- Paz-Miguel, J. E., F. Pardo-Manuel de Villena, P. Sanchez-Velasco and F. Leyva-Cobian, 2001. H2-haplotype-dependent unequal transmission of the 17(16) translocation chromosome from Ts65Dn females. Mamm. Genome 12: 83–85. [DOI] [PubMed] [Google Scholar]
- Reeves, R. H., N. G. Irving, T. H. Moran, A. Wohn, C. Kitt et al., 1995. A mouse model for Down syndrome exhibits learning and behaviour deficits. Nat. Genet. 11: 177–184. [DOI] [PubMed] [Google Scholar]
- Reeves, R. H., L. L. Baxter and J. T. Richtsmeier, 2001. Too much of a good thing: mechanisms of gene action in Down syndrome. Trends Genet. 17: 83–88. [DOI] [PubMed] [Google Scholar]
- Richtsmeier, J. T., L. L. Baxter and R. H. Reeves, 2000. Parallels of craniofacial maldevelopment in Down syndrome and Ts65Dn mice. Dev. Dyn. 217: 137–145. [DOI] [PubMed] [Google Scholar]
- Roizen, N. J., and D. Patterson, 2003. Down's syndrome. Lancet 361: 1281–1289. [DOI] [PubMed] [Google Scholar]
- Rueda, N., R. Mostany, A. Pazos, J. Florez and C. Martinez-Cue, 2005. Cell proliferation is reduced in the dentate gyrus of aged but not young Ts65Dn mice, a model of Down syndrome. Neurosci. Lett. 380: 197–201. [DOI] [PubMed] [Google Scholar]
- Spencer, K., 2001. What is the true fetal loss rate in pregnancies affected by trisomy 21 and how does this influence whether first trimester detection rates are superior to those in the second trimester? Prenat. Diagn. 21: 788–789. [DOI] [PubMed] [Google Scholar]
- Verloes, A., Y. Gillerot, L. Van Maldergem, R. Schoos, C. Herens et al., 2001. Major decrease in the incidence of trisomy 21 at birth in south Belgium: Mass impact of triple test? Eur. J. Hum. Genet. 9: 1–4. [DOI] [PubMed] [Google Scholar]
- Villar, A. J., P. V. Belichenko, A. M. Gillespie, H. M. Kozy, W. C. Mobley et al., 2005. Identification and characterization of a new Down syndrome model, Ts[Rb(12.1716)]2Cje, resulting from a spontaneous Robertsonian fusion between T(171)65Dn and mouse chromosome 12. Mamm. Genome 16: 79–90. [DOI] [PubMed] [Google Scholar]
- Wright, A., Y. Zhou, J. F. Weier, E. Caceres, M. Kapidzic et al., 2004. Trisomy 21 is associated with variable defects in cytotrophoblast differentiation along the invasive pathway. Am. J. Med. Genet. A 130: 354–364. [DOI] [PubMed] [Google Scholar]