Abstract
We established the mutant mouse line, B6;CB-SktGtAyu8021IMEG (SktGt), through gene-trap mutagenesis in embryonic stem cells. The novel gene identified, called Sickle tail (Skt), is composed of 19 exons and encodes a protein of 1352 amino acids. Expression of a reporter gene was detected in the notochord during embryogenesis and in the nucleus pulposus of mice. Compression of some of the nuclei pulposi in the intervertebral discs (IVDs) appeared at embryonic day (E) 17.5, resulting in a kinky-tail phenotype showing defects in the nucleus pulposus and annulus fibrosus of IVDs in SktGt/Gt mice. These phenotypes were different from those in Danforth's short tail (Sd) mice in which the nucleus pulposus was totally absent and replaced by peripheral fibers similar to those seen in the annulus fibrosus in all IVDs. The Skt gene maps to the proximal part of mouse chromosome 2, near the Sd locus. The genetic distance between them was 0.95 cM. The number of vertebrae in both [Sd +/+ SktGt] and [Sd SktGt/+ +] compound heterozygotes was less than that of Sd heterozygotes. Furthermore, the enhancer trap locus Etl4lacZ, which was previously reported to be an allele of Sd, was located in the third intron of the Skt gene.
THE notochord is an integral component of the axial structure of vertebrates, functions as a signaling center during embryogenesis, and plays essential roles in patterning of both somites and the neural tube (Ang and Rossant 1994; Wilson et al.1995; Chiang et al.1996). In addition, the notochord has major roles in vertebral column formation. In the mouse, the notochord is a continuous rod of constant diameter extending from the hypophysis to almost the tip of the tail at embryonic day (E) 9.5. At E10.5–E11.5, signals from the notochord induce the migration, proliferation, and fusion of the sclerotome to form a continuous and unsegmented perichordal tube around the notochord and neural tube. At 12.5, mesenchyme acquires a characteristic metameric pattern of densely packed areas caudally and loosely packed areas cranially. Some densely packed cells move cranially and give rise to the annulus fibrosus of the future intervertebral disc (IVD). The remaining densely packed cells fuse with the immediately caudal loosely packed cells to form the cartilaginous primordia of the vertebral bodies. Notochord cells located in the vertebral body of cranial regions start to relocate into intervertebral regions (Paavola et al. 1980; Rufai et al. 1995; Aszodi et al. 1998). At E13.5, the vertebral regions are enlarged and chondrified. The notochord proliferates and undergoes hypertrophy to form the gelatinous center of the intervertebral disc, called the nucleus pulposus. This nucleus is surrounded by the circularly arranged fibers of the annulus fibrosus. These two structures together constitute the IVD (Langman 1969; Theiler 1988). At E14.5, nearly all chondrocytes are hypertrophied. Starting from E14.5, the annulus fibrosus can be subdivided into a fibrous outer annulus and a cartilaginous inner annulus. At E16.0, notochord cells complete relocation from vertebral regions into intervertebral regions. Failures in somite, neural tube, and notochord formation are closely correlated with vertebral malformations. However, the mechanisms that underlie the formation of IVDs are largely unknown.
In the mouse, several mutations are known to affect the formation of the vertebral column due to functional defects in the notochord, including Danforth's short tail (Sd) and the enhancer trap line (Etl4lacZ). Sd, located on chromosome 2 (Lane and Birkenmeiser 1993; Alfred et al. 1997), is a semidominant mutation affecting the development of the vertebral column and the urogenital system (Dunn et al. 1940; Gruneberg 1953, 1958). At E9.5, the notochord shows discontinuities. At E11.5, mesenchymal organization around the notochord is abnormal. At E13.5, the notochord is fragmented and does not show proliferation and dilatation. Chondrification is much reduced in vertebral regions. Thus, an early reduction of the notochord results in cellular degeneration in the sclerotome, leading to reduced vertebral bodies and a characteristic short tail due to a reduced number of caudal vertebrae. Homozygous Sd animals show a similar but much more severe tailless phenotype. In the enhancer trap line, Etl4lacZ, a reporter (lacZ) gene was inserted near the Sd locus and was expressed in the notochord, mesonephric mesenchyme, and apical ectoderm ridge (Gossler et al. 1989; Korn et al. 1992; Maatman et al. 1997; Zachgo et al. 1998). Etl4lacZ homozygotes exhibited kinks in the caudal region of their tails and a synergistic genetic interaction between Etl4lacZ and Sd was observed. Genetically, Etl4lacZ and Sd are separated by 0.75 cM. Interestingly, attenuation or enhancement of the Sd phenotype was observed when the Etl4lacZ insertion was in a cis- or trans-conformation, respectively. This suggests that Etl4lacZ is an allele of Sd, presumably by trapping a cis-regulatory element of the Sd gene, and that Sd is a gain-of-function mutation (Zachgo et al. 1998). Nevertheless, neither the cis-element nor the trapped gene has been identified.
In this study, we report a new mutant mouse line, SktGt, obtained by gene-trap mutagenesis in embryonic stem (ES) cells. SktGt/Gt mice exhibit a kinky tail in the caudal vertebral columns due to malformation of the IVDs. The gene identified, Sickle tail (Skt), was expressed in the notochord, its derivative nucleus pulposus, and in the mesonephros. Interestingly, the Skt gene maps to the proximal part of mouse chromosome 2, near the locus for Sd, and the lacZ insertion site in Etl4lacZ was found to be located in the third intron of the Skt gene. Furthermore, a cumulative effect of the SktGt mutation on the Sd mutant was observed.
MATERIALS AND METHODS
Generation and genotyping of mutant mice:
The gene-trap method using the pU-8 trap vector was previously described (Araki et al. 1999). Chimeric mice were produced by the aggregation method using ES gene-trap clones and morulas of ICR (Charles River, Wilmington, MA) mice, and the chimeric mice were mated with C57BL/6 (CLEA) females to obtain F1 heterozygotes. Sd mice were purchased from the Jackson Laboratory (Bar Harbor, ME) and propagated by in vitro fertilization. Sd +/+ SktGt mice were generated by mating Sd +/+ + mice (C57BL/6 genetic background) to SktGt/Gt mice with a C57BL/6 genetic background. One heterozygote carrying the Sd mutation and the SktGt insertion on the same chromosome (Sd SktGt/+ +; cis-configuration) was obtained through mating between trans-heterozygotes and wild-type C57BL/6 mice. Sd mice were distinguished by external inspection. Genotyping for SktGt alleles was done with PCR using tail genomic DNA as a template. For the wild-type allele, the 5′ primer, GTS (5′-CCACCCCTACATGTGTCTTT-3′), and the 3′ primer, GTA (5′-CGAGTAAGTAACATCCCTCC-3′) located in the 14th intron, were used to generate a 339-bp wild-type fragment. To detect the trapped allele, the 5′ primer, called Z1 (5′-GCGTTACCCAACTTAATCG-3′), and the Z2 (5′-TGTGAGCGAGTAACAACCCG-3′) located in lacZ gene, were used to generate a 320-bp fragment.
Skeletal preparations:
After tail skins were peeled off, tails were fixed in 95% ethanol for 3 days. Tails were cleared by placing in 1% KOH for 1 day and were stained in alizarin red for 1 day until the bone was red. Excess stain was removed with 2% KOH. After removing excessive alizarin red stain, tails were transferred to glycerol (Hogan et al. 1994).
To count the number of vertebral bodies, we examined all mice by X-ray photography. We counted the number of normal vertebral bodies, that is, those without obvious malformations, but did not consider the size reduction observed in the vertebral bodies of Sd mutants.
Cloning of genomic DNA and cDNA:
Plasmid rescue to obtain flanking genomic DNA was performed as described (Araki et al. 1999). The 5′-end of the cDNA of the trapped gene was isolated by 5′-rapid amplification of cDNA ends (5′-RACE) using the 5′-RACE system (Invitrogen, Carlsbad, CA). Total RNA from a B6;CB-SktGtAyu8021IMEG gene-trapped ES clone was extracted by using Sepasol-RNA I (NACALAI TESQUE, Kyoto, Japan), and then poly(A)+ RNA was isolated with an oligo(dT) column (Takara Biomedicals, Shiga, Japan). First-strand cDNA synthesis from 1 μg of poly(A)+ RNA was performed with reverse transcriptase from ReverScript (Wako, Osaka, Japan) and with the primer SA13 (5′-TCTGAAACTCAGCCTTGAGC-3′) in the splice acceptor (SA) sequence. After dCTP tailing with terminal deoxynucleotidyl transferase (Invitrogen), cDNA was purified using a QIAquick nucleotide removal kit (QIAGEN, Chatsworth, CA). The initial PCR was performed using the primer SA10 (5′-AGCAGTGAAGGCTGTGCGA-3′) in the SA sequence and the anchor primer (5′-GGCCACGCGTCGACTAGTACGGGiiGGGiiGGGiiG-3′) (Invitrogen). Then, nested PCR was performed using primer 63 (5′-GCTTGTCCTCTTTGTTAGGG-3′) in the SA sequence and the amplification primer (5′-GGCCACGCGTCGACTAGTAC-3′) in the anchor primer sequence. Amplified fragments were then sequenced directly by the dideoxy-chain termination method using Big Dye terminator cycle sequencing (Perkin-Elmer, Foster City, CA).
RT–PCR analysis:
RT–PCR was performed using the Thermoscript RT–PCR system (Invitrogen) according to the manufacturer's instructions. The PCR was performed using the primers a–f in the sense and antisense sequences in the Skt gene. The sequences of the primers used are as follows: primer-a, 5′-TCACCATGAAGATGCTGGAG-3′; primer-b, 5′-CTACAGTAAGCACTCGCTGAC-3′; primer-c, 5′-ACTCCTCAGCCTTGATGAAC-3′; primer-d, 5′-GTGGTGGTAAGTCCTGATCC-3′; primer-e, 5′-GCCACCTTAAAGACACTAGG-3′; and primer-f, 5′-TGAGGAGGAAGAGGTAGTAG-3′. The PCR conditions were 94° for 1 min, 55° for 2 min, and 72° for 2 min using 0.5 units of Taq polymerase for 30 cycles.
Northern blot analysis:
Total RNA and poly(A)+ mRNA isolated from ES cells and embryo and adult tissues were electrophoresed on a 0.7% denaturing formaldehyde-MOPS-containing agarose gel and transferred to a positively charged nylon membrane (Roche). After baking at 80° for 1 hr, the membrane was prehybridized and then hybridized using the Skt gene-specific RNA probes and the lacZ RNA probes prepared using DIG RNA labeling and detection kit (Roche).
Detection of β-galactosidase (lacZ) activities:
Whole-mount X-gal staining was performed according to the method of Allen et al. (1988). Samples were fixed for 30 min at room temperature in fix solution [1% formaldehyde, 0.2% glutaraldehyde, and 0.02% NP-40 in phosphate-buffered saline (PBS)]. Fixed samples were washed two times in PBS and incubated overnight at 30° in staining solution (5 mm potassium ferricyanide, 5 mm potassium ferrocyanide, 2 mm MgCl2, 0.5% X-gal in PBS). Samples were rinsed twice in PBS, postfixed in 4% paraformaldehyde, and made transparent using benzylalcohol/benzylbenzoate (1:2) after dehydration with a series of ethanol steps (25, 50, 70, 100, and 100%, 1 hr each). Adult tissues were fixed in 4% paraformaldehyde in PBS. Tissue sections of 10 μm were prepared and stained overnight at 30° with X-gal in staining solution. After staining, sections were counterstained with Fast Red. For the section of the intervertebral discs, after X-gal stained tails were refixed in 3.7% formaldehyde/PBS, the caudal vertebral bones were demineralized in Plank–Rychlo solution and embedded in paraffin according to standard procedures. Sections of 8 μm were prepared and counterstained with Nuclear Fast red.
Construction and transfection of the Skt expression vector:
To clone the full open reading frame (ORF) of the Skt gene, RT–PCR was performed using the Thermoscript RT–PCR system. The initial PCR was performed using the primer ORFS1 in the sense strand upstream from the start codon in the Skt gene (5′- ACCGGAGTGGAGACTAGTTG-3′) and primer ORFA1 in the antisense strand downstream from the stop codon in the Skt gene (5′-TGCATGAGGCCTTGAACGATACAG-3′). Then, nested PCR was performed using the sense-strand primer ORFS2 (5′-TTTCTGCGAGCTTTCCGAAC-3′) and the antisense-strand primer ORFA2 (5′-ACCTTGGTCCTAATAGGATCTGGC-3′). The PCR conditions used were 94° for 1 min, 58° for 1 min, and 72° for 3 min using 1.0 unit of LA Taq polymerase (Takara Biomedicals) for 25 cycles. The 4.1-kb PCR products were cloned into the pGEM-T vector (Promega, Madison, WI). This cDNA ORF was confirmed by sequencing and cloned into the pCAGGS expression vector (Niwa et al. 1991). Transfection into BMT10 cells (Gerard and Gluzman 1985) was carried out by the lipofection method using LipofectAMINE reagent (Invitrogen).
Western blot analysis:
BMT10 cells and 40 pieces of the nucleus pulposus of caudal IVDs of adult mice were homogenized in 2× sample buffer (100 mm Tris HCl pH 6.8, 4% SDS, 12% β-mercaptoethanol, 20% glycerol). Extracts were electrophoresed on a 6.0% polyacrylamide gel, transferred to a nitrocellulose filter (Immobilon, Millipore, Bedford, MA), and detected using anti-Skt antibodies with the ECL detection system (Amersham, Arlington Heights, IL).
Immunohistochemistry:
Tails were fixed in 4% paraformaldehyde in PBS. The caudal vertebral bones were demineralized in 0.24 m EDTA·2Na, 0.22 m EDTA·4Na solution for 48 hr and embedded in paraffin blocks. Sections of the IVDs were immunostained with anti-Skt antibodies by the avidin–biotin complex method (Vector Laboratories, Burlingame, CA). Sections were counterstained with hematoxylin.
RESULTS
Generation of Sickle tail mutant mice:
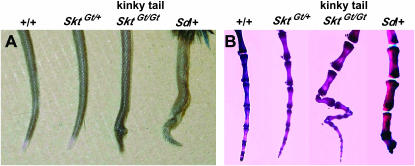
A gene-trap ES clone was isolated by the exchangeable gene-trap method using the trap vector pU-8 (Araki et al. 1999). We obtained eight chimeric mice of which three were germline chimeras. Heterozygous animals appeared normal and were fertile. About half (35/66) of the homozygotes, however, showed a peculiar kinky-tail phenotype (Figure 1A and Table 1). This mutant mouse line was designated as B6;CB-SktGtAyu8021IMEG, in which Skt means Sickle tail because of the characteristic shape of the tail. Shortened and curved caudal vertebrae were apparent by the age of 2 weeks and were restricted to the 20–25th caudal vertebrae (Figure 1B). In contrast, heterozygous Sd mice showed short tails with truncation of vertebral columns at the 6th caudal vertebral body on average (Figures 1B and 7A) as reported previously. In SktGt/Gt mice, no other skeletal abnormality was observed by bone X-ray examination (data not shown).
Figure 1.
(A) Tail phenotype of 8-week-old mice. SktGt/Gt mice had kinked tails compared to wild-type, SktGt/+, and heterozygous Sd mice. (B) Alizarin red whole-mount preparations of the tails of 8-week-old mice. SktGt/Gt mice confirmed this kinky-tail phenotype. Sd mice showed decreased numbers of vertebrae with truncation at the caudal vertebrae.
TABLE 1.
Summary of genotyping of 4-week-old mice from SktGt heterozygote matings
|
Gt/Gt
|
|||
|---|---|---|---|
| +/+ | Gt/+ | Kinked | Normala |
| 60 | 151 | 35 (53%) | 31 |
All normal-looking homozygotes showed deformity of the caudal discs by histological analysis.
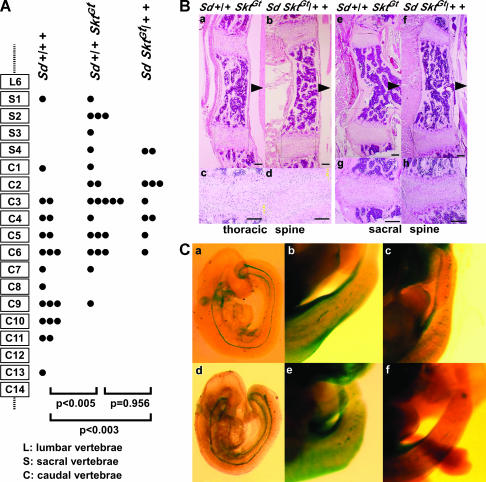
Figure 7.
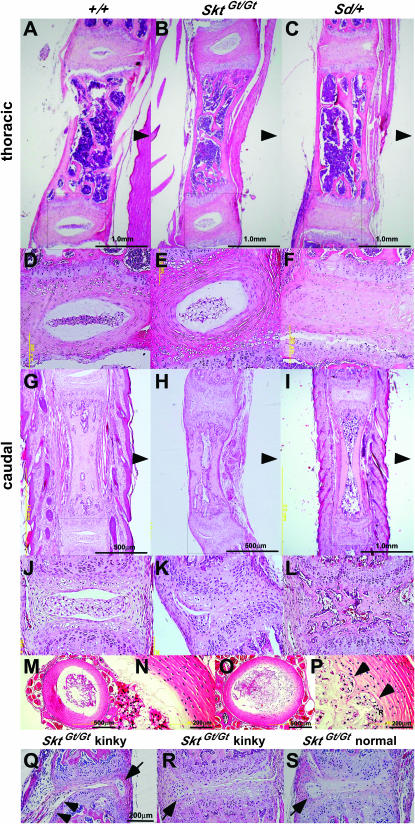
(A) Schematic of axial levels and severity of vertebral malformations in compound mutant 8-week-old mice. A single solid circle indicates the level of the terminal vertebral body for a single mouse. In both trans and cis compound mutant mice, the degree of vertebral malformation is more severe than that in heterozygous Sd mice (trans, P < 0.005; cis, P < 0.003, Mann–Whitney U-test). There was no significant difference between trans [Sd +/+ SktGt] (n = 23) and cis [Sd SktGt/+ +] (n = 10) mice (P = 0.956, Mann–Whitney U-test). (B) Histological analyses of vertebral columns in the Sd +/+ SktGt and Sd SktGt/+ + mutant mice. HE staining of midsagittal sections in thoracic spines of Sd +/+ SktGt (a and c) and Sd SktGt /+ + (b and d) mice and in sacral spines of Sd +/+ SktGt (e and g) and Sd SktGt/+ + (f and h) mice. Arrowheads indicate dorsal sides. (c, d, g, and h) Higher magnification of the areas indicated by the boxes in a, b, e, and f, respectively. (C) Whole-mount X-gal staining in the Sd +/+ SktGt and Sd SktGt/+ + mutant embryos. The β-gal expression in the tail notochord of trans-heterozygous embryos [Sd +/+ SktGt] at E9.5 (a) and E13.5 (b and c) and of cis-heterozygous embryos [Sd SktGt/+ +] at E9.5 (d) and E13.5 (e and f).
Characterization of the integration site of the trap vector:
To characterize the gene-trap locus, we cloned and sequenced genomic DNA fragments flanking the gene-trap vector. A single copy of the vector was integrated into the genome as determined by Southern blot analysis using genomic DNA samples extracted from SktGt mice. Three base pairs of genomic DNA were deleted at the integration site of the trap vector. Using PCR amplification on genomic DNA samples, the genotype of offspring from the heterozygous intercross was easily determined (data not shown).
Identification of the Sickle tail gene:
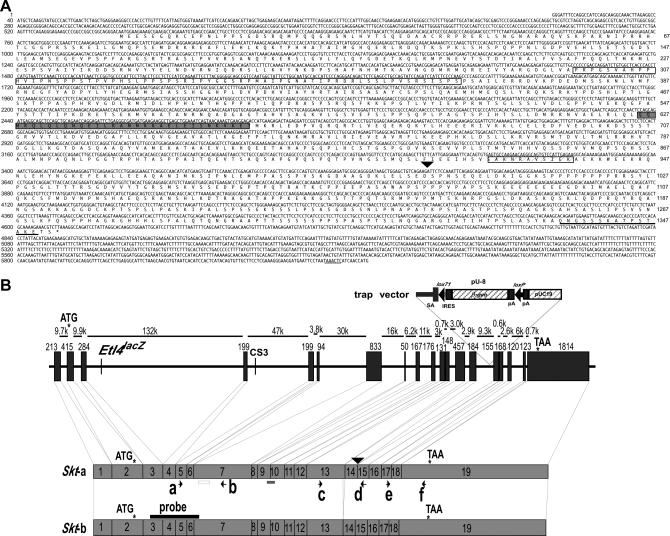
To identify the gene trapped, we performed 5′-and 3′-RACE. The sequence of the ORF of the trapped gene was determined by compiling sequences of 5′- and 3′-RACE products and of EST that showed 100% homology to the RACE products. We thus obtained a cDNA sequence comprising 5930 nucleotides (accession no. AB125594) that encodes a putative protein of 1352 amino acids with a predicted molecular weight of 147 kDa (Figure 2A). This gene was termed Skt. The protein contains a proline-rich region (amino acid residues 298–364) and a coiled-coil region (amino acid residues 626–656) as determined by analysis using Lupas's algorithm (Lupas et al. 1991). An ATG codon is located at nucleotide positions 559–561. This codon is most likely the initiation codon, because there is a Kozak sequence surrounding this ATG codon (Kozak 1996): i.e., there is a G residue following the ATG codon and an A residue three nucleotides upstream. A BLAST search of the amino acid sequence deduced from the Skt cDNA sequence revealed 80.6% homology with an uncharacterized human protein, KIAA 1217 (accession no. AB033043), and no other evolutionarily conserved protein was identified.
Figure 2.
Identification of the trapped gene Skt. (A) The nucleotide sequence of the Skt cDNA and predicted amino acid sequence. The open box indicates the pro-rich region at the N terminus and the shaded box indicates the coiled-coil region in the middle. The striped box indicates the sequence deleted in Skt-b by alternative splicing. The 15-amino-acid peptide used for the production of anti-Skt antibodies is shown by underlining. The polyadenylation signal is underlined at the 3′-end of the nucleotide sequence. The nucleotide sequence is numbered on the left side and the amino acid sequence is numbered on the right side. (B) Genomic structure of the Skt gene. (Top) Exon-intron structure of the Sickle tail gene. The trap vector, pU-8, was inserted into the 14th intron. Sizes of exons and introns are given. (Bottom) Two transcripts produced from the Skt allele. There are at least two types of Skt transcripts: one contains all the exons (termed Skt-a) and the other lacks 33 bp of the 13th exon (termed Skt-b). Arrows (a–f) indicate the location of the primers used for RT–PCR analyses in Figure 3, A–C, to detect the expression of each part of the Skt transcripts. The solid bar represents a probe used for Northern blotting. The open and shaded boxes indicate the pro-rich region and the coiled-coil region, respectively. The start and stop codons of the Skt gene are shown by asterisks. A sequence with high homology to the CS3 in node/notochord enhancers is located in the fourth intron of the Skt gene 106 kb downstream of the insertion site of Etl4lacZ.
Sequence comparison of Skt cDNA with the murine genome sequence in the public Mouse Genome Resources (http://www.ncbi.nlm.nih.gov/genome/guide/mouse/) revealed that the Skt gene consists of 19 exons spanning >300 kb and that the ATG codon is located in the second exon (Figure 2B). The trap vector was integrated in the 14th intron, resulting in the disruption of the Skt protein at position 998 (arrowhead in Figure 2, A and B). Sequence analyses of fusion transcripts revealed the presence of two fusion transcripts with the β-geo sequence: Skt-a, containing the 1st–14th exons, and Skt-b, lacking 33 bp of the 13th exon from Skt-a (Figure 2B). In any case, a truncated protein lacking 355 amino acids encoded by exons 15–19 in the C-terminal region is expected to be produced from the SktGt allele.
RT–PCR and Northern blot analyses:
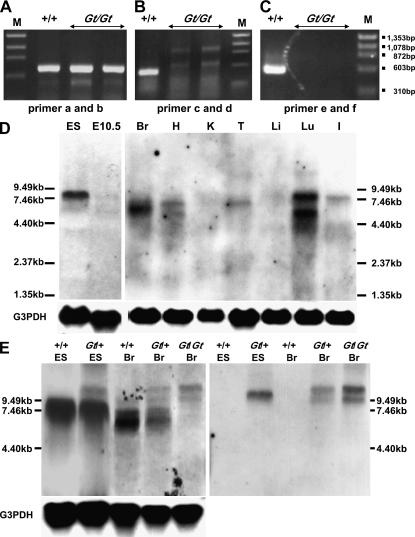
We performed RT–PCR using wild-type and SktGt/Gt embryos at E10.5. RT–PCR with primers a and b located within the 5′-region of the integration site and detected the expected band in both embryos (Figure 3A). Using RT–PCR with two primer pairs—c in the 5′-region and d in the 3′-region of the integration site and primers e and f within the 3′-region of the integration site—we could not detect any product in SktGt/Gt embryos (Figure 3, B and C), indicating the absence of Skt mRNA containing exons 15–19 in SktGt/Gt embryos.
Figure 3.
Analyses of Skt transcripts. (A–C) RT–PCR analyses using E10.5 embryos to detect Skt transcripts in wild-type (+/+) and SktGt/Gt embryos. The transcripts containing nucleotide sequences upstream of the insertion site of the trap vector were detected in both wild-type and SktGt/Gt embryos (A). The transcripts containing nucleotide sequences downstream of the Skt sequence were not detected in SktGt/Gt embryos (B and C). M, molecular marker. (D) Northern blot analyses to detect Skt mRNA in the wild-type ES cells, E10.5 embryo, and 8-week-old mice, using the Skt-specific probe in the 5′-region (see Figure 2B). Total RNA (10 μg) from TT2 ES cells, wild-type E10.5 embryos, mRNA (5 μg) from wild-type organs, and a Skt RNA probe were used for Northern blotting. (E) Northern blot analyses to detect Skt and β-geo fusion transcripts. Total RNA (20 μg) from TT2 and heterozygous (Gt/+) ES cells, wild-type (+/+), heterozygous (Gt/+), and homozygous (Gt/Gt) adult brains was used for Northern blotting. The Skt RNA probe or lacZ RNA probe was used in the left and right panels, respectively. Br, brain; H, heart; K, kidney; T, testis; Li, liver; Lu, lung; I, intestine.
To analyze the size and expression pattern of Skt mRNA, we performed Northern blot analysis using the Skt-specific probe in the 5′-region (see Figure 2B). As shown in Figure 3D, a major band of 8 kb and a minor band of 6.5 kb were detected in wild-type ES cells. In wild-type whole embryos at E10.5, the 8- and 6.5-kb bands were also detected, although the expression levels were low. Surprisingly, Northern blot analysis using mRNA from wild-type adult organs revealed the presence of four different mRNA transcripts, 5.5, 6.5, 7.0, and 8.0 kb. The faint 7.0-kb and strong 6.5-kb bands in the brain, the 7.0- and 5.5-kb bands in the heart, the 7.0-kb band in the testis, the 8.0- and the 5.5-kb bands in the lung, and the 8.0-kb band in the intestine were detected (Figure 3D). We then examined the presence of the Skt and β-geo fusion mRNA (Figure 3E). In SktGt/+ ES cells, an expected 10.5-kb band representing the fusion transcript containing β-geo was detected with both the Skt and lacZ probes, although the intensity was weaker than that of the 8-kb band of the endogenous transcript. In the SktGt/+ and SktGt/Gt adult brains, two bands of 10.5 and 9.5 kb were detected with the Skt probe (Figure 3E, left), and these bands were also hybridized with the lacZ probe (Figure 3E, right), confirming that both transcripts corresponded to the fusion mRNA containing β-geo. This result indicates the existence of alternative splicing in the upstream region of the insertion site. Northern blot analysis using mRNA from wild-type adult heart revealed the presence of two different transcripts, the 7- and 5.5-kb bands when the Skt probe was used. However, in the SktGt adult heart, one 9.5-kb band was detected with the lacZ probe (data not shown). This result indicates that alternative splicing in the heart may occur in the downstream region of the insertion site, leading to the production of the 7- and 5.5-kb bands. The largest size of mRNA is 8 kb, which is larger than that of the predicted cDNA sequence comprising 5930 nucleotides (accession no. AB125594). As described below, the anti-Skt antibodies recognized only an ∼150-kDa protein corresponding to the predicted molecular weight of 147 kDa in extracts from the nucleus pulposus of the caudal IVDs (see lane 4 in Figure 6A). Thus, the 8-kb mRNA may contain untranslated regions at both the 5′- and 3′-ends and splicing may occur in each untranslated region. Further study will be required to determine the untranslated regions.
Figure 6.
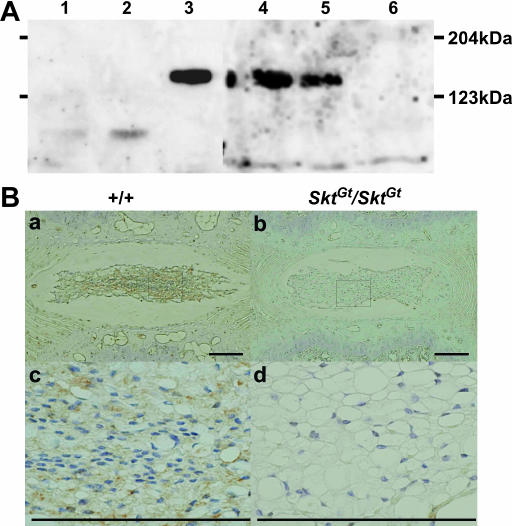
Detection of the Skt protein. (A) Western blot analysis to detect Skt protein using extracts from untreated BMT10 cells (lane 1), BMT10 cells transfected with vector (lane 2), BMT10 cells transfected with the Skt expression vector (lane 3), and extracts of the nucleus pulposus of caudal IVDs from 8-week-old wild-type mice (lane 4), SktGt/+ mice (lane 5), and SktGt/Gt mice (lane 6). An ∼150-kDa protein corresponding to the predicted molecular weight of 147 kDa was detected in lanes 3, 4, and 5, but not in lane 6. The amount of Skt protein was reduced in the SktGt/+ mutant (lane 5) and was below the detectable level in SktGt/Gt (lane 6). (B) Immunohistochemistry of frontal sections of the nucleus pulposus in upper caudal IVDs from adult 8-week-old mice using purified anti-Skt antibodies. Skt protein was detected in the cytoplasm of nucleus pulposus cells in wild-type (a and c), but not SktGt/Gt, mice (b and d). (c and d) Higher magnification of the area indicated by the boxes in a and b, respectively. Bars, 200 μm.
Formation of vertebral column and β-geo expression in SktGt mice:
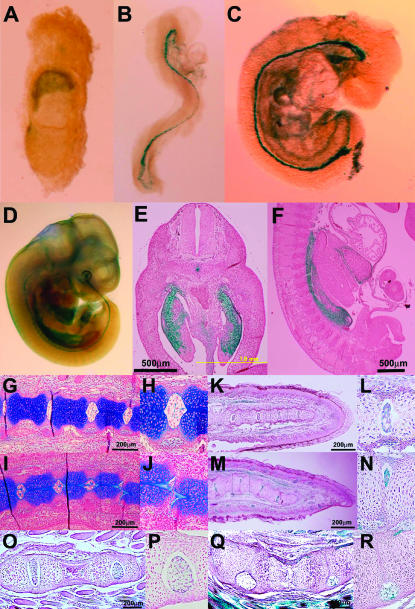
We examined formation of the vertebral column in SktGt mice during the embryonic, fetal, and postnatal periods (E8.0, E8.5, E9.0, E9.5, E10.5, E11.5, E12.5, E13.5, E14.5, 15.5, E16.5, E17.5, E18.5, E19.5, newborn, and 2 weeks of age) by histological analysis with and without X-gal staining. Before E8.0, the β-geo gene was expressed in the chorion, but not in the embryo (Figure 4A). At E8.5 (Figure 4B) and E9.0 (Figure 4C), intense staining was detected in the notochord. At E11.5, the notochord and the mesonephros expressed β-geo strongly (Figure 4, D–F). In addition, sections of E11.5 embryos showed β-geo expression in the epithalamus sulcus, roof of the neopallial cortex, lens vesicle, inner layer of retina, heart (atrium and ventricle), surface of hepatic primordium, infundiblum, surface ectoderm, hind gut, and mesenchyme of the limb bud (data not shown). No abnormality was found in both vertebral and intervertebral regions up to E16.5. In the SktGt/Gt embryo at E17.5, compression was observed in some IVDs (Figure 4, I and J) of the SktGt/Gt embryo, but not in the wild-type embryo (Figure 4, G and H). In the SktGt/Gt neonate, the X-gal-positive cells in the nucleus pulposus were shifted to the periphery and the alignment of the vertebral bodies was undulated (Figure 4, M and N). At this stage, sizes of IVDs are the same as those of wild-type mice. In 2-week-old SktGt/Gt mice, the X-gal-positive notochord cells were dislocated to the left or right side and the nuclei pulposi were smaller and eccentric, resulting in the disappearance of normal IVDs (Figure 4, Q and R). In the SktGt/+ embryo and mice, the X-gal-positive cells were positioned in the center of the vertebrae at any stage (Figure 4, G, K, and O). Interestingly, the expression of the reporter gene was much lower in the upper IVDs than in the fifth to seventh caudal IVD, coinciding with a relationship between the phenotype of the SktGt/Gt and strong expression in the caudal region of the tail.
Figure 4.
β-gal expression and histological analyses in SktGtmice. At E7.5 (A), the chorion was stained with X-gal, but the midline region of the embryo was not stained. At E8.5 (B) and E9.0 (C) intense staining was detected in the notochord. At E11.5 (D–F), the notochord and the mesonephros expressed β-geo strongly in whole-mount X-gal staining (D), the frontal section (E), and the sagittal section (F). Sagittal sections of the tail bud of SktGt/+ (G and H) and SktGt/Gt (I and J) embryos at E17.5. At E17.5, some IVDs were compressed in the tail bud of SktGt/Gt (J). Sagittal sections of the tail tips of newborn SktGt/+ (K and L) and SktGt/Gt (M and N) mice. In the SktGt/Gt neonate, the vertebral body alignment was undulated (M and N). Sagittal sections of the tail tips of 2-week-old SktGt/+ (O and P) and SktGt/Gt (Q and R) mice. In the 2-week-old SktGt/Gt mice, the X-gal-positive nuclei pulposi were dislocated to the periphery (Q and R). (H, J, L, N, P, and R) Higher magnification of the area indicated by the boxes in G, I, K, M, O, and Q, respectively. Sections (G–J) were stained with alcian blue and sections of X-gal staining (E, F, and K–R) were counterstained with Nuclear Fast red. Bars, 200 μm.
Histological analyses of IVDs of SktGt/Gt and Sd mutant adult mice:
In SktGt/Gt adults, the β-geo gene was also expressed in many tissues such as the corpus callosum in the brain, uriniferous tubules in kidney, cardiac muscle, Sertoli's cells in testes, and basal cells and outer root sheaths of hair follicles in skin (data not shown).
In normal adult mice, the nucleus pulposus was located in the center of IVDs at all levels of the vertebral column (Figure 5, A, D, G, and J). However, the nucleus pulposus in the IVDs of the tail region of SktGt/Gt mice contained an aggregation of notochord-like cells with fewer vacuoles than normal and were dislocated to the periphery (Figure 5, H and K). The nucleus pulposus in the upper regions appeared normal (Figure 1, B and E). Although histochemical analyses of embryos and newborns in the Sd mutant were reported by Paavola et al. (1980) and Theiler (1988), the histochemical analyses of the IVDs in Sd mutant adult mice have not previously been performed in detail. Thus, we analyzed the IVDs of the whole spine in Sd +/+ + mutant mice. Surprisingly, the IVDs were totally occupied by peripheral fibers similar to those seen in the annulus fibrosus and no nucleus pulposus was found within the IVDs (Figure 5, C and F). The degeneration of the nucleus pulposus in the center of caudal IVDs was occasionally observed (Figure 5, I and L). Although tail kinks of SktGt/Gt mice were restricted to the 20–25th caudal vertebrae (Figure 1B), an irregular boundary with direct contact between the nucleus pulposus and annulus fibrosus (Figure 5, O and P) was observed in the 5–25th caudal IVDs of SktGt/Gt mice, compared to the sharp boundary and fixed space observed between the nucleus pulposus and the fibrous layers of the annulus fibrosus in normal mice (Figure 5, M and N). In addition to the abnormalities of the nucleus pulposus in IVDs, the annulus fibrosus development was also impaired in SktGt/Gt mice as demonstrated by the thin fibrous layers of annulus fibrosus and the failure of fibrous adhesion in the vertebral bodies (Figure 5, Q and R). We also examined whether similar abnormalities were observed in the regions that did not have kinks in SktGt/Gt mice (Table 1). Surprisingly, histological analysis of externally normal tails of SktGt/Gt mice revealed the presence of similar IVD abnormalities such as dislocation of the nucleus pulposus and thin fibrous layers of the annulus fibrosus (Figure 5S) as found in kinked tails (Figure 5, Q and R). This IVD phenotype was observed over 10 generations in the progeny backcrossed to C57BL/6. Thus, the primary phenotype of SktGt/Gt mice is the deformity of IVDs in the tail region. These histological pictures were clearly distinct from those of Sd mice.
Figure 5.
Histological analyses of IVDs of SktGt/Gt and Sd mutant adult mice. Sagittal sections of the thoracic and caudal IVD from 8-week-old adult wild-type (A, D, G, and J), SktGt/Gt (B, E, H, and K), and heterozygous Sd mice (C, F, I, and L). (D, E, F, J, K, and L) Higher magnification of the area indicated by the boxes in A, B, C, G, H, and I, respectively. Arrowheads indicate the dorsal side. Axial sections of the upper caudal IVD in wild-type (M and N) and SktGt/Gt (O and P) 8-week-old mice. (N and P) Higher magnification of the area indicated by the boxes in M and O, respectively. The arrowheads in P indicate an irregular boundary with close contact between the nucleus pulposus and annulus fibrosus. (Q–S) The 20–25th caudal IVDs of SktGt/Gt 8-week-old mice. Impaired development of the annulus fibrosus in SktGt/Gt mice was demonstrated by the thin fibrous layers of annulus fibrosus (Q and R) and the failure of fibrous adhesion to the vertebral bodies (arrowhead in Q). Similar IVD abnormalities such as dislocation of the nucleus pulposus (arrow in Q–S) and impaired growth of the annulus fibrosus were observed in the nonkinked regions (S) and in the kinked regions (Q and S). Haematoxylin and eosin (HE) staining was used. Bars, 200 μm.
Analysis of Skt protein using anti-Skt antibody:
The anti-Skt antibodies recognized an ∼150-kDa protein corresponding to the predicted molecular weight of 147 kDa in the lysates of BMT10 cells transiently expressing the Skt cDNA by the CAG promoter as well as in extracts from the nucleus pulposus of the caudal IVDs. No band was detected in extracts from untreated BMT10 cells and BMT10 cells transfected with mock expression vector (Figure 6A). This is consistent with the notion that ATG at positions 559–561 and TAA at positions 4615–4617 in the Skt cDNA are the start codon and termination codon, respectively. As expected, the amount of Skt protein was decreased in SktGt/+ mice and was below the detectable level in SktGt/Gt mice (Figure 6A). After immunohistochemical staining, the nucleus pulposus cells from upper caudal vertebral discs were positively stained in wild-type mice (Figure 6B, a and c), but not in SktGt/Gt mice (Figure 6B, b and d). At higher magnification, the staining was observed mainly in the cytoplasm of the nucleus pulposus cells, indicating cytoplasmic localization of the Skt protein. Since we could not produce antibodies against the N terminus, the expression of the truncated protein was not confirmed in SktGt/Gt mice.
Relationship of Skt with Etl4lacZ or Sd loci:
Both the expression pattern of the Skt gene and the phenotype of SktGt mutant mice were quite similar to those of Etl4lacZ mutant mice (Zachgo et al. 1998). Furthermore, by examining the reported primer sequences of the Etl4lacZ locus (Maatman et al. 1997), we found that the Etl4lacZ locus is located in the third intron of the Skt gene (Figure 2B) and that the distance between the integration sites of Etl4lacZ and SktGt was 237 kb.
To examine the genetic distance and interaction between the SktGt and Sd locus, we crossed SktGt mice with Sd mice to produce the compound heterozygote (Sd +/+ SktGt; trans-configuration). Then, these compound heterozygotes were used to analyze the recombination rate between the two loci. As shown in Table 2, one compound heterozygote carrying the Sd mutation and the SktGt insertion on the same chromosome (Sd SktGt/+ +; cis-configuration) and two wild-type mice were obtained among 249 mice obtained by mating the trans compound heterozygote with the wild-type C57BL/6 mice, demonstrating that the Sd and SktGt mutations were genetically separated. The genetic distance was calculated to be ∼0.95 cM by combining the data from mating the [Sd SktGt/+ +] (cis) compound heterozygote with the wild-type mice (Table 2).
TABLE 2.
Distribution of the haplotypes among the 315 offspring of the backcross Sd +/+ SktGt or Sd SktGt /+ + × C57BL/6
|
Sd +/+ SktGt × C57 BL/6
|
Sd SktGt/+ + × C57 BL/6
|
||||||
|---|---|---|---|---|---|---|---|
| + +/+ SktGt | Sd +/+ + | Sd SktGt/+ + | + +/+ + | + SktGt/+ + | Sd +/+ + | Sd SktGt/+ + | + +/+ + |
| 144 | 102 | 1 | 2 | 0 | 0 | 24 | 42 |
Sd, SktGt compound heterozygotes were generated from crosses with an inbred laboratory strain (C57BL/6)
To evaluate the effect of the SktGt mutation on the tail phenotype in heterozygous Sd mice in either the trans- or the cis-configuration, the number of vertebrae was determined by X-ray analysis. Heterozygous Sd [Sd +/+ +] mice (n = 22) had a variable number of vertebrates and the vertebral columns were truncated at the sixth caudal vertebral body on average. Both trans [Sd +/+ SktGt] (n = 23) and cis [Sd SktGt/+ +] (n = 10) compound heterozygous mice had shorter tails, in which the vertebral columns were truncated at the second and third caudal vertebral body on average (Figure 7A). We examined whether the phenotype of [Sd SktGt/+ SktGt] neonatal mice is more severe than those of trans [Sd +/+ SktGt] and cis [Sd SktGt/+ +] compound heterozygous mice. Approximately 80% of [Sd SktGt/+ SktGt] neonatal mice died within 48 hr of birth and all mutant mice died within 2 weeks. In addition, as shown by making alcian blue/alizarin red whole-mount preparations of [Sd SktGt/+ SktGt] neonatal mice, [Sd SktGt/+ SktGt] neonatal mice had shorter tails than those of trans [Sd +/+ SktGt] and cis [Sd SktGt/+ +] compound heterozygous mice, in which the vertebral columns were truncated at the fourth sacral vertebral body on average, suggesting a cumulative effect of the SktGt mutation on the Sd mutant.
To examine the pathologic effect of the Skt mutation on the Sd phenotype, we carried out histological analyses on the IVDs of Sd +/+ SktGt and Sd SktGt/+ + mutant mice. Both compound heterozygous mice showed IVD histology similar to that seen in Sd +/+ + mutant mice. The nucleus pulposus was totally absent and replaced by peripheral fibers similar to those seen in the annulus fibrosus in all IVDs (Figure 7B, a–h). These results suggest that the Skt mutation did not affect the histological picture of IVD in the Sd mutation. In addition, the expression pattern of the β-geo gene in the tail notochord was the same in both trans [Sd +/+ SktGt] and cis [Sd SktGt/+ +] compound heterozygous embryos at E9.5 and E13.5 (Figure 7C). The notochord of both trans and cis compound heterozygous embryos was thin and clearly stained at E9.5 (Figure 7C, a and d), and the X-gal-positive notochord was similarly fragmented in both trans and cis compound heterozygous embryos at E13.5 (Figure 7C, b, c, e, and f). This suggested that the Sd mutation did not affect Skt expression. In addition, this trans–cis test demonstrated that the double heterozygotes exhibited indistinguishable phenotypes in regard to notochord degradation whether Sd and SktGt were in a trans- or in a cis-configuration.
DISCUSSION
We established a new recessive trap line, B6;CB-SktGtAyu8021IMEG, which had a deformity in caudal IVDs. The insertion site of the trap vector is in the 14th intron of the novel gene Skt, located on chromosome 2, near the locus for Danforth's short tail. In addition, we found that the enhancer trap locus Etl4lacZ, which was previously reported to be an allele of Sd, was located in the third intron of the Skt gene.
Structure, expression, and function of the Skt gene:
The sequence of the trapped gene, Skt, was obtained using the gene-trap ES clone. The Skt gene contains 19 exons encoding a novel protein of 1352 amino acids with a proline-rich region in the N terminus (amino acid positions 298–364) and a coiled-coil domain in the middle (amino acid positions 626–656). Although the role of the proline-rich region in many proteins is not clear yet, a proline-rich region located in the amino-terminal region has been shown to be important for proper folding in cytochrome P450s (mitochondrial, microbial, and microsomal P450s) (Kusano et al. 2001a,b). Thus, the proline-rich region in the Skt protein may have a similar function in protein folding. The coiled-coil motif was first described by Crick (1952) and by Pauling and Corey (1953) as the main structural element of a large class of fibrous proteins that included keratin, myosin, and fibrinogen, mediating dimerization, heterodimer formation, or trimerization (for a review see Lupas 1996). These proteins provide a scaffold for regulatory complexes such as tropomyosin and a protective surface for pathogens. As the Skt protein is localized in the cytoplasm, it is conceivable that the Skt protein may provide structural elements or act as a scaffold for regulatory complexes.
Insertion of the gene-trap vector into the 14th intron of the Skt gene, downstream of the coiled-coil domain and the proline-rich region, resulted in production of two fusion transcripts with the β-geo sequence: one, Skt-a, containing only exons 1–14 and the other, Skt-b, lacking 33 bp of the 13th exon from Skt-a (see Figure 2B). Northern blot analysis showed that the amount of fusion transcripts from the trapped allele was lower than that of the wild-type allele. Therefore, it is also possible that the insertion of the gene-trap vector resulted in decreased mRNA stability. As tail phenotypes are recessive, insertion of the trap vector may cause a hypomorphic or null mutation, but not a dominant-negative mutation. Although we detected four types of mRNA transcribed from the wild-type Skt allele, further analysis will be required to elucidate the function of these mRNA.
The expression patterns of the Skt gene can be monitored by X-gal staining, because the expression of the reporter gene, β-geo, is under control of the regulatory region of the Skt gene. At E11.5, β-geo was expressed mainly in the notochord and mesonephros at high levels, and in other tissues as described in the results. In the Etl4lacZ line, the lacZ reporter gene was also expressed in two main tissues, the notochord and the mesonephros (Zachgo et al. 1998). In both lines, lacZ expression was first detected at E8.5 in the presumptive notochord cells. In the Etl4lacZ line, lacZ expression was detected in the future IVDs at E13.5 and persisted up to E14.5. However, no lacZ expression was detected in the IVDs of newborn and adult mice. On the other hand, lacZ expression in the IVDs persisted to adult stage in the SktGt line. These results suggest that the enhancer located near the Etl4lacZ locus is not sufficient to express the lacZ gene in the IVDs of adult mice.
Histochemical analyses of the vertebral column revealed the differences between Sd mice and SktGt mice. As histological data on Etl4lacZ mice were not described by Zachgo et al. (1998), it is not clear whether the IVD histology is similar to that seen in Sd or SktGt mice. As reported previously, the development of both the vertebral column and the urogenital system is affected in Sd mutation, suggesting that the Sd gene is required for formation of derivatives from both the paraxial and the intermediate mesoderm. In addition, both vertebral bodies and IVDs are affected in Sd mice. The notochord shows discontinuities as early as E9.5, resulting in the total absence of the nucleus pulposus at all levels and is replaced by peripheral fibers similar to those of the annulus fibrosus in Sd adult mice. All vertebral bodies are reduced in a dorso-ventral direction and the number of tail vertebrae is reduced, leading to shortening or absence of the tail. However, in SktGt mice, the compression and dislocation of the nucleus pulposus were first observed at E17.5 and were restricted to the tail region. Interestingly, the size of the nucleus pulposus was similar to that in wild-type mice until birth. After birth, the nucleus pulposus did not expand and was dislocated to the periphery, resulting in a kinky-tail phenotype in adults. These observations suggest that Sd acts at an early stage of mesoderm development involving both sclerotome and notochord development and that Skt acts during the fetal period and at the later stage involving growth and hypertrophy of the nucleus pulposus. Although massive apoptosis in notochord cells was observed in embryos with targeted disruption of Jun (Behrens et al. 2003) or Sox5−/−/Sox6−/− (Smits and Lefebvre 2003), no apoptotic notochord cells were observed in the SktGt/Gt embryos at E16.5 and in neonatal mice. Thus, apoptosis is not the cause of compression or dislocation of the nucleus pulposus in SktGt mice. As the Skt protein contains the coiled-coil motif that is involved in the formation of mechanically rigid structures, it is possible that the nucleus pulposus lacking Skt protein may not be capable of sustaining mechanical loads, leading to compression or dislocation of the nucleus pulposus.
Although the IVD is formed from two components of developmentally different origins, the nucleus pulposus and the annulus fibrosus, interaction of the nucleus pulposus and annulus fibrosus is not clear yet. In Sd mice, disappearance of the notochord cells occurs at early stages of development. Theiler (1988) reported that the fibers of the annulus fibrosus are reduced in Sd heterozygous embryos and newborns. However, our results suggest that the annuli fibrosi are not reduced in Sd heterozygous adult mice. Thus, the annulus fibrosus may be able to completely compensate for the loss of the nucleus pulposus during growth after birth. In SktGt mice, the thin annulus fibrosus was observed together with abnormalities in the nucleus pulposus. At present, it is not known whether the thin annulus fibrosus is caused by the direct effect of Skt deficiency or indirectly caused by defects in the nucleus pulposus.
The relationship among the SktGt, Etl4lacZ, and Sd mutations:
Our breeding studies revealed that the genetic distance between the Skt and Sd loci was 0.95 cM. We believe that the Skt gene is distinct from the Sd gene for the following reasons. First, anti-Skt antibody detected the predicted size of protein deduced from the Skt gene. If the Skt was part of the Sd gene, we should have detected a larger protein by Western blot analysis. But, we could not detect any such protein. Second, our trans–cis test demonstrated that both double heterozygotes exhibited indistinguishable phenotypes, regardless of whether Sd and SktGt were in a trans- or a cis-configuration. On the basis of the results by Zachgo et al. (1998), Sd is a gain-of-function mutation. As the Skt is a recessive mutation, producing a hypomorphic or null allele, the Sd phenotype is expected to be attenuated in the cis-configuration. Therefore, the phenotype in double heterozygotes in the trans-configuration might be more severe than that in the cis-configuration if the Skt gene is part of the Sd gene.
We have shown that the Etl4lacZ locus is located in the third intron of the Skt gene. Interestingly, a sequence that has 93% homology to consensus sequence 3 (CS3) in node/notochord enhancers of Hnf3β (Nishizaki et al. 2001) was found in the fourth intron of the Skt gene, located 106 kb downstream of the insertion site of Etl4lacZ. As the expression patterns of Etl4lacZ are quite similar to those in SktGt mice and as Etl4lacZ mice have similar kinks in the tail region (Zachgo et al. 1998), the lacZ gene in the enhancer trap vector may be expressed under the control of this possible node/notochord enhancer. Zachgo et al. (1998) reported that the Etl4lacZ locus is localized ∼0.75 cM distal to Sd and that the Sd phenotype is attenuated when Etl4lacZ is present in cis. These results suggest that the possible enhancer sequence in the fourth intron of the Skt gene functions also as the enhancer for the Sd gene and that the insertion of an enhancer trap vector in the third intron, as found in the Etl4lacZ line, may eliminate the enhancer function for the Sd gene, resulting in the attenuation of the Sd phenotype.
Future study on the functions of the Skt and Sd proteins may provide important clues for understanding the mechanisms for the development of notochordal cells and their differentiation into the nucleus pulposus cells.
Acknowledgments
We thank T. Iwamura, K. Miike, K. Haruna, and H. Hino for helpful and critical discussions and comments on the manuscript, and Michiyo Nakata and Ikuyo Kawasaki for technical assistance. This work was supported in part by a Grant-in-Aid on Priority Areas from the Ministry of Education, Science, Culture, and Sports of Japan and a grant from the Osaka Foundation for Promotion of Clinical Immunology.
References
- Alfred, J. B., K. Rance, B. A. Taylor, S. J. Phillips, C. M. Abbott et al., 1997. Mapping in the region of Danforth's short tail and the localization of tail length modifiers. Genome Res. 7: 108–117. [DOI] [PubMed] [Google Scholar]
- Allen, N. D., D. G. Cran, S. C. Barton, S. Hettel, W. Reik et al., 1988. Transgenes as probes for active chromosomal domains in mouse development. Nature 333: 852–855. [DOI] [PubMed] [Google Scholar]
- Ang, S., and J. Rossant, 1994. HNF-3β is essential for node and notochord formation in mouse development. Cell 78: 561–574. [DOI] [PubMed] [Google Scholar]
- Araki, K., T. Imaizumi, T. Sekimoto, K. Yoshinobu, J. Yoshimuta et al., 1999. Exchangeable gene trap using the Cre/mutated lox system. Cell. Mol. Biol. 45: 737–750. [PubMed] [Google Scholar]
- Aszodi, A., D. Chan, E. Hunziker, J. F. Bateman and R. Fassler, 1998. Collagen II is essential for the removal of the notochord and the formation of intervertebral discs. J. Cell Biol. 143: 1399–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens, A., J. Haigh, F. Mechta-Grigoriou, A. Nagy, M. Yaniv et al., 2003. Impaired intervertebral disc formation in the absence of Jun. Development 130: 103–109. [DOI] [PubMed] [Google Scholar]
- Chiang, C., Y. Litingtung, E. Lee, K. E. Young, J. Corden et al., 1996. Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature 383: 407–413. [DOI] [PubMed] [Google Scholar]
- Crick, F. H., 1952. Is alpha-keratin a coiled coil? Nature 170: 882–883. [DOI] [PubMed] [Google Scholar]
- Dunn, L. C., S. Gluecksohn-Schoenheimerl and V. Bryson, 1940. A new mutation in the mouse affecting spinal column and urogenital system. J. Hered. 31: 343–348. [Google Scholar]
- Gerard, R. D., and Y. Gluzman, 1985. New host cell system for regulated simian virus 40 DNA replication. Mol. Cell. Biol. 5: 3231–3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossler, A., A. L. Joyner, J. Rossant and W. C. Skarnes, 1989. Mouse embryonic stem cells and reporter constructs to detect developmentally regulated genes. Science 244: 463–465. [DOI] [PubMed] [Google Scholar]
- Gruneberg, H., 1953. Genetical studies on the skeleton of the mouse. VI. Danforth's short-tail. J. Genet. 51: 317–326. [Google Scholar]
- Gruneberg, H., 1958. Genetical studies on the skeleton of the mouse. XXII. The development of Danforth's short-tail. J. Embryol. Exp. Morphol. 6: 124–148. [PubMed] [Google Scholar]
- Hogan, B., R. Beddington, F. Costantini and E. Lacy, 1994. Manipulating the Mouse Embryo: A Laboratory Manual, pp. 379–381. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Korn, R., M. Schoor, H. Neuhaus, U. Henseling, R. Sonininen et al., 1992. Enhancer trap integrations in mouse embryonic stem cells give rise to staining patterns in chimaeric embryos with a high frequency and detect endogenous genes. Mech. Dev. 39: 95–109. [DOI] [PubMed] [Google Scholar]
- Kozak, M., 1996. Interpreting cDNA sequences: some insights from studies on translation. Mamm. Genome 7: 563–574. [DOI] [PubMed] [Google Scholar]
- Kusano, K, N. Kagawa, M. Sakaguchi, T. Omura and M. R. Waterman, 2001. a Importance of a proline-rich sequence in the amino-terminal region for correct folding of mitochondrial and soluble microbial p450s. J. Biochem. 129: 271–277. [DOI] [PubMed] [Google Scholar]
- Kusano, K., M. Sakaguchi, N. Kagawa, M. R. Waterman and T. Omura, 2001. b Microsomal p450s use specific proline-rich sequences for efficient folding, but not for maintenance of the folded structure. J. Biochem. 129: 259–269. [DOI] [PubMed] [Google Scholar]
- Lane, P. W., and C. S. Birkenmeiser, 1993. Urogenital syndrome (us): a developmental mutation on chromosome 2 of the mouse. Mamm. Genome 4: 481–484. [DOI] [PubMed] [Google Scholar]
- Langman, J., 1969. Medical Embryology, pp. 134–135. Williams & Wilkins, Baltimore.
- Lupas, A., 1996. Coiled coils: new structures and new functions. Trends Biochem. Sci. 21: 375–382. [PubMed] [Google Scholar]
- Lupas, A., M. Van Dyke and J. Stock, 1991. Predicting coiled coils from protein sequences. Science 252: 1162–1164. [DOI] [PubMed] [Google Scholar]
- Maatman, R., J. Zachgo and A. Gossler, 1997. The Danforth's short tail mutation acts cell autonomously in notochord cells and ventral hindgut endoderm. Development 124: 4019–4028. [DOI] [PubMed] [Google Scholar]
- Nishizaki, Y., K. Shimazu, H. Kondof and H. Sasaki, 2001. Identification of sequence motifs on the node/notochord enhancer of Foxa2 (Hnf3β) gene that are conserved across vertebrate species. Mech. Dev. 102: 57–66. [DOI] [PubMed] [Google Scholar]
- Niwa, H., K. Yamamura and J. Miyazaki, 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108: 193–200. [DOI] [PubMed] [Google Scholar]
- Paavola, L. G., D. B. Wilson and E. M. Center, 1980. Histochemistry of the developing notochord, perichordal sheath and vertebrae in Danforth's short-tail (Sd) and normal C57BL/6 mice. J. Emboryol. Exp. Morphol. 55: 227–245. [PubMed] [Google Scholar]
- Pauling, L., and R. B. Corey, 1953. Compound helical configurations of polypeptide chains: structure of proteins of the alpha-keratin type. Nature 171: 59–61. [DOI] [PubMed] [Google Scholar]
- Rufai, A., M. Benjamin and J. R. Ralphs, 1995. The development of fibrocartilage in the rat intervertebral disc. Anat. Embryol. 192: 53–62. [DOI] [PubMed] [Google Scholar]
- Smits, P., and V. Lefebvre, 2003. Sox5 and Sox6 are required for notochord extracellular matrix sheath formation, notochord cell survival and development of the nucleus pulposus of intervertebral discs. Development 130: 1135–1148. [DOI] [PubMed] [Google Scholar]
- Theiler, K., 1988. Vertebral malformations. Adv. Anat. Embryol. Cell Biol. 112: 1–99. [DOI] [PubMed] [Google Scholar]
- Wilson, V., L. Manson, W. C. Skarnes and R. S. Beddington, 1995. The T gene is necessary for normal mesodermal morphogenetic cell movements during gastrulation. Development 121: 877–886. [DOI] [PubMed] [Google Scholar]
- Zachgo, J., R. Korn and A. Gossler, 1998. Genetic interactions suggested that Danforth's short tail (Sd) is a gain-of-function mutation. Dev. Genet. 23: 86–96. [DOI] [PubMed] [Google Scholar]